Abstract
Comparison of the ratio of nonsynonymous to synonymous polymorphisms within species with the ratio of nonsynonymous to synonymous substitutions between species has been widely used as a supposed indicator of positive Darwinian selection, with the ratio of these 2 ratios being designated as a neutrality index (NI). Comparison of genome-wide polymorphism within 12 species of bacteria with divergence from an outgroup species showed substantial differences in NI among taxa. A low level of nonsynonymous polymorphism at a locus was the best predictor of NI < 1, rather than a high level of nonsynonymous substitution between species. Moreover, genes with NI < 1 showed a strong tendency toward the occurrence of rare nonsynonymous polymorphisms, as expected under the action of ongoing purifying selection. Thus, our results are more consistent with the hypothesis that a high relative rate of between-species nonsynonymous substitution reflects mainly the action of purifying selection within species to eliminate slightly deleterious mutations rather than positive selection between species. This conclusion is consistent with previous results highlighting an important role of slightly deleterious variants in bacterial evolution and suggests caution in the use of the McDonald–Kreitman test and related statistics as tests of positive selection.
Keywords: bacterial evolution, homologous recombination, McDonald–Kreitman test, neutrality index, nucleotide substitution
Introduction
Numerous studies have compared the patterns of synonymous and nonsynonymous polymorphisms within species with the patterns of synonymous and nonsynonymous divergences between species in order to obtain information regarding the action of natural selection on protein-coding genes; but the interpretation of such data is complicated (Hughes 2007). McDonald and Kreitman (1991) suggested that a higher ratio of nonsynonymous to synonymous divergence than of nonsynonymous to synonymous polymorphism might provide evidence of positive Darwinian selection that favored adaptive amino acid changes between species; and they proposed a statistical test (the McDonald–Kreitman [MK] test) of this hypothesis based on a contingency table. Several other authors have used similar reasoning to define as a “neutrality index” (NI), the ratio of nonsynonymous to synonymous polymorphism divided by the ratio of nonsynonymous to synonymous divergence, which may or may not be used in conjunction with the MK test (Rand and Kann 1996; Bazin et al. 2006). Alternatively, the inverse of NI may be defined as a “fixation index” (Shapiro et al. 2007).
It has been argued that the MK test and related methods may be subject to distortions caused by the presence of slightly deleterious nonsynonymous mutations in natural populations. As has frequently been suggested, these methods may not be able to distinguish clearly between the fixation of nonsynonymous substitutions due to positive selection and the fixation of slightly deleterious nonsynonymous mutations during a bottleneck accompanying speciation (Ohta 1993; Eyre-Walker 2002; Hughes et al. 2006; Hughes 2007). Furthermore, slightly deleterious nonsynonymous polymorphisms can confound the MK test and related methods in other ways. An abundance of slightly deleterious polymorphisms, subject to ongoing purifying selection, within a species might mask the effect of between-species divergence (Hughes 2007). Conversely, effective removal of slightly deleterious variants by purifying selection might lead to a false inference of positive selection between species because within-species nonsynonymous polymorphism is reduced.
There are 2 aspects of purifying selection that leave signatures detectable by nucleotide sequence analysis: 1) certain deleterious mutations have previously been eliminated from the population, thereby contributing the fact that dN is less than dS in most pairwise comparisons of coding sequences and; 2) certain slightly deleterious mutations, still present in populations, are subject to ongoing purifying selection that acts to lower their frequencies in comparison to neutral variants (Hughes et al. 2003; Hughes AL and Hughes MA 2007a, 2007b; Hughes and Piontkivska 2008; Irausquin and Hughes 2008). Because nonsynonymous mutations are more likely to be slightly deleterious than synonymous mutations, a signature of ongoing purifying selection is that gene diversity (“heterozygosity”) is reduced at nonsynonymous polymorphic sites in comparison to that at synonymous polymorphic sites (Hughes et al. 2003). Likewise, ongoing purifying selection will influence statistics that compare the pairwise nucleotide difference with the number of segregating sites and thereby identify the presence of rare variants (Tajima 1989).
Several lines of evidence suggest that slightly deleterious variants, subject to ongoing purifying selection, are widespread in protein-coding genes of many bacterial species. A survey of 149 data sets of bacterial sequence polymorphism revealed an excess of rare nonsynonymous variants but not of synonymous variants; the former is likely to represent slightly deleterious alleles whose frequency in the population has decreased as a result of ongoing purifying selection (Hughes 2005). Similar results were reported in a study of Escherichia coli and Salmonella enterica, even when singletons (possibly due to sequencing errors) were removed from the data (Charlesworth and Eyre-Walker 2006). Moreover, Rocha et al. (2006) presented evidence that the ratio of the number of nonsynonymous substitutions per nonsynonymous site (dN) to the number of synonymous substitutions per synonymous site (dS) tends to decrease as the evolutionary time between 2 related bacterial genomes increases.
The efficiency with which slightly deleterious variants are removed is correlated with effective population size. Although there is evidence that effective population sizes of bacteria are in general much greater than those of multicellular eukaryotes (Lynch and Conery 2003), little is known about variations among bacterial species with respect to effective population size. Another factor that will affect these analyses is the rate of homologous recombination because recombination is necessary to purge deleterious variants from a population (“Mueller's ratchet”; Lynch et al. 1993; Lynch 2007); and recombination rates are known to vary among bacterial species (Feil et al. 2001). If nonsynonymous polymorphism involves mainly slightly deleterious variants subject to ongoing purifying selection, species with large effective population sizes and/or efficient recombination may be expected to have reduced within-species nonsynonymous polymorphism relative to within-species synonymous polymorphism. As a result, the MK test and related methods may tend to identify positive selection in such species. Moreover, within a given species, the MK test and related methods may tend to infer positive selection on individual genes subject to strong ongoing purifying selection and thus having low ratios of nonsynonymous to synonymous polymorphism.
Here we address this question by examining within-species polymorphism in protein-coding genes in the completely sequenced genomes of 12 species of bacteria, including important pathogens of humans, domestic animals, and plants. Using the closest available outgroup species, we estimate between-species synonymous and nonsynonymous substitutions and compare the pattern of net between-species divergence with that of within-species polymorphism. In particular, we examine the NI, defined as the ratio of nonsynonymous to synonymous polymorphism divided by the ratio of nonsynonymous to synonymous divergence (Rand and Kann 1996). In addition, in the case of 3 species for which polymorphism data were available on a substantial number of genes in 4 or more genomes (Burkholderia pseudomallei, Staphylococcus aureus, and Streptococcus pyogenes), we test for the presence of rare nonsynonymous variants likely to be subject to ongoing purifying selection. By comparing the patterns of within-species purifying selection within species with those of between-species divergence, we test for the role of slightly deleterious nonsynonymous variants on the MK and related methods.
Methods
Sequences Analyzed
We analyzed polymorphism within 12 species of Bacteria for which at least 3 complete genome sequences were available and for which the complete genome sequence of a congeneric outgroup species was available (for GenBank accession numbers, see supplementary table S1, Supplementary Material online). The species (with numbers of sequences) and outgroups were as follows: B. pseudomallei (4) and Burkholderia thailandensis, Campylobacter jejuni (3) and Campylobacter fetus, Chlamydophila pneumoniae (4) and Chlamydophila felis, Ehrlichia ruminantium (3) and Ehrlichia chaffeensis, Helicobacter pylori (3) and Helicobacter acinonychis, Pseudomonas syringae (3) and Pseudomonas fluorescens, Shigella flexneri (3) and Shigella sonnei, S. aureus (9) and Staphylococcus epidermidis, Streptococcus pneumoniae (3) and Streptococcus sanguinis, S. pyogenes (12) and Streptococcus agalactiae, and Yersinia pestis (5) and Yersinia pseudotuberculosis. We designate the species for which multiple genomes were analyzed as the ingroup species. Gene families in a species and its outgroup were identified by applying the Blastclust software (Altschul et al. 1997) to predicted protein translations. Putative orthologs were identified as families with exactly one representative per genome. The parameters L (minimum length coverage) and S (similarity threshold measured as Blast similarity score divided by the alignment length) were chosen for each pair of species so as to yield as many orthologs as possible, given the level of sequence divergence between ingroup and outgroup species (supplementary table S1, Supplementary Material online). Orthologs were aligned at the amino acid level using ClustalW (Thompson et al. 1994), and the alignment was imposed on the DNA sequences.
The species concept in Bacteria has been controversial because genetic exchange can occur beyond the boundaries of named species (Vulić et al. 1997; Majewski 2001; Cohan 2002; Fraser et al. 2007). Furthermore, named bacterial species do not always correspond to monophyletic groups. We used preliminary phylogenetic analyses to ensure that each of the 12 ingroup species used here constituted a monophyletic group, at least as regards the majority of its protein-coding genes. Likewise, we used phylogenetic analyses to test the hypothesis that each of the outgroup species constituted an outgroup to the ingroup species with which it was compared, at least as regards the majority of its protein-coding genes. These phylogenetic analyses were based on the aligned sets of orthologous amino acid sequences using the following methods: 1) maximum parsimony (MP) using branch-and-bound search (Swofford 2002), 2) Neighbor-Joining (Saitou and Nei 1987) based on the JTT model (Jones et al. 1992) with the gamma correction for rate variation among sites, using the MEGA 3 program (Kumar et al. 2004), and 3) the quartet maximum likelihood method using the Tree-Puzzle 5.2 program (Schmidt et al. 2002) based on the JTT model with gamma correction for rate variation among sites. We only included in our analyses species for which these analyses supported monophyly and the validity of the outgroup.
Nucleotide Diversity and Divergence
The number of synonymous nucleotide substitutions per synonymous site (dS) and the number of nonsynonymous nucleotide substitutions per nonsynonymous site (dN) were estimated for all pairwise comparisons of orthologs by the method of Yang and Nielsen (2000). This method takes into account nucleotide content and mutational biases (Yang and Nielsen 2000). For each ortholog, we computed the mean of dS values in all pairwise comparisons within the ingroup species (i.e., the synonymous nucleotide diversity, symbolized πS). Likewise, for each ortholog, we computed the mean of dN values in all pairwise comparisons within the ingroup species (i.e., the nonsynonymous nucleotide diversity, symbolized πA). For each ortholog, we computed the net synonymous (kS) and nonsynonymous (kA) nucleotide divergences between the ingroup and outgroup species following Nei and Jin (1989; see also Nei and Kumar 2000). To compute kS, we first computed the mean of dS for all comparisons between ingroup and outgroup (dSb), then kS = dSb − πS. Similarly, to compute kA, we first computed the mean of dN for all comparisons between ingroup and outgroup (dNb), then kA = dNb − πA.
Different authors have used different methods based on the original idea of McDonald and Kreitman (1991) to compare synonymous and nonsynonymous polymorphisms and divergences (e.g., Rand and Kann 1996; Bustamante et al. 2002; Smith and Eyre-Walker 2002; Bazin et al. 2006). Some of these methods have been developed because counting of synonymous and nonsynonymous differences between species may underestimate the amount of substitution if the evolutionary time has been long and because, when there are multiple substitutions per codon, the numbers of synonymous and nonsynonymous substitutions depend on the pathway taken by evolution (Whittam and Nei 1991). In preliminary analyses, we found that the results of all these methods are highly correlated. For example, we reconstructed the ancestral sequences of the set of orthologs for selected ingroup species by MP and estimated dS and dN between the outgroup species and the reconstructed ancestor of the ingroup species. In addition, we used the maximum likelihood method to reconstruct internal branches within the trees. The resulting values of these preliminary analyses were highly correlated with kS and kA computed as described above. Therefore, we used the latter as measures of between-species divergence because they involve fewer assumptions than methods dependent on ancestral reconstruction, while taking into account both multiple hits and the probability of different evolutionary pathways (Yang and Nielsen 2000).
Because of homologous recombination, certain loci may have evolutionary histories that contrast markedly with those of other genes in the genomes. We used a 2-pronged approach to exclude these genes from our data set. First, we used a k-means clustering algorithm to identify genes with unusual patterns of dS in pairwise comparisons among ingroup members (Hughes and Friedman 2004, 2005; Hughes and French 2007; Hughes and Langley 2007). Of 16,776 orthologs identified by homology search, 73 (0.4%) showed highly unusual patterns and were excluded from further analyses. Second, we excluded all genes with πS > dSb (162 such genes or 1.0% of the total) and all genes with πA > dNb (232 such genes or 1.4% of the total). Note that the fact that such a small proportion of genes showed greater within-species polymorphism than between-species divergence provides further support for the conclusion that the outgroup species chosen constituted genuine outgroups to the ingroup species.
We computed the following quantity:
| (1) |
NI (Rand and Kann 1996) is greater than 1 when the ratio of nonsynonymous to synonymous polymorphism exceeds the ratio of nonsynonymous to synonymous divergence; this is the condition of the absence of positive selection increasing between-species divergence at nonsynonymous sites according to the assumptions of McDonald and Kreitman (1991). By contrast, NI < 1 is taken to be indicative of such selection (McDonald and Kreitman 1991), although other interpretations are possible (Ohta 1993; Eyre-Walker 2002; Hughes et al. 2006; Hughes 2007). In the present analyses, we excluded genes for which NI was undefined; 5,956 (35.5%) of orthologs identified by homology search were excluded for this reason. NI was defined for relatively few genes when the available genomes for a given ingroup species were very closely related because in those species, πS = 0 in the case of many genes. Chlamydophila pneumoniae and Y. pestis were the genomes that had the highest numbers of genes excluded for this reason.
Codon Usage
Within each genome, we computed for each predicted protein-coding gene 5 quantities summarizing codon usage in that gene. For a given gene, let nTC2f = the number of 2-fold degenerate sites using T or C, nAG2f = the number of 2-fold degenerate sites using A or G, and n4f = the number of 4-fold degenerate sites. Let nC2f = the number of occurrences of C at 2-fold degenerate sites; nG2f = the number of occurrences of G at 2-fold degenerate sites; and nC4f, nG4f, and nA4f designate, respectively, the numbers of occurrences of C, G, and A at 4-fold degenerate sites. Then define T2 = nT2f/nTC2f, A2 = nA2f/nAG2f, T4 = nT4f/n4f, C4 = nC4f/n4f, and A4 = nA4f/n4f. Taken together, these 5 codon usage variables (pC2f, pG2f, pC4f, pG4f, and pA4f) provide a measure of nucleotide usage, including both nucleotide content and content skewness, at almost all synonymous sites in the gene (excluding only the 3-fold degenerate sites in isoleucine codons and the very rare C/A synonymous mutations in the first positions of certain arginine codons). The advantage of this approach is that it describes codon usage in only 5 linearly independent variables that are not dependent on amino acid composition (Hughes and Langley 2007). Moreover, these 5 variables are not subject to the stochastic errors due to small sample size seen in variables measured on each amino acid or each codon.
In order to compare these 5 variables across species, we computed standard normal deviates for each of these quantities within each species. We computed the standard normal deviate for a given value by subtracting from that value the mean for the species and dividing that difference by the standard error for the species. In order to identify genes with unusual codon usage for the species, we first computed the absolute value of the standard normal deviates. Then we used principal components, extracted from the correlation matrix, to reduce dimensionality. The first principal component (PC1) accounted for 35.1% of the variance in the correlation matrix, and PC1 was strongly positively correlated with the absolute values of all 5 standard normal deviates. We therefore used PC1 as an overall measure of atypical codon usage patterns. In preliminary analyses, separate analyses of all 5 standard normal deviates yielded similar results to those based on PC1 (data not shown); for simplicity, we report below only the results based on PC1.
Within-Species Polymorphism
For selected genomes, we compared the pattern of synonymous and nonsynonymous polymorphisms within individual genes. There were 3 species for which we had sequences from 4 or more genomes and on a substantial number of genes including both synonymous and nonsynonymous polymorphic sites: B. pseudomallei (4 genomes; 1,710 genes), S. aureus (9 genomes; 1,317 genes), and S. pyogenes (11 genomes; 875 genes). For these genes, we computed the single-locus gene diversity (heterozygosity) independently for each polymorphic site by the following formula:
| (2) |
where n is the number of alleles and xi is the population frequency of the ith allele (Nei 1987, p. 177). Single nucleotide polymorphisms were classified either as synonymous or as nonsynonymous depending on their effect of the encoded nucleotide sequence; we excluded ambiguous sites at which both synonymous and nonsynonymous variants occurred or at which the polymorphism could be considered synonymous or nonsynonymous depending on the pathway taken by evolution. There were 4,316 such ambiguous polymorphic sites out of 177,038 total polymorphic sites (2.4%).
In order to examine the relative frequency of rare alleles at synonymous and nonsynonymous sites, we compared the average number of nucleotide differences and the number of segregating sites (Tajima 1989) separately for synonymous and nonsynonymous sites (Rand and Kann 1996; Hughes 2005; Hughes AL and Hughes MA 2007a, 2007b). For each gene, we computed the difference Ks − S*s. Ks is the mean number of synonymous nucleotide differences for all pairwise comparisons among the n allelic sequences in the data set. If Ss is the number of synonymous segregating sites, then
| (3) |
The divisor in equation (3) is a factor providing an adjustment for sample size (n) and is given by the following (Tajima 1989):
| (4) |
Similarly, for each gene, we computed the difference Kn − S*n. Kn is the mean number of nonsynonymous nucleotide differences for all pairwise comparisons among the n allelic sequences in the data set; and, if Sn is the number of segregating nonsynonymous sites
| (5) |
The differences Ks − S*s and Kn − S*n constitute the numerator of Tajima's (1989) D statistic computed separately for synonymous and nonsynonymous polymorphisms, respectively. We then computed the ratio of this difference to the absolute value of the minimum possible value of the difference, which would occur if all polymorphisms were singletons (Schaeffer 2002). We designate this ratio Qsyn in the case of synonymous polymorphisms and Qnon in the case of nonsynonymous polymorphisms. Comparing Qsyn and Qnon provides an index of the relative abundance of rare alleles at synonymous and nonsynonymous sites, with a strongly negative value indicating an abundance of rare alleles (Hughes AL and Hughes MA 2007a, 2007b). Note that, unlike Tajima's D, these ratios are independent of sample size and thus can be compared between data sets of different size.
Tajima's D was designed to provide a test of neutrality but that test depends on the assumption of mutation-drift equilibrium (Tajima 1989; Nei and Kumar 2000). For example, a recent population bottleneck can yield a negative value of D in the absence of purifying selection. However, in the present analyses, we did not test for neutrality but merely used Qsyn and Qnon as indices of the relative abundance of rare variants at synonymous and nonsynonymous sites, respectively. Note also that, because a bottleneck should affect both synonymous and nonsynonymous polymorphisms equally (Tajima 1989), a marked and consistent difference between Qsyn and Qnon is likely to be due to selection on nonsynonymous sites.
In order to examine the relationships among NI, the action of purifying selection, and the results of the MK tests (McDonald and Kreitman 1991), we constructed for each gene in our data sets for B. pseudomallei, S. aureus, and S. pyogenes, a 2 × 2 contingency table including the following quantities: Ss and Sn; the estimated number of synonymous differences between the ingroup and outgroup species and the estimated number of nonsynonymous differences between the ingroup and outgroup species. The latter 2 quantities were estimated by the method of Yang and Nielsen (2000) and rounded to the nearest integer. Note that this approximation is probably reasonably valid in the case of these 3 species because in each of these species, the outgroup was relatively close to the ingroup (see Results). For each contingency table, we conducted G-tests of independence using Williams’ correction (Sokal and Rohlf 1981).
Other Statistical Methods
In order to compare πS, πA, kS, and kA across species, we computed standard normal deviates for each of these quantities within each species. We computed the standard normal deviate for a given value by subtracting from that value the mean for the species and dividing that difference by the standard error for the species; we designate the standard normal deviates as πSdev, πAdev, kSdev, and kAdev, respectively.
Because the variables analyzed were not normally distributed, we used for hypothesis testing nonparametric methods of statistical analysis, which make no assumptions regarding the form of the underlying distribution (Hollander and Wolfe 1973). However, in preliminary analyses, methods assuming a normal distribution generally yielded essentially identical results.
Results
Comparisons among Taxa
Table 1 summarizes means of synonymous (πS) and nonsynonymous (πA) nucleotide diversity for sets of orthologous genes from 12 bacterial species, along with mean net synonymous (kS) and nonsynonymous (kA) divergence from an outgroup species. The synonymous nucleotide diversity within species ranged from less than 1% to over 50% (in P. syringae; table 1). Likewise, available outgroups covered a wide range of divergences. For example, C. fetus was very distant from the 3 C. jejuni genomes analyzed, with mean kS estimated at nearly 3 substitutions per site (table 1). By contrast, Y. pseudotuberculosis was very close to the 5 Y. pestis genomes analyzed (mean kS = 0.0186; table 1).
Table 1.
Synonymous (πS) and Nonsynonymous (πA) Nucleotide Diversity within 12 Bacterial Species and Mean Net Numbers of Synonymous Substitutions per Synonymous Site (kS) and of Nonsynonymous Substitutions per Nonsynonymous Site (kA) between Each Species and an Outgroup Species
| Species | Outgroup | No. of Genes | πS ± SE | πA ± SE | kS ± SE | kA ± SE |
| Burkholderia pseudomallei | Burkholderia thailandensis | 1,711 | 0.01591 ± 0.00023 | 0.00104 ± 0.00003 | 0.3436 ± 0.0030 | 0.0180 ± 0.0003 |
| Campylobacter jejuni | Campylobacter fetus | 934 | 0.06937 ± 0.00177 | 0.00477 ± 0.00022 | 2.8491 ± 0.0295 | 0.3818 ± 0.0053 |
| Chlamydophila pneumoniae | Chlamydophila felis | 158 | 0.00232 ± 0.00011 | 0.00047 ± 0.00006 | 1.9790 ± 0.0527 | 0.2557 ± 0.0109 |
| Ehrlichia ruminantium | Ehrlichia chaffeensis | 633 | 0.02913 ± 0.00078 | 0.00252 ± 0.00016 | 1.9871 ± 0.0314 | 0.1364 ± 0.0026 |
| Helicobacter pylori | Helicobacter acinonychis | 1,130 | 0.11903 ± 0.00099 | 0.02118 ± 0.00049 | 0.1609 ± 0.0023 | 0.0336 ± 0.0009 |
| Neisseria meningitidis | Neisseria gonorrhoeae | 942 | 0.07336 ± 0.00193 | 0.00806 ± 0.00026 | 0.0843 ± 0.0022 | 0.0117 ± 0.0003 |
| Pseudomonas syringae | Pseudomonas fluorescens | 1,005 | 0.54810 ± 0.00883 | 0.01169 ± 0.00030 | 0.9906 ± 0.0160 | 0.0477 ± 0.0007 |
| Shigella flexneri | Shigella sonnei | 562 | 0.00295 ± 0.00011 | 0.00081 ± 0.00006 | 0.0380 ± 0.0014 | 0.0040 ± 0.0002 |
| Staphylococcus aureus | Staphylococcus epidermidis | 1,463 | 0.03415 ± 0.00094 | 0.00273 ± 0.00010 | 2.6932 ± 0.0267 | 0.1646 ± 0.0026 |
| Streptococcus pneumoniae | Streptococcus sanguinis | 819 | 0.02362 ± 0.00149 | 0.00307 ± 0.00019 | 1.6791 ± 0.0242 | 0.1793 ± 0.0040 |
| Streptococcus pyogenes | Streptococcus agalactiae | 931 | 0.02924 ± 0.00114 | 0.00407 ± 0.00019 | 2.1018 ± 0.0335 | 0.2014 ± 0.0040 |
| Yersinia pestis | Yersinia pseudotuberculosis | 65 | 0.00119 ± 0.00011 | 0.00051 ± 0.00014 | 0.0186 ± 0.0027 | 0.0033 ± 0.0006 |
NOTE.—SE, standard error.
As predicted by Rocha et al. (2006), mean πA/πS for the 12 taxa was negatively correlated with mean πS (rS = −0.699; P = 0.011; fig. 1A). Pseudomonas syringae provided an influential point in this relationship, with the lowest mean πA/πS (0.0232) and by far the highest mean πS (0.5481; fig. 1A). Nonetheless, even when Pseudomonas was excluded from the analysis, the negative relationship between mean πA/πS and mean πS remained (rS = −0.609; P = 0.047). Besides Pseudomonas, the next lowest value of πA/πS was that seen in Campylobacter (0.0800; fig. 1A).
FIG. 1.—

(A) Plot of mean πA/πS versus mean πS for 12 bacterial taxa (rS = −0.699; P = 0.011). Abbreviations for taxa: Burk (Burkholderia), Camp (Campylobacter), Chlam (Chlamydophila), Ehr (Ehrlichia), Helic (Helicobacter), Neis (Neisseria), Pseud (Pseudomonas), Shig (Shigella), Staph (Staphylococcus), Streppn (Streptococcus pneumoniae), Streppy (Streptococcus pyogenes), and Yers (Yersinia pestis). (B) Numbers of genes with NI < 1 and NI ≥ 1. There was a significant difference among taxa with respect to the portion of genes with NI ≥ 1 (χ2 = 1158.5; 11 df; P < 0.001).
There was a highly significant difference among taxa with respect to the portion of genes with NI ≥ 1 (χ2 = 1158.5; 11 degrees of freedom [df]; P < 0.001; fig. 1B). The proportion of orthologs with NI ≥ 1 exceeded the proportion with NI < 1 in 4 of the 12 taxa, whereas in other 8 taxa, the proportion of orthologs with NI < 1 exceeded the proportion with NI ≥ 1 (fig. 1B). There were 2 taxa in which the proportion of genes with NI ≥ 1 was strikingly low. In Campylobacter, there were only 150 of 934 orthologs (16.1%) with NI ≥ 1; and in Pseudomonas, there were only 94 of 1,005 orthologs (9.4%) with NI ≥ 1 (fig. 1B). In the remaining 10 taxa, the lowest proportion of orthologs with NI ≥ 1 was 38.5% (363 of 942) in Neisseria (fig. 1A). Combining data for all taxa, there were 4,551 (44.0%) of 10,353 orthologs with NI ≥ 1 (fig. 1A). But when Campylobacter and Pseudomonas were excluded, 4,307 of the remaining 8,414 genes (51.2%) had NI ≥ 1 (fig. 1A).
If values of NI < 1 are due to positive selection leading to amino acid sequence divergence between species, it would be expected that species with NI < 1 would be species with unusually high kA values. As an initial test of this prediction, we computed for each species, the number of genes falling in the top quartile (25%) of values of πA/πS across all species, then we compared this value with the number of genes expected to be in the top quartile if the distribution of πA/πS were uniform across species (i.e., 25% of the number of genes for the species; fig. 2A). Likewise, we computed for each species, the number of genes falling in the top quartile (25%) of values of kA/kS across all species, then we compared this value with the number of genes expected to be in the top quartile if the distribution of kA/kS was uniform across species (i.e., 25% of the number of genes for the species; fig. 2B).
FIG. 2.—
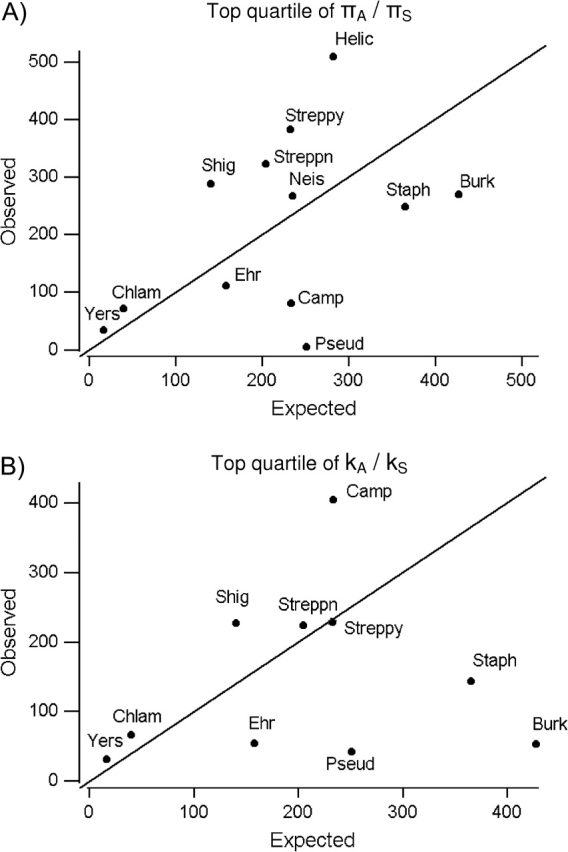
Observed versus expected numbers of genes in the top quartile of values of (A) πA/πS and (B) kA/kS. Abbreviations for taxa are as in figure 1A.
Pseudomonas and Campylobacter had by far the lowest ratios of observed to expected gene numbers in the top quartile of πA/πS values (fig. 2A). In the case of Pseudomonas, only 4 of 1,005 genes (0.4%) were in the top quartile of πA/πS values; and in Campylobacter, only 80 of 934 (8.6%) were in the top quartile of πA/πS values (fig. 2A). Burkholderia (52 of 1,711 or 3.1%) had the lowest proportion of genes in the top quartile of kA/kS values (fig. 2B). Pseudomonas also had a very low proportion of genes (42 of 1,005 or 4.2%) in the top quartile of kA/kS values (fig. 2B). By contrast, the 2 highest proportions of genes in the top quartile of kA/kS values were seen in Yersinia (31 of 65 or 47.7%) and Campylobacter (405 of 934 or 43.4%; fig. 2B).
Thus, contrary to the usual interpretation of the NI statistic, Pseudomonas, the taxon with the highest proportions of genes with NI < 1 (fig. 1B), showed exceptionally few genes with high kA/kS values (fig. 2B). On the other hand, Pseudomonas also showed exceptionally few genes with high πA/πS values (fig. 2A). Because NI is a ratio, these results suggest that low NI values in the case of Pseudomonas were due to low numerators rather than to high denominators.
Campylobacter, the taxon with the second highest proportions of genes with NI < 1 (fig. 1B), did show a high proportion of genes with high kA/kS values (fig. 2B). But, like Pseudomonas, Campylobacter also showed exceptionally few genes with high of πA/πS values (fig. 2A). Thus, the factors contributing to values of NI < 1 were not consistent between the 2 taxa with the highest proportions of such genes.
Codon Usage
Synonymous codon usage is one factor that might be expected to influence the pattern of nucleotide substitution (Sharp 1991); but the method we used to estimate synonymous and nonsynonymous substitutions is designed to take into account differing patterns of synonymous codon usage (Yang and Nielsen 2000). Moreover, there was no obvious relationship between codon usage and the frequency of genes with NI < 1. Campylobacter jejuni has an AT-rich genome, whereas P. syringae is GC-rich (supplementary table S2, Supplementary Material online). However, the species with lower frequencies of genes with NI < 1 included both very AT-rich species (e.g., E. ruminantium) and GC-rich species (e.g., B. pseudomallei; supplementary table S2, Supplementary Material online).
Rank Partial Correlations
In order to examine the factors influencing the value of NI for individual genes, we used standard normal deviates as a measure of the deviation of πA, πS, kA, and kS for a given gene from the mean values for the species. We designated the standard normal deviates as πAdev, πSdev, kAdev, and kSdev, respectively. We used partial rank correlation to analyze the relationship between these variables and NI. We also tested for an independent effect of atypical patterns of codon usage, using the first PC1 extracted from the correlation matrix of absolute values of the standard normal deviates of 5 variables describing nucleotide content at synonymous sites (see Methods). We computed partial rank correlation coefficients between NI and each of 5 predictor variables (πAdev, πSdev, kAdev, kSdev, and PC1) simultaneously controlling in each case for the other 4 predictor variables (table 2). Because certain ingroup species showed much higher kS values in comparison with the outgroup than did others, we analyzed the taxa with close outgroups (Burkholderia, Helicobacter, Neisseria, Shigella, and Yersinia; table 1) separately from the other taxa (table 2). However, the results were similar when all species were considered together (data not shown).
Table 2.
Partial Rank Correlation Coefficients between 5 Predictor Variables and NI, Computed Separately for Genes from Species with Close Outgroups and Species with Distant Outgroupsa
| Predictor Variable | Close Outgroup (N = 4,410) | Distant Outgroup (N = 5,943) |
| πAdev | 0.747b | 0.684b |
| πSdev | 0.340bc | −0.523bc |
| kAdev | −0.412bc | −0.422bc |
| kSdev | 0.305bc | 0.400bc |
| PC1 | −0.020c | −0.102bc |
Each value is a fourth-order partial rank correlation between the predictor variable and NI simultaneously controlling for the other 4 predictor variables.
Tests of the hypothesis that a given partial rank correlation coefficient equals zero: P < 0.001.
Tests of the hypothesis that a given partial rank correlation coefficient equals that between πAdev and NI: P < 0.001.
Given that NI is a ratio of 2 ratios, it was unsurprising that there were significant positive partial rank correlation coefficients both between πAdev and NI and between kSdev and NI, a pattern seen both in genes from species with close outgroups and in genes from species with distant outgroups (table 2). Likewise, it was unsurprising that πSdev and kAdev showed significant negative partial rank correlations with NI both for genes from species with close outgroups and for genes from species with distant outgroups (table 2). However, in the case of both close and distant outgroups, the strongest partial rank correlation between NI and any of the predictor variables was that with πAdev (table 2). In fact, whether positive or negative, all the other partial rank correlations differed significantly in absolute value from that between NI and any of the predictor variables was that with πAdev (table 2). Thus, πAdev, rather than kAdev, was the strongest predictor of NI, implying that low NI was primarily a consequence of unusually low within-species nonsynonymous nucleotide diversity rather than unusually high nonsynonymous between-species divergence. This result is contrary to the expectation that a low value of NI is evidence of positive selection favoring between-species divergence at the amino acid level.
In the case of species with close outgroups, there was not a significant partial rank correlation between NI and PC1 (table 2). By contrast, when the outgroup was distant, there was a highly significant negative partial rank correlation between NI and PC1. Thus, in these species, NI tended to be decreased when codon usage was unusual, independently of the estimates of nucleotide sequence polymorphism and divergence. It seems plausible that this result may have occurred because of the difficulty of estimating kS in the case of distant outgroups, where synonymous sites approached saturation (table 1), which may be especially problematic when codon usage is highly unusual.
We tested this interpretation by computing rank partial correlations between PC1 and kSdev, simultaneously controlling for NI, πAdev, πSdev, and kAdev. In the case of species with close outgroups, the partial rank correlation between PC1 and kSdev was −0.034 (P = 0.025), whereas in the case of species with distant outgroups, the partial rank correlation between PC1 and kSdev was −0.143 (P < 0.001). The difference between these 2 correlation coefficients was highly significant (P < 0.001). Thus, unusual nucleotide content tended to cause a reduction in the estimate of kS, and this effect was much more pronounced when the outgroup was distant.
Within-Species Polymorphism
For genes of B. pseudomallei, S. aureus, and S. pyogenes, we computed mean gene diversity separately for synonymous and nonsynonymous polymorphic sites (table 3). In each species, the median of the gene diversity values at synonymous sites was significantly greater than that at nonsynonymous sites (sign test; P < 0.001 in each case; table 3). The fact that gene diversities at nonsynonymous polymorphic sites tended to be lower than those at synonymous polymorphic sites in the same genes was evidence that nonsynonymous variants in these species include many that are subject to ongoing purifying selection. Additional evidence in support of this interpretation was provided by comparisons of Qsyn and Qnon (table 3). In all 3 species, median Qnon was significantly lower than median Qsyn (sign test; P < 0.001 in each case; table 3). In all 3 species, median Qnon was strongly negative (table 3). Median Qsyn was negative in Burkholderia and in Staphylococcus, although less negative than Qnon, whereas in S. pyogenes, median Qsyn was actually slightly positive (table 3). Thus, in all 3 species, nonsynonymous polymorphic sites showed a greater bias toward rare polymorphisms than did synonymous sites, implying the presence of stronger ongoing purifying selection on the former than on the latter.
Table 3.
Median Gene Diversity at Polymorphic Nonsynonymous and Synonymous Sites and Median Qnon and Qsyn in 3 Bacterial Species
| Species (no. of genes) | Gene Diversity |
Qnon | Qsyn | |
| Nonsynonymous | Synonymous | |||
| Burkholderia pseudomallei (1,710) | 0.375 | 0.377a | −0.501 | −0.368b |
| Staphylococcus aureus (1,317) | 0.272 | 0.294a | −0.438 | −0.145b |
| Streptococcus pyogenes (875) | 0.254 | 0.299a | −0.289 | 0.039b |
Tests of the hypothesis that median gene diversity at nonsynonymous sites equals median gene diversity at synonymous sites: P < 0.001 (sign test).
Tests of the hypothesis that median Qnon equals median Qsyn: P < 0.001 (sign test).
In order to examine the relationship between the value of NI and purifying selection on nonsynonymous polymorphic variants, we compared Qsyn and Qnon between genes with NI < 1 and genes with NI ≥ 1 separately for the 3 species. In each species, median Qnon values differed significantly between these 2 categories of genes (fig. 3). For all 3 species, median Qnon was lower in genes with NI < 1 than in genes with NI ≥ 1. Median Qsyn also differed significantly between the 2 categories of genes in all 3 species (fig. 3). However, in the case of Qsyn, median values for genes with NI < 1 were consistently higher than those for genes with NI ≥ 1 (fig. 3). Thus, genes with NI < 1 were characterized by an excess of rare nonsynonymous variants but not of synonymous variants.
FIG. 3.—

Median Qsyn and Qnon in genes with NI < 1 and NI ≥ 1 in (A) Burkholderia, (B) Staphylococcus, and (C) Streptococcus pyogenes. Mann–Whitney tests of the hypothesis that the median value for genes with NI < 1 equals that for genes with NI ≥ 1: *P < 0.05; ***P < 0.001.
MK Tests
In order to examine the relationship between within-species polymorphism and the results of the MK test, we compared numbers of polymorphic synonymous and nonsynonymous sites within B. pseudomallei, S. aureus, and S. pyogenes with the estimated numbers of synonymous and nonsynonymous substitutions between each of these species and the appropriate outgroup species. According to the usual interpretation of the MK test, genes with NI ≥ 1 and a significant G-test represent genes with a significant excess of nonsynonymous polymorphisms, whereas genes with NI < 1 and a significant G-test represent cases of positive selection between species. In the present data, the latter was much less frequent than the former. Of 1,580 genes with NI < 1, the G-test was significant at the 5% level in only 50 cases (3.2%). By contrast, of 2,322 genes with NI ≥ 1, the G-test was significant at the 5% level in 499 cases (21.5%).
Among genes with NI ≥ 1, Qnon was greater than zero in 103 of 499 genes with significant G-tests (20.6%). By contrast, of 1,823 genes with NI ≥ 1 and nonsignificant G-tests, Qnon was greater than zero in only 276 (15.1%) genes. The difference between proportions was highly significant (χ2 = 8.7; 1 df; P < 0.001). This difference is expected because a significant G-test with NI ≥ 1 implies an excess of within-species nonsynonymous polymorphisms. Among genes with NI < 1, Qnon was greater than zero in 15 of 50 (30.0%) genes with significant G-tests (30%) but in only 146 of 1,530 (9.5%) genes with nonsignificant G-tests. Again, the difference in proportions was highly significant (χ2 = 22.1; 1 df; P < 0.001). The latter result is surprising because a significant G-test for genes with NI < 1 is typically interpreted as indicating positive selection between species. Our results suggest on the contrary that the test may be influenced by the pattern of within-species polymorphism. In particular, because a high value of Qnon implies few nonsynonymous variants of low frequency, our results suggest that the absence of such variants is likely to lead to a significant result in the MK test.
We used discriminant analysis to examine further the relationship between Qnon and the outcome of the G-test. For genes with NI < 1, a linear discriminant function using Qnon as the sole predictor correctly predicted the outcome of the G-test (significant or nonsignificant) in 80.6% of cases. By contrast, in genes with NI ≥ 1, a linear discriminant function with Qnon as the sole predictor successfully predicted the outcome of the G-test in only 36.2% of cases. This analysis further supported the conclusion that the pattern of within-species nonsynonymous polymorphism can strongly influence the MK G-test when NI < 1.
Discussion
Comparison of the ratio of nonsynonymous to synonymous nucleotide diversity within species (πA/πS) with the ratio of nonsynonymous to synonymous substitutions between species (kA/kS) has been widely used as an indicator of positive Darwinian selection favoring amino acid changes between species. Comparison of genome-wide polymorphism within 12 species of bacteria with divergence from an outgroup species yielded results that cannot easily be explained on this supposition. Two species, C. jejuni and P. syringae, showed extremely high frequencies (83.9% and 90.6%, respectively) of genes for which NI (the ratio of πA/πS to kA/kS) was less than 1 (fig. 1B). By contrast, in the 10 other species analyzed, NI was less than 1 in only about 48% of genes. According to the usual interpretation of NI, these results might be taken to indicate an extraordinarily high frequency of between-species positive selection in Campylobacter and Pseudomonas, but there is no obvious reason to expect that these 2 taxa are subject to unusually strong positive selection.
As an alternative to the hypothesis of positive selection, it might be proposed that the differences among taxa are artifacts of the estimation of synonymous and nonsynonymous substitutions. For example, it might be suggested that factors such as an unusual pattern of codon usage or a high rate of synonymous substitution have biased the estimates of synonymous and nonsynonymous substitutions in certain taxa. However, the method used here to estimate the numbers of synonymous and nonsynonymous substitutions per site is designed to take into account factors such as nucleotide usage and mutational bias and appears to be robust even when the rate of synonymous substitution is high (Yang and Nielsen 2000). In the present data, the level of synonymous substitution between ingroup and outgroup species was high in Campylobacter and Pseudomonas but not unusually so in comparison to the other species analyzed (table 1). Moreover, Campylobacter and Pseudomonas did not share any atypical patterns of codon usage that set them apart from the other species analyzed.
On the other hand, our results showed that Campylobacter and Pseudomonas shared a high proportion of genes with an unusually low πA/πS (fig. 2A). Moreover, Pseudomonas had by far the lowest overall mean πA/πS of the 12 taxa analyzed (fig. 1A). Thus, contrary to the usual interpretation of NI < 1 as evidence of positive selection favoring amino acid sequence divergence between species, our results suggested that in our data this statistic mainly reflected a low πA/πS within species rather than a high kA/kS.
This interpretation was strongly supported by partial correlation analyses showing that an unusually low value of πA was a significantly better predictor of a low NI than was an unusually high kA (table 2). The same pattern was seen both in species with close outgroups and in species with distant outgroups. Thus, it seemed unlikely that problems in estimating kS when synonymous sites were near saturation had a major impact on NI. The fact that, in the case of distantly related outgroups, there was a significant negative correlation between NI and a measure of unusual codon usage suggested that there may have been some problems with estimating kS between distantly related species, particularly in the case of genes with unusual patterns of codon usage. But this effect was quite minor in comparison with the strong positive relationship between πA and NI.
Factors that might cause relatively low πA in a given gene include the effects of purifying selection acting to eliminate or reduce in frequency slightly deleterious nonsynonymous variants. In order to examine purifying selection further, we analyzed synonymous and nonsynonymous polymorphisms in 3 species (B. pseudomallei, S. aureus, and S. pyogenes) for which we had sequences of at least 3 genomes with numerous polymorphic sites. All these species had a majority of genes with NI ≥ 1 but a substantial number with NI < 1 (fig. 1B). All 3 of these genomes showed an excess of rare nonsynonymous polymorphisms, indicative of ongoing purifying selection acting to eliminate slightly deleterious variants (Hughes et al. 2003; Hughes AL and Hughes MA 2007a, 2007b; Hughes and Piontkivska 2008; Irausquin and Hughes 2008). Moreover, the tendency toward rare nonsynonymous variants was strongest in genes with NI < 1.
Across the 12 taxa in our data set, there was a negative correlation between mean πA/πS and mean πS (fig. 1A). Taking πS as a proxy for average time since the common ancestor of a set of sequences, these results are consistent with those of Rocha et al. (2006) and support those authors’ hypothesis that this effect is due to a time lag in the removal of slightly deleterious nonsynonymous variants by purifying selection. In our data set, Pseudomonas showed by far the highest mean πS, the lowest mean πA/πS, and the highest proportion of genes with NI < 1.
Our results thus imply that the major factor in causing NI < 1 was not positive selection between species but effective purifying selection within species, lowering πA/πS and thus lowering NI. Moreover, certain results suggested that the MK test (G-test) is sensitive to the effectiveness of purifying selection acting to eliminate slightly deleterious nonsynonymous variants within populations. In the usual interpretation of the MK test, genes with NI < 1 and a significant G-test are considered to be subject to positive selection causing the fixation of nonsynonymous differences between species. However, we found an association in genes with NI < 1 between a significant G-test and a positive Qnon. Because a strongly negative Qnon characterizes a gene containing numerous nonsynonymous variants of low frequency, this result implies that the G-test is more likely to be significant when such low-frequency nonsynonymous variants are lacking. The absence of rare nonsynonymous polymorphisms evidently causes the level of nonsynonymous between-species divergence to be relatively large. Because rare nonsynonymous variants are often slightly deleterious (Hughes et al. 2003), their absence reflects the action of purifying selection. An ironic aspect of the MK test and related statistics thus appears to be that they mistakenly identify as positively selected between species the very genes that are most stringently negatively selected within species.
These considerations can explain, without resorting to the hypothesis of positive selection, the finding of lower NI in the mitochondrial genomes of invertebrates than of vertebrates (Bazin et al. 2006). On the assumption that invertebrates tend to have larger effective population sizes than vertebrates, this result is easily explained if low NI is generally due to purifying selection within a species reducing πA/πS because a species with a larger effective population size will in general be more efficient in eliminating slightly deleterious variants. The same reasoning may also explain the detection of more “adaptive evolution” by the MK test in regions of the Drosophila genome with normal recombination than in those with low recombination (Shapiro et al. 2007) because purifying selection is expected to be more effective in removing deleterious variants in the former regions than in the latter.
There are additional factors affecting NI that could not be addressed easily by our analyses. One such factor might be the fixation, during a population bottleneck, of slightly deleterious nonsynonymous variants, thereby increasing kA (Ohta 1993; Eyre-Walker 2002; Hughes et al. 2006; Hughes 2007). On the other hand, it is possible that given the presumably very large effective population sizes of the geographically widespread bacterial species analyzed here that no extreme bottleneck occurred in speciation. And of course, it is possible that certain nonsynonymous substitutions between species in our data set were indeed fixed by positive selection (Charlesworth and Eyre-Walker 2006). Indeed, if fixation of slightly deleterious mutations is a widespread phenomenon, one might expect it to be accompanied by a certain degree of positive selection favoring “compensatory mutations” that ameliorate the effect of deleterious alleles (Charlesworth and Eyre-Walker 2007; Sawyer et al. 2007).
Nonetheless, our results suggest that fixation of nonsynonymous substitutions between species—whether by drift or by positive selection—has a minor impact on the MK test and related methods. Rather, the strongest effect is that of ongoing purifying selection on within-species nonsynonymous polymorphism. Thus, our results suggest caution regarding the use of the MK test and related methods in testing for positive selection. Moreover, our results provide support for the nearly neutral theory of Ohta (1973) and its prediction that slightly deleterious variants play a significant role in evolution.
Supplementary Material
Supplementary tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health to A.L.H.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The rate of adaptive evolution in enteric bacteria. Mol Biol Evol. 2006;23:1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A. The other side of the nearly neutral theory, evidence of slightly advantageous back-mutations. Proc Natl Acad Sci USA. 2007;104:16992–16997. doi: 10.1073/pnas.0705456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. What are bacterial species? Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. Changing effective population size and the McDonald-Kreitman test. Genetics. 2002;162:2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil EJ, Holmes EC, Bessen DE, et al. (12 co-authors) Recombination within natural populations of pathogenic bacteria: Short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG. Recombination and the nature of bacterial speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA. Nonparametric statistical methods. New York: Wiley; 1973. [Google Scholar]
- Hughes AL. Evidence for abundant slightly deleterious polymorphisms in bacterial populations. Genetics. 2005;169:533–538. doi: 10.1534/genetics.104.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: The misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL, French JO. Homologous recombination and the pattern of nucleotide substitution in Ehrlichia ruminantium. Gene. 2007;387:31–37. doi: 10.1016/j.gene.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Patterns of sequence divergence in 5′ intergenic spacers and linked coding regions in 10 species of pathogenic Bacteria reveal distinct recombinational histories. Genetics. 2004;168:1795–1803. doi: 10.1534/genetics.104.032979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J Bacteriol. 2005;187:2698–2704. doi: 10.1128/JB.187.8.2698-2704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R, Glenn NL. The future of data analysis in evolutionary genomics. Curr Genomics. 2006;7:227–234. [Google Scholar]
- Hughes AL, Hughes MA. More effective purifying selection on RNA viruses than in DNA viruses. Gene. 2007a;404:117–125. doi: 10.1016/j.gene.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Hughes MA. Coding sequence polymorphism in avian mitochondrial genomes reflects population histories. Mol Ecol. 2007b;16:1369–1376. doi: 10.1111/j.1365-294X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Langley KL. Nucleotide usage, synonymous substitution pattern, and past recombination in genomes of Streptococcus pyogenes. Infect Genet Evol. 2007;7:188–196. doi: 10.1016/j.meegid.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Piontkivska H. Nucleotide sequence polymorphism in circoviruses. Infect Genet Evol. 2008;8:130–138. doi: 10.1016/j.meegid.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Packer B, Welsch R, Bergen AW, Chanock SJ, Yeager M. Widespread purifying selection at polymorphic sites in human protein-coding loci. Proc Natl Acad Sci USA. 2003;100:15754–15757. doi: 10.1073/pnas.2536718100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irausquin SJ, Hughes AL. Distinctive pattern of sequence polymorphism in the NS3 protein of hepatitis C virus type 1b reflects conflicting evolutionary pressures. J Gen Virol. 2008;89:1921–1929. doi: 10.1099/vir.0.2008/000992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer; 2007. [Google Scholar]
- Lynch M, Conery S. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lynch M, Bürger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- Majewski J. Sexual isolation in bacteria. FEMS Microbiol Lett. 2001;199:161–169. doi: 10.1111/j.1574-6968.2001.tb10668.x. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:114–116. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M, Jin L. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 1989;6:290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Ohta T. Amino acid substitution at the Adh locus of Drosophila is facilitated by small population size. Proc Natl Acad Sci USA. 1993;90:4548–4551. doi: 10.1073/pnas.90.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Excess amino acid polymorphism in mitochondrial DNA: Contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Maynard Smith J, Hurst LD, Holden MT, Cooper JE, Smith NH, Feil EJ. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Parsch J, Zhang Z, Hart DL. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. Proc Natl Acad Sci USA. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW. Molecular population genetics of sequence length diversity in the Adh region of Drosophila pseudoobscura. Genet Res. 2002;80:163–175. doi: 10.1017/s0016672302005955. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Shapiro J, Huang W, Zhang C, et al. (12 co-authors) Adaptive genic evolution in Drosophila genomes. Proc Natl Acad Sci USA. 2007;104:2271–2276. doi: 10.1073/pnas.0610385104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A. Adaptive evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. 2nd ed. New York: W.H. Freeman; 1981. [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods) Sunderland (MA): Sinauer; 2002. [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improvement of the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulić M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: dNA polymorphism and the control of genetic exchange in enterobacteria. Proc Natl Acad Sci USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam TS, Nei M. Neutral mutation hypothesis test. Nature. 1991;354:115–116. doi: 10.1038/354114e0. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


