Abstract
Various lectins have attracted attention as potential microbicides to prevent HIV transmission. Their capacity to bind glycoproteins has been suggested as a means to block HIV binding and entry into susceptible cells. The previously undescribed lectin actinohivin (AH), isolated by us from an actinomycete, exhibits potent in vitro anti-HIV activity by binding to high-mannose (Man) type glycans (HMTGs) of gp120, an envelope glycoprotein of HIV. AH contains 114 aa and consists of three segments, all of which need to show high affinity to gp120 for the anti-HIV characteristic. To generate the needed mechanistic understanding of AH binding to HIV in anticipation of seeking approval for human testing as a microbicide, we have used multiple molecular tools to characterize it. AH showed a weak affinity to Manα(1–2)Man, Manα(1–2)Manα(1–2)Man, of HMTG (Man8 or Man9) or RNase B (which has a single HMTG), but exhibited a strong and highly specific affinity (Kd = 3.4 × 10−8 M) to gp120 of HIV, which contains multiple Man8 and/or Man9 units. We have compared AH to an alternative lectin, cyanovirin-N, which did not display similar levels of discrimination between high- and low-density HMTGs. X-ray crystal analysis of AH revealed a 3D structure containing three sugar-binding pockets. Thus, the strong specific affinity of AH to gp120 is considered to be due to multivalent interaction of the three sugar-binding pockets with three HMTGs of gp120 via the “cluster effect” of lectin. Thus, AH is a good candidate for investigation as a safe microbicide to help prevent HIV transmission.
Keywords: action mechanism, anti-HIV, gp120, high-mannose type glycan, cyanovirin-N
The global public health community has become acutely aware of the difficulties and obstacles that still remain in the path of HIV vaccine development (1). To find alternative means of blocking HIV infection, multiple groups (such as the Alliance for Microbicide Development) have promoted research on compounds that can be formulated for vaginal or rectal delivery, and which would bind to the infectious virus to block its association with cellular receptors. These compounds, termed microbicides, offer the tantalizing promise of a new strategy for global public health intervention to prevent AIDS. Unfortunately, human clinical trials have so far failed to demonstrate efficacy of broad-spectrum microbicides against vaginal HIV-1 transmission (2). The failure in human trials has been stated to be, at least in part, a lack of underpinning scientific understanding of how candidate microbicides work (2). The current study was conducted to provide a detailed basis for the mode of action of actinohivin (AH) before its entry into human clinical trials.
The HIV-1 envelope, which is responsible for both receptor binding and membrane fusion that permit viral entry into susceptible cells, consists of a gp120 surface subunit that is noncovalently associated with a gp41 membrane-spanning subunit. In the first step of viral entry, the surface subunit of gp120 binds to the target cell and determines viral tropism through interaction with the cell surface receptor CD4, and is one of several coreceptors that are members of the chemokine receptor family (3).
The HIV-1 gp120 is highly glycosylated, and half of its molecular weight is due to its carbohydrate side chain (glycan). As an example, the recombinant gp120 produced in CHO cells was found to contain 13 complex-type glycans and 11 high-mannose (Man) or hybrid-type glycans (4). Such a high-density, clustering oligomannose structure as that present on HIV-1 gp120 has not been found in mammalian glycoproteins or envelope proteins of different virus families. Therefore, a specific oligomannose-binding agent that binds only to glycoproteins with a clustering oligomannose structure may be a specific microbicide (virucide) for prevention of HIV transmission and infection by blocking viral entry into the cell.
We previously sought HIV entry inhibitors using a syncytium formation assay that uses T cell line-tropic and macrophage-tropic HIV env expressed cell lines (5), and found a previously undescribed anti-HIV lectin, AH, in a cultured actinomycete, Longispora albida K97-0003T (6–8). AH exhibits potent anti-HIV activity against various strains of T-tropic and M-tropic HIV-1 and HIV-2 (IC50 = 2–110 nM) and inhibits viral entry to cells by binding to the high-Man type glycans (HMTGs) of gp120 (9). AH has a unique sequence consisting of 114 aa and a highly conserved internal sequence triplication (comprising amino acids 1–38, 39–77, and 78–114; segments 1, 2, and 3, respectively) (7). These three segments of AH are necessary for potent anti-HIV activity (10).
In recent years, carbohydrate-binding agents have received specific attention as microbicides, because their binding to HIV could prevent the initial step in viral infection by blocking association with cell receptors. Several plant lectins with specificity for Man and/or N-acetylglucosamine have been reported to possess favorable properties to qualify as potential microbicidal drugs (11, 12).
Also, several carbohydrate-binding agents of prokaryotic origin or from invertebrates have been isolated and characterized. Among them, cyanovirin (CV)-N, an 11-kDa protein derived from the cyanobacterium Nostoc ellipsosporum, has received the most attention and has already been thoroughly investigated to determine its structural properties, carbohydrate-binding potential, and antiviral activity (13–19). CV-N was recently shown to inhibit SHIV infection in a vaginal transmission model (female Macaca fascicularis infected with a pathogenic recombinant chimerical SIV 89.6P), which suggests that CV-N is a good candidate for testing in humans as an anti-HIV topical microbicide (20). Therefore, we aimed to clarify which structures of HMTGs of gp120 are necessary for AH binding, and compared the specificities of AH and CV-N. The investigation was performed by ELISA and analysis with a resonance mirror biosensor and a frontal affinity chromatography (FAC) system, which enabled us to analyze lectin–oligosaccharide interactions in a high-throughput manner (21, 22). Consequently, we found that target units of AH are a HMTG with Manα(1–2)Man units, that AH exhibits a very specific, high affinity solely to glycoproteins having many HMTGs (unlike CV-N), and that AH is much more specific to HIV than CV-N.
Also, X-ray crystal analysis revealed the 3D structure of AH, which contains three similar sugar-binding pockets.
Results
AH Recognizes α(1–2)Man Residues of N-Linked Glycan on the Surface of gp120.
As shown in a previous article (9), AH–gp120 binding is strongly inhibited by yeast mannan (IC50 = 3.0 μg/mL), an α-type mannan, but not by coffee mannan, a β-type mannan, or many other saccharides tested, including monosaccharides and disaccharides. AH binds to glycoproteins having N-linked HMTGs, such as thyroglobulin, but not to other glycoproteins having hybrid or complex type N-linked glycans alone. To investigate the saccharide structure recognized by AH, we examined the effects of mannosaccharides, which are commonly contained in the structures of yeast mannan, and a HMTG on AH–gp120 binding by ELISA.
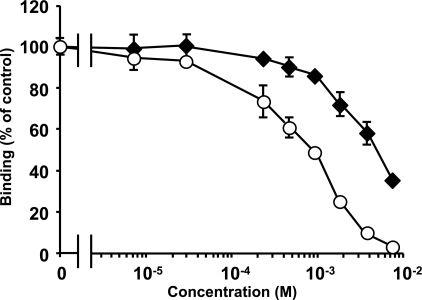
The results (Fig. 1) showed that Manα(1–2)Man and Manα(1–2)Manα(1–2)Man exhibit low inhibitory activities (IC50 = 5.1 and 0.92 mM, respectively) but Man, Manα(1–3)Manα, Manα(1–6)Man, and Manα(1–3)Manα (1–6)Man(1–6)Manα(1–3)Man showed no inhibition, even at 10 mM.
Fig. 1.
Inhibition of AH binding to gp120 by α(1–2)Man oligomers. Open circle indicates Manα(1–2)Manα(1–2)Man, and solid square indicates Manα(1–2)Man.
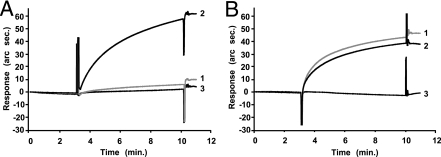
We then tested whether Manα(1–2)Man residues without a reducing end of N-linked HMTG participated in binding of AH to gp120. After the purified gp120 was digested with glycosidase, AH–gp120 binding activity was checked with an IAsys resonant mirror biosensor. HIV gp120 contains many N-linked glycans, including complex, hybrid, and HM types, among which endoglycosidase H (Endo H) digests both HM and hybrid types. When purified gp120 was treated with Endo H, the molecular mass of the treated gp120 decreasing to ≈80 kDa, with the AH binding activity also decreasing to a control level. Also, when gp120 was treated with 1,2α-mannosidase, which specifically removes the nonreducing end of 1,2α-linked Man residues, but not those of 1,3α- or 1,6α-linked Man residues, AH interaction with the treated gp120 dramatically decreased to a background level (Fig. 2A). Conversely, when gp120 was treated with sialidase or β-galactosidase, both of which digest hybrid- and complex-type sugar chains of gp120, no effect on AH–gp120 binding was observed (Fig. 2B).
Fig. 2.
AH-binding activity of gp120 after being treated with glycosidase. gp120 was treated with or without 1,2α-mannosidase from Aspergillus saltol (A), sialidase, or β-galactosidase (B). Association and dissociation curves for the interaction of gp120 treated or untreated with glycosidase to AH-immobilized carboxylate cuvette for IAsys: 1, with glycosidase; 2, without glycosidase; and 3, glycosidase alone without gp120. When treated with sialidase, a similar profile to that of β-galactosidase was observed.
Some decrease in molecular mass of gp120 was observed after digestion with 1,2α-mannosidase, sialidase, and β-galactosidase. These results strongly indicate that AH recognizes α(1–2)Man residues of N-linked HMTGs on the surface of gp120.
Analysis of Carbohydrate-Binding Properties of AH by FAC.
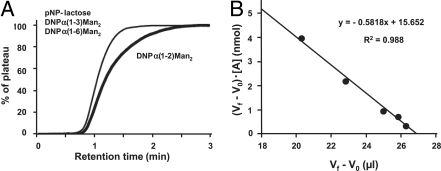
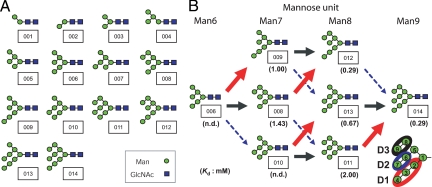
AH was immobilized on NHS-activated Sepharose, and the resulting resin was packed into a miniature column, as described in Materials and Methods. For evaluation of the AH column, it was necessary to determine the effective ligand content (Bt). For this purpose, we used 2,4-dinitrophenyl (DNP) derivative [DNP-α(1–2)Man2] of Manα(1–2)Man, which possesses AH-binding affinity as described above. Concentration dependence analysis was performed with DNP-α(1–2)Man2 at various concentrations ranging from 5 to 200 μM. As a result, Bt and Kd values were determined to be 15.7 nM and 0.58 mM, respectively, although the Kd value was relatively large, which is discussed below (Fig. 3). DNP-α(1–3)Man2 and DNP-α(1–6)Man2 had no affinity toward the AH-binding column, which coincided with the results obtained by ELISA. We next tested affinities of various types of HMTGs to the AH-immobilized column. HMTGs and their Kd values are shown in Fig. 4. The relatively strong affinities were obtained for 014 and 012 (designated Man9 and Man8, Kd = 2.9 × 10−4 and 2.1 × 10−4 M, respectively), followed by 013 (6.7 × 10−4 M), 009 (1.0 × 10−3 M), 008 (1.4 × 10−3 M), and 011 (2.0 × 10−3 M). However, 001-007 and 010 showed no affinity to the AH column. These results showed that among three domains (D1–D3) of α(1–2)Man structures of HMTG, the Μanα(1–2)Manα(1–2)Man structure (D1) is the most effective, followed by D3 and D2, and that the combination of D1 and D3 (or D2) in Man8 and Man9 are necessary for high affinity to AH (Fig. 4). Consequently, among three kinds of Man8, Man8 (D1D3) having D1 and D3 domains has the highest affinity.
Fig. 3.
Analysis for Bt value determination of AH column. For evaluation of AH column, DNP-α(1–2)Man2 was diluted to various concentrations (5–200 μM) and applied to an AH-immobilized column. The thick and thin lines indicate elution profiles of DNP-α(1–2)Man2 and control sugar (pNP-lactose), respectively (A). Woolf–Hofstee type plots were made by using V − V0 values (B). For details, see main text.
Fig. 4.
Analysis of affinity of AH to α(1–2)Man glycans. (A) HMTGs used for FAC analysis. For numbering of glycans in square boxes, see ref. 22. (B) Structures of HMTGs and Kd values of AH toward the glycans. Figures in parentheses indicate Kd values. ND, no affinity to AH was detected. D1, Manα(1–2)Manα(1–2)Man red; D2 and D3, Manα(1–2)Man blue and black, respectively.
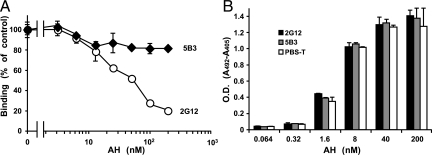
Effect of AH on 2G12-gp120 Binding.
The antibody 2G12 is a broadly neutralizing human monoclonal antibody against HIV-1. The epitope of 2G12 might consist of several α(1–2)Man linked moieties contributed by the HMTGs that form a unique cluster on HIV-1 gp120 at sites N295, N332, and N392 (23, 24). To confirm whether AH affects 2G12 binding to gp120, 2G12 or AH was added to a gp120-coated plate that was pretreated with AH or 2G12, and the amounts of 2G12 or AH bound to gp120 were measured by ELISA. We found that the 2G12 binding to gp120 was inhibited by AH in a concentration-dependent manner, but another gp120 antibody, 5B3, which recognizes gp120 peptides at the C1 region of N-terminus, was not inhibited by AH (Fig. 5A). Contrastingly, the AH binding to gp120 was not inhibited even by 1.0 μg/mL 2G12, which was the maximum concentration added to the gp120-coated ELISA plate (Fig. 5B). These results indicate that AH can bind to the binding sites of 2G12 on gp120, as well as to HMTGs other than at the 2G12 binding sites.
Fig. 5.
Effects of AH on gp120-gp120 antibody binding (A) and of gp120 antibody on AH–gp120 binding (B).
Comparative Studies of Binding Affinities to Glycoproteins Between AH and CV-N.
We previously reported that AH shows affinity to glycoproteins, such as bovine RNase B and bovine thyroglobulin, which have only one and four HMTG(s), respectively, but that those binding affinities are much lower than to gp120 (9). However, CV-N (13), an anti-HIV lectin, binds strongly to N-linked HMTGs of gp120 (14, 15) and to RNase B. To clarify how the mechanism of binding specificity of AH differs from that of CV-N, we analyzed the affinity of AH and CV-N with gp120 or RNase B using a resonant mirror biosensor (IAsys). A carboxylate cuvette surface was coated with gp120 or RNase B, and the binding of AH or CV-N was monitored in real time with various concentrations of AH or CV-N. Table 1shows a comparison between AH and CV-N in affinity to glycans and glycoproteins. AH and CV-N interacted strongly with HIV-1NL4-3 gp120 and HIV-1SF162 gp120 (SI Materials and Methods); their Kd values were 1.7 × 10−9 ≈ 3.4 × 10−8 M. CV-N also interacted strongly with RNase B and had a similar Kd value to those of gp120 and Man9. The affinities of AH to glycans were weak (Table 1), as described above, and an AH interaction with RNase B was not detected. These data indicate that CV-N can bind strongly to a free HMTG and to a glycoprotein with a single HMTG. However, AH binds strongly only to glycoproteins having many HMTGs, such as HIV-1 gp120. These results were confirmed by the ELISA experiments using various glycoproteins that have different numbers of HMTGs (Fig. S1).
Table 1.
Comparison between AH and CV-N in affinity to glycans and glycoproteins
Crystal Structure of AH.
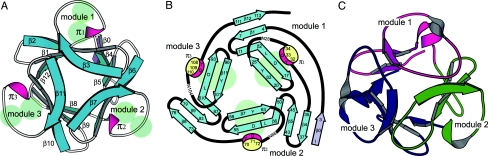
In this study, before X-ray analysis, the tertiary structure of AH has been predicted by the computer modeling program FAMS (25) using the small domain of xylanase from Streptomyces olivaceoviridis (26), to which AH has high sequence homology (30% identical and 40% similar). In parallel, the crystal structure of AH has also been determined at high (1.2 Å) resolution (SI Materials and Methods and Table S1). The two proteins found in the asymmetric unit of the crystal adopt a similar structure so that a rmsd between the corresponding Cα atoms is only 0.17 Å when superimposed on each other. The overall structure of AH revealed is shown in Fig. 6A. The structure strongly reinforces the accuracy of the predicted structure. Both structures support our previous discussions (6, 9, 10) and the above results.
Fig. 6.
Structural features of AH. (A) Tertiary structure of AH, (B) secondary-structure topology, and (C) tertiary structure of the small domain of xylanase (26). (B) Ellipsoids colored in light green indicate the three Man-binding pockets composed of the LD-QXW motifs. Arrows and disks show β-strands and π-helices, respectively, with the amino acid sequence numbers.
The structure appears composed of three modules that are similar to each other, as speculated from the tandem repeats in the sequence. However, the folding is not so simple. Although each module contains a β-sheet with four antiparallel strands and a short 310 helix (π), the module 2 has an additional β-strand.
The secondary-structure topologies are slightly different between the modules. As seen in Fig. 6B, the N-terminal β-strand (β0) starts in the module 2. The subsequent three β-strands (β1, β2, and β3), a long loop (L), and a π-helix form the module 1, which is followed by the second module (β4, β5, β6, β7, L, and π2) and the third module (β8, β9, β10, β11, L, and π3). The last β12-strand completes the first module 1 so as to close the cyclic assembly. Only the module 2 is structurally stabilized by forming a larger β-sheet with the five β-strands. In the xylanase small domain (26) shown in Fig. 6C, three long Ls with short α-helices are inserted between β2 and β3, between β6 and β7, and between β10 and β11, respectively.
The three modules are associated with a pseudo 3-fold symmetry, in which the three β-sheets form a triangular barrel. Inside the barrel, hydrophobic residues form a stable core. On the outer surface, a long L with a π-helix of each module is running from the top to the bottom of the barrel. This L and the preceding β-strands containing the carbohydrate-binding motifs LD-QXW form a valley with a pocket for carbohydrate-binding, except W, which participates in the central-core formation. The size of a valley is ≈10 Å in length, 15 Å in width, and 3 Å in depth, which creates enough space to accept two units of Manα(1–2)Man of an HMTG. The end of the D1 branch (Man-4; for the Man numbering system, see Fig. 4) could be accommodated in a pocket, and the end of D2 or D3 (Man-7 or Man-9) might contact with a valley. Therefore, it is possible to speculate that each carbohydrate-binding pocket of the three modules of AH accepts the D1 and D2/D3 branches of Man8 and Man9; the combination of D1 and D3 may be preferred, because both are closely located in their conformations (17, 27, 28). The three pockets are located to form an almost regular triangle at a distance of 17 Å between the aspartate residues in the three conserved motifs on AH. This separation might allow acceptance of three HMTGs on a gp120 at the same time.
Discussion
As reported previously, AH is an anti-HIV lectin of CBM family 13, which has three segments, each containing a sugar-binding pocket (three LD-QXW motifs) (29, 30).
AH binds to HMTGs on gp120, and the binding requires all three segments of AH (9, 10). The QXW motif mutants of AH, His-AH(Q33A), His-AH(Q71A), and His-AH(Q109A), in which glutamine residues were substituted into alanine, showed much reduced anti-syncytium formation activities (1/20, 1/10, and 1/30, respectively) compared with that of His-AH (10). In the present article, it has been shown by analyses of the affinity of AH to various HMTGs with ELISA (Fig. 1), resonant mirror sensor IAsys (Fig. 2), and FAC (Figs. 3 and 4) that AH targets Manα(1–2)Manα(1–2)Man of the D1 arm and Manα(1–2)Man of the D3 arm of the HMTGs Man8(D1D3) and Man9 on gp120, indicating that the target of AH resembles that of CV-N (17).
A comparison between AH and CV-N affinities to glycans and glycoproteins (Table 1) revealed several differences. CV-N showed strong affinities to both HMTGs and glycoproteins having a HMTG(s) including RNase B. However, AH showed a weak or no affinity to HMTG(s) and RNase B, but showed a strong affinity to gp120.
In summary, AH exhibits a strong affinity only to glycoproteins with many HMTGs, whereas CV-N can bind to all glycoproteins having Man8(D1D3) or Man9, regardless of the number of glycans on a glycoprotein. CV-N contains two carbohydrate binding sites that binds to Man8(D1D3) and Man9 with nanomolar affinity (17), and a possible cross-linking binding model for CV-N and trimeric gp120 has been proposed (31). Consequently, AH has a much more specific affinity to glycoproteins with many HMTGs than CV-N.
Also, although CV-N proved effective in preventing SHIV transmission in macaques in the absence of toxic side-effects (20), CV-N can also cause a broad variety of side effects such as a cytopathic effect and mitogenic activity (32, 33). AH showed no cytotoxicity, even at 100 μM in MT-4 cells (6). It is considered that the basis of the side effects of CV-N may be due to its strong binding to a single HMTG in contrast with AH, or by protein–protein interaction of CV-N with cellular factions.
As described above, the crystal structure (Fig. 6A) of AH showed that AH has three sugar-binding pockets that are constructed by the LD-QXW motifs. The ends of D1 and D3 branches of Man8(D1D3) and Man9 could be bound in the pocket and its nearby valley. It appears that the amino acid sequence of the motif determines the kind of glycans that lectin binds to. Interestingly, amino acid homology of three LD-QXW motifs of AH is extremely high among known lectins of CBM 13. Seventeen of 22 aa (77%) in the LD-QXW motif are conserved. Practically, the structures of the three pockets are very similar to each other with high symmetry. In other words, it suggests that these pockets have an almost equal binding affinity to Mans in particular. However, in the small domain of xylanase of Streptomyces olivaceoviridis E-86 (26) and the ricin B chain of Ricinus communis (34), which belong to CBM 13, only 41 and 3% have conserved between the three LD-QXW motifs, respectively. Therefore, the three pockets of the small domain are deformed, suggesting that they are not equivalent to each other, as shown in Fig. 6C. Consistently with the structural differences, the specificities of these lectins to sugars are not very high; for example, the xylan-binding domain accepts glucose, galactose, and lactose in addition to xylose and xylooligosaccharides (26). The extremely high specificity of AH to HMTGs is likely due to the high homology of the three-pocket structures. Consequently, the binding of D1 and D3 domains of three HMTGs on one molecule of gp120 to the three equivalent sugar-binding pockets of AH might underlie the highly specific, strong affinity via the so-called cluster effect of lectin (35).
Human monoclonal antibody 2G12 against HIV-1 recognizes a unique oligomannose cluster that is mainly contributed by the HMTG at glycosylation sites N295, N332, and N392 (23, 24). The results depicted in Fig. 5 indicate that AH can also bind to HMTGs other than the epitope of 2G12. Incidentally, the inhibitory activity of AH against gp120-soluble CD4 binding is very weak (IC50 = 1.2 mM by ELISA) (6). It seems to show that HMTGs are not involved in gp120 binding to CD4 on susceptible cells, and that AH interferes with the entry steps after gp120-CD4 binding occurs. The Man specific lectins from Galanthus nivalis and Hippeastrum hybrid also do not interfere with HIV binding to CD4 (36).
A typical HIV-1 gp120 is highly glycosylated and has many HMTGs. Although a Man oligosaccharide moiety exists in some human glycoproteins, a high-density, clustering oligomannose structure similar to the one present on gp120 has not yet been found in any other human glycoproteins. Therefore, it seems that AH does not bind to such human glycoproteins. The above results support AH being developed as a potential topical microbicide to prevent HIV transmission and infection.
Materials and Methods
Materials.
AH was prepared from a cultured broth of L. albida K97-0003T as described previously (4). Manα(1–2)Man, Manα(1–3)Man, Manα(1–6)Man, Manα(1–3)Manα(1–6)Manα(1–6)Manα(1–3)Man, bovine thyroglobulin, and bovine RNase B were purchased from Sigma. Manα(1–2)Manα(1–2)Man was donated by Y. Ito and I. Matsuo (RIKEN Advanced Science Institute). CV-N and its polyclonal antibody were a gift from M. R. Boyd (University of South Carolina, Columbia, SC). Pyridylamine (PA)-labeled Man type N-glycans (001-014) were purchased from Takara and Seikagaku Kogyo. DNP oligosaccharides, DNP-α-d-Manp-α (1–2)d-Manp (DNP-α(1–2)Man2), DNP-α-d-Manp-α (1–3)d-Manp (DNP-α(1–3)Man2), and DNP-α-d-Manp-α (1–6)d-Manp (DNP-α(1–6)Man2) were generous gifts from T. Nakamura (Obihiro University of Agriculture and Veterinary Medicine, Obihiro, Japan). Anti-AH polyclonal antibody was prepared as previously described (9). Human anti-HIV-1 gp120 mAb 2G12 was provided by Hermann Katinger through the National Institutes of Health (NIH) AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: HIV-1 gp120 mAb 2G12. Anti-HIV gp120 antibody 5B3 and HIVΙΙΙΒgp120 were generous gifts from Genentech.
Effects of Man Oligomers on AH–gp120 Interaction.
We used the ELISA method as follows. Each well of a 96-well protein-adsorbing plate (Maxisorp Immunoplate; Nalge NINC) was treated with 50 μL of AH solution (3.0 μg/mL) for 18 h at 4 °C. The plate was blocked with 3.0% nonfat dry milk (50 μL) for 1 h, washed with PBS (10 mM phosphate buffer, 2.7 mM potassium chloride and 137 mM sodium chloride, pH 7.4) containing 0.05% Tween 20 (PBS-T), and incubated for 2 h with a solution containing 0.18 μg/mL sgp120IIIB and various concentrations of Man oligomers [Manα(1–2)Man,Manα(1–3)Man, Manα(1–6)Man, Manα(1–2)Manα(1–2)Man, and Manα(1–3)Manα(1–6)Manα(1–6)Manα(1–3)Man]. Each well was rinsed four times with PBS-T.
A solution (50 μL) of anti-gp120 monoclonal antibody 5B3 (0.3 μg/mL) was added to the wells and incubated at room temperature for 30 min, followed by washing with PBS-T three times.
A solution (50 μL) of HRP-conjugated anti-mouse IgG (1/2,000 dilution) was added to each well, and the plate was incubated at room temperature for 30 min. The plate was washed four times with PBS-T, followed by incubation with 100 μL of o-phenylenediamine dihydrochloride (OPD) solution (0.45 mg/mL OPD/16 mM citrate/50 mM disodium hydrogenphosphate, pH 5.0) for 8 min at room temperature. The color reaction was stopped by adding 2.0 M sulfuric acid, and the optical density was measured at 405 nm.
Glycosidase Digestion.
Endo H, 1,2α-mannosidase (from Aspergillus saitoi), and β-galactosidase were purchased from Seikagaku, and sialidase was purchased from Roche Diagnostics. The gp120 from HeLa/T-env/Tat cells was digested with a glycosidase in a glycosidase digestion buffer as specified by the manufacturer.
Assay of Protein–Protein Interaction Activity Using a Resonant Mirror Biosensor.
Analysis of protein–protein interaction was performed using an IAsys resonant mirror biosensor (Affinity Sensors). For preparation of an AH-coated cuvette, we used dual-well IAsys amino cuvette. Polymerized glutaraldehyde (PG) was prepared by taking 5.0 mL of 5% (vol/vol) glutaraldehyde, adding 500 μL of 0.1 M NaOH, and leaving it for 30 min to polymerize before neutralizing with 500 μL of 0.1 M HCl [final 4.2% (vol/vol) PG]. A dual-well IAsys amino cuvette was washed with 10 mM sodium phosphate (PB, pH 7.7) and activated with PG for 30 min. One well was blocked with BSA as a control surface. The other one was treated with 10 μL of 80 μM solution of AH in PB (0.24 ng of AH bound/mm2). A gp120 or RNase B-coated cuvette was prepared using a dual-well IAsys carboxylate cuvette. The IAsys carboxylate cuvette was activated with a mixture of 0.2 M N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride and 0.05 M NHS as described in IAsys protocols. The purified gp120 or RNase B was immobilized on a well of the cuvette (4.13 ng/mm2 of HIV-1NL4-3 gp120, 4.88 ng/mm2 of HIV-1SF162 gp120, or 4.4 ng/mm2 of RNase B). The other well of each cuvette was blocked with β-casein as a control surface. Remaining activated groups were blocked with 1.0 M 2-aminoethanol (pH 8.5). Binding reaction was carried out in PBS (total volume of 50 μL) containing 0.02% Tween20 at 25 °C.
Functional Analysis by FAC.
Kd of AH for PA-labeled HMTGs (001-014; Fig. 4) were determined by FAC (21, 22). All FAC data were obtained in 10 mM Tris-Cl buffer, pH 7.4, containing 0.7% NaCl (TBS) at a flow rate of 0.125 mL/min with the FAC-1 machine, which had been specially designed and manufactured by Shimadzu. Purified AH (1.0 mg) was immobilized on 100 μL of NHS-activated Sepharose resin (Amersham Bioscience). The slurry of the AH-immobilized Sepharose was packed into a capsule-type miniature column (inner diameter, 2 mm; length, 10 mm; bed volume, 31.4 μL). The resulting column was slotted into a stainless steel holder, and connected to the FAC-1 machine. For the determination of effective ligand content, Bt of the AH-immobilized column, concentration-dependence analysis and subsequent Woolf–Hofstee type plots were performed. Here, various concentrations ([A]0) of DNP-α(1,2)Man2 dissolved in TBS were applied to the miniature column, and the elution was monitored by absorbance at 304 nm. Woolf–Hofstee type plots, i.e., (Vf – V0) vs. [A]0 · (Vf − V0), were made to determine Bt and Kd values from the intercept and the slope, respectively, of the fitted curve (Fig. 3). Vf is the elution volume of A and V0 is the elution volume of pNP-β-lactose, which has no affinity toward AH.
Next, PA-labeled HMTGs were applied to the column, and the elution was monitored by fluorescence (excitation and emission wavelength 320 and 400 nm, respectively). As in the assays used in this study, V0 is the elution volume of PA-lactose, which has no affinity toward AH and [A]0, the initial concentration of the PA-labeled HMTGs of interest (A), was 2.5 nM, which was negligible compared with Kd. Thus, Vf approached its maximum value, which was independent of [A]0, and we used the following equation to obtain the values of Kd of AH toward HM type N-glycan derivatives.
Effects of AH on Human mAb 2G12-gp120 Binding and of 2G12 on AH–gp120 Binding.
Each well of a 96-well protein-adsorbing plate was coated with purified HIV-1NL4-3 gp120 for 18 h at 4 °C and then washed three times with PBS-T. Various concentrations of AH were added to the gp120-coated wells and held for 1 h at room temperature. Each well was then washed four times with PBS-T.
Human anti-HIV-1 gp120 mAb 2G12 (1.0 μg/mL) or anti-HIV-1 gp120 antibody 5B3 (as a control) was added to the appropriate wells and incubated for 1 h at room temperature. The plate was washed four times with PBS-T, and then antibody bound to gp120 was detected, as described above. To determine whether AH–gp120 binding is inhibited by 2G12 antibody, the gp120-coated plate was next incubated with 1.0 μg/mL 2G12 or 5B3 antibody for 1 h. The plate was washed four times with PBS-T and various concentrations of AH were added to the wells. After incubation for 1 h, the plate was washed with PBS-T four times and AH bound to gp120 was detected by anti-AH polyclonal antibody (9).
Supplementary Material
Acknowledgments.
We thank Dr. M. R. Boyd (University of South Carolina, Columbia, SC) for generous gifts of samples of CV-N and its polyclonal antibody; Genentech Inc. for generous gifts of HIV-1IIIB gp120 and anti-gp120 mouse mAb 5B3; Ms. E. Kanda and Mr. T. Sagara for their helpful assistance; and Drs Y. Yamada, N. Matsugaki, and N. Igarashi (Photon Factory, Tsukuba, Japan) for their assistance in data collection with the synchrotron facility. HIV-1 gp120 mAb 2G12 was obtained through the National Institutes of Health AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH, courtesy of Dr. Hermann Katinger. This work was supported by Japan Ministry of Education, Culture, Sports, Science, and Technology grants from the 21st Century Program (to H.T. and S.Ō.) and the Grant-in-Aid for Young Scientists B, 14771313, 2004 (to H.C.); the Japan Ministry of Health, Labor, and Welfare Research on Japan Health Science Foundation on Drug Innovation (H.T.); and the Iwaki City, Japan Strategic Industry Producing Project (H.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3A07).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907572106/DCSupplemental.
References
- 1.Cohen J. Did Merck's failed HIV vaccine cause harm? Science. 2007;318:1048–1049. doi: 10.1126/science.318.5853.1048. [DOI] [PubMed] [Google Scholar]
- 2.Grant RM, et al. Whither or wither microbicides? Science. 2008;321:532–534. doi: 10.1126/science.1160355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapham PR, McKnight A. Cell surface receptors, virus entry and tropism of primate lentiviruses. J Gen Virol. 2002;83:1809–1829. doi: 10.1099/0022-1317-83-8-1809. [DOI] [PubMed] [Google Scholar]
- 4.Leonard CK, et al. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type-1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 5.Chiba H, et al. A simple screening system for anti-HIV drugs: Syncytium formation assay using T-cell line tropic and macrophage tropic HIV env expressing cell lines—establishment and validation. J Antibiot. 2001;54:818–826. doi: 10.7164/antibiotics.54.818. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, et al. Actinohivin, a novel anti-HIV protein from an actinomycete that inhibits syncytium formation: Isolation, characterization, and biological activities. Biochem Biophys Res Commun. 2001;282:595–601. doi: 10.1006/bbrc.2001.4495. [DOI] [PubMed] [Google Scholar]
- 7.Inokoshi J, et al. Molecular cloning of actinohivin, a novel anti-HIV protein from an actinomycete, and its expression in Escherichia coli. Biochem Biophys Res Commun. 2001;281:1261–1265. doi: 10.1006/bbrc.2001.4496. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto A, et al. Longispora albida gen nov, sp nov, a novel genus of the family Micromonosporaceae. Int J Syst Evol Microbiol. 2003;53:1553–1559. doi: 10.1099/ijs.0.02595-0. [DOI] [PubMed] [Google Scholar]
- 9.Chiba H, et al. Actinohivin, a novel anti-human immunodeficiency virus protein from an actinomycete, inhibits viral entry to cells by binding high-mannose type sugar chains of gp120. Biochem Biophys Res Commun. 2004;316:203–210. doi: 10.1016/j.bbrc.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi A, et al. Essential regions for antiviral activities of actinohivin, a sugar-binding anti-human immunodeficiency virus protein from an actinomycete. Arch Biochem Biophys. 2005;437:233–240. doi: 10.1016/j.abb.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Balzarini J. Targeting the glycans of glycoproteins: A novel paradigm for antiviral therapy. Nat Rev Microbiol. 2007;5:583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balzarini J. Targeting the glycans of gp120: A novel approach at the Achilles heel of HIV. Lancet. 2005;5:726–731. doi: 10.1016/S1473-3099(05)70271-1. [DOI] [PubMed] [Google Scholar]
- 13.Boyd MR, et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: Potential applications to microbicide development. Antimicrob Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shenoy SR, et al. Selective interactions of the human immunodeficiencyvirus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J Pharmacol Exp Ther. 2001;297:704–710. [PubMed] [Google Scholar]
- 15.Bolmstedt AJ, et al. Cyanovirin-N defines a new class of antiviral agent targeting N-linked, high-mannose glycans in an oligosaccharide-specific manner. Mol Pharmacol. 2001;59:949–954. doi: 10.1124/mol.59.5.949. [DOI] [PubMed] [Google Scholar]
- 16.Bewley CA. Solution structure of a cyanovirin-N:Man alpha 1–2Man alpha complex: Structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure. 2001;9:931–940. doi: 10.1016/s0969-2126(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 17.Bewley CA, Otero-Quintero S. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man(8) D1D3 and Man(9) with nanomolar affinity: Implications for binding to the HIV envelope protein gp120. J Am Chem Soc. 2001;123:3892–3902. doi: 10.1021/ja004040e. [DOI] [PubMed] [Google Scholar]
- 18.Bewley CA. Rapid validation of the overall structure of an internal domain-swapped mutant of the anti-HIV protein cyanovirin-N using residual dipolar couplings. J Am Chem Soc. 2001;123:1014–1015. doi: 10.1021/ja005714o. [DOI] [PubMed] [Google Scholar]
- 19.Bewley CA, Kiyonaka S, Hamachi I. Site-specific discrimination by cyanovirin-N for alpha-linked trisaccharides comprising the three arms of Man(8) and Man(9) J Mol Biol. 2002;322:881–889. doi: 10.1016/s0022-2836(02)00842-2. [DOI] [PubMed] [Google Scholar]
- 20.Tsai CC, et al. Cyanovirin-N inhibits AIDS virus infections in vaginal transmission models. AIDS Res Hum Retroviruses. 2004;20:11–18. doi: 10.1089/088922204322749459. [DOI] [PubMed] [Google Scholar]
- 21.Hirabayashi J, Arata Y, Kasai K. Frontal affinity chromatography as a tool for elucidation of sugar recognition properties of lectins. Methods Enzymol. 2003;362:353–368. doi: 10.1016/s0076-6879(03)01025-5. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura-Tsuruta S, Uchiyama N, Hirabayashi J. High-thoughtput analysis of lectin-oligosaccharide interactions by automated frontal affinity chromatography. Methods Enzymol. 2006;415:311–325. doi: 10.1016/S0076-6879(06)15019-3. [DOI] [PubMed] [Google Scholar]
- 23.Sanders RW, et al. The mannose-dependent epitope for neutralizing antibody 2g12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlan CN, et al. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2g12 recognizes a cluster of α1->2 mannose residues on the outer face of gp120. J Virol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umeyama H, Iwadate M. In: Current Protocols in Bioinformatics. Baxevanis AD, editor. Hoboken, NJ: Wiley; 2004. Chap 6, Unit 5.2. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto Z, et al. Crystal structure of Streptomyces olivaceoviridis E-86 beta-xylanase containing xylan-binding domain. J Mol Biol. 2000;300:575–585. doi: 10.1006/jmbi.2000.3877. [DOI] [PubMed] [Google Scholar]
- 27.Shenoy SR, et al. Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: Calorimetric and NMR characterization. Chem Biol. 2002;10:1109–1118. doi: 10.1016/s1074-5521(02)00237-5. [DOI] [PubMed] [Google Scholar]
- 28.Woods RJ, Pathiaseril A, Wormald MR, Edge CJ, Dwek RA. The high degree of internal flexibility observed for an oligomannose oligosaccharide does not alter the overall topology of the molecule. Eur J Biochem. 1998;258:372–386. doi: 10.1046/j.1432-1327.1998.2580372.x. [DOI] [PubMed] [Google Scholar]
- 29.Hazes B. The (QXW)3 domain: A flexible lectin scaffold. Protein Sci. 1996;5:1490–1501. doi: 10.1002/pro.5560050805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilkes NR, et al. Domains in microbial β-1,4-glycanases: Sequence conservation, function, and enzyme families. Microbiol Rev. 1991;55:303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bewley CA. Solution structure of a cyanovirin-N:Manα1–2Manα complex: Structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure. 2001;9:931–940. doi: 10.1016/s0969-2126(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 32.Balzarini J, et al. Mutational pathways, resistance profile, and side effects of cyanovirin relative to human immunodeficiency virus type 1 strains with N-glycan deletions in their gp120 envelopes. J Virol. 2006;80:8411–8421. doi: 10.1128/JVI.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huskens D, et al. Safety concerns for the potential use of cyanovirin-N as a microbicidal anti-HIV agent. Int J Biochem Cell Biol. 2008;40:2802–2814. doi: 10.1016/j.biocel.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 34.Rutenber E, Robertus JD. Structure of ricin B-chain at 2.5 Α resolution. Proteins. 1991;10:260–269. doi: 10.1002/prot.340100310. [DOI] [PubMed] [Google Scholar]
- 35.Lee YC, Lee RT. Carbohydrate–protein interactions: Basis of glycobiology. Acc Chem Res. 1995;28:321–327. [Google Scholar]
- 36.Balzarini J, et al. Mannose-specific plant lectins from the Amaryllidaceae family qualify as efficient microbicides for prevention of human immunodeficiency virus infection. Antimicrob Agents Chemother. 2004;48:3858–3870. doi: 10.1128/AAC.48.10.3858-3870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.