Abstract
Summary: Massively parallel sequencing technologies hold incredible promise for the study of DNA sequence variation, particularly the identification of variants affecting human disease. The unprecedented throughput and relatively short read lengths of Roche/454, Illumina/Solexa, and other platforms have spurred development of a new generation of sequence alignment algorithms. Yet detection of sequence variants based on short read alignments remains challenging, and most currently available tools are limited to a single platform or aligner type. We present VarScan, an open source tool for variant detection that is compatible with several short read aligners. We demonstrate VarScan's ability to detect SNPs and indels with high sensitivity and specificity, in both Roche/454 sequencing of individuals and deep Illumina/Solexa sequencing of pooled samples.
Availability and Implementation: Source code and documentation freely available at http://genome.wustl.edu/tools/cancer-genomics implemented as a Perl package and supported on Linux/UNIX, MS Windows and Mac OSX.
Contact: dkoboldt@genome.wustl.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Massively parallel sequencing technologies have supplanted traditional capillary-based methods as the predominant approach to identifying sequence variation. The relatively low cost and incredible throughput of Roche/454, Illumina/Solexa and other platforms have made it possible to sequence complete human genomes to high coverage in a matter of months. The genomes of a Nobel laureate (Wheeler et al., 2008), anonymous African and Asian individuals (Bentley et al., 2008; Wang et al., 2008), and both tumor and normal cells of a cancer patient (Ley et al., 2008) have been fully sequenced; ambitious efforts like the 1,000 Genomes Project aim to extend whole genome sequencing (WGS) to a thousand or more individuals (http://www.1000genomes.org). New platforms have also enabled rapid sequencing and variant discovery for numerous plants (Ossowski et al., 2008), animals (Hillier et al., 2008) and microbes (Holt et al., 2008).
There is considerable interest in applying high-throughput sequencing to targeted regions of the genome (Harismendy et al., 2009), particularly to identify sequence variants affecting human disease. In such studies, samples may be pooled together and deeply sequenced to minimize costs. The extraordinary read depth from massively parallel targeted resequencing makes it feasible to detect rare variants and accurately estimate allele frequencies in pools containing many samples (Brockman et al., 2008).
Analysis of massively parallel sequencing data generally requires alignment to a reference sequence, a bioinformatics problem made particularly challenging by the relatively short read lengths and sheer volume of data. Many algorithms have been adapted or newly developed for short read alignment, including Maq (Li et al., 2008), Newbler (Roche), Bowtie (Langmead et al., 2009) and many others. Yet variant detection tools that make use of these alignments are often limited to a single platform or aligner. For example, Newbler's alignment and assembly tools, while powerful, are limited to the Roche/454 platform. The SHORE pipeline (Ossowski et al., 2008) calls SNPs and small indels, but uses its own alignment algorithm and operates only on Illumina/Solexa data. Very few analysis tools are compatible with multiple data and aligner types.
Here, we describe VarScan, an open source tool for detecting SNPs, insertions, and deletions and assessing their frequencies in massively parallel sequencing data. Unlike currently available variant detection tools, VarScan is compatible with several read aligners (BLAT, Newbler, cross_match, Bowtie and Novoalign), and calls variants in both individual and pooled samples.
2 METHODS
The VarScan package includes a complete workflow for variant detection from alignments of next-generation sequencing data (Supplementary Fig. 1). Given an alignments file, VarScan scores and sorts the alignments on a per-read basis, discarding reads that aligned with low identity or to multiple (ambiguous) locations in the reference sequence. Next, the single best alignment for each read is screened for sequence changes. Variants detected in multiple reads are then combined together into unique SNPs and indels. For each predicted variant, VarScan determines the overall coverage, as well as the number of supporting reads, average base quality, and number of strands observed for each allele. Thresholds for coverage, quality, variant frequency, and/or number of reads required to call a variant are set automatically with the easyrun command, but can be manually adjusted by the user. VarScan reports SNPs, insertions, and deletions with their chromosomal coordinates, alleles, flanking sequence, and read counts.
The software was implemented in Perl and inline C. The package installer, source code, and documentation are freely available for non-commercial use at http://genome.wustl.edu/tools/cancer-genomics.
3 RESULTS
We examined VarScan's performance on real data from targeted resequencing of 1000 PCR amplicons (∼250 kb) on the Roche/454 and Illumina/Solexa platforms. Forty-two samples were sequenced individually to ∼ 70 × coverage on the Roche 454 XLR platform. The same samples were also pooled together, and sequenced to ∼ 6000 × (∼125× per sample) on the Illumina GAII platform.
We aligned 454 reads (average length 230.2 bp) from individual sequencing to the Hs36 reference sequence with BLAT (v32x1). Reads that aligned with a score of <50, less than 95% identity, or to multiple competing locations in the genome were discarded by VarScan. SNPs were called at positions with >10× coverage where >25% of reads supported the variant allele (see Supplementary Data and Supplementary Fig. 1). For the pooled samples, we aligned Illumina reads (average length 36 bp) to the Hs36 reference sequence using Bowtie (v0.9.8) with the −m option, which returns only reads with a single best match in the genome. Average runtime was 1423 s per individual 454 sample and 1625 s per lane of Illumina/Solexa data (Supplementary Table 1).
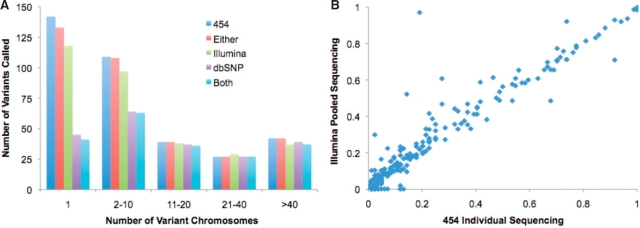
To evaluate sensitivity and specificity, we compared the SNP calls from individual 454 sequencing to those from pooled Illumina sequencing as well as dbSNP build 129 (Fig. 1A). We limited our analysis to positions with >10× 454 coverage in every individual and >100× Illumina coverage in the pool. Of 359 SNPs called in 454 data, 215 (59.78%) were present in dbSNP. VarScan detected 344 of the SNPs (95.82%) with Illumina data including 118 of 142 singleton heterozygotes (83%) present on just 1 of the 84 pooled chromosomes. Overall, 349 of 359 SNPs (97.21%) were either confirmed in the Illumina data or present in dbSNP.
Fig. 1.
Variant detection results from individual and pooled sequencing. (A) Overlap of variants called between 454, Illumina, and dbSNP build 129, by allele frequency in the sample pool. (B) Allele frequency from Illumina pooled sequencing versus known values from 454 individual sequencing.
To compare VarScan's performance with that of other tools, we also applied canonical variant calling algorithms to our sequencing datasets. Of the 359 SNPs detected by VarScan in 454 individual sequencing, Newbler identified 80 (22.28%) in the 454 data. Maq identified 340/359 SNPs (94.71%) in the Illumina/Solexa data, but only three passed Maq's SNPfilter (Supplementary Table 2).
To assess how well variant chromosomes were represented in the pooled samples, we compared the estimated allele frequency by Illumina read count to the known allele frequency from individual 454 sequencing results. We limited our analysis to 344 SNPs that were detected in both data sets (Supplementary Table 3). Variant allele frequency estimates from pooled sequencing tracked well with expected values from 454 results (Fig. 1B). Correlation between frequencies from each platforms was 0.962 (Pearson). At higher variant frequencies, estimates from pooled sequencing were slightly lower than expected, most likely because variant-containing short reads are at a mapping disadvantage during alignment to the reference sequence.
Detection of insertion/deletion (indel) variants remains a challenging area of bioinformatics, but the longer reads generated by 454 sequencing are well suited to this purpose. Even after filtering indel calls near homopolymers, VarScan predicted over 200 indels in the 454 data ranging in size from 1 to 97 bp. To validate these predictions, we isolated a set of 77 high-confidence indels that were small enough (1−5 bp) to be spanned by Illumina reads. Next, we remapped the Illumina reads with Novoalign, which aligns short reads across gaps in single-end mode, and predicted indels in the resulting alignments using VarScan. Of 77 high-confidence small indels (1−5 bp) from the 454 data, some 46 (59.74%) were also detected in Illumina/Solexa reads (Supplementary Table 4).
We have shown that VarScan efficiently calls variants with data from multiple sequencing platforms, in both individual and pooled samples. Comparisons between 454/Illumina data and dbSNP suggest 97% specificity in individual 454 data, and 93% sensitivity in pooled Illumina data. Even SNPs present in ∼1% of pooled chromosomes were called with 83% sensitivity. Of course, sensitivity and specificity of variant calls are dependent upon the accuracy of the sequence alignments provided to VarScan. For this reason, the VarScan documentation includes recommended parameters for every compatible aligner.
By accepting multiple data and aligner types, VarScan offers a central, platform-independent tool for variant detection. It can also detect variants at low (1%) frequencies, which is advantageous for sequencing of pooled samples. These capabilities, coupled with the incredible throughput of massively parallel sequencing platforms, offer a powerful system for large-scale targeted studies of genetic variation by deep resequencing.
Technologies for high-throughput sequencing and the algorithms for data processing continue to evolve. For this reason, we developed VarScan as an open-source, modular tool whose underlying functions can be easily expanded. We hope to make subsequent releases of the software that accommodate additional aligner outputs and data types. This aligner-independent, open-source approach to variant detection ensures that VarScan will continue to be useful to the research community.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephen Daiger and colleagues at the University of Texas HSC Houston; David Dooling, Scott Smith, Erica Sodergren, Bob Fulton and Rachel Abbott of the Washington University Genome Center; and the Foundation Fighting Blindness.
Funding: National Human Genome Research Institute [grant number HG003079, PI R.K.W.].
Conflict of Interest: none declared.
REFERENCES
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, et al. Whole-genome sequencing and variant discovery in C. elegans. Nat Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat. Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.