Abstract
The major forms of diabetes are characterized by pancreatic islet β-cell dysfunction and decreased β-cell numbers, raising hope for cell replacement therapy. Although human islet transplantation is a cell-based therapy under clinical investigation for the treatment of type 1 diabetes, the limited availability of human cadaveric islets for transplantation will preclude its widespread therapeutic application. The result has been an intense focus on the development of alternate sources of β cells, such as through the guided differentiation of stem or precursor cell populations or the transdifferentiation of more plentiful mature cell populations. Realizing the potential for cell-based therapies, however, requires a thorough understanding of pancreas development and β-cell formation. Pancreas development is coordinated by a complex interplay of signaling pathways and transcription factors that determine early pancreatic specification as well as the later differentiation of exocrine and endocrine lineages. This review describes the current knowledge of these factors as they relate specifically to the emergence of endocrine β cells from pancreatic endoderm. Current therapeutic efforts to generate insulin-producing β-like cells from embryonic stem cells have already capitalized on recent advances in our understanding of the embryonic signals and transcription factors that dictate lineage specification and will most certainly be further enhanced by a continuing emphasis on the identification of novel factors and regulatory relationships.
Keywords: Diabetes, insulin, islet, pancreas
Diabetes is rapidly becoming a global epidemic, with a staggering health, societal, and economic impact. Recent estimates by the American Diabetes Association suggest that the lifetime risk of developing diabetes for Americans born in the year 2000 is one in three. Diabetes results when insulin production by the pancreatic islet β cell is unable to meet the metabolic demand of peripheral tissues such as liver, fat, and muscle.
A reduction in β-cell function and mass leads to hyperglycemia (elevated blood sugar) in both type 1 and type 2 diabetes. In type 1 diabetes, autoimmune destruction of the β cell itself severely reduces β-cell mass, resulting in marked hypoinsulinemia and potentially life-threatening ketoacidosis. In contrast, during the progression to type 2 diabetes, impaired β-cell compensation in the setting of insulin resistance (impaired insulin action) eventually leads to β-cell failure and a modest but significant reduction in β-cell mass (Maclean and Ogilvie 1955; Butler et al. 2003; Yoon et al. 2003). More recently, autoimmunity has been detected in a subset of patients with type 2 diabetes, which has led to a revision of the classification to include LADA, latent autoimmune diabetes of adulthood, underscoring the continuum between type 1 and type 2 diabetes, and raising questions as to the role of immunity and inflammation in β-cell dysfunction and death in type 2 diabetes (Syed et al. 2002; Pozzilli and Buzzetti 2007). Conversely, forms of ketosis prone diabetes due to severe β-cell dysfunction but without evidence of autoimmunity are now recognized (Balasubramanyam et al. 2008). Decreased β-cell function also underlies early-onset monogenic forms of diabetes termed MODY (maturity-onset diabetes of the young) resulting from mutations in transcription factors that regulate β-cell development and differentiation. Mutations in MODY genes are also found in patients with common late-onset type 2 diabetes (Fajans et al. 2001). Thus, reduced β-cell number and function underlie the progression of the full spectrum of diabetes, prompting intense effort to develop new sources of insulin-producing β cells for replacement therapies.
Islet transplantation is a cell replacement approach whose widespread applicability as a treatment for diabetes has been curtailed by limited donor islet availability and the toxicity of immunosuppressive drug regimens. The success of islet transplantation was markedly improved by the introduction of the Edmonton protocol, which involved the transplantation of greater numbers of islets as well as a glucocorticoid-free immunosuppressant regimen that reduces but does not eliminate β-cell toxicity (Shapiro et al. 2000; Shapiro et al. 2006). However, the long-term success of this approach is still limited by a gradual reduction in β-cell function, resulting in a return to insulin dependence in a majority of subjects, as well as by the ongoing severe mismatch between the numbers of cadaveric donor islets available and those needed to treat patients with type 1 diabetes.
As a result, basic research has continued to focus on methods of generating β cells from alternative sources, such as transdifferentiation from related cell types or differentiation from stem and progenitor cells (Hogan et al. 2008). Although success in achieving stem cell differentiation into insulin expressing cells has been highly variable and even controversial, recent reports indicate highly promising results in the derivation of endoderm-like cells from embryonic stem (ES) cells and subsequent in vitro or in vivo differentiation into insulin-expressing β-like cells (D'Amour et al. 2005, 2006; J. Jiang et al. 2007; W. Jiang et al. 2007; Phillips et al. 2007; Shim et al. 2007; Kroon et al. 2008). This research was directly guided by fundamental advances in our understanding of the extracellular signals and transcription factors that dictate the embryonic development of the pancreas. This review emphasizes recent advances in our knowledge of both extracellular signals/signaling pathways and transcription factors that govern the formation of β cells during pancreas development. Although tremendous progress in delineating the molecular underpinnings of β development has been made, there is still much to be learned regarding the regulatory relationships among signaling pathways and transcription factors and how these processes guide pancreatic specification, endocrine differentiation, and β-cell maturation.
Overview of mouse pancreas development
The definitive endoderm, from which the pancreas arises, begins as a flat sheet of cells that is specified during gastrulation (Fig. 1). Genes required for definitive endoderm formation include Wnt/β-catenin, Nodal, GATA4/6, FoxA2, Sox17, and Mix (Grapin-Botton and Constam 2007; Zorn and Wells 2007). At the beginning of somitogenesis, closure of the gut tube starts as the anterior region of this sheet folds posteriorly, and eventually joins the anteriorly moving posterior fold. Specification of the pancreatic domain within the gut endoderm is mediated by a combination of mesodermally derived signals, including but not limited to TGFβ (transforming growth factor) superfamily members, retinoic acid (RA), and fibroblast growth factors (Fgf) (Grapin-Botton and Constam 2007; Spence and Wells 2007). Pancreatic specification becomes evident around embryonic day 8.5 (E8.5, eight to 10 somites) with the expression of pancreatic duodenal homeobox 1 (Pdx1) in two ventral domains, preceding expression of Pdx1 in the single dorsal domain at E8.5–E8.75. At this time, mesenchyme accumulates around the prospective dorsal and ventral pancreatic anlagen, with one of the two ventral domains eventually disappearing.
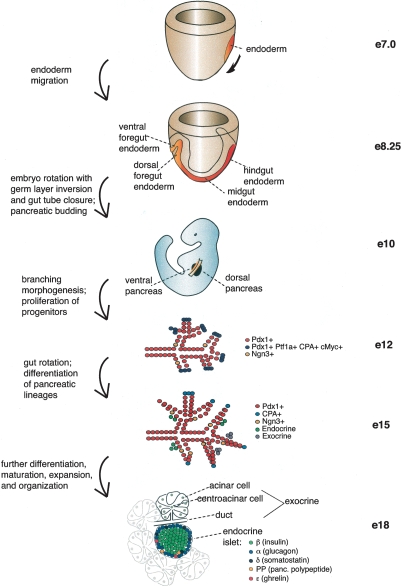
Figure 1.
Overview of mouse pancreatic organogenesis. Drawing depicts mouse embryonic development from early primitive streak stage (E7) through endoderm migration and specification, pancreatic budding, branching morphogenesis, and differentiation of pancreatic lineages. Multipotent progenitors that give rise to all pancreatic lineages express Pdx1, Ptf1a, CPA, and cMyc in early pancreas development, while exocrine progenitors express CPA during mid-pancreatic development. At all stages, endocrine progenitors express Ngn3.
Around E9.5, epithelial budding into the surrounding mesenchyme occurs with subsequent branching morphogenesis. Recent evidence indicates that multipotent progenitors that give rise to hormone-expressing endocrine cells, ductal cells, and digestive enzyme-producing acinar cells of the exocrine compartment are located at the tips of the branching network and are marked by Pdx1, Ptf1a, cMyc, and carboxypeptidase A (CPA) (Zhou et al. 2007). Although some endocrine cells, mostly glucagon+, are evident at E9.5 during the primary transition, most of the hormone-expressing cells that will comprise the endocrine islets begin to rapidly emerge around E13.5, a period called the secondary transition. Reports on the precise appearance of the different lineages vary, but by E13.5–E14.5 all five hormone-expressing endocrine lineages (α, β, δ, ε, and PP) are detectable. Emergence of amylase-expressing acinar cells commences around this time as well. The development of the ventral bud is generally delayed compared with the dorsal bud, except with regard to the differentiation of the acinar lineage. By this time, the gut tube has rotated, bringing the two buds into proximity. As embryogenesis proceeds, organ growth and differentiation continue by forming digestive enzyme-producing acinar cell clusters that empty into the ductal network leading into the duodenum, interspersed with endocrine cells that organize by delaminating from the epithelium and forming islet clusters (for comprehensive reviews, see Pictet et al. 1972; Slack 1995; Jorgensen et al. 2007).
Signaling pathways
Extracellular factors that modulate signaling pathways are required for distinct aspects of pancreas formation. The early specification of the pancreatic domain within the gut endoderm requires escape from Hedgehog signaling as well as regulation by endothelially derived signals and Fgf. The Fgf, Notch, Wnt, TGFβ superfamily and EGF pathways influence the growth of the early pancreas. TGFβ family members and EGF are also important in lineage specification and differentiation. Little is known about how these pathways cross-talk and how they regulate transcription factor cascades during pancreatic development (Kim and Hebrok 2001). Recent advances in signaling pathway cross-talk and their impingement on the transcriptional hierarchy will be particularly highlighted throughout this review.
Hedgehog signaling
Three members of the Hedgehog family, Sonic (Shh), Indian (Ihh), and Desert (Dhh), are expressed in the developing gut (for a comprehensive review, see Lau et al., 2006). All three proteins have similar binding affinities and function by interacting with the Patched receptor, relieving its repression of the G-protein-coupled receptor Smoothened, thereby initiating a signaling cascade that results in nuclear translocation of Gli transcription factors. Gli1 and Gli3 function as transcriptional activators and repressors, respectively, while both functions have been attributed to Gli2. Shh and Ihh are expressed throughout the early foregut endoderm but are excluded from the pancreatic domain.
Hedgehog signaling inhibits pancreas formation and early Pdx1 expression (Apelqvist et al. 1997; Kim and Melton 1998; Hebrok et al. 2000). Conversely, inhibition of Shh signaling is a critical step for the early specification of both pancreatic buds and for permitting Pdx1 expression. In the chick embryo, evagination of the dorsal bud occurs in response to mesodermal notochord-derived signals, including activin-βB, a TFGβ family ligand, which represses Shh, thereby enabling Pdx1 expression and inducing a pancreatic fate (Kim et al. 1997; Hebrok et al. 1998; Kim and Melton 1998). Consistent with the idea that activin signaling is necessary for inhibition of Shh and pancreatic specification, activin receptor II loss of function mouse mutants have decreased pancreatic size with a concomitant misexpression of Shh and reduced Isl1 expression (Kim et al. 2000). In contrast, ventral pancreas development is initiated when presumptive pancreatic endoderm migrates away from the high Fgf signaling environment of the cardiac mesoderm that up-regulates Shh and evades septum transversum mesoderm-derived bone morphogenetic protein (BMP) signaling, both of which normally specify a liver fate (Deutsch et al. 2001; Rossi et al. 2001).
RA-mediated inhibition of Shh has also been suggested, although its role in mammals is unclear. In Xenopus and zebrafish, RA signaling inhibits Shh and induces both pancreas gene expression in dorsal endoderm and endocrine differentiation (Stafford and Prince 2002; Chen et al. 2004; Stafford et al. 2004; Pan et al. 2007). In contrast, the phenotype of mouse embryos deficient for the RA-synthesizing enzyme Raldh2 suggests a Shh-independent pathway mediating RA effects on pancreas development. Raldh2 is expressed in mouse dorsal mesenchyme adjacent to pancreatic progenitors until E12.5, and RA signaling is detected in both mesenchyme and pancreatic epithelium. Raldh2-deficient mice do not develop the dorsal pancreatic bud and have fewer mesenchymal cells and decreased expression of the transcription factors, Hb9, Isl1, Pdx1, and Prox1 (Martin et al. 2005; Molotkov et al. 2005). However, this was not associated with ectopic expression of Ihh or Shh in the pancreas. These studies suggest that RA signaling in the mouse might influence early dorsal pancreatic development in a hedgehog-independent manner, by regulating expression of Isl1, Pdx1, or other factors, directly or indirectly.
Hedgehog signaling may also regulate pancreatic size and endocrine mass by its activity later in pancreas development. Expression of Ihh and Patched1 can be detected in the pancreas by E13.5 (Hebrok et al. 2000). Transgenic overexpression of Shh or Ihh under the control of the Pax4 promoter at mid-development dramatically inhibits formation of epithelial cells, acinar cells, and Neurogenin3 (Ngn3)-expressing endocrine precursors and decreases β-cell proliferation (Kawahira et al. 2005), suggesting that Hh signaling could regulate the number of pancreatic progenitors by modulating survival, proliferation, or differentiation state. Similarly, increased Hh activity in hedgehog inhibitor Hhip-null mice leads to defects in pancreas morphogenesis, reductions in pancreatic size, and endocrine cell formation likely in part due to delayed expression of Fgf10 in the mesenchyme, which normally promotes proliferation of Pdx1+ progenitors (Bhushan et al. 2001; Hart et al. 2003; Kawahira et al. 2003; Norgaard et al. 2003). This study provides the first indication that hedgehog signaling influences mesenchymal function.
Endothelial signals
Mesodermally derived aortic endothelial cells are important in dorsal pancreatic specification following initiation of Pdx1 expression. Aortic endothelial cells in close proximity to the dorsal Pdx1+ endoderm induce this region to express the pancreatic transcription factor Ptf1a and to maintain Pdx1 expression (Lammert et al. 2001; Yoshitomi and Zaret 2004). Moreover, the dorsal bud fails to emerge in Flk-1−/− embryos, which lack these endothelial cells likely due to the apoptotic loss of the overlying dorsal mesenchyme, thereby revealing a second role for the endothelium (Jacquemin et al. 2006). Even following specification of the dorsal pancreatic bud, interaction with overlying dorsal mesenchyme is necessary for further development, at least in part due to its expression of Fgf10 beginning at 15–20 somites (E9.5), which together with the transcription factor Hepatocyte Nuclear Factor 6 (HNF6), contributes to initial induction of Pdx1 expression and enhances aorta-induced Ptf1a expression.
Intraislet endothelial cells also play an essential role in endocrine development. Both the developing endoderm and islets express vascular endothelial growth factor (VEGF) and its receptor Flk-1 (Christofori et al. 1995; Dumont et al. 1995; Kuroda et al. 1995; Vasir et al. 2001). In mature islets, β cells synthesize VEGF-A, which may attract endothelial cells, evidenced by the marked reduction in intraislet endothelial cell numbers in mice with β-cell-specific deletion of VEGF-A (Nikolova et al. 2006). Conversely, transgenic overexpression of VEGF-A driven by the Pdx1 promoter induces formation of hypervascular hyperplastic islets (Lammert et al. 2001). Endothelial cells in turn synthesize the basement membrane laminin chains α4 and α5, which in culture studies up-regulate insulin gene expression and β-cell proliferation by interacting with β1-integrin containing laminin receptors on the β cell (Nikolova et al. 2006). The role of the endothelium in β-cell proliferation is further supported by the observation that vascular proliferation is closely associated temporally with the peak of β-cell proliferation in rat pancreatic development (Johansson et al. 2006). Altogether, these studies demonstrate the critical role of the endothelium in both pancreas specification and β-cell differentiation and proliferation.
Fgf signaling
In addition to the role of Fgf2 in initiating Pdx1 expression in the pancreatic primordium, multiple members of the Fgf family have also been described to regulate expansion and differentiation of the pancreatic epithelium. Expression of multiple Fgf ligands and receptors, including alternatively spliced isoforms, has been detected in the early rodent pancreas (Le Bras et al. 1998; Cras-Meneur and Scharfmann 2002; Elghazi et al. 2002; Dichmann et al. 2003; Hart et al. 2003). FgfR1c is detected in both the epithelium and mesenchyme, while FgfR1b, FgfR2b, and FgfR4 are detected exclusively in the epithelium and FgfR2c is present in the mesenchyme. Expression of the ligands Fgf1, Fgf7, Fgf9, Fgf10, Fgf11, and Fgf18 is also detected early in the developing pancreas (Dichmann et al. 2003). The role for Fgf9 is inferred from genetic mouse models with targeted disruption of its receptor, FgfR3, that reveal FgfR3 as a negative regulator of pancreatic epithelial cell expansion (Arnaud-Dabernat et al. 2007). In contrast, the Fgfr2b ligands Fgf1, Fgf7, and Fgf10, have all been shown to induce epithelial cell proliferation and switch the default fate of pancreatic epithelial explants from endocrine differentiation toward an exocrine fate (Miralles et al. 1999; Bhushan et al. 2001; Elghazi et al. 2002; Dichmann et al. 2003; Pulkkinen et al. 2003).
The most well-characterized of the Fgf ligands involved in pancreas development is Fgf10. Fgf10 is expressed by the dorsal and ventral mesenchyme from E9.5 to E12.5. Transgenic overexpression or targeted deletion of Fgf10 in mice indicates that Fgf10 regulates bud growth and morphogenesis by promoting proliferation of Pdx1+ epithelial pancreatic progenitors after its initial role to induce Pdx1 expression and maintain Ptf1a expression (Bhushan et al. 2001; Hart et al. 2003; Norgaard et al. 2003; Jacquemin et al. 2006). Fgf10 also maintains Notch signaling and Hes1 expression, thus preventing differentiation of progenitors (Hart et al. 2003; Norgaard et al. 2003; Miralles et al. 2006). These studies have begun to clarify the mechanism behind the well-observed phenomenon that mesenchyme is required for pancreatic bud growth and morphogenesis as well appropriate lineage differentiation (Golosow and Grobstein 1962; Wessels and Cohen 1967; Gittes et al. 1996; Miralles et al. 1998).
Notch signaling
In the classic Notch signaling pathway, interaction of the membrane-bound ligands Delta or Serrate with the Notch receptor on the adjacent cell leads to cleavage of the Notch intracellular domain (ICD) (for a comprehensive review of this pathway, see Louvi and Artavanis-Tsakonas 2006). The cleaved ICD translocates to the nucleus and interacts with the transcription factor RPB-Jκ (recombining binding protein suppressor of hairless), thereby activating expression of basic helix–loop–helix (bHLH) proteins of the hairy and enhancer of split family (Hes). Hes transcription factors, in turn, actively repress differentiation factors. Multiple Notch ligands and receptors are expressed in the early rodent pancreas, including Notch1-4, Delta-like 1 (Dll1), Dll3, Jagged 1 (Jag1), Jag2, Serrate 1, Serrate 2, and the Notch signaling target Hes1 (Apelqvist et al. 1999; Jensen et al. 2000b; Lammert et al. 2000). Promoter reporter assays indicate that Hes1 represses expression of both the bHLH factor Ngn3, a marker of endocrine precursors, and the cyclin-dependent kinase inhibitor p57, thus regulating cell cycle exit (Lee et al. 2001; Georgia et al. 2006). Mice deficient for Notch signaling pathway components such as RBP-Jκ, Dll1, or Hes1, display increased numbers of Ngn3+ cells, premature endocrine differentiation and a hypoplastic pancreas that is likely the result of premature depletion of pancreatic precursor cells (Apelqvist et al. 1999; Jensen et al. 2000b; Fujikura et al. 2006). Similarly, transgenic overexpression of Notch3 ICD, a Notch signaling inhibitor, leads to decreased epithelial branching and decreased bud size, low Ptf1a and Hes1 expression, increased Ngn3+ and hormone+ cells, and decreased numbers of mature acinar cells. Conversely, mice expressing a constitutively active Notch1 (Notch1-ICD) in the Pdx1 domain display reduced Ngn3+ and endocrine cell number as well as impaired acinar differentiation (Hald et al. 2003; Murtaugh et al. 2003). Expressing Notch1-ICD in the Ngn3+ population also prevents differentiation (Murtaugh et al. 2003). HES family members Hes1, Hey1, and Hey2 directly bind Ptf1 and inhibit its activity in promoter reporter assays, suggesting that Notch signaling also regulates exocrine differentiation by regulating Ptf1 activity (Esni et al. 2004; Ghosh and Leach 2006). All together, these studies indicate that Notch signaling in the early developing pancreas is responsible for maintaining the undifferentiated state of pancreatic progenitors, enabling proper growth and morphogenesis, and suppressing exocrine and endocrine differentiation by inhibiting Ptf1 activity and Ngn3 expression, respectively. Further investigation will be required to assess the specific roles of individual Notch signaling components in pancreas development.
Wnt signaling
Wnts are secreted glycoproteins that bind to Frizzled receptors and LRP5–LRP6 coreceptors in the plasma membrane, thereby recruiting Dishevelled and inhibiting the Axin/APC (Adenomatous Polyposis coli)/GSK-3β (glycogen synthase kinase 3β)/CK1α (casein kinase 1α) complex, leading to stabilization of β-catenin. Upon nuclear translocation, β-catenin interacts with transcription factors such as TCF/LEF to activate gene transcription. In the absence of active Wnt signaling, β-catenin is targeted for proteosomal degradation by CK1α- and GSK-3β-mediated phosphorylation and subsequent ubiquitination. In addition to its role in the canonical Wnt signaling pathway, β-catenin also binds E-cadherin in adherens junctions (for a comprehensive review of this pathway, see Klaus and Birchmeier 2008). Expression of multiple Wnt ligands (1, 2b, 5a,7b, 11, and 16), Frizzled receptors (2, 3, 4, 5, 6, 7, 8, and 9), Wnt coreceptors LRP5 and LRP6, and Wnt inhibitors Fsrp (1–4) has been detected in the developing pancreas, in the mesenchyme, and in the epithelium (Heller et al. 2002).
Multiple genetic mouse models have been derived to elucidate the role of Wnt/β-catenin signaling in vivo, including transgenic mouse models of Wnt or Wnt inhibitor Frz8CRD-igG overexpression, as well as Cre-Lox-mediated deletion of APC and stabilization or deletion of β-catenin using various Pdx1-Cre lines (Heller et al. 2002; Dessimoz et al. 2005; Murtaugh et al. 2005; Papadopoulou and Edlund 2005; Heiser et al. 2006; Strom et al. 2007; Wells et al. 2007). Taken as a whole, these studies suggest that β-catenin mediates overall pancreas growth, by regulating both early proliferation of pancreatic epithelial progenitor cells as well as acinar cell differentiation and proliferation.
Recently, single-nucleotide polymorphisms in the TCF7L2 locus have been linked to type 2 diabetes in humans and associated with decreased insulin secretion, implying an important role for Wnt/β-catenin signaling in β cells (Cauchi et al. 2007; Salonen et al. 2007; Sladek et al. 2007; Loder et al. 2008). In mice, Wnt3a treatment of cultured β cells or islets stimulates proliferation, and in vivo expression of active β-catenin in β cells postnatally leads to β-cell expansion (Rulifson et al. 2007). Genetic studies manipulating this pathway through conditional ablation or overexpression of a stabilized form of β-catenin, however, have given conflicting phenotypes with regard to the developing endocrine compartment. In agreement with the adult role of Wnt signaling, expression of the Wnt inhibitor Axin in the Pdx1 domain during development decreases β-cell mass (Rulifson et al. 2007). Expression of stabilized β-catenin also increases pancreatic proliferation (Heiser et al. 2006; Rulifson et al. 2007). By contrast, slightly earlier expression of stabilized β-catenin in the epithelium, driven by a different Pdx1-Cre recombinase strain, modulates mesenchymal Fgf10 and Hh signaling, reducing Pdx1 expression in pancreatic progenitors and decreasing the number of insulin+ cells (Heiser et al. 2006). Similarly, conditional ablation of β-catenin in the Pdx1 domain gave divergent results in three studies, two of which showed no effect on the developing endocrine compartment while the third showed a decrease in endocrine islet cell number (Dessimoz et al. 2005; Murtaugh et al. 2005; Wells et al. 2007). While these discrepancies may be due in part to methodological differences in the quantitative assessment of the endocrine compartment, the precise timing of perturbation of the β-catenin signal also has a profound impact on the phenotypic result, likely reflecting highly dynamic, temporally, and spatially distinct roles for β-catenin in the developing endocrine compartment.
TGF-β signaling
The secreted growth factors in the TGFβ superfamily include TGFβ, Inhibin, Activin, BMP, and growth differentiation factors (GDF). Expression of the TGFβ signaling pathway components has been detected in the pancreatic epithelium and mesenchyme and in mature and developing islets. These include the ligand subunits of TGF-β1-3, activinβB, inhibin-α, BMP2, BMP4, BMP5, BMP6, BMP7, GDF11, ligand antagonists such as follistatin, the receptors TβR-II, ActR-IA, ActR-IB, ActR-IIA, ActR-IIB, BMPR-IA, BMPR-IB, BMPR-II, and intracellular mediators such as Smad2 (Ogawa et al. 1993; Feijen et al. 1994; Furukawa et al. 1995; Verschueren et al. 1995; Crisera et al. 1999; Tremblay et al. 2000; Dichmann et al. 2003).
Activin receptors (ActRII-A and ActRII-B), which may be bound by ligands in multiple TGFβ subclasses, are expressed early in pancreatic epithelium and later in islets (Yamaoka et al. 1998; Shiozaki et al. 1999; Kim et al. 2000; Goto et al. 2007). Mice harboring null or dominant-negative mutations in these receptors as well as the downstream mediator Smad2, display decreased endocrine–hormone expression and islet hypoplasia, supporting a role for activin/TGFβ signaling in islet development and differentiation. Mice overexpressing Smad7, a potent inhibitor of TFGβ superfamily signaling, in the Pdx1 domain have β-cell hypoplasia (Smart et al. 2006). Together, these studies underscore the role of TGFβ family signaling in endocrine development, but whether signaling influences formation, survival, or expansion of endocrine progenitors or of differentiated endocrine cells has not been established.
In addition to its previously mentioned role in notochord-mediated specification of the early pancreatic bud and in regulating early branching morphogenesis, activin signaling has other important functions in lineage specification (Ritvos et al. 1995). For example, the ligand Activin B is detected at E12.5 in glucagon+ cells and by E18.5 it is localized to islets (Maldonado et al. 2000). The inhibitor follistatin is present in the mesenchyme prior to E12.5 and reappears in islets at E18.5. Cultured explant data indicate that follistatin mimics mesenchyme by inhibiting endocrine differentiation and promoting exocrine development (Miralles et al. 1998). Consistent with this, adult mice with a targeted disruption of Follistatin like 3 (FSTL3) exhibit increased islet size (Mukherjee et al. 2007). Thus, activin may be permissive for endocrine differentiation, such that inhibition by follistatin may inhibit endocrine differentiation and redirect it toward an exocrine lineage.
Another ligand member of the TGFβ superfamily, GDF11, regulates development of the exocrine and endocrine compartments (Harmon et al. 2004; Dichmann et al. 2006). GDF11−/− mice display increased numbers of Ngn3+ endocrine precursors, suggesting an inhibitory role for GDF11 in endocrine development (Harmon et al. 2004). The increase in islet precursor number does not result in a corresponding increase in mature islet cells, implying a distinct role for GDF11 in terminal islet cell differentiation. GDF11 can bind type II activin receptors (ActRII-A, ActRII-B), thereby eliciting Smad2 phosphorylation (Oh et al. 2002). Not surprisingly, Smad2+/− mice display similar pancreatic defects to GDF11−/− mice, thereby identifying an additional TGFβ family pathway that influences endocrine development. Additionally, the acinar compartment is reduced in GDF11−/− mice, attributing an additional function to GDF11 in exocrine development, possibly by influencing progenitor cell fate decision between endocrine and exocrine lineages (Harmon et al. 2004; Dichmann et al. 2006).
Other than their role in the early inductive events leading to pancreas specification, the function of BMP ligands within the developing pancreas is unclear. Culture studies suggest that BMPs 4–6 may promote proliferation of pancreatic epithelial cells and endocrine differentiation (Jiang et al. 2002; Jiang and Harrison 2005; Hua et al. 2006). In contrast, mouse embryos expressing BMP6 under the control of the Pdx1 promoter display pancreatic agenesis (Dichmann et al. 2003). The source of the BMPs within the pancreas that could mediate these effects in vivo has not been established. In adult animals, studies of β-cell deletion of BMPR1A as well as overexpression of its ligand BMP4 indicate that this pathway positively regulates insulin expression and processing as well as glucose-stimulated insulin secretion (Goulley et al. 2007).
In summary, the TGFβ family of ligands regulates multiple processes during pancreas development after pancreatic specification, including both exocrine and endocrine development. Elucidation of the mechanisms by which each of the individual ligands regulates these processes will require further investigation of the signaling pathways and genetic targets affected by specific ligand and receptor isoforms.
Transcription factors
Over the past 15 years, targeted gene disruption in mice has provided enormous insight into the role that transcription factors play during embryonic development of the mouse pancreas. In particular, conditional gene ablation has revealed that many transcription factors have early roles in pancreas development as well as later distinct roles in specific lineage formation, differentiation, and function (Table 1). Most of the known pancreatic transcription factors, with few exceptions, are expressed prior to or near the onset of pancreas development. Further, many of these factors regulate one another through positive feedback loops; as a result, earlier models of the transcriptional hierarchy have evolved into less linear schemes (Fig. 2). As development proceeds, networks regulating specific lineages segregate from one another and are stabilized. In addition to suggesting pathways that can be manipulated to differentiate progenitor cells into β cells, understanding these cross-regulatory networks may also provide insights into how fully differentiated cells might transdifferentiate; e.g., how a mature acinar cell might be reprogrammed to become a fully differentiated insulin-producing β cell.
Table 1.
Transcription factors that regulate pancreatic β-cell development and function
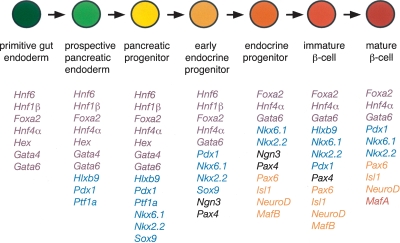
Figure 2.
Transcription factor profile during stages of β cell formation from endodermal derivatives. Diagram indicates transcription factors expressed at each stage of differentiation. Factors initially expressed at a particular stage are color-coded as follows: gut endoderm (purple), pancreatic endoderm progenitor (blue), early endocrine progenitor (black), endocrine progenitor (orange), β cell (red).
This review will emphasize transcription factors with recently discovered or newly expanded roles in pancreatic progenitors (Ptf1, Pdx1, Sox9), islet progenitors (HNF1β, Ngn3, and Pax4), lineage specification (Nkx2.2, Nkx6.1) and β-cell differentiation (Maf A/B, HNF4α, Foxa2). Studies elucidating the regulation of these factors by signaling pathways will be highlighted. Due to space constraints and the growing list of transcription factors expressed during pancreas development, this review is necessarily incomplete; however, a concise summary of the transcription factors with roles in pancreas development and human disease is provided in Table 1. For more comprehensive reviews of the entire hierarchy, of the distinct aspects of the hierarchy governing dorsal versus ventral pancreas development and of the embryonic pancreas development of other model organisms, the reader is referred to several excellent recent reviews (Jensen 2004; Servitja and Ferrer 2004; Jorgensen et al. 2007).
Pancreatic progenitors
Ptf1
Ptf1 is a heterooligomeric transcription factor composed of bHLH p48, p64, and p75 subunits that together control exocrine pancreas gene expression, including elastase 1(Ela1) (Cockell et al. 1989; Roux et al. 1989; Sommer et al. 1991; Krapp et al. 1996; Liu et al. 2001). Ptf1a (p48) is detected throughout the early pancreatic epithelium and later becomes restricted to acinar cells. The complete absence of exocrine cells in Ptf1a-null mice led to the initial interpretation that Ptf1a was exclusively required for the formation of the exocrine pancreas (Krapp et al. 1998). This concept was revised after elegant lineage tracing experiments revealed that Ptf1a is expressed in the pancreatic progenitors that give rise to endocrine, ductal, as well as acinar lineages (Kawaguchi et al. 2002). In the absence of Ptf1a, these progenitors adopt an intestinal fate. Thus, Ptf1a is important for both pancreatic specification and the development of all pancreatic lineages. More recent studies in Ptf1a-null mice indicate that Ptf1a is not absolutely required for formation of Ngn3+ endocrine progenitor cells or endocrine β cells, which are present in the remaining dorsal pancreatic rudiment (Burlison et al. 2008).
A substantial emerging literature indicates significant interactions between Ptf1 and the Notch signaling pathway. Notch signaling components modulate Ptf1 activity and therefore exocrine differentiation, in Notch signaling-dependent and -independent manners. Hes1, Hey1, and Hey2 directly interact with p48/Ptf1a, inhibiting its activity (Esni et al. 2004; Ghosh and Leach 2006).The Notch mediator RBP-J can bind Ptf1a in the Ptf1 complex and enhance Ptf1 transactivation of the Area III enhancer of the Pdx1 promoter, possibly a Notch signaling-independent effect (Obata et al. 2001; Beres et al. 2006; Miyatsuka et al. 2007; Wiebe et al. 2007). Chromatin immunoprecipitation (ChIP) experiments demonstrate in vivo binding of Ptf1 to this Pdx1 promoter region at E11.5. Consistent with this observation, deletion of RBP-J in the Ptf1 domain starting at E10.5 leads to decreased Pdx1 expression, epithelial growth and acinar cell differentiation (Fujikura et al. 2007). Masui et al. (2007) suggest that the RBP-J/Ptf1a complex during the secondary transition activates transcription of its paralog RBP-JL, which can replace RBP-J in this complex and mediate activation of acinar digestive enzymes. Taken together, these studies suggest that Ptf1 is important in the specification of early pancreatic progenitors, in the regulation of Pdx1 expression and in acinar cell differentiation and that these functions occur in part through interaction with RBP proteins and Hes family members.
Pdx1
Pdx1 is a homedomain transcription factor whose gene is mutated in early onset monogenic human diabetes (MODY4) (Stoffers et al. 1997a). Pdx1 is expressed as early as E8.5 in the dorsal and ventral endoderm regions that give rise to pancreatic buds, as well as in the common bile duct, distal stomach, Brunner's glands, and duodenal epithelium (Ohlsson et al. 1993; Guz et al. 1995; Stoffers et al. 1999). Pancreatic expression continues in the pancreatic epithelium and high level expression progressively becomes restricted to pancreatic β cells and a small subpopulation of δ and PP cells (Miller et al. 1994; Guz et al. 1995). Low levels are detected in the nuclei of acinar cells.
Lineage tracing studies reveal that Pdx1 expressing cells, like those expressing Ptf1a, are pancreatic progenitor cells, since all pancreatic lineages develop from these cells (Herrera 2000; Gu et al. 2002). Pdx1-null mice display pancreatic agenesis, as well as an abnormal duodenal epithelium and decreased numbers of enteroendocrine cells (Jonsson et al. 1994; Offield et al. 1996). Similarly, human congenital pancreatic agenesis is caused by homozygous deletion of IPF1, the human ortholog of Pdx1 (Stoffers et al. 1997b). Close examination of the pancreatic endoderm in Pdx1−/− mice reveals the presence of small rudimentary buds with some glucagon+ cells, indicating that Pdx1 is not essential for initial pancreatic induction but is required for pancreatic growth and development of the full spectrum of pancreatic lineages. In pancreatic explants, growth of Pdx1−/− epithelium does not occur in the presence of wild-type mesenchyme, indicating that Pdx1 imparts competence to the epithelium to respond to mesenchymally derived growth signals (Ahlgren et al. 1996). Furthermore, Pdx1-null mice do not contain Ngn3+ pancreatic endocrine progenitors after E9.5 (Burlison et al. 2008). The specific requirement for Pdx1 in the formation, survival, or proliferation of endocrine progenitor cells remains to be determined.
A variety of genetic models that permit spatially and temporally regulated expression of Pdx1 reveal additional roles for Pdx1 in endocrine and exocrine development. In vivo transgenic reporter mice reveal specific conserved regions in the Pdx1 promoter that confer endocrine and β-cell-specific expression (Gannon et al. 2001). Areas I and II impart endocrine expression, while Area III confers β-cell specificity. Mice homozygous for deletion of the Area I, II, and III containing enhancer region recapitulate the pancreatic phenotype of Pdx1-null mice, including the absence of a ventral bud, a hypoplastic dorsal bud, and impaired endocrine differentiation (Fujitani et al. 2006). In vivo Tet-off-mediated down-regulation of Pdx1 throughout embryonic development also recapitulates the Pdx1-null phenotye (Holland et al. 2002). In contrast, Pdx1 down-regulation starting at E12.5–E13.5 leads to a selective defect in acinar cell differentiation and markedly reduced levels of Ptf1a, indicating a later role for Pdx1 in the exocrine pancreas (Hale et al. 2005). Cre-lox-mediated deletion of Pdx1 in the β cell in late development leads to a defect in β-cell proliferation (Gannon et al. 2008). All together, these studies indicate that Pdx1 plays crucial roles in pancreatic growth and in the differentiation of both exocrine and endocrine lineages.
Postnatally, high level Pdx1 expression becomes restricted to islet β cells and β-cell growth and function become exquisitely sensitive to Pdx1 protein level. Pdx1 heterozygosity, cre-lox-mediated deletion of Pdx1 in the β cell, as well as Tet-off-mediated down-regulation of Pdx1 in adult mice all lead to overt hyperglycemia, highlighting the importance of Pdx1 in mature β-cell differentiation (Ahlgren et al. 1998; Brissova et al. 2002; Holland et al. 2005). In addition to its role in glucose-stimulated insulin secretion, Pdx1 is also important in β-cell survival and in the functional and morphological compensatory response in genetic models of insulin resistance (Brissova et al. 2002, 2005; Johnson et al. 2003; Kulkarni et al. 2004).
The specificity whereby Pdx1 regulates gene transcription is dictated in part by its interaction with other transcription factors and cofactors. One set of partners are the atypical TALE (three-amino-acid loop extension) homeodomain containing factors, including Pbx and Meis. In a heterotrimeric complex with Pdx1 and MEIS, Pbx regulates expression of Elastase I in acinar cells and CK19 in ductal cells (Swift et al. 1998; Liu et al. 2001; Deramaudt et al. 2006). In partnership with Prep1 and Pdx1, Pbx regulates expression of somatostatin and with Prep1 alone Pbx silences glucagon expression in non-α cells (Goudet et al. 1999; Herzig et al. 2000). Heterodimer formation with Pdx1 requires the FPWMK pentapeptide motif conserved in Pdx 1. To determine the role of Pdx1–Pbx complexes in vivo, Dutta et al. (2001) performed rescue experiments of null Pdx1 embryos with a wild-type Pdx1 transgene or a Pbx interaction motif-deficient Pdx1 mutant. As compared with wild-type, animals with the mutant transgene have a hypoplastic pancreas with small disorganized islets and hormone+ ductal cells. Decreased proliferation of all cell lineages is observed at birth, suggesting that Pdx1–Pbx complexes are required for pancreatic growth, cell migration, and expansion of all lineages during development. The development of diabetes in transheterozygous Pdx1+/− Pbx1+/− mice provides genetic evidence for the importance of this interaction in β cells (Kim et al. 2002). Remarkably, no β-cell transcriptional targets of the Pdx1–Pbx heterodimer have been reported.
Sox
Twelve members of the Sox gene family of high-mobility group (HMG) transcription factors are expressed in the pancreas during development (Lioubinski et al. 2003). Sox4 transcript is detectable in the pancreas by E10.5 and peaks at E12.5 (Wilson et al. 2005). By E18.5, Sox4 is present in endocrine islets and subset of acinar cells (Lioubinski et al. 2003). Sox4−/− embryos show grossly normal morphology at E12.5 (Wilson et al. 2005). Pancreatic explants from these mice develop fewer endocrine cells, indicating a specific role for Sox4 in islet development. More recently, Sox4 has been implicated in the regulation of insulin secretion in adult β cells (Goldsworthy et al. 2008).
An exciting recent discovery is the critical role of Sox9 in the maintenance of the pancreatic progenitor pool. Sox9 is first detected in the pancreatic epithelium in Pdx1+ progenitors at E9.0–E9.5, and is later coexpressed with Hes1 cells (Seymour et al. 2007). Consistent with the idea that Sox9 expression marks progenitor cells, the numbers of Sox9+ cells are unchanged in Ngn3 or Nkx6.1-null animals, whereas in transgenic Pdx1 promoter–Fgf10 embryos, where ectopic expression of Fgf10 maintains Pdx1+ cells in their progenitor state, Sox9 expression is abnormally maintained at E18.5. Deletion of Sox9 in the Pdx1 domain results in increased apoptosis and decreased proliferation of Pdx1+ progenitors. An increase in glucagon+ and Isl1+ cells indicates premature differentiation, and decreased percentages of Pdx1+ cells that coexpress Hes1 in these mice suggests that Sox9 is required to maintain Notch signaling in the progenitor pool.
Sox9 also participates in a cross-regulatory network during early pancreas development. Sox9 is coexpressed with Ngn3 at E14.5, and in vitro and cell based reporter studies suggest that Sox9 positively regulates Ngn3 (Lynn et al. 2007). To determine whether Sox9 regulates Ngn3 via HNF1β, HNF6, and Foxa2, factors that have been implicated in Ngn3 regulation (Fig. 3), EMSA, ChIP, and siRNA-mediated gene silencing studies were performed, which indicated that Sox9 directly regulates expression of these factors (Lynn et al. 2007). Conversely, knockdown of Foxa2 also decreases Sox9 levels in ductal cells, suggesting a cross-regulatory relationship between these factors. Moreover, Sox9 binds a site in its own promoter, suggesting autoregulation. The relevance of these coregulatory relationships during the transition from Sox9-mediated maintenance of a progenitor pool to endocrine differentiation warrant further exploration.
Figure 3.
Regulation of endocrine progenitor marker Ngn3. Schematic depicts the factors and pathways within the epithelium and the mesenchyme that regulate Ngn3 expression. Large arrows indicate regulation that has been demonstrated by in vivo mouse models, while small arrows denote regulation supported by cell culture and in vitro evidence.
Islet progenitors
HNF1β
HNF1β plays early roles in visceral endoderm differentiation and overall pancreas development, as well as a specific role in the precursor population for Ngn3-expressing islet progenitor cells. HNF1β is expressed in the primitive endoderm and later in the epithelium of both pancreatic buds, eventually becoming relatively restricted to ductal cells by E14, although expression in endocrine cells continues at very low but detectable levels (Barbacci et al. 1999; Coffinier et al. 1999a, b; Maestro et al. 2003). In HNF1β-null embryoid bodies, activation of HNF4α, HNF1α, and Foxa3 is reduced, suggesting direct or indirect regulatory relationships among these factors. Further studies have confirmed that HNF1β likely directly activates Foxa3 (Hiemisch et al. 1997). Rescue of the visceral endoderm defect using tetraploid aggregation chimeric embryos reveals that HNF1β is also required for pancreatic organogenesis (Haumaitre et al. 2005). Mice lacking HNF1β have a small pancreatic dorsal bud rudiment at E9.5 that expresses Pdx1 and Hb9 and reduced HNF6, but no detectable Ptf1a and Ngn3. Altogether these data suggest that HNF1β may be important in directly or indirectly activating pancreatic (Ptf1a, HNF6) and endocrine (Ngn3) gene expression. HNF1β activates HNF6 in the embryo (Poll et al. 2006). In turn, HNF6−/− animals have reduced HNF1β, suggesting that HNF6 and HNF1β regulate one another during early pancreas development; this could be an underlying mechanism that contributes to diabetes development in humans with MODY5, which results from mutations in the HNF1β locus (Horikawa et al. 1997; Maestro et al. 2003). Moreover, morphologic studies imply that HNF6+/HNF1β+ cells may be precursors to the Ngn3+ endocrine progenitor population (Maestro et al. 2003).
Ngn3
The bHLH transcription factor Ngn3 is first detected in the embryonic pancreatic epithelium at E9.5, peaks by E15.5, and declines after birth, such that little if any expression is present in the adult (Gradwohl et al. 2000; Gu et al. 2003). Unlike humans where presumptive null mutations result in endocrine cell loss in the gut but not in the pancreas, Ngn3-null mice are completely devoid of both intestinal and pancreatic endocrine lineages and pancreatic acinar morphogenesis is abnormal (Gradwohl et al. 2000; Jenny et al. 2002; Lee et al. 2002a; Schonhoff et al. 2004; Bjerknes and Cheng 2006; Wang et al. 2006; Jensen et al. 2007). Conversely, overexpression of Ngn3 in the Pdx1 domain leads to premature endocrine differentiation toward the glucagon lineage at the expense of the exocrine lineage (Apelqvist et al. 1999). These studies, together with lineage tracing and morphological studies, indicate that Ngn3+ cells are endocrine progenitors (Jensen et al. 2000a; Schwitzgebel et al. 2000; Gu et al. 2002).
The mechanisms by which Ngn3+ cells give rise to the different endocrine lineages are poorly understood. However, a recent study using inducible reexpression of Ngn3 in Ngn3-null pancreas at various times throughout development suggests that the context of different developmental time windows dictates lineage allocation. Early Ngn3 induction (E8.7) produces glucagon+ cells, whereas later induction additionally leads to insulin+ and PP+ cells, and even later induction (E14.5) contributes somatostatin+ cells while decreasing glucagon+ cells (Johansson et al. 2007). The specific signals that influence competence of Ngn3+ progenitors to generate specific endocrine lineages at each developmental stage remain to be elucidated.
Various Ngn3 targets have been identified by in vitro, cell-based, and in vivo Xenopus studies, including NeuroD/BETA2, IA-1, Pax4, Nxk2.2, and Ngn3 itself (Huang et al. 2000; Smith et al. 2003; Watada et al. 2003; Smith et al. 2004; Mellitzer et al. 2006). More recently, microarrays using E13 and E15 pancreata of wild-type and Ngn3-null mice were performed and a new list of potential Ngn3 targets was generated (Petri et al. 2006). Furthermore, gene expression profiles of isolated Ngn3+ cells and their descendants from E13.5–E17.5 embryos will aid in identifying developmentally determined gene regulatory relationships of high potential importance to in vitro differentiation of mature β cells (Gu et al. 2004; White et al. 2008).
Insulinoma-associated antigen 1 (IA-1), a zinc finger transcription factor that may function as a transcriptional repressor, has been identified as a direct target of Ngn3 (Goto et al. 1992; Breslin et al. 2002). IA-1-null mice have reduced numbers of endocrine cells and they accumulate endocrine precursors without hormone expression (Gierl et al. 2006; Mellitzer et al. 2006).
The zinc finger transcription factor Myt1, which encodes for two isoforms, is expressed in Ngn3 expressing endocrine progenitors cells during development and appears to be downstream from Nkx6.1 (Henseleit et al. 2005). In chick, ectopic expression of the Notch inhibitor, Manic Fringe, promotes expression of Ngn3 and cMyt1/3 as well as endocrine differentiation (Xu et al. 2006). Expression of a dominant-negative form of Myt1 in the Ngn3 domain leads to a reduction in the number of insulin+ and glucagon+ cells (Gu et al. 2004). Surprisingly, pancreas-specific and global Myt1-null mice have a mild endocrine defect consisting of the presence of multihormone expressing cells (Wang et al. 2007). Compensation by Myt1 paralogs may be masking a critical role for this gene family; targeted disruption of the other paralogs, individually and in combination, may be required to fully appreciate the importance of this gene family in endocrine differentiation.
Pax4
The paired domain transcription factor Pax4 is expressed by E9.5 in the pancreas, becomes restricted to insulin+ cells, and is absent in the adult (Sosa-Pineda et al. 1997; Wang et al. 2004). Homozygous disruption of Pax4 leads to a marked reduction in the number of β and δ cells, along with increased numbers of α cells. Interestingly, these mice also contain increased number of ghrelin+ ε cells, as does the Nkx2.2 knockout, suggesting a genetic interaction between these two factors (Prado et al. 2004). Lineage tracing reveals that Pax4+ cells contribute to all endocrine lineages, indicating that, similar to Ngn3, Pax4 may mark endocrine precursors (Greenwood et al. 2007; Wang et al. 2008). Interestingly, expression of the Notch1-ICD in this population switches the fate of Pax4-expressing cells to ductal cells, consistent with Notch signaling preventing commitment to an endocrine fate (Greenwood et al. 2007).
Lineage specification and β-cell differentiation
Nkx2.2
Nkx2.2 is an NK type of homeodomain transcription factor whose expression is first detected at E9.5 in the pancreatic epithelium, and by E15.5 becomes restricted to endocrine cells, specifically α, β, and PP cells (Sussel et al. 1998). Nkx2.2-null mice lack β cells and are hyperglycemic at birth. Decreased numbers of α and PP cells are observed as well. Interestingly, a group of PC1/3-expressing cells that are negative for other hormones is expanded within the islets. These cells were later identified as ghrelin+ ε endocrine cells, a fifth endocrine cell type that derives from Ngn3+ progenitor cells and normally disappears postnatally (Prado et al. 2004). Whether these cells represent an alternative lineage to β cells derived from a common intermediate progenitor or are themselves β-cell precursors remains to be determined.
Nkx2.2 may function as both a transcriptional activator and transcriptional repressor (Doyle et al. 2007; Doyle and Sussel 2007). The phenotypes of transgenic mice expressing fusion proteins containing the Nkx2.2 homeodomain (HD) fused to either the VP16 activation domain or the Engrailed repressor domain (Engr) suggest that Nkx2.2 functions in part as a repressor for α- and β-cell formation during development and as an activator for β-cell maturation and postnatal function. The Nkx2.2HD-Engr transgene completely rescues the reduction in α-cells and partially rescues β-cell number in Nkx2.2-null embryos, while Nkx2.2HD-VP16 has no effect, implying at least in part a repressive role for Nkx2.2 in development. However, expression of β-cell maturation factors MafA and Glut2 is not recovered. Postnatally, the presence of the Nkx2.2HD-Engr transgene on a wild-type background decreases endogenous Nkx2.2 levels as well as expression of MafA, Glut-2, and insulin, suggesting a requirement for Nkx2.2 activator function in this context. This example demonstrates the markedly different roles that transcription factors may play at different times during development and highlights the need to confirm transcriptional targets in vivo at the appropriate time point in order to elucidate relevant regulatory relationships.
Nkx6.1
Nkx6.1 is another NK-type homeodomain transcription factor that is first detected in the pancreatic epithelium at E10.5. As development progresses, Nkx6.1 is expressed in Ngn3+ endocrine precursors and becomes restricted to the pancreatic β cell, where it is maintained postnatally (Rudnick et al. 1994; Sander et al. 2000). Nkx6.1−/− embryos have normal pancreatic size at E18.5, with reduced islet size due to a marked reduction of the β-cell lineage. The early population of insulin+ cells at E12.5 is unaffected, suggesting that Nkx6.1 is only necessary for the second wave of β-cell differentiation starting at the secondary transition. A role for Nkx6.1 in α-cell development has also been suggested (Henseleit et al. 2005; Nelson et al. 2007).
Similar to Nkx2.2, Nkx6.1 may function as a transcriptional repressor (Mirmira et al. 2000). Cell line-based evidence suggests that Nkx6.1 represses glucagon expression and regulates insulin secretion in the mature β cell (Schisler et al. 2005; Gauthier et al. 2007). Nkx6.1 also appears to function as a transcriptional activator by positively regulating its own transcription (Iype et al. 2004). Moreover, there is some evidence suggesting postranscriptional regulation of Nkx6.1 (Watada et al. 2000).
A highly related gene, Nkx6.2, is also expressed in the pancreatic epithelium beginning at E10.5 (Nelson et al. 2005). Transgenic expression of Nkx6.2 rescues β-cell differentiation in an Nkx6.1-null as effectively as an Nkx6.1 transgene, suggesting a high degree of functional redundancy between these family members (Nelson et al. 2007). Although Nkx6.2-null mice have no obvious pancreatic phenotype, double Nkx6.1/Nkx6.2-null mice have a further reduction in the number of insulin+ cells compared with Nkx6.1-null animals, and an additional reduction in glucagon+ cells, which is not observed in either Nkx6.1- or Nkx6.2-null mice (Henseleit et al. 2005; Alanentalo et al. 2006). This result suggests an additional role for Nkx6 factors in α cell development.
Mafs
The basic leucine zipper proteins of the large Maf family, MafA, MafB, and c-Maf, are expressed in pancreatic islets. MafA was independently identified by three groups as the β-cell-specific RIPE3b1 factor that binds the well characterized RIPE3b1 element of the insulin promoter (Kataoka et al. 2002; Olbrot et al. 2002; Matsuoka et al. 2003). MafA directly interacts with Pdx1 and BETA2 to synergistically activate insulin gene transcription (Aramata et al. 2005; Zhao et al. 2005). Interestingly, all three Mafs are capable of activating the insulin gene, although direct regulation of glucagon by MafB and c-Maf has been observed as well (Matsuoka et al. 2003; Kataoka et al. 2004; Artner et al. 2006).
Onset of MafA expression occurs at E13.5 in the first insulin producing cells and remains in this population until adulthood (Matsuoka et al. 2004). Expression is markedly reduced in Nkx6.1−/− mice positioning MafA downstream from Nkx6.1. Surprisingly, MafA-null mice do not exhibit defects in pancreatic or β-cell development but become glucose intolerant postnatally, revealing a role for MafA in glucose-stimulated insulin secretion (Zhang et al. 2005).
In contrast, MafB is expressed in both β and α cells starting at E12.5 and becomes restricted to α cells postnatally (Artner et al. 2006). MafB-null mice have reduced numbers of both insulin+ and glucagon+ cells during development, with a delay in the emergence of insulin+ cells until after the onset of MafA expression at E13.5, suggesting a role for MafB in both α- and β-cell differentiation and/or maturation (Artner et al. 2007). Decreased expression of Pdx1, Nkx6.1, and Glut-2 is observed in insulin+ cells after E15.5, suggesting al regulatory role for MafB in β-cell differentiation. Consistent with this, MafB occupies the MafA, Glut2, and Nkx6.1 promoters in vivo. Developmental studies suggest that insulin+ cells switch from a MafB+ to a MafA+ state during development, and that this process is associated with up-regulation of Pdx1 expression (Nishimura et al. 2006). This idea is consistent with the recent identification of MafA as a direct transcriptional target of Pdx1 (Raum et al. 2006). Another study suggests that Pdx1 is itself a target of MafA (Samaras et al. 2003), emphasizing the cross-regulatory nature of the maturing β-cell transcription factor network.
Forkheads
The Forkhead family of transcription factors includes Foxa1 (HNF3α), Foxa2 (HNF3β), and Foxa3 (HNF3γ). Foxa2 is the first of these to be expressed in the embryo at E6.5 in the anterior primitive streak and node and becomes expressed in the foregut endoderm, notochord, and floor plate, prior to pancreas formation (Ang et al. 1993; Kaestner et al. 1993, 1994; Monaghan et al. 1993; Sasaki and Hogan 1993, 1994). Foxa2-null mice die at E11 from multiple defects (Ang and Rossant 1994; Weinstein et al. 1994); thus, the role of Foxa2 in pancreas development was determined by conditional gene ablation. Interestingly, pancreatic specification and growth are unaltered in Foxa3-Cre;Foxa2 loxP/loxP mice, but α-cell differentiation is impaired, resulting in a reduced number of α cells and reduced glucagon gene expression (Lee et al. 2005b). In the mature β cell, Foxa2 regulates insulin secretion (Sund et al. 2001; Lantz et al. 2004; Gao et al. 2007). Mice with Foxa2 deletion in β cells using RIP-Cre transgene display increased insulin secretion in response to amino acids consistent with the observed reduction the Foxa2 targets Sur1, Kir6.2, and Hadhsc, which are mutated in various forms of human congenital hyperinsulinism (Sund et al. 2001; Lantz et al. 2004). More recently, derivation of adult Tam-inducible Pdx1-Cre ERT2; Foxa2 loxP/loxP mice reveals additional novel roles for Foxa2 in β-cell calcium responses and in the formation and docking of insulin containing secretory granules (Gao et al. 2007).
In vitro and cell line studies identify Pdx1 and Ngn3 as direct Foxa2 targets (Wu et al. 1997; Gerrish et al. 2000; Marshak et al. 2000; Lee et al. 2001). Interestingly, dysregulation of Ngn3 has not been observed in the absence of Foxa2 in vivo, and dysregulation of Pdx1 by Foxa2 deletion has only been observed in the differentiated β cell and not during early pancreas development, suggesting that this regulatory relationship depends on developmental context (Lee et al. 2002b, b; Gao et al. 2007).
Foxa1 is first detected at E7.0 in midline endoderm cells and Foxa3 expression begins at E8.5 in the midgut to hindgut region (Ang et al. 1993; Monaghan et al. 1993). Both Foxa1- and Foxa3-null mice appear to have normal pancreas organogenesis (Kaestner et al. 1998, 1999; Shih et al. 1999). Foxa1−/− mice have impaired insulin secretion and decreased proglucagon expression, revealing glucagon as a novel target of Foxa1 (Kaestner et al. 1999; Shih et al. 1999; Vatamaniuk et al. 2006).
Surprisingly, despite the early expression and important role for forkhead factors in early endoderm, Foxa1, Foxa2 and Foxa3 are not individually essential for overall pancreas development. Similarly, these factors are not individually required for liver development, but simultaneous deletion of Foxa1 and Foxa2 in the endoderm leads to absence of hepatic induction, indicating redundancy of these factors during liver development (Lee et al. 2005a) and leaving open the possibility of a critical early role for these factors in pancreas development as well.
HNF4α
HNF4α is nuclear hormone receptor that is mutated in MODY1 (Yamagata et al. 1996). It is expressed in the primitive endoderm at E4.5, in the gut endoderm at E8.5, the pancreatic epithelium at E9.5, and postnatally in both endocrine and exocrine cells (Duncan et al. 1994; Nammo et al. 2008). Isoforms result from both alternative splicing and expression driven by alternative promoters, P1 and P2. HNF4α-null mice are embryonic lethal by E6.5 (Chen et al. 1994), requiring conditional gene ablation to define the role of this factor during β-cell development. β-cell-specific deletion of HNF4α in RIP-Cre; HNF4α loxP/loxP adult mice leads to impaired glucose tolerance, glucose-stimulated insulin secretion, and calcium responses (Gupta et al. 2005; Miura et al. 2006). Moreover, β-cell mass expansion during pregnancy is impaired in these animals due to reduced HNF4α dependent-transcription of ST5 (suppression of tumorigenicity 5), a novel regulator of Ras/ERK signaling (Gupta et al. 2007). This animal model has proven useful in the identification of multiple HNF4α targets, including the K-channel subunit Kir6.2 (Gupta et al. 2005). Interestingly, no change in HNF1α mRNA is observed, in contradiction to the previously proposed model of HNF4α-mediated regulation of HNF1α. Other studies using islet ChIP/promoter arrays, expression of dominant-negative or wild-type HNF4α in cell lines or ES cell-derived visceral endoderm have identified a multitude of putative transcriptional targets, although the in vivo regulation of many of these are still to be determined (Stoffel and Duncan 1997; Wang et al. 2000; Bartoov-Shifman et al. 2002; Odom et al. 2004; Thomas et al. 2004; Gupta et al. 2005).
Recently, the regulatory relationships among HNF4α and other factors including HNF1α, HNF1β, Foxa2, and HNF6 were examined throughout liver development (Kyrmizi et al. 2006). ChIP assays for HNF1α, HNF1β, Foxa2, HNF4α, HNF6, LRH, C/EBPα, COUP-TFII, GATA6, and RNA pol-II, were performed using liver tissue from E14.5, E18.5, P2 (postnatal day 2), and P45. The results revealed a dynamic recruitment of transcription factors to individual promoters that varies depending on the developmental time point. For instance, occupancy of the HNF1α promoter by HNF1β is only detected at E18.5. Moreover, six of these factors appear to regulate one another and downsteam targets as part of a cross-regulatory network. Furthermore, hepatocyte-specific deletion of HNF4α in adult liver had little effect on expression of these other hepatic factors, while deletion in the embryo had a marked effect on expression HNF1α, HNF1β, Foxa2, and HNF6, among others, indicating a developmental stage-specific requirement of these factors for cross-regulation. This study and others highlight the importance of identifying promoter occupancy and regulatory relationships in the appropriate developmental context.
Directed differentiation of ES cells
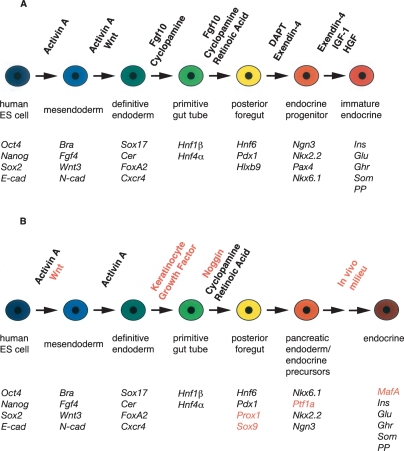
These advances in our understanding of the signaling pathways and transcription factors that regulate the embryonic development of the pancreas have informed efforts to derive glucose-responsive β-like cells from human and mouse ES cells (Lumelsky et al. 2001; Hori et al. 2002; Kim et al. 2003; Blyszczuk et al. 2004; Vaca et al. 2008). For a detailed review of directed differentiation of ES cells to pancreatic endocrine cells, see Spence and Wells (2007). The use of the Nodal mimic Activin A to generate an endodermal intermediate has significantly enhanced the efficiency of generating β-like cells in a stepwise fashion by sequential treatment with a variety of factors (Kubo et al. 2004; D'Amour et al. 2005, 2006; J. Jiang et al. 2007; W. Jiang et al. 2007; Phillips et al. 2007; Shim et al. 2007; Kroon et al. 2008). One notable example was published by D'Amour et al. (2006) (Fig. 4). Using an in vitro differentiation protocol, treatment of human ES cells with Activin A and Wnt followed by Activin A alone resulted in the generation of definitive endoderm, marked by expression of Sox17, FoxA2, the mouse Cerberus homolog, CER, and the chemokine receptor, CXCR4. Then, addition of Fgf10 and the hedgehog signaling inhibitor cyclopamine and the subsequent addition of RA led to the generation of primitive gut tube-like cells marked by HNF1β and HNF4α, followed by posterior foregut-like cells that expressed Pdx1, HNF6, and Hb9. Finally, treatment with the Notch signaling inhibitor DAPT and the glucagon-like peptide 1 (GLP-1) receptor agonist Exendin-4 with the subsequent addition of insulin-like growth factor 1 (IGF-1) and hepatocyte growth factor (HGF), sequentially generated pancreatic/endocrine precursors marked by expression of Nkx6.1, Ngn3, Pax4, Nkx2.2, and finally cells expressing endocrine hormones, including insulin. GLP-1 promotes fetal β-cell maturation in culture, and HGF is a β-cell mitogen (Otonkoski and Hayek 1995; Otonkoski et al. 1996; Garcia-Ocana et al. 2000; Movassat et al. 2002), whereas IGF-1 plays postnatal roles in β-cell differentiation and survival (Ueki et al. 2006). Thus, by attempting to recapitulate the signaling cascades governing embryonic development and β-cell differentiation, cells expressing a transcription factor signature resembling that of β cells were generated from ES cells. Although the importance of this contribution cannot be overstressed, the efficiency of this protocol in generating insulin-expressing cells was still low (7%), and the cells themselves were poorly glucose-responsive, indicating incomplete differentiation, further evident by the absence of MafA expression. These cells may more closely resemble “primary transition” endocrine cells.
Figure 4.
Differentiation of β-like cells from human ES cells. Diagrams depicts protocols used by D'Amour et al. (2006) (A) and Kroon et al. (2008) (B). Exogenous factors added to induce differentiation into particular lineages are indicated above, and transcription factors expressed at each stage are denoted in italics below. Differences between the protocols are indicated in red. Adapted by permission from Macmillan Publishers Ltd.: (Nature Biotechnology, http://www.nature.com/nbt) from D'Amour et al. (2006) (© 2006) (A) and Kroon et al. (2008) (© 2008) (B).
In a more recent study, the same group modified their protocol to generate pancreatic endoderm-like cells from hES cells (Kroon et al. 2008). The resulting cell mixture, which included Pdx1, Foxa2, HNF6, and Nkx6.1 expressing pancreatic endoderm-like cells as well as nonpancreatic endoderm, mesoderm, and ectoderm-like cells, was implanted into epididymal fat pads of immunocompromised SCID-Bg mice. Within a 3-mo period, endocrine cells were generated within the implants in vivo, a proportion of which were insulin/Pdx1/Nkx6.1/MafA expressing β-like cells. These cells were glucose-responsive, as measured by serum human insulin and C-peptide levels that approached the levels achieved by a curative dose of human islets transplanted into a parallel set of recipients. The presence of hES-derived pancreatic endoderm implants prevented development of overt hyperglycemia upon streptozotocin treatment, which selectively targets mouse pancreatic β cells. This study clearly demonstrated the potential to generate glucose-responsive β-like cells from hES cells in vivo. However, the occurrence of a teratoma in one graft could impact the clinical utility of this approach. Since the endogenous β cells of the host were not destroyed until after the implants were engrafted, it is also unclear whether the generation of fully functional β-like cells from pancreatic endoderm-like cells would be impaired under the hyperglycemic conditions that might be present in diabetic human recipients.
Concluding remarks
The last decade has been marked by tremendous progress in the elucidation of the basic transcriptional hierarchy governing β-cell development, the identification of critical signaling pathways that impinge on this hierarchy and the application of this fundamental knowledge toward the development of β-cell replacements to complement the inadequate supply of human islets available for transplantation into patients with diabetes. Parallel progress in human islet transplantation to achieve at least short-term insulin independence raises considerable hope for cell-based therapy for diabetes. Significant challenges, however, remain to be overcome before ES- or other progenitor-derived β-cell replacements can be used in a clinical setting. The efficiency of differentiation into a stable mature β-cell phenotype, whether from ES cells, adult progenitor cells, or even differentiated cells of other lineages, must be considerably improved, and the generation of unwanted cell types (including potentially cancerous cells) suppressed. Complementary progress in addressing immunological issues of rejection and modulation of recipient autoimmunity must also continue.
Improvements in both efficiency and fidelity of guided ES cell differentiation will rely on new advances in our understanding of the extracellular signals governing each lineage choice along the path of β-cell development. One notable gap is the identity of the extracellular signals that promoted ES cell differentiation toward a mature β-cell fate in vivo (Kroon et al. 2008). The inability to achieve the same outcome in the culture dish reflects the paucity of knowledge of the factors that normally regulate β-cell maturation during late embryonic and neonatal development in vivo. Similarly, our knowledge of the bidirectional signaling between pancreatic mesenchyme and epithelium during early pancreas development remains limited. Other emerging areas include the critical role of cell–cell contact in limiting cellular plasticity (Minami et al. 2008) and the ability of severe injury and inflammation, perhaps through the destruction of cell–cell interactions and the production of cytokines, to unleash previously unappreciated regenerative pathways in vivo (Xu et al. 2008).
Knowledge of the transcription signature characteristic of each stage of β-cell development has been critical to the advances thus far, but a refinement of the in vivo transitions between pancreatic progenitor and endocrine progenitor and between endocrine progenitor and mature β cell will further extend the efficiency and fidelity of β-cell development ex vivo. Already a distinct transcription factor profile has begun to emerge for immature β cells and similarly for early endocrine progenitors (Fig. 2). Recent advances in high-throughput technology will greatly assist in this endeavor. A thorough comparative study of the gene expression profiles of cells isolated from definitive and visceral endoderm, pancreatic progenitors including single-cell preparations, Ngn3+ endocrine progenitors at different stages, and islets, has already enabled the determination of a “genetic identity” of different cell types at different stages of development and the identification of novel regulatory elements (Chiang and Melton 2003; Gu et al. 2004; Sherwood et al. 2007; White et al. 2008).
Many of the regulatory relationships that have been studied in the mature β cell have not been directly examined during embryonic development of the pancreas. Further, until recently, the regulation of targets by specific transcription factors has been determined primarily using in vitro and cell culture systems in the form of electrophoretic mobility shift assays (EMSA) and promoter/reporter studies. With the advent of ChIP to examine transcription factor occupancy in vivo, siRNA gene-silencing techniques and more widespread application of conditional gene ablation, the relevance of specific target gene regulation can now be more easily established in vivo.
Combining high-throughput ChIP and promoter arrays (Odom et al. 2004; Keller et al. 2007) and more recently high-throughput sequencing (Johnson et al. 2007), using material from developing pancreas along with gene expression profiles of specific pancreatic cell types from mice with targeted genetic mutations (Petri et al. 2006; Svensson et al. 2007) will advance our knowledge of the regulatory relationships among transcription factors.
Ultimately, the integration of these new advances in the extracellular signals and a refined transcription factor network governing each stage of β-cell development will redefine our understanding of how insulin-producing β cells are derived from primitive endoderm in vivo. There is considerable hope that the application of this knowledge will optimize efforts to generate β cells from precursor or stem cells ex vivo, thereby providing a potentially limitless source of β cells for transplantation into patients with diabetes as well as providing the first tractable cell-based model of islet β-cell development, which will find wide application among basic scientists who endeavor to understand this process.
Acknowledgments
We thank James Wells (University of Cincinnati) for his critical review of this manuscript. The Stoffers' laboratory is supported by grants from the National Institutes of Health (R01 DK068157, R01 DK062965, and P01 DK49210 to D.A.S. and individual NIH predoctoral NRSA 5F31HL071273 to J.O.K.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1670808.
References
- Ahlgren U., Jonsson J., Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes & Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanentalo T., Chatonnet F., Karlen M., Sulniute R., Ericson J., Andersson E., Ahlgren U. Cloning and analysis of Nkx6.3 during CNS and gastrointestinal development. Brain Res. Gene Expr. Patterns. 2006;6:162–170. doi: 10.1016/j.modgep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Ang S.L., Rossant J. HNF-3 β is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Ang S.L., Wierda A., Wong D., Stevens K.A., Cascio S., Rossant J., Zaret K.S. The formation and maintenance of the definitive endoderm lineage in the mouse: Involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Apelqvist A., Ahlgren U., Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr. Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T., Hrabe de Angelis M., Lendahl U., Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Aramata S., Han S.I., Yasuda K., Kataoka K. Synergistic activation of the insulin gene promoter by the β-cell enriched transcription factors MafA, Beta2, and Pdx1. Biochim. Biophys. Acta. 2005;1730:41–46. doi: 10.1016/j.bbaexp.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Arnaud-Dabernat S., Kritzik M., Kayali A.G., Zhang Y.Q., Liu G., Ungles C., Sarvetnick N. FGFR3 is a negative regulator of the expansion of pancreatic epithelial cells. Diabetes. 2007;56:96–106. doi: 10.2337/db05-1073. [DOI] [PubMed] [Google Scholar]
- Artner I., Le Lay J., Hang Y., Elghazi L., Schisler J.C., Henderson E., Sosa-Pineda B., Stein R. MafB: An activator of the glucagon gene expressed in developing islet α- and β-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- Artner I., Blanchi B., Raum J.C., Guo M., Kaneko T., Cordes S., Sieweke M., Stein R. MafB is required for islet β cell maturation. Proc. Natl. Acad. Sci. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam A., Nalini R., Hampe C.S., Maldonado M. Syndromes of ketosis-prone diabetes mellitus. Endocr. Rev. 2008;29:292–302. doi: 10.1210/er.2007-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci E., Reber M., Ott M.O., Breillat C., Huetz F., Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- Bartoov-Shifman R., Hertz R., Wang H., Wollheim C.B., Bar-Tana J., Walker M.D. Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4 α. J. Biol. Chem. 2002;277:25914–25919. doi: 10.1074/jbc.M201582200. [DOI] [PubMed] [Google Scholar]
- Beres T.M., Masui T., Swift G.H., Shi L., Henke R.M., MacDonald R.J. PTF1 is an organ-specific and Notch-independent basic helix–loop–helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan A., Itoh N., Kato S., Thiery J.P., Czernichow P., Bellusci S., Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Bjerknes M., Cheng H. Neurogenin 3 and the enteroendocrine cell lineage in the adult mouse small intestinal epithelium. Dev. Biol. 2006;300:722–735. doi: 10.1016/j.ydbio.2006.07.040. [DOI] [PubMed] [Google Scholar]
- Blyszczuk P., Asbrand C., Rozzo A., Kania G., St-Onge L., Rupnik M., Wobus A.M. Embryonic stem cells differentiate into insulin-producing cells without selection of nestin-expressing cells. Int. J. Dev. Biol. 2004;48:1095–1104. doi: 10.1387/ijdb.041904pb. [DOI] [PubMed] [Google Scholar]
- Breslin M.B., Zhu M., Notkins A.L., Lan M.S. Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: Identification of consensus IA-1 binding sequence. Nucleic Acids Res. 2002;30:1038–1045. doi: 10.1093/nar/30.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissova M., Shiota M., Nicholson W.E., Gannon M., Knobel S.M., Piston D.W., Wright C.V., Powers A.C. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J. Biol. Chem. 2002;277:11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- Brissova M., Blaha M., Spear C., Nicholson W., Radhika A., Shiota M., Charron M.J., Wright C.V., Powers A.C. Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 2005;288:E707–E714. doi: 10.1152/ajpendo.00252.2004. [DOI] [PubMed] [Google Scholar]
- Burlison J.S., Long Q., Fujitani Y., Wright C.V., Magnuson M.A. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev. Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. β-Cell deficit and increased β-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- Cauchi S., El Achhab Y., Choquet H., Dina C., Krempler F., Weitgasser R., Nejjari C., Patsch W., Chikri M., Meyre D., et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: A global meta-analysis. J. Mol. Med. 2007;85:777–782. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- Chen W.S., Manova K., Weinstein D.C., Duncan S.A., Plump A.S., Prezioso V.R., Bachvarova R.F., Darnell J.E., Jr Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes & Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Chen Y., Pan F.C., Brandes N., Afelik S., Solter M., Pieler T. Retinoic acid signaling is essential for pancreas development and promotes endocrine at the expense of exocrine cell differentiation in Xenopus. Dev. Biol. 2004;271:144–160. doi: 10.1016/j.ydbio.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Chiang M.K., Melton D.A. Single-cell transcript analysis of pancreas development. Dev. Cell. 2003;4:383–393. doi: 10.1016/s1534-5807(03)00035-2. [DOI] [PubMed] [Google Scholar]
- Christofori G., Naik P., Hanahan D. Vascular endothelial growth factor and its receptors, flt-1 and flk-1, are expressed in normal pancreatic islets and throughout islet cell tumorigenesis. Mol. Endocrinol. 1995;9:1760–1770. doi: 10.1210/mend.9.12.8614412. [DOI] [PubMed] [Google Scholar]
- Cockell M., Stevenson B.J., Strubin M., Hagenbuchle O., Wellauer P.K. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol. Cell. Biol. 1989;9:2464–2476. doi: 10.1128/mcb.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C., Barra J., Babinet C., Yaniv M. Expression of the vHNF1/HNF1β homeoprotein gene during mouse organogenesis. Mech. Dev. 1999a;89:211–213. doi: 10.1016/s0925-4773(99)00221-x. [DOI] [PubMed] [Google Scholar]
- Coffinier C., Thepot D., Babinet C., Yaniv M., Barra J. Essential role for the homeoprotein vHNF1/HNF1β in visceral endoderm differentiation. Development. 1999b;126:4785–4794. doi: 10.1242/dev.126.21.4785. [DOI] [PubMed] [Google Scholar]
- Cras-Meneur C., Scharfmann R. FGFR1-IIIb is a putative marker of pancreatic progenitor cells. Mech. Dev. 2002;116:205–208. doi: 10.1016/s0925-4773(02)00138-7. [DOI] [PubMed] [Google Scholar]
- Crisera C.A., Rose M.I., Connelly P.R., Li M., Colen K.L., Longaker M.T., Gittes G.K. The ontogeny of TGF-β1, -β2, -β3, and TGF-β receptor-II expression in the pancreas: Implications for regulation of growth and differentiation. J. Pediatr. Surg. 1999;34:689–693. doi: 10.1016/s0022-3468(99)90357-3. [DOI] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- D'Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Deramaudt T.B., Sachdeva M.M., Wescott M.P., Chen Y., Stoffers D.A., Rustgi A.K. The PDX1 homeodomain transcription factor negatively regulates the pancreatic ductal cell-specific keratin 19 promoter. J. Biol. Chem. 2006;281:38385–38395. doi: 10.1074/jbc.M605891200. [DOI] [PubMed] [Google Scholar]
- Dessimoz J., Bonnard C., Huelsken J., Grapin-Botton A. Pancreas-specific deletion of β-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr. Biol. 2005;15:1677–1683. doi: 10.1016/j.cub.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Deutsch G., Jung J., Zheng M., Lora J., Zaret K.S. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Dichmann D.S., Miller C.P., Jensen J., Scott Heller R., Serup P. Expression and misexpression of members of the FGF and TGFβ families of growth factors in the developing mouse pancreas. Dev. Dyn. 2003;226:663–674. doi: 10.1002/dvdy.10270. [DOI] [PubMed] [Google Scholar]
- Dichmann D.S., Yassin H., Serup P. Analysis of pancreatic endocrine development in GDF11-deficient mice. Dev. Dyn. 2006;235:3016–3025. doi: 10.1002/dvdy.20953. [DOI] [PubMed] [Google Scholar]
- Doyle M.J., Sussel L. Nkx2.2 regulates β-cell function in the mature islet. Diabetes. 2007;56:1999–2007. doi: 10.2337/db06-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M.J., Loomis Z.L., Sussel L. Nkx2.2-repressor activity is sufficient to specify α-cells and a small number of β-cells in the pancreatic islet. Development. 2007;134:515–523. doi: 10.1242/dev.02763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont D.J., Fong G.H., Puri M.C., Gradwohl G., Alitalo K., Breitman M.L. Vascularization of the mouse embryo: A study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev. Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- Duncan S.A., Manova K., Chen W.S., Hoodless P., Weinstein D.C., Bachvarova R.F., Darnell J.E., Jr Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proc. Natl. Acad. Sci. 1994;91:7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S., Gannon M., Peers B., Wright C., Bonner-Weir S., Montminy M. PDX:PBX complexes are required for normal proliferation of pancreatic cells during development. Proc. Natl. Acad. Sci. 2001;98:1065–1070. doi: 10.1073/pnas.031561298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elghazi L., Cras-Meneur C., Czernichow P., Scharfmann R. Role for FGFR2IIIb-mediated signals in controlling pancreatic endocrine progenitor cell proliferation. Proc. Natl. Acad. Sci. 2002;99:3884–3889. doi: 10.1073/pnas.062321799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esni F., Ghosh B., Biankin A.V., Lin J.W., Albert M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., et al. Notch inhibits Ptf1 function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- Fajans S.S., Bell G.I., Polonsky K.S. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- Feijen A., Goumans M.J., van den Eijnden-van Raaij A.J. Expression of activin subunits, activin receptors and follistatin in postimplantation mouse embryos suggests specific developmental functions for different activins. Development. 1994;120:3621–3637. doi: 10.1242/dev.120.12.3621. [DOI] [PubMed] [Google Scholar]
- Fujikura J., Hosoda K., Iwakura H., Tomita T., Noguchi M., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K. Notch/Rbp-j signaling prevents premature endocrine and ductal cell differentiation in the pancreas. Cell Metab. 2006;3:59–65. doi: 10.1016/j.cmet.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Fujikura J., Hosoda K., Kawaguchi Y., Noguchi M., Iwakura H., Odori S., Mori E., Tomita T., Hirata M., Ebihara K., et al. Rbp-j regulates expansion of pancreatic epithelial cells and their differentiation into exocrine cells during mouse development. Dev. Dyn. 2007;236:2779–2791. doi: 10.1002/dvdy.21310. [DOI] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Boyer D.F., Gannon M., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes & Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., Eto Y., Kojima I. Expression of immunoreactive activin A in fetal rat pancreas. Endocr. J. 1995;42:63–68. doi: 10.1507/endocrj.42.63. [DOI] [PubMed] [Google Scholar]
- Gannon M., Gamer L.W., Wright C.V. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev. Biol. 2001;238:185–201. doi: 10.1006/dbio.2001.0359. [DOI] [PubMed] [Google Scholar]
- Gannon M., Tweedie Ables E., Crawford L., Lowe D., Offield M.F., Magnuson M.A., Wright C.V. pdx-1 function is specifically required in embryonic β cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev. Biol. 2008;314:406–417. doi: 10.1016/j.ydbio.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N., White P., Doliba N., Golson M.L., Matschinsky F.M., Kaestner K.H. Foxa2 controls vesicle docking and insulin secretion in mature β cells. Cell Metab. 2007;6:267–279. doi: 10.1016/j.cmet.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Garcia-Ocana A., Takane K.K., Syed M.A., Philbrick W.M., Vasavada R.C., Stewart A.F. Hepatocyte growth factor overexpression in the islet of transgenic mice increases β cell proliferation, enhances islet mass, and induces mild hypoglycemia. J. Biol. Chem. 2000;275:1226–1232. doi: 10.1074/jbc.275.2.1226. [DOI] [PubMed] [Google Scholar]
- Gauthier B.R., Gosmain Y., Mamin A., Philippe J. The β-cell specific transcription factor Nkx6.1 inhibits glucagon gene transcription by interfering with Pax6. Biochem. J. 2007;403:593–601. doi: 10.1042/BJ20070053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgia S., Soliz R., Li M., Zhang P., Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev. Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Gerrish K., Gannon M., Shih D., Henderson E., Stoffel M., Wright C.V., Stein R. Pancreatic β cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3β sites. J. Biol. Chem. 2000;275:3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- Ghosh B., Leach S.D. Interactions between hairy/enhancer of split-related proteins and the pancreatic transcription factor Ptf1-p48 modulate function of the PTF1 transcriptional complex. Biochem. J. 2006;393:679–685. doi: 10.1042/BJ20051063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierl M.S., Karoulias N., Wende H., Strehle M., Birchmeier C. The zinc-finger factor Insm1 (IA-1) is essential for the development of pancreatic β cells and intestinal endocrine cells. Genes & Dev. 2006;20:2465–2478. doi: 10.1101/gad.381806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes G.K., Galante P.E., Hanahan D., Rutter W.J., Debase H.T. Lineage-specific morphogenesis in the developing pancreas: Role of mesenchymal factors. Development. 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- Goldsworthy M., Hugill A., Freeman H., Horner E., Shimomura K., Bogani D., Pieles G., Mijat V., Arkell R., Bhattacharya S., et al. The role of the transcription factor Sox4 in insulin secretion and impaired glucose tolerance. Diabetes. 2008 doi: 10.2337/db07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golosow N., Grobstein C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Dev. Biol. 1962;4:242–255. doi: 10.1016/0012-1606(62)90042-8. [DOI] [PubMed] [Google Scholar]
- Goto Y., De Silva M.G., Toscani A., Prabhakar B.S., Notkins A.L., Lan M.S. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J. Biol. Chem. 1992;267:15252–15257. [PubMed] [Google Scholar]
- Goto Y., Nomura M., Tanaka K., Kondo A., Morinaga H., Okabe T., Yanase T., Nawata H., Takayanagi R., Li E. Genetic interactions between activin type IIB receptor and Smad2 genes in asymmetrical patterning of the thoracic organs and the development of pancreas islets. Dev. Dyn. 2007;236:2865–2874. doi: 10.1002/dvdy.21303. [DOI] [PubMed] [Google Scholar]
- Goudet G., Delhalle S., Biemar F., Martial J.A., Peers B. Functional and cooperative interactions between the homeodomain PDX1, Pbx, and Prep1 factors on the somatostatin promoter. J. Biol. Chem. 1999;274:4067–4073. doi: 10.1074/jbc.274.7.4067. [DOI] [PubMed] [Google Scholar]
- Goulley J., Dahl U., Baeza N., Mishina Y., Edlund H. BMP4-BMPR1A signaling in β cells is required for and augments glucose-stimulated insulin secretion. Cell Metab. 2007;5:207–219. doi: 10.1016/j.cmet.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapin-Botton A., Constam D. Evolution of the mechanisms and molecular control of endoderm formation. Mech. Dev. 2007;124:253–278. doi: 10.1016/j.mod.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Greenwood A.L., Li S., Jones K., Melton D.A. Notch signaling reveals developmental plasticity of Pax4+ pancreatic endocrine progenitors and shunts them to a duct fate. Mech. Dev. 2007;124:97–107. doi: 10.1016/j.mod.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Gu G., Brown J.R., Melton D.A. Direct lineage tracing reveals the ontogeny of pancreatic cell fates during mouse embryogenesis. Mech. Dev. 2003;120:35–43. doi: 10.1016/s0925-4773(02)00330-1. [DOI] [PubMed] [Google Scholar]
- Gu G., Wells J.M., Dombkowski D., Preffer F., Aronow B., Melton D.A. Global expression analysis of gene regulatory pathways during endocrine pancreatic development. Development. 2004;131:165–179. doi: 10.1242/dev.00921. [DOI] [PubMed] [Google Scholar]
- Gupta R.K., Vatamaniuk M.Z., Lee C.S., Flaschen R.C., Fulmer J.T., Matschinsky F.M., Duncan S.A., Kaestner K.H. The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. J. Clin. Invest. 2005;115:1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K., Gao N., Gorski R.K., White P., Hardy O.T., Rafiq K., Brestelli J.E., Chen G., Stoeckert C.J., Jr, Kaestner K.H. Expansion of adult β-cell mass in response to increased metabolic demand is dependent on HNF-4α. Genes & Dev. 2007;21:756–769. doi: 10.1101/gad.1535507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guz Y., Montminy M.R., Stein R., Leonard J., Gamer L.W., Wright C.V., Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in β cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Hald J., Hjorth J.P., German M.S., Madsen O.D., Serup P., Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev. Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Hale M.A., Kagami H., Shi L., Holland A.M., Elsasser H.P., Hammer R.E., MacDonald R.J. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev. Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Harmon E.B., Apelqvist A.A., Smart N.G., Gu X., Osborne D.H., Kim S.K. GDF11 modulates NGN3+ islet progenitor cell number and promotes β-cell differentiation in pancreas development. Development. 2004;131:6163–6174. doi: 10.1242/dev.01535. [DOI] [PubMed] [Google Scholar]
- Hart A., Papadopoulou S., Edlund H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev. Dyn. 2003;228:185–193. doi: 10.1002/dvdy.10368. [DOI] [PubMed] [Google Scholar]
- Haumaitre C., Barbacci E., Jenny M., Ott M.O., Gradwohl G., Cereghini S. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc. Natl. Acad. Sci. 2005;102:1490–1495. doi: 10.1073/pnas.0405776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M., Kim S.K., Melton D.A. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes & Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M., Kim S.K., St Jacques B., McMahon A.P., Melton D.A. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- Heiser P.W., Lau J., Taketo M.M., Herrera P.L., Hebrok M. Stabilization of β-catenin impacts pancreas growth. Development. 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- Heller R.S., Dichmann D.S., Jensen J., Miller C., Wong G., Madsen O.D., Serup P. Expression patterns of Wnts, Frizzleds, sFRPs, and misexpression in transgenic mice suggesting a role for Wnts in pancreas and foregut pattern formation. Dev. Dyn. 2002;225:260–270. doi: 10.1002/dvdy.10157. [DOI] [PubMed] [Google Scholar]
- Henseleit K.D., Nelson S.B., Kuhlbrodt K., Hennings J.C., Ericson J., Sander M. NKX6 transcription factor activity is required for α- and β-cell development in the pancreas. Development. 2005;132:3139–3149. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- Herrera P.L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- Herzig S., Fuzesi L., Knepel W. Heterodimeric Pbx-Prep1 homeodomain protein binding to the glucagon gene restricting transcription in a cell type-dependent manner. J. Biol. Chem. 2000;275:27989–27999. doi: 10.1074/jbc.M003345200. [DOI] [PubMed] [Google Scholar]
- Hiemisch H., Schutz G., Kaestner K.H. Transcriptional regulation in endoderm development: Characterization of an enhancer controlling Hnf3g expression by transgenesis and targeted mutagenesis. EMBO J. 1997;16:3995–4006. doi: 10.1093/emboj/16.13.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan A., Pileggi A., Ricordi C. Transplantation: Current developments and future directions; The future of clinical islet transplantation as a cure for diabetes. Front. Biosci. 2008;13:1192–1205. doi: 10.2741/2755. [DOI] [PubMed] [Google Scholar]
- Holland A.M., Hale M.A., Kagami H., Hammer R.E., MacDonald R.J. Experimental control of pancreatic development and maintenance. Proc. Natl. Acad. Sci. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.M., Gonez L.J., Naselli G., Macdonald R.J., Harrison L.C. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of β-cells in the adult pancreas. Diabetes. 2005;54:2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- Hori Y., Rulifson I.C., Tsai B.C., Heit J.J., Cahoy J.D., Kim S.K. Growth inhibitors promote differentiation of insulin-producing tissue from embryonic stem cells. Proc. Natl. Acad. Sci. 2002;99:16105–16110. doi: 10.1073/pnas.252618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y., Iwasaki N., Hara M., Furuta H., Hinokio Y., Cockburn B.N., Lindner T., Yamagata K., Ogata M., Tomonaga O., et al. Mutation in hepatocyte nuclear factor-1 β gene (TCF2) associated with MODY. Nat. Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- Hua H., Zhang Y.Q., Dabernat S., Kritzik M., Dietz D., Sterling L., Sarvetnick N. BMP4 regulates pancreatic progenitor cell expansion through Id2. J. Biol. Chem. 2006;281:13574–13580. doi: 10.1074/jbc.M600526200. [DOI] [PubMed] [Google Scholar]
- Huang H.P., Liu M., El-Hodiri H.M., Chu K., Jamrich M., Tsai M.J. Regulation of the pancreatic islet-specific gene BETA2 (neuroD) by neurogenin 3. Mol. Cell. Biol. 2000;20:3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iype T., Taylor D.G., Ziesmann S.M., Garmey J.C., Watada H., Mirmira R.G. The transcriptional repressor Nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic β-cell differentiation: Evidence for feedback activation of the nkx6.1 gene by Nkx6.1. Mol. Endocrinol. 2004;18:1363–1375. doi: 10.1210/me.2004-0006. [DOI] [PubMed] [Google Scholar]
- Jacquemin P., Yoshitomi H., Kashima Y., Rousseau G.G., Lemaigre F.P., Zaret K.S. An endothelial-mesenchymal relay pathway regulates early phases of pancreas development. Dev. Biol. 2006;290:189–199. doi: 10.1016/j.ydbio.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Jenny M., Uhl C., Roche C., Duluc I., Guillermin V., Guillemot F., Jensen J., Kedinger M., Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev. Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jensen J., Heller R.S., Funder-Nielsen T., Pedersen E.E., Lindsell C., Weinmaster G., Madsen O.D., Serup P. Independent development of pancreatic α- and β-cells from neurogenin3-expressing precursors: A role for the notch pathway in repression of premature differentiation. Diabetes. 2000a;49:163–176. doi: 10.2337/diabetes.49.2.163. [DOI] [PubMed] [Google Scholar]
- Jensen J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000b;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jensen J.N., Rosenberg L.C., Hecksher-Sorensen J., Serup P.2007Mutant neurogenin-3 in congenital malabsorptive diarrhea N Engl J Med 3561781-1782 [DOI] [PubMed] [Google Scholar]
- Jiang F.X., Harrison L.C. Convergence of bone morphogenetic protein and laminin-1 signaling pathways promotes proliferation and colony formation by fetal mouse pancreatic cells. Exp. Cell Res. 2005;308:114–122. doi: 10.1016/j.yexcr.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Jiang F.X., Stanley E.G., Gonez L.J., Harrison L.C. Bone morphogenetic proteins promote development of fetal pancreas epithelial colonies containing insulin-positive cells. J. Cell Sci. 2002;115:753–760. doi: 10.1242/jcs.115.4.753. [DOI] [PubMed] [Google Scholar]
- Jiang J., Au M., Lu K., Eshpeter A., Korbutt G., Fisk G., Majumdar A.S. Generation of insulin-producing islet-like clusters from human embryonic stem cells. Stem Cells. 2007;25:1940–1953. doi: 10.1634/stemcells.2006-0761. [DOI] [PubMed] [Google Scholar]
- Jiang W., Shi Y., Zhao D., Chen S., Yong J., Zhang J., Qing T., Sun X., Zhang P., Ding M., et al. In vitro derivation of functional insulin-producing cells from human embryonic stem cells. Cell Res. 2007;17:333–344. doi: 10.1038/cr.2007.28. [DOI] [PubMed] [Google Scholar]
- Johansson M., Mattsson G., Andersson A., Jansson L., Carlsson P.O. Islet endothelial cells and pancreatic β-cell proliferation: Studies in vitro and during pregnancy in adult rats. Endocrinology. 2006;147:2315–2324. doi: 10.1210/en.2005-0997. [DOI] [PubMed] [Google Scholar]
- Johansson K.A., Dursun U., Jordan N., Gu G., Beermann F., Gradwohl G., Grapin-Botton A. Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev. Cell. 2007;12:457–465. doi: 10.1016/j.devcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Johnson J.D., Ahmed N.T., Luciani D.S., Han Z., Tran H., Fujita J., Misler S., Edlund H., Polonsky K.S. Increased islet apoptosis in Pdx1+/− mice. J. Clin. Invest. 2003;111:1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jorgensen M.C., Ahnfelt-Ronne J., Hald J., Madsen O.D., Serup P., Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr. Rev. 2007;28:685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- Kaestner K.H., Lee K.H., Schlondorff J., Hiemisch H., Monaghan A.P., Schutz G. Six members of the mouse forkhead gene family are developmentally regulated. Proc. Natl. Acad. Sci. 1993;90:7628–7631. doi: 10.1073/pnas.90.16.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner K.H., Hiemisch H., Luckow B., Schutz G. The HNF-3 gene family of transcription factors in mice: Gene structure, cDNA sequence, and mRNA distribution. Genomics. 1994;20:377–385. doi: 10.1006/geno.1994.1191. [DOI] [PubMed] [Google Scholar]
- Kaestner K.H., Hiemisch H., Schutz G. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol. Cell. Biol. 1998;18:4245–4251. doi: 10.1128/mcb.18.7.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner K.H., Katz J., Liu Y., Drucker D.J., Schutz G. Inactivation of the winged helix transcription factor HNF3α affects glucose homeostasis and islet glucagon gene expression in vivo. Genes & Dev. 1999;13:495–504. doi: 10.1101/gad.13.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K., Han S.I., Shioda S., Hirai M., Nishizawa M., Handa H. MafA is a glucose-regulated and pancreatic β-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- Kataoka K., Shioda S., Ando K., Sakagami K., Handa H., Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet α- and β-cells. J. Mol. Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kawahira H., Ma N.H., Tzanakakis E.S., McMahon A.P., Chuang P.T., Hebrok M. Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development. 2003;130:4871–4879. doi: 10.1242/dev.00653. [DOI] [PubMed] [Google Scholar]
- Kawahira H., Scheel D.W., Smith S.B., German M.S., Hebrok M. Hedgehog signaling regulates expansion of pancreatic epithelial cells. Dev. Biol. 2005;280:111–121. doi: 10.1016/j.ydbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Keller D.M., McWeeney S., Arsenlis A., Drouin J., Wright C.V., Wang H., Wollheim C.B., White P., Kaestner K.H., Goodman R.H. Characterization of pancreatic transcription factor Pdx-1 binding sites using promoter microarray and serial analysis of chromatin occupancy. J. Biol. Chem. 2007;282:32084–32092. doi: 10.1074/jbc.M700899200. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Hebrok M. Intercellular signals regulating pancreas development and function. Genes & Dev. 2001;15:111–127. doi: 10.1101/gad.859401. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Melton D.A. Pancreas development is promoted by cyclopamine, a hedgehog signaling inhibitor. Proc. Natl. Acad. Sci. 1998;95:13036–13041. doi: 10.1073/pnas.95.22.13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Hebrok M., Melton D.A. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Hebrok M., Li E., Oh S.P., Schrewe H., Harmon E.B., Lee J.S., Melton D.A. Activin receptor patterning of foregut organogenesis. Genes & Dev. 2000;14:1866–1871. [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Selleri L., Lee J.S., Zhang A.Y., Gu X., Jacobs Y., Cleary M.L. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat. Genet. 2002;30:430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- Kim D., Gu Y., Ishii M., Fujimiya M., Qi M., Nakamura N., Yoshikawa T., Sumi S., Inoue K. In vivo functioning and transplantable mature pancreatic islet-like cell clusters differentiated from embryonic stem cell. Pancreas. 2003;27:e34–e41. doi: 10.1097/00006676-200308000-00021. [DOI] [PubMed] [Google Scholar]
- Klaus A., Birchmeier W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Frutiger S., Hughes G.J., Hagenbuchle O., Wellauer P.K. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix–loop–helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J., et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:553–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Kubo A., Shinozaki K., Shannon J.M., Kouskoff V., Kennedy M., Woo S., Fehling H.J., Keller G. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- Kulkarni R.N., Jhala U.S., Winnay J.N., Krajewski S., Montminy M., Kahn C.R. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J. Clin. Invest. 2004;114:828–836. doi: 10.1172/JCI21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Oka T., Oka Y., Yamochi T., Ohtsubo K., Mori S., Watanabe T., Machinami R., Ohnishi S. Colocalization of vascular endothelial growth factor (vascular permeability factor) and insulin in pancreatic islet cells. J. Clin. Endocrinol. Metab. 1995;80:3196–3200. doi: 10.1210/jcem.80.11.7593426. [DOI] [PubMed] [Google Scholar]
- Kyrmizi I., Hatzis P., Katrakili N., Tronche F., Gonzalez F.J., Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes & Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert E., Brown J., Melton D.A. Notch gene expression during pancreatic organogenesis. Mech. Dev. 2000;94:199–203. doi: 10.1016/s0925-4773(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Lammert E., Cleaver O., Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Lantz K.A., Vatamaniuk M.Z., Brestelli J.E., Friedman J.R., Matschinsky F.M., Kaestner K.H. Foxa2 regulates multiple pathways of insulin secretion. J. Clin. Invest. 2004;114:512–520. doi: 10.1172/JCI21149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J., Kawahira H., Hebrok M. Hedgehog signaling in pancreas development and disease. Cell. Mol. Life Sci. 2006;63:642–652. doi: 10.1007/s00018-005-5357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S., Miralles F., Basmaciogullari A., Czernichow P., Scharfmann R. Fibroblast growth factor 2 promotes pancreatic epithelial cell proliferation via functional fibroblast growth factor receptors during embryonic life. Diabetes. 1998;47:1236–1242. [PubMed] [Google Scholar]
- Lee J.C., Smith S.B., Watada H., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Perreault N., Brestelli J.E., Kaestner K.H. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes & Dev. 2002a;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Sund N.J., Vatamaniuk M.Z., Matschinsky F.M., Stoffers D.A., Kaestner K.H. Foxa2 controls Pdx1 gene expression in pancreatic β-cells in vivo. Diabetes. 2002b;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Friedman J.R., Fulmer J.T., Kaestner K.H. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005a;435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- Lee C.S., Sund N.J., Behr R., Herrera P.L., Kaestner K.H. Foxa2 is required for the differentiation of pancreatic α-cells. Dev. Biol. 2005b;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Lioubinski O., Muller M., Wegner M., Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Dev. Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- Liu Y., MacDonald R.J., Swift G.H. DNA binding and transcriptional activation by a PDX1.PBX1b.MEIS2b trimer and cooperation with a pancreas-specific basic helix–loop–helix complex. J. Biol. Chem. 2001;276:17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- Loder M.K., Xavier Gda S., McDonald A., Rutter G.A. TCF7L2 controls insulin gene expression and insulin secretion in mature pancreatic β-cells. Biochem. Soc. Trans. 2008;36:357–359. doi: 10.1042/BST0360357. [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lumelsky N., Blondel O., Laeng P., Velasco I., Ravin R., McKay R. Differentiation of embryonic stem cells to insulin-secreting structures similar to pancreatic islets. Science. 2001;292:1389–1394. doi: 10.1126/science.1058866. [DOI] [PubMed] [Google Scholar]
- Lynn F.C., Smith S.B., Wilson M.E., Yang K.Y., Nekrep N., German M.S. Sox9 coordinates a transcriptional network in pancreatic progenitor cells. Proc. Natl. Acad. Sci. 2007;104:10500–10505. doi: 10.1073/pnas.0704054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean N., Ogilvie R.F. Quantitative estimation of the pancreatic islet tissue in diabetic subjects. Diabetes. 1955;4:367–376. doi: 10.2337/diab.4.5.367. [DOI] [PubMed] [Google Scholar]
- Maestro M.A., Boj S.F., Luco R.F., Pierreux C.E., Cabedo J., Servitja J.M., German M.S., Rousseau G.G., Lemaigre F.P., Ferrer J. Hnf6 and Tcf2 (MODY5) are linked in a gene network operating in a precursor cell domain of the embryonic pancreas. Hum. Mol. Genet. 2003;12:3307–3314. doi: 10.1093/hmg/ddg355. [DOI] [PubMed] [Google Scholar]
- Maldonado T.S., Kadison A.S., Crisera C.A., Grau J.B., Alkasab S.L., Longaker M.T., Gittes G.K. Ontogeny of activin B and follistatin in developing embryonic mouse pancreas: Implications for lineage selection. J. Gastrointest. Surg. 2000;4:269–275. doi: 10.1016/s1091-255x(00)80075-x. [DOI] [PubMed] [Google Scholar]
- Marshak S., Benshushan E., Shoshkes M., Havin L., Cerasi E., Melloul D. Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3β transcription factors mediate β-cell-specific expression. Mol. Cell. Biol. 2000;20:7583–7590. doi: 10.1128/mcb.20.20.7583-7590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Gallego-Llamas J., Ribes V., Kedinger M., Niederreither K., Chambon P., Dolle P., Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev. Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Masui T., Long Q., Beres T.M., Magnuson M.A., MacDonald R.J. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes & Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T.A., Zhao L., Artner I., Jarrett H.W., Friedman D., Means A., Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet β cells. Mol. Cell. Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T.A., Artner I., Henderson E., Means A., Sander M., Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc. Natl. Acad. Sci. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer G., Bonne S., Luco R.F., De Van Casteele M., Lenne-Samuel N., Collombat P., Mansouri A., Lee J., Lan M., Pipeleers D., et al. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25:1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.P., McGehee R.E., Jr, Habener J.F. IDX-1: A new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami K., Okano H., Okumachi A., Seino S. Role of cadherin-mediated cell–cell adhesion in pancreatic exocrine-to-endocrine transdifferentiation. J. Biol. Chem. 2008;283:13753–13761. doi: 10.1074/jbc.M710034200. [DOI] [PubMed] [Google Scholar]
- Miralles F., Czernichow P., Scharfmann R. Follistatin regulates the relative proportions of endocrine versus exocrine tissue during pancreatic development. Development. 1998;125:1017–1024. doi: 10.1242/dev.125.6.1017. [DOI] [PubMed] [Google Scholar]
- Miralles F., Czernichow P., Ozaki K., Itoh N., Scharfmann R. Signaling through fibroblast growth factor receptor 2b plays a key role in the development of the exocrine pancreas. Proc. Natl. Acad. Sci. 1999;96:6267–6272. doi: 10.1073/pnas.96.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Lamotte L., Couton D., Joshi R.L. Interplay between FGF10 and Notch signalling is required for the self-renewal of pancreatic progenitors. Int. J. Dev. Biol. 2006;50:17–26. doi: 10.1387/ijdb.052080fm. [DOI] [PubMed] [Google Scholar]
- Mirmira R.G., Watada H., German M.S. β-Cell differentiation factor Nkx6.1 contains distinct DNA binding interference and transcriptional repression domains. J. Biol. Chem. 2000;275:14743–14751. doi: 10.1074/jbc.275.19.14743. [DOI] [PubMed] [Google Scholar]
- Miura A., Yamagata K., Kakei M., Hatakeyama H., Takahashi N., Fukui K., Nammo T., Yoneda K., Inoue Y., Sladek F.M., et al. Hepatocyte nuclear factor-4α is essential for glucose-stimulated insulin secretion by pancreatic β-cells. J. Biol. Chem. 2006;281:5246–5257. doi: 10.1074/jbc.M507496200. [DOI] [PubMed] [Google Scholar]
- Miyatsuka T., Matsuoka T.A., Shiraiwa T., Yamamoto T., Kojima I., Kaneto H. Ptf1a and RBP-J cooperate in activating Pdx1 gene expression through binding to Area III. Biochem. Biophys. Res. Commun. 2007;362:905–909. doi: 10.1016/j.bbrc.2007.08.076. [DOI] [PubMed] [Google Scholar]
- Molotkov A., Molotkova N., Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev. Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- Monaghan A.P., Kaestner K.H., Grau E., Schutz G. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 α, β and γ genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Movassat J., Beattie G.M., Lopez A.D., Hayek A. Exendin 4 up-regulates expression of PDX 1 and hastens differentiation and maturation of human fetal pancreatic cells. J. Clin. Endocrinol. Metab. 2002;87:4775–4781. doi: 10.1210/jc.2002-020137. [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Sidis Y., Mahan A., Raher M.J., Xia Y., Rosen E.D., Bloch K.D., Thomas M.K., Schneyer A.L. FSTL3 deletion reveals roles for TGF-β family ligands in glucose and fat homeostasis in adults. Proc. Natl. Acad. Sci. 2007;104:1348–1353. doi: 10.1073/pnas.0607966104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh L.C., Stanger B.Z., Kwan K.M., Melton D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh L.C., Law A.C., Dor Y., Melton D.A. β-Catenin is essential for pancreatic acinar but not islet development. Development. 2005;132:4663–4674. doi: 10.1242/dev.02063. [DOI] [PubMed] [Google Scholar]
- Nammo T., Yamagata K., Tanaka T., Kodama T., Sladek F.M., Fukui K., Katsube F., Sato Y., Miyagawa J., Shimomura I. Expression of HNF-4α (MODY1), HNF-1β (MODY5), and HNF-1α (MODY3) proteins in the developing mouse pancreas. Brain Res. Gene Expr. Patterns. 2008;8:96–106. doi: 10.1016/j.modgep.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Nelson S.B., Janiesch C., Sander M. Expression of Nkx6 genes in the hindbrain and gut of the developing mouse. J. Histochem. Cytochem. 2005;53:787–790. doi: 10.1369/jhc.5B6619.2005. [DOI] [PubMed] [Google Scholar]
- Nelson S.B., Schaffer A.E., Sander M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting β-cell fate specification in Pdx1+ pancreatic progenitor cells. Development. 2007;134:2491–2500. doi: 10.1242/dev.002691. [DOI] [PubMed] [Google Scholar]
- Nikolova G., Jabs N., Konstantinova I., Domogatskaya A., Tryggvason K., Sorokin L., Fassler R., Gu G., Gerber H.P., Ferrara N., et al. The vascular basement membrane: A niche for insulin gene expression and β cell proliferation. Dev. Cell. 2006;10:397–405. doi: 10.1016/j.devcel.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Nishimura W., Kondo T., Salameh T., El Khattabi I., Dodge R., Bonner-Weir S., Sharma A. A switch from MafB to MafA expression accompanies differentiation to pancreatic β-cells. Dev. Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgaard G.A., Jensen J.N., Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev. Biol. 2003;264:323–338. doi: 10.1016/j.ydbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Obata J., Yano M., Mimura H., Goto T., Nakayama R., Mibu Y., Oka C., Kawaichi M. p48 subunit of mouse PTF1 binds to RBP-Jκ/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger N., Gordon D.B., Bell G.W., Rinaldi N.J., Murray H.L., Volkert T.L., Schreiber J., Rolfe P.A., Gifford D.K., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offield M.F., Jetton T.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L., Wright C.V. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ogawa K., Abe K., Kurosawa N., Kurohmaru M., Sugino H., Takahashi M., Hayashi Y. Expression of α, β A and β B subunits of inhibin or activin and follistatin in rat pancreatic islets. FEBS Lett. 1993;319:217–220. doi: 10.1016/0014-5793(93)80549-a. [DOI] [PubMed] [Google Scholar]
- Oh S.P., Yeo C.Y., Lee Y., Schrewe H., Whitman M., Li E. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes & Dev. 2002;16:2749–2754. doi: 10.1101/gad.1021802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson H., Karlsson K., Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrot M., Rud J., Moss L.G., Sharma A. Identification of β-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc. Natl. Acad. Sci. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otonkoski T., Hayek A. Constitution of a biphasic insulin response to glucose in human fetal pancreatic β-cells with glucagon-like peptide 1. J. Clin. Endocrinol. Metab. 1995;80:3779–3783. doi: 10.1210/jcem.80.12.8530635. [DOI] [PubMed] [Google Scholar]
- Otonkoski T., Cirulli V., Beattie M., Mally M.I., Soto G., Rubin J.S., Hayek A. A role for hepatocyte growth factor/scatter factor in fetal mesenchyme-induced pancreatic β-cell growth. Endocrinology. 1996;137:3131–3139. doi: 10.1210/endo.137.7.8770939. [DOI] [PubMed] [Google Scholar]
- Pan F.C., Chen Y., Bayha E., Pieler T. Retinoic acid-mediated patterning of the pre-pancreatic endoderm in Xenopus operates via direct and indirect mechanisms. Mech. Dev. 2007;124:518–531. doi: 10.1016/j.mod.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Papadopoulou S., Edlund H. Attenuated Wnt signaling perturbs pancreatic growth but not pancreatic function. Diabetes. 2005;54:2844–2851. doi: 10.2337/diabetes.54.10.2844. [DOI] [PubMed] [Google Scholar]
- Petri A., Ahnfelt-Ronne J., Frederiksen K.S., Edwards D.G., Madsen D., Serup P., Fleckner J., Heller R.S. The effect of neurogenin3 deficiency on pancreatic gene expression in embryonic mice. J. Mol. Endocrinol. 2006;37:301–316. doi: 10.1677/jme.1.02096. [DOI] [PubMed] [Google Scholar]
- Phillips B.W., Hentze H., Rust W.L., Chen Q.P., Chipperfield H., Tan E.K., Abraham S., Sadasivam A., Soong P.L., Wang S.T., et al. Directed differentiation of human embryonic stem cells into the pancreatic endocrine lineage. Stem Cells Dev. 2007;16:561–578. doi: 10.1089/scd.2007.0029. [DOI] [PubMed] [Google Scholar]
- Pictet R.L., Clark W.R., Williams R.H., Rutter W.J. An ultrastructural analysis of the developing embryonic pancreas. Dev. Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- Poll A.V., Pierreux C.E., Lokmane L., Haumaitre C., Achouri Y., Jacquemin P., Rousseau G.G., Cereghini S., Lemaigre F.P. A vHNF1/TCF2-HNF6 cascade regulates the transcription factor network that controls generation of pancreatic precursor cells. Diabetes. 2006;55:61–69. [PubMed] [Google Scholar]
- Pozzilli P., Buzzetti R. A new expression of diabetes: Double diabetes. Trends Endocrinol. Metab. 2007;18:52–57. doi: 10.1016/j.tem.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Prado C.L., Pugh-Bernard A.E., Elghazi L., Sosa-Pineda B., Sussel L. Ghrelin cells replace insulin-producing β cells in two mouse models of pancreas development. Proc. Natl. Acad. Sci. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen M.A., Spencer-Dene B., Dickson C., Otonkoski T. The IIIb isoform of fibroblast growth factor receptor 2 is required for proper growth and branching of pancreatic ductal epithelium but not for differentiation of exocrine or endocrine cells. Mech. Dev. 2003;120:167–175. doi: 10.1016/s0925-4773(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Raum J.C., Gerrish K., Artner I., Henderson E., Guo M., Sussel L., Schisler J.C., Newgard C.B., Stein R. FoxA2, Nkx2.2, and PDX-1 regulate islet β-cell-specific mafA expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Mol. Cell. Biol. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritvos O., Tuuri T., Eramaa M., Sainio K., Hilden K., Saxen L., Gilbert S.F. Activin disrupts epithelial branching morphogenesis in developing glandular organs of the mouse. Mech. Dev. 1995;50:229–245. doi: 10.1016/0925-4773(94)00342-k. [DOI] [PubMed] [Google Scholar]
- Rossi J.M., Dunn N.R., Hogan B.L., Zaret K.S. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes & Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux E., Strubin M., Hagenbuchle O., Wellauer P.K. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes & Dev. 1989;3:1613–1624. doi: 10.1101/gad.3.10.1613. [DOI] [PubMed] [Google Scholar]
- Rudnick A., Ling T.Y., Odagiri H., Rutter W.J., German M.S. Pancreatic β cells express a diverse set of homeobox genes. Proc. Natl. Acad. Sci. 1994;91:12203–12207. doi: 10.1073/pnas.91.25.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson I.C., Karnik S.K., Heiser P.W., ten Berge D., Chen H., Gu X., Taketo M.M., Nusse R., Hebrok M., Kim S.K. Wnt signaling regulates pancreatic β cell proliferation. Proc. Natl. Acad. Sci. 2007;104:6247–6252. doi: 10.1073/pnas.0701509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen J.T., Uimari P., Aalto J.M., Pirskanen M., Kaikkonen J., Todorova B., Hypponen J., Korhonen V.P., Asikainen J., Devine C., et al. Type 2 diabetes whole-genome association study in four populations: The DiaGen consortium. Am. J. Hum. Genet. 2007;81:338–345. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaras S.E., Zhao L., Means A., Henderson E., Matsuoka T.A., Stein R. The islet β cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J. Biol. Chem. 2003;278:12263–12270. doi: 10.1074/jbc.M210801200. [DOI] [PubMed] [Google Scholar]
- Sander M., Sussel L., Conners J., Scheel D., Kalamaras J., Dela Cruz F., Schwitzgebel V., Hayes-Jordan A., German M. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of β-cell formation in the pancreas. Development. 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hogan B.L. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hogan B.L. HNF-3 β as a regulator of floor plate development. Cell. 1994;76:103–115. doi: 10.1016/0092-8674(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Schisler J.C., Jensen P.B., Taylor D.G., Becker T.C., Knop F.K., Takekawa S., German M., Weir G.C., Lu D., Mirmira R.G., et al. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet β cells. Proc. Natl. Acad. Sci. 2005;102:7297–7302. doi: 10.1073/pnas.0502168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Servitja J.M., Ferrer J. Transcriptional networks controlling pancreatic development and β cell function. Diabetologia. 2004;47:597–613. doi: 10.1007/s00125-004-1368-9. [DOI] [PubMed] [Google Scholar]
- Seymour P.A., Freude K.K., Tran M.N., Mayes E.E., Jensen J., Kist R., Scherer G., Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl. Acad. Sci. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro A.M., Lakey J.R., Ryan E.A., Korbutt G.S., Toth E., Warnock G.L., Kneteman N.M., Rajotte R.V. Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 2000;343:230–238. doi: 10.1056/NEJM200007273430401. [DOI] [PubMed] [Google Scholar]
- Shapiro A.M., Ricordi C., Hering B.J., Auchincloss H., Lindblad R., Robertson R.P., Secchi A., Brendel M.D., Berney T., Brennan D.C., et al. International trial of the Edmonton protocol for islet transplantation. N. Engl. J. Med. 2006;355:1318–1330. doi: 10.1056/NEJMoa061267. [DOI] [PubMed] [Google Scholar]
- Sherwood R.I., Jitianu C., Cleaver O., Shaywitz D.A., Lamenzo J.O., Chen A.E., Golub T.R., Melton D.A. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev. Biol. 2007;304:541–555. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Shih D.Q., Navas M.A., Kuwajima S., Duncan S.A., Stoffel M. Impaired glucose homeostasis and neonatal mortality in hepatocyte nuclear factor 3α-deficient mice. Proc. Natl. Acad. Sci. 1999;96:10152–10157. doi: 10.1073/pnas.96.18.10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.H., Kim S.E., Woo D.H., Kim S.K., Oh C.H., McKay R., Kim J.H. Directed differentiation of human embryonic stem cells towards a pancreatic cell fate. Diabetologia. 2007;50:1228–1238. doi: 10.1007/s00125-007-0634-z. [DOI] [PubMed] [Google Scholar]
- Shiozaki S., Tajima T., Zhang Y.Q., Furukawa M., Nakazato Y., Kojima I. Impaired differentiation of endocrine and exocrine cells of the pancreas in transgenic mouse expressing the truncated type II activin receptor. Biochim. Biophys. Acta. 1999;1450:1–11. doi: 10.1016/s0167-4889(99)00022-1. [DOI] [PubMed] [Google Scholar]
- Slack J.M. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D., Boutin P., Vincent D., Belisle A., Hadjadj S., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Smart N.G., Apelqvist A.A., Gu X., Harmon E.B., Topper J.N., MacDonald R.J., Kim S.K. Conditional expression of Smad7 in pancreatic β cells disrupts TGF-β signaling and induces reversible diabetes mellitus. PLoS Biol. 2006;4:e39. doi: 10.1371/journal.pbio.0040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.B., Gasa R., Watada H., Wang J., Griffen S.C., German M.S. Neurogenin3 and hepatic nuclear factor 1 cooperate in activating pancreatic expression of Pax4. J. Biol. Chem. 2003;278:38254–38259. doi: 10.1074/jbc.M302229200. [DOI] [PubMed] [Google Scholar]
- Smith S.B., Watada H., German M.S. Neurogenin3 activates the islet differentiation program while repressing its own expression. Mol. Endocrinol. 2004;18:142–149. doi: 10.1210/me.2003-0037. [DOI] [PubMed] [Google Scholar]
- Sommer L., Hagenbuchle O., Wellauer P.K., Strubin M. Nuclear targeting of the transcription factor PTF1 is mediated by a protein subunit that does not bind to the PTF1 cognate sequence. Cell. 1991;67:987–994. doi: 10.1016/0092-8674(91)90371-5. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B., Chowdhury K., Torres M., Oliver G., Gruss P. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- Spence J.R., Wells J.M. Translational embryology: Using embryonic principles to generate pancreatic endocrine cells from embryonic stem cells. Dev. Dyn. 2007;236:3218–3227. doi: 10.1002/dvdy.21366. [DOI] [PubMed] [Google Scholar]
- Stafford D., Prince V.E. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr. Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Stafford D., Hornbruch A., Mueller P.R., Prince V.E. A conserved role for retinoid signaling in vertebrate pancreas development. Dev. Genes Evol. 2004;214:432–441. doi: 10.1007/s00427-004-0420-6. [DOI] [PubMed] [Google Scholar]
- Stoffel M., Duncan S.A. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc. Natl. Acad. Sci. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers D.A., Ferrer J., Clarke W.L., Habener J.F. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 1997a;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997b;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- Stoffers D.A., Heller R.S., Miller C.P., Habener J.F. Developmental expression of the homeodomain protein IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional reporter. Endocrinology. 1999;140:5374–5381. doi: 10.1210/endo.140.11.7122. [DOI] [PubMed] [Google Scholar]
- Strom A., Bonal C., Ashery-Padan R., Hashimoto N., Campos M.L., Trumpp A., Noda T., Kido Y., Real F.X., Thorel F., et al. Unique mechanisms of growth regulation and tumor suppression upon Apc inactivation in the pancreas. Development. 2007;134:2719–2725. doi: 10.1242/dev.02875. [DOI] [PubMed] [Google Scholar]
- Sund N.J., Vatamaniuk M.Z., Casey M., Ang S.L., Magnuson M.A., Stoffers D.A., Matschinsky F.M., Kaestner K.H. Tissue-specific deletion of Foxa2 in pancreatic β cells results in hyperinsulinemic hypoglycemia. Genes & Dev. 2001;15:1706–1715. doi: 10.1101/gad.901601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L., Kalamaras J., Hartigan-O'Connor D.J., Meneses J.J., Pedersen R.A., Rubenstein J.L., German M.S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Svensson P., Williams C., Lundeberg J., Ryden P., Bergqvist I., Edlund H. Gene array identification of Ipf1/Pdx1−/− regulated genes in pancreatic progenitor cells. BMC Dev. Biol. 2007;7:129. doi: 10.1186/1471-213X-7-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift G.H., Liu Y., Rose S.D., Bischof L.J., Steelman S., Buchberg A.M., Wright C.V., MacDonald R.J. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2) Mol. Cell. Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed M.A., Barinas-Mitchell E., Pietropaolo S.L., Zhang Y.J., Henderson T.S., Kelley D.E., Korytkowski M.T., Donahue R.P., Tracy R.P., Trucco M., et al. Is type 2 diabetes a chronic inflammatory/autoimmune disease? Diabetes Nutr. Metab. 2002;15:68–83. [PubMed] [Google Scholar]
- Thomas H., Senkel S., Erdmann S., Arndt T., Turan G., Klein-Hitpass L., Ryffel G.U. Pattern of genes influenced by conditional expression of the transcription factors HNF6, HNF4α and HNF1β in a pancreatic β-cell line. Nucleic Acids Res. 2004;32:e150. doi: 10.1093/nar/gnh144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay K.D., Hoodless P.A., Bikoff E.K., Robertson E.J. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development. 2000;127:3079–3090. doi: 10.1242/dev.127.14.3079. [DOI] [PubMed] [Google Scholar]
- Ueki K., Okada T., Hu J., Liew C.W., Assmann A., Dahlgren G.M., Peters J.L., Shackman J.G., Zhang M., Artner I., et al. Total insulin and IGF-I resistance in pancreatic β cells causes overt diabetes. Nat. Genet. 2006;38:583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- Vaca P., Berna G., Araujo R., Carneiro E.M., Bedoya F.J., Soria B., Martin F. Nicotinamide induces differentiation of embryonic stem cells into insulin-secreting cells. Exp. Cell Res. 2008;314:969–974. doi: 10.1016/j.yexcr.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Vasir B., Jonas J.C., Steil G.M., Hollister-Lock J., Hasenkamp W., Sharma A., Bonner-Weir S., Weir G.C. Gene expression of VEGF and its receptors Flk-1/KDR and Flt-1 in cultured and transplanted rat islets. Transplantation. 2001;71:924–935. doi: 10.1097/00007890-200104150-00018. [DOI] [PubMed] [Google Scholar]
- Vatamaniuk M.Z., Gupta R.K., Lantz K.A., Doliba N.M., Matschinsky F.M., Kaestner K.H. Foxa1-deficient mice exhibit impaired insulin secretion due to uncoupled oxidative phosphorylation. Diabetes. 2006;55:2730–2736. doi: 10.2337/db05-0470. [DOI] [PubMed] [Google Scholar]
- Verschueren K., Dewulf N., Goumans M.J., Lonnoy O., Feijen A., Grimsby S., Vandi Spiegle K., ten Dijke P., Moren A., Vanscheeuwijck P., et al. Expression of type I and type IB receptors for activin in midgestation mouse embryos suggests distinct functions in organogenesis. Mech. Dev. 1995;52:109–123. doi: 10.1016/0925-4773(95)00395-h. [DOI] [PubMed] [Google Scholar]
- Wang H., Maechler P., Antinozzi P.A., Hagenfeldt K.A., Wollheim C.B. Hepatocyte nuclear factor 4α regulates the expression of pancreatic β-cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J. Biol. Chem. 2000;275:35953–35959. doi: 10.1074/jbc.M006612200. [DOI] [PubMed] [Google Scholar]
- Wang J., Elghazi L., Parker S.E., Kizilocak H., Asano M., Sussel L., Sosa-Pineda B. The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic β-cell differentiation. Dev. Biol. 2004;266:178–189. doi: 10.1016/j.ydbio.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Wang J., Cortina G., Wu S.V., Tran R., Cho J.H., Tsai M.J., Bailey T.J., Jamrich M., Ament M.E., Treem W.R., et al. Mutant neurogenin-3 in congenital malabsorptive diarrhea. N. Engl. J. Med. 2006;355:270–280. doi: 10.1056/NEJMoa054288. [DOI] [PubMed] [Google Scholar]
- Wang S., Zhang J., Zhao A., Hipkens S., Magnuson M.A., Gu G. Loss of Myt1 function partially compromises endocrine islet cell differentiation and pancreatic physiological function in the mouse. Mech. Dev. 2007;124:898–910. doi: 10.1016/j.mod.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Elghazi L., Martin S., Martins I., Srinivasan R.S., Geng X., Sleeman M., Collombat P., Houghton J., Sosa-Pineda B. ghrelin is a novel target of Pax4 in endocrine progenitors of the pancreas and duodenum. Dev. Dyn. 2008;237:51–61. doi: 10.1002/dvdy.21379. [DOI] [PubMed] [Google Scholar]
- Watada H., Mirmira R.G., Leung J., German M.S. Transcriptional and translational regulation of β-cell differentiation factor Nkx6.1. J. Biol. Chem. 2000;275:34224–34230. doi: 10.1074/jbc.M004981200. [DOI] [PubMed] [Google Scholar]
- Watada H., Scheel D.W., Leung J., German M.S. Distinct gene expression programs function in progenitor and mature islet cells. J. Biol. Chem. 2003;278:17130–17140. doi: 10.1074/jbc.M213196200. [DOI] [PubMed] [Google Scholar]
- Weinstein D.C., Ruiz i Altaba A., Chen W.S., Hoodless P., Prezioso V.R., Jessell T.M., Darnell J.E., Jr The winged-helix transcription factor HNF-3 β is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Wells J.M., Esni F., Boivin G.P., Aronow B.J., Stuart W., Combs C., Sklenka A., Leach S.D., Lowy A.M. Wnt/β-catenin signaling is required for development of the exocrine pancreas. BMC Dev. Biol. 2007;7:4. doi: 10.1186/1471-213X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels N., Cohen J. Early pancreas organogenesis: Morphogenesis, tissue interactions, and mass effects. Dev. Biol. 1967;15:237–270. doi: 10.1016/0012-1606(67)90042-5. [DOI] [PubMed] [Google Scholar]
- White P., Lee May C., Lamounier R.N., Brestelli J.E., Kaestner K.H. Defining pancreatic endocrine precursors and their descendants. Diabetes. 2008;57:654–668. doi: 10.2337/db07-1362. [DOI] [PubMed] [Google Scholar]
- Wiebe P.O., Kormish J.D., Roper V.T., Fujitani Y., Alston N.I., Zaret K.S., Wright C.V., Stein R.W., Gannon M. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol. Cell. Biol. 2007;27:4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.E., Yang K.Y., Kalousova A., Lau J., Kosaka Y., Lynn F.C., Wang J., Mrejen C., Episkopou V., Clevers H.C., et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54:3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- Wu K.L., Gannon M., Peshavaria M., Offield M.F., Henderson E., Ray M., Marks A., Gamer L.W., Wright C.V., Stein R. Hepatocyte nuclear factor 3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Wang S., Zhang J., Zhao A., Stanger B.Z., Gu G. The fringe molecules induce endocrine differentiation in embryonic endoderm by activating cMyt1/cMyt3. Dev. Biol. 2006;297:340–349. doi: 10.1016/j.ydbio.2006.04.456. [DOI] [PubMed] [Google Scholar]
- Xu X., D'Hoker J., Stange G., Bonne S., De Leu N., Xiao X., de Van Casteele M., Mellitzer G., Ling Z., Pipeleers D., et al. β Cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Furuta H., Oda N., Kaisaki P.J., Menzel S., Cox N.J., Fajans S.S., Signorini S., Stoffel M., Bell G.I. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- Yamaoka T., Idehara C., Yano M., Matsushita T., Yamada T., Ii S., Moritani M., Hata J., Sugino H., Noji S., et al. Hypoplasia of pancreatic islets in transgenic mice expressing activin receptor mutants. J. Clin. Invest. 1998;102:294–301. doi: 10.1172/JCI2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K.H., Ko S.H., Cho J.H., Lee J.M., Ahn Y.B., Song K.H., Yoo S.J., Kang M.I., Cha B.Y., Lee K.W., et al. Selective β-cell loss and α-cell expansion in patients with type 2 diabetes mellitus in Korea. J. Clin. Endocrinol. Metab. 2003;88:2300–2308. doi: 10.1210/jc.2002-020735. [DOI] [PubMed] [Google Scholar]
- Yoshitomi H., Zaret K.S. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131:807–817. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
- Zhang C., Moriguchi T., Kajihara M., Esaki R., Harada A., Shimohata H., Oishi H., Hamada M., Morito N., Hasegawa K., et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell. Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Guo M., Matsuoka T.A., Hagman D.K., Parazzoli S.D., Poitout V., Stein R. The islet β cell-enriched MafA activator is a key regulator of insulin gene transcription. J. Biol. Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Law A.C., Rajagopal J., Anderson W.J., Gray P.A., Melton D.A. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Zorn A.M., Wells J.M. Molecular basis of vertebrate endoderm development. Int. Rev. Cytol. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]