Abstract
Maintenance of the shape and diameter of biological tubules is a critical task in the development and physiology of all metazoan organisms. We have cloned the exc-9 gene of C. elegans, which regulates the diameter of the single-cell excretory canal tubules. exc-9 encodes a homologue of the highly expressed mammalian intestinal LIM-domain protein CRIP, whose function has not previously been determined. A second well-conserved CRIP homologue functions in multiple valves of C. elegans. EXC-9 shows genetic interactions with other EXC proteins, including the EXC-5 guanine exchange factor that regulates CDC-42 activity. EXC-9 and its nematode homologue act in polarized epithelial cells that must maintain great flexibility at their apical surface; our results suggest that CRIPs function to maintain cytoskeletal flexibility at the apical surface.
Keywords: Tubulogenesis, LIM domain, Cysteine-Rich Intestinal Protein, FGD1, epithelia
Introduction
Tubular structures play important biological roles in all metazoan organisms. Maintaining the structure of tubules such as vasculature, lung sacs, and kidney nephrons is critical for many biological functions (Lubarsky and Krasnow, 2003). Most tubules are constructed from a series of linked epithelial cells. Tubules such as the Caenorhabditis elegans excretory canal and the single-cell tips of branches of the Drosophila trachea provide simplified models in which a single epithelial cell forms a long tubular structure (Buechner, 2002; Casanova, 2007; Ghabrial and Krasnow, 2006; Kerman et al., 2006; Tonning et al., 2005).
The C. elegans excretory canal regulates the osmolarity of the nematode (Nelson and Riddle, 1984). The excretory canal cell body forms and extends a tubule from each side of the cell body on the ventral surface of the nematode; the two tubules bifurcate when they reach the lateral side, to form an H-shaped structure that extends the entire length of the animal (Nelson et al., 1983; White, 1987). Guidance and full extension of the canals to the anterior and posterior ends of the animal are compromised in mutants affecting basolateral receptors and basement membrane proteins such as the UNC-6 netrin, EPI-1 laminin, and UNC-52 heparan sulfate proteoglycan (Buechner et al., 1999; Hedgecock et al., 1987; Huang et al., 2003). In contrast, apically expressed proteins affect the morphology of the lumen of the canals. These proteins include ion channels and pumps (Berry et al., 2003; Liegeois et al., 2007), cytoskeletal proteins (Gao et al., 2001; Gobel et al., 2004; McKeown et al., 1998; Praitis et al., 2005; Suzuki et al., 2001), an apically secreted protein (Jones and Baillie, 1995), and proteins that affect trafficking of apical cytoskeletal components (Fujita et al., 2003). The functions of a large number of other genes that alter the apical cytoskeleton to affect excretory canal shape remain to be identified (Buechner et al., 1999). A single cell must closely regulate the relative growth of the apical and basal surfaces in order to create a lumen of uniform diameter that extends to the full length of the tubule (Buechner, 2002). In addition, outgrowth of the excretory canals occurs while the animal is growing and bending, so the cytoskeleton at both the apical and basal surfaces of these narrow tubules must exhibit significant flexibility during and after growth.
We have identified the EXC-9 protein as a small LIM-domain protein homologous to the mammalian Cysteine-Rich Intestinal Protein (CRIP) (Blackshaw et al., 2004; Davis et al., 1998; Lanningham-Foster et al., 2002). A second CRIP homologue found in multiple valve tissues of C. elegans can substitute for EXC-9 if expressed in the excretory canal, and its absence inhibits the function of these valves. The function of CRIP proteins in mammals is presently not known. Overexpression of exc-9 in other exc backgrounds indicates that EXC-9/CRIP acts upstream of the EXC-5 FGD-like guanine exchange factor (GEF). We suggest that CRIPs modulate stability of the actin cytoskeleton to affect cell morphology.
Materials and methods
Nematode genetics
All mutants were derived from the N2 Bristol strain background. The exc-9(gk395) deletion allele was generously supplied by the C. elegans Knockout Consortium (Vancouver, http://ko.cigenomics.bc.ca). Other strains were generously supplied by the Caenorhabditis Genetics Center (Minneapolis; http://biosci.umn.edu/CGC). All strains were maintained as described (Brenner, 1974).
We refined the map position of exc-9 on linkage group IV (Buechner et al., 1999) by use of snip-SNP mapping. bli-6(sc16) exc-9(n2669) and exc-9(n2669) fem-3(q20) double mutants were crossed to the Hawaiian strain CB4856. From the heterozygous F1 progeny, Bli non-Exc and Exc non-Fem recombinant F2 progeny were picked and allowed to self-fertilize. Homozygous F3 progeny were tested for the presence of CB4856 SNP markers on LG IV. SNP site C06A6:2004 was tested on Bli non-Exc recombinants, and SNP site B0218:15870 was tested on Exc non-Fem recombinants; the results indicated that these two sites are the boundary of exc-9. Cosmids in this region, provided by the Sanger Center, Cambridge, UK, were prepared and microinjected into worms as described (Mello and Fire, 1995). The plasmid or cosmid DNA was microinjected together with pRF4, which contains the dominant mutation rol-6(su1006) as marker (Kramer et al., 1990), into the gonadal syncytia of hermaphrodites. 100ng/μl was the concentration used for injection unless otherwise specified.
DNA
Transcriptional exc-9 construct: Primers specific to the 2.2kb upstream of predicted gene F20D12.5 (5’-CTGCAGTGTTGGCTCTCTGAAATGGA-3’ and 5’-AAGCTTCAACTTCGGGTCCTGGCACGA-3’) were used to amplify the exc-9 promoter from N2 genomic DNA. Translational construct: The same upstream primer was used in combination with a primer specific to the last predicted exon of exc-9 (5’-AAGCTTCAACTTCGGGTCCTGGCACGA-3’). For both constructs, amplified DNA was first ligated into pCR®-XL-TOPO® vector (Invitrogen), then digested with Pst I and Hind III and cloned into the multi-cloning site of pPD95.75 (gift of A. Fire), upstream and in-frame with the gfp gene.
Construction of Pvalv-1∷gfp, in which the valv-1 (B0496.7) promoter drives gfp expression, was done in a similar way as was the exc-9 transcriptional vector. The primers used are: 5’-CTGCAGGATTATTACGATGGTTTTG -3’; 5’-AAGCTTATCGTAATATCGTTCATTTT -3’.
The Pvha-1∷exc-9∷gfp construct, incorporating the exc-9∷gfp translational fusion under the control of the vha-1 promoter, was made by ligating the exc-9 coding region into the MscI and SacI restriction sites of the vector pCV01 (Oka et al., 2001). The primers used are:
exc-9 upstream: 5’-TGGCCACATTTCAGAGAGCCAACA-3’
exc-9 downstream: 5’-GAGCTCGGAACCTGAAAATGTTGAGAAT-3’
valv-1 upstream: 5’-TGGCCAATGCCAAACTGTCCAAAT-3’
valv-1 downstream: 5’-GAGCTCGGATTTCCAGTAGTTCCTTGAA-3’
RNAi
Double-stranded RNA was synthesized by use of the MEGAscript T7 kit (Ambion) with an exc- 9 PCR product (primers: 5’-TAATACGACTCACTATAGGAAGTCATTCGGAGGTTTGTTTAG-3’ and 5’-TAATACGACTCACTATAGGTCTTACCTGTCCAGTTTGTC-3’) from N2 genomic DNA as template. For valv-1, primers 5’-TAATACCACTCACTATAGGGTAAGTTGTTTTTTCAAGTT-3’ and 5’-TAATACCACTCACTATAGGTTTCCAGTAGTTCCTTGA-3’ were used. The dsRNA was microinjected directly into N2 animals without further treatment (Fire et al., 1998), along with plasmid pRF4 containing the rol-6(su1006) dominant allele as marker.
Sequence analysis
The exc-9 (n2669) amber mutation was identified via PCR amplification of the F20D12.5 sequence from DNA purified from exc-9 (n2669) mutant animals. The amplified fragment was ligated into pCR®2.1-TOPO® vector (Invitrogen). Five positive colonies with the insert were sequenced to make sure the error was not introduced randomly by the PCR. DNA sequencing of the amplified region was performed on both strands.
Alignment of the exc-9 gene and homologues was performed using ClustalW analysis on Vector NTI (Invitrogen), followed by the use of TreeView to draw the phylogenetic tree. Homologous sequences used for analysis (and their GenBank accession numbers) include: Nematode proteins EXC-9 (NP_501326), VALV-1 (NP_501187), CBG17716 (CAE70917), and CBG05866 (CAE61880); vertebrate proteins CRIP and CRP1 (AAP36964, XP_001333107, AAI35403, NP_031789), CRP2 (NP_001005968, NP_001303, NP_077185), and CRP3 (NP_996805, Q6Q6R3, XP_696083), insect CRP1 (XP_967808, XP_001122454); leech CRP1 (AAN73075); and distant CRP homologues from insect (XP_001120788, NP_651126, EAT43197, XP_310992, XP_969829) and mushroom (EAU87616). Additional vertebrate proteins (accession numbers XP_001501981, XP_001094052, XP_001137689, NP_001102773, XP_855529, Q5R7Y1, AAI35603, CAF93748, CAG13149, XP_001507066, XP_001364171, NP_001020520, EDL18557, EDL93791, XP_001171359, XP_001507066, NP_001087303, NP_001087271) mapped to the same branches in Fig. 3 as did the included genes, but were omitted for clarity. A full tree (including homologues from echinoderm and trematode, accession numbers XP_001188306, AAX30636) is included as Supplementary Fig. S1.
Fig. 3.
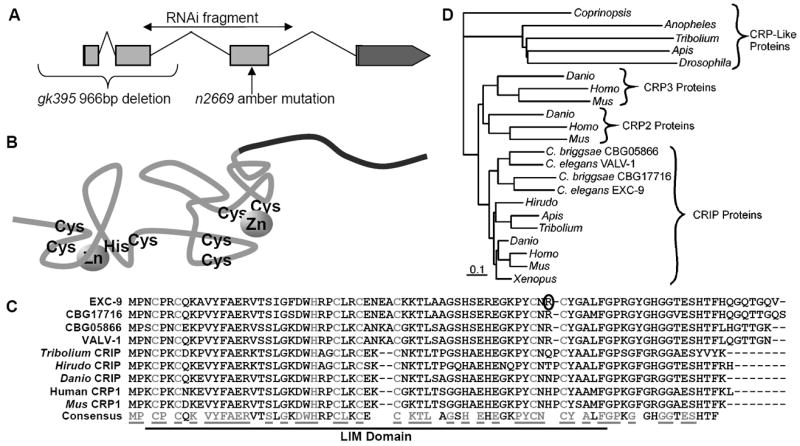
EXC-9 is a conserved small LIM-domain protein, CRIP. (A) Structure of the exc-9 gene, indicating exons, introns, and UTRs. The position of the n2669 and gk395 mutations are indicated, as is the amplified region used for PCR in order to perform RNAi. (B) exc-9 encodes a small protein containing a single LIM domain with 2 Zinc-finger domains, plus a short variable-length and -shape tail (in black) at the C-terminus. Structure (adapted from (Perez-Alvarado et al., 1996)) shows approximate positions of conserved cysteine and histidine residues used to co-ordinate zinc binding. (C) Alignment of EXC-9 and other CRIP/CRP1 proteins shows well-conserved homology. The Zn-binding cysteine and histidine residues of the LIM domain are shown in grey, as are all residues in the consensus sequence that are 100% conserved between these proteins. The arginine residue of EXC-9 that is mutated to a stop codon in the n2669 allele is circled. (D) Phylogenetic tree of EXC-9 and CRIP homologues suggest that EXC-9 and VALV-1 result from a duplication found only in nematodes. EXC-9 and VALV-1 show closest homology to the CRIP/CRP1 family of vertebrate proteins.
Microscopy
Worms and embryos were observed at high magnification on 2% agarose pads in M9 buffer, with 1% of 1-phenoxy-2-propanol as anaesthetic added as needed (Sulston and Hodgkin, 1988). DIC and/or fluorescence microscopy (Sulston and Hodgkin, 1988) images were taken on a Zeiss Axioskop Microscope with a MagnaFire Electronic Camera (Optronics), and images merged with Corel PhotoPaint software. GFP-positive cells were identified according to their position and morphology (www.wormatlas.org) (Sulston and Horvitz, 1977).
Canal Measurements
Excretory canals of living animals expressing a GFP canal marker were observed through a Zeiss Axioskop Microscope. Shortening of the lumen due to cyst formation was graded as either: completely shortened with no lumen visible past cysts at the cell body (0); significantly shortened to less than half-length (1); lumen reaching only midlength to the area around the vulva (2); lumen slightly shortened, reaching to between the vulva and tail (3); or no shortening/full length to or past the anus (4). While many animals had 1 or 2 large cysts, we instead graded the overall general canal cyst size either as: large - over half the width of the animal (0); medium – ¼ to ½ the width of the animal (1.3); small – less than ¼ the width of the animal (2.7); or no cysts (4). Wild-type canals were graded as (4) for both canal length and cyst size, while highly affected cystic mutants have lower scores. “Convoluted canals,” in which canal extension fails because the basolateral surface is affected (Suzuki et al., 2001) rather than due to shortening via cyst formation and failure of the apical surface, were graded as full-length (4) for length, and 4 (no cysts) for cyst size. An overall score was calculated by averaging all scores together. Statistical analysis of canal length was conducted by binning similar categories: numbers of canals with poor growth (no or short outgrowth, scores 0 or 1); numbers of canals with partial growth (halfway or ¾ length, scores 1 or 2); and number of canals with full outgrowth (score 4). The binned results were then analysed via a 3x2 Fisher’s Exact Test to measure the presence of any full or partial rescue. Canal cyst lumen size was similarly evaluated with a 3x2 Fisher’s Exact Test by binning numbers of canals with large or very large cysts (score 0 or 1.3); vs. number of canals with small cysts (score 2.7); and number of canals with no cysts (score 4). A p-value of 0.0001% or lower was regarded as strong statistical significance of rescue; p-values between 0.0001% and 0.01% or less were regarded as partial rescue.
Yeast 2-Hybrid Assay
A cDNA clone of exc-9 was purified and sent to the Molecular Interaction Facility of the University of Wisconsin, Madison, where the clone was ligated into a bait vector (encoding GAL4 downstream of EXC-9) and tested via 2-hybrid assay at low stringency with the Facility’s library of 36 million C. elegans RNAs derived from mixed-stage hermaphrodites. Five putative interactors were isolated; one was eliminated as a false positive because it also interacted with empty bait vector. The four validated prey clones all encoded CSN-5, the nematode homologue to subunit 5 of the COP9 signalosome.
Results
exc-9 mutation alters epithelial structure
Mutation in exc-9 was first identified through changes in the shape and size of the lumen of the excretory canal. Normally, the canal tubules extend the length of the animal and have a lumen that is only several microns wide (Buechner et al., 1999) (Fig. 1A, B). In contrast, both exc-9 alleles (n2669 and gk335) exhibit penetrant defects in canal morphology; the canal lumen is variably wide and short, often with septations between cystic areas (Fig. 1C). In n2669, almost half or the animals exhibit at least one very large fluid-filled cyst that is greater than half the diameter of the animal (Fig. 1D), and all animals have multiple smaller cysts (Table). In addition to canal defects, mutations in exc-9 affect the fine structure of the thin tail whip of hermaphrodite worms (Fig. 1E); 40% of hermaphrodite exc-9 mutants show tail defects (Fig. 1F). Finally, male exc-9 mutants show defects in genital development. Wild-type males form 9 rays on each side of the tail (Fig. 1G); each ray is composed of a single epithelial cell surrounding a pair of neurons (Emmons, 2005). In 40% of exc-9 mutant male animals, ray development is abnormal and leads to abnormally shaped or fused rays (Fig. 1H). As a result, exc-9 male worms show impaired mating efficiency, but overall are capable of mating.
Fig. 1.
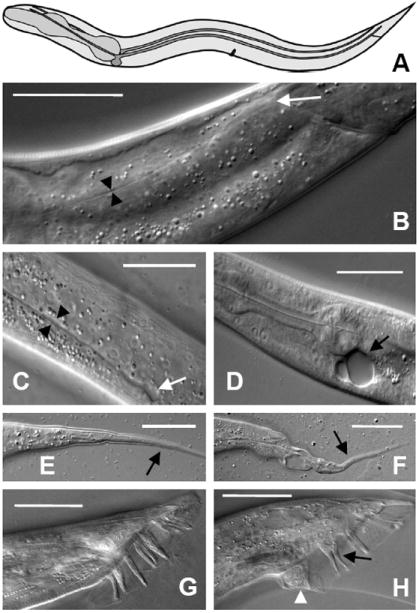
exc-9 Mutant Phenotype. (A) Schematic drawing of excretory canals (in blue) in adult wild-type worm. Anterior to the left. For relative position, the pharynx is indicated in dark grey, vulva at the center of the animal, and anus at the posterior. The cell body beneath the pharynx sends processes right and left, which bifurcate to create four canals that extend the length of the animal. (B) DIC micrograph of canal lumen in wild-type young adult. Black arrowheads indicate the canal lumen diameter. The tip of the canal lumen (white arrow) is located past the anal opening near the tail of the worm. (C) Section near center of exc-9(n2669) mutant shows wider lumen (arrowheads) than in wild-type worm and fluid-filled cyst (white arrow) at end of shortened canal. (D) Another exc-9(n2669) animal shows large cysts at the cell body and no canal extension. (E) Tail whip (arrow) in wild-type hermaphrodite tapers smoothly to a point. (F) Tail whip (arrow) of exc-9(n2669) animal has a poorly formed tip. (G) Wild-type male has 9 sensory rays at one side of the tail, while exc-9(n2669) animal (H) has malformed ray (white arrowhead) and attached rays (black arrow). Scale bars: 50μm.
Table 1.
Interactions of EXC-9 with other exc mutants.
| Canal lumen length, % |
Cyst size, % |
p-value (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full-length | |||||||||||||||
| Genotype | Injected with | n | None | Short | Midway (vulva) |
3/4 | WT | Convoluted | Large | Medium | Small | None | Score | Lumen length |
Cyst size |
| Wild-type | - | 100 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 100 | 4.0 | ||
| Wild-type | exc-9∷gfp | 44 | 0 | 0 | 0 | 0 | 77 | 23 | 0 | 0 | 0 | 100 | 4.0 | 100 | 100 |
| exc-9 (n2669) | - | 104 | 1 | 75 | 11 | 14 | 0 | 0 | 2 | 18 | 80 | 0 | 1.9 | ||
| exc-9 (n2669) | exc-9∷gfp | 41 | 0 | 27 | 34 | 15 | 0 | 24 | 0 | 10 | 56 | 34 | 2.7 | 2.75×10-8 | 6.76×10-7 |
| exc-9 (gk395) | - | 119 | 0 | 82 | 15 | 3 | 0 | 0 | 13 | 52 | 35 | 0 | 1.4 | ||
| exc-9 (gk395) | exc-9∷gfp | 161 | 0 | 17 | 34 | 34 | 6 | 9 | 0 | 1 | 86 | 13 | 2.7 | 1.86×10-25 | 5.07×10-37 |
| exc-9 (gk395) | exc-5∷gfp | 32 | 13 | 0 | 3 | 3 | 0 | 81 | 0 | 0 | 16 | 84 | 3.6 | 3.85×10-22 | 2.33×10-25 |
| exc-9 (gk395) | valv-1∷gfp | 34 | 0 | 32 | 62 | 6 | 0 | 0 | 0 | 18 | 82 | 0 | 2.1 | 1.30×10-5 | 1.06×10-4 |
| exc-5 (rh232) | - | 87 | 63 | 28 | 7 | 2 | 0 | 0 | 20 | 32 | 44 | 5 | 1.1 | ||
| exc-5 (rh232) | exc-9∷gfp | 382 | 16 | 54 | 22 | 7 | 1 | 1 | 34 | 38 | 28 | 1 | 1.2 | 0.0115 | 0.0116 |
| exc-2 (rh90) | - | 125 | 3 | 94 | 2 | 0 | 0 | 0 | 44 | 29 | 27 | 0 | 1.1 | ||
| exc-2 (rh90) | exc-9∷gfp | 117 | 0 | 33 | 52 | 9 | 0 | 7 | 2 | 16 | 76 | 7 | 2.2 | 1.49×10-25 | 4.01×10-17 |
| exc-4 (rh133) | - | 69 | 57 | 44 | 0 | 0 | 0 | 0 | 41 | 28 | 32 | 0 | 0.8 | ||
| exc-4 (rh133) | exc-9∷gfp | 29 | 16 | 76 | 0 | 0 | 0 | 8 | 14 | 21 | 45 | 21 | 1.7 | 8.54 | 0.161 |
| exc-1 (rh26) | - | 91 | 0 | 18 | 19 | 48 | 15 | 0 | 9 | 17 | 66 | 9 | 2.5 | ||
| exc-1 (rh26) | exc-9∷gfp | 119 | 14 | 16 | 24 | 24 | 22 | 0 | 13 | 7 | 63 | 17 | 2.3 | 2.05 | 20.7 |
| sma-1 (ru18) | - | 56 | 0 | 79 | 13 | 9 | 0 | 0 | 0 | 9 | 91 | 0 | 1.9 | ||
| sma-1 (ru18) | exc-9∷gfp | 132 | 0 | 59 | 19 | 3 | 1 | 19 | 1 | 2 | 79 | 19 | 2.4 | 0.0218 | 4.41×10-3 |
n, number of canals examined
EXC-9 is homologous to a mammalian intestinal protein with a conserved LIM domain
We used snip-SNP mapping to narrow the position of exc-9 to a region of 13 cosmids. Injection of cosmid F20D12 rescued the canal and tail whip phenotypes of exc-9(n2669) animals (Fig. 2A). Injections of dsRNA corresponding to the predicted gene F20D12.5 encoded on this cosmid caused defects both in the canals (Fig. 2B) and in the tail whip (Fig. 2C) identical to those of exc-9 mutation. Injection of a PCR fragment corresponding to the entire F20D12.5 gene rescued exc-9 mutants. A strain (VC976) carrying a deletion of the first two exons of F20D12.5 showed similar or somewhat more severe canal, tail whip, and male ray phenotypes as does exc-9(n2669), and failed to complement that allele. Finally, sequencing of the n2669 allele showed an amber mutation in the center of the predicted coding region. Since the canal morphology of the gk395 is somewhat more severe than that of n2669 (Table), this amber allele may allow a low level of EXC-9 function. We conclude that F20D12.5 encodes exc-9.
Fig. 2.
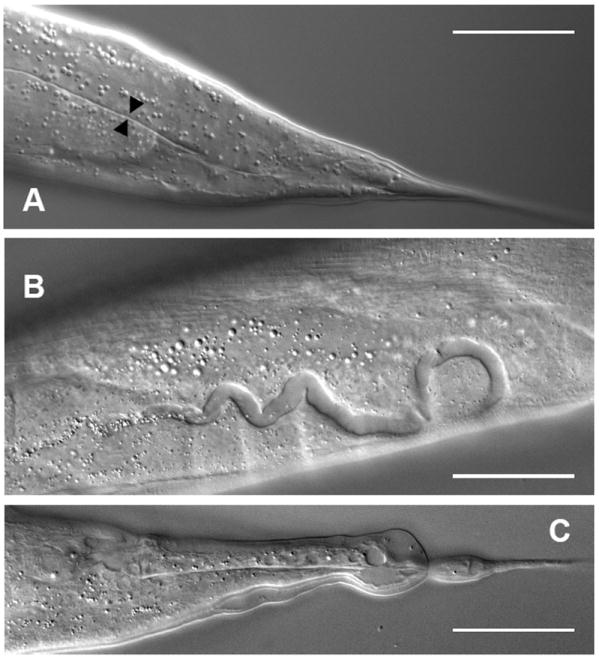
F210D12.5 encodes exc-9. (A) Canal in progeny of exc-9(n2669) animal microinjected with F20D12.5 cosmid DNA. Canal lumen is restored to normal diameter (black arrowheads), and extends all the way to the tail. (B, C) dsRNA corresponding to the exc-9 2nd and 3rd exons (364 bp, see Fig. 3A) was injected into wild-type worms. Progeny shows wider canals (B) and malformed tail whip (C), similar to the phenotype of exc-9 mutants. Scale bars: 50μm.
The exons of exc-9 cDNA as determined from 5’- and 3’-RACE (data not shown) match the GeneFinder algorithm predictions obtained from WormBase (www.wormbase.org) (Fig. 3A). This gene produces a single transcript that encodes an 85-aa protein. The protein contains a single LIM domain at the N-terminus (Fig. 3B) and shows 63% identity and over 70% similarity with both human and mouse CRIP (Cysteine-rich intestinal protein) (Lanningham-Foster et al., 2002), both in the LIM domain and in the remaining unique C-terminal end of the protein (Fig. 3C). LIM domains use cysteine and histidine in two zinc fingers to mediate protein-protein binding (Schmeichel and Beckerle, 1994). The function of CRIP is not presently known, but it is enriched in the mammalian gut, heart, lung, skin, and immune system (Levenson et al., 1993; Tsui et al., 1994) as well as in regenerating invertebrate neurons (Emes et al., 2003), and overexpression of CRIP in mice affects immune system function in the gut (Davis et al., 1998; Lanningham-Foster et al., 2002).
There is additionally another CRIP homologue in C. elegans homologue, B0496.7, which we have named valv-1. Since the gene structure and amino acid sequence of exc-9 and valv-1 are highly homologous (81% identity, 91% similarity) and the two genes are located close to each other on linkage group IV, valv-1 and exc-9 likely arose through a duplication of an ancestral gene in nematodes. There are similarly two copies of these CRIP homologues in C. briggsae (Fig. 3C). Different duplications occurred in mammals (Fig. 3D, Fig. S1), which have only a single CRIP orthologue, but also contain two other genes, CRP2 and CRP3, that each contain 2 complete CRIP sequences (both LIM domain and tail) held together in tandem (van Ham et al., 2003). Homology to CRIP is found in multiple animal phyla and in the basidiomycte fungus Coprinopsis. Several insect species have both a CRIP orthologue and a much larger more distant protein containing a CRIP motif, but a CRIP orthologue was not found in the fully sequenced Dipteran genomes Drosophila, Aedes, or Anopheles, and is presumably lost in this order.
EXC-9 and VALV-1 act cell-autonomously
To determine the expression pattern of EXC-9, we made a transcriptional construct carrying the 2.2kb exc-9 promoter region linked to gfp. Expression was found in the excretory canal cell, tail spike, uterine seam cell (UTSE), distal tip cells, intestine, ALM and PLN neurons, and nerve ring (Fig. 4A-E). For male worms, although ray development is affected, no GFP was detected in the mature rays (Fig. 4F, arrows), although GFP is weakly expressed in the tail spike in larval stages (Fig. 4G). The rescuing translational construct, containing the entire exc-9 promoter and coding regions linked to gfp, showed the same expression pattern, and expression was seen throughout the cytoplasm (FIG. 4H).
Fig. 4.
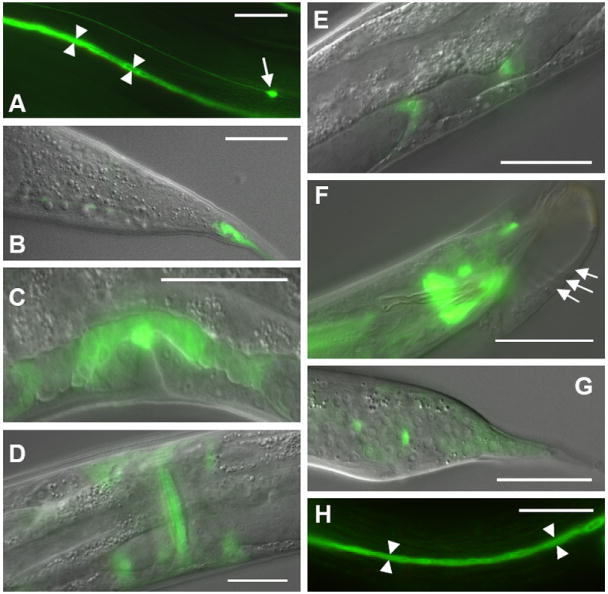
exc-9 is expressed in multiple epithelia and neurons. The 2.2kb exc-9 promoter was linked to gfp and microinjected into N2 worms. Strong expression was consistently found in the excretory canal cytoplasm (arrowheads) (A) and tail whip (B), anchor cell (C), uterine seam cell (ventral view) (D), distal tip cells (oblique dorsal view) (E), as well as in the ALM neurons (arrow in A) and PLN neurons (not shown). In males, a few neurons near the tail expressed the construct (F), and weaker expression was found throughout the developing larval male tail (G). Arrows in (F) indicate Rays 4-6 in newly molted male. A translational exc-9∷gfp construct was expressed throughout the cytoplasm of the canals and substantially rescued the cystic canal phenotype of exc-9(n2669) animals (H). Anterior to the left in all panels; scale bars 50 μm.
The translational GFP construct is lost spontaneously during some cell divisions. Mosaic animals that expressed this construct in other tissues where exc-9 is normally expressed, but not in the excretory canals, still show large cysts. In addition, we found that an exc-9∷gfp translational construct driven by the vha-1 promoter, which is expressed only in the excretory canal and head mesodermal cell (Oka et al., 2001), is capable of rescuing the exc-9 mutant’s cystic canal phenotype (Table), but cannot rescue the tail whip defects. We conclude from these results that EXC-9 acts cell-autonomously to regulate cell structure solely in the tissues in which it is expressed.
We additionally examined the role of the C. elegans EXC-9 homologue VALV-1. A construct containing the valv-1 promoter linked to gfp was expressed in the cytoplasm of multiple cells that control passage of material from tissue to another, such as the pharyngeal-intestinal valve (Fig. 5A) spermathecal-uterine valves, vulva (Fig. 5B), and the tissue of the rectal valve (Fig. 5C). Expression was also seen in tissue surrounding the pharynx (Fig. 5A), in the seam cells of the hypodermis that synthesize the cuticular alae (Fig. 5D), and in the gonadal sheath that surrounds the oocytes (Fig. 5E). No expression was found in the excretory canals. Since no knockout mutant of this gene is available, we reduced function of VALV-1 via injection of dsRNA specific to this gene. Progeny of injected animals that expressed the co-injected marker rol-6(su1006) were noticeably affected in the function of the valves where this protein normally is expressed. The animals appeared starved and had reduced ability to pass food into the intestine (not shown), even though pumping of the pharynx appeared to be normal (not shown). At the same time, they seemed unable to excrete properly. Finally, the animals often had fertilized oocytes trapped in the spermatheca, unable to continue into the uterus (Fig. 5F). We conclude that the VALV-1 protein is necessary for the ability of these valves to function efficiently.
Fig. 5.
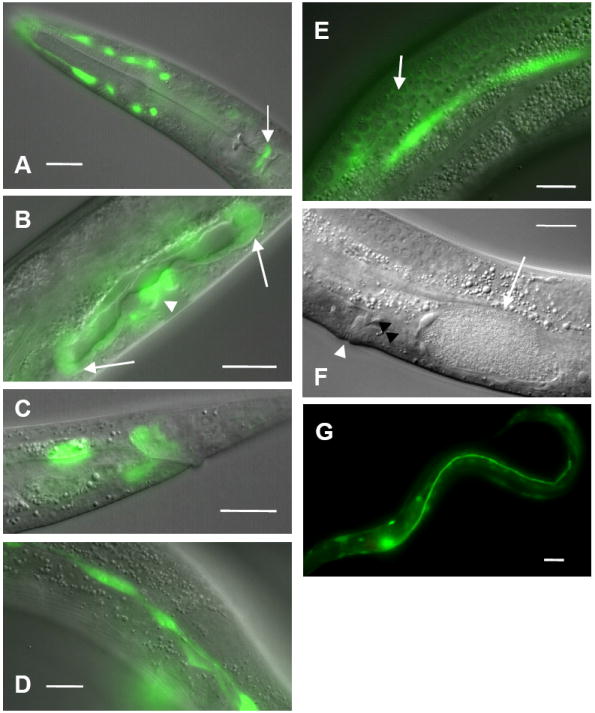
VALV-1 CRIP is expressed in multiple valves. The promoter for the valv-1 (B0496.7) gene was linked to gfp and microinjected into N2 worms. Strong expression was consistently found in the pharyngeal-intestinal valve (arrow) and tissue surrounding the pharynx, possibly the amphid sheath (A), spermathecal-uterine valves (arrows) and vulval epithelial cells (arrowhead) (B), intestinal-rectal valve (C), hypodermal seam cells (D), and gonadal sheath cell that surrounds developing oocytes (e.g. arrow; bright green is hypodermal seam cell out of focus) (E). (F) Wild-type animal injected with RNAi to valv-1 shows a fertilized egg (arrow) trapped in the spermatheca. Spermatocytes (black arrowhead) are visible between the oocyte and the uterus above the vulval opening (white arrowhead). (G) Expression of valv-1∷gfp in the canals largely rescues the Exc-9 mutant phenotype; the canal extends most of the way to the anus. Anterior to the left in all panels; scale bars 25 μm.
As noted, valv-1 is not expressed in the excretory canals. We linked this gene to gfp and placed it under the control of the exc-9 promoter. Expression of a valv-1∷gfp translational construct in the excretory canals greatly improved the Exc-9 mutant phenotype (Fig. 5G, Table), extending canal length and preventing the formation of large cystic canals, although complete restoration was not observed. We conclude that EXC-9 and VALV-1 perform substantially similar cellular tasks in different tissues, and that this task affects the structural integrity of the luminal surface of epithelial cells.
Overexpression of exc-9 also affects cell shape
High levels of expression of either functional EXC-9 or EXC-9∷GFP in the excretory canals caused a phenotype similar to that caused by overexpression of an exc-5 translational construct. The exc-5 gene encodes the nematode homologue to the FGD1 GEF that activates CDC-42 in mammalian cells (Gao et al., 2001; Olson et al., 1996; Suzuki et al., 2001). When EXC-5 is overexpressed in nematodes, the canals polarize normally and create a normal lumen from the apical surface, but the structure of the basolateral surface necessary for extension of the canals along the hypodermal basement membrane is not maintained (Huang et al., 2003; Suzuki et al., 2001). As for exc-5 overexpression, nematodes with strong levels of exc-9∷gfp expression exhibited canals that were incapable of extending to their full length, but which contained canal lumena of normal diameter circling inside the enlarged cell body (Fig. 6A). This “convoluted” canal phenotype was never seen in animals microinjected with the GFP vector alone (not shown), and could be seen both in N2 worms and in some exc-9 worms injected with high levels of the exc-9∷gfp construct under the expression of either the exc-9 or vha-1 promoter. The frequency of convoluted canals decreased when N2 worms were fed a bacterial strain expressing RNAi specific to gfp (data not shown). Finally, N2 nematodes expressing high levels of exc-9∷gfp also showed abnormal tail whip morphology (Fig. 6B). We conclude that, as for exc-5, not only does loss of EXC-9 activity result in disruption of the apical surface of the canals, but overexpression of EXC-9 disrupts structure at the basolateral surface of these tubules.
Fig. 6.
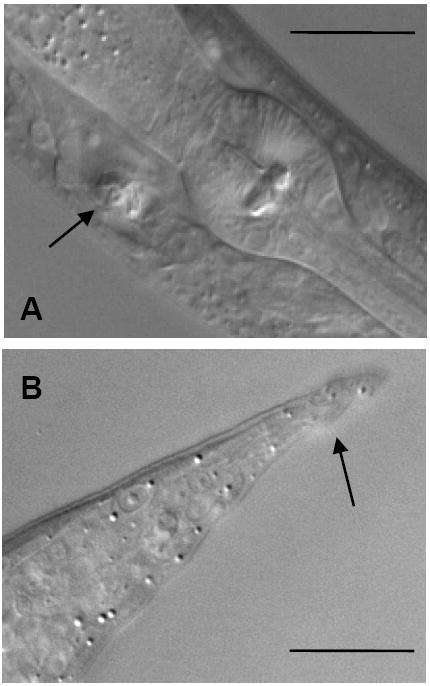
CRIP overexpression also affects epithelial shape. A) An animal with high levels of exc-9∷gfp expression exhibiting a convoluted canal phenotype, with the narrow lumen visible (arrow) wound up inside the excretory cell body (anterior to the right). (B) Abnormal tailspike in N2 animal containing high levels of the exc-9∷gfp transgene. Scale bars 25 μm.
EXC-9 expression rescues other exc Mutant phenotypes
Since overexpression of exc-9 behaves similarly to that of exc-5-overexpressing constructs, we tested whether exc-9 exhibited genetic interactions with the EXC-5 GEF and with other exc genes (Table). The exc-9∷gfp construct was injected into various other mutants with cystic canals to see if exc-9 overexpression could rescue the cystic phenotype.
We assessed the effects of injection by measuring the approximate length of the canals and general size of cysts, if any, to form an overall measure of canal morphology (see Materials and Methods). The exc-5 mutants form very large cysts predominantly at the tips of the shortened canals. Expression of exc-9∷gfp caused no statistically significant improvement on this phenotype. In contrast, in the reciprocal experiment, expression of exc-5∷gfp rescued the phenotype of exc-9(gk395) deletion mutants about as well as did injection of exc-9∷gfp (Table), yielding almost complete loss of cysts, much longer canals, and frequent formation of convoluted canals. The ability of EXC-5 to restore canal morphology to exc-9 mutants, while EXC-9 cannot restore the structure of exc-5 canals, strongly indicates that EXC-5 functions downstream of EXC-9 to maintain the structure of the epithelial apical surface.
In contrast to its inability to rescue exc-5, the expression of exc-9∷gfp was able to improve canal morphology in other exc mutants (Table). Mutants in exc-2, which has not yet been cloned, show severe cystic swelling of severely shortened canals. Expression of exc-9∷gfp in this mutant showed clear improvement of canal morphology, with animals presenting much smaller cysts, longer canals, and occasional convoluted canals. Weaker rescue by exc-9∷gfp was found for sma-1 mutants (encoding βH-spectrin (McKeown et al., 1998)), mutants of which have short wide canals with unseptate cysts. Finally, mutants in exc-1 (uncloned) present defects similar to those of exc-5, with large cysts predominantly at the terminus of the canals. Injection of exc-9∷gfp was unable to rescue the canal defects of exc-1 mutants (which has not been cloned). Mutants in exc-4 (encoding a CLC ion channel(Berry et al., 2003)) show severe cystic swelling similar to that of exc-2 mutants. Injection of exc-9∷gfp into these exc-4 mutants did cause the formation of some animals with wild-type canals, but not at a statistically significant level.
A yeast two-hybrid assay was performed to look for interactions between EXC-9 and other proteins. Out of 36 million clones tested from a mixed-stage hermaphrodite cDNA library, 4 yielded positive interactions. All 4 positive clones encoded CSN-5, the nematode homologue of subunit 5 of the COP9 signalosome which acts in conjunction with E3 ubiquitin ligases to degrade proteins (Schwechheimer, 2004). The C. elegans Interactome Project has previously found that VALV-1 also shows a yeast 2-hybrid interaction with CSN-5 (Li et al., 2004). However, we did not see strong GFP expression of a csn-5 promoter construct either in the excretory canals or in tissues where valv-1 is expressed (data not shown). The addition of 11 amino acids to the 5’ end of the exc-9 gene (the side of the gene where the Gal4 bait was attached) substantially reduces the gene’s ability to rescue exc-9 mutants and renders it unable to create animals with convoluted canals (data not shown). We suggest that the assay therefore indicated that EXC-9 and VALV-1 proteins can themselves degraded by the COP9 signalosome.
Discussion
EXC-9 is a nematode CRIP homologue
The Cysteine-Rich Intestinal Proteins were originally discovered as developmentally-regulated zinc-binding proteins in the mammalian gut (Birkenmeier and Gordon, 1986; Hempe and Cousins, 1991). They have since been found to be expressed in multiple tissues, including gut, heart, lung, skin, and the immune system (Levenson et al., 1993; Tsui et al., 1994), and more recently found as a component of the nervous system in annelids (Emes et al., 2003), but their function has remained obscure. No mutants have previously been discovered in CRIP-encoding genes. Overexpression of mouse CRIP in the intestine increased the ratio of Th2:Th1 immune response (Lanningham-Foster et al., 2002), and reduced overall white blood cell count (Davis et al., 1998), but the biochemical basis for this response is unidentified. The strong level of homology between human CRIP and nematode EXC-9 suggests that these proteins play a conserved role in cell biology. Our expression and functional studies indicate that EXC-9 acts in the excretory canal cytoplasm in a cell-autonomous manner to prevent failures of the lumenal surface. Mammalian CRIP may therefore have a similar role in regulating cell structure in those tissues where it is expressed.
LIM domains can bind to DNA or to other proteins. Expression of our exc-9 translational construct was predominantly cytoplasmic (Fig. 4H). In addition, the related mouse protein CRP2 was found to be expressed at in the cytoplasm and cell surface when expressed in COS cells (van Ham et al., 2003). We conclude that EXC-9 and VALV-1 likely bind to other proteins in the cytoplasm of the cells where they are expressed.
VALV-1 is a functional homologue of EXC-9
A second CRIP homologue in C. elegans has maintained a similar biological function as has EXC-9, and can substitute for EXC-9 if expressed in the excretory canals. VALV-1 is normally expressed, among other tissues, in junctions between several epithelial tubes, such as the pharyngeal-intestinal valve, spermathecal valve, and intestinal-anal valve. While no mutant of VALV-1 has yet been isolated, our RNAi studies suggest that VALV-1 function is necessary for function of these tissues. While these epithelia polarize normally and initially form a superficially normal structure, they have difficulty opening enough to pass material. Isolation of a mutant and further study of the ultrastructure of these tissues will be necessary to confirm the location and placement of VALV-1 protein and its function in cell contractility.
The tissues where EXC-9 and VALV-1 are expressed in C. elegans include: A) many particularly long processes, such as the excretory canals and PLN neuron, which extend nearly the entire length of the animal; B) tissues with thin protrusions or highly sculpted thin shapes, such as the tail whip and distal tip cells, and; C) valves and other tissues, such as the uterine seam cell, in which the apical surface must change shape rapidly in order to function. In the mammalian intestine, CRIP is expressed most highly in villus during weaning, a period of rapid morphological differentiation (Birkenmeier and Gordon, 1986). Villus cells, like the C. elegans canals, have highly sculpted surfaces, with a thick terminal web of actin and intermediate filaments anchoring thin characteristic structures: canalicular invaginations in the case of the excretory canals (Nelson et al., 1983), protruding microvilli in the case of the intestine (Heintzelman and Mooseker, 1992; Hirokawa et al., 1982). In annelids, CRIP is upregulated during injured nerve regeneration (Blackshaw et al., 2004). As an animal moves, long neuronal process as well as the long thin hollow canal processes will be required to bend repeatedly, which will greatly strain the surface of the canals. A common feature of all of these cell types is the necessity for rapid cytoskeletal remodeling during cell movement. A possible function for CRIP homologues may be to provide flexibility to thick cytoskeletal surfaces. Loss of CRIP function results in an apical surface that cannot move rapidly, and therefore breaks. Indeed, in exc mutants, individual cysts appear instantly and stochastically along the length of the canals as the animal moves and pumps liquid through the canals, as if the apical surface is brittle; and electron microscopy of some exc mutants clearly shows that the thick apical cytoskeleton is broken and fragmented around the cysts (Buechner et al., 1999).
EXC-9 acts with other EXC proteins to regulate tubule shape
exc-9 exhibits genetic interactions with other exc mutants. Results here show that expression of a functional exc-9∷gfp construct significantly rescues exc-2 mutants, and improves the phenotype of sma-1 mutants (and perhaps exc-4 mutants), but cannot rescue exc-1 or exc-5 mutants. In contrast, expression of exc-5 prevents formation of cysts in exc-9 mutants. Our results strongly suggest that EXC-9 CRIP functions in a pathway upstream of EXC-5 to regulate apical cytoskeletal structure (Fig. 7). Yeast two-hybrid data taken by themselves could indicate that EXC-9 and VALV-1 potentially act on CSN-5-mediated protein degradation, and suggest that perhaps EXC-9 prevents turnover of EXC-5. Since csn-5 is not strongly expressed in larval or adult excretory canals or neurons, however, it is more likely that the yeast two-hybrid interactions resulted from binding of EXC-9 degradation products to CSN-5. The presence of the GAL4 DNA-binding domain at the N-terminus of the small CRIP protein may prevent it from binding to its normal partner in a yeast two-hybrid assay.
Fig. 7.
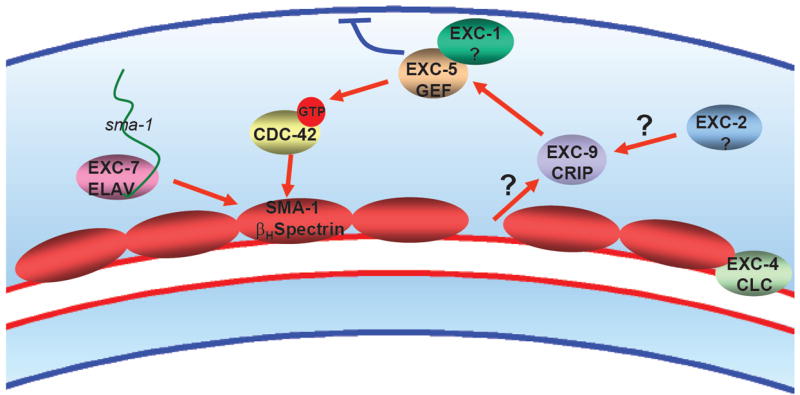
Model of EXC protein interactions. Diagram depicts a section of the excretory canals. sma-1 mRNA is transported via EXC-7 ELAV and is translated along the length of the canals. We hypothesize that bending of the tube stretches or causes a gap at the apical cytoskeleton that sends a signal to EXC-9, which activates EXC-1 and EXC-5 to cause CDC-42 to be phosphorylated and active, and direct the growth and stability of the apical cytoskeleton, while inhibiting the basolateral cytoskeleton. In this model, placement of spectrin at the apical surface would repress activation of EXC-9.
exc-5 encodes a homologue of the FGD1 (Facial-Genital Dysplasia) GEF defective in Aarskog-Scott syndrome (Gao et al., 2001; Suzuki et al., 2001). Mammalian FGD1 activates the small GTPase CDC-42 in mammalian tissue culture to regulate apical cytoskeletal morphology (Zheng et al., 1996), and also uses an SH3 domain to stimulate cortactin to polymerize actin via Arp 2/3 (Kim et al., 2004), Nematode EXC-5 lacks this SH3 domain, and therefore should affect cytoskeletal structure predominantly through GEF activity. EXC-5 and EXC-9 are both highly expressed in the excretory canals and tailspike. In addition, EXC-5 is found in some valve cells and is highly expressed in the muscular nematode pharynx, while EXC-9 is not found in this tissue. Interestingly, mutations in another mammalian homologue, FGD4, result in Charcot-Marie-Tooth syndrome, a form of muscular dystrophy (Delague et al., 2007). The differences in nematode tissue expression patterns suggests that VALV-1 could activate EXC-5 in some tissues, and that EXC-5 has separate functions unregulated by either EXC-9 or VALV-1 in muscle tissue.
The identity of EXC-2 and EXC-1 are not yet known, although since the phenotype of exc-1 mutants resembles that of exc-5 mutants, and since overexpression of exc-9 is similarly unable to rescue exc-1 mutants, EXC-1 may function together with EXC-5 downstream of EXC-9 in a pathway that activates CDC-42 at the canal apical surface. EXC-4 encodes a CLC chloride channel (Berry et al., 2003); it is possible that the function of this channel may provide an ionic environment necessary for EXC-9 to function normally.
Based on these results, we suggest a model of tubule maintenance in the excretory canals as presented in Figure 7. The presence of the EXC-4 CLC channel and EXC-2 allow EXC-9 to function. EXC-9 signals through EXC-5 (and presumably EXC-1) to activate CDC-42 and possibly other small GTPases to maintain the flexibility of the apical epithelial surface. The cytoskeletal terminal web is anchored to the luminal surface through the β-heavy spectrin SMA-1. Since exc-9 overexpression partially rescues sma-1 mutation, we hypothesize that placement of spectrin at the apical surface may send a signal to EXC-9 to modulate the activity of this pathway in a feedback loop, while stretching or lack of spectrin at the apical surface could increase EXC-9 activity. Future studies should determine other potential binding partners and the biochemical role of CRIP proteins in signaling to the apical surface, how this signaling may repress cytoskeletal maintenance at the basolateral surface, as well as the identity of EXC-1 and other proteins that form and maintain the structure and malleability of apical cytoskeleton in tubules and other epithelia.
Supplementary Material
Supplementary Figure 1. Complete phylogenetic tree of EXC-9 and all CRIP homologues (shown in abbreviated form in Fig. 3D), including echinoderm, trematode, and additional vertebrate homologues.
Acknowledgments
We thank Drs. Andrew Fire, T. Oka, and M. Futai for vectors used to make exc-9 and exc-5 constructs; Dr. Andrew Chisholm for first supplying the n2669 allele of exc-9; Lisa Timmons for a bacterial strain expressing dsRNA to gfp; the Caenorhabditis Genetics Center for strains; the C. elegans Knockout Project for isolation of the gk395 deletion allele of exc-9; Stuart MacDonald for help in performing statistical analyses, Lena Hileman for help with evolutionary trees, and Lisa Timmons, Erik Lundquist, Robert S. Cohen, Victoria Corbin, William S. Dentler, and Kathy Suprenant for helpful discussions. This work has been supported by NIH R01 DK55526. Dr. Tong was supported in part through the Kansas City Area Life Science Institute and the NSF EPSCoR for Ecological Genomics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berry KL, et al. A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science. 2003;302:2134–7. doi: 10.1126/science.1087667. [DOI] [PubMed] [Google Scholar]
- Birkenmeier EH, Gordon JI. Developmental regulation of a gene that encodes a cysteine-rich intestinal protein and maps near the murine immunoglobulin heavy chain locus. Proc Natl Acad Sci U S A. 1986;83:2516–20. doi: 10.1073/pnas.83.8.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw SE, et al. Identifying genes for neuron survival and axon outgrowth in Hirudo medicinalis. J Anat. 2004;204:13–24. doi: 10.1111/j.1469-7580.2004.00260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechner M. Tubes and the single C. elegans excretory cell. Trends Cell Biol. 2002;12:479–84. doi: 10.1016/s0962-8924(02)02364-4. [DOI] [PubMed] [Google Scholar]
- Buechner M, et al. Cystic canal mutants in Caenorhabditis elegans are defective in the apical membrane domain of the renal (excretory) cell. Developmental Biology. 1999;214:227–241. doi: 10.1006/dbio.1999.9398. [DOI] [PubMed] [Google Scholar]
- Casanova J. The emergence of shape: notions from the study of the Drosophila tracheal system. EMBO Rep. 2007;8:335–9. doi: 10.1038/sj.embor.7400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BA, et al. Structural characterization of the rat cysteine-rich intestinal protein gene and overexpression of this LIM-only protein in transgenic mice. DNA Cell Biol. 1998;17:1057–64. doi: 10.1089/dna.1998.17.1057. [DOI] [PubMed] [Google Scholar]
- Delague V, et al. Mutations in FGD4 encoding the Rho GDP/GTP exchange factor FRABIN cause autosomal recessive Charcot-Marie-Tooth type 4H. Am J Hum Genet. 2007;81:1–16. doi: 10.1086/518428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes RD, et al. HmCRIP, a cysteine-rich intestinal protein, is expressed by an identified regenerating nerve cell. FEBS Lett. 2003;533:124–8. doi: 10.1016/s0014-5793(02)03741-9. [DOI] [PubMed] [Google Scholar]
- Emmons SW. The C elegans Research Community. Wormbook; 2005. Male development. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fujita M, et al. The role of the ELAV homologue EXC-7 in the development of the Caenorhabditis elegans excretory canals. Dev Biol. 2003;256:290–301. doi: 10.1016/s0012-1606(03)00040-x. [DOI] [PubMed] [Google Scholar]
- Gao J, et al. The Caenorhabditis elegans homolog of FGD1, the human Cdc42 GEF gene responsible for faciogenital dysplasia, is critical for excretory cell morphogenesis. Hum Mol Genet. 2001;10:3049–62. doi: 10.1093/hmg/10.26.3049. [DOI] [PubMed] [Google Scholar]
- Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–9. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- Gobel V, et al. Lumen morphogenesis in C. elegans requires the membrane-cytoskeleton linker erm-1. Dev Cell. 2004;6:865–73. doi: 10.1016/j.devcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, et al. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- Heintzelman MB, Mooseker MS. Assembly of the intestinal brush border cytoskeleton. Curr Top Dev Biol. 1992;26:93–122. doi: 10.1016/s0070-2153(08)60442-1. [DOI] [PubMed] [Google Scholar]
- Hempe JM, Cousins RJ. Cysteine-rich intestinal protein binds zinc during transmucosal zinc transport. Proc Natl Acad Sci U S A. 1991;88:9671–4. doi: 10.1073/pnas.88.21.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, et al. Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J Cell Biol. 1982;94:425–43. doi: 10.1083/jcb.94.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, et al. Laminin alpha subunits and their role in C. elegans development. Development. 2003;130:3343–58. doi: 10.1242/dev.00481. [DOI] [PubMed] [Google Scholar]
- Jones SJ, Baillie DL. Characterization of the let-653 gene in Caenorhabditis elegans. Mol Gen Genet. 1995;248:719–26. doi: 10.1007/BF02191712. [DOI] [PubMed] [Google Scholar]
- Kerman BE, et al. From fate to function: the Drosophila trachea and salivary gland as models for tubulogenesis. Differentiation. 2006;74:326–48. doi: 10.1111/j.1432-0436.2006.00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, et al. Effect of Fgd1 on cortactin in Arp2/3 complex-mediated actin assembly. Biochemistry. 2004;43:2422–7. doi: 10.1021/bi036173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, et al. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–9. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanningham-Foster L, et al. Overexpression of CRIP in transgenic mice alters cytokine patterns and the immune response. Am J Physiol Endocrinol Metab. 2002;282:E1197–203. doi: 10.1152/ajpendo.00508.2001. [DOI] [PubMed] [Google Scholar]
- Levenson CW, et al. Regulation of cysteine-rich intestinal protein by dexamethasone in the neonatal rat. Proc Natl Acad Sci U S A. 1993;90:712–5. doi: 10.1073/pnas.90.2.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegeois S, et al. Genes required for osmoregulation and apical secretion in Caenorhabditis elegans. Genetics. 2007;175:709–24. doi: 10.1534/genetics.106.066035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubarsky B, Krasnow MA. Tube morphogenesis: making and shaping biological tubes. Cell. 2003;112:19–28. doi: 10.1016/s0092-8674(02)01283-7. [DOI] [PubMed] [Google Scholar]
- McKeown C, et al. sma-1 encodes a βH-spectrin homolog required for Caenorhabditis elegans morphogenesis. Development. 1998;125:2087–2098. doi: 10.1242/dev.125.11.2087. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA Transformation. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Vol. 48. Academic Press; San Diego: 1995. pp. 451–482. [Google Scholar]
- Nelson FK, et al. Fine structure of the Caenorhabditis elegans secretory-excretory system. J Ultrastructural Res. 1983;82:156–171. doi: 10.1016/s0022-5320(83)90050-3. [DOI] [PubMed] [Google Scholar]
- Nelson FK, Riddle DL. Functional study of the Caenorhabditis elegans secretory-excretory system using laser microsurgery. J Exp Zool. 1984;231:45–56. doi: 10.1002/jez.1402310107. [DOI] [PubMed] [Google Scholar]
- Oka T, et al. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem. 2001;276:33079–85. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- Olson MF, et al. Faciogenital dysplasia protein (FGD1) and Vav, two related proteins required for normal embryonic development, are upstream regulators of Rho GTPases. Curr Biol. 1996;6:1628–33. doi: 10.1016/s0960-9822(02)70786-0. [DOI] [PubMed] [Google Scholar]
- Perez-Alvarado GC, et al. Structure of the cysteine-rich intestinal protein, CRIP. J Mol Biol. 1996;257:153–74. doi: 10.1006/jmbi.1996.0153. [DOI] [PubMed] [Google Scholar]
- Praitis V, et al. SMA-1 spectrin has essential roles in epithelial cell sheet morphogenesis in C. elegans. Dev Biol. 2005;283:157–70. doi: 10.1016/j.ydbio.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–9. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta. 2004;1695:45–54. doi: 10.1016/j.bbamcr.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Press; Cold Spring Harbor, New York: 1988. pp. 587–606. [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic Cell Lineages of the Nematode, Cænorhabditis elegans. Developmental Biology. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Suzuki N, et al. A putative GDP-GTP exchange factor is required for development of the excretory cell in Caenorhabditis elegans. EMBO Rep. 2001;2:530–5. doi: 10.1093/embo-reports/kve110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonning A, et al. A transient luminal chitinous matrix is required to model epithelial tube diameter in the Drosophila trachea. Dev Cell. 2005;9:423–30. doi: 10.1016/j.devcel.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Tsui SK, et al. Isolation and characterization of a cDNA that codes for a LIM-containing protein which is developmentally regulated in heart. Biochem Biophys Res Commun. 1994;205:497–505. doi: 10.1006/bbrc.1994.2693. [DOI] [PubMed] [Google Scholar]
- van Ham M, et al. Cloning and characterization of mCRIP2, a mouse LIM-only protein that interacts with PDZ domain IV of PTP-BL. Genes Cells. 2003;8:631–44. doi: 10.1046/j.1365-2443.2003.00660.x. [DOI] [PubMed] [Google Scholar]
- White J. The Anatomy. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Vol. 17. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1987. pp. 81–122. [Google Scholar]
- Zheng Y, et al. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs- specific guanine-nucleotide exchange factor. J Biol Chem. 1996;271:33169–72. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Complete phylogenetic tree of EXC-9 and all CRIP homologues (shown in abbreviated form in Fig. 3D), including echinoderm, trematode, and additional vertebrate homologues.


