Abstract
Pathogenic bacteria continuously encounter multiple forms of stress in their hostile environments, which leads to DNA damage. With the new insight into biology offered by genome sequences, the elucidation of the gene content encoding proteins provides clues toward understanding the microbial lifestyle related to habitat and niche. Campylobacter jejuni, Haemophilus influenzae, Helicobacter pylori, Mycobacterium tuberculosis, the pathogenic Neisseria, Streptococcus pneumoniae, Streptococcus pyogenes and Staphylococcus aureus are major human pathogens causing detrimental morbidity and mortality at a global scale. An algorithm for the clustering of orthologs was established in order to identify whether orthologs of selected genes were present or absent in the genomes of the pathogenic bacteria under study. Based on the known genes for the various functions and their orthologs in selected pathogenic bacteria, an overview of the presence of the different types of genes was created. In this context, we focus on selected processes enabling genome dynamics in these particular pathogens, namely DNA repair, recombination and horizontal gene transfer. An understanding of the precise molecular functions of the enzymes participating in DNA metabolism and their importance in the maintenance of bacterial genome integrity has also, in recent years, indicated a future role for these enzymes as targets for therapeutic intervention.
Keywords: genome sequences, gene profile, DNA repair, recombination, competence, transformation
Introduction
Continuously, whole genome sequences of bacterial pathogens are being completed, allowing a comparative genomic analysis of the adaptation of different species to their natural habitats. We selected the genomes of nine pathogens and two model organisms for analysis of their gene complements related to genome maintenance and horizontal gene transfer (HGT). In this context, the aim was to focus on major pathogens with relatively small genomes exhibiting vivid genome dynamics, related to competence for transformation, and also include Gram-positive and Gram-negative representatives and model organisms, without covering a wall-to-wall panel for all infectious diseases. Among the major microbial pathogens dominating the human infectious disease scenario at the global level, Neisseria meningitidis, Haemophilus influenzae and Streptococcus pneumoniae are the causative agents of meningitis and airway-related infections. Neisseria gonorrhoeae is the causative agent of gonorrhoea, Helicobacter pylori is the cause of gastric and duodenal ulcers and precancerous gastric lesions, and Campylobacter jejuni is a main source of diarrhea. Streptococcus pyogenes, ‘the flesh-eating bug’ or group A streptococcus, is a major tissue destructor and the cause of a number of disease types including tonsillitis, serious skin infections with tissue damage, erysipelas, scarlatina, rheumatic fever and puerperal fever. Staphylococcus aureus is a typical abscess-forming agent including the methicillin-resistant S. aureus (MRSA), which is an emerging and feared multiresistant opportunist. Mycobacterium tuberculosis is the cause of tuberculosis, infecting one-third of the world's population, making it the most widespread pathogen known. Thus, C. jejuni, H. influenzae, H. pylori, M. tuberculosis, pathogenic Neisseria, S. pneumoniae, S. pyogenes and S. aureus all contribute to frequent infectious disease cases of mild to grave severity, as well as a large numbers of deaths each year. Most of these pathogens are opportunistic, mucosal surface or skin organisms, while M. tuberculosis is an intracellular parasite.
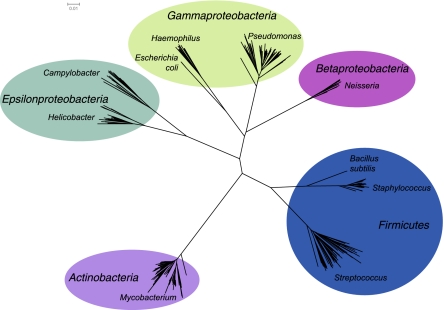
Representing members of the phylae Proteobacteriae, Actinobacteria and Firmicutes (Fig. 1), each of these microbial pathogens exhibits a lifestyle and survival strategy relevant for their respective niches (Table 1): N. meningitidis (Parkhill et al., 2000a; Tettelin et al., 2000), N. gonorrhoeae, H. influenzae (Fleischmann et al., 1995), S. pneumoniae (Tettelin et al., 2001), S. pyogenes (Ferretti et al., 2001; Hidalgo-Grass et al., 2002), C. jejuni (Parkhill et al., 2000b) and H. pylori (Alm et al., 1999) are all fastidious and face up to their environments in their exclusive human host with small, but hyperdynamic genomes, while they are naturally competent for transformation. The genome of the versatile pathogen S. pyogenes contains all the predicted ORFs required to be competent for transformation (Ferretti et al., 2001) and has also been shown to be competent (Hidalgo-Grass et al., 2002). The noncompetent M. tuberculosis (Cole et al., 1998) and S. aureus (Herron-Olson et al., 2007) have larger genomes, reflecting their lifestyles and fitness for survival also in versatile environments outside the human body.
Fig. 1.
Phylogenetic map based on 16S rRNA gene sequences of the bacterial species under study.
Table 1.
Characteristics of major human pathogens and reference bacteria included in the study
| Species (Classification) No. of genomes analyzed | Genome size (mb) | Habitat niche | Disease | References |
|---|---|---|---|---|
| Neisseria meningitidis (Betaproteobacteria) 5 | 2.2 | Upper airway colonizer | Meningitis, septicemia, Waterhouse–Friderichsen syndrome | Parkhill et al. (2000a), Tettelin et al. (2000) |
| Neisseria gonorrhoeae (Betaproteobacteria) 1 | 2.1 | Urethra | Gonorrhea | Oklahoma University Website |
| Haemophilus influenzae (Gammaproteobacteria) 6 | 1.9 | Upper airway colonizer | Pneumonia, meningitis, bacteremia, otitis media and more | Fleischmann et al. (1995) |
| Streptococcus pneumoniae (Bacilli) 3 | 2.0 | Upper airway colonizer | Pneumonia, septicemia, otitis media, peritonitis, pericarditis, meningitis, brain abscess and more | Tettelin et al. (2001) |
| Helicobacter pylori (Epsilonproteobacteria) 3 | 1.6 | Stomach and duodenum | Peptic ulcer and gastric cancer | Alm et al. (1999) |
| Campylobacter jejuni (Epsilonproteobacteria) 6 | 1.6 | Gastrointestinal | Main cause of diarrhea in Europe, inducer of the Guillain Barre syndrome | Parkhill et al. (2000b) |
| Staphylococcus aureus/MRSA (Bacilli) 14 | 2.8+ | Nose and skin | Pneumonia, meningitis, toxic shock syndrome, septicemia and skin infections | Herron-Olson et al. (2007) |
| Streptococcus pyogenes (Bacilli) 14 | 1.9 | Throat and skin | Diverse pathogenic outcomes/diseases – flesh-eating bacteria | Ferretti et al. (2001) |
| M. tuberculosis (Actinobacteria) 5 | 4.2+ | Lungs, extrapulmonary locations | Tuberculosis | Cole et al. (1998) |
| Bacillus subtilis (Bacilli) 1 | 4.2 | Soil | Reference bacterium | Kunst et al. (1997) |
| Escherichia coli (Gammaproteobacteria) 18 | 4.6+ | Intestine, sewer | Reference bacterium | Blattner et al. (1997) |
Here, we summarize comparative genomic characteristics of this subset of human pathogens and studies that have contributed to our understanding of how they adapt to different environments, combat antibiotics and acquire increased virulence. We address selected parts of the total gene content of these major pathogens in order to elucidate how these reflect their major traits and enable them to persist in their respective environments. As such, gene complements shed new light on the basis for genome dynamics in microbial pathogens. In the context of genome maintenance and HGT, major emphasis will be placed on DNA repair, type IV secretion and transformation processes.
Methods
Identification of orthologs by use of the DNA Repair Gene Orthologs system
Initially, genes known to be involved in different types of DNA repair, replication and recombination as well as genes responsible for secretion systems (mainly type II and IV secretion), pilus biogenesis and DNA uptake were identified in representative bacterial species on the basis of a combination of manual selection, databases (COG), lists in review papers (Cascales & Christie, 2004; Chen et al., 2005) and other sources. A system for the identification of gene orthologs was then used in order to see whether orthologs of the selected genes were present or absent in the genomes of the selected pathogenic bacteria under the study listed in Table 1. Based on the known genes of the various functions and their orthologs in the selected pathogenic bacteria under study, an overview of the presence of the different types of genes was created.
In brief, the ortholog system DNA Repair Gene Orthologs system (T. Rognes, O. Aussedat & B. Eliassen, unpublished data) identifies orthologous genes based on similarity of sequence. Initially, all protein sequences in Refseq were compared using an all-vs.-all blast search, and all significant matches were identified (E<1e-7). Genes were linked using single-linkage clustering based on the protein sequence alignment scores, starting with the highest-scoring pairs of sequences and progressing to gradually lower-scoring pairs. Genes belonging to the same organisms were not allowed to be clustered, unless all genes in the cluster belonged to that same organism (inparalogs). Access to the clustering information was provided through a web interface, where organisms and groups of genes could be selected. In addition to DNA Repair Gene Orthologs, the KEGG pathway database (http://www.genome.jp/kegg/pathway.html) (Kanehisa et al., 2008) was used for identifying orthologs. Positive hits, and lack thereof, were checked against homology identifications from http://www.microbesonline.org/ (Alm et al., 2005) and blast searches.
DNA repair and recombination
DNA repair
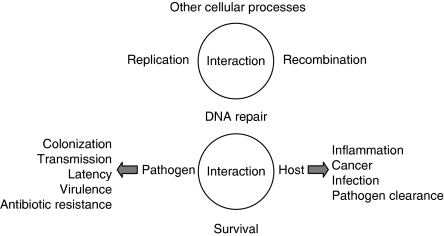
DNA repair is essential to all organisms (Fig. 2). In the context of this review, we will focus particularly on the DNA repair machinery of the pathogens C. jejuni, H. influenzae, H. pylori, M. tuberculosis, the pathogenic Neisseria, S. pneumoniae, S. pyogenes and S. aureus, and how DNA repair contributes to the ability of these bacteria to colonize, transmit and survive inside their host.
Fig. 2.
DNA repair, recombination and replication (3R) are essential processes in living cells. These processes are interconnected, often sharing components to restore or replicate genetic information. A growing body of evidence is pointing at the necessity of harboring 3R mechanisms for pathogens to effectively colonize their human host, which exerts, among others, oxidative stress on the bacterial genomes through the oxidative burst (described in the main text). Also, the host relies on 3R mechanisms to survive an invasion of potentially deadly organisms. Bacteria or bacterial components, as well as the inflammation process triggered by the bacteria, may induce host DNA damage (Box 1). The outcome of a bacterial invasion depends on both the host and the pathogen: they are not static players independent of each other. This interaction is best described as an interplay where the actions of one affects the other, for better or for worse. One example illustrating this scenario is the effect of antibiotics; although helping the host to clear the invading pathogen, induction of bacterial DNA repair mechanisms triggered by the antibiotic may lead to the dissemination of bacterial virulence determinants (Box 2).
Box 1 Host DNA repair induced by pathogens.
DNA repair is critical for the survival of pathogens inside the host because of the DNA lesions introduced in the genome of the pathogen by harmful agents released from the host (e.g. through the oxidative burst). What happens in the host, related to DNA repair, when colonized by a pathogen? Recent literature point at several mechanisms for host DNA damage during pathogen colonization (this list is not exhaustive):
Factors released from the bacteria cause DNA damage: Campylobacter jejuni cytolethal distending toxin (CDT), a protein toxin affecting the cell cycle of the host, can induce DNA damage responses in human cells. This has been identified by the presence of Rad50 foci near sites of DNA damage and the formation of the phosphoproteinγ-H2AX that mediates recruitment of repair complexes to sites where double-strand breaks occur (Hassane et al., 2003).
Reactive oxygen species (ROS) induce DNA damage: During chronic infection, ROS produced by factors of the host immune system can directly induce host DNA damage as has been shown by Obst et al. (2000). After incubation of gastric cells with Helicobacter pylori extracts, an induced synthesis of ROS and DNA fragmentation in host cells were identified.
Host levels of DNA repair enzymes are altered: Yao et al. (2006) recently demonstrated a reduction of mismatch repair protein levels in cells cocultured with H. pylori compared with cells not cocultured with this pathogen. The reduction of repair protein levels was associated with an increased number of frameshift and point mutations in the cells cocultured with H. pylori.
Host DNA repair components are selectively mutated: Shibata et al. (2002) found that H. pylori infection with strains harboring the pathogenicity island cag is associated with a higher prevalence of p53 mutation in gastric adenocarcinoma.
Box 2 Unintended consequences of antibiotic treatment.
Antibiotic treatment is the major ingredient in the battle against pathogenic bacteria. Resistance against such drugs has been a major concern since penicillin resistance was first discovered in Staphylococcus aureus in 1947. However, other important unintended side effects in the microorganisms have recently been described: several antimicrobial agents induce bacterial DNA responses, like the SOS system. In S. aureus, this induction results in replication and high-frequency transfer of a pathogenicity island, thereby promoting the spread of staphylococci virulence factors (Ubeda et al., 2005). In bacteria not harboring an SOS system, like Streptococcus pneumoniae, antibiotic stress has been shown to induce genetic exchange through transformation (Prudhomme et al., 2006), a mechanism that may contribute to the acquisition of new survival or virulence determinants.
Nearly 60 years ago, the studies of Escherichia coli DNA repair systems were initiated (Friedberg, 2008), and this organism now represents the most well-characterized bacterium of all. However, there is a growing body of evidence showing that not all bacteria function as E. coli, and in order to gain a wider genome-based perspective beyond the E. coli paradigm, a thorough analysis of different groups of bacteria is warranted. Campylobacter jejuni, H. influenzae, H. pylori, M. tuberculosis, pathogenic Neisseria, S. pneumoniae, S. pyogenes and S. aureus contribute to the majority of morbidity and mortality caused by bacteria worldwide. At the same time, E. coli and Bacillus subtilis serve as Gram-negative and Gram-positive model organisms, respectively. When comparing the DNA repair, recombination and replication (3R) enzymes in these bacteria with those present in E. coli (Table 2), a general theme seems to be the occurrence of a reduced number of genes in each class of DNA repair as compared with E. coli: in base excision repair, which normally removes subtle base damages (Seeberg et al., 1995), nei, alkA, nfi, nfo, tag and xthA are often not present in these pathogens. In the postreplication mismatch repair (MMR) pathway, base–base mismatches and insertion/deletion loops (IDLs) are recognized and excised (Schofield & Hsieh, 2003). The key enzymes of this pathway, MutS and MutL, are absent in some of the pathogens, as is MutH. A more detailed description of this pathway and some interesting features are given below. Direct repair, in which DNA lesions are chemically reversed (Mishina et al., 2006), and especially for enzymes handling alkylation damage (ada and alkB), the pathogens often show an absence of genes. On the one hand, the apparent lack of function might allow adapted genome dynamics. On the other hand, one needs to bear in mind that there might exist genes encoding products that perform identical functions, but that lack sequence homology. In this context, new protein-encoding and RNA genes remain to be discovered. Also, a number of (error-prone) DNA polymerases and the SOS response regulator, LexA, are often not present in these bacteria. Likewise, when considering helicases, which are important proteins involved in various aspects of 3R activities, E. coli is the organism studied most and is generously equipped (Table 3). The only 3R pathways that appear to be ubiquitous for most organisms are nucleotide excision repair, recombinational repair and replication (Table 3). Nucleotide excision repair removes many types of bulky lesions from DNA, often of exogenous origin, while recombinational repair is crucial for the repair of DNA strand breaks that occur during recombination (de Laat et al., 1999). Replication is fundamental for the perpetuation of the genome, and this process is tightly coupled to most of the DNA repair pathways (Friedberg et al., 2006). Our observations (Table 3) corroborate the results of Eisen & Hanawalt (1999), who performed a phylogenomic study of DNA repair genes, proteins and processes in, among others, 11 bacterial species. An immediate question arising from these findings is that how representative for each species is the number of DNA repair genes found in one strain, considering the variable level conservation and diversity in clonal and polyphyletic species? In a larger context, more central questions arising from the discussion above are as follows: how does the DNA repair enzyme repertoire affect the lifestyle of the bacteria? For instance, what does it mean for Neisseria sp. not to host an SOS response? Or for M. tuberculosis not to encode conventional mismatch repair? How do DNA repair enzymes from different pathways interact? How do DNA repair enzymes interact with enzymes from other cellular systems? And most importantly, how are colonization, transmission and virulence of the pathogenic bacteria affected by the presence or the absence of specific DNA repair enzymes?
Table 2.
Selected genes in major DNA repair pathways, recombination and replication that are present in the genome sequences of the human pathogens Campylobacter jejuni NCTC 11168 (C.j.), Haemophilus influenzae Rd KW20 (H.i.), Helicobacter pylori J99 (H.p.), Neisseria gonorrhoeae FA1090 (N.g.), Neisseriameningitidis MC58 (N.m.), Mycobacterium tuberculosis H37Rv (M.t.), Staphylococcusaureus MRSA 252 (S.a.), Streptococcuspneumoniae TIGR4 (S.p.) and Streptococcuspyogenes SF370 (S.py.) and reference bacteria Bacillus subtilis ssp. subtilis str. 168 (B.s.) and Escherichia coli K12 (E.c.)
| Symbol | Description | B.s. | E.c. | C.j. | H.i. | H.p. | N.g. | N.m. | M.t. | S.a. | S.p. | S.py. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Direct repair† | ||||||||||||
| ada | Methyltransferase, transcriptional regulator | * | * | * | * | |||||||
| alkB | Oxidative demethylase | * | * | |||||||||
| ogt | Methyltransferase | * | * | * | * | * | ||||||
| phr/spl | Photolyase | * | * | * | * | * | * | |||||
| Base excision repair† | ||||||||||||
| mutY | Adenine DNA glycosylase | * | * | * | * | * | * | * | * | * | * | * |
| mutM | Formamidopyrimidine DNA glycosylase | * | * | * | * | * | * | * | * | * | ||
| nei | Endonuclease VIII | * | * | |||||||||
| nth | Endonuclease III | * | * | * | * | * | * | * | * | * | * | * |
| tag | 3-Methyl-adenine DNA glycosylase I | * | * | * | * | * | * | * | ||||
| alkA | 3-Methyl-adenine DNA glycosylase II | * | * | |||||||||
| ung | Uracil-DNA glycosylase | * | * | * | * | * | * | * | * | * | * | * |
| xth | Exonuclease III | * | * | * | * | * | * | * | * | |||
| mpg | 3-Methylpurine DNA glycosylase | * | * | * | ||||||||
| ygjF | Thymine-DNA-glycosylase | * | ||||||||||
| nfo | Endonuclease IV | * | * | * | ||||||||
| Nucleotide excision repair† | ||||||||||||
| uvrA | Damage recognition, ATPase | * | * | * | * | * | * | * | * | * | * | * |
| uvrB | Exinuclease | * | * | * | * | * | * | * | * | * | * | * |
| uvrC | Exinuclease | * | * | * | * | * | * | * | * | * | * | * |
| uvrD | Helicase II, DNA-dependent ATPase | * | * | * | * | * | * | * | * | * | * | * |
| mfd | Transcription-repair coupling factor | * | * | * | * | * | * | * | * | * | * | * |
| Mismatch repair† | ||||||||||||
| mutS | MutS1, mismatch recognition | * | * | * | * | * | * | * | * | |||
| mutL | Endonuclease/recruitment of MutS | * | * | * | * | * | * | * | * | |||
| mutH | Endonuclease | * | * | |||||||||
| Recombinational repair† | ||||||||||||
| recA | DNA strand exchange and recombination protein | * | * | * | * | * | * | * | * | * | * | * |
| recB | Exonuclease V, β subunit | * | * | * | * | * | ||||||
| recC | Exonuclease V, γ chain | * | * | * | * | * | ||||||
| recD | Exonuclease V, α chain | * | * | * | * | * | ||||||
| recF | Gap repair protein | * | * | * | * | * | * | * | ||||
| recO | Gap repair protein | * | * | * | * | * | * | * | * | * | ||
| recR | Gap repair protein | * | * | * | * | * | * | * | * | * | * | * |
| ruvA | RuvABC resolvasome, regulatory subunit | * | * | * | * | * | * | * | * | * | * | * |
| ruvB | RuvABC resolvasome, DNA helicase | * | * | * | * | * | * | * | * | * | * | * |
| ruvC/recU | RuvABC resolvasome, endonuclease | * | * | * | * | * | * | * | * | |||
| ssb | Single-stranded-binding protein | * | * | * | * | * | * | * | * | * | * | |
| Other repair† | ||||||||||||
| lexA | Transcriptional repressor of SOS regulon | * | * | * | * | * | ||||||
| polB | DNA polymerase II | * | ||||||||||
| umuC | DNA polymerase V, subunit C | * | * | |||||||||
| umuD | DNA polymerase V, subunit D | * | ||||||||||
| dinB | DNA polymerase IV | * | * | * | * | * | * | * | * | * | ||
| ligA | DNA ligase | * | * | * | * | * | * | * | * | * | * | * |
| mutT | Pyrophosphohydrolase | * | * | * | * | * | ||||||
| Replication† | ||||||||||||
| dnaA | Chromosomal replication initiator protein | * | * | * | * | * | * | * | * | * | * | * |
| dnaB | Replicative DNA helicase | * | * | * | * | * | * | * | * | * | * | * |
| dnaG | DNA primase | * | * | * | * | * | * | * | * | * | * | * |
| gyrA | DNA gyrase, subunit A | * | * | * | * | * | * | * | * | * | * | * |
| gyrB | DNA gyrase, subunit B | * | * | * | * | * | * | * | * | * | * | * |
| parC | DNA topoisomerase IV, subunit A | * | * | * | * | * | * | * | * | |||
| parE | DNA topoisomerase IV, subunit B | * | * | * | * | * | * | * | * | |||
| priA | Primosome assembly protein | * | * | * | * | * | * | * | * | * | * | * |
| rep | DNA helicase | * | * | * | * | * | * | * | ||||
| topA | DNA topoisomerase I | * | * | * | * | * | * | * | * | * | * | * |
| polA | DNA polymerase I | * | * | * | * | * | * | * | * | * | * | * |
A complete list of the genes associated with DNA repair, recombination and replication can be found at http://cmr.jcvi.org/cgi-bin/CMR/shared/Genomes.cgi.
At least one sequence homolog is present; whether the gene contains point mutations and authentic frameshifts or whether the enzyme is active or not is not considered.
Results based on the presence of genes as identified through the DNA Repair Gene orthologs database (T. Rognes et al., unpublished data) and TIGR genome sequences, role category: DNA metabolism, DNA replication, recombination and repair (http://cmr.jcvi.org/cgi-bin/CMR/shared/Genomes.cgi).
Table 3.
Distribution of helicases involved in DNA repair, replication and recombination among the bacteria examined
| Symbol | Description | B.s. | E.c. | C.j. | H.i. | H.p. | N.g. | N.m. | M.t. | S.a. | S.p. | S.py. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dnaB | Replicative DNA helicase | * | * | * | * | * | * | * | * | * | * | * |
| uvrD | DNA-dependent ATPase I and helicase II | * | * | * | * | * | * | * | ** | * | * | * |
| dinG | ATP-dependent DNA helicase | * | * | * | * | * | * | * | ||||
| lhr | Predicted ATP-dependent helicase | * | * | |||||||||
| recG | ATP-dependent DNA helicase | * | * | * | * | * | * | * | * | * | * | * |
| recQ | ATP-dependent DNA helicase | * | * | * | * | * | * | |||||
| ruvB | ATP-dependent DNA helicase, component of RuvABC resolvasome | * | * | * | * | * | * | * | * | * | * | * |
| ruvA | Component of RuvABC resolvasome, regulatory subunit | * | * | * | * | * | * | * | * | * | * | * |
| ercc3 | Excision repair cross-complementing rodent repair | * | ||||||||||
| mfd | Transcription-repair coupling factor | * | * | * | * | * | * | * | * | * | * | * |
B.s., Bacillus subtilis; E.c., Escherichia coli; C.j., Campylobacter jejuni; H.i., Haemophilus influenzae; H.p., Helicobacter pylori; N.g., Neisseria gonorrhoea; N.m., Neisseria meningitidis; M.t., Mycobacterium tuberculosis; S.a., Staphylococcus aureus; S.p., Streptococcus pneumoniae; S.py., Streptococcus pyogenes.
Although substantial wet-lab analysis regarding DNA repair enzymes in these pathogens is not available, some studies have been conducted. The readers are referred to recent summaries (Davidsen & Tønjum, 2006; Wang et al., 2006; Davidsen et al., 2007a) and the present review (dos Vultos et al., 2009) on DNA repair in N. meningitidis, H. influenzae, S. pneumoniae, H. pylori and M. tuberculosis. For instance, in N. meningitidis mutants inactivated in genes representing all the main DNA repair pathways, Davidsen et al. (2007b) have demonstrated that the highest spontaneous mutation frequency among the N. meningitidis single mutants are found in MutY-deficient strains, as opposed to mutS mutants in E. coli, indicating a possible role for meningococcal MutY in antibiotic resistance development. In general, distinct differences between N. meningitidis and established DNA repair characteristics in E. coli have been found. Interestingly, an increasing number of studies are focusing on the in vivo survival of DNA repair mutants in animal models. In M. tuberculosis, it has been shown that a nucleotide excision repair mutant and a DNA polymerase E2 mutant are attenuated in mice (Boshoff et al., 2003; Darwin & Nathan, 2005). In H. pylori, the base excision glycosylases MutY and Nth, as well as recombinational repair, are required for effective colonization of the stomach of mice (O'Rourke et al., 2003; Eutsey et al., 2007; Amundsen et al., 2008; Wang & Maier, 2008). These findings suggest that the host induces DNA lesions in the genomes of the infectious agents, and therefore effective DNA repair is crucial for the pathogen to be able to colonize its host (Fig. 2). The observations are also supported by gene expression analysis of the pathogens upon contact with host cells. In several studies, DNA repair components have been found to be upregulated upon interaction with human cells (Vriesema et al., 2000; Morelle et al., 2005; Tala et al., 2008). In contrast to the importance of functional DNA repair mechanisms for effective host colonization stands the putative correlation between the lack of DNA repair genes associated with transmission and virulence of the pathogens. Neisseria meningitidis strains harboring mutated mutS or mutL gene copies have been identified at high prevalence in epidemic serogroup A isolates, while mutT and ogt mutations have been characterized in strains belonging to the hypervirulent M. tuberculosis W-Beijing family (Richardson et al., 2002; Rad et al., 2003). Such DNA repair deficiencies are often accompanied by a hypermutator phenotype that may be beneficial under specific selective pressures (Giraud et al., 2001). In a clinical setting, this is highly relevant concerning the development of antibiotic resistance. In S. pneumoniae, MMR mutants may have a selective advantage in the setting of antibiotic pressure (Gould et al., 2007), while Schaaff et al. (2002) recently showed that an elevated mutation frequency favors development of vancomycin resistance in S. aureus. Likewise, high mutation frequencies have been suggested as the cause of the frequently acquired antibiotic resistance during treatment of H. pylori infections (Bjorkholm et al., 2001). However, a general role for hypermutators in the emergence of clinically relevant antibiotic resistance and disease remains to be elucidated (Woodford & Ellington, 2007). Thus, the absence of certain DNA repair activities may be beneficial and allow adaptation during specific stages of the pathogen's life cycle, but intact repair machineries are vital for long-term colonization. This scenario highlights the importance for bacteria to possess mechanisms for reacquiring genes encoding DNA repair functions at some stage, for example through HGT.
Focus on MMR
MMR functions
The DNA MMR pathway is conserved from prokaryotes/bacteria to eukaryotes including humans. Defects in MMR increase mutation rates and cause genome instability, which in turn may expand the fitness landscape of bacterial pathogens. In humans, impaired MMR may cause a range of cancers, and the documented association to hereditary nonpolyposis colorectal cancer (or Lynch syndrome) has been studied intensively (Lynch & Lynch, 1985). MMR is a postreplicative process and provides an efficient way of repairing both base mismatches and IDLs that are generated during DNA synthesis. In essence, MMR allows degradation of error-containing DNA and resynthesis of unimpaired DNA. High-fidelity DNA replication is central to genome maintenance, and evidence for a close spatio-temporal association between replication, recombination and MMR is growing (Simmons et al., 2008).
MMR has been extensively investigated in E. coli and much of our current knowledge is obtained from studies of this model organism. In short, the process of MMR can be summarized as follows: MutS binds the mismatch and recruits MutL, which orchestrates several interactions including the activation of MutH – an endonuclease nicking the unmethylated strand of newly synthesized DNA at GATC sites. A piece of the nascent DNA that has received a nick is degraded by exonucleases with the aid of the DNA helicase UvrD before DNA polymerase III accurately resynthesizes DNA, and the remaining nick is sealed by DNA ligase. The process also depends on single-strand-binding proteins and the initial methylation of GATC sites by Dam methylase. The strand containing the mismatch is degraded in the 5′-to-3′ or 3′-to-5′ direction, depending on the location of the mismatch relative to the nick. The minimal human MMR has also been reconstituted in vitro by the following elements: MutSα, MutLα, ExoI, proliferating cell nuclear antigen (PCNA), replication factor C (which loads PCNA onto DNA), the single-strand-binding factor replication protein A, polδ and DNA ligase I (Constantin et al., 2005; Zhang et al., 2005). It is now clear, however, that many bacteria differ from E. coli in their basic MMR machinery.
The conundrum of MutH vs. MutL
The absence of MutH in many bacteria and all eukaryotes is particularly striking, because it suggests that strand discrimination and the initiation of excision may have a basis different from that of the methyl-directed process in E. coli and in certain other Gram-negative microorganisms. How can MMR discriminate between the template strand and the newly synthesized strand if it is not methyl-directed? It has been proposed that MMR can be directed to the newly synthesized strand by interacting with strand termini during replication. As such, MMR would constitute a part of the replisome and could direct repair activity from the termini between okazaki fragments on the lagging strand or from the 3′-terminus on the leading strand, linking the two processes tightly (Jiricny, 2006). Indeed, the interaction between MutS from B. subtilis, which does not belong to the methyl-directed MMR group, and the β-clamp (PCNA in eukaryotes) was recently described in detail and supported that the MMR complex acts at, or in association with, the replication fork (Simmons et al., 2008). A breakthrough came from the laboratory of Paul Modrich when they found that human MutLα itself is an endonuclease that is able to produce a nick in nascent DNA (Kadyrov et al., 2006). A model where MutL can introduce random nicks on both sides of the mismatch has helped explain how a bidirectional DNA repair process was able to operate with a single exonuclease (EXOI in the human model) that degrades only in the 5′-3′ direction. Based on the finding that endonucleolytic hydrolysis of DNA depends on one or two divalent cations as the metal required, a binding site with a motif [DQHA(X)2E(X)4E] was identified, which is conserved in archaeal, eukaryotic (PMS2 and MLH3) and eubacterial MutL homologs. Convincingly, this motif was absent in all MutL homologs from those Gram-negative organisms known to have MutH and the methyl-directed MMR pathway (Kadyrov et al., 2006). We compiled a list of organisms containing a MutH homolog (Supporting Information, Table S1) and found that the distribution of MutH-dependent MMR is very limited. With a few exceptions, MutH is primarily found in bacterial species sorting under the class of Gammaproteobacteria. In our selection of pathogens, only H. influenzae belonging to the Pasteurellaceae (class Gammaproteobacteria) contains the mutH gene whereas the Neisseria (class Betaproteobacteria) and the staphylococci and streptococci (both class Bacilli) contain the MutH-less MMR (Table 2 and Fig. 1). The Epsilonproteobacteria member C. jejuni and the Actinobacterium M. tuberculosis lack MMR altogether. Helicobacter pylori contains a gene encoding a homolog of MutS-2, but this protein is likely to be involved in processes other than MMR as we know it (Table 2). Taken together, these bacteria may experience elevated mutation rates because they lack a functional MMR system.
An alignment based on the MutL pfam entries from 822 organisms was used to investigate sequence conservation of the metal-binding motif. We note that the motif in Neisseria sp. has a Q/M substitution in the second position DQ/MHA(X)2E(X)4E. As Fig. S1 shows, the Q/M substitution is one of the most common substitutions. Whether this substitution from a polar to a nonpolar amino acid is functionally important for MutL activity and function remains to be investigated. It has, however, been established that MutL knock-outs in N. meningitidis produce a mutator phenotype as expected for MMR malfunction (Richardson et al., 2002).
MutS sequence diversity
Genome comparisons have also revealed differences in the distribution of the mutS gene and also great diversity within the mutS-group. Phylogenomic analysis has revealed that the mutS lineage split early in evolution and gave several distinct lines, where, importantly, only one belongs to the MMR pathway (Eisen, 1998; Lin et al., 2007).
When MMR is absent
One of the striking characteristics of the M. tuberculosis and partially for H. pylori DNA repair system is the absence of recognized MMR homologs, which might suggest that these bacteria do not perform MMR activity. The consequent reduced fidelity in genome maintenance might add to the adaptive ability of M. tuberculosis, which otherwise seems to exist in genetic isolation. On the other hand, MMR activity could exist without sequence homology to recognized MMR components, and the search for components exerting MMR activity in M. tuberculosis should still be pursued.
Distribution of helicases in pathogenic bacteria
Helicases are ubiquitous enzymes vital to all living organisms. They are motor proteins that move directionally along the nucleic acid phosphodiester backbone separating two annealed nucleic acid strands using energy from NTP hydrolysis. Helicases are involved in various aspects of cellular processes including replication, repair, recombination, transcription and RNA processing (Schmid & Linder, 1992; Matson et al., 1994). The vital role(s) that these enzymes play has been underscored by a number of genetic discoveries. Mutations in three out of the five human recQ homologs have been identified as causes of Werner (WRN), Bloom (BLM) or Rothmund–Thomson syndrome (RECQ4), respectively (Ellis et al., 1995; Yu et al., 1996; Kitao et al., 1999). Mutations interfering with the proper function of XPB and XPD helicases in humans have been linked to disorders such as Xeroderma Pigmentosum (XP), Cockayne syndrome (CS) and trichothiodystrophy (TTD) (Hoeijmakers, 1994; Vermeulen et al., 1994; de Boer & Hoeijmakers, 2000).
First discovered in E. coli as a ‘DNA-unwinding enzyme’ more than 32 years ago, the number of helicases identified and characterized has since then increased tremendously (Abdel-Monem & Hoffmann-Berling, 1976). Most organisms host multiple helicases; for example the E. coli genome encodes at least 12 helicases (Matson et al., 1994). When examining the pathogens under study (Table 3), some helicases that are essential to cellular functions, such as DnaB and UvrD, are distributed across all the organisms. In addition, RecG, RuvA and RuvB helicases that participate in recombinational repair, as well as Mfd involved in nucleotide excision repair, are found in all the pathogens. On the other hand, some helicases, such as RecQ, Ercc3, DinG and Lhr, are not universally distributed in our selected organisms (Table 3). The recQ gene homolog is present in H. influenzae, the Neisseria and S. aureus while it is missing in C. jejuni, H. pylori, M. tuberculosis, S. pneumoniae and S. pyogenes. The E. coli RecQ DNA helicase has served as a paradigm for the RecQ family and has been proposed to have multiple functions in the initiation of recombination, resolution of recombination intermediates and suppression of illegitimate recombination also required for proper induction of the ‘SOS’ response to stalled replication forks (Hishida et al., 2004; Chow & Courcelle, 2007).
The helicase- and RNAse-like C-terminal (HRDC) domain is characteristic of many members of the RecQ helicase clade (Bernstein & Keck, 2005; Wu et al., 2005; Killoran & Keck, 2006a). Interestingly, RecQ, which usually contains a single HRDC domain in most organisms, is identified with three HRDC domains in N. meningitidis and N. gonorrhoeae and plays a critical role in determining pilin antigenic variation and also participates in DNA repair (Mehr & Seifert, 1998; Killoran & Keck, 2006b, 2008; Stohl & Seifert, 2006). This might indicate that the multiplicity of HRDC domains can represent one specialized way to exert specificity in RecQ activities (Killoran et al., 2009). Even though RecQ is absent in M. tuberculosis, the HRDC domain is identified in its UvrD2 helicase, which is one out of the two UvrD-like paralogs found in mycobacteria (Morozov et al., 1997; Sinha et al., 2008). However, the HRDC domain appeared not to be essential for enzymatic activity of UvrD2, suggesting that it might be involved in DNA binding (Sinha et al., 2008). It was proposed that the HRDC domain might target RecQ-family proteins to specific DNA structures (Bernstein & Keck, 2005).
Another feature noted among the distribution of the helicases in the pathogenic bacteria under study is the presence of XPB/ERCC3 homolog, which is found only in M. tuberculosis (Table 3) (Poterszman et al., 1997). XPB/ERCC3/RAD25 in eukaryotes is an integral subunit of the transcription factor TFIIH, which is involved in transcription initiation and nucleotide excision repair (Weeda et al., 1990; Schaeffer et al., 1993). Even though well studied in humans, the role of ERCC3 helicase in bacteria is not yet known. However, the occurrence of the ercc3 gene in prokaryotes seems to be limited to mycobacteria and Kineococcus radiotolerans (Biswas et al., 2009), which might have acquired the gene through infrequent HGT that might occur from eukaryotes to certain bacterial species (Poterszman et al., 1997; Aravind et al., 1999).
The DinG helicase in E. coli is a damage-inducible, SOS-regulated, strucure-specific enzyme, related to the human helicases XPD and BACH1, Rad3 from Saccharomyces cerevisiae and Rad15 from Schizosaccharomyces pombe (see Voloshin & Camerini-Otero, 2007 and references therein) (Voloshin & Camerini-Otero, 2007). Similar to XPB, XPD is also a part of the multisubunit complex TFIIH that plays a dual role in the transcription initiation and nucleotide excision repair (de Boer & Hoeijmakers, 2000).
Another less-distributed helicase, the long helicase-related protein (Lhr), which is the longest protein identified in E. coli, is also found in M. tuberculosis (Table 3) (Reuven et al., 1995). However, its exact function is not yet known. The fact that the M. tuberculosis genome encodes the eukaryotic DNA repair proteins such as ERCC3 and Mpg might reflect past HGT events and enable this bacterium to survive in the hostile environment inside human macrophages.
HGT
Natural transformation in selected pathogens
Transformation, type IV pili and type II and IV secretions
The strong selective advantage of HGT has in several instances driven the evolution of complex machineries in favor of transformation. These divergent competence vehicles can ultimately cause the acquisition of novel traits such as antibiotic resistance, while they still allow homologous recombination that in turn could facilitate DNA repair and the fixation of beneficial alleles. Because different bacteria have solved their sex drives in various ways, the study of their strategies provides an exemplary case of convergent evolution. An interesting feature of all these systems is that they are based on structures already present in the cell that have become modified to facilitate and control genetic flux. Neisseria meningitidis, H. influenzae and the two streptococcal species S. pneumoniae and S. pyogenes all host systems composed of partners involved in the assembly of type IV pili and proteins with homology to type II secretion systems (Table 4) (Woodbury et al., 2006). Type IV pili are important virulence factors in many pathogens, are required for transformation and are also associated with many other functions including cell adhesion, twitching motility and biofilm formation (Tønjum & Koomey, 1997; Mattick, 2002). In order to identify the entire complement of proteins driving the transformation machinery, the complete set of neisserial DNA-binding proteins should be defined (Lång et al., 2009) (Fig. 3). Interestingly, the pilus biogenesis component PilQ binds DNA (Assalkhou et al., 2007), thus contributing to the transformation process directly and indirectly, through pilus biogenesis. Type II secretion is also important for pathogenesis in facilitating the release of toxins and hydrolytic enzymes (Sandkvist, 2001). Helicobacter pylori and C. jejuni make use of a different transformation system that is evolutionarily related to type IV secretion systems. This is a secretion system that runs in reverse and resembles the conjugation systems of Agrobacterium tumefaciens (see separate section). Staphylococcus aureus, including MRSA, are not competent for transformation, but do contain a few of the known competence genes (ComC, ComGA, ComGB, ComGC) (Table 4). These are likely to be involved in other transport processes in this organism (Sibbald et al., 2006). Strains of S. aureus display up to 20% variability in their genome sequence, and virulence and evolution of S. aureus are influenced by frequently occurring prophages and pathogenicity islands. It seems that prophages in moderately virulent S. aureus strains contribute important properties to pathogenesis, as fewer virulence factors in these cases are found outside of the prophages than for the highly virulent strains identified (Baba et al., 2008). Similarly, M. tuberculosis is nontransformable and nonconjugative, although (remnants of) a few transducing phages are found. Despite its relative genetic/sexual isolation, this intensively studied organism is a most successful pathogen, and ends up with a clonal lifestyle (for the 3R and recombinational history of Mycobacterium, see review dos Vultos, in this issue; Jang et al., 2008; Stinear et al., 2008).
Table 4.
The competence gene profile and presence and absence of genes related to competence in major microbial pathogens and reference bacteria
| Organism |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B.s. | E.c | C.j. | H.i. | H.p. | N.g | N.m | M.t | S.a | S.p. | S.py. | ||
| Symbol | Description/function | G+ | G− | G− | G− | G− | G− | G− | G+ | G+ | G+ | G+ |
| Competence/type II secretion/pilus biogenesis proteins | ||||||||||||
| ComEA | Competence protein, helix–hairpin–helix region | * | * | * | * | * | * | * | * | * | * | |
| ComEC | Hydrolase, Rec2 | * | * | * | * | * | * | * | * | * | * | * |
| ComFA | Late competence protein | * | * | * | * | |||||||
| ComFB | Competence protein FB | * | ||||||||||
| ComFC | Amidophosphoribosyltransferase, | * | * | * | * | * | * | * | * | * | * | * |
| ComGA | ATPase, PulE/PilB/PilF/PilT/PilU | * | * | * | * | * | * | * | * | * | * | |
| ComGB | Type II secretory pathway, PulF/PilG | * | * | * | * | * | * | * | * | * | ||
| ComGC-GG | Pilin/pseudopilin, PilA, PilE | * | * | * | * | * | * | * | * | * | ||
| ComC | Type IV prepilin peptidase, PilD | * | * | * | * | * | * | * | * | * | ||
| Smf | DNA-processing chain A, DprA | * | * | * | * | * | * | * | * | * | * | * |
| CinA | Competence/damage-inducible protein | * | * | * | * | * | * | * | * | * | * | |
| ComL | Competence lipoprotein | * | * | * | * | * | * | * | ||||
| PilQ | Secretin, HofQ, ComE, PulD | * | * | * | * | * | ||||||
B.s., Bacillus subtilis; E.c., Escherichia coli; C.j., Campylobacter jejuni; H.i., Haemophilus influenzae; H.p., Helicobacter pylori; M.t., Mycobacterium tuberculosis; N.m., Neisseria meningitidis; N.g., Neisseria gonorrhoeae; S.p., Streptococcus pneumoniae; S.py., Streptococcuspyogenes; S.a., Staphylococcus aureus.
Fig. 3.

Model of the meningococcal transformation machinery based on the current information on the components involved in this process. DNA is predicted to enter the meningococcal cell through the PilQ pore, which, when it is wound around the pilus rod, sterically just allows the DNA to enter the cell. This hypothesis needs to be biologically verified.
Repeat sequences promoting transformation
HGT is associated with the risk of allowing entry of alien and potentially harmful DNA from other organisms such as viruses. Even similar and only slightly diverged DNA from other species may be disadvantageous in a new host with separate sets of adaptations and fine-tuned processes. Various strategies have therefore been used in many bacteria to control the entry and persistence of DNA, including ecological isolation such as competence induction by quorum sensing, restriction modification systems and stringent homologous recombination. The Neisseria sp. and members of the Pasteurellaceae, such as H. influenzae, discriminate between homologous and alien DNA by recognizing a short specific sequence in DNA from their own genus. These sequences are known as DNA uptake sequence (DUS) and uptake signal sequence (USS). DUS/USS are found in exceptionally high numbers throughout the genomes of these species, ensuring that almost any piece of the chromosome of a certain length will contain such a signal and hence be recognized, taken up and finally constitute a substrate for homologous recombination (Goodman & Scocca, 1991; Ambur et al., 2007). In contrast, the pneumococci and streptococci use quorum sensing of a competence-stimulating peptide and fratricide to ensure that most of the DNA available in their surroundings is homologous (Johnsborg et al., 2007). Again, the positive effects of allowing HGT has driven the evolution of different strategies that ultimately produce the same result, namely homologous recombination between closely related alleles.
The identification of the positive effects that have influenced the evolution of transformation has not been straightforward, and many selective pressures have been proposed, most of which are not mutually exclusive. Firstly, it has been proposed that transformation evolved from its ability to provide nutrients for the recipient organism, also known as the sex-for-food hypothesis. The rationale behind this notion is that in many organisms, transformation is induced upon starvation and would provide a good system to ensure uptake of high-energy compounds that could promote survival (Redfield et al., 1997; Palchevskiy & Finkel, 2006). Other hypotheses are based on incoming DNA providing a benefit after having been recombined into the chromosome and include, for example, sex for repair and in response to oxidative stress (Nedelcu & Michod, 2003; Nedelcu et al., 2004; Michod et al., 2008) and innovation (Ochman et al., 2000; Narra & Ochman, 2006; Jeon et al., 2008). An in-depth discussion and evaluation of these hypotheses are beyond the scope of this review and the debate on the evolution of transformation and bacterial sex in general is still ongoing (for excellent reviews, please see Johnsborg et al., 2007; Michod et al., 2008). Our own studies of DUS, itself a sign of transformation, have shown that that the complex DUS-mediated control of transformation is not likely to have evolved for its ability to import completely novel sequences. The genomic distribution of these sequences showed that DUS were over-represented in the conserved common core genome, under-represented in regions under diversification, and absent in both recently acquired genes and recently lost core genes (Treangen et al., 2008). Previously, we have found that DUS and USS in Neisseria sp. and H. influenzae, respectively, are biased toward 3R genes, suggesting that a functional relationship between genome maintenance and transformation exists (Davidsen et al., 2004). In addition, DUS occurrences correlate with the size of conversion fragments. We have therefore proposed that transformation has evolved from its ability to incorporate homologous sequences, either for the regeneration of damaged DNA or from benefits associated with the reassortment of alleles, or both (Treangen et al., 2008). We hypothesize that the core genomes in species that have no means of biasing their DNA uptake, such as S. pneumoniae, H. pylori and C. jejuni, still could have experienced more recombination during the evolutionary time than the more variable parts of their respective genomes. Although speculative, such frequent nonrandom allelic replacements could be generated by biases at the level of recombination by, for example, repeats. We have no indication that nondiscriminatory transformation differs from the signature-discriminating DUS/USS system of Neisseria/Pasteurellaceae in its regenerative properties or that these species experience fundamental differences in DNA damage or the need to reassort alleles. On the contrary, we suspect that these diverse transformation systems have been shaped by the same evolutionary forces. Thus, the study of the genomics of DUS/USS represents an example of convergent evolution (Davidsen et al., 2004), which may also be helpful in generating new testable hypotheses regarding transformation and its history in other organisms. Comparative genomics in a multispecies approach could elaborate on this hypothesis and increase our understanding of the selective advantages of transformation. The hypothesis that transformation evolved due to its ability to provide substrate for recombination has also been strengthened by the observation that these two processes are physically linked in space and time (Kidane & Graumann, 2005).
Conjugation-related genes: type IV secretion in bacterial DNA transfer/sex
The versatile type IV secretion (T4S) systems
T4S systems are involved in the transport of macromolecules such as proteins and DNA across the outer envelope of bacteria. These systems are primarily known from Gram-negative bacteria, where they transport components over both the cytoplasmic and the outer membranes. Interestingly, the conjugation systems that transfer plasmid DNA are one form of the T4S system and are also described for Gram-positive bacteria (Grohmann et al., 2003). While proteins are secreted by the T4S systems and can also be transferred through the plasma membrane of a target cell, DNA can also be secreted and even imported by the T4S system (Ding et al., 2003; Economou et al., 2006). Bacterial conjugation systems represent a prominent subfamily of the T4S systems, while the VirB/D4 system in A. tumefaciens is the prototype example of a T4S system. A subclassification of the T4S systems was established based on ancestral lineage, building on two main groups (Christie & Vogel, 2000), followed by a subsequent systematic organization/grouping of all known T4S systems based on their function in conjugation, DNA uptake and release, and effector translocation (Cascales & Christie, 2004). Further analysis of the evolution of the T4S systems based on a protein homology-network defined them into four groups (Medini et al., 2006). Core T4S proteins are part of all T4S systems and can be complemented with independently recruited subunits or proteins to gain a system-specific function (Ding et al., 2003; Medini et al., 2006). It was also suggested that those T4S systems particularly involved in HGT between species led to a functional divergence of these systems (Frank et al., 2005). The secretion of DNA has probably evolved from protein secretion systems (Cascales & Christie, 2003, 2004). The DNA-binding relaxases recognize and translocate DNA, which is suggested to lead to an only coincidental ‘hitch-hiking’ of DNA together with the protein secreted (Cascales & Christie, 2004; Lybarger & Sandkvist, 2004; Chen et al., 2005).
Among the pathogenic bacteria assessed in this review, C. jejuni, H. pylori, H. influenzae and N. meningitidis as well as N. gonorrhoeae possess one or more T4S systems (Table 5) (Hofreuter et al., 1998, 2001; Bacon et al., 2000, 2002; Cascales & Christie, 2004; Snyder et al., 2005). The C. jejuni strain 81-176 pTet plasmid is a true conjugative plasmid. It coexists with the smaller pVir plasmid, which also encodes a T4S system, and influences C. jejuni virulence (Bacon et al., 2000; Batchelor et al., 2004). Other conjugative plasmids in C. jejuni had, as opposed to pVir, no influence on the invasiveness of this bacterium, but often encode antibiotic resistance (Schmidt-Ott et al., 2005; Dasti et al., 2007). Furthermore, other conjugative plasmids are found in other Campylobacter species (Boosinger et al., 1990; Fouts et al., 2005).
Table 5.
The presence and absence of genes related to T4S systems in microbial genomes
| Organism |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Description | B.s. | E.c. (IncF) | C.j. (Vir) | H.i. (Tra) | H.p. (Com) | N.g. (GGI) | N.m. (GGI) | M.t. | S.a. | S.p. | S.py. |
| VirB1 | * | |||||||||||
| VirB2 | pilin | * | * | * | * | |||||||
| VirB3 | * | * | * | * | ||||||||
| VirB4 | ATPase | * | * | * | * | * | * | |||||
| VirB5 | * | * | * | * | ||||||||
| VirB6 | * | * | * | * | ||||||||
| VirB7 | lipoprotein | * | * | * | * | * | * | |||||
| VirB8 | pore protein | * | * | * | ||||||||
| VirB9 | pore protein | * | * | * | * | * | * | |||||
| VirB10 | pore protein | * | * | * | * | * | * | |||||
| VirB11 | ATPase | * | * | |||||||||
| VirD4 | T4CP | * | * | * | * | * | ||||||
B.s., Bacillus subtilis; E.c., Escherichia coli; C.j., Campylobacter jejuni; H.i., Haemophilus influenzae; H.p., Helicobacter pylori; M.t., Mycobacterium tuberculosis; N.m., Neisseria meningitidis; N.g., Neisseria gonorrhoeae; S.p., Streptococcus pneumoniae; S.py., Streptococcuspyogenes; S.a., Staphylococcus aureus.
A detailed description of the single components is given by Christie et al. (2005).
Another pathogenic member of the order Campylobacteriales is H. pylori, which has three T4S systems: the Com-system, the Cag- or HP-system and the Tsf3-system (Chen et al., 2005; Zhong et al., 2007). While the Cag- or HP-system is used for exotoxin effector translocation and the function of the Tsf3-system is unknown, the Com-system has evolved for DNA transport (Hofreuter et al., 2001). The conjugation-like Com system is special in that it is used for the uptake of DNA, thus translocating DNA in a direction opposite to that for secretion. This might also be the case for the C. jejuni Vir system. These ‘competence’ systems have probably evolved to increase the possibilities for genetic variation or renewal, leading to enhanced cellular fitness, survival and invasion of the eukaryotic host (Ding et al., 2003). In addition, the transfer of chromosomally encoded properties by a conjugation-like mechanism may contribute to horizontal DNA transfer between different members of the Campylobacteriales group (Oyarzabal et al., 2007). Little is known about the T4S systems in H. influenzae. There are two Tra-like plasmid-encoded systems (Smoot et al., 2002; McGillivary et al., 2005).
T4S systems on genomic islands (GIs)
Recently, a GI containing a T4S system, which is evolutionarily distant from the plasmid-based systems and a vector for antibiotic resistance, was discovered (Juhas et al., 2007a, b). This system belongs to a new type of T4S systems found in a wide number of bacteria. They were named GI-like T4S systems and allow GIs encoding many different properties to mobilize and spread (Juhas et al., 2008).
The gonococcal genetic island (GGI) was first identified in N. gonorrhoeae. The T4S system encoded by GGI is related to the conjugational F plasmid system of E. coli and is used by the bacteria for secretion of chromosomal DNA (Ding et al., 2003; Hamilton et al., 2005). Later, complete and partial forms of the GGI were also found in N. meningitidis (Snyder et al., 2005). Approximately 80% of gonococcal strains and some N. meningitidis strains carry the GGI, probably inserted by the site-specific recombinase XerCD into the dif site (Dillard & Hamilton, 2002; Hamilton et al., 2005). The sequence of the GGI is characterized by a low G+C and low DUS content, suggesting that it is not of neisserial origin, but the amelioration of some regions to a typical neisserial composition indicate an already long-term existence of GGIs in neisserial genomes. As chromosomal DNA is secreted by the T4S system encoded by GGI, no direct contact between the donor and recipient of DNA is needed. This may be so because Neisseria species are naturally competent throughout their life cycle and preferentially take up DUS-containing DNA (Lie, 1965; Sparling, 1966; Mathis & Scocca, 1982; Goodman & Scocca, 1991) The GGI contains only one DUS per 10 kb, which is only about 10% of the average DUS density found in the whole genome, but, in addition, it contains several incomplete DUS with one mutation showing that the DUS may be on the way to establish itself in the GGI. For the stable maintenance of GGI in the neisserial genome, the imperfections of one dif site were shown to be responsible, because reversion to a perfect site led to significant loss of the GGI (Dillard & Dominguez, 2008). The two parts of the GGI missing in N. meningitidis serogroup H and Z strains are flanked by DUS (Snyder et al., 2005). These sites may be the sites of recombination that led to an excision of the sequence blocks (Treangen et al., 2008). Because surrounding sequences of the GGI are still available, they may serve as a target for reintroduction of chunks of DNA by recombination with GGI DNA taken up by the bacterium through the DUS-specific uptake/recombination system. The effects of the GGI are still mostly obscure. It was shown that for the peptidoglycan fragment release in culture, neither the T4S system components nor the GGI-encoded lytic transglycosylases AtlA and LtgX are required for this process, but that the presence of the GGI can bypass the TonB-dependent iron acquisition of intracellular gonococci (Hagen et al., 2006; Cloud-Hansen et al., 2008). On the other hand, the high number of strains of N. gonorrhoeae that host a GGI, which can support efficient conjugation, may explain why plasmids, and the consequent antibiotic resistance when selective pressures exert their action, are more prevalent in gonococcal than in meningococcal strains.
Conclusions
Acquisition and loss of genetic material are essential forces in bacterial microevolution, also challenging functions involved in DNA repair, recombination and HGT. These functions have been repeatedly linked with adaptation of lineages to new lifestyles, and in particular to pathogenicity. Comparative genomics has the potential to elucidate this genetic flux, but there are many methodological challenges involved in inferring gene content and evolutionary events from collections of genome sequences. Here, we have described a method for detecting the presence or the absence of genes in whole genome sequences to elucidate the impact of gene content on microbial lifestyle. Our approach is purely sequence based and relies on gene identification. We have demonstrated its use on datasets from the genomes of C. jejuni, H. influenzae, H. pylori, M. tuberculosis, pathogenic Neisseria, S. pneumoniae, S. pyogenes and S. aureus. In all these examples, we found interesting variations in the presence and absence/gain and loss of genetic material, which correlate with their niches and fitness for survival.
Competence for transformation, according to the gene content detected in many genomes, might be under-rated in many microbial species, including pathogens. At the same time, the different strategies in Gram-negative and Gram-positive organisms to achieve the net result of competence for transformation, leading to the same outcome, namely preferential uptake of its own DNA, represent an exciting diversity in biology. In this context, the presence of repeats, such as DUS and USS, has a tendency to accumulate in the core genome (Treangen et al., 2008), emphasizing their importance. Recently, the linkage between transformation and recombination, including the close proximity of the recombination process to the cytoplasmatic side of the inner membrane (Kidane & Graumann, 2005), has been elucidated. Taken together, this study of the presence and the absence of genes related to DNA metabolism and HGT enlightens how the gene profile affects the lifestyle of microbial pathogens in their respective niches.
Acknowledgments
We are grateful to Ophélie Aussedat for her work with the DNA Repair Genes Orthologs database. This work was supported by grants from the Research council of Norway.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Sequence conservation of the MutL metal-binding motif DQH/MA(X)2E(X)4E based on a MutL alignment of entries from 822 organisms.
Table S1. Bacteria containing a MutH homolog.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abdel-Monem M, Hoffmann-Berling H. Enzymic unwinding of DNA. 1. Purification and characterization of a DNA-dependent ATPase from Escherichia coli. Eur J Biochem. 1976;65:431–440. doi: 10.1111/j.1432-1033.1976.tb10358.x. [DOI] [PubMed] [Google Scholar]
- Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Ling LS, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Ambur OH, Frye SA, Tønjum T. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol. 2007;189:2077–2085. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase–nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assalkhou R, Balasingham S, Collins RF, et al. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology. 2007;153:1593–1603. doi: 10.1099/mic.0.2006/004200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon DJ, Alm RA, Burr DH, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon DJ, Alm RA, Hu L, et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81-176. Infect Immun. 2002;70:6242–6250. doi: 10.1128/IAI.70.11.6242-6250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology. 2004;150:3507–3517. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Biswas T, Pero J, Joseph C, Tsodikov O. DNA-dependent ATPase activity of bacterial XPB helicases. Biochemistry. 2009 doi: 10.1021/bi8022416. DOI 10.1021/bi8022416. [DOI] [PubMed] [Google Scholar]
- Bjorkholm B, Sjolund M, Falk PG, Berg OG, Engstrand L, Andersson DI. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. P Natl Acad Sci USA. 2001;98:14607–14612. doi: 10.1073/pnas.241517298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Boosinger TR, Blevins WT, Heron JV, Sunter JL. Plasmid profiles of six species of Campylobacter from human beings, swine, and sheep. Am J Vet Res. 1990;51:718–722. [PubMed] [Google Scholar]
- Boshoff HI, Reed MB, Barry CE, III, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KH, Courcelle J. RecBCD and RecJ/RecQ initiate DNA degradation on distinct substrates in UV-irradiated Escherichia coli. Radiat Res. 2007;168:499–506. doi: 10.1667/RR1033.1. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud-Hansen KA, Hackett KT, Garcia DL, Dillard JP. Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J Bacteriol. 2008;190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Nathan CF. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect Immun. 2005;73:4581–4587. doi: 10.1128/IAI.73.8.4581-4587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasti JI, Gross U, Pohl S, Lugert R, Weig M, Schmidt-Ott R. Role of the plasmid-encoded tet(O) gene in tetracycline-resistant clinical isolates of Campylobacter jejuni and Campylobacter coli. J Med Microbiol. 2007;56:833–837. doi: 10.1099/jmm.0.47103-0. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Tønjum T. Meningococcal genome dynamics. Nat Rev Microbiol. 2006;4:11–22. doi: 10.1038/nrmicro1324. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Rødland EA, Lagesen K, Seeberg E, Rognes T, Tønjum T. Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res. 2004;32:1050–1058. doi: 10.1093/nar/gkh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen T, Koomey M, Tønjum T. Microbial genome dynamics in CNS pathogenesis. Neuroscience. 2007a;145:1375–1387. doi: 10.1016/j.neuroscience.2007.01.059. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Tuven HK, Bjørås M, Rødland EA, Tønjum T. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J Bacteriol. 2007b;189:5728–5737. doi: 10.1128/JB.00161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Dominguez NM. Factors Affecting Excision of the Gonococcal Genetic Island from the Chromosome. the Netherlands: Rotterdam; 2008. pp. 106–107. [Google Scholar]
- Dillard JP, Hamilton HL. Sequence and mutational analysis of the gonococcal genetic island. In: Caugant DA, Wedege E, editors. Proceedings of the 13th IPNC meeting. Oslo: Nordberg Aksidenstrykkeri AS; 2002. p. 79. [Google Scholar]
- Ding Z, Atmakuri K, Christie PJ. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 2003;11:527–535. doi: 10.1016/j.tim.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Vultos T, Mestre O, Tønjum T, Gicquel B. DNA repair in Mycobacterium tuberculosis revisited. FEMS Microbiol Rev. 2009;33:471–487. doi: 10.1111/j.1574-6976.2009.00170.x. [DOI] [PubMed] [Google Scholar]
- Economou A, Christie PJ, Fernandez RC, Palmer T, Plano GV, Pugsley AP. Secretion by numbers: protein traffic in prokaryotes. Mol Microbiol. 2006;62:308–319. doi: 10.1111/j.1365-2958.2006.05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA. A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 1998;26:4291–4300. doi: 10.1093/nar/26.18.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, et al. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Eutsey R, Wang G, Maier RJ. Role of a MutY DNA glycosylase in combating oxidative DNA damage in Helicobacter pylori. DNA Repair. 2007;6:19–26. doi: 10.1016/j.dnarep.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. P Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Fouts DE, Mongodin EF, Mandrell RE, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Alsmark CM, Thollesson M, Andersson SG. Functional divergence and horizontal transfer of type IV secretion systems. Mol Biol Evol. 2005;22:1325–1336. doi: 10.1093/molbev/msi124. [DOI] [PubMed] [Google Scholar]
- Friedberg EC. A brief history of the DNA repair field. Cell Res. 2008;18:3–7. doi: 10.1038/cr.2007.113. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 2006. [Google Scholar]
- Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- Goodman SD, Scocca JJ. Factors influencing the specific interaction of Neisseria gonorrhoeae with transforming DNA. J Bacteriol. 1991;173:5921–5923. doi: 10.1128/jb.173.18.5921-5923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CV, Sniegowski PD, Shchepetov M, Metlay JP, Weiser JN. Identifying mutator phenotypes among fluoroquinolone-resistant strains of Streptococcus pneumoniae using fluctuation analysis. Antimicrob Agents Ch. 2007;51:3225–3229. doi: 10.1128/AAC.00336-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol R. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen TA, Dominguez NM, Dillard JP, Cornelissen CN. The Gonococcal Genetic Island Provides a Bypass Mechanism to TonB-Dependent Intracellular Survival of Neisseria Gonorrhoeae Within Cervical Epithelial Cells. P2.1.02. Cairns, Australia: Cambridge Publishing; 2006. [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Hassane DC, Lee RB, Pickett CL. Campylobacter jejuni cytolethal distending toxin promotes DNA repair responses in normal human cells. Infect Immun. 2003;71:541–545. doi: 10.1128/IAI.71.1.541-545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Grass C, Ravins M, Dan-Goor M, Jaffe J, Moses AE, Hanski E. A locus of group A Streptococcus involved in invasive disease and DNA transfer. Mol Microbiol. 2002;46:87–99. doi: 10.1046/j.1365-2958.2002.03127.x. [DOI] [PubMed] [Google Scholar]
- Hishida T, Han YW, Shibata T, Kubota Y, Ishino Y, Iwasaki H, Shinagawa H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. Human nucleotide excision repair syndromes: molecular clues to unexpected intricacies. Eur J Cancer. 1994;30A:1912–1921. doi: 10.1016/0959-8049(94)00381-e. [DOI] [PubMed] [Google Scholar]
- Hofreuter D, Odenbreit S, Henke G, Haas R. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol Microbiol. 1998;28:1027–1038. doi: 10.1046/j.1365-2958.1998.00879.x. [DOI] [PubMed] [Google Scholar]
- Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- Jang J, Becq J, Gicquel B, Deschavanne P, Neyrolles O. Horizontally acquired genomic islands in the tubercle bacilli. Trends Microbiol. 2008;16:303–308. doi: 10.1016/j.tim.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Jeon B, Muraoka W, Sahin O, Zhang Q. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob Agents Ch. 2008;52:2699–2708. doi: 10.1128/AAC.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J. MutLalpha: at the cutting edge of mismatch repair. Cell. 2006;126:239–241. doi: 10.1016/j.cell.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Johnsborg O, Eldholm V, Havarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol. 2007;158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Dimopoulou ID, Lunter G, Harding RM, Ferguson DJ, Hood DW. Novel type IV secretion system involved in propagation of genomic islands. J Bacteriol. 2007a;189:761–771. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Power PM, Harding RM, et al. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007b;8:R237. doi: 10.1186/gb-2007-8-11-r237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Crook DW, Hood DW. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane D, Graumann PL. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell. 2005;122:73–84. doi: 10.1016/j.cell.2005.04.036. [DOI] [PubMed] [Google Scholar]
- Killoran MP, Keck JL. Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J Biol Chem. 2006a;281:12849–12857. doi: 10.1074/jbc.M600097200. [DOI] [PubMed] [Google Scholar]
- Killoran MP, Keck JL. Sit down, relax and unwind: structural insights into RecQ helicase mechanisms. Nucleic Acids Res. 2006b;34:4098–4105. doi: 10.1093/nar/gkl538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killoran MP, Keck JL. Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ. Nucleic Acids Res. 2008;36:3139–3149. doi: 10.1093/nar/gkn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killoran MP, Kohler PL, Dillard JP, Keck JL. RecQ DNA helicase HRDC domains are critical determinants in Neisseria gonorrhoeae pilin antigenic variation and DNA repair. Mol Microbiol. 2009;71:158–171. doi: 10.1111/j.1365-2958.2008.06513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund–Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Lång E, Haugen K, Fleckenstein B, Homberset H, Frye SA, Ambur OH, Tønjum T. Identification of neisserial DNA binding components. Microbiology. 2009;155:852–862. doi: 10.1099/mic.0.022640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie S. Studies on the phenotypic expression of competence in Neisseria meningitidis. Acta Pathol Mic Sc. 1965;64:119–129. doi: 10.1111/apm.1965.64.1.119. [DOI] [PubMed] [Google Scholar]
- Lin Z, Nei M, Ma H. The origins and early evolution of DNA mismatch repair genes – multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 2007;35:7591–7603. doi: 10.1093/nar/gkm921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybarger SR, Sandkvist M. Microbiology. A hitchhiker's guide to type IV secretion. Science. 2004;304:1122–1123. doi: 10.1126/science.1098806. [DOI] [PubMed] [Google Scholar]
- Lynch HT, Lynch JF. Hereditary nonpolyposis colorectal cancer (Lynch syndromes I and II): a common genotype linked to oncogenes? Med Hypotheses. 1985;18:19–28. doi: 10.1016/0306-9877(85)90115-x. [DOI] [PubMed] [Google Scholar]
- Mathis LS, Scocca JJ. Haemophilus influenzae and Neisseria gonorrhoeae recognize different specificity determinants in the DNA uptake step of genetic transformation. J Gen Microbiol. 1982;128:1159–1161. doi: 10.1099/00221287-128-5-1159. [DOI] [PubMed] [Google Scholar]
- Matson SW, Bean DW, George JW. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- McGillivary G, Tomaras AP, Rhodes ER, Actis LA. Cloning and sequencing of a genomic island found in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect Immun. 2005;73:1927–1938. doi: 10.1128/IAI.73.4.1927-1938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D, Covacci A, Donati C. Protein homology network families reveal step-wise diversification of Type III and Type IV secretion systems. PLoS Comput Biol. 2006;2:e173. doi: 10.1371/journal.pcbi.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehr IJ, Seifert HS. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol. 1998;30:697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- Michod RE, Bernstein H, Nedelcu AM. Adaptive value of sex in microbial pathogens. Infect Genet Evol. 2008;8:267–285. doi: 10.1016/j.meegid.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Mishina Y, Duguid EM, He C. Direct reversal of DNA alkylation damage. Chem Rev. 2006;106:215–232. doi: 10.1021/cr0404702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle S, Carbonnelle E, Matic I, Nassif X. Contact with host cells induces a DNA repair system in pathogenic Neisseriae. Mol Microbiol. 2005;55:853–861. doi: 10.1111/j.1365-2958.2004.04426.x. [DOI] [PubMed] [Google Scholar]
- Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- Narra HP, Ochman H. Of what use is sex to bacteria? Curr Biol. 2006;16:R705–R710. doi: 10.1016/j.cub.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Michod RE. Sex as a response to oxidative stress: the effect of antioxidants on sexual induction in a facultatively sexual lineage. Proc Biol Sci. 2003;270(suppl 2):S136–S139. doi: 10.1098/rsbl.2003.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu AM, Marcu O, Michod RE. Sex as a response to oxidative stress: a twofold increase in cellular reactive oxygen species activates sex genes. Proc Biol Sci. 2004;271:1591–1596. doi: 10.1098/rspb.2004.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst B, Wagner S, Sewing KF, Beil W. Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis. 2000;21:1111–1115. [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- O'Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, Radicella JP. Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization. P Natl Acad Sci USA. 2003;100:2789–2794. doi: 10.1073/pnas.0337641100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzabal OA, Rad R, Backert S. Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J Clin Microbiol. 2007;45:402–408. doi: 10.1128/JCM.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchevskiy V, Finkel SE. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol. 2006;188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Achtman M, James KD, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000a;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000b;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Poterszman A, Lamour V, Egly JM, Moras D, Thierry JC, Poch O. A eukaryotic XPB/ERCC3-like helicase in Mycobacterium leprae? Trends Biochem Sci. 1997;22:418–419. doi: 10.1016/s0968-0004(97)01124-9. [DOI] [PubMed] [Google Scholar]
- Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- Rad ME, Bifani P, Martin C, et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg Infect Dis. 2003;9:838–845. doi: 10.3201/eid0907.020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield RJ, Schrag MR, Dean AM. The evolution of bacterial transformation: sex with poor relations. Genetics. 1997;146:27–38. doi: 10.1093/genetics/146.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven NB, Koonin EV, Rudd KE, Deutscher MP. The gene for the longest known Escherichia coli protein is a member of helicase superfamily II. J Bacteriol. 1995;177:5393–5400. doi: 10.1128/jb.177.19.5393-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Yu Z, Popovic T, Stojiljkovic I. Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. P Natl Acad Sci USA. 2002;99:6103–6107. doi: 10.1073/pnas.092568699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M. Biology of type II secretion. Mol Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- Schaaff F, Reipert A, Bierbaum G. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Ch. 2002;46:3540–3548. doi: 10.1128/AAC.46.11.3540-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer L, Roy R, Humbert S, et al. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- Schmid SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott R, Pohl S, Burghard S, Weig M, Gross U. Identification and characterization of a major subgroup of conjugative Campylobacter jejuni plasmids. J Infect. 2005;50:12–21. doi: 10.1016/j.jinf.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Seeberg E, Eide L, Bjørås M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- Shibata A, Parsonnet J, Longacre TA, et al. CagA status of Helicobacter pylori infection and p53 gene mutations in gastric adenocarcinoma. Carcinogenesis. 2002;23:419–424. doi: 10.1093/carcin/23.3.419. [DOI] [PubMed] [Google Scholar]
- Sibbald MJ, Ziebandt AK, Engelmann S, et al. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol Mol Biol R. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Davies BW, Grossman AD, Walker GC. Beta clamp directs localization of mismatch repair in Bacillus subtilis. Mol Cell. 2008;29:291–301. doi: 10.1016/j.molcel.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha KM, Stephanou NC, Unciuleac MC, Glickman MS, Shuman S. Domain requirements for DNA unwinding by mycobacterial UvrD2, an essential DNA helicase. Biochemistry. 2008;47:9355–9364. doi: 10.1021/bi800725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot LM, Franke DD, McGillivary G, Actis LA. Genomic analysis of the F3031 Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization. Infect Immun. 2002;70:2694–2699. doi: 10.1128/IAI.70.5.2694-2699.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LA, Jarvis SA, Saunders NJ. Complete and variant forms of the ‘gonococcal genetic island’ in Neisseria meningitidis. Microbiology. 2005;151:4005–4013. doi: 10.1099/mic.0.27925-0. [DOI] [PubMed] [Google Scholar]
- Sparling PF. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92:1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, Seemann T, Harrison PF, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl EA, Seifert HS. Neisseria gonorrhoeae DNA recombination and repair enzymes protect against oxidative damage caused by hydrogen peroxide. J Bacteriol. 2006;188:7645–7651. doi: 10.1128/JB.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tala A, De Stefano M, Bucci C, Alifano P. Reverse transcriptase-PCR differential display analysis of meningococcal transcripts during infection of human cells: up-regulation of priA and its role in intracellular replication. BMC Microbiol. 2008;8:131. doi: 10.1186/1471-2180-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Tønjum T, Koomey M. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships – a review. Gene. 1997;192:155–163. doi: 10.1016/s0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Ambur OH, Tønjum T, Rocha EP. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol. 2008;9:R60. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Maiques E, Knecht E, Lasa I, Novick RP, Penades JR. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- Vermeulen W, Scott RJ, Rodgers S, et al. Clinical heterogeneity within xeroderma pigmentosum associated with mutations in the DNA repair and transcription gene ERCC3. Am J Hum Genet. 1994;54:191–200. [PMC free article] [PubMed] [Google Scholar]
- Voloshin ON, Camerini-Otero RD. The DinG protein from Escherichia coli is a structure-specific helicase. J Biol Chem. 2007;282:18437–18447. doi: 10.1074/jbc.M700376200. [DOI] [PubMed] [Google Scholar]
- Vriesema AJ, Beekhuizen H, Hamdi M, et al. Altered gene expression in Staphylococcus aureus upon interaction with human endothelial cells. Infect Immun. 2000;68:1765–1772. doi: 10.1128/iai.68.4.1765-1772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Maier RJ. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect Immun. 2008;76:153–160. doi: 10.1128/IAI.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Alamuri P, Maier RJ. The diverse antioxidant systems of Helicobacter pylori. Mol Microbiol. 2006;61:847–860. doi: 10.1111/j.1365-2958.2006.05302.x. [DOI] [PubMed] [Google Scholar]
- Weeda G, van Ham RC, Vermeulen W, Bootsma D, van der Eb AJ, Hoeijmakers JH. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990;62:777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- Woodbury RL, Wang X, Moran CP., Jr Sigma X induces competence gene expression in Streptococcus pyogenes. Res Microbiol. 2006;157:851–856. doi: 10.1016/j.resmic.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infec. 2007;13:5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- Wu L, Chan KL, Ralf C, et al. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Tao H, Park DI, Sepulveda JL, Sepulveda AR. Demonstration and characterization of mutations induced by Helicobacter pylori organisms in gastric epithelial cells. Helicobacter. 2006;11:272–286. doi: 10.1111/j.1523-5378.2006.00408.x. [DOI] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Shao SH, Cui LL, Mu RH, Ju XL, Dong SR. Type IV secretion system in Helicobacter pylori: a new insight into pathogenicity. Chinese Med J. 2007;120:2138–2142. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.