SUMMARY
HP1 proteins are a highly conserved family of eukaryotic proteins, which bind to methylated histone H3 lysine 9 (H3K9) and are required for heterochromatic gene silencing. In fission yeast, two HP1 homologs, Swi6 and Chp2, function in heterochromatic gene silencing, but their relative contribution to silencing remains unknown. Here we show that Swi6 and Chp2 exist in non-overlapping complexes and make distinct contributions to silencing. Chp2 associates with the SHREC histone deacetylase complex (SHREC2), is required for histone H3 lysine 14 (H3K14) deacetylation, and mediates transcriptional repression by limiting RNA polymerase II access to heterochromatin. In contrast, Swi6 associates with a different set of nuclear proteins and with noncoding centromeric transcripts, and is required for efficient RNAi-dependent processing of these transcripts. Our findings reveal an unexpected role for Swi6 in RNAi-mediated gene silencing and suggest that different HP1 proteins ensure full heterochromatic gene silencing through largely non-overlapping inhibitory mechanisms.
Introduction
Silent or heterochromatic DNA domains play important roles in maintenance of chromosome integrity and regulation of gene expression in eukaryotes ranging from fission yeast to human. A core characteristic of heterochromatin is its association with heterochromatin protein 1 (HP1) proteins, a highly conserved family of chromosomal proteins that bind to di- and tri-methylated H3K9 via a conserved N-terminal domain called the chromodomain (CD) (Grewal and Moazed, 2003; Jacobs and Khorasanizadeh, 2002; Jacobs et al., 2001; Lomberk et al., 2006; Maison and Almouzni, 2004; Richards and Elgin, 2002). HP1 proteins contain a C-terminal chromo-shadow domain (CSD) that is thought to mediate protein-protein interactions, including self-association (Brasher et al., 2000; Lomberk et al., 2006). The fission yeast Schizosaccharomyces pombe contains two HP1 homologs, Swi6 and Chp2, and a third chromodomain protein, Chp1, all of which bind to methylated H3K9 and are involved in heterochromatic gene silencing (Bannister et al., 2001; Halverson et al., 2000; Partridge et al., 2002; Thon and Verhein-Hansen, 2000). Chp1 is associated with Tas3, Ago1, and repeat-associated short interfering RNAs (siRNAs) in the RITS complex and links heterochromatin to RNA interference (RNAi) (Motamedi et al., 2004; Verdel et al., 2004). However, despite their conservation and importance, the precise role(s) of Swi6 and Chp2 in heterochromatin assembly and function has remained unclear.
HP1 proteins specifically bind to methylated H3K9 (Jacobs and Khorasanizadeh, 2002; Jacobs et al., 2001), and are believed to recruit the H3K9 methyltransferase, KMT1 (Clr4 in S. pombe, Su(var)3–9 in Drosophila, and Suv39H in mammals), directly to indirectly to chromatin (Aagaard et al., 1999; Lachner et al., 2001; Rea et al., 2000; Schotta et al., 2002; Stewart et al., 2005). Sequential cycles of HP1/Swi6 binding and KMT1 recruitment have been proposed to mediate the spreading of H3K9 methylation along the chromatin fiber (Bannister et al., 2001; Grewal and Moazed, 2003; Nakayama et al., 2001). However, recent studies indicate that in fission yeast neither Swi6 nor Chp2 is required for the spreading of H3K9 methylation at centromeres (Sadaie et al., 2004). Instead spreading requires the RITS-associated Chp1 protein and the RNAi machinery (Verdel et al., 2004; Volpe et al., 2002). Swi6 and Chp2 act downstream of H3K9 methylation to promote heterochromatic gene silencing by mechanisms that appear to involve the recruitment of additional chromatin modifying activities. In this regards, previous studies have suggested a role for H3K9 methylation and Swi6/HP1 in recruitment of the SHREC complex, containing Clr1, Clr2, the Clr3 histone deacetylase (HDAC), and the Mit1 chromatin remodeling protein. SHREC mediates the deacetylation of histone H3 lysine 14 (H3K14) and promotes transcriptional gene silencing (Sugiyama et al., 2007).
Most eukaryotes contain multiple HP1 proteins. For example, Drosophila, mouse, and human cells possess three closely related HP1 proteins (referred to as HP1, HP1b, and HP1c in Drosophila, and HP1α, HP1β, and HPγ in human and mouse). The Drosophila HP1 and HP1b (and mammalian HP1α and β) localize predominantly to heterochromatin, whereas HP1c and HP1γ localize to euchromatin and function in gene-specific silencing. In addition to methylated H3K9 and KMT1, HP1 proteins interact with diverse groups of non-histone chromosomal proteins (Lomberk et al., 2006). The Drosophila HP1 associates with the nuclear lamin B receptor (LBR), subunits of the Origin Recognition Complex, and chromatin remodeling proteins (Pak et al., 1997; Shareef et al., 2001; Ye and Worman, 1996). In human, HP1 α interacts with Ku70 DNA repair protein (Song et al., 2001), HP1β associates with Dnmt1 and Dnmt3a DNA methyltransferses (Fuks et al., 2003), and HP1 and the human KMT1 (SUV39H1) associate with Rb, and mediate cell cycle dependent repression of Rb targets in euchromatin (Nielsen et al., 2001). In addition to the interaction between the fission yeast Swi6/HP1 and the Clr3 HDAC (Yamada et al., 2005), interactions between the mouse HP1α class II HDACs have been reported (Zhang et al., 2002).
HP1 proteins also localize to actively transcribed genes in flies and mammals (Cryderman et al., 2005; Vakoc et al., 2005). The significance of this localization is not understood but has been proposed to reflect an HP1 affinity for RNA, and possible involvement in RNA processing during transcription elongation. In fact, HP1 binds to RNA and DNA in vitro, with preference for the single-stranded form in the case of DNA (Muchardt et al., 2002). The latter binding activity may contribute to the telomere capping function of HP1 in Drosophila, which is independent of its ability to bind to methylated H3K9 via its chromodomain (Perrini et al., 2004). Although no RNA-binding activity has been described for the fission yeast HP1 proteins, Swi6 is required for efficient accumulation of centromeric siRNAs, suggesting that it may interact directly with the RNAi machinery (Buhler et al., 2006; Motamedi et al., 2004).
It has been unclear why heterochromatic gene silencing requires multiple HP1 proteins and whether these proteins play distinct or largely redundant roles within heterochromatin. Here we report studies aimed at understanding the relative contributions of each Swi6, Chp2, and RNAi to heterochromatic gene silencing in fission yeast. We perform biochemical purifications of Chp2 and Swi6, and use quantitative assays to determine the degree to which each of the above pathways contributes to silencing. We find that the Chp2 and Swi6 proteins associate with non-overlapping sets of proteins. Chp2 is associated with the SHREC deacetylase complex (which we call SHREC-Chp2 or SHREC2), is required for deacetylation of H3K14, and mediates transcriptional gene silencing by limiting RNA polymerase II (Pol II) access to heterochromatin. On the other hand, Swi6 associates with a large number of chromosomal proteins, including transcription elongation proteins, chromatin remodeling complexes, and DNA-binding and DNA replication proteins. Swi6 plays a minor role in limiting Pol II access to heterochromatin and a major role in processing of centromeric transcripts into siRNAs. Consistent with the latter result, Swi6 localizes to noncoding centromeric RNAs in a Clr4-dependent manner and is required for efficient localization of RNAi complexes (RITS and the RNA-dependent RNA polymerase complex - RDRC) to heterochromatic transcripts, suggesting that it may tether nascent transcripts to heterochromatin for the RNAi-dependent heterochromatic gene silencing. Overall, our results reveal the existence of two tiers of silencing mechanisms that act together to ensure that transcription within heterochromatic regions is properly regulated. In one tier, RNAi and an unknown RNAi-independent pathway act together to recruit Clr4 to mediate H3K9 methylation of heterochromatic nucleosomes. In the second tier, Chp2 and Swi6 act downstream of H3K9 methylation and ensure efficient silencing by limiting Pol II access and processing of heterochromatic transcripts into siRNAs, respectively.
RESULTS
Chp2 associates with the SHREC complex
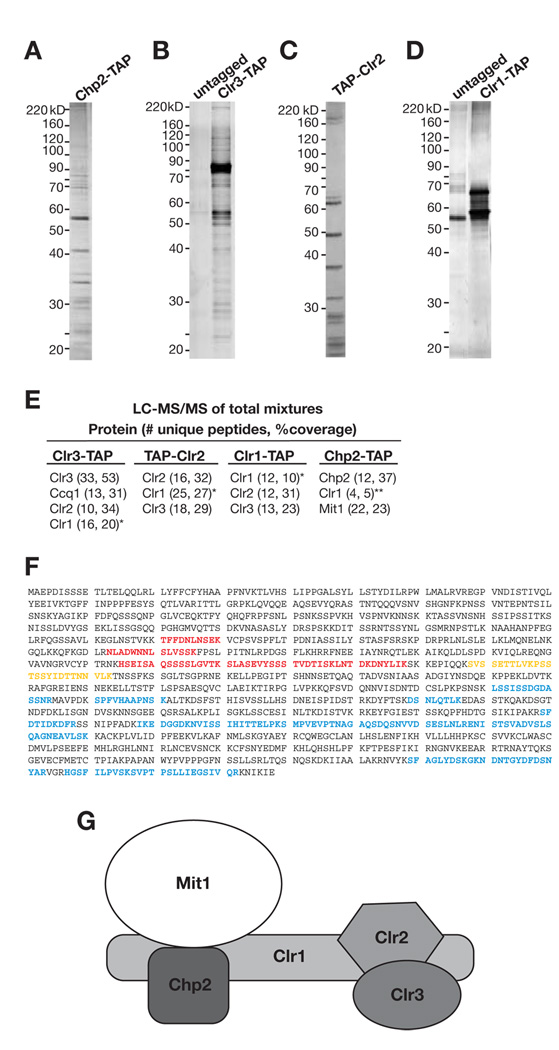
The fission yeast HP1 homolog Chp2 is required for the transcriptional silencing of reporter genes inserted at all known S. pombe heterochromatic loci including the pericentromeric repetitive innermost (imr) and outer (otr) DNA elements, telomeres, rDNA and the mating-type loci (Halverson et al., 2000; Thon and Verhein-Hansen, 2000). In order to gain insight into how Chp2 contributes to silencing, we constructed a strain in which a functional C-terminally TAP-tagged Chp2 protein was expressed from its endogenous promoter, and purified this protein using a tandem affinity purification scheme (Motamedi et al., 2004; Rigaut et al., 1999). Polyacrylamide gel electrophoresis and liquid chromatography coupled to tandem mass spectromertry (LC-MS/MS) analyses revealed that Chp2 interacted with two previously characterized proteins, Clr1 and Mit1 (Figure 1A, E, Table S1), both of which are components of the Snf2/Hdac-containing Repressor Complex (SHREC)(Figure 1B–E and Sugiyama et al., 2007). Examination of the Clr1 peptide spectra from two independent Chp2-TAP purifications revealed a non-uniform distribution of peptides, mapping only to the N terminus of the Clr1 protein (Figure 1F), suggesting that only the N-terminal portion of Clr1 was present in the Chp2-TAP purifications. These data suggested that Chp2 was a component of the SHREC complex, interacting with Clr2 and Clr3 via the N terminus of Clr1.
Figure 1. Purification of Chp2 and identification of the SHREC2 complex.
Representative silver-stained gels depicting the tandem affinity purifications of (A) Chp2-TAP, (B) Clr3-TAP, (C) TAP-Clr2, and (D) Clr1-TAP proteins. (E) Proteins identified by tandem mass spectrometry sequencing of mixture of proteins (LC-MS/MS) for the indicated purification. The numbers in parentheses correspond to the number of unique peptides and protein coverage based on total number of amino acids. (F) Clr1 peptide spectra in SHREC2 purifications. Red, residues represent peptides only identified in Chp2-TAP purification. Blue residues represent peptides identified in Clr3-TAP, TAP-Clr2 or Clr1-TAP purifications. Yellow residues show peptides common to both Chp2-TAP and TAP-Clr2 purifications. (G) SHREC2 complex is composed of Chp2 and Mit1, which interact with the N-terminal region of Clr1, and Clr2 and Clr3, which interact with the C-terminal region of Clr1.
In order to further analyze the composition of the SHREC complex, we constructed functional C-terminally TAP-tagged Clr3 and Clr1, and N-terminally TAP-tagged Clr2 expressed under the control of their endogenous promoters. Several tandem affinity purifications were performed and the resulting purified proteins were subjected to LC-MS/MS analysis (Table S1). Results from three independent Clr3-TAP, two independent TAP-Clr2, and two independent Clr1-TAP purifications revealed a largely non-overlapping Clr1 peptide spectra compared to those found in the two independent Chp2-TAP purifications (Figure 1A, F). In contrast to Chp2-TAP purifications in which only peptides from the N terminus of Clr1 were identified, in all Clr1-TAP, TAP-Clr2, and Clr3-TAP purifications (seven purifications in total), peptides only from the C terminus of the Clr1 protein were identified (Figure 1F). The only exception was a single peptide, mapping to the middle of Clr1 protein, which was shared among the Chp2-TAP and TAP-Clr2 purifications. Western blot analysis of Clr1-TAP revealed a 160KD protein compatible with the predicted full-length TAP-tagged protein, which is rapidly degraded after cell lysis, especially during purification (Supplemental Figure S1A–C). Based on these data, we conclude that Chp2, and probably Mit1, interact with the N-terminus, and Clr2 and Clr3 interact with the C-terminus of Clr1. This shows that Chp2 is a component of the SHREC complex, which we named SHREC associated with Chp2 complex, or SHREC2 complex (Figure 1G). The SHREC complex previously has been shown to mediate deacetylation of H3K14 (via Clr3) and chromatin remodeling (via Mit1)(Bjerling et al., 2002; Sugiyama et al., 2007). Our results suggest that these activities are recruited to heterochromatin via the association of the Chp2 subunit of SHREC2 with H3K9me.
The requirement for SHREC2 in heterochromatic gene silencing
Previous reports (Bjerling et al., 2004; Ekwall and Ruusala, 1994; Sugiyama et al., 2007; Thon and Verhein-Hansen, 2000) have shown that components of SHREC2 are required for the silencing of a reporter construct (e.g. ura4+) inserted at various S. pombe heterochromatic loci, including centromeres, telomeres, mating-type region, and rDNA. In agreement with these findings, we found that deletion of any of the SHREC2 components led to the loss of silencing in strains carrying a ura4+ reporter gene inserted at the innermost (imr1::ura4+) or outer (otr1::ura4+) pericentromeric DNA regions, the mating-type (mat::ura4+) or the rDNA (rDNA::ura4+) loci (Figure S2A–D). Even though these growth-based assays serve as an indicator for silencing defects, they are not quantitative, and therefore do not reveal differences in expression levels above the minimum RNA level required to generate the growth phenotype.
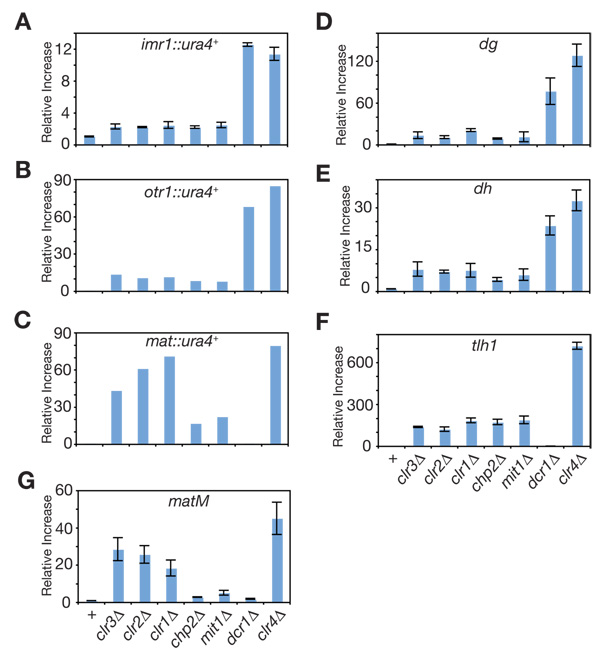
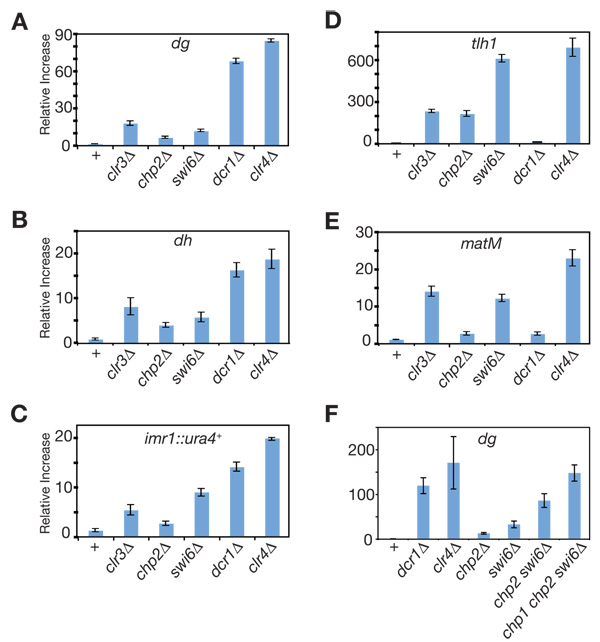
In order to quantify the extent to which the components of SHREC2 contribute to heterochromatic gene silencing, we measured the steady-state levels of the inserted ura4+ reporter gene (imr1::ura4+, otr1::ura4+, and mat::ura4+), or endogenous heterochromatic transcripts originating from the dg and dh elements of centromeres, the matM gene of the mating-type locus, and tlh1+ gene of the sub-telomeres using reverse-transcriptase real-time PCR. Compared to clr4Δ cells, in which heterochromatin is completely disrupted, we found that deletion of SHREC2 subunits resulted in only partial derepression of the centromeric ura4+ reporter genes (Figure 2A, B), the endogenous centromeric dg and dh, and telomeric tlh1+ transcripts (Figure D–F). These results indicated that the contribution of SHREC2 to heterochromatic gene silencing at these loci is a fraction (10–20%) of the full silencing levels observed in clr4Δ cells (Table S2). Similar to the 5-FOA silencing results (Figure S2C), the contribution of clr1, clr2, and clr3 genes to silencing of mat::ura4+ as well as matM (Figure 2C) was greater than that of chp2 and mit1, suggesting a Chp2-Mit1-independent mechanism for Clr1, Clr2, and Clr3 recruitment to this locus. Overall, these results indicate a partial requirement for SHREC2 in heterochromatic gene silencing.
Figure 2. Contribution of SHREC2 to heterochromatic gene silencing.
Quantitative real-time RT-PCR depicting the steady-state levels of (A) imr1::ura4+, (B) otr1::ura4+, (C) mat::ura4+, (D) dg, (E) dh, (F) tlh1+, and (G) matM transcripts in SHREC2 mutant cells compared to wild-type, dcr1Δ and clr4Δ cells. Error bars represent the standard deviations from three independent biological experiments. All values were normalized to act1+ transcript levels.
SHREC2 couples H3K14 deacetylation to H3K9me binding via Chp2 and limits Pol II access to heterochromatin
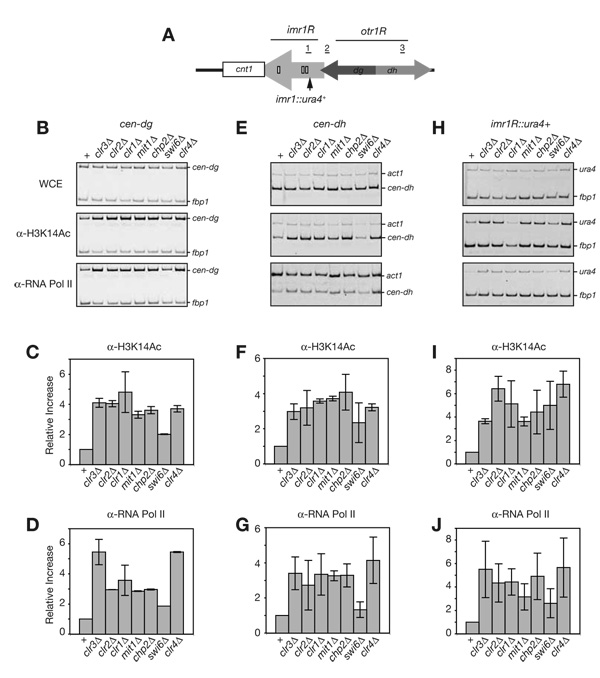
As mentioned above, the composition of the SHREC2 complex suggests that the Clr3-mediated H3K14 deacetylation activity is recruited to heterochromatin via Chp2 binding to H3K9me. In order to test this, we performed chromatin immunoprecipitation (ChIP) experiments using H3K14Ac antibody in cells lacking SHREC2 components. At the centromeric dg and dh repeats and the imr1R::ura4+ transgene (Figure 3A), we found that SHREC2Δ and clr4Δ mutants display a similar increase in H3K14Ac levels, compared to wild type (Figure 3B–I, lanes 1–6 and 8, respectively). In comparison, we observed a smaller increase in H3K14Ac in swi6Δ cells (Figure 3B–I, lane 7), suggesting that majority of Clr3-dependent H3K14 deacetylation occur in a Swi6-independent but Chp2-dependent manner.
Figure 3. SHREC2 is required for H3K14 deacetylation and limits RNA polymerase II (Pol II) access to heterochromatin.
(A) Schematic diagram of the pericentromeric DNA repeats immediately to the right of the S. pombe centromere 1 central core region (cnt1 – white box), including the imr1R (light grey arrow), and otr1R elements. The dh and dg elements of otr1R are shown as light grey and dark grey arrows, respectively. The site of the ectopic ura4+ insert, three tRNA genes which act as barrier elements to heterochromatin spreading, and the DNA segments amplified in the chromatin immunoprecipitation (ChIP) experiments are shown as a black arrow, small white boxes, and underlined numbers 1 (corresponding to imr1R::ura4+), 2 (corresponding to cen-dg), and 3 (corresponding to cen-dh), respectively. ChIP experiments showing that H3K14 deacetylation and Pol II occupancy increase in the absence of SHREC2 components at cen-dg (B–D), cen-dh (E–G), and imr1R::ura4+ (H–J). In swi6Δ cells, the increase in H3K14 deacetylation and Pol II occupancy is smaller compared to SHREC2 mutant cells. Error bar represent variation from the mean from two independent biological experiments. fbp1+ was used as an internal control for the ChIP experiments.
Previous work had shown that SHREC components limit the access of Pol II machinery to heterochromatin (Sugiyama et al., 2007). We performed ChIP experiments using an antibody against Pol II to assess the role of SHREC2 components, including Chp2, in regulating Pol II access to centromeric heterochromatin. We found that chp2Δ, clr4Δ as well as clr3Δ, clr2Δ, clr1Δ, and mit1Δ mutants displayed a similar level of increase in Pol II occupancy compared to wild-type cells (Figure 3B–H, lower panels, and Figure 3D–J). In swi6Δ cells however, the increase in Pol II occupancy (similar to H3K14Ac levels) was smaller than that in SHREC2 mutant cells (Figure 3B–H, lower panels, and Figure 3D–J). Because Clr4-dependent H3K9 methylation creates the binding substrate for Chp2 and Swi6, these data suggest that all H3K9me-dependent control of Pol II access to heterochromatin is SHREC2-mediated.
Even though we observed a comparable level of increase in Pol II occupancy in cells lacking Clr4 or any of the SHREC2 components, the increase in the accumulation of dg, dh, and imr1R::ura4+ RNA levels in SHREC2 mutants (Figure 2A, D, E) was only a fraction (10–20%) of the total clr4Δ increase (Table S2). These results suggest that SHREC2-dependent transcriptional gene silencing contributes to only a portion of the total heterochromatic gene silencing that prevents the accumulation of endogenous and transgene centromeric RNAs. The rest appears to involve an RNAi-dependent post-transcriptional silencing mechanism that operates in cis (Buhler et al., 2007; Buhler et al., 2006).
Swi6 associates with a distinct set of nuclear proteins
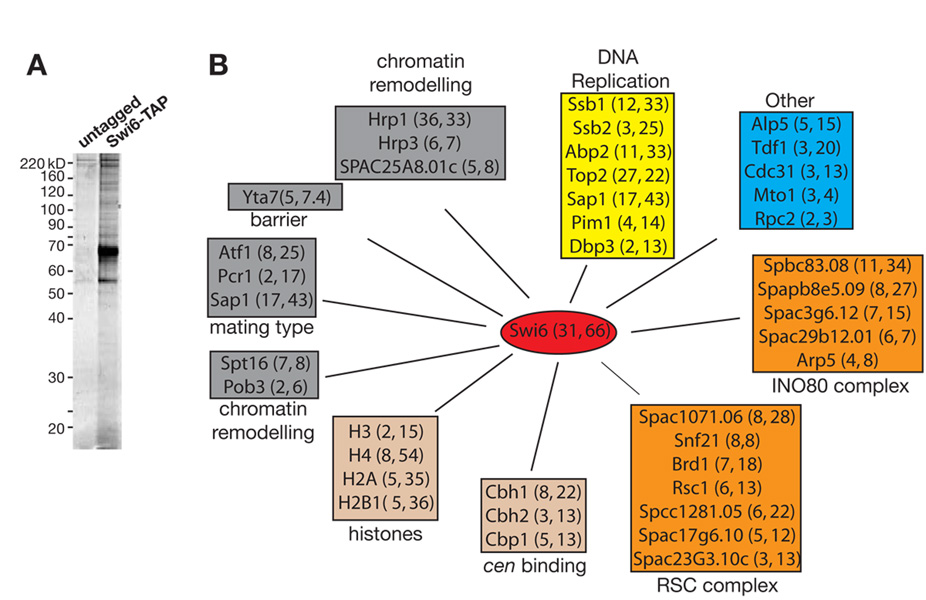
Previous reports have shown that, similar to Chp2, Swi6 is required for silencing at all S. pombe heterochromatic loci (Allshire et al., 1995; Ekwall et al., 1995; Klar and Bonaduce, 1991). However, Swi6 also is required for the efficient processing of heterochromatic transcripts into siRNAs (Buhler et al., 2006; Motamedi et al., 2004) suggesting a role for Swi6 in the RNAi-dependent heterochromatic gene silencing pathway. In order to better understand the contribution of Swi6 to this pathway, we purified a C-terminally TAP-tagged version of Swi6 using the previously described tandem affinity purification scheme (Figure 4A), and analyzed the purified proteins using tandem mass spectrometry. The purifications yielded three main classes of nuclear proteins, none of which overlapped with any of the SHREC2 or other known RNAi complexes (Figure 4B, Table S1). DNA binding, chromatin remodeling/histone modifying, and DNA replication proteins represent the three main groups of proteins that co-purified with Swi6. From among these, several proteins with previously characterized roles in silencing were identified. Three members of the chromo-ATP/helicase-DNA-binding (CHD) remodeling proteins, two of which, Hrp1 and Hrp3, previously have been shown to be involved in silencing (Jae Yoo et al., 2002; Walfridsson et al., 2005), co-purified with Swi6. Chp2 also associates with another member of this family, Mit1, and mammalian HP1 has been previously shown to interact with the Brg1 subunit of the SWI/SNF chromatin remodeling complex (Nielsen et al., 2002). Thus interactions between HP1s and chromatin remodelers appear to be conserved.
Figure 4. Swi6 interacts with a diverse set of nuclear proteins involved in a variety of nuclear functions.
(A) Silver-stained gel of Swi6-TAP and untagged control purifications. (B) Schematic summary of the network of nuclear proteins that co-purify with Swi6. Numbers in the parentheses next to each protein indicate the number of unique peptides and percent coverage based on total number of amino acids, respectively. Proteins are grouped in colored boxes based on previously defined roles in heterochromatin function, histone/centromere binding, chromatin remodeling, DNA replication and other nuclear functions. Sap1 has been implicated in mating-type switching and DNA replication, and is included in groups corresponding to both nuclear functions.
Other Swi6-associated proteins with known heterochromatin silencing functions include the FACT chromatin remodeling members, Spt16 and Pob3 (Lejeune et al., 2007), ATF/CREB proteins Atf1/Pcr1, which are involved in silencing at the mating-type locus (Jia et al., 2004), and Sap1, a general replication factor (Krings and Bastia, 2005; Noguchi and Noguchi, 2007) also required for mating-type switching (Arcangioli and Klar, 1991). The S. pombe homolog of the S. cerevisiae Yta7 protein, which has been suggested to restrict spreading of silencing proteins at euchromatin-heterochromatin boundaries (Jambunathan et al., 2005), also co-purified with Swi6. Furthermore, the three centromere binding proteins (CENP-B) homologs Cbh1, Cbh2, and Cbp1 with known functions in centromeric silencing (Nakagawa et al., 2002), the core histones H3, H4, H2A and H2B1, and several protein involved in DNA replication co-purified with Swi6 (Figure 4B and Table S1). These data suggest that Swi6 is at the center of a complex network of chromatin-associated proteins, whose activities contribute to a number of different chromosomal functions, including transcriptional silencing.
Contribution of Swi6 to heterochromatic gene silencing
The current evidence on the requirement of Swi6 in heterochromatic gene silencing is largely based on the assessment of the expression of transgenes inserted at various heterochromatic loci using growth-based assays (Allshire et al., 1995; Ekwall et al., 1995; Klar and Bonaduce, 1991). Although very sensitive, as shown above, these assays are not quantitative. Here, we quantified the relative contribution of Swi6 to heterochromatic gene silencing using real-time PCR, measuring the accumulation of transgene (imr1::ura4+) and endogenous heterochromatic RNAs originating from dg and dh of centromeric repeats, matM gene of the mating-type region, and tlh1+ gene of sub-telomeres. Because Clr4 is the sole H3K9 methyltransferase in S. pombe, we used RNA levels in clr4Δ cells as our reference (1.00) for complete loss of H3K9me-dependent silencing and present the RNA levels in SHREC2 or swi6 mutant cells relative to this maximal derepression in clr4Δ cells. Our analysis revealed that similar to Chp2, Swi6 contributed to a fraction of the total H3K9me-mediated silencing at centromeres (Figure 5A, B, E, and Table S3), as the levels of centromeric dg, dh, and imr1R::ura4+ transcripts in swi6Δ cells were 0.14, 0.32, and 0.44 compared to clr4Δ, respectively. Unlike Chp2, Swi6 was required for the majority of H3K9me-dependent silencing at the sub-telomeric tlh1+ gene (Figure 5D, Table S3). Also, at the mating-type gene matM, Swi6 contribution to silencing was greater than Chp2 and similar to the Clr3 contribution (Figure 5C and Table S3). These results show that centromeric gene silencing is only partially disrupted in swi6Δ cells.
Figure 5. Swi6 and SHREC2 components make non-redundant contributions to heterochromatic gene silencing.
Quantitative real-time RT-PCR showing the steady-state levels of (A) dg, (B) dh, (C) imr1::ura4+ (D) tlh1+, and (E) matM transcripts in swi6Δ cells compared to wild-type, clr3Δ, chp2Δ, dcr1Δ and clr4Δ cells. (F) The steady-state levels of the dg transcript in chp2Δ swi6Δ double, and chp1Δ chp2Δ swi6Δ triple mutant cells were compared to single mutants. See Figure 4SA for additional data, including chp1Δ, chp1Δ chp2Δ, and chp1Δ swi6Δ cells at dg dh, imr1::ura4+, and tlh1+. Error bars represent standard deviations for three independent biological experiments. All values were normalized to act1+ transcript levels.
The residual silencing in each swi6Δ or chp2Δ single mutant cells is likely to be mediated by the remaining HP1 protein (Chp2 or Swi6, respectively) or the Chp1-containing RITS complex. We therefore tested whether this residual silencing is lost in double-mutant cells lacking both Swi6 and Chp2, or triple-mutant cells lacking Swi6, Chp2, and Chp1. We used real time RT-PCR to measure the accumulation of transgene (imr1::ura4+) and endogenous (dg, dh, and tlh1+) heterochromatic RNAs in double chp2Δ swi6Δ and triple chp1Δ chp2Δ swi6Δ mutant cells and compared these values to those obtained from swi6Δ, chp2Δ and clr4Δ cells. We found that the double and triple mutant cells displayed a larger defect in silencing (See Figure 5F, Figure 4S and Table S3) than any of the single swi6Δ, chp2Δ mutants. In particular, deletion of all three chromodomain proteins resulted in accumulation of heterochromatic transcripts to levels that were similar to that found in clr4Δ cells, which lack any H3K9 methylation. These data support a model in which Chp2 and Swi6 make distinct non-overlapping contribution to heterochromatic gene silencing in S. pombe.
Swi6 associates with noncoding centromeric transcripts
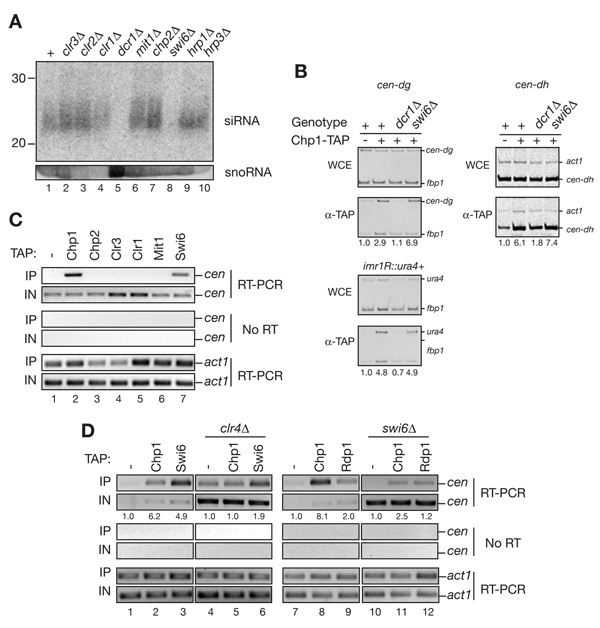
The biochemical purifications of Chp2 and Swi6, their relative contributions to heterochromatic gene silencing, and their distinct roles in regulating Pol II access to heterochromatin show that these proteins contribute to silencing by non-redundant mechanisms. Previously, it has been shown that Swi6 is required for efficient accumulation of centromeric siRNAs (Buhler et al., 2007; Motamedi et al., 2004; also see Figure 6A, lane 8), whereas siRNA levels are slightly increased or unaffected in cells that lack the Clr3 component of SHREC2 (Figure 6A lanes 1 and 2; Irvine et al., 2006; Sugiyama et al., 2007). Consistent with its association with Clr3 in the SHREC complex, we found that deletion of chp2+ or other subunits of the SHREC complex had little or no effect on the levels of centromeric siRNA (Figure 6A, compare lanes 1–4 and 6,7 with lanes 5 and 8). These results further support distinct roles for Swi6 and Chp2 in mediating RNAi-dependent RNA processing and transcriptional gene silencing, respectively. The modest increase in siRNA levels observed in SHREC2 mutant cells is likely to result from an increase in the transcription of siRNA precursors.
Figure 6. Swi6, but not SHREC2, associates with noncoding cenRNAs and is required for their processing into siRNAs.
(A) Northern blot showing that Swi6 is required for accumulation of centromeric siRNAs. Size fractionated total small RNAs purified from the indicated cells were probed with end-labeled DNA oligos complementary to centromeric siRNAs. The membrane was then re-probed with snoR69, serving as a loading control. Unlike swi6Δ, SHREC2 mutant cells exhibited no defect in siRNA production. (B) ChIP experiments showing that RITS recruitment to centromeric dg, dh, and imr1::ura4+ is Swi6 independent. Immunoprecipitations were performed using wild-type, dcr1Δ, and swi6Δ cells carrying a chp1-TAP gene. Untagged cells were used as a control. Values below each lane represent enrichment compared to the untagged control fbp1+ and act1+ were used as internal controls. (C) RNA immunoprecipitation (RNA-IP) experiments showing that centromeric transcripts (cen) associate specifically with Swi6, but not with SHREC2 components Chp2, Clr3, Mit1 or Clr1. Chp1 is used as a positive control. Actin mRNA (act1) was used as a control to show that the interaction is specific to heterochromatic transcripts. (D) RNA-IP experiments showing that Chp1 and Swi6 associate with centromeric transcripts in a Clr4-dependent manner (lanes 1–6); efficient Chp1 and Rdp1 interaction with cen transcripts requires Swi6 (lanes 7–12). Actin mRNA (act1) and the untagged sample were used as specificity controls. The number shown below each lane is the average enrichment calculated by comparing the enrichment values to the untagged control in two independent experiments.
The significant reduction in siRNA levels in swi6Δ mutants (Figure 6A and Buhler et al., 2006; Motamedi et al., 2004) gives rise to the possibility that Swi6 may be required for the recruitment of the RNAi effector complex RITS, to centromeric heterochromatin. We used chromatin immunoprecipitation (ChIP) experiments to test this hypothesis and found that the Chp1 subunit of RITS associated with centromeric dg and dh repeats independently of Swi6 (Figure 6B), but, as shown previously (Verdel et al., 2004), this interaction required Dcr1-generated siRNAs(Figure 6B). Also, consistent with the MS/MS profile of proteins identified in Swi6 purifications, using co-immunoprecipitation assays, we did not detect an interaction between C-terminally tagged Rdp1, Tas3, Chp1, Cid14, Dcr1 or an N-terminally FLAG-tagged Ago1 with Swi6 (data not shown), suggesting that Swi6 did not interact with RDRC/Dcr1, RITS, TRAMP or ARC complexes. The Swi6 contribution to RNAi-mediated processing of heterochromatic transcripts therefore is unlikely to be mediated by the recruitment of any of the above RNAi complexes to heterochromatin.
Prior work has shown that HP1 proteins are capable of binding to RNA in vivo and in vitro via their basic patch region, located in between the chromo- and chormo-shadow domains (Maison and Almouzni, 2004; Muchardt et al., 2002). We hypothesized that Swi6 may contribute to the RNAi-dependent pathway by binding to heterochromatic RNAs, transcribed within nucleosomal regions enriched for H3K9me, the binding substrate for Swi6. In our previous work, we used an RNA immunoprecipitation assay to show that RITS and RDRC complexes interact with noncoding centromeric transcripts in a Dcr1- and Clr4-dependent manner (Motamedi et al., 2004). Here we examined the ability of SHREC2 components Chp2, Clr3, Clr1 and Mit1, and Swi6 to interact with cen transcripts. We found that similar to the Chp1 subunit of the RITS complex (Motamedi et al., 2004), Swi6 specifically cross-linked to noncoding cen transcripts in a manner that was mostly Clr4-dependent (Figure 6C, lanes 2 and 7; Figure 6D, compare lanes 1–3 to lanes 4–6). In contrast, the Chp2, Clr1, Clr3, and Mit1 subunits of SHREC did not cross-link to cen transcripts (Figure 6C, lanes 3–6). Together with previous observations, these results suggest that Swi6 is a bifunctional protein that associates with both H3K9 methylated nucleosomes and noncoding transcripts within heterochromatic domains.
The association of Swi6 with cen transcripts and its requirement for siRNA generation suggest that Swi6 may contribute to the stable assembly of RNAi complexes on heterochromatic transcripts by tethering the transcripts to sites of RNA synthesis. To test this possibility, we used RNA-IP experiments to ask whether Swi6 was required for localization of Chp1 and Rdp1 (components of RITS and RDRC, respectively) to cen transcripts. As shown in Figure 6D, we observed a marked decrease in the association of Chp1 and a smaller decrease in the association of Rdp1 with cen transcripts in swi6Δ compared to the wild-type cells (compare lanes 7–9 with lanes 10–12), suggesting that Swi6 was required for efficient association of RITS and RDRC with centromeric transcripts. Together with the ChIP data in Figure 6B, these results reveal a role for Swi6 in promoting the association of RNAi complexes with heterochromatic transcripts by a mechanism that is independent of their association with H3K9 methylated chromatin.
DISCUSSION
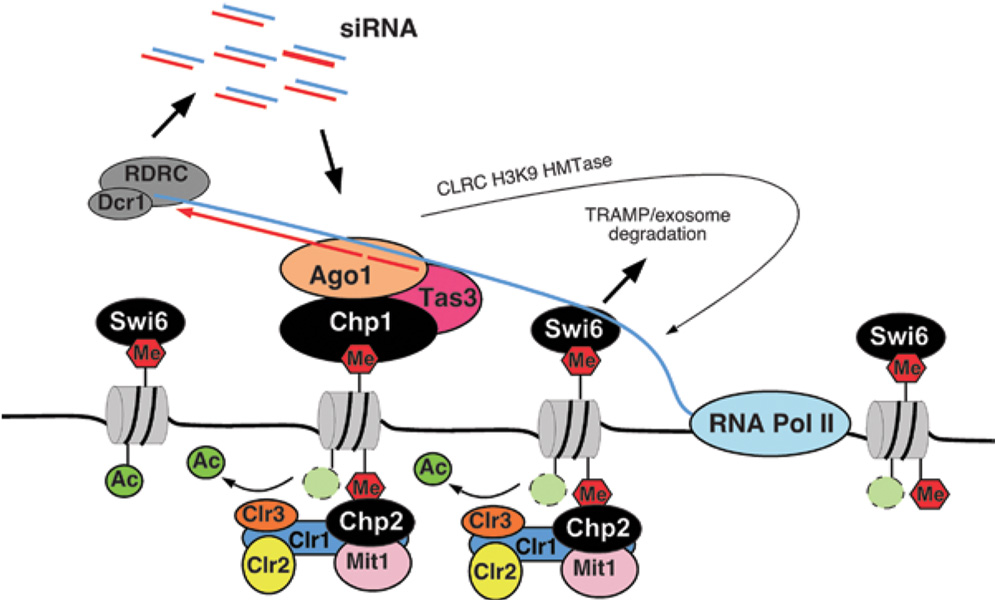
HP1 proteins play a critical role in H3K9 methylation-dependent heterochromatic gene silencing in eukaryotes, however the nature of their contribution to silencing has remained elusive. In this study, we show that the two S. pombe HP1 proteins contribute to heterochromatin-dependent gene silencing by non-redundant mechanisms. Chp2 associates with the SHREC complex and recruits the H3K14 deacetylase Clr3, whose activity is required for restricting RNA Pol II access to heterochromatin. Swi6, on the other hand, interacts with noncoding centromeric transcripts, similar to the RITS chromodomain subunit Chp1, and contributes to silencing primarily through RNAi-dependent cis-processing of heterochromatic transcripts (Figure 6D, Figure 7). These findings demonstrate that efficient heterochromatic gene silencing requires the recruitment of distinct silencing mechanisms by different HP1 proteins.
Figure 7. Model highlighting the contributions of HP1 proteins to heterochromatic gene silencing in S. pombe.
Swi6 promotes the stable association of nascent heterochromatic transcripts to sites of RNA synthesis by its ability to bind to H3K9me and RNA. The association of Swi6 with the nascent transcript is required for the subsequent assembly of RNAi complexes (RITS and RDRC/Dcr1) on the RNA, probably by helping to tether the RNA to chromatin. Swi6-mediated tethering may also be required for targeting of heterochromatic transcripts for degradation by the TRAMP/exosome pathway. SHREC2 binds to H3K9me via its Chp2 component, targeting the H3K14 deacetylase Clr3 and CHD ATPase Mit1 to heterochromatin, which are required for limiting Pol II access to heterochromatin.
SHREC2 links H3K9me binding to H3K14 deacetylation
Previous purifications of Clr3 (Sugiyama et al., 2007) as well as our Clr3-TAP, TAP-Clr2 and Clr1-TAP purifications (Figure 1B–E) failed to reveal SHREC interaction with Chp2. This is likely due to rapid degradation of Clr1 during purification (Figure S1A–C). Our LC-MS/MS analyses revealed that full length Clr1 forms an N- and a C-terminal fragment, which interact with Chp2 and Mit 1 or Clr2 and Clr3, respectively (Figure 1A–F). The largely non-overlapping peptide spectra of Clr1 in the Clr1-TAP, TAP-Clr2 and Clr3-TAP versus Chp2-TAP purifications (Figure 1F) demonstrate that Clr1 serves as a connector between Chp2 (and possibility Mit1) and Clr2 and Clr3 subunits of SHREC (Figure 1G).
The association of Chp2 with the SHREC complex demonstrates a coupling between two highly conserved post-translational modifications of histone H3 tails (lysine 9 methylation and lysine 14 deacetylation), both of which are critical for heterochromatic gene silencing. What is the biological significance of this coupling? Previous work (Sadaie et al., 2004) and data present here (Figure S3A–G) show that Clr4-mediated H3K9 methylation of pericentromeric repeats occurs independently of Chp2 or Swi6. This combined with the requirement of these proteins for silencing suggests that these HP1 proteins associate with heterochromatin and recruit downstream activities that are critical for heterochromatin maturation. Consistent with this hypothesis, we found that the deacetylation of H3K14 by Clr3 at centromere is H3K9me- and Chp2-dependent (Figure 3). Clr3-dependent H3K14 deacetylation regulates the transcriptional state of chromatin (Bjerling et al., 2002; Sugiyama et al., 2007), and the importance of H3K14 in transcriptional gene silencing has been demonstrated in a wide range of organisms including S. cerevisiae (Braunstein et al., 1996; Suka et al., 2001; Thompson et al., 1994) and S. pombe (Mellone et al., 2003; Yamada et al., 2005). Conversely, H3K14 acetylation is correlated with active transcription (Pokholok et al., 2005) and in human cells acetylated H3K14 is critical for the recruitment of TFIID to active promoters (Agalioti et al., 2002). Our results show that deacetylation of H3K14 by the SHREC complex occurs downstream of H3K9 methylation by Clr4, and is largely or entirely mediated by H3K9-binding HP1 protein, Chp2. Conservation of HP1 proteins and their mode of association with heterochromatin, as well as histone hypoacetylation in heterochromatic gene silencing, raise the possibility that recruitment of homologous deacetylase complexes in other eukaryotes may be HP1-dependent.
Consistent with previous work (Yamada et al., 2005), our data suggest that Clr3 is recruited to the mating-type locus by a distinct and parallel mechanism that does not involve the Chp2 and Mit1 subunits of the SHREC2 complex. We found that the silencing defect observed at matM or mat::ura4+ transgene in chp2Δ or mit1Δ cells is 3- to 6-fold less severe compared to clr1Δ, clr2Δ, and clr3Δ cells (Figure 2C and G, Figure 5E and S2C; see Table S1 for quantification). Previous work has shown that Atf1/Pcr1 proteins recruit Clr3 to its nucleation site at the mat locus within the REIII silencer element. We propose that in the absence of Mit1 and Chp2, Clr3 can be recruited to the mat region via the Atf1/Pcr1-dependent pathway, perhaps as a Clr1/2/3 subcomplex. In addition, the association of Swi6 with Clr3 (Yamada et al., 2005) and with Atf1/Pcr1 proteins (Figure 5C)(Jia et al., 2004) may provide a parallel, Chp2-independent, mechanism for the recruitment of Clr1/2/3 to the mating-type locus. Alternative mechanisms for the recruitment of SHREC have been reported for other loci. For example, at telomeres the telomere binding protein, Taz1, and the HEAT domain protein, Ccq1, cooperate with the RNAi machinery to recruit SHREC (Sugiyama et al., 2007). Overall, these results demonstrate the existence of distinct, partially redundant, pathways for targeting of the SHREC complex to different heterochromatic domains.
Association of Swi6 with noncoding heterochromatic transcripts
Recent studies suggest that chromatin and RNA silencing pathways intersect at several levels (Bernstein and Allis, 2005). Our analysis of the role of Swi6 in heterochromatic gene silencing suggests that this HP1 protein plays a central role in coordinating histone H3K9 methylation with RNA processing. Purification of Swi6 revealed a complex network of nuclear proteins involved in a wide range of cellular processes, including transcriptional repression and elongation, chromosome biogenesis, and chromatin remodeling. Remarkably, our purifications revealed no overlapping peptides in Chp2 and Swi6 purifications, suggesting that these closely related HP1 proteins make distinct non-overlapping contributions to gene silencing. Consistent with the biochemical purifications, we found that Swi6 and Chp2 operate via different mechanisms to effect silencing within heterochromatic regions. While Chp2 functions to limit Pol II access to heterochromatin, and thereby mediate transcriptional gene silencing, the contribution of Swi6 to limiting Pol II access to heterochromatin is minimal (Figure 3D, G, J lanes 7), suggesting an unexpected silencing role for an HP1 protein in a step downstream of transcription initiation. Together with the requirement for Swi6 in siRNA accumulation (Buhler et al., 2006; Motamedi et al., 2004), Figure 6A), our data suggest that the primary function of Swi6 in heterochromatin involves co-transcriptional processing of heterochromatic transcripts.
How does Swi6 contribute to the RNAi–mediated processing of heterochromatic transcripts? HP1 proteins have been shown to bind to RNA in vitro, and in addition to heterochromatin, co-localize with transcriptional elongation complexes of highly transcribed genes in Drosophila and human (Cryderman et al., 2005; Vakoc et al., 2005), suggesting that they play a role in RNA processing. Our observation that Swi6 specifically interacts with noncoding cen transcripts, suggests a possible model for the involvement of Swi6 in RNAi-mediated processing of heterochromatic transcripts. We propose that Swi6 is a bifunctional protein that tethers heterochromatic transcripts to the chromosome (Figure 7). In this model, the Swi6 chromodomain binds to H3K9 methylated nucleosomes and its basic hinge region, which in other HP1 proteins has been implicated in RNA binding, associates with nascent RNAs. This tethers nascent RNAs to heterochromatin and allows RNAi complexes to assemble on the transcript. In the absence of Swi6, centromeric siRNA levels are greatly diminished (∼20-fold below swi6+ cells) and RITS and RDRC localization to cen transcripts is reduced compared to wild type (Figure 6D), suggesting that Swi6 is required for the efficient association of RITS/RDRC/Dicer with their target centromeric transcripts. The actual Swi6-mediated decrease in the levels of heterochromatic transcripts may result from the conversion of the RNA to dsRNA and its degradation by Dicer or by the slicer activity of siRNA-programmed RITS complexes (Figure 7). Recent studies in Drosophila have uncovered a direct physical interaction between HP1 and Piwi, an Argonaute family protein that binds to small RNAs that originate from repetitive DNA elements and transposons (Brower-Toland et al., 2007). HP1 proteins therefore appear to play direct but distinct roles in distantly related chromatin-associated RNA silencing mechanisms. Finally, any model for the role of Swi6 in heterochromatic gene silencing must take into account the requirement for Swi6 in RNAi-independent gene silencing at the silent mating-type locus. In addition to Swi6 and H3K9 methylation, efficient silencing at the mating-type locus requires the TRAMP polyadenylation complex and exosome-mediated RNA degradation (Buhler et al., 2007). We propose that the RNA association activity of Swi6 serves a general function in retention of heterochromatic RNAs on chromatin that contributes to the processing of these RNAs by either the RNAi or the TRAMP/exosome pathways (Figure 7).
Experimental Procedures
Strain construction and Protein Purification
Strain construction was performed following standard procedures as described in Supplemental Data. (Bahler et al., 1998). 10–20 grams of TAP-clr2, clr3-TAP and, and 80–110 grams of clr1-TAP and chp2-TAP of logarithmically growing cells were harvested and frozen in liquid nitrogen for later use. All tandem affinity TAP purifications were preformed as described previously (Motamedi et al., 2004), except that all TEV cleavage steps were done at 4°C overnight. Proteins were identified as described previously (Verdel and Moazed, 2005).
Silencing Assays
All silencing assays were performed as described previously (Verdel et al., 2004). Briefly, cells were grown in 5 ml of YEA (Yeast extract supplemented with adenine) to logarithmic phase, after which five-fold dilutions of each culture were spotted on non-selective (N/S) YEA plates, and YEA plates supplemented with 5-fluoro-oratic acid (5-FOA). 5FOA is used as a counter selection against cells expressing ura4+.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described in detail previously (Huang and Moazed, 2003). Cells were grown to an OD600 of 1.5–2, crosslinked for 15 minutes with 1% formaldehyde. Immunoprecipitation was performed with the following antibodies: 3ul of α-H3K14Ac (Upstate, 07–353), 3ul of α-H3K9Me2 (Abcam ab1220), 4ul of α-RNA Polymerase II (Covance, 8WG16) per 400 ul reaction. PCR amplifications were performed to determine the linear range for each reaction and a final non-radioactive PCR set was performed according to the linear range determination on all samples. PCR products were run on 6% acrylamide gels and stained with Ethidium bromide (EtBr) at 2.5mg/l. Image ReaderLAS-3000 was used to capture the image and the PCR bands were quantified using ImageGauge V4.22 (Fuji Film Life Science, Stanford, Connecticut).
RNA Analysis
RNA immunoprecipitation (RNA-IP) experiments were performed as described previously (Motamedi et al., 2004). Cells were grown to OD600 of 1.5–2, crossed-linked with formaldehyde for 30 minutes and frozen in liquid nitrogen. Whole cell extracts were prepared from frozen pellets as described previously (Gilbert et al., 2004; Hurt et al., 2004), and treated with DNAse I (Sigma, 700 units) for one hour at 30 deg in the buffer supplemented with 25mM MgCl2 and 5mM CaCl2. RNA was precipitated and used to perform semi-quantitative RT-PCR reactions (Motamedi et al., 2004).
RNA preparation, semi-quantitative and real-time RT-PCR, and northern blot analysis were performed as described previously (Buhler et al., 2007, 2006; Leeds et al., 1991; Motamedi et al., 2004).
Western blots
Western blots were performed as described previously (Motamedi et al., 2004). For detection of Clr1-TAP cell lysis was performed in buffer containing 30% trichloroacetic acid as described in the Supplemental Data section.
Supplementary Material
Acknowledgments
We thank Charles Hoffman, Shiv Grewal, and Karl Ekwall for S. pombe strains, Tessi Iida, and Marc Buhler for primer sets, and members of the Moazed lab for helpful discussions. M.R.M. was supported by a postdoctoral fellowship from NSERC and CIHR; E.E.H. is a Leukemia and Lymphoma Society fellow. D.M. was a Leukemia and Lymphoma Scholar and is an HHMI Investigator. This work was supported by grants from the NIH (GM72805 to D.M.). M.R.M. dedicates this paper to the memory of his beloved father, Fazlollah Motamedi, who passed away on Sept 24, 2008.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. Embo J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Arcangioli B, Klar AJ. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 1991;10:3025–3032. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bjerling P, Ekwall K, Egel R, Thon G. A novel type of silencing factor, Clr2, is necessary for transcriptional silencing at various chromosomal locations in the fission yeast Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:4421–4428. doi: 10.1093/nar/gkh780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerling P, Silverstein RA, Thon G, Caudy A, Grewal S, Ekwall K. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol Cell Biol. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232:767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136:53–64. doi: 10.1093/genetics/136.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Halverson D, Gutkin G, Clarke L. A novel member of the Swi6p family of fission yeast chromo domain-containing proteins associates with the centromere in vivo and affects chromosome segregation. Mol Gen Genet. 2000;264:492–505. doi: 10.1007/s004380000338. [DOI] [PubMed] [Google Scholar]
- Huang J, Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. Embo J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae Yoo E, Kyu Jang Y, Ae Lee M, Bjerling P, Bum Kim J, Ekwall K, Hyun Seong R, Dai Park S. Hrp3, a chromodomain helicase/ATPase DNA binding protein, is required for heterochromatin silencing in fission yeast. Biochem Biophys Res Commun. 2002;295:970–974. doi: 10.1016/s0006-291x(02)00797-0. [DOI] [PubMed] [Google Scholar]
- Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics. 2005;171:913–922. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304:1971–1976. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- Klar AJ, Bonaduce MJ. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics. 1991;129:1033–1042. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem. 2005;280:39135–39142. doi: 10.1074/jbc.M508996200. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Ball L, Suka N, Grunstein MR, Partridge JF, Allshire RC. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr Biol. 2003;13:1748–1757. doi: 10.1016/j.cub.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, Grewal SI, Tanaka K, Murakami Y. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Sanchez C, Ichinose H, Cervino M, Lerouge T, Chambon P, Losson R. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1alpha. Embo J. 2002;21:5797–5806. doi: 10.1093/emboj/cdf560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Noguchi C, Noguchi E. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics. 2007;175:553–566. doi: 10.1534/genetics.106.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, Turano C, Ferraro A, Pimpinelli S. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–476. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. Embo J. 2004;23:3825–3835. doi: 10.1038/sj.emboj.7600401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3–9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (hp1)/origin recognition complex (orc) protein is associated with hp1 and orc and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12:1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Jung Y, Jung D, Lee I. Human Ku70 interacts with heterochromatin protein 1alpha. J Biol Chem. 2001;276:8321–8327. doi: 10.1074/jbc.M008779200. [DOI] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol. 2005;25:2525–2538. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- Thon G, Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155:551–568. doi: 10.1093/genetics/155.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Moazed D. Labeling and characterization of small RNAs associated with the RNA interference effector complex RITS. Methods Enzymol. 2005;392:297–307. doi: 10.1016/S0076-6879(04)92017-4. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Walfridsson J, Bjerling P, Thalen M, Yoo EJ, Park SD, Ekwall K. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell. 2005;20:173–185. doi: 10.1016/j.molcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.