Abstract
Cell surface proteins have been shown to be effective therapeutic targets. In addition, shed forms of these proteins and secreted proteins can serve as biomarkers for diseases, including cancer. Thus, identification of cell surface and secreted proteins has been a prime area of interest in the proteomics field. Most cell surface and secreted proteins are known to be glycosylated and therefore, a proteomics strategy targeting these proteins was applied to obtain proteomic profiles from various thyroid cancer cell lines that represent the range of thyroid cancers of follicular cell origin. In this study, we oxidized the carbohydrates of secreted proteins and those on the cell surface with periodate and isolated them via covalent coupling to hydrazide resin. The glycoproteins obtained were identified from tryptic peptides and N-linked glycopeptides released from the hydrazide resin using 2-dimensional liquid chromatography-tandem mass spectrometry in combination with the gas phase fractionation. Thyroid cancer cell lines derived from papillary thyroid cancer (TPC-1), follicular thyroid cancer (FTC-133), Hürthle cell carcinoma (XTC-1), and anaplastic thyroid cancer (ARO and DRO-1) were evaluated. An average of 150 glycoproteins were identified per cell line, of which more than 57 percent are known cell surface or secreted glycoproteins. The usefulness of the approach for identifying thyroid cancer associated biomarkers was validated by the identification of glycoproteins (e.g. CD44, galectin 3 and metalloproteinase inhibitor 1) that have been found to be useful markers for thyroid cancer. In addition to glycoproteins that are commonly expressed by all of the cell lines, we identified others that are only expressed in the more well-differentiated thyroid cancer cell lines (follicular, Hürthle cell and papillary), or by cell lines derived from undifferentiated tumors that are uniformly fatal forms of thyroid cancer (i.e. anaplastic). Based on the results obtained, a set of glycoprotein biomarker candidates for thyroid cancer is proposed.
Introduction
Growth of abnormal cells that form nodules in the thyroid gland is the most common endocrine problem in the United States.1–4 Most nodules are benign, however a significant number (<10%) are malignant and form thyroid tumors.1, 2, 4, 5 Each year more than 25,000 women and 8,000 men are diagnosed with thyroid cancer in the United States.3, 4 There are several histologic types of thyroid cancer including the more common types, papillary thyroid cancer (PTC) and follicular thyroid cancer (FTC), and those that are less common, medullary thyroid cancer (MTC) and anaplastic thyroid cancer (ATC).4 PTC, FTC and ATC originate from follicular cells of the thyroid, whereas MTC originates from the parafollicular or calcitonin-producing (C) cells in the thyroid gland. PTC and FTC (including a subtype of Hürthle cell carcinoma, HCC) are composed of cancer cells that are considered well differentiated and account for 80 and 15 percent of all thyroid cancer in US, respectively.4, 6 In contrast, the cells of MTC and ATC are considered less differentiated, or dedifferentiated and account for about 3–5 percent each of all thyroid cancer. Individuals diagnosed with PTC, FTC and MTC generally have a good prognosis, whereas ATC is the most aggressive and the least likely form of thyroid cancer to respond to treatment, with a median survival of 4–12 months from the time of diagnosis.4, 7
Fine needle aspiration (FNA) is currently the principle technique used to obtain samples of thyroid tissue for diagnostic evaluation.4, 5, 8 The type of thyroid tumor is determined by evaluating the cytological features of cells obtained at biopsy. However, results from FNA can be inconclusive. Moreover this diagnostic method is unable to distinguish between benign and malignant follicular or Hürthle cell neoplasm.5 Although ultrasound guided fine needle aspiration improves thyroid cancer diagnostic sensitivity, up to 30 percent of the FNA results are still indeterminate or nondiagnostic. Most patients with inconclusive thyroid nodule FNA biopsy results are subjected to at least a diagnostic hemithyroidectomy to exclude a thyroid cancer diagnosis. Commonly, only 20% are found to be malignant on permanent histology and this means about 80% of patients are subjected to an unnecessary surgical procedure.5, 8 Thus, there is a need to establish more specific methods to distinguish benign nodules from malignant nodules, and to develop improved approaches to identify various types of thyroid cancer.
Several gene expression profiling and immunohistochemical studies of thyroid tissue and cell lines have been conducted with the aim to identifying diagnostic biomarkers for thyroid cancer. Gene expression studies comparing cancer versus non-cancer thyroid cells or tissues have identified a group of up-regulated genes including MET, SERPINA, FN1, CD44 and DPP4, whereas the expression of many other genes, including TFF3, are down-regulated.9 Proteomics approaches (2D-gel with MALDI- or ESI-MS analysis) have been used recently to search for biomarkers of thyroid cancer.10–12 These biomarker discovery-based studies identified several, mostly cytosolic, proteins differentially expressed in various subtypes of PTC, FTC and follicular thyroid adenoma. However, none of these candidate markers for malignant thyroid tumors have been validated or clinically applied to make management decision.
In this work, we used a glycoproteomic approach to identify cell surface and secreted candidate biomarkers for thyroid cancer. To achieve this objective, we used a modification of the periodate oxidation/hydrazide method13 in which intact cells are treated with periodate to selectively oxidize the carbohydrate moieties of integral plasma membrane glycoproteins and secreted glycoproteins.14 We identified a total of 333 glycoproteins, including a substantial number of integral plasma membrane glycoproteins and secreted glycoproteins from each thyroid cancer cell line. Analysis of the glycoproteins isolated by this method also identified an average of 224 N-linked glycopeptides per cell line. In addition to glycoproteins that were identified in all of the cell lines, each thyroid cancer cell line expressed a set of unique glycoproteins.
Experimental Methods
Cell Culture
Five thyroid cancer cell lines, (follicular, FTC-133; papillary, TPC-1; Hürthle, XTC-1; and anaplastic, ARO (ARO-82-1) and DRO-1(DRO-90-1)) were grown in 10cm culture dishes with 10ml of culture medium (50/50 Dubelco’s Modified Eagle’s Medium/Ham’s F-12 Medium supplemented with 10% fetal bovine serum, 10μg/ml insulin, 0.01U/ml thyroid stimulating hormone, 2mM L-glutamine, 50μg/ml penicillin/streptomycin and 250ng/ml fungizone). Cells were grown at 37°C with 5% CO2 until roughly 90% confluent. Each sample for analysis was prepared from 30 culture dishes (10cm), yielding 1.2–2.8mg of total protein. Because there has been some question in the literature of whether some thyroid cancer cell lines used for research are not of thyroid origin, we determined the gene expression levels of markers specific to follicular thyroid cells in the cell lines studied.15 In all of the thyroid cancer cell lines used in this study, at least 2 of the 3 genes were expressed confirming that the cell lines are of follicular thyroid cell origin (data not shown).
Periodate Oxidation
Thyroid cancer cell lines were grown to ~90% confluence. The growth media was aspirated from each plate of cells and the cells were rinsed once with 5ml of PBS. Cells were oxidized with 3ml of 10mM NaIO4 in oxidation buffer (20mM sodium acetate, 150mM NaCl, pH 5) in the dark at 25°C for 1 hour with gentle rocking, and then the periodate solution was removed by aspiration. The cells were incubated with 3ml of coupling buffer (100mM sodium acetate, 1.5M NaCl, pH 4.5) with gently rocking plates for 2 min, then the coupling buffer was removed.
FTC-133 cells grew as a loosely adherent monolayer and were partially dislodged with coupling buffer after periodate oxidation. Therefore, in some instances FTC-133 cells were detached from the dish with 4mM EDTA (ethylenediaminetetraacetic acid) and recovered by centrifugation prior to the removal of the wash solution. The results obtained from cells processed by either approach were not significantly different.
Cell Lysate Preparation
Cell lysates were prepared as previously described.14 Briefly, cells were treated with 0.5ml of lysis buffer containing 1% the non-ionic detergent octyl-β-D-1-thioglucopyranoside (BTGP), 1% protease inhibitor cocktail, 20mM Tris-HCl, and 150mM NaCl, pH 7.5, and allowed to incubate at room temperature for 10 minutes. BTGP has been shown to result in complete solubilization of whole membranes, including lipid rafts.16 Cells were scraped from the dishes and homogenized by multiple passing through a syringe with 27 ½ gauge needle. Homogenized lysates were clarified by centrifugation at 14,000 rpm for 5 minutes to pellet insoluble material, the supernatant was carefully collected, and total protein content was determined by Bradford assay and either immediately subjected to glycoprotein enrichment by hydrazide resin coupling or frozen at −20°C before the coupling process was carried out.
Glycoprotein Enrichment via Hydrazide Resin Binding
Protein lysates were spiked with 5μg of periodate oxidized chicken ovalbumin (Sigma-Aldrich, St. Louis, MO), which served as an internal control for the coupling process, and well mixed. Hydrazide Sepharose resin (3.2 ml) (BioRad, Hercules, CA) in a 50% slurry of ethanol storage buffer was rinsed with 7ml of coupling buffer (100mM sodium acetate, 1.5M NaCl, pH 4.5) then centrifuged at 5,000 rpm for 3 minutes to pellet the resin. The buffer was discarded and the wash process repeated twice. The spiked lysate was mixed with the rinsed hydrazide resin and incubated for at least 8 hours at 25ºC, with end-over-end tumbling.
Glycoprotein Reduction, Alkylation, Denaturation and Trypsin Digestion
The hydrazide resin was transferred to a disposable plastic column using 7ml of 8M urea in 100mM ammonium bicarbonate pH 8.3 buffer, which also was used to denatured the bound glycoproteins. The immobilized proteins were washed with 5ml of 50mM ammonium bicarbonate to remove the remaining urea. Subsequently, the bound proteins were reduced with 5ml of 50mM dithiothreitol (DTT) for 60 minutes at 37°C. The DTT was discarded and the resin rinsed with 5ml of 50mM ammonium bicarbonate buffer. The denatured glycopeptides were alkylated with 5ml of 65mM iodoacetamide (IAM) for 30 minutes at 25ºC in the dark using a thermo-mixing apparatus set to medium shaking. The IAM was discarded, and the immobilized glycoproteins rinsed with 5ml of ammonium bicarbonate buffer. To ensure complete protein denaturation, the resin was incubated in 5ml of 6M urea for 5 minutes at 25ºC, and subsequently rinsed 3 times with 5ml of 1.5M sodium chloride to remove non-specific binding. The resin was rinsed 3 times with 5ml of 50mM ammonium bicarbonate to remove the remaining sodium chloride. The glycoproteins were digested with 40ng/μl trypsin (Promega, Madison, WI) in 50mM ammonium bicarbonate buffer at a final concentration of at 37ºC for more than 8 hours in a water bath. After digestion, the tryptic fraction was collected, and the hydrazide column was washed with 3–5ml of 50mM ammonium bicarbonate to collect any remaining tryptic peptides. The combined eluate was processed by solid phase extraction (SPE).
N-Glycopeptide Release from the Hydrazide Resin with N-Glycosidase F
The hydrazide resin containing N-linked glycopeptides was washed 3 times with 3ml of 1.5M sodium chloride, and 3 times with 3ml of 80% acetonitrile in HPLC grade water (Fisher Scientific) to remove non-specifically bound peptides. The column was further washed with 9ml of HPLC grade water and finally with 9ml of 50mM ammonium bicarbonate. The N-linked glycopeptides were released from the resin with N-glycosidase F (Glyco N-Glycanase 200mU, Prozyme) (4μl in 1ml of 50mM ammonium bicarbonate buffer) overnight at 37°C. The N-linked glycopeptide fraction was collected, and the resin was rinsed with 3ml of 50mM ammonium bicarbonate buffer. The ammonium bicarbonate rinse solution was collected and combined with the PNGase F released fraction, stored at 4°C, and subsequently processed by SPE.
Solid Phase Extraction (SPE)
The tryptic peptides and N-linked glycopeptides were each subjected to SPE (Strata-X reversed phase, Phenomenex). The SPE resin was activated with 1ml of HPLC grade methanol, then rinsed with 1ml of HPLC grade water to remove the remaining methanol. The tryptic peptides or PNGase F treated N-linked glycopeptides were bound to an SPE column using a flow rate of 0.5 ml/min. The resin-bound peptides were rinsed with 2ml of HPLC grade water to remove salts, eluted with 1ml of 65% methanol/water, dried using a Speed-Vac apparatus, and stored at 4°C prior to mass spectrometric analysis.
Peptide/Protein Identifications by ESI-MS/MS Analysis
The tryptic peptides and the PNGase F treated N-linked glycopeptides were analyzed by liquid chromatography/electrospray ionization-tandem mass spectrometry (LC/ESI-MS/MS) using an LTQ ion trap mass spectrometer (Thermo Fisher, San Jose, CA). NanoLC and on-line 2D-LC/ESI-MS/MS analyses were conducted using a dual pump Thermo Surveyor HPLC system. For on-line 2D-LC/ESI-MS/MS analysis peptide mixtures were chromatographically separated (Thermo Fisher, BioBasic SCX column, 320μm x 100mm) into seven fractions using various concentrations (0, 20, 40, 60, 80, 200, 400mM) of NH4Cl, followed by in-line desalting with a C18 trap column (300μm x 5mm, Agilent), and finally reverse phase (C18, 75μm x 130mm) nanoLC separation. The mobile phases for the reverse phase chromatography were (mobile phase A) 0.1% HCOOH/water and (mobile phase B) 0.1% HCOOH in acetonitrile. A three-step, linear gradient was used for the nanoLC separation (5% to 30% B in the first 65 min, followed by 30% to 80% B in the next 10 min, and 80% B in the last 10 min). The ESI-MS/MS data acquisition was set up to collect ion signals from the eluted peptides using an automatic, data-dependent scan procedure in which a cyclical series of three different scan modes (1 full scan, 4 zoom scans, and 4 MS/MS scans for top four abundant ions) was performed. The full scan mass range was set from m/z 400 to 1800. The MS/MS analysis exclusion rule for the same precursor ion was set to a value of 2 during a 30 s period. For gas phase fractionation experiments, three sample injections were performed with full mass range settings of either m/z 400–800, m/z 700–1100, or m/z 1100–1500 followed by the automatic, data-dependent scan procedure described previously. This gas phase fractionation data acquisition procedure was repeated twice for each mass range, once with an MS/MS analysis exclusion rule for the same precursor ion of 2 during a 30 s period and once with the exclusion value set at 1. The resulting MS/MS spectra were searched against the SwissProt human protein database using the Mascot and X!Tandem algorithms to identify the peptides.17, 18 Scaffold (Proteome Software) was used to merge and summarize the files from Mascot and X!Tandem. Protein identifications were based on a minimum of 2 peptide hits, with a probability score of 95% or greater using the Peptide and Protein Prophet algorithms in Scaffold.19, 20 The false positive rate in this study was 0.57% based on results obtained using the Mascot program with a decoy database in which the sequences in the SwissProt human protein database were reversed. The ProteinID Finder (Proteome Solutions) program was used to extract protein information such as subcellular location from the UniProt database for each identified protein.
Results
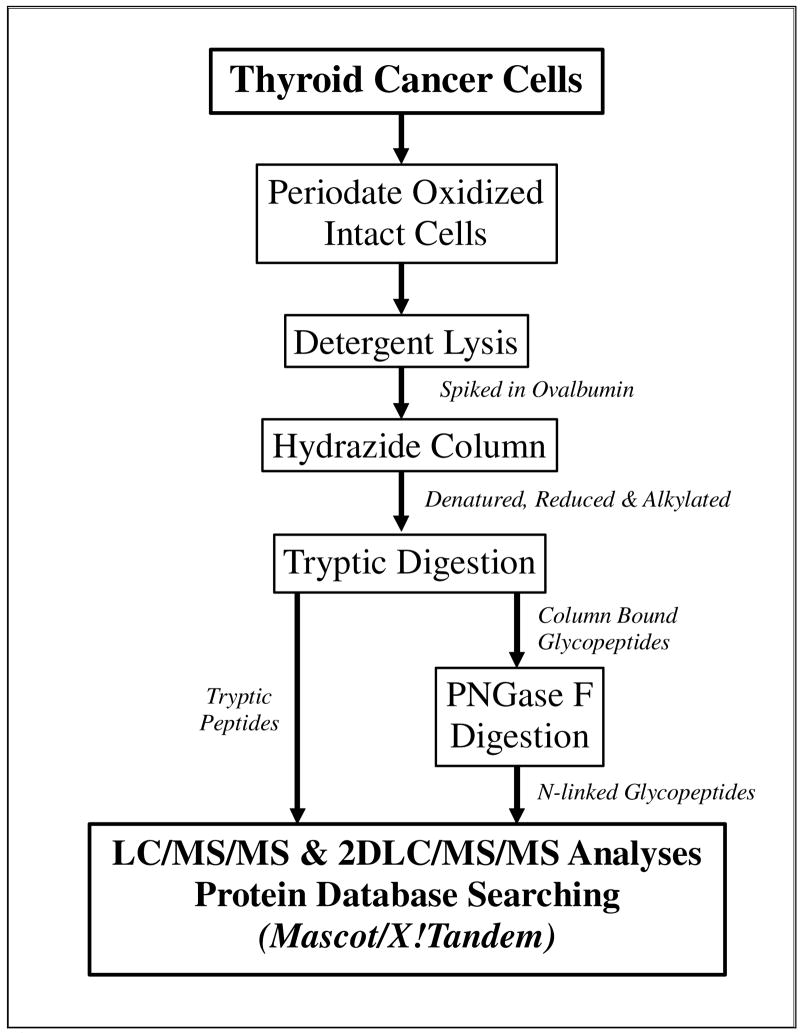
Figure 1 presents a diagram of the experimental approach14 used to enrich for secreted and plasma membrane glycoproteins from five thyroid cancer cell lines. Total protein content in the lysates from cells harvested from 30 plates for each thyroid cancer cell line ranged from 1.22 mg for DRO-1 cells to 2.79 mg for ARO cells and varied by 2–13% among replicates (data not shown). To analyze the overall glycoprotein recovery rate in each sample, a known amount of the periodate-oxidized glycoprotein, ovalbumin (5μg), was spiked into each cell lysate sample prior to the hydrazide resin coupling step. Covalently linked glycoproteins were denatured, reduced, alkylated and digested into tryptic peptides for bottom-up proteomics analysis. Tryptic peptides were collected and analyzed by nanoLC/ESI-MS/MS using an LTQ ion trap. Gas phase fractionation, using a narrow mass range for the precursor ion (see Experimental Methods), was employed to increase the number of protein identifications.21 2D-LC/ESI-MS/MS analysis 22 was used to separate and detect the complex mixture of tryptic peptides obtained, and to overcome the limitation of the loading capacity of the nanoLC column. The hydrazide resin-bound, N-linked glycopeptides were subsequently released using PNGase F, and converting the glycosylated Asn residues to Asp. The released glycopeptides were analyzed by nanoLC/ESI-MS/MS, in combination with the gas phase fractionation.
Figure 1.
Workflow diagram of the glyco-capture (periodate oxidation/hydrazide resin) method for profiling the cell surface membrane and secreted glycoproteins.
Glycoproteins Identified in Thyroid Cancer Cell Lines
Protein identifications were obtained using a combination of nanoLC, 2D-LC/ESI-MS/MS and gas phase fractionation MS/MS analyses, and were based on a minimum of 2 peptide hits, with a probability score of 95% or greater using the Peptide and Protein Prophet algorithms in the Scaffold program.19, 20 A script program (Proteome Solutions) was used to obtain basic information (subcellular location, the number of the transmembrane domains, N-linked and O-linked sites) about the identified proteins. This program utilizes the UniProt primary access number for each protein to extract the available information from the UniProtKB website and summarize it in an Excel spreadsheet format.
LC/MS/MS analyses of the tryptic peptides and the PNGase F released glycopeptides from the five thyroid cancer cell lines resulted in the identification of a total of 862 unique proteins (see Supplemental Table 1). Each cell line (TPC-1, FTC-133, XTC-1, ARO and DRO-1) was analyzed twice using two different cell culture passage samples and the results were merged using the Scaffold program (see Supplemental Table 2). Ovalbumin, the spiked internal glycoprotein standard, was identified by at least three unique peptides in each 2D-LC/ESI-MS/MS analysis, demonstrating that the recovery of glycoproteins for each sample was consistent. Run-to-run reproducibility from the same cell lysate was also very good as measured by the intra-sample variation of the number of proteins identified. For example, replicates of 2D-LC/ESI-MS/MS analyses from an ARO sample identified an average of more than 84% of the same proteins. As expected, the inter-sample variation of proteins identified between replicates was lower. For example, when two sets of ARO cells (passage 65 and 67) were prepared as describe in Figure 1 and analyzed, 62.8% of the same proteins were identified. Thus, it is clear that multiple sample preparations run in replicates produce a maximum number of protein identifications with the highest confidence.
Analyses of the two of the well-differentiated thyroid cancer cell lines resulted in the identification of 397 proteins in the TPC-1 cell lysates, and 363 in cell lysates from FTC-133 cells (see Table 1). Among the proteins identified in the TPC-1 cell line, 42% (166) are glycoproteins, compared to the 29% (107) for the FTC-133 cell line. The largest number (477) of proteins was identified in the other well differentiated cell line XTC-1 or Hürthle cell carcinoma line. Glycoproteins accounted for 38% (181) of the total proteins identified in the XTC-1 cell line. Analyses of the undifferentiated cancer cell line lysates yielded similar numbers of proteins (402 in ARO and 409 proteins in DRO-1), of which 173 (43%) and 121 (30%) were glycoproteins in ARO and DRO-1, respectively (see Table 1).
Table 1.
Total Numbers of Proteins and Glycoproteins Identified in Thyroid Cancer Cell Lines
| Cell Lines | Glycoproteins Identified | Non-Glycoproteins Identified | Total Proteins Identified |
|---|---|---|---|
| FTC-133 | 107 (29%) | 256 (71%) | 363 |
| TPC-1 | 166 (42%) | 231 (58%) | 397 |
| XTC-1 | 181 (38%) | 296 (62%) | 477 |
| ARO | 173 (43%) | 229 (57%) | 402 |
| DRO-1 | 121 (30%) | 288 (70%) | 409 |
The numbers listed in columns 2 and 3 are the total number of glycoproteins and non-glycoproteins identified in each thyroid cancer cell line. The percentile listed in the parenthesis in columns 2 and 3 indicate the percentage of glycoproteins and non-glycoproteins identified in each cell line.
In total, 313 unique glycoproteins were identified in the lysates prepared from the five thyroid cancer cell lines (see Supplemental Table 3). Among the glycoproteins identified, 134 are classified as integral membrane proteins and are predicted to have from 1 to 12 transmembrane regions. These results demonstrate the effectiveness of our method for solubilizing glycoproteins from cells. The glycoproteins identified include cell surface receptors, antigens, and adhesion molecules.
On average, glycoproteins accounted for 36.4% of the total proteins identified in the five thyroid cancer cell lines; similar to the result of 32.4% observed in our previous analysis of HeLa cells.14 As found in the HeLa cell study, the non-glycoprotein identified in the thyroid cells are highly abundant cellular proteins including protein disulfide-isomerase, enolase, GRP 78, HSP90, histones and actin. We previously demonstrated that these proteins non-specifically bind to the hydrazide resin and are incompletely removed by washing the resin with 8M urea, organic solvents (methanol and acetonitrile), and high salt washes (1.5M NaCl).14
Subcellular Location of Glycoproteins Identified
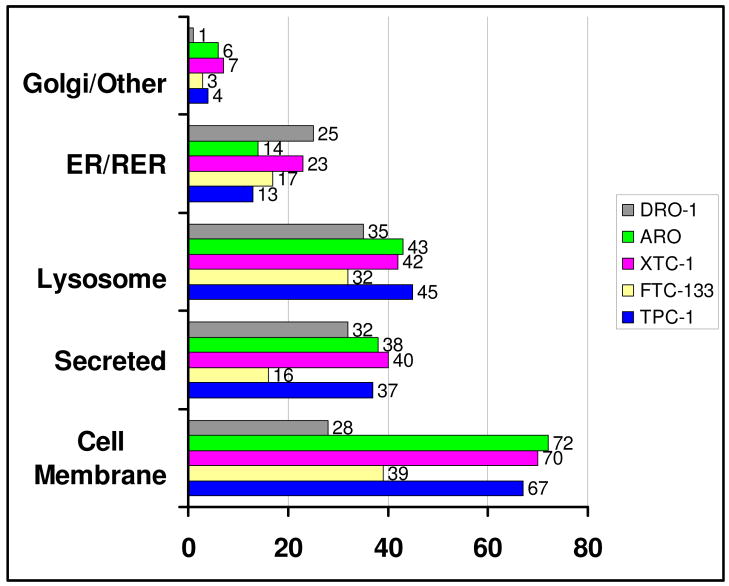
Based on the UniProtKB database, the subcellular locations of the glycoproteins identified in the thyroid cancer cell lines are the plasma membrane, the endoplasmic reticulum (ER), the golgi apparatus, lysosomes, secreted proteins and other proteins (cytosol, unknown) as shown in Figure 2. Among all of the glycoproteins identified within the papillary thyroid cancer cell line (Figure 2) TPC-1, 67 (40%) were identified as plasma membrane glycoproteins, including those designated as cell surface receptors, CD antigens (such as CD44, CD147 and CD222), membrane transport proteins and cell adhesion proteins. A significant number of lysosomal glycoproteins, 45 (27%), were also identified in TCP-1 cells. Several of these lysosomal proteins are known to be located on the plasma membrane during some point in their cellular trafficking. For example, the cation-independent mannose-6-phosphate receptor precursor binds lysosomal enzymes that have been secreted from the cell and directs them to endosomes in the cell. Of the proteins identified in TPC-1, 37 proteins (22%) were categorized as secreted proteins. Only a small fraction of glycoproteins identified in TPC-1, 17 proteins (10%), were classified as being resident in the ER/golgi or elsewhere.
Figure 2.
Subcellular location of glycoproteins detected from the tryptic digests and PNGase F-released glycopeptides from TPC-1, FTC-133, XTC-1, ARO and DRO-1 cells. The horizontal axis represents the number of glycoproteins identified.
A similar subcelluar distribution was found for the glycoproteins identified in the other differentiated thyroid cancer cell lines FTC-133 and XTC-1. Among the glycoproteins found in the follicular thyroid cancer cell line (Figure 2) FTC-133, 36% were classified as plasma membrane. Although fewer (107 vs. 166 for TPC-1) glycoproteins were identified in lysates from FTC-133, the ratio of plasma membrane glycoproteins identified was similar. Nearly 40 percent or 70 out of 181 glycoproteins identified in the Hürthle cell carcinoma cell line, XTC-1 (Figure 2), are classified as plasma membrane proteins. Glycoproteins in other subcellular locations include 40 secreted proteins, 42 lysosomal proteins and 30 ER/golgi or elsewhere proteins.
Of the glycoproteins identified in the anaplastic cell line ARO, 42% or 72 are classified as plasma membrane glycoproteins (Figure 2), whereas the percentage of plasma membrane proteins identified in DRO-1 cells was significantly lower, 23% or 28 glycoproteins (Figure 2). In contrast, comparable numbers of secreted proteins (32 in DRO vs. 38 in ARO) and lysosomal proteins (35 in DRO-1 vs. 43 in ARO) were identified in the two cell lines. Among the MS/MS spectra obtained from the DRO-1 cell lysate, a very large number corresponded to tryptic peptides from alpha-1-anti-chymotrypsin. More than 2400 spectra counts were obtained for peptides derived from alpha-1-anti-chymotrypsin in each of two samples, which is 3–5 times higher than for any other peptide from a glycoprotein in any other cell lysate. The large number of spectra for the alpha-1-anti-chymotrypsin tryptic peptides would overwhelm the ion signal for any co-eluted peptides derived from other glycoproteins, resulting in a relatively low number of glycoprotein identifications for the DRO-1 cell line.
Additional Glycoprotein identifications, and N-linked Glycosylation Site identifications from peptides released by PNGase F treatment of the hydrazide resin
In the case of low abundant glycoproteins, their tryptic peptides are less likely to be identified due to the presence of other more abundant tryptic peptides. However, the detection of glycopeptides can be substantially enhanced when the less complex, PNGase F-released samples are analyzed. The N-linked glycopeptides are collected, concentrated by SPE chromatography, and analyzed by nanoLC/MS/MS in combination with gas phase fractionation.
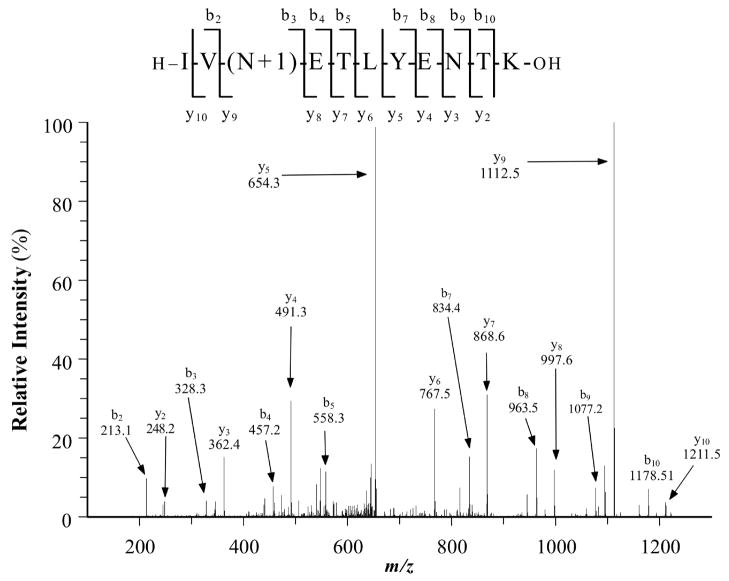
Sites of N-linked glycosylation were identified by locating peptides with an N-linked glycosylation consensus sequence (NX(S/T), where X is not P) in which the Asn residue was converted to Asp. To assure the accuracy of peptide sequence assignments, the MS/MS spectrum for each identified glycopeptide was manually evaluated for i) the expected mass deviation between the mass of the detected ion and the calculated mass of the deamidated peptide, ii) the Mascot ion score, iii) the X!Tandem –log(e) score, and iv) fragment ion matches. This extensive validation process eliminated several false positives. As a result, the average Mascot ion score for the N-linked glycopeptides in our list is more than 70, reflecting the high quality of the spectra. Most of N-linked glycopeptides identified in the list are doubly charged peptides. Any peptides with a charge greater than 3 were discarded due to the lack of fragments generated within the N-linked consensus sequence. Figure 3 shows an example of the MS/MS spectrum of an N-linked glycopeptide derived from tetraspanin 8 (AA# 116–126) derived from ARO cells. The fragments shown in Figure 3 demonstrate that N118 was converted to Asp, whereas N124 was not. Thus, the mass difference of 115 Da between fragments y8 and y9 confirms that a conversion of N to D occurred at N118, but not at N124, showing that tetraspanin 8 is N-glycosylated on N118. Using this approach, 20 additional glycoproteins were identified from peptides released by PNGase F treatment of the hydrazide resin (Supplemental Tables 4 and 5), bringing the total identified glycoproteins to 333 from the five thyroid cancer cell lines. These 20 glycoproteins were identified based on the spectra for a single N-linked glycopeptide (Supplemental Figures 1). In addition, 37 glycoproteins were identified in some cells based on a single N-linked glycopeptide (MS/MS spectra presented in Supplemental Figures 1). However, the latter glycoproteins were identified in at least one other thyroid cancer cell line by more than one peptide. For example, fibrillin-1 was identified in the TPC-1 cell line based on 2 unique peptides, whereas it was identified by a single N-linked glycopeptide in XTC-1 cells.
Figure 3.
MS/MS spectrum of the N-linked glycopeptide containing amino acids 116–126 of tetraspanin 8 from ARO cells. Asn118 has been converted to Asp by PNGase F as denoted by N+1 with the mass difference of 115 Da between fragments y8 and y9.
In addition to the glycoproteins uniquely identified from the PNGase F released glycopeptides, the PNGase F released peptide also contained a large number of N-linked glycopeptides from glycoproteins identified in the tryptic peptide fractions. For the TPC-1 cell line, 315 unique N-linked glycopeptides, occurring within 150 glycoproteins, were identified (see Table 2 and Supplemental Table 4 for lists of glycopeptides identified). Of the N-linked glycopeptides identified, 152 sites (see Table 2) have not been previously verified as being glycosylated (i.e. these sites are listed as potential N-linked sites based on the presence of the NXT/S consensus sequence). 23–26
Table 2.
Total N-linked Glycopeptides, Newly Verified N-linked Glycosylation Sites and the Total Number of Glycoproteins Identified via N-linked Glycopeptides in the Thyroid Cancer Cell Lines
| Cell Lines | Unique N-linked Glycopeptides Identified | Newly Verified N-linked Sites | Total # of Glycoproteins Identified |
|---|---|---|---|
| FTC-133 | 72 | 30 | 41 (3) |
| TPC-1 | 315 | 152 | 150 (20) |
| XTC-1 | 320 | 165 | 146 (17) |
| ARO | 259 | 142 | 126 (8) |
| DRO-1 | 152 | 64 | 84 (9) |
The values listed in column 2 are the total number of N-linked glycopeptides identified in the samples released from the hydrazide resin by PNGase F treatment. The values listed in column 3 represent the number of newly verified N-linked sites (listed in UniProt as potential N-linked sites based on the presence of the NXT/S consensus sequence, but not previously verified as being glycosylated by experimental data).23–26 The values in parentheses in column 4 is the number of glycoproteins identified based on a single N-linked glycopeptide.
Compared to the number N-linked glycopeptides identified for TPC-1, far fewer N-linked glycopeptides (72 N-glycopeptides within 41 proteins) were identified for the FTC-133 cell line. This observation is consistent with the finding that the total glycoproteins identified from tryptic digests of FTC-133 samples was the smallest among all samples analyzed. Among the N-linked glycopeptides, a few glycopeptides were identified only when the data was analyzed using X!Tandem (see Supplemental Table 4). This demonstrates the usefulness of carrying out data analysis with more than one algorithm (i.e., Mascot and X!Tandem). Table 2 provides a summary of the numbers of glycoproteins and glycopeptides identified, as well as the number of potential N-linked sites that were verified as being glycosylated in glycoproteins from the thyroid cancer cell lines. Our data verify that more than 400 potential N-linked glycosylation sites are actually glycosylated in more than 240 glycoproteins. Five or more N-linked glycosylation sites have been verified in several proteins (e.g. integrin alpha-3, laminin alpha-3 chain, plexin-B2).
Glycoproteins Patterns in the Five Thyroid Cancer Cell Lines
To systematically study the distribution of the glycoproteins identified among the five thyroid cancer cell lines (Supplemental Table 3) a series of Venn diagrams were generated in which the glycoproteins found in each cell line were compared with those found in two other cell lines. Ten Venn diagrams (Figures 4A, 4B and Supplemental Figures 2) representing all of the possible combinations of each of three cell lines were prepared. Figures 4A and 4B show two examples of these results and demonstrate that in each case the three cell lines evaluated had more than 50 glycoproteins in common. When the glycoprotein patterns for the five cell lines were compared, a total of 52 commonly expressed glycoproteins were found (Supplemental Table 6). More than 60% of these commonly expressed proteins are classified as lysosomal and represent proteins that are likely expressed in most cell types (see Discussion). The second most abundant (~20%) class of commonly expressed glycoproteins are plasma membrane proteins and include GPI-anchored and single pass membrane proteins with a range of functions (e.g. adhesion, amino acid transport).
Figure 4.
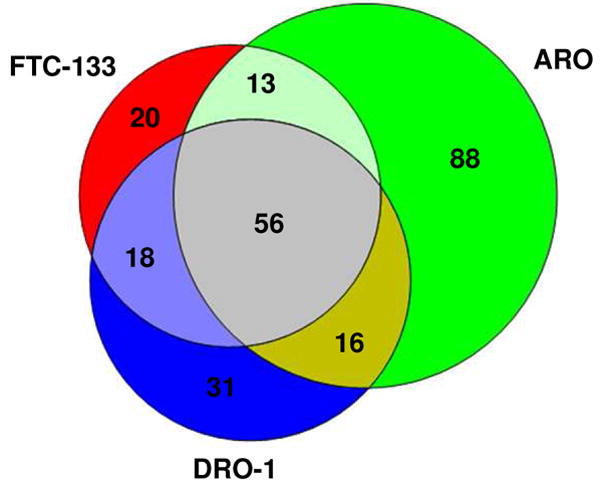
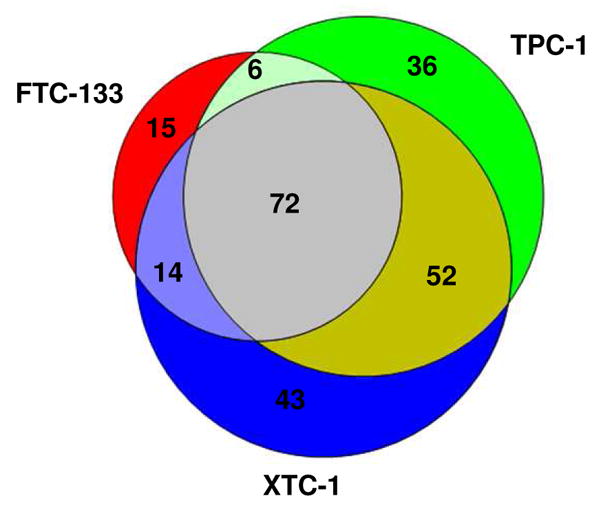
Figure 4A: Venn Plot of Glycoproteins Profiling of FCT-133, ARO and DRO-1 cells.
Figure 4B: Venn Plot of Glycoproteins Profiling of FCT-133, TPC-1 and XTC-1 cells
Distinct Glycoprotein Patterns Among the Thyroid Cancer Cell Lines
Comparisons of the glycoproteins identified in the various thyroid cancer cell lines revealed sets of glycoproteins that were found in only (1) the well-differentiated cell lines (TPC-1, FTC-133 and XTC-1), (2) the cell lines derived from thyroid cancer subtypes with poor prognosis (ARO and DRO-1), or (3) one of the cell lines.
Five glycoproteins were detected only in the well-differentiated thyroid cancer cell lines TPC-1, FTC-133 and XTC-1 (Supplemental Table 3). Cell membrane proteins, vasorin, neural cell adhesion molecule 1 (NCAM-1), trophoblast glycoprotein, discoidin and the integrin alpha-5 chain were exclusively detected in these cell lines, with average spectra counts for the identifying peptides ranging from 7.8 to 25.7.
Alpha-2-macroglobulin, UDP-glucose ceramide glucosyltransferase-like 1, and lactadherin were exclusively identified in the anaplastic thyroid cancer cells (ARO and DRO-1). Alpha-2-macroglobulin is a secreted protein and lactadherin is a plasma membrane glycoprotein, whereas UDP-glucose ceramide glucosyltransferase-like 1 is classified as being located in the endoplasmic reticulum. Alpha-2-macroglobulin was confidently detected in all ARO and DRO-1 cell line samples with average spectral counts of 17.8 and 13.8, respectively. Lactadherin was found in both ARO samples, and in one DRO-1 sample, with average spectral counts of 20.3. Four unique tryptic peptides were detected from alpha-2-macroglobulin, and 8 tryptic peptides were found for lactadherin. Tryptic peptides for UDP-glucose ceramide glucosyltransferase-like 1 were detected in both ARO and DRO-1 samples with average spectra counts of 5 and 22.5, respectively.
Analyses of each of the five cell lines resulted in lists of uniquely identified glycoproteins including 7 for FTC-133, 17 for TPC-1, 26 for XPC-1, 34 for ARO and 19 for DRO-1 (Supplemental Table 6). Most of these proteins are classified as membrane proteins (65–85%) in all of the cell lines except DRO-1 where secreted or extracellular proteins were found to predominate.
Among the glycoproteins uniquely expressed by the various thyroid cancer cell lines are two transmembrane heparan sulfate proteoglycans, syndecan-1 (FTC-133) and syndecan-4 (XTC-1). These proteins are involved in cell adhesion and signaling process.27 In contrast to the cell specific expression of these syndecans, a GPI-anchored proteoglycan, glypican-1, was identified in three of the thyroid cancer cells (FTC-133, XTC-1 and DRO-1). Finally, an extracellular heparan sulfate proteoglycan basement membrane-specific core protein or perlecan was only detected in the ARO cell line.
Cadherin-6 was found only in lysates from XTC-1, the Hürthle cell carcinoma cell line, whereas cadherin-13 was observed only in lysates from FTC-133 cells. These are two of several cadherin proteins (E-cadherin, N-cadherin or cadherin-2, H-cadherin or cadherin-13, desmoglein 2 and desmocolin 2) identified in the various thyroid cancer cell lines. Cadherins are transmembrane or GPI-anchored plasma membrane glycoproteins and mediate Ca2+-dependent cell-cell adhesion through homophilic interaction of their extracellular domains. Members of this protein family, particularly E-cadherin, have been suggested to be useful diagnostic markers for thyroid cancer (see Potter for a review).28
A number of other glycoproteins that have been linked to cancer were identified in the various thyroid cancer cell lines including proteins that are known to interact with the cadherin family of proteins, WNT, and proteins that regulate WNT signaling (Dickkopf-related protein 3 inhibitor of signaling pathway and secreted frizzled-related protein 1). Other glycoproteins that were either identified in a single cell line or identified in only a few of the thyroid cancer cell lines and which have been linked to cancer include receptor-type tyrosine-protein phosphatases and tyrosine-protein kinase receptors.
Discussion
The objectives of this study were to qualitatively evaluate the cell surface and secreted glycoprotein profiles of five human thyroid cancer cell lines that were derived from papillary, follicular, Hürthle cell carcinoma, and anaplastic thyroid cancer tumors, and identify potential biomarkers for various forms of thyroid cancer. The majority of the 333 glycoproteins identified are classified as either cell surface or secreted proteins. Many of our identifications are supported by other independent studies such as thyroid cancer gene expression profiling. For example, a recent review of 21 thyroid cancer gene expression studies by Griffith et al.9 showed that 23 genes are upregulated in thyroid cancer cells. Among these 23 upregulated genes, 9 encode glycoproteins (MET, SERPINA1, TIMP1, PROS1, FN1, SDC4, CD44, DPP4 and P4HA2) that were identified in our analysis, demonstrating the usefulness of the methodology employed in our study.
Glycoproteins Found in all of the Thyroid Cancer Cell Lines
Proteins commonly found among the thyroid cell lines may be useful as biomarkers for thyroid cancer in general. Of the 333 glycoproteins identified, 52 (16%) were detected in the five thyroid cancer cell lines (Supplemental Table 6). Of these common proteins, 12 proteins are cell surface, CD antigens (CD44, CD51, CD55, CD59, CD73, CD107A, CD107B, CD146, CD147, CD222, CD276 and CD315). Many of these CD antigens have been identified in several cancer types and have been suggested to be involved in tumor progression.29, 30 Recently, Tan et al. reported that CD147, in conjunction with MMP-2, are useful markers for differentiated thyroid carcinomas.31 Previous studies have shown that the message levels for three of these proteins, CD44, galectin-3-binding protein (LGALS3) and metalloproteinase inhibitor 1 (TIMP1) are up-regulated in thyroid cancer.9 Two other CD antigens, CD59 and CD73, are strongly expressed on the cell surface of benign and malignant thyroid follicular cells,32 and on the apical cell membrane of papillary thyroid cancer cells.33
Two lysosomal enzymes, cathepsin D and proactivator polypeptide, are among the most abundant glycoproteins (based on the spectra counts) identified in all of the thyroid cancer cell lines. Cathepsin D, with spectra counts of ~200-1,400 for the various cell lines, has been reported to have a higher concentration in thyroid neoplastic tissues than normal tissues.34 Proactivator polypeptide, with spectra counts of ~300-800 for the various cell lines, has been proposed to act as a mitogenic, survival, and anti-apoptotic factor for prostate cancer cells.35
In a previous study, we identified glycoproteins expressed by the cervical cancer cell line, HeLa14, and recently we have characterized the glycoproteins expressed by a group of human breast cancer cell lines (unpublished). Comparison of the glycoproteins that were found to be expressed by all five thyroid cancer cell lines with those identified in HeLa cells and the breast cancer cell lines demonstrates that only one protein, aspartylglucosaminidase (AGA) is exclusively identified in thyroid cancer cells. AGA cleaves the GlcNAc-Asn bond that joins oligosaccharides to the peptide of asparagine-linked glycoproteins and functions in the catabolism of glycoproteins. Deficiency of the AGA enzyme causes the lysosomal storage disease, aspartylglycosaminuria.36
Differential Expression of Glycoproteins among various thyroid cancer cell lines
Various subtypes of thyroid cancers have been distinguished primarily by differences in tumor tissue morphology and cytology. Our results demonstrate clear differences in glycoprotein expression patterns among the various thyroid cancer cell lines (Supplemental Table 6).
Comparing the glycoproteins identified in the three cell lines (TPC-1, FTC-133 and XTC-1) derived from tumors described as being well differentiated cancers to those in cell lines defined as dedifferentiated (anaplastic, ARO and DRO-1) resulted in a list of five glycoproteins that were exclusively identified in the differentiated cell lines. Cell membrane proteins, vasorin, neural cell adhesion molecule 1 (NCAM-1), trophoblast glycoprotein, the integrin alpha-5 subunit, and discoidin were exclusively detected in TPC-1, FTC-133 and XTC-1. Interestingly, NCAM-1 expression has been shown to be down regulated in papillary thyroid cancer compared to normal thyroid tissue and adenomas, but has been detected in papillary thyroid cancer and TPC-1 cells.37, 38 Trophoblast glycoprotein, an oncofetal antigen, has been found in many cancers including colorectal, gastric and ovarian.39 Hoffmann et al.40 evaluated integrin expression in a range of follicular, papillary and anaplastic cell lines and demonstrated that the alpha-5 subunit is expressed at high levels in follicular cell lines, including FTC-133, and papillary cell lines. They also reported the presence of the alpha-5 subunit in two anaplastic cell lines, C643 and Hth74.40
Three glycoproteins, alpha-2-macroglobulin, UDP-glucose ceramide glucosyltransferase-like 1, and lactadherin, were exclusively identified in anaplastic thyroid cancer cells (ARO and DRO-1). Alpha-2-macroglobulin is a secreted protein and lactadherin is a plasma membrane glycoprotein, whereas UDP-glucose ceramide glucosyltransferase-like 1 is classified as an endoplasmic reticulum protein. Interestingly, lactadherin has also been previously reported to promote tumor growth.41
Glycoproteins Found Uniquely in Specific Thyroid Cancer Cell lines
Of the 333 glycoproteins identified in the five thyroid cancer cell lines, nearly one third, 105, were exclusively found in a single cell line. FTC-133 had the fewest, 7, uniquely identified glycoproteins, ARO the most, 34, whereas the other cell lines had between 19 and 26 uniquely identified glycoproteins (TPC-1 and DRO-1, 19, and XTC-1, 26).
Most (22 of 34 or 65%) of the glycoproteins uniquely identified in ARO cells are integral membrane glycoproteins, including eight multi-pass membrane proteins (Supplemental Table 6). Tumor-associated calcium signal transducer 1(CD326), and tetraspanin-8 appeared to be the most abundant (i.e. largest number of spectra counts) of these ARO cell glycoproteins. Interestingly, five of the multi-pass membrane glycoproteins (tetraspanin-1, tetraspanin-3, tetraspanin-8, tetraspanin-15 and CD82) uniquely identified in ARO cells belong to the tetraspanin superfamily, a family of plasma membrane proteins, with four membrane spanning segments, that are thought to mediate signal transduction processes in the regulation of cell development, activation, growth and motility.42–44 Additional members of the tetraspanin superfamily (CD9, CD63, CD81 and CD151) were identified in ARO cells and TPC-1 cells. Tetraspanin-8 has been found to be over-expressed in many cancer cells.45, 46 Anti-CD9 pull-down experiments with human colon cancer cells demonstrated that many proteins including CD9, CD44, CD46, CD81, CD151,CD315, CD326, tetraspanin-1, tetraspanin-8, tetraspanin-15 are involved in forming a tetraspanin web. Interesting most of these proteins were detected in ARO cells.47 The unique detection of an extensive range of the tetraspanin family members, and members of the tetraspanin web such as integrin alpha-6, CD326 and Notch 2 in ARO cells suggests that the tetraspanin web may have a critical role in anaplastic thyroid cancer.
In addition to the tetraspanin family of glycoproteins, ARO cells were found to be unique among the thyroid cell lines in expressing ephrin type-B receptor 3 and its ligands, ephrin-B1 and ephrin-B2, which are reported to involve in the tumor neovascularization and the regeneration of intestine cells.48–50 Another member of the ephrin receptors, ephrin type-B receptor 2, was detected in ARO, XTC-1 and TPC-1 cells. Ephrin-B2 mRNA is over expressed in differentiated thyroid cancer (papillary and follicular thyroid cancer) and is associated with more aggressive disease. 51, 52
Of the 19 uniquely identified glycoproteins in the DRO-1 cell line, 15 are secreted or plasma membrane proteins (see Supplemental Table 6). Serotransferrin precursor (24 tryptic peptides) and HPLN1 (10 tryptic peptides) were identified by approximately 40% sequence coverage and had the highest spectra counts. Transferrin is a secreted glycoprotein that is involved in binding and transporting iron ions. It has been reported that serum transferrin is upregulated in lung cancer patients with brain metastases.53 HPLN1 is found in the extracellular matrix of cells and involved in the development of the heart.54 It also stabilizes the interactions of the proteoglycans of versican and aggrecan with hyaluronic acid.55
Hepatitis A virus cellular receptor 1, follistatin-related protein 1, and protein-tyrosine phosphatase mu were the most abundant, uniquely identified glycoproteins in TPC-1 cells based on spectra counts. Follistatin-related protein 1 promotes revascularization56, hepatitis A virus cellular receptor 1 is a member of the TIM family, and is involved in immune system regulation57, and protein-tyrosine phosphatase mu is a member of the protein tyrosine phosphatase family and is over-expressed in ovarian cancer.58
Hürthle cell carcinoma (HCC) has the highest risk of metastasis among the well differentiated thyroid cancer subtypes. Twenty-six glycoproteins were uniquely identified in XTC-1 cells including CD14, which was identified by 7 tryptic peptides (31% sequence coverage). CD14 is often found on the cell surface of monocytes or macrophages, but has not been reported to be expressed in normal thyroid tissue or thyroid cancer. Bone marrow stromal antigen 1 (CD157) was also confidently identified and has been reported to be overexpressed in circulating endothelial cells of patients with metastatic carcinomas.59
Only seven glycoproteins were uniquely identified in FTC-133 cells. In contrast to the other cell lines used in this study that grow in tightly adherent monolayers, FTC-133 cells loosely adhere to the culture dish. Consistent with this cell trait, few cell surface adhesion glycoproteins were detected in FTC-133 relative to the number identified in the other cell lines. For example, intercellular adhesion molecule 1 (CD54) and integrin alpha-2 were identified in most of the thyroid cancer cell lines, whereas neither was detected in FTC-133 cells. However, FTC-133 cells did uniquely express syndecan-1 and cadherin-13. Syndecan-1 has been found to be expressed in lower levels in the de-differentiated thyroid cancer cell lines compared to the well-differentiated thyroid gland carcinomas.60 Cadherin-13 is a unique member of the cadherin family that lacks the transmembrane and cytoplasmic domains found in other cadherins. Instead Cadherin-13 is anchored to the plasma membrane through a GPI anchor. The functions of cadherin-13 are not well understood61, but recent studies have shown that it interacts with Grp78 (a protein detected in all of the thyroid cancer cell lines, see Supplemental Table 1) and this interaction associated with cell survival signal transduction via AKT in endothelial cells.
Thyroid Cancer Biomarker Candidates
Distinguishing normal thyroid cells from various types of thyroid cancer cells is generally based on cytological features observed in cells obtained from fine needle aspiration biopsies. The cytological features for papillary and medullary thyroid cancer are unique and usually allow accurate diagnosis, but it is almost impossible to distinguish between benign and malignant follicular and Hürthle cell neoplasm fine needle aspiration biopsies. Although, cytologic examination is useful in the diagnosis of thyroid cancer, inconclusive cases occur in up to 30% of biopsy requiring the patient to undergo an operation to exclude a thyroid cancer diagnosis. Most patients with differentiated thyroid cancer do well, but 10-15% of patients have aggressive disease. Better predictors of disease recurrence are needed to tailor the treatment patients receive to reduce unnecessary aggressive therapy for the majority of patients and to identify those patients who would benefit from an aggressive therapy. The differences in cell surface glycoproteins observed in our analysis could provide candidate diagnostic and prognostic biomarkers, and therapeutic targets for differentiated thyroid cancer if validated in future studies.
Several of the glycoproteins identified in the five thyroid cancer cell lines have been implicated in cancer and/or tumor progression. Thus, they have potential as biomarkers for a specific type of thyroid cancer or as general thyroid cancer biomarkers. In Table 3 we lists a panel of biomarker candidates based on the following criteria; the glycoproteins are classified as secreted or located in the plasma membrane, their identification was verified by manual inspection of the MS/MS spectra, and whenever available the immunohistochemical data in the Human Protein Atlas62 indicates that the glycoprotein's expression level in thyroid cancer tissue is greater than that in normal thyroid gland tissue. In some cases glycoproteins are included in the table even though data is unavailable in the Human Protein Atlas or the antibody used does not stain either normal or cancer tissue from thyroid. MS/MS spectra of peptides identified for each of the glycoproteins listed in Table 3 were manually evaluated for the expected fragmentations, and information on these peptides is summarized in Supplemental Figures 3.
Table 3.
Glycoprotein Biomarker Candidates for Thyroid Carcinomas
| Biomarker Candidates | Thyroid Carcinoma Subtypes | ||||||
|---|---|---|---|---|---|---|---|
| Protein Name (SwissProt Accession Number) | Papillary Thyroid Carcinoma (TPC-1) | Follicular Thyroid Carcinoma (FTC-133) | Hürthle cell Thyroid Carcinoma (XPC-1) | Anaplastic Thyroid Carcinoma (ARO) | Anaplastic Thyroid Carcinoma (DRO-1) | Expression Level in Normal Thyroida | Expression Level in Thyroid Cancera |
| CD59 (P13987) | ☑ | ☑ | ☑ | ☑ | ☑ | − | ++ |
| Basigin, CD147 (P35613) | ☑ | ☑ | ☑ | ☑ | ☑ | − | + to +++ |
| Epididymal secretory protein E1 (P61916) | ☑ | ☑ | ☑ | ☑ | ☑ | ++ | +++ |
| Galectin-3-binding protein (Q08380) | ☑ | ☑ | ☑ | ☑ | ☑ | ||
| Neutral cell adhesion molecule 1, CD56, NCAM-1 (P13591) | ☑ | ☑ | ☑ | − | + to +++ | ||
| Lactadherin (Q08431) | ☑ | ☑ | +++b +c | ++b + to + to ++c | |||
| Tetraspanin-8 (P19075) | ☑ | ||||||
| Tumor-associated calcium signal transducer 1, CD326 (P16422) | ☑ | ++ | + to +++ | ||||
| Ephrin type-B receptor 3 (P54753) | ☑ | ++d | ++ to +++d | ||||
| Ephrin-B1 (P98172) | ☑ | ||||||
| Hyaluronan and proteoglycan link protein 1(P10915) | ☑ | ||||||
| Elastin microfibril interfacer 1 (Q9Y6C2) | ☑ | − | − | ||||
| Syndecan-1 (P18827) | ☑ | − | + | ||||
| Cadherin-13 (P55290) | ☑ | − | − | ||||
| Dickkopf-3 (Q9UBP4) | ☑ | + | + to ++ | ||||
| Follistatin-related protein 1 (Q12841) | ☑ | ||||||
| Receptor-type tyrosine-protein phosphatase mu (P28827) | ☑ | ||||||
| Bone marrow stromal antigen 1, CD157 (Q10588) | ☑ | ||||||
| G-protein coupled receptor 64 (Q8IZP9) | ☑ | − | − | ||||
Results for Protein Atlas (http://www.proteinatlas.org/index.php) Immunohistochemical results are listed as (−) no staining or negative, (+) weak staining, (++) moderate staining, (+++) strong staining. A blank cell indicates that no information on antibody staining is available.
Results for antibody CAB002753
Results for antibody HPA002807
Results for antibody HPA007698
The glycoproteins listed in Table 3 include those detected in all of the thyroid cancer cell lines, and thus could be used as markers of thyroid cancer tissue vs. normal or possibly benign thyroid tissue. Other glycoproteins listed in Table 3 have a more limited distribution and may serve as markers for the more aggressive forms of thyroid cancer vs. those types that have more promising treatment outcomes. Still others may serve to distinguish among the various thyroid cancer types. Even if these glycoproteins do not have utility as thyroid cancer biomarkers, evaluation of their distribution in normal, benign and thyroid cancer tissues has the potential to add to our understanding of the origin of the various thyroid cancer types.63
Cellular Origin of ARO and DRO-1 cell lines
A recent genetic study of 40 thyroid cancer cell lines provided evidence that TPC-1 and FTC-133 cell lines are of thyroid origin, but indicated that ARO (a different cell line than used in our study) was identical to colon cancer HT-29 cell line, and that a DRO-1 was identical to melanoma A-375 cell line.15. We did, however, perform quantitative gene expression analysis of markers that are specific for thyroid follicular cells to confirm a thyroid origin of the cell lines we used in our studies.
Unfortunately, no comprehensive proteomic analyses have been done on HT-29 or A-375. Therefore, we cannot do a comparison with the results obtained here to determine what overlap, if any, exists. Le Naour et al.47 used pull-down experiments, Western blotting and mass spectrometry to evaluate the tetraspanin web of several human colon cell lines, including HT-29. Comparing the list of glycoproteins identified by Le Naour and coworkers with those obtained from the ARO cell line used in our study does show some overlap in proteins identified. Since the number of glycoproteins available from the tetraspanin web study is limited it is difficult to draw conclusions about the relationship between the HT-29 and ARO cell lines. Previous proteomic studies on human melanoma cell lines are even more limited than those on colon cancer cell lines, therefore comparisons of glycoproteomic profiles are not possible
Conclusion
In the current study, we demonstrate that the glyco-capture method is very effective for profiling secreted and cell surface glycoproteins of thyroid cancer cells. Of 333 glycoproteins identified, more than 50 were detected in all five thyroid cancer cell lines, while many others were exclusively identified in a particular subtype of thyroid cancer cells. Most of the common glycoproteins also are found in HeLa and human breast cancer cell lines, indicating that these glycoproteins are commonly expressed in many human cell lines. A panel of biomarker candidates was derived from the glycoproteins identified among the thyroid cancer cell lines. In future studies, antibodies to these glycoproteins will be used to evaluate their distribution in normal thyroid and cancer tissues and to determine whether they would serve as diagnostic and prognostic biomarkers, and therapeutic target genes.
Supplementary Material
Acknowledgments
Support for this work was provided by grants from the National Institutes of Health, Grant P20 MD000544 and GM048972, and the National Science Foundation, Grant CHE-0619163. Venn diagrams were plotted used Venn Diagram Plotter program by Kyle Littlefield and Mattew Monroe at PNNL, Richland WA. The authors thank Ms. Julie Wang for the gene expression work.
Abbreviations
- TC
thyroid cancer
- PTC
papillary thyroid cancer
- FTC
follicular thyroid cancer
- MTC
medullary thyroid cancer
- ATC
anaplastic thyroid cancer
- FNA
fine needle aspiration
- BTGP
octyl-β-D-1-thioglucopyranoside
- DTT
dithiothriol
- IAM
iodoacetamide
- EDTA
ethylenediaminetetraacetic acid
- PNGase F
Peptide-N-Glycosidase F
- SPE
solid phase extraction
- ER
endoplasmic reticulum
- CD
cluster of differentiation
- GPI
glycosylphosphatidylinositol
Footnotes
Supporting Information Available: This information is available free of charge via the Internet at http://pubs.acs.org
References
- 1.ATA. Thyroid Nodules the American Thyroid Association. ATA Publications; 2005. http://www.thyroid.org/patients/patient_brochures/nodules.html. [Google Scholar]
- 2.ATA. Cancer of the Thyroid the American Thyroid Association. ATA Publications; 2005. http://www.thyroid.org/patients/patient_brochures/cancer_of_thyroid.html. [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 4.NCI. National Cancer Institute, NIH Publication No. 07-4994. 2007. What You Need To Know About Thyroid Cancer. [Google Scholar]
- 5.de Micco C, Savchenko V, Giorgi R, Sebag F, Henry JF. Utility of malignancy markers in fine-needle aspiration cytology of thyroid nodules: comparison of Hector Battifora mesothelial antigen-1, thyroid peroxidase and dipeptidyl aminopeptidase IV. Br J Cancer. 2008;98(4):818–23. doi: 10.1038/sj.bjc.6604194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slough CM, Randolph GW. Workup of well-differentiated thyroid carcinoma. Cancer Control. 2006;13(2):99–105. doi: 10.1177/107327480601300203. [DOI] [PubMed] [Google Scholar]
- 7.Are C, Shaha AR. Anaplastic Thyroid Carcinoma: Biology, Pathogenesis, Prognostic Factors, and Treatment Approaches. Ann Surg Oncol. 2006;13(4):453–464. doi: 10.1245/ASO.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 8.Sakorafas GH, Peros G, Farley DR. Thyroid nodules: Does the suspicion for malignancy really justify the increased thyroidectomy rates? Surgical Oncology. 2006;15(1):43–55. doi: 10.1016/j.suronc.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Griffith OL, Melck A, Jones SJM, Wiseman SM. Meta-Analysis and Meta-Review of Thyroid Cancer Gene Expression Profiling Studies Identifies Important Diagnostic Biomarkers. J Clin Oncol. 2006;24(31):5043–5051. doi: 10.1200/JCO.2006.06.7330. [DOI] [PubMed] [Google Scholar]
- 10.Giusti L, Iacconi P, Ciregia F, Giannaccini G, Donatini GL, Basolo F, Miccoli P, Pinchera A, Lucacchini A. Fine-needle aspiration of thyroid nodules: proteomic analysis to identify cancer biomarkers. J Proteome Res. 2008;7(9):4079–88. doi: 10.1021/pr8000404. [DOI] [PubMed] [Google Scholar]
- 11.Netea-Maier RT, Hunsucker SW, Hoevenaars BM, Helmke SM, Slootweg PJ, Hermus AR, Haugen BR, Duncan MW. Discovery and validation of protein abundance differences between follicular thyroid neoplasms. Cancer Res. 2008;68(5):1572–80. doi: 10.1158/0008-5472.CAN-07-5020. [DOI] [PubMed] [Google Scholar]
- 12.Srisomsap C, Subhasitanont P, Otto A, Mueller EC, Punyarit P, Wittmann-Liebold B, Svasti J. Detection of cathepsin B up-regulation in neoplastic thyroid tissues by proteomic analysis. Proteomics. 2002;2(6):706–12. doi: 10.1002/1615-9861(200206)2:6<706::AID-PROT706>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Aebersold R, Zhang H. Isolation of N-linked glycopeptides from plasma. Anal Chem. 2007;79(15):5826–37. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 14.McDonald CA, Yang JY, Marathe V, Yen T-Y, Macher BA. Combining Results from Lectin Affinity Chromatography and Glycocapture Approaches Substantially Improves the Coverage of the Glycoproteome. Mol Cell Proteomics. 2009;8(2):287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland JA, Smallridge RC, Haugen BR. Deoxyribonucleic Acid Profiling Analysis of 40 Human Thyroid Cancer Cell Lines Reveals Cross-Contamination Resulting in Cell Line Redundancy and Misidentification. J Clin Endocrinol Metab. 2008;93(11):4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garner AE, Smith DA, Hooper NM. Visualization of detergent solubilization of membranes: implications for the isolation of rafts. Biophys J. 2008;94(4):1326–40. doi: 10.1529/biophysj.107.114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17(20):2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 18.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal Chem. 2002;74(20):5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 21.Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23(18):3205–16. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17(7):676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 23.Lewandrowski U, Moebius J, Walter U, Sickmann A. Elucidation of N-glycosylation sites on human platelet proteins: a glycoproteomic approach. Mol Cell Proteomics. 2006;5(2):226–33. doi: 10.1074/mcp.M500324-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4(6):2070–80. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5(6):1493–503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21(6):660–6. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 27.Fears CY, Woods A. The role of syndecans in disease and wound healing. Matrix Biology. 2006;25(7):443–456. doi: 10.1016/j.matbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Potter E, Bergwitz C, Brabant G. The Cadherin-Catenin System: Implications for Growth and Differentiation of Endocrine Tissues. Endocr Rev. 1999;20(2):207–239. doi: 10.1210/edrv.20.2.0362. [DOI] [PubMed] [Google Scholar]
- 29.Bogenrieder T, Herlyn M. Axis of evil: molecular mechanisms of cancer metastasis. Oncogene. 2002;22(42):6524–6536. doi: 10.1038/sj.onc.1206757. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Park YW, Schiff BA, Doan DD, Yazici Y, Jasser SA, Younes M, Mandal M, Bekele BN, Myers JN. An orthotopic model of anaplastic thyroid carcinoma in athymic nude mice. Clin Cancer Res. 2005;11(5):1713–21. doi: 10.1158/1078-0432.CCR-04-1908. [DOI] [PubMed] [Google Scholar]
- 31.Tan H, Ye K, Wang Z, Tang H. Clinicopathologic evaluation of immunohistochemical CD147 and MMP-2 expression in differentiated thyroid carcinoma. Japanese journal of clinical oncology. 2008;38(8):528–33. doi: 10.1093/jjco/hyn065. [DOI] [PubMed] [Google Scholar]
- 32.Yamakawa M, Yamada K, Tsuge T, Ohrui H, Ogata T, Dobashi M, Imai Y. Protection of thyroid cancer cells by complement-regulatory factors. Cancer. 1994;73(11):2808–17. doi: 10.1002/1097-0142(19940601)73:11<2808::aid-cncr2820731125>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 33.Kondo T, Nakazawa T, Murata SI, Katoh R. Expression of CD73 and its ecto-5'-nucleotidase activity are elevated in papillary thyroid carcinomas. Histopathology. 2006;48(5):612–4. doi: 10.1111/j.1365-2559.2005.02277.x. [DOI] [PubMed] [Google Scholar]
- 34.Metaye T, Millet C, Kraimps JL, Aubouin B, Barbier J, Begon F. Estrogen receptors and cathepsin D in human thyroid tissue. Cancer. 1993;72(6):1991–6. doi: 10.1002/1097-0142(19930915)72:6<1991::aid-cncr2820720633>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 35.Lee TJ, Sartor O, Luftig RB, Koochekpour S. Saposin C promotes survival and prevents apoptosis via PI3K/Akt-dependent pathway in prostate cancer cells. Mol Cancer. 2004;3:31. doi: 10.1186/1476-4598-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikonen E, Aula P, Gron K, Tollersrud O, Halila R, Manninen T, Syvanen AC, Peltonen L. Spectrum of mutations in aspartylglucosaminuria. Proc Natl Acad Sci USA. 1991;88(24):11222–6. doi: 10.1073/pnas.88.24.11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laco J, Ryska A. The use of immunohistochemistry in the differential diagnosis of thyroid gland tumors with follicular growth pattern. Cesk Patol. 2006;42(3):120–4. [PubMed] [Google Scholar]
- 38.Scarpino S, Di Napoli A, Melotti F, Talerico C, Cancrini A, Ruco L. Papillary carcinoma of the thyroid: low expression of NCAM (CD56) is associated with downregulation of VEGF-D production by tumour cells. J Pathol. 2007;212(4):411–9. doi: 10.1002/path.2183. [DOI] [PubMed] [Google Scholar]
- 39.Smyth LJ, Elkord E, Taher TEI, Jiang H-R, Burt DJ, Clayton A, Veelen PAv, Ru Ad, Ossendorp F, Melief CJM, Drijfhout JW, Dermime S, Hawkins RE, Stern PL. Cd8 T-cell recognition of human 5T4 oncofetal antigen. Int J Cancer. 2006;119(7):1638–1647. doi: 10.1002/ijc.22018. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann S, Maschuw K, Hassan I, Reckzeh B, Wunderlich A, Lingelbach S, Zielke A. Differential pattern of integrin receptor expression in differentiated and anaplastic thyroid cancer cell lines. Thyroid. 2005;15(9):1011–20. doi: 10.1089/thy.2005.15.1011. [DOI] [PubMed] [Google Scholar]
- 41.Neutzner M, Lopez T, Feng X, Bergmann-Leitner ES, Leitner WW, Udey MC. MFG-E8/lactadherin promotes tumor growth in an angiogenesis-dependent transgenic mouse model of multistage carcinogenesis. Cancer Res. 2007;67(14):6777–85. doi: 10.1158/0008-5472.CAN-07-0165. [DOI] [PubMed] [Google Scholar]
- 42.Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. 2003;19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609. [DOI] [PubMed] [Google Scholar]
- 43.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–11. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 44.Lazo PA. Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 2007;98(11):1666–77. doi: 10.1111/j.1349-7006.2007.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gesierich S, Paret C, Hildebrand D, Weitz J, Zgraggen K, Schmitz-Winnenthal FH, Horejsi V, Yoshie O, Herlyn D, Ashman LK, Zoller M. Colocalization of the Tetraspanins, CO-029 and CD151, with Integrins in Human Pancreatic Adenocarcinoma: Impact on Cell Motility. Clin Cancer Res. 2005;11(8):2840–2852. doi: 10.1158/1078-0432.CCR-04-1935. [DOI] [PubMed] [Google Scholar]
- 46.Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, Weitz J, Zoller M. A Complex of EpCAM, Claudin-7, CD44 Variant Isoforms, and Tetraspanins Promotes Colorectal Cancer Progression. Mol Cancer Res. 2007;5(6):553–567. doi: 10.1158/1541-7786.MCR-06-0384. [DOI] [PubMed] [Google Scholar]
- 47.Le Naour F, Andre M, Greco C, Billard M, Sordat B, Emile J-F, Lanza F, Boucheix C, Rubinstein E. Profiling of the Tetraspanin Web of Human Colon Cancer Cells. Mol Cell Proteomics. 2006;5(5):845–857. doi: 10.1074/mcp.M500330-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Holmberg J, Genander M, Halford MM, Annerén C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisén J. EphB Receptors Coordinate Migration and Proliferation in the Intestinal Stem Cell Niche. Cell. 2006;125(6):1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 49.Merlos-Suarez A, Batlle E. Eph-ephrin signalling in adult tissues and cancer. Curr Opin Cell Biol. 2008;20(2):194–200. doi: 10.1016/j.ceb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Hughes SE. Role of the ephrin and Eph receptor tyrosine kinase families in angiogenesis and development of the cardiovascular system. J Pathol. 2006;208(4):453–461. doi: 10.1002/path.1937. [DOI] [PubMed] [Google Scholar]
- 51.Kebebew E, Peng M, Reiff E, Duh QY, Clark OH, McMillan A. Diagnostic and prognostic value of angiogenesis-modulating genes in malignant thyroid neoplasms. Surgery. 2005;138(6):1102–9. doi: 10.1016/j.surg.2005.05.025. discussion 1109–10. [DOI] [PubMed] [Google Scholar]
- 52.Kebebew E, Peng M, Reiff E, McMillan A. Diagnostic and extent of disease multigene assay for malignant thyroid neoplasms. Cancer. 2006;106(12):2592–7. doi: 10.1002/cncr.21922. [DOI] [PubMed] [Google Scholar]
- 53.Marchi N, Mazzone P, Fazio V, Mekhail T, Masaryk T, Janigro D. ProApolipoprotein A1. Cancer. 2008;112(6):1313–1324. doi: 10.1002/cncr.23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirrig EE, Snarr BS, Chintalapudi MR, O'Neal JL, Phelps AL, Barth JL, Fresco VM, Kern CB, Mjaatvedt CH, Toole BP, Hoffman S, Trusk TC, Argraves WS, Wessels A. Cartilage link protein 1 (Crtl1), an extracellular matrix component playing an important role in heart development. Dev Biol. 2007;310(2):291–303. doi: 10.1016/j.ydbio.2007.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsumoto K, Kamiya N, Suwan K, Atsumi F, Shimizu K, Shinomura T, Yamada Y, Kimata K, Watanabe H. Identification and Characterization of Versican/PG-M Aggregates in Cartilage. J Biol Chem. 2006;281(26):18257–18263. doi: 10.1074/jbc.M510330200. [DOI] [PubMed] [Google Scholar]
- 56.Ouchi N, Oshima Y, Ohashi K, Higuchi A, Ikegami C, Izumiya Y, Walsh K. Follistatin-like 1, a Secreted Muscle Protein, Promotes Endothelial Cell Function and Revascularization in Ischemic Tissue through a Nitric-oxide Synthase-dependent Mechanism. J Biol Chem. 2008;283(47):32802–32811. doi: 10.1074/jbc.M803440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuchroo VK, Dardalhon V, Xiao S, Anderson AC. New roles for TIM family members in immune regulation. Nat Rev Immunol. 2008;8(8):577–80. doi: 10.1038/nri2366. [DOI] [PubMed] [Google Scholar]
- 58.Gyorffy B, Dietel M, Fekete T, Lage H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int J Gynecol Cancer. 2008 doi: 10.1111/j.1525-1438.2007.01169.x. [DOI] [PubMed] [Google Scholar]
- 59.Smirnov DA, Foulk BW, Doyle GV, Connelly MC, Terstappen LW, O'Hara SM. Global gene expression profiling of circulating endothelial cells in patients with metastatic carcinomas. Cancer Res. 2006;66(6):2918–22. doi: 10.1158/0008-5472.CAN-05-4003. [DOI] [PubMed] [Google Scholar]
- 60.Mitselou A, Ioachim E, Peschos D, Charalabopoulos K, Michael M, Agnantis NJ, Vougiouklakis T. E-cadherin adhesion molecule and syndecan-1 expression in various thyroid pathologies. Exp Oncol. 2007;29(1):54–60. [PubMed] [Google Scholar]
- 61.Cavallaro U, Liebner S, Dejana E. Endothelial cadherins and tumor angiogenesis. Exp Cell Res. 2006;312(5):659–67. doi: 10.1016/j.yexcr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 62.Bjorling E, Lindskog C, Oksvold P, Linne J, Kampf C, Hober S, Uhlen M, Ponten F. A web-based tool for in silico biomarker discovery based on tissue-specific protein profiles in normal and cancer tissues. Mol Cell Proteomics. 2008;7(5):825–44. doi: 10.1074/mcp.M700411-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Zhang P, Zuo H, Ozaki T, Nakagomi N, Kakudo K. Cancer stem cell hypothesis in thyroid cancer. Pathology International. 2006;56(9):485–489. doi: 10.1111/j.1440-1827.2006.01995.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.