Abstract
Adenosine deaminases acting on RNA (ADARs) are best known for altering the coding sequences of mRNA through RNA editing, as in the GluR-B Q/R site. ADARs have also been shown to affect RNA interference (RNAi) and microRNA processing by deamination of specific adenosines to inosine. Here, we show that ADAR proteins can affect RNA processing independently of their enzymatic activity. We show that ADAR2 can modulate the processing of mir-376a2 independently of catalytic RNA editing activity. In addition, in a Drosophila assay for RNAi deaminase-inactive ADAR1 inhibits RNAi through the siRNA pathway. These results imply that ADAR1 and ADAR2 have biological functions as RNA-binding proteins that extend beyond editing per se and that even genomically encoded ADARs that are catalytically inactive may have such functions.
Keywords: Drosophila, DSH1, microprocessor, RNA editing, RNA interference
Introduction
The most common RNA editing event in human mRNA is deamination of adenosine to inosine. Proteins of the adenosine deaminases acting on RNA (ADAR) family catalyse this editing event. ADAR activity has been implicated in cancer, development, innate immunity and RNA interference (RNAi) (Patterson et al, 1995; George and Samuel, 1999; Beghini et al, 2000; Maas et al, 2001; Scadden and Smith, 2001; Hartwig et al, 2004; Luciano et al, 2004; Scadden, 2005; Scadden and O'Connell, 2005; Yang et al, 2005, 2006; Blow et al, 2006; Takahashi et al, 2006; Ohman, 2007; Paz et al, 2007; Kawahara et al, 2007a, 2007b; Cenci et al, 2008). In humans, the two most studied proteins of the ADAR family are ADAR1 and ADAR2. The ADAR1−/− mice are embryonic lethal with liver disintegration and a deficiency in erythropoiesis (Wang et al, 2000, 2004; Hartner et al, 2004). ADAR2 has been shown to be necessary for the editing of the GluR-B transcript at the critical Q/R site, which limits calcium entry through AMPA receptors. ADAR2−/− mice die around day p20 with seizures (Higuchi et al, 2000).
ADAR1 and ADAR2 have similar domain structures (Figure 1D) but mainly display distinct editing site specificities—although their editing activities do overlap at some sites. There are two major isoforms of ADAR1 because of alternative promoter usage, the longer shuttling p150 isoform and the shorter nuclear p110 isoform. ADAR1 p150 is upregulated by interferon and can compete with PKR for double-stranded (ds)RNA. At the amino terminus ADAR1 p150 has two Z-DNA-binding domains and a nuclear export signal, which allows it to shuttle in and out of the nucleus, however it is predominantly found in the cytoplasm. Both isoforms have three dsRNA-binding domains, the third one overlaps with a nuclear localization signal (NLS) and the catalytic deaminase domain is at the carboxy terminus (Strehblow et al, 2002). ADAR1 is widely expressed throughout the body with the exception of skeletal muscle. ADAR2, on the other hand, is most highly expressed in the central nervous system, has two dsRNA-binding domains and a carboxy-terminal deaminase domain. It is localized to the nucleus by an NLS present in the N-terminal region (Desterro et al, 2003; Wong et al, 2003). To what extent each of the different domains of ADAR1 and ADAR2 contributes to their biological activities is an open question. Two other members of the ADAR family in vertebrates are catalytically inactive, ADAR3 that is brain specific and TENR that is expressed in the testis (Melcher et al, 1996; Connolly et al, 2005). Drosophila has a single Adar gene encoding a deaminase with the same domain structure as ADAR2. Adar mutant flies are viable with severe locomotion defects and loss of RNA editing in transcripts expressed in the CNS (Palladino et al, 2000a, 2000b).
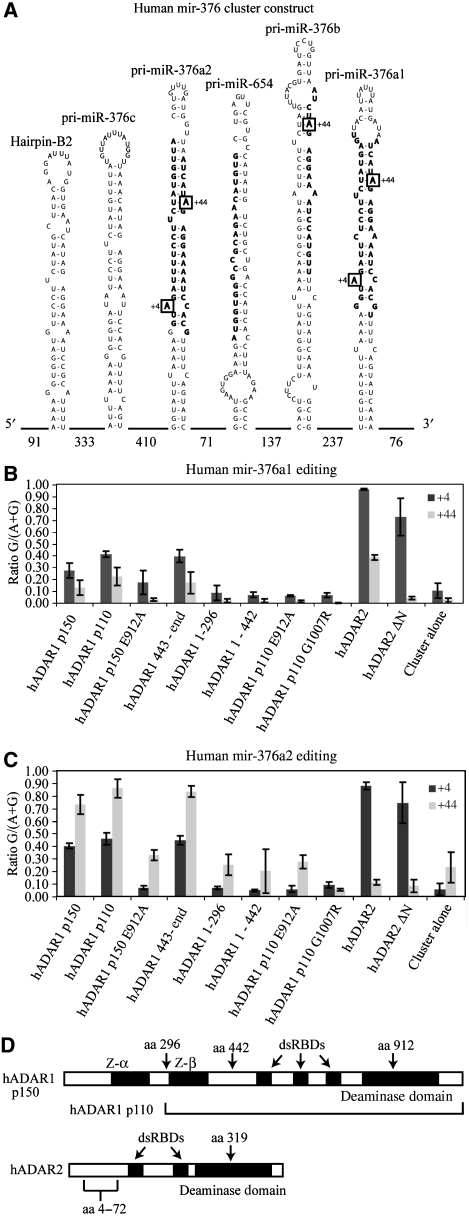
Figure 1.
Editing levels in the human mir-376 cluster. (A) Diagram of the mir-376 cluster; nomenclature is from miRBASE http://microrna.sanger.ac.uk/sequences. Numbers at bottom indicate intervening lengths of sequences. Bold text regions are the mature microRNAs. Boxes indicate major sites of editing. There is no evidence of editing in mir-654. (B) Editing in mir-376a1 expressed from a complete mir-376 cluster. (C) Editing in mir-376a2. Ratio G/(A+G) indicates the ratio of the guanosine signal to the total (adenosine+guanosine) chromatogram signal. +4 and +44 are positions within mir-376a2 indicated in A. (D) ADAR1 and ADAR2 protein domains showing the positions and features used in the construction of the hADAR expression vectors. hADAR1 p150 contains two Z-DNA-binding domains, three dsRBDs and one deaminase domain. hADAR1 p110 begins at amino-acid position 296 of hADAR1 p150. hADAR2 has two dsRBDs and one deaminase domain. hADAR2 ΔN is a deletion mutant of hADAR2 with amino acids from positions 4–72 removed. A key glutamate residue in the deaminase domain is amino acid 912 for ADAR1 and 319 for ADAR2. Mutation of this residue to alanine renders the ADAR catalytically inactive.
Mutations in human ADAR1 are associated with Dyschromatosis Symmetrica Hereditaria (DSH1) (Tojo et al, 2006), an autosomal dominant hyperpigmentation of the hands and feet occurring in Chinese and Japanese families. The majority of DSH1 mutations are associated with single allele truncations of ADAR1 and the dominant phenotype seems to be due to haploinsufficiency for ADAR1. The effect may arise in migrating neural crest cells that develop into melanocytes in the affected skin areas. The point mutant ADAR1 G1007R is an unusual example of a DSH1-associated variant that produces a full-length ADAR1 protein. On the basis of the known crystal structure of the ADAR2 deaminase domain (Macbeth et al, 2005) this mutant should not affect deaminase domain folding and the protein may be capable of RNA binding. However, the mutation introduces an additional positively charged arginine residue on the RNA-binding face of the deaminase domain very close to the active site and the DSH1 phenotype suggests that the ADAR1 G1007R protein may be catalytically inactive.
dsRNA is required for both RNA editing by ADARs and RNAi. Thus, it is not surprising that there have been several reports of effects of ADARs in RNAi. Most of these studies have focused on the effect of the editing base change on processing of dsRNA or edited microRNA precursors (Scadden and Smith, 2001; Luciano et al, 2004; Scadden, 2005; Scadden and O'Connell, 2005; Yang et al, 2005, 2006; Blow et al, 2006; Ohman, 2007; Kawahara et al, 2007a, 2007b). More recent studies focused on the downstream effects on target selection by edited miRNAs. For example, it was recently shown that an miRNA predicted to target the PRPS1 transcript is edited by ADAR2 and, interestingly, levels of PRPS1 protein are elevated in ADAR2-deficient mice (Kawahara et al, 2007b). This result is the only demonstration so far that miRNA modulation of protein levels is altered by ADARs.
There are several key steps in the miRNA pathway, which could be affected by ADARs (for review see Tomari and Zamore, 2005; Valencia-Sanchez et al, 2006). First, primary transcripts (pri-miRNA) are processed in the nucleus by the microprocessor complex, which contains Drosha and its partner DGCR8, to pre-miRNA hairpins (Gregory et al, 2004). Nuclear ADARs could therefore affect this step through steric hindrance of Drosha/DGCR8 binding or by inhibiting Drosha action through the alteration at the edited base in the primary miRNA transcript. As pre-miRNA hairpins can be bound by ADARs it is possible that their export from the nucleus, which is mediated by Exportin 5 (Yi et al, 2003), can be disrupted by binding. The next step in miRNA processing is the Dicer complex-mediated cleavage of the pre-miRNA hairpin into a 21–23 nucleotide RNA duplex in the cytoplasm. Cytoplasmic ADARs could also influence this step through sequestering the pre-miRNA or editing the pre-miRNA. Ultimately, the mature miRNA guide strand guides RISC to mRNAs that have complementarity with the guide strand, resulting in translation inhibition or mRNA degradation. ADAR antagonism of miRNA or siRNA production should result in alteration in the functional activity elicited by miRNAs or siRNAs.
In this work, we sought to determine which properties of ADARs are responsible for influencing miRNA function. Specifically, we have examined the ability of several truncation and point mutations of ADAR1 and ADAR2 to alter the function of mir-376a2 in a cell culture system optimized for analysis of the activity of a single miRNA. Our results indicate that editing can result in retargeting of human mir-376a2 as shown earlier for murine mir-376 (Kawahara et al, 2007b). Interestingly, our results also show that even in the absence of editing, ADARs can affect the processing of miRNAs at the Drosha cleavage level step, thereby altering the functional activity of miRNAs. This suggests that ADARs may influence a larger set of miRNAs than would be predicted from the numbers that are known to be edited. We have also examined the ability of the human ADARs to alter silencing of the white+ eye colour gene in Drosophila by a 300plus base-pair hairpin, which is processed into siRNA duplexes (Vagin et al, 2006). We again find that a deaminase-inactive ADAR can antagonize siRNA activity against white+. Therefore, RNA binding by itself is an important function of ADAR biology.
Results
Editing of the mir-376 cluster by ADAR isoforms shows asymmetric editing in pri-mir-376a2
The human mir-376 cluster comprises several miRNA hairpins encoding mir-376c, mir-376a2, mir-654, mir-376b and mir-376a1 and others (Figure 1A). It was reported earlier that in human medulla several of the adenosines in this cluster are edited (Kawahara et al, 2007b). To elucidate whether particular domains of human ADAR1 or ADAR2 have prominent roles in the effect of ADARs on miRNA function, we cotransfected HEK 293T cells with constructs that express the mir-376 cluster and with constructs that express individual human ADAR1 or ADAR2 isoforms (Figure 1D summarizes key features of ADAR1 and ADAR2). All mutant proteins were expressed to the same level as the respective wild-type protein, as detected on immunoblots probed with anti-FLAG monoclonal antibody (data not shown). HEK 293T cells do not express detectable levels of the endogenous cluster. RNA editing levels were examined initially, before endeavouring to examine the effects of these ADAR variants on precursor processing and effects on downstream microRNA targets.
RNA was extracted from transfected cells, reverse transcribed with random hexamers and cDNA corresponding to individual pri-mir-376 hairpins were amplified using primers located around a 100 nucleotides before and after each hairpin within the long intervening sequences between the hairpins. RNA editing levels at sites within the pri-mir-376 hairpins were measured from mixed sequence chromatograms on the amplified sequence pools for each pri-mir-376 hairpin. We found that sites in pri-mir-376 hairpins were highly edited, as reported earlier for human medulla (Kawahara et al, 2007b).
The different specificities of ADAR1 and ADAR2 account for the different patterns of editing in the pri-mir-376a1 (Figure 1B), -376a2 (Figure 1C) and -376b hairpins (Kawahara et al, 2007b). For instance, ADAR2 edits pri-mir-376a1 at the +4 and +44 site more efficiently than ADAR1 (Figure 1B). In contrast, the two naturally occurring isoforms of ADAR1, p150 and p110, showed similar efficiencies at a range of sites within pri-miR-376a1, with the nuclear ADAR1 p110 editing slightly more efficiently in all cases (Figure 1B). The larger isoform has an extended amino terminus that encodes Z-DNA-binding domains that can bind to dsRNA (Oh et al, 2002; Koeris et al, 2005; Placido et al, 2007).
The editing sites that showed the strongest preferences for editing by a particular ADAR occurred in pri-mir-376a2, where hADAR2 preferentially edited the +4 site of the mature 5′ miRNA with 80–90% efficiency. ADAR1 edits this site with 40% efficiency (Figure 1C). By contrast, hADAR1 edits the +44 site within the mature 3′ miRNA with 80–90% efficiency whereas ADAR2 edits this site with only 10% efficiency. This is in contrast with the editing of miR-376a1 at the +44 site as described above (Figure 1B). RNA editing sites in other microRNA precursors within the cluster show less specificity for ADARs, the +44 site of pri-mir-376b was edited by both hADAR1 and hADAR2 (data not shown). As observed earlier in human medulla RNA, mir-654 was not edited in HEK 293T cells cotransfected with ADAR.
As anticipated, cotransfection of the ADAR1 1-296 construct that expresses only the first 296 residues, containing the first Z-DNA-binding domain of ADAR or the ADAR1 1-442 construct containing both Z-DNA-binding domains did not increase RNA editing. On the other hand, cotransfection with the ADAR1 443-end construct that expresses a protein lacking both Z DNA-binding-domains gave levels of RNA editing similar to intact ADAR1 p110.
To determine whether editing activity is critical for effects of ADARs on microRNA processing and/or function we tested the effects of two ADAR1 mutant proteins. First, a construct with a glutamate to alanine change in an essential catalytic site residue (ADAR1 E912A) was shown to cause only background levels of RNA editing similar to that seen with cotransfection of the control pcDNA3.1 construct. We also cotransfected a construct expressing the naturally occurring G1007R mutation in ADAR1 that has been linked to DSH1 with the construct expressing the mir-376 cluster and found that this protein is incapable of editing any sites in the microRNA precursors. This protein, expressed in and purified from the yeast Pichia pastoris binds long dsRNA similarly to wild-type ADAR1 as assayed by filter binding but does not deaminate long dsRNA (Supplementary Figure S1). ADAR2 ΔN is a deletion of ADAR2 amino acids 4–72 that removes the nuclear localization sequence and causes active ADAR2 to accumulate in the cytoplasm (Wong et al, 2003). Surprisingly, the ADAR2 ΔN isoform edited the pri-miRNA, which is normally found only in the nucleus (Figure 1B and C). The observed editing by the cytoplasmic ADAR2 ΔN is due to editing of the pri-miRNA transcripts that escaped to the cytoplasm without being cleaved by the Drosha complex in the nucleus (Supplementary Figure S2).
ADAR2 shows strong inhibition of miRNA activity
As the RNA editing sites in mir-376a2 show the highest levels of preference for editing by ADAR1 or ADAR2 within the entire miR-376 cluster, we generated a construct that expresses pri-mir-376a2 alone. This allows the effects of ADAR2 binding and editing at the +4 site and ADAR1 binding and editing at the +44 site to be studied in the absence of other microRNAs that have strong sequence similarities. We verified that the editing of mir-376a2 alone was the same as editing of mir-376a2 expressed in the intact cluster (Supplementary Figure S3A).
Processing of pre-mir-376a2 by the miRNA processing machinery produces mature 5′ and 3′ microRNAs. To determine how the production and the specific targeting of the mature microRNAs is affected by the presence of ADARs we measured RNA-silencing activities with two sets of luciferase constructs containing full-length microRNA target sites in the 3′ UTR regions of the luciferase transcripts (Supplementary Figure S3B). The 5′ miR luciferase reporter and the 3′ miR luciferase reporters measure the luciferase-silencing activity of the corresponding wild-type mature unedited microRNAs. The use of target sites with perfect homology extending beyond the seed match may lead to site-directed cleavage of the target transcript as well as translational inhibition. When ADARs edit within miRNAs they have the potential of retargeting miRNAs to another transcript. The RNA-silencing activities of the mature microRNAs retargeted as a result of editing are measured using a second pair of luciferase reporters with single base changes in the target sites (Supplementary Figure S3B).
We found that mir-376a2 expressed and processed in the absence of cotransfected ADAR constructs generated substantial silencing activities corresponding to the mature, unedited 5′ and 3′ miRs, as expected, (Supplementary Figure S3B), reducing luciferase levels 10-fold to approximately 0.1 of the value obtained in the absence of the miRNA. We optimized the level of mir-376a2-expressing construct so that it was sufficient to obtain this maximal level of silencing for both microRNAs.
Expression of the mir-376a2 construct in the absence of ADARs produced much less silencing activity directed against the reporters for the edited microRNAs, reducing the luciferase levels by only approximately two-fold, to approximately 0.5 of the value obtained in the absence of the miRNA (Supplementary Figure S3B). This five-fold discrimination by the mature unedited microRNAs between the unedited and the edited target sites is highly specific, considering that the edited target sites in each case differ by only 1 nucleotide in the 22 base sequences.
Having established conditions to maximize silencing activities of mature miRNAs from the transfected pri-mir-376a2 construct and to determine their target site preferences we cotransfected the mir-376a2 reporter with constructs expressing ADARs and ADAR mutations to assess how ADARs affect the silencing activities of resulting miRNAs. Consistent with their editing of pri-mir-376a2 at position +44, both hADAR1 p110 and hADAR1 p150 increased the silencing activity corresponding to the edited form of the 3′ miR (Figure 2B). This change was largely editing dependent, as the deaminase mutant ADARs (ADAR p150 E912A and ADAR p110 E912A) did not have this effect (Figure 2B); the effect was not dependent on the Z-DNA-binding domains of ADAR1 (ADAR1 443-end). Further, the Z-DNA-binding domains of ADAR1 (ADAR1 1-442) were not sufficient on their own to significantly increase the silencing activity of edited 3′ miRNA.
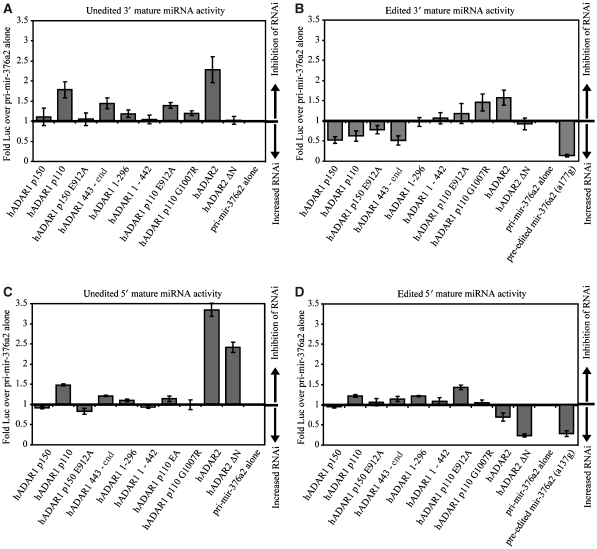
Figure 2.
Effects of ADARs on functional activities of mature miRNAs processed from pri-mir-376a2. (A) Inhibition of 3′ miRNA activity. A value more than 1 means that the miRNA is less active when the ADAR is present than when the pri-mir-376a2 is expressed alone. (B) Retargeting of 3′ miRNA activity. A value less than 1 means that the miRNA is more active in the presence of the ADAR than when the pri-mir-376a2 is expressed alone. Pre-edited mir-376a2 (a177g) indicates a construct expressing pri-mir-376a2 with the +44 site (position 177) mutated from an adenosine to a guanosine. (C) Inhibition of 5′ miRNA activity. (D) Retargeting of 5′ miRNA activity. The activity for pre-edited mir-376a2 (a137g) is from a construct expressing pri-mir-376a2 with the +4 site (position 137) mutated from an adenosine to a guanosine.
Notably, it was ADAR2 that produced the most striking inhibitory effects on miRNA function, particularly on the activity of the unedited miRNAs (Figure 2A and C). ADAR2 edits the pri-mir-376a2 at position +4 and thereby increases the silencing activity of edited 5′ miR (Figure 2D), at the expense of unedited 5′ mir activity (Figure 2C). As ADAR2 edits the +4 site in pri-mir-376a2 with 90% efficiency it is surprising that the retargeting effect was not more complete. Silencing of the reporters for edited microRNAs was maximized by cotransfecting equivalent amounts of mir-376a2 constructs bearing A–G point mutations at the RNA editing sites so that the mature miRNAs were ‘pre-edited' to mimic the inosine-containing product. Cotransfecting a pri-mir-376a2 construct pre-edited at the +4 site gives a silencing activity corresponding to the edited 5′ miRNA that was almost twice as great as seen with ADAR2 (Figure 2D). A possible explanation is that ADAR2 edits pri-mir-376a2 very efficiently but that it also slows Drosha processing of the pri-mir-376a2 so that less mature miRNA is produced.
The hypothesis that ADAR2 inhibits Drosha processing of pri-mir-376a2 is supported, first, by the fact that ADAR2 inhibits the activity of the 3′ miRNA even though it edits the +44 site within this miRNA with only 10% efficiency. Second, the cytoplasmic protein ADAR2 ΔN has no effect either on activity or on retargeting of the 3′ micro RNA (Figure 2A and B). This is consistent with a nuclear inhibition of Drosha being responsible for the ADAR2-mediated inhibition of 3′ miR activity.
ADAR2 ΔN did affect the activity of the 5′ miRNA and increased the activity of the edited form. This appears to be due to base deamination and it does not suggest further cytoplasmic influence of ADAR2 on cytoplasmic processing steps. ADAR2 ΔN, which is cytoplasmic, edits the Drosha product, the pre-miRNA, after it is exported to the cytoplasm (Figure 2 and Supplementary Figure S2).
Drosha processing of pri-mir-376a2 is inhibited by ADAR2 protein and not by the RNA editing base changes
As mentioned earlier ADAR interactions with miRNAs can occur at all levels of the miRNA processing pathway. We examined the processing of pri-mir-376a2 in transfected cells by northern analysis (Figure 3A and B; Supplementary Figure S4). ADARs were expressed in HEK293T cells in the presence of the mir-376a2 expression construct and the appropriate reporter construct. Only ADAR2 caused a significant reduction in the mature ∼22-nucleotide miRNA and in the ∼64-nucleotide pre-miRNA, with a reduction of over 65% in both cases. This indicates that ADAR2 activity on the pri-mir-376a2 hairpin disrupts Drosha processing in the nucleus. In further support of this hypothesis, ADAR2 ΔN, a cytoplasmically localized mutant of ADAR2, did not cause a significant reduction in the levels of either the pre-miRNA or the mature miRNA.
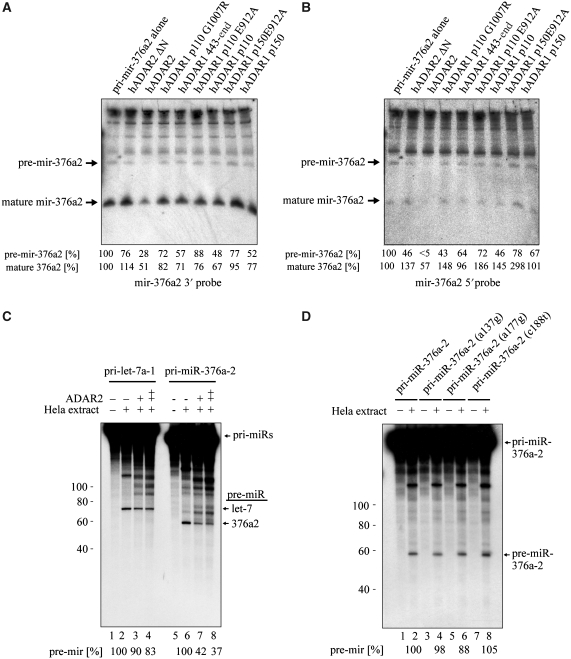
Figure 3.
Effects of ADARs on processing of pri-mir-376a2. (A, B) Effects of ADARs on processing in HEK 293T cells. Small RNA northern on RNA from cotransfected HEK 293T cells probed with antisense probes to (A) the 3′ miRNA and (B) the 5′ miRNA. Nothern blots were also probed with a 5S rRNA-specific probe as a loading control (data not shown). ADAR2 expression reduced both the pre-miRNA and the mature 3′ and 5′ miRNA signals. The numbers below the blot indicate the amount of both pre-mir-376a2 or mature 376a2 signal in each lane relative to that in pri-mir-376a2 expressed alone. (C) Recombinant ADAR2 inhibits Drosha cleavage in HeLa extracts in vitro. Drosha cleavage assays performed in the presence of recombinant ADAR2 and either pri-let-7a-1 or pri-mir-376a2. A double plus in the ADAR2 table indicates that a higher amount of ADAR2 was added. Cleavage of pri-mir-376a2 but not pri-let-7a-1 was inhibited by ADAR2, indicating that ADAR2 directly and specifically effects Drosha processing of mir-376a2. The percentage of pre-miRNA and mature 376a2 signal relative to cleavage in the absence of ADAR2 is indicated below the blot. (D) Lack of effect of RNA editing base changes in pri-mir-376a2 on Drosha cleavage in HeLa extracts in vitro. Drosha cleavage was not impaired by point mutations mimicking RNA editing events in mir-376a2. a137g mimics editing of the +4 site. a177g mimics editing of the +44 site. c188t closes the A–C mismatch at the +4 site in pri-mir-376a2. The amount of pre-miRNA signal relative to wild-type pri-mir-376a2 cleavage is indicated below the blot.
By performing in vitro pri-miRNA processing assays with HeLa nuclear extract we found that the addition of purified ADAR2 specifically inhibited Drosha processing of pri-mir-376a2 but not pri-let-7a-1 (Figure 3C), though some pre-mir-376a2 is still produced. To determine whether Drosha cleavage in the HeLa nuclear extract is inhibited by the RNA editing base changes or by binding of ADAR2, in vitro transcribed mutant pri-mir-376a2 transcripts bearing adenosine to guanosine changes at the edited positions were assayed. Specifically, in mir-376a2 a137g, a guanosine replaced adenosine at the +4 position and in mir-376a2 a177g, a guanosine replaced adenosine at the +44 site that is the +6 position of the 3′ miR. Importantly, Drosha cleavage was not affected by either mutation (Figure 3D), clearly showing that the nucleotide changes mediated by editing are not themselves sufficient to inhibit the processing of mir-376a2. The c188t construct that removes the A-C mismatch at the +4 site also had no effect.
Catalytically inactive ADAR2 disrupts pri-mir-376a2 processing
To confirm that inhibition of Drosha cleavage is due to ADAR2 binding rather than because of the base change at the edited site an active site-mutant ADAR2 E319A was co-expressed. ADAR2 E319A retained the ability to reduce the silencing activity of unedited 5′ miR but, as expected, it was unable to retarget the silencing activity (Figure 4A). Furthermore, ADAR2 E319A caused inhibition of the processing of pri-miRNA-376a2, as detected on small RNA northerns with RNA from cotransfected cells (Figure 4B). ADAR2 E319A disruption of Drosha processing verifies the conclusion that ADAR2 interference with miRNA processing is independent of editing. ADAR2 ΔN E319A, a deaminase domain mutant localized to the cytoplasm, did not inhibit 3′ miR activity and only weakly affected 5′ miR activity (Figure 4A). This shows further that it is nuclear Drosha cleavage that is affected and not cytoplasmic Dicer activity.
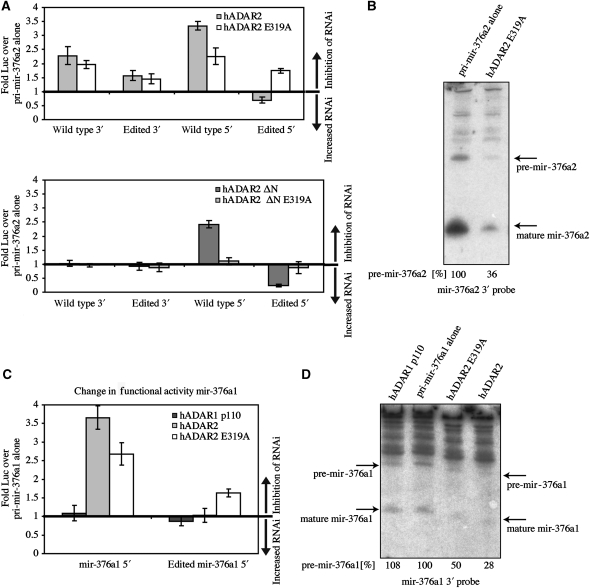
Figure 4.
Deaminase-inactive ADAR2 fails to retarget miRNAs but still inhibits their processing. (A) Top panel: effects of normal versus deaminase mutant ADAR2 on production of mature unedited and edited miRNA activities. Inactive ADAR2 E319A retained the inhibitory effect of ADAR2 on the unedited 5′ miRNA activity but was unable to retarget the activity by editing. Bottom panel: effects of cytoplasmic ADAR2 ΔN and ADAR2 ΔN E319A on the activity of mir-376a2. ADAR2 ΔN E319A did not affect the 5′ reporter, indicating a loss in retargeting. Neither hADAR2 ΔN nor hADAR2 ΔN deaminase mutant affected the 3′ mature miRNA activity. (B) Effect of ADAR2 E319A on processing of mir-376a2 in cotransfected HEK 293T cells. Small RNA northerns of RNA from cotransfected cells probed with antisense probes to the 3′ miRNA. Blots were also probed with a 5S rRNA-specific probe as a loading control (data not shown). Deaminase mutant ADAR2 E319A expression reduced both pre-miRNA and mature miRNA levels. Percentage of pre-miRNA signal relative to pri-mir-376a2 expressed alone is noted below the blot. (C) Effects of main isoforms of ADAR1 and ADAR2 on functional activities of 5′ and 3′ miRNAs expressed from a construct bearing mir-376a1 alone. As in the case of mir-376a2, ADAR2 was able to inhibit the activity of mir-376a1 independently of deaminase activity. (D) The ADAR2 E319A deaminase domain mutant reduced the level of pre-mir-376a1 in cotransfected HEK 293T cells. Small RNA northern blot of RNA from transfected cells probed with antisense probe to the 3′ miRNA. The percentage of pre-mir-376a1 signal relative to pri-mir-376a1 expressed alone is listed below the blot.
Another pri-miRNA within the cluster is edited by ADAR2. We wanted to determine whether binding of ADAR2 to pri-mir-376a1 also affected processing. Similar to pri-mir-376a2, expression of pri-mir-376a1 alone showed that its activity was inhibited by ADAR2 E319A (Figure 4C). In addition, a northern analysis was performed to determine the processing of pri-mir-376a1 (Figure 4D). It was affected by the binding of either active or inactive ADAR2 but not by ADAR1 (Figure 4D). This shows that inhibition of Drosha cleavage by the ADAR2 deaminase-inactive mutant is not restricted to pri-mir-376a2.
Human ADAR1 protein expressed in Drosophila inhibits RNAi through a mechanism that is partially independent of RNA editing activity
We generated a series of Drosophila strains that express different human ADAR proteins using the GAL4/UAS binary expression system. Interactions between expressed human ADARs and the RNAi system were examined by crossing these UAS-ADAR-bearing strains to a fly strain in which silencing of the wild-type white+ gene in the eye by a cytoplasmic white RNA hairpin construct is also directed by the GAL4/UAS binary expression system.
Silencing of the white+ gene in the eye is directed by expressing a UAS-white hairpin construct under the control of the eye-specific GMR-GAL4 driver (Kalidas and Smith, 2002). Silencing of white+ results in flies with white eyes and this has been used successfully in a screen to find genes that antagonize the siRNA pathway (Lee and Carthew, 2003). The white hairpin primary transcript contains an intron and has been designed to be efficiently processed, polyadenylated and exported to the cytoplasm. Silencing is mediated by the Dicer processing of the resulting cytoplasmic RNA hairpin, which is over 300-nucleotides long (Vagin et al, 2006). After Dicer cleavage, the white hairpin-derived siRNA targets the white+ mRNA for silencing by RISC.
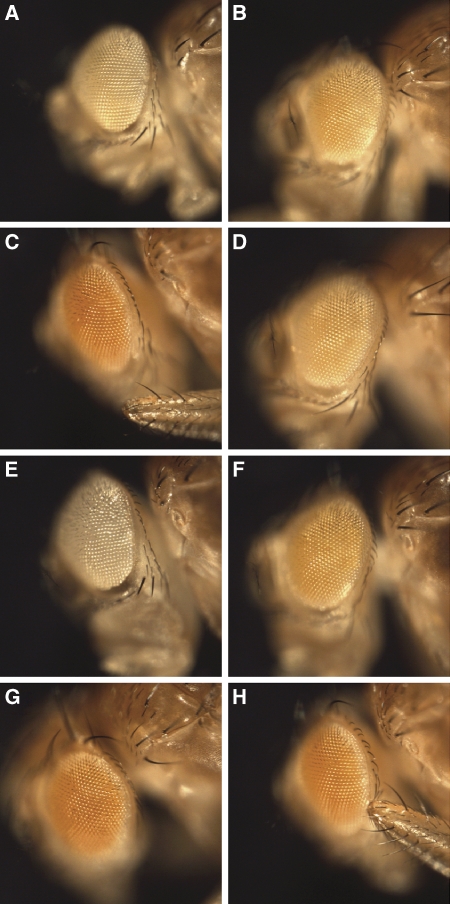
When strains containing UAS-ADAR are crossed with the silencing reporter strain (homozygous w+; GMR>wIR), ADAR is co-expressed with the white hairpin in the eye. The interaction of ADAR with RNAi is observed as an alteration in the silenced white eye colour. The strongest effect of a human ADAR on white hairpin-mediated silencing was caused by ADAR1 p150, the shuttling isoform of ADAR1 that accumulates in cytoplasm (Figure 5C). This ADAR1 isoform antagonized RNAi strongly to restore the eye colour to red by allowing expression of the intact wild-type white+ gene in the strain background. This effect on the eye colour was not variegated but was absolutely consistent across the eyes and between flies.
Figure 5.
Human ADAR1 antagonizes white hairpin-mediated RNA interference in Drosophila. Effects of human ADAR proteins on silencing of white+ gene expression in Drosophila eyes by a co-expressed cytoplasmic white RNA hairpin. These flies are female progeny of homozygous w+; GMR-GAL4, UAS-whiteIR (GMR>wIR) crossed to w1118; UAS construct lines. (A) w+; GMR>wIR alone, (B) ADAR p110 (line 19 on Chr III), (C) ADAR p150 (line 25 on Chr III), (D) ADAR2 (line 22 on Chr II), (E) Dicer-2 (line D2 on Chr II), (F) Dicer-2 (line D2) and ADAR1 p150 (line 25), (G) ADAR1 p150 E319A (line 34 on Chr III), (H) ADAR1 p150 E319A (line 35 on Chr II).
Neither nuclear ADAR1 p110 nor ADAR2 had any effect on white hairpin-mediated silencing (Figure 5B and D). The human ADARs are expressed in addition to the normal level of endogenous Drosophila ADAR in the flies shown here. Drosophila ADAR, which is also nuclear, behaved like ADAR2 in this assay: neither increasing the expression with a UAS-dADAR construct nor deleting the single endogenous Drosophila ADAR gene on the X chromosome affected silencing (data not shown). When editing of the white hairpin over a stretch of several hundred base pairs was analysed in these flies there was some editing even in the absence of any human ADAR that is presumably because of the endogenous fly protein (Supplementary Figure S5A). Of the human proteins only the cytoplasmic ADAR1 p150 showed a substantially increased level of editing. This suggests that cytoplasmic localization is required for efficient editing and for full inhibition of siRNA activity in this assay.
Combining UAS-Dicer-2 with UAS-ADAR1 p150 in crosses to the homozygous w+; GMR>wIR strain showed that increasing DICER-2 expression above normal levels increases the background level of RNAi in the absence of ADAR1p150 in the progeny (Figure 5, compare panels E and A) and also partly but not completely suppresses the antagonism of RNAi by ADAR1 (Figure 5, compare panels C and F). This suggests that the antagonism of RNAi by ADAR1p150 is partly attributable to competition with DICER-2 for long dsRNA, although ADAR1p150 does also bind the shorter dsRNA products of DICER cleavage (Yang et al, 2005) and changing dosages of R2D2 and ARGONAUTE-2 might have similar effects.
The visual assay is very consistent, especially because groups of ADAR-expressing flies can be compared with sibling progeny of the same age, which have white silencing but lack ADAR. This gives sensitivity to the assay and testing further ADAR1 derivatives shows that all the domains of ADAR1, including the Z-DNA-binding domains, have perceptible effects on RNAi in this assay. The truncation mutant, ADAR1 1-442, containing two Z-DNA-binding domains showed an effect in this assay and removing the Z-DNA-binding domains in the ADAR1 443-end construct weakened the inhibition (data not shown).
When human ADAR1 p150EA, the deaminase-inactive mutant, was expressed in this system it was found to still antagonize the RNAi activity of the white hairpin (Figure 5G and H). Again the effect was very consistent between flies. The red eye colour was 40% as strong as ADAR1 p150, based on quantization of red pigment in ethanol-extracted heads and in digital images of eyes (Supplementary Figure S5B). This result suggests that, as in the case of miRNA processing, a significant part of the antagonism is independent of deaminase activity. Very surprisingly, expressing the ADAR2 ΔN protein that lacks the nuclear localization signal increased the antagonism by ADAR2 only very slightly (not shown). This suggests that the specificity of the different ADARs is also important for this effect.
Discussion
Earlier work on the interaction between ADARs and the miRNA/siRNA pathways in mammals has focused on the effects of the base changes introduced by RNA editing. This editing was shown to affect microRNA processing or produce retargeting (Scadden and Smith, 2001; Luciano et al, 2004; Scadden, 2005; Yang et al, 2005, 2006; Blow et al, 2006; Landgraf et al, 2007; Kawahara et al, 2007a, 2007b). Our data show, for the first time, that catalytically inactive ADAR2 can inhibit the processing and decrease the downstream function of miRNA, independent of base deamination. In addition, we show that deaminase-inactive ADAR1 p150 inhibits siRNA activity in a Drosophila test system. ADAR1 p150 also inhibits siRNA in ADAR1−/− MEFs (Yang et al, 2005). As ADARs may bind to a wider range of pre-miRNAs than they successfully edit, the influence of ADARs on miRNA function may also extend beyond the subset of miRNA, which are edited. Modulation of miRNA biogenesis by binding Lin-28 protein (Newman et al, 2008; Piskounova et al, 2008) or hnRNP A1 (Guil and Caceres, 2007; Michlewski et al, 2008) to miRNA precursors has also been reported recently. Therefore, regulation of pri-miRNA processing by accessory RNA-binding proteins may be more common than appreciated earlier.
There is specificity in how different ADARs affect different RNAi phenomena as only ADAR2 robustly affects the processing of pri-mir-376a2. Despite overexpressing other ADAR proteins with dsRNA-binding domains, they did not radically inhibit processing of this pri-miRNA by Drosha. What is really surprising is that ADAR1 binds to and edits the +4 position in mir-376a2 to between 40 and 50% yet its affect on Drosha processing is not as strong as ADAR2 (Figure 3). The +4 editing site is a mere three base pairs from the Drosha cleavage site.
The specificity of ADAR effects is also clear in relation to Dicer processing. It was recently shown that pre-mir-151 could be edited by ADAR1 p110 and ADAR p150 in vitro, and that this editing led to disruption of Dicer processing (Kawahara et al, 2007a). However, cytoplasmic ADAR2 ΔN did not result in disruption of Dicer processing of the pre-mir-376a2 in this study. Neither was there degradation of miRNA duplexes by Tudor-SN but instead there was retargeting of 5′ miRNA activity by ADAR2 ΔN because of editing. In addition, ADAR1 p150 did not have a major impact on mir-376a2 processing. Therefore, not all editing events in pre-miRNA in the cytoplasm necessarily disrupt Dicer processing just as not all editing events in pri-miRNA in the nucleus affect Drosha cleavage.
We found that single nucleotide changes in miRNA target sites impair the ability of a particular miRNA to target the mRNA. Several publications have documented the decrease in the ability of miRNA to silence an mRNA when there are nucleotide changes in the miRNA responsive site (reviewed in Tomari and Zamore, 2005). It seems that siRNA activity is even more sensitive to nucleotide changes. Therefore, editing may have a more dramatic effect on silencing of a transcript if it occurs within a miRNA responsive site in the 3′UTRs of specific mRNAs rather than if the miRNA itself is edited. This may be important in some species like Caenorhabditis elegans where there is significant editing of 3′UTRs.
We have shown here that one DSH1-associated variant of ADAR1 (Miyamura et al, 2003; Zhang et al, 2004, 2008; Liu et al, 2006) produces an ADAR1 protein that binds dsRNA with a similar Kd to wild type but is catalytically inactive (Supplementary Figure S1). Members of the family having this variant show the same dermatological symptoms of DSH1, which are associated with protein truncations of ADAR1 and that probably reflect decreased RNA editing activity in other families. However, affected members of the Japanese family having the ADAR1 G1009R allele also show additional symptoms including progressive neuromotor defects and calcium accumulation in basal ganglia. On the basis of our demonstration of efficient RNA binding by this protein we suggest that these additional symptoms arise because ADAR1 G1007R partially antagonizes normal RNA editing. Inhibition of mir-376 cluster processing in the brain may be relevant but the mutant protein would also probably bind RNA editing target sites such as the Q/R editing site in the GluR-B transcript and reduce editing by ADAR2 and ADAR1 at this site. This would lead to calcium-permeable AMPA receptors and possibly contribute to the brain calcium accumulations and to the slowly progressing neurological symptoms in this family.
In conclusion, we report a number of different and very selective effects of ADARs on RNAi processes that appear to depend significantly on the RNA-binding activities of ADARs. It has been shown earlier that ADARs can bind to many transcripts without editing them (Klaue et al, 2003). A biological function can now be ascribed to this binding activity. Binding of ADAR2 to pri-mir-376a2 is sufficient to inhibit processing by Drosha, an inactive form of ADAR1 p150 is sufficient to inhibit RNAi in a Drosophila assay, and disease symptoms in a Japanese family are probably explained by production of an ADAR1 protein that binds RNA but lacks catalytic activity. This implies a broader functional role for ADARs as RNA-binding proteins in different cellular compartments interacting with a range of dsRNA-based phenomena.
Materials and methods
Expression plasmids
miRNA-expressing plasmids contained mir-376 cluster genomic DNA were amplified with PCR primers and cloned using BamHI and HindIII restriction enzyme sites in the primers into the pcDNA3. 1 vector. All constructs had 76–100 nucleotides of genomic sequence on both the 5′ and 3′ sides of the encoded miRs depicted in Figure 1A (see Supplementary Table 1 for sequences of primers). Site-directed mutagenesis was performed to incorporate the a137g, a177g and c188t mutations into pri-mir-376a2.
Human ADAR-expressing plasmids, containing ADAR coding sequences with N-terminal Flag and C-terminal His epitope tags, were constructed by Gateway Cloning (Invitrogen) with pc3D as the destination vector. pc3D was derived by blunt-end ligation of the gateway cloning cassette-B from Invitrogen into pcDNA3.1. Therefore, the vector for all expression constructs was pcDNA3.1.
miRNA reporter plasmids
Target sites for each miRNA were subcloned into the 3′ UTR of Renilla luciferase in psicheck 2.0 (Promega) using the XhoI and NotI restriction enzyme sites. As a transfection control the vector also contains humanized Firefly luciferase under its own promoter. This is also a control for any general effects on translation. (see Supplementary Figure 3 for sequences of target sites).
Editing assays
RNA was purified from transiently transfected cell cultures and subjected to reverse transcription with random hexamers (Invitrogen) and MMLV-RnaseH (Invitrogen). Editing was determined by direct sequencing of PCR product pools with primers located substantially downstream and upstream of the relevant mir-hairpin, as performed by Blow et al (2006) and Kawahara et al (2007b). Primers used were originally described in Kawahara et al (2007b) and their sequences are listed in Supplementary Table 1. The primers are designed to amplify a sequence from the pri-miRNA before Drosha cleavage. Our notation in Figure 1 for the +4 and +44 site follows Kawahara et al, although new information in miRBASE suggests that the Drosha cleavage site is one base closer to the editing site.
In vitro RNA binding and editing
In vitro RNA-binding assay was performed as described by Ohman (Ohman et al, 2000). In vitro RNA editing assays was performed as described by O'Connell (O'Connell and Keller, 1994).
Cell culture
HEK 293T cells were cultured in DMEM supplemented with 10% FCS.
Transient transfections
Transient transfections were performed in 24-well plates seeded 1 day before transfection with 0.14 × 106 cells, with 1.005 μg DNA and 1.01 μl Lipofectamine 2000. The amounts of miRNA-expressing plasmid, ADAR-expressing plasmid and luciferase reporter target construct plasmid were 500, 500 and 5 ng, respectively. Cells were collected on the third day after the transfection.
Luciferase assays
The miRNA target plasmid, derived from psicheck 2.0, contains humanized Firefly and Renilla luciferase. The miRNA responsive site is in the 3′UTR of Renilla luciferase. Assays for Firefly and Renilla luciferase were performed with the Dual-Glo luciferase kit from Promega. Luciferase was measured on a Perkin-Elmer Victorlite. To ascertain the activity of mir-376a2, pri-mir-376a2 was co-expressed with the luciferase reporters so that the amount of DNA was constant. The resulting luciferase level was compared to that obtained in a cotransfection of pcDNA3.1 and the luciferase reporter without the pri-mir-376a2 expression vector. Fold Luc activity is the activity remaining in the presence of the miRNA.
Fold Luc=(luciferase activity with the pri-mir-376a2 construct)/(luciferase activity without the pri-mir-376a2 construct).
Assays on ADARs were performed by co-expressing the ADAR, pri-mir-376a2 and the appropriate luciferase reporter. The resulting luciferase level was compared to co-expression of ADAR, pcDNA3.1 and the appropriate reporter without pri-mir-376a2. For the ADAR constructs.
Fold Luc=(luciferase level with pri-mir-376a2 and ADAR construct)/(luciferase level without pri-mir-376a2 and with the relevant ADAR construct).
The activity was then normalized to the ratio obtained with pcDNA3.1 and the pri-miRNA expression construct alone. Fold Luc over pri-mir-376a2 alone (labelled on the y-axis in Figure 2) is the fold change in luciferase activity compared with expression of the pri-miRNA alone. This indicates how the ability of the microRNAs to direct silencing of luciferase activity from the different reporters has been affected by the presence of ADAR proteins. A value greater than 1 indicates inhibition of the miRNA activity, a value less than 1 indicates increased miRNA activity.
Small RNA northerns
RNA samples were electrophoresed on a denaturing 12% PAGE 8 M urea gel. The RNA was then transferred to a Nylon+ membrane (Amersham) and hybridized with the appropriate probe. Radioactive probes that were internally labelled were made with the Mirvana probe construction kit (Ambion) with the primers listed in Supplementary Table 1. The probes used in the northerns were antisense to either mature mir-376a2 5′ miR, mature mir-376a2 3′ miR or to the 5S rRNA. The sizes of the resulting bands were verified by electrophoresis next to the Ambion decade marker. The northerns were visualized with a phosphoimager and their density was measured. All experiments were performed in triplicate. Probe for the loop specific primer, used in Supplementary Figure S4, is GTTACGTGTTTGATGGTTAATCCTGTCTC.
In vitro Drosha cleavage assays
Drosha cleavage assays were performed as described earlier (Guil and Caceres, 2007; Michlewski et al, 2008). pri-miRNA-376a2, which was transcribed in vitro, was added to HeLa extracts competent for Drosha cleavage. After incubation at 37°C, reactions were stopped and electrophoresed on a denaturing PAGE gel.
For Drosha cleavage assays in the presence of hADAR2, the recombinant hADAR2 had been expressed and purified from P. pastoris (Ring et al, 2004).
Drosophila transgenic lines
ADAR coding sequences were cloned in the pUAST plasmid bearing GAL4-binding sites. Constructs were injected in Drosophila eggs and transgenic lines were obtained and balanced by standard methods. Female flies bearing UAS-ADAR constructs on chromosomes II or III were crossed to males homozygous for both the GMR-GAL4 and UAS-whiteIR constructs (w+/Y; ; GMR-GAL4, UAS-wIR/ GMR-GAL4, UAS-wIR). Levels of red pigment in the eyes of female progeny flies in the RNAi assay were quantitated by ethanol extraction of batches of 50 heads followed by optical absorption measurements (Pal-Bhadra et al, 2004). The maximal level of red pigment in the presence of UAS-ADAR1 p150 is 12% of the wild-type level.
The images in Figure 5 were made with an imaging system comprising a Nikon AZ100 macroscope with 2 × plan fluor objective at 3 × zoom setting (Nikon UK Ltd, Kingston-on-Thames, UK), a Qimaging Micropublisher 5 cooled colour camera (Qimaging, Burnaby, BC, Canada). Image capture and control of the filterwheel and image analysis were performed using IPLab Spectrum (Scanalytics Corp, Fairfax, VA). White balance for the illumination level used was set on a white background, and all images were captured in a single session using identical capture settings.
The data for Supplementary Figure 5B were imaged on a Leica MZFLIII stereo-microscope with 0.5 × , 0.63 × , 1 × and 1.6 × objectives. Images were captured with a Hamamatsu Orca AG CCD camera (Hamamatsu Photonics (UK) Ltd, Welwyn Gardens City, UK) and CRI liquid crystal RGB filter (Cambridge Research & Instrumentation, Woburn, MA). Image capture and analysis were performed with in-house scripts written for IPLab Spectrum (Scanalytics Corp). The pixel intensity was measured on both eyes from 10 different flies from each genotype.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure Legends
Supplementary Table 1
Review Process File
Acknowledgments
We thank David W Lazinski for the gift of an ADAR2 ΔN expression construct, David Finnegan, Dean Smith, and the Bloomington Drosophila Stock Center for fly strains. In addition, we thank Roberto Marcucci for recombinant ADAR2, Marion Hogg for discussion, Anne Leroy for sequencing white hairpin, Paul Perry and Matthew Pearson for assistance with photography and Craig Nicol for assistance with figures. MO'C is supported by core funding from the Medical Research Council (U.1275.01.005.00001.01). BH is supported by a Fellowship from the Marie Curie Foundation (PIIF-GA-2008-220317).
Footnotes
The authors declare that they have no conflict of interest.
References
- Beghini A, Ripamonti CB, Peterlongo P, Roversi G, Cairoli R, Morra E, Larizza L (2000) RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 9: 2297–2304 [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR (2006) RNA editing of human microRNAs. Genome Biol 7: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A (2008) Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 283: 7251–7260 [DOI] [PubMed] [Google Scholar]
- Connolly CM, Dearth AT, Braun RE (2005) Disruption of murine Tenr results in teratospermia and male infertility. Dev Biol 278: 13–21 [DOI] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M (2003) Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci 116: 1805–1818 [DOI] [PubMed] [Google Scholar]
- George CX, Samuel CE (1999) Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 229: 203–213 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Guil S, Caceres JF (2007) The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol 14: 591–596 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH (2004) Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279: 4894–4902 [DOI] [PubMed] [Google Scholar]
- Hartwig D, Schoeneich L, Greeve J, Schutte C, Dorn I, Kirchner H, Hennig H (2004) Interferon-alpha stimulation of liver cells enhances hepatitis delta virus RNA editing in early infection. J Hepatol 41: 667–672 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP (2002) Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron 33: 177–184 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K (2007a) RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K (2007b) Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaue Y, Kallman AM, Bonin M, Nellen W, Ohman M (2003) Biochemical analysis and scanning force microscopy reveal productive and nonproductive ADAR2 binding to RNA substrates. RNA 9: 839–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeris M, Funke L, Shrestha J, Rich A, Maas S (2005) Modulation of ADAR1 editing activity by Z-RNA in vitro. Nucleic Acids Res 33: 5362–5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW (2003) Making a better RNAi vector for Drosophila: use of intron spacers. Methods 30: 322–329 [DOI] [PubMed] [Google Scholar]
- Liu Q, Jiang L, Liu WL, Kang XJ, Ao Y, Sun M, Luo Y, Song Y, Lo WH, Zhang X (2006) Two novel mutations and evidence for haploinsufficiency of the ADAR gene in dyschromatosis symmetrica hereditaria. Br J Dermatol 154: 636–642 [DOI] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S (2004) RNA editing of a miRNA precursor. RNA 10: 1174–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA 98: 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL (2005) Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH (1996) RED2, a brain specific member of the RNA-specific adenosine deaminase family. J Biol Chem 271: 31795–31798 [DOI] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Semple CA, Caceres JF (2008) Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol Cell 32: 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, Tomita Y (2003) Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet 73: 693–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MA, Keller W (1994) Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc Natl Acad Sci USA 91: 10596–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DB, Kim YG, Rich A (2002) Z-DNA-binding proteins can act as potent effectors of gene expression in vivo. Proc Natl Acad Sci USA 99: 16666–16671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman M (2007) A-to-I editing challenger or ally to the microRNA process. Biochimie 89: 1171–1176 [DOI] [PubMed] [Google Scholar]
- Ohman M, Kallman AM, Bass BL (2000) In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA 6: 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC (2004) Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA (2000a) dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6: 1004–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MJ, Keegan LP, O'Connell MA, Reenan RA (2000b) A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 102: 437–449 [DOI] [PubMed] [Google Scholar]
- Patterson JB, Thomis DC, Hans SL, Samuel CE (1995) Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology 210: 508–511 [DOI] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, Krupsky M, Ben-Dov I, Cazacu S, Mikkelsen T, Brodie C, Eisenberg E, Rechavi G (2007) Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 17: 1586–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283: 21310–21314 [DOI] [PubMed] [Google Scholar]
- Placido D, Brown BA II, Lowenhaupt K, Rich A, Athanasiadis A (2007) A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure 15: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring GM, O'Connell MA, Keegan LP (2004) Purification and assay of recombinant ADAR proteins expressed in the yeast Pichia pastoris or in Escherichia coli. Methods Mol Biol 265: 219–238 [DOI] [PubMed] [Google Scholar]
- Scadden AD (2005) The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 12: 489–496 [DOI] [PubMed] [Google Scholar]
- Scadden AD, O'Connell MA (2005) Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites. Nucleic Acids Res 33: 5954–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD, Smith CW (2001) RNAi is antagonized by A-->I hyper-editing. EMBO Rep 2: 1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehblow A, Hallegger M, Jantsch MF (2002) Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell 13: 3822–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yoshimoto T, Shimoda M, Kono T, Koizumi M, Yazumi S, Shimada Y, Doi R, Chiba T, Kubo H (2006) Loss of function of the candidate tumor suppressor prox1 by RNA mutation in human cancer cells. Neoplasia 8: 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo K, Sekijima Y, Suzuki T, Suzuki N, Tomita Y, Yoshida K, Hashimoto T, Ikeda S (2006) Dystonia, mental deterioration, and dyschromatosis symmetrica hereditaria in a family with ADAR1 mutation. Mov Disord 21: 1510–1513 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19: 517–529 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K (2000) Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768 [DOI] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279: 4952–4961 [DOI] [PubMed] [Google Scholar]
- Wong SK, Sato S, Lazinski DW (2003) Elevated activity of the large form of ADAR1 in vivo: very efficient RNA editing occurs in the cytoplasm. RNA 9: 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K (2006) Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13: 13–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Wang Q, Howell KL, Lee JT, Cho DS, Murray JM, Nishikura K (2005) ADAR1 RNA deaminase limits short interfering RNA efficacy in mammalian cells. J Biol Chem 280: 3946–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Liu H, Jiang D, Tian H, Wang C, Yu L (2008) Six novel mutations of the ADAR1 gene in Chinese patients with dyschromatosis symmetrica hereditaria. J Dermatol Sci 50: 109–114 [DOI] [PubMed] [Google Scholar]
- Zhang XJ, He PP, Li M, He CD, Yan KL, Cui Y, Yang S, Zhang KY, Gao M, Chen JJ, Li CR, Jin L, Chen HD, Xu SJ, Huang W (2004) Seven novel mutations of the ADAR gene in Chinese families and sporadic patients with dyschromatosis symmetrica hereditaria (DSH). Hum Mutat 23: 629–630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure Legends
Supplementary Table 1
Review Process File