Abstract
Staphylococcus aureus is the leading cause of bacterial infections in developed countries and produces a wide spectrum of diseases, ranging from minor skin infections to fatal necrotizing pneumonia. Although S. aureus infections were historically treatable with common antibiotics, emergence of drug-resistant organisms is now a major concern. Methicillin-resistant S. aureus (MRSA) was endemic in hospitals by the late 1960s, but it appeared rapidly and unexpectedly in communities in the 1990s and is now prevalent worldwide. This Review focuses on progress made toward understanding the success of community-associated MRSA as a human pathogen, with an emphasis on genome-wide approaches and virulence determinants.
Introduction
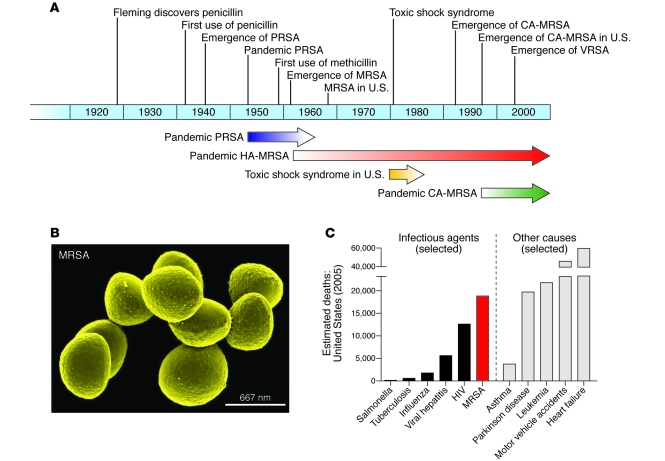
Staphylococcus aureus is the leading cause of bacterial infections involving the bloodstream, lower respiratory tract, and skin and soft tissue in many developed countries, including the United States (1). Penicillin was initially highly effective against staphylococcal infections, but penicillinase-producing S. aureus emerged in the mid-1940s, and its prevalence increased dramatically within a few years (2, 3) (Figure 1A). This increase was entirely attributable to widespread use of penicillin, which selected for bacteria containing resistance genes. Several “epidemic waves” of antibiotic-resistant S. aureus have occurred since then (Figure 1A). Penicillin-resistant S. aureus was pandemic in the 1950s and early 1960s (4). These infections, both in hospitals and in the community, were caused primarily by one S. aureus clone known as phage-type 80/81 (4). Pandemic phage-type 80/81 S. aureus infections declined after the introduction of methicillin in 1959 (5). Within 2 years, however, Jevons and colleagues reported the emergence of methicillin-resistant S. aureus (MRSA) (Figure 1B) (6), and the first cluster of cases was reported shortly thereafter (7).
Figure 1. Emergence of antibiotic-resistant S. aureus, epidemic waves, and estimated deaths caused by MRSA in the United States.
(A) Timeline indicates the year in which an event occurred or was reported. Arrows indicate approximate length of time for each pandemic/epidemic. (B) Scanning electron micrograph of strain COL, one of the first MRSA isolates from Colindale, United Kingdom. Original magnification, ×45,000. (C) Comparison of estimated deaths in the United States in 2005 due to individual infectious agents or other causes. Data are CDC estimates from National Vital Statistics Reports (12) and Klevens et al. (11). Deaths associated with MRSA infection are based on the estimated number of in-hospital deaths rather than attributable mortality, whereas data for all other causes of mortality are based on US Standard Certificate of Death. Note also that mortality due to MSSA is not included, and thus estimated mortality associated with all S. aureus infections is not shown.
MRSA has since spread and is endemic in most hospitals worldwide. In the United States, S. aureus is the number one cause of hospital-associated infections, and a high percentage of these are caused by MRSA (8–10). The mortality rate associated with invasive MRSA infections is approximately 20% (11), and in the United States these infections are probably the leading cause of death by any single infectious agent; fatalities resulting from these infections are estimated to surpass those caused by HIV/AIDS (Figure 1C) (11, 12). Thus, it is clear that S. aureus, especially MRSA, remains a major problem for health care facilities.
The rapid emergence of community-associated MRSA (CA-MRSA) infections (i.e., those that occur outside of health care facilities in otherwise healthy people) is one of the most surprising events in infectious diseases in recent years (13–17). This is in part because community-associated S. aureus infections were traditionally caused by methicillin-susceptible S. aureus (MSSA), rather than the antibiotic-resistant strains so prevalent in hospitals (13). Thus, it remains a mystery why these antibiotic-resistant bacteria emerged in a niche (the community) not obviously under the relatively high selective pressure exerted by antibiotics in the hospital setting. CA-MRSA was first reported in Western Australia in the 1990s (18) and has since emerged worldwide (19) and become epidemic in the United States (16, 17). In addition to the increased prevalence of infections, CA-MRSA strains can cause severe or fatal disease. In this Review, we discuss key components of the recent emergence of CA-MRSA, including transmission and virulence, and we highlight some of the genome-wide approaches used to understand the success of this pathogen. Due to space constraints, we focus mainly, but not exclusively, on the epidemic in the United States.
Recent emergence of CA-MRSA

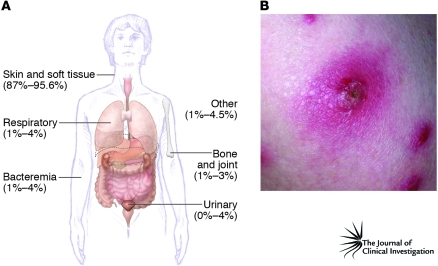
The molecular epidemiology and evolution of MRSA is a topic too expansive for this Review, but several key features of CA-MRSA strains merit mention. Based on molecular typing and microarray approaches, S. aureus can be grouped broadly into at least 10 major lineages, named clonal complexes (Table 1) (20–22). Pulsed-field gel electrophoresis is commonly used to identify S. aureus isolates in the United States, and for simplicity, here we use this typing system to describe S. aureus strains (Sidebar 1) (23). In 2003, pulsed-field gel electrophoresis types USA100, USA200, USA500, USA600, and USA800 were the predominant health care–associated strains, whereas USA300 and USA400 were the most abundant CA-MRSA strains (23). MRSA strains can be characterized further by at least five distinct staphylococcal cassette chromosome mec (SCCmec) types (I–V), mobile genetic elements that encode methicillin resistance. Health care–associated MRSA strains historically contain predominantly SCCmecI–III, whereas CA-MRSA typically carries SCCmecIV (24, 25). SCCmecIV, which is smaller than SCCmecI–III, lacks other antibiotic resistance elements and appears to impart little or no fitness cost to the organism (24). Consistent with this idea, CA-MRSA strains in general have faster growth rates than health care–associated MRSA strains (24). In addition to SCCmecIV, CA-MRSA strains carry genes encoding a toxin known as Panton-Valentine leukocidin (PVL, discussed below), and both are often used as markers for CA-MRSA.
Table 1 .
Major clonal complexes of S. aureus
CA-MRSA in the clinic
Clinical epidemiology and disease.
CA-MRSA infections appear to have emerged first among indigenous people of the remote Kimberley region of Western Australia in the early 1990s (18, 26). The first well-documented cases of CA-MRSA infection in the United States occurred in four otherwise healthy children from rural Minnesota and North Dakota in 1997–1999 (15). Lacking any risk factors for MRSA, e.g., contact with a health care facility, they presented with severe infections, septic arthritis, bacteremia and septic shock, and necrotizing pneumonia. A retrospective study performed in Chicago suggested an earlier beginning (1993–1995) of the CA-MRSA epidemic in the United States (14). CA-MRSA has subsequently been identified in numerous populations, including high school, college, and professional athletes; prison inmates; men who have sex with men; military personnel; and children in day care centers (17, 27–32). CA-MRSA strains are so prevalent in the United States (17, 33) and abroad (19) (Table 1), and such an important cause of hospital infections (34, 35), that no group can be considered not at risk. This idea is underscored by the surprising finding that most MRSA clinical isolates in Denmark, a country of very low MRSA prevalence, were CA-MRSA isolates (36).
The CA-MRSA epidemic in the United States is actually two sequential, overlapping epidemics. USA400 — the original CA-MRSA strain commonly known as MW2 — predominated prior to 2001 (15). This strain may be a descendant of an early Western Australian CA-MRSA strain (WA-1) (Table 1). Although USA400 is still an important cause of community-associated disease in some regions (e.g., Alaska and the Pacific Northwest), USA300 is now responsible for most CA-MRSA infections in the United States (17, 33). Importantly, the complete genomes of USA300 and USA400 have been sequenced, and these data have been fundamental for approaches directed toward better understanding the epidemiology and virulence of CA-MRSA infections and developing diagnostic tools (see below).
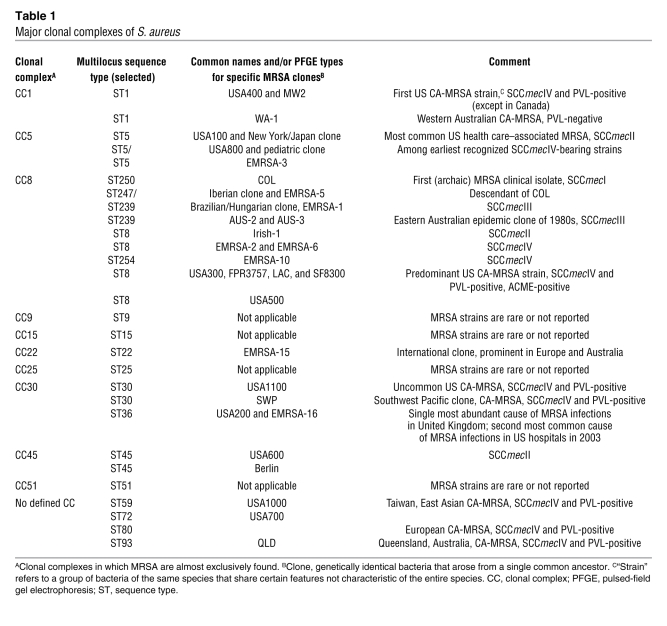
The emergence of CA-MRSA has led to an increase in the overall burden of staphylococcal disease (37, 38). Skin and soft tissue infections, the most common types of CA-MRSA infection, constitute approximately 90% of cases (Figure 2). However, CA-MRSA strains can cause virtually any infection, ranging from mild skin infections to fatal necrotizing pneumonia (Figure 2) (14, 16, 37, 39–48).
Figure 2. CA-MRSA infections.
(A) Distribution of all CA-MRSA infections (indicated as a percentage) (16, 34, 37, 139). Studies by Kaplan et al. (37) and Purcell et al. (139) reported distribution in children. (B) Human CA-MRSA skin infection with a typical “spider bite” appearance. Photo credit: Kelly Cardoro, Department of Dermatology, UCSF.
Approximately a third of healthy noninstitionalized humans are asymptomatically colonized by S. aureus (49, 50), and thus it can be considered normal flora. In this respect, CA-MRSA should be no different from other S. aureus strains. However, most individuals are colonized by MSSA rather than MRSA (28.6% were colonized with S. aureus in the United States in 2003–2004, of which 1.5% were colonized with MRSA), and only a fraction (15.6%) of these MRSA isolates are USA300 or USA400 (50). It should be noted, however, that this represents a doubling of the MRSA nasal colonization rate compared with 2001–2002 (before USA300 became so prevalent) and that much of this increase is attributable to USA300 and USA400 strains, which doubled in percentage of all isolates. Nevertheless, given the high prevalence of CA-MRSA infections in the United States, these findings suggest that CA-MRSA strains cause infection in the absence of nasal colonization, a notion that merits further investigation. CA-MRSA strains may be more easily transmitted than other stains for reasons that are not known (51). Skin-to-skin contact with a colonized or infected individual is the predominant mode of transmission, although contaminated fomites (e.g., towels and razors) may play a role in some settings (33, 52, 53). The CDC has articulated “Five Cs” of transmission: first, contact with an infected or colonized person; second, cleanliness (actually, a lack thereof); third, compromised skin integrity; fourth, contaminated objects, surfaces, and items (e.g., towels and sauna benches); and fifth, crowded living conditions. Infection occurs when S. aureus, either as a component of the resident flora or when introduced from the outside by a contaminated agent, exploits a breach in host defenses, producing local infection, invasive disease, or both. Being from an ethnic minority, young age (in particular, less than 2 years of age), low socioeconomic status, and drug use (especially injection drug use) are risk factors for CA-MRSA infection (16, 27, 32, 42, 54–58).
Diagnosis, treatment, and prevention.
Diagnosis of S. aureus infection is easily made by culturing organisms from a focal site of infection, blood, or other sterile body fluid. Methicillin resistance is determined by standard antimicrobial susceptibility test methods, including disk diffusion, broth dilution, and automated methods. Molecular typing to determine clonal type (23, 59–61) (Sidebar 1), presence of SCCmecIV (62), and/or presence of PVL genes (19) are useful for differentiating between CA-MRSA and classical health care–associated MRSA clones, but these methods are relatively labor intensive for routine use and of unproven clinical benefit.
The CA-MRSA epidemic has had a profound impact on empirical therapy of suspected staphylococcal infection. Given the prevalence of CA-MRSA throughout the United States, if antimicrobial therapy is indicated for treatment of staphylococcal infection, it should be active against MRSA strains. Unfortunately, this requires the use of second-line agents, such as trimethoprim-sulfamethoxazole and clindamycin, for which efficacy is not well defined, or vancomycin. Despite its being a drug of choice, there are major issues with the use of vancomycin, including problems with prolonged, persistent, and/or recurrent bacteremia during therapy (63), high rates of microbiological and clinical failure (64), nephrotoxicity (65), and increasing prevalence of nonsusceptible strains (66).
S. aureus genomics
CA-MRSA emerged coincident with completion of the first whole genome sequences of bacterial pathogens. There are at least 14 published S. aureus genomes, and reports of more are on the way. The first genomes of S. aureus to be sequenced were the genomes of MRSA strains N315 and Mu50, each of which is a pulsed-field gel electrophoresis type USA100 strain (67). Mu50 is also a vancomycin-resistant S. aureus (VRSA) strain. These studies, which were conducted by Kuroda et al. to better understand virulence and multidrug resistance, revealed that most of the antibiotic resistance genes and many of the virulence genes reside on mobile genetic elements, such as plasmids, transposons, and prophages, indicating substantial horizontal gene transfer from other bacteria (67). This work was followed by the completion of the genome sequences of several key S. aureus clinical isolates, including MW2 and MSSA476 (USA400 strains) (68, 69), MRSA252 (a USA200 strain) (69), and FPR3747 (a USA300 strain) (70). MW2 was found to be a distant relative of the previously sequenced USA100 strains and, surprisingly, to show PBP2a resistance only to β-lactam antibiotics (antibiotics that contain a β-lactam ring in their molecular structure) (68). This resistance was conferred by blaZ (which encodes a penicillinase) and mecA (which encodes PBP2a, a low-affinity penicillin-binding protein that mediates resistance to β-lactam antibiotics such as methicillin). The novel SCCmec element identified within MW2, SCCmecIVa, is much smaller than the SCCmec element found in USA100 strains and other hospital-associated MRSA clones (69) and appears to impart no fitness cost to the organism (24). Another major finding from the MW2 genome sequence was the presence of novel virulence determinants encoded by prophages, including the genes encoding PVL and the enterotoxin G and K homologs. However, the relative contribution of these molecules to human disease remains undetermined. In addition to identifying novel virulence and resistance determinants, the availability of complete S. aureus genome sequences served as a springboard for comparative genomics analyses and expression microarray studies.
Whole genome sequencing as a tool to understand the evolution of antibiotic resistance.
VRSA strains are rare (there are 7 reported isolates from the United States) (71), but vancomycin-intermediate resistant S. aureus (VISA) are an emerging problem in the major S. aureus clones worldwide (66). A key difference between VRSA and VISA is the minimum inhibitory concentration of vancomycin required to inhibit growth in vitro (≥16 μg/ml for VRSA and 4–8 μg/ml for VISA). The mechanisms for resistance are also quite distinct: VRSA isolates carry resistance on a specific mobile genetic element, whereas VISA isolates acquire resistance as a gradual adaptive process, and the molecular genetic basis for resistance was unknown until recently (72). Mwangi et al. used whole-genome sequencing to follow the evolution of the VISA phenotype in S. aureus isolates recovered from the same patient over time (72). These researchers sequenced an S. aureus isolate at the start of vancomycin therapy that was fully susceptible to vancomycin and then sequenced a nonsusceptible isolate recovered 3 months later. This interesting approach identified 35 point mutations in S. aureus that conferred a VISA phenotype and defined loci critical for multidrug resistance (72). These studies provide an illustration of how whole genome sequencing can be used to elucidate the molecular genetic basis of emerging antibiotic resistance.
Using microarray-based approaches to understand bacterial epidemics.
There is not enough space here to highlight all the comparative genomics analyses of S. aureus performed to date (22, 73–75). However, the study by Fitzgerald et al. was the first to use a comparative microarray approach to study the evolution of MRSA (76). This work demonstrated that the mecA gene was acquired by distinct S. aureus genotypes multiple times, indicating that MRSA has evolved from nonresistant S. aureus strains independently on several occasions, a finding consistent with more recent work (77). Importantly, these studies revealed that the epidemic of toxic shock syndrome in the 1970s and 1980s (Figure 1A) was caused by a change in host environment rather than an emerging hypervirulent strain of S. aureus (76). Although the outbreak of toxic shock syndrome is unrelated to the current problem with CA-MRSA, the studies demonstrate how genome-wide approaches can be used to investigate S. aureus outbreaks/epidemics.
The first whole genome sequence of USA300.
To better understand epidemic CA-MRSA in the United States, Diep et al. determined the first complete genome sequence of a USA300 strain in 2006 (70). It is much more closely related to S. aureus strain COL, one of the first MRSA clinical isolates, than it is to many of the other prominent contemporary MRSA strains, including the USA400 prototype CA-MRSA strain (70, 78). For example, the core genomes (excluding mobile genetic elements) of USA300 and strain COL differ by only 678 SNPs. By comparison, USA300 differs from the sequenced USA100 and USA200 strains by 21,313 and 44,964 SNPs, respectively (70). This finding is remarkable, because COL has limited virulence in mice (73) and is not a prominent cause of human disease, whereas USA300 is rapidly lethal in mice (73, 79–82) and is the leading cause of CA-MRSA infections in the United States (16, 17). However, although the core genomes of COL and USA300 are similar, there are differences in the mobile genetic elements that they harbor (70). COL carries SCCmecI, whereas USA300 carries SCCmecIV, the same SCCmec type as MW2. Immediately adjacent to SCCmecIV in USA300 is a novel mobile genetic element that is known as type I arginine catabolic mobile element (ACME) (70). ACME is present in most (more than 99%) PVL-positive USA300 clinical isolates but not in MW2 or the published genomes of any other S. aureus strains. Three other regions of difference between COL and USA300 correspond to prophages, and two of these mobile genetic elements are of interest here. Prophage ϕSa2 contains the PVL-encoding genes lukS-PV and lukF-PV, which are epidemiologically associated with emergence of CA-MRSA. Prophage ϕSa3 encodes staphylokinase and two modulators of the human innate immune system, staphylococcal complement inhibitor (SCIN) and S. aureus chemotaxis inhibitory protein (CHIPS) (83). Thus, both ϕSa3 and ϕSa2 carry genes that could contribute to the hypervirulence of USA300. S. aureus pathogenicity island (SaPI) 5 in USA300 is similar to SaPI3 of COL (84). Although SaPI5 lacks the toxic shock toxin gene (tst), it carries seq and sek, genes that encode two novel pyrogenic toxin superantigens, enterotoxin Q and K, respectively; these are also present in the MW2 genome. Acquisition of seq and sek is epidemiologically associated with emergence of USA300 and related strains (85), and therefore SaPI5 is of interest in our efforts to understand the success of USA300.
Clonal expansion of USA300: identifying the epidemic USA300 clone.
Our understanding of emerging pathogens and bacterial epidemics has traditionally relied on approaches that index genomic variation at a relatively limited number of nucleotides. Although significant progress has been made using these methods, whole genome analysis of strains within a species is essential to fully understand key aspects of pathogen success, including emergence, virulence, and epidemics. To determine whether USA300 arose from a recent common ancestor or by convergent evolution toward a hypervirulent phenotype, Kennedy et al. performed comparative whole genome sequencing of ten USA300 isolates recovered from patients hospitalized in several regions across the United States (80). Whole genome sequencing and comparative whole genome sequencing identifies all SNPs and non-SNP regions of difference within an organism and yields a much higher resolution of the genetic variability of strains that appear to be closely related by other methods. Although the ten USA300 isolates were relatively similar, as determined by analysis of SNPs, some were clearly much more closely related than others and had descended from a recent common ancestor. The studies demonstrated that there has been recent clonal expansion of a subset of USA300 isolates that are now known as the epidemic USA300 clone. This work (80) and that of Mwangi et al. (72) provide insight into the potential utility of whole genome sequencing on a much greater scale. As more whole genome sequences for S. aureus isolates become available, it may be possible to pinpoint precise evolutionary events in the emergence of new successful clones such as USA300. With such precise understanding of microbe evolution, it should be possible to alter the selective pressures (e.g., antibiotic use or specific environmental conditions) under which these microorganism evolve to become successful pathogens.
CA-MRSA virulence and pathogenesis
Is CA-MRSA more virulent than traditional health care–associated MRSA?
Klevens et al. found that 85% of severe invasive MRSA infections either occur in hospitals or can be traced to the health care system (11). The death rate was approximately ten times higher for health care–associated MRSA infections than for CA-MRSA infections, 5.7 versus 0.5 deaths per 100,000 infections, respectively (11). Bacteremia and pneumonia accounted for 75.2% and 13.3% of these health care–associated infections, respectively. In contrast, Fridkin and colleagues found that 77% of CA-MRSA infections were of skin and soft tissue (mainly abscesses and cellulitis), with only 6% causing invasive disease (16). In the United States, USA100, USA200, and USA500 constitute the most abundant isolates from health care–associated infections, whereas USA300 and USA400 account for the greatest number of CA-MRSA isolates (11, 17, 23, 47, 86). These differences in invasive disease and mortality for health care–associated MRSA versus CA-MRSA infections could suggest that health care–associated strains are more virulent than those causing community infections. However, there is strong evidence to suggest the contrary.
First and foremost is the host. Health care–associated infections occur in individuals with predisposing risk factors — including surgery, dialysis, history of hospitalization, substance abuse, indwelling devices, and/or residence in a long-term care facility — that make them susceptible to invasive disease (11, 43). In contrast, CA-MRSA infections occur in otherwise healthy individuals lacking such risk factors. Notably, USA300 and USA400 have the capacity to cause severe and fatal infections in healthy subjects, albeit relatively infrequently (15, 40).
Although transmission and spread of the organism are not a component of disease pathogenesis per se, they contribute to the overall burden of disease and thus are an important component of virulence. USA300 has an extraordinary ability to spread among groups of individuals (53), and thus it disseminated rapidly throughout the United States (11, 17, 47). USA300 has invaded the health care system and is an abundant cause of bloodstream infections in hospitals (11, 35, 87, 88). In contrast, typical health care–associated MRSA strains (e.g., USA100 and USA200), although highly prevalent among health care facilities, are an infrequent cause of community-associated infections (11, 23, 55). This indirect evidence suggests that prominent CA-MRSA strains have enhanced virulence compared with traditional health care–associated MRSA.
There is also direct evidence that prominent CA-MRSA strains (i.e., USA300 and USA400) are more virulent than representative health care–associated strains (73). For example, MRSA252, a USA200 strain representative of the EMRSA-16 epidemic hospital clone, is essentially avirulent in a mouse model of sepsis, whereas USA300 and USA400 cause rapid death (73, 79). There is also considerable variance in virulence among CA-MRSA strains in mouse models of disease (79, 80, 82). For example, although USA300 and USA400 each cause fatal sepsis in a mouse bacteremia model, USA300 is substantially more virulent than USA400 in mouse and rat models of pneumonia (79, 82, 89). This observation is interesting because MW2 was notorious for causing fatal necrotizing pneumonia (15) and sepsis (40). Therefore, the mouse or rat lung infection models may not fully mimic human CA-MRSA necrotizing pneumonia, and/or the human disease may occur under specific preexisting conditions, such as antecedent infection with influenza.
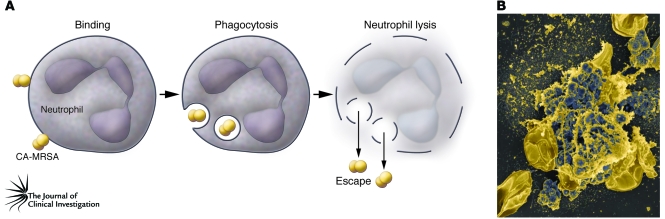
The success of S. aureus as a human pathogen depends largely on its ability to circumvent destruction by neutrophils. Compared with representative health care–associated MRSA strains, CA-MRSA strains have substantially enhanced ability to evade killing by human neutrophils (73). This ultimately results in lysis of the neutrophils, which occurs more rapidly with CA-MRSA strains than with prominent health care–associated MRSA strains (73). Early studies by Rogers and colleagues demonstrated that S. aureus is readily phagocytosed by neutrophils and that a substantial number of these bacteria remain viable and either serve as a potential reservoir for infection or ultimately cause neutrophil lysis (90). Consistent with these early studies, most S. aureus strains we have tested cause eventual neutrophil lysis after phagocytosis, albeit with marked variance among strains (ref. 73 and our unpublished observations). The important point here is that the most prominent CA-MRSA strains excel at killing neutrophils after phagocytosis (Figure 3). Thus, there is little doubt that the most prominent CA-MRSA strains are more virulent than traditional health care–associated MRSA strains.
Figure 3. Destruction of neutrophils after phagocytosis of CA-MRSA.
(A) A schematic model illustrating neutrophil lysis by CA-MRSA and subsequent escape of the pathogen. (B) Scanning electron micrograph of human neutrophils that have been destroyed after phagocytosis of USA400. Original magnification, ×5000.
As a first step toward understanding this differential virulence, Voyich et al. used a microarray-based approach to elucidate changes in the transcriptomes of S. aureus strains COL, USA200, USA300, and USA400 during phagocytosis by human neutrophils (73). The studies revealed changes common among all S. aureus strains during phagocytosis and those specific to USA300 and USA400, including many genes encoding proteins of unknown or uncharacterized function (73). Although progress has been made, understanding the molecular basis for the enhanced virulence of USA300 remains an area of intense research.
Is CA-MRSA more virulent than CA-MSSA?
Historically, community-associated S. aureus infections were almost always caused by MSSA. However, USA300 isolates are now the most abundant cause of community-associated S. aureus infections in many regions of the United States (17, 47). CA-MRSA and CA-MSSA each can cause disease in otherwise healthy individuals. The ability of CA-MRSA, and USA300 strains in particular, to displace CA-MSSA suggests no loss, and perhaps enhancement, of virulence or fitness. Indeed, one retrospective study found higher rates of hospitalization, higher rates of recurrent infection, and higher rates of infections in household contacts for patients with CA-MRSA compared with CA-MSSA infection (91). In other studies, clinical outcomes for CA-MRSA and CA-MSSA infections were similar (56, 92, 93). There is no a priori reason to expect that methicillin resistance, although an issue for treatment, would enhance virulence per se. Consistent with this idea was the demonstration that deletion of SCCmecIV in the USA300 epidemic clone had no impact on the ability of bacteria to colonize vital organs in a rabbit model of bacteremia (94). Infections caused by community-associated methicillin-resistant USA300 strains (CA-MRSA) or methicillin-susceptible variants of USA300 (CA-MSSA), which account for approximately 4.2%–31.0% of the CA-MSSA clinical isolates (17, 56, 95), have similar clinical outcomes. Thus, if CA-MRSA strains are more virulent than those that are CA-MSSA, factors other than antibiotic resistance must account for this difference.
Virulence determinants
S. aureus has an extensive repertoire of virulence factors, which includes both freely secreted and surface-bound factors (96). These molecules promote host colonization, alter leukocyte recruitment or function (81, 97), inhibit complement (97) and antimicrobial peptides (98–101), and cause destruction of leukocytes (81, 102–104). Also, the enhanced growth rate of prominent CA-MRSA strains could play a role in establishing infection or contribute to disease severity (24). Although many types of host cells and molecular processes are involved in S. aureus–host interaction, for the purposes of this Review we focus on those for which there is information in the context of CA-MRSA (19, 68, 70, 79, 81, 82, 85, 94, 105, 106).
One hundred years of PVL.
The role of PVL in the emergence of CA-MRSA is hotly debated, and the relative contribution of PVL to severe invasive CA-MRSA syndromes remains to be determined. The ability of S. aureus to produce molecules cytolytic for leukocytes has been known for more than a century. In 1894, van de Velde identified such a substance, which he named “substance leucocide” or leukocidin (107). Panton and Valentine then discovered a leukocidin that was nonlethal for rabbits and distinct from the hemolytic activity associated with a lethal toxin (103). However, Panton and Valentine showed a correlation between presence of the leukocidin and severe skin infections, especially carbuncles (large abscesses), some of which were ultimately fatal in the absence of antibiotics (103). PVL has been studied extensively over the past 70 years. This leukocidin consists of two subunits, LukS-PV and LukF-PV (108), which are encoded by lukS-PV and lukF-PV, respectively, within specific bacteriophages (68, 70, 109). Thus, acquisition of PVL by S. aureus may occur by horizontal gene transfer. PVL assembles as a pore-forming octamer containing LukS-PV/LukF-PV heterodimers on the surface of susceptible host cells (110), although the plasma membrane binding site and/or receptor is unknown (111). PVL is highly specific for myeloid cells, especially granulocytes, and there are differences in susceptibility among granulocytes and macrophages from various animal species and humans (112). In general, PVL damages the plasma membrane of myeloid cells; this may ultimately cause cell lysis depending on the concentration of toxin. Sublytic concentrations of PVL trigger neutrophil apoptosis within 6 hours of treatment, whereas higher toxin concentrations cause cell lysis within 1 hour (113). Woodin and colleagues made the unexpected discovery that PVL induces immediate changes in neutrophils that mimic priming or activation, including degranulation (114). PVL causes release of granulocyte leukotriene B4 and IL-8 (115), prominent mediators of inflammation. In addition, sublytic concentrations of PVL prime neutrophils for enhanced release of ROS after stimulation with N-formylpeptide (fMLP) (116). Therefore, it is reasonable to speculate that PVL might contribute to pathogenesis by recruiting and activating neutrophils, thereby eliciting an exaggerated inflammatory response.
Notably, PVL is nonlethal even at high concentrations. Jeljaszewicz and colleagues found that although intravenous injection of partially or highly purified PVL in rabbits or mice caused transient granulocytopenia followed by marked granulocytosis, animal health remained unaffected (117, 118). This transient granulocytopenia probably was caused by redistribution of granulocytes from blood into tissues, rather than direct lysis in the bloodstream, which would dramatically affect animal health. Subsequent work demonstrated that purified PVL causes dermonecrosis in rabbits (119), and these findings were confirmed by Cribier et al., who described the ability of the toxin to cause edema, erythema, and tissue necrosis (120). This effect was limited to administration of purified PVL, as there was no difference in skin lesions between animals infected with PVL-producing and -nonproducing bacteria (120). Even with these caveats, there is no question that PVL is an S. aureus virulence factor. The prevailing question is to what degree does PVL contribute to emergence of CA-MRSA and to its pathogenesis?
Early epidemiologic studies were at variance in correlating PVL-producing S. aureus strains and severe skin infections (120, 121). However, Lina and colleagues subsequently provided strong evidence that PVL is associated with severe furunculosis and severe necrotic hemorrhagic pneumonia (122). In 2002, Gillet et al. reported that PVL-producing S. aureus strains, mostly MSSA strains, were associated with fatal necrotizing pneumonia in otherwise healthy individuals (123). These reports coincided roughly with the emergence of CA-MRSA (14, 15, 19). CA-MRSA strains, including USA300 and USA400, contain the genes that encode PVL (19, 68, 70). Given that USA300 is a leading cause of skin and soft tissue infections and that PVL is associated with severe skin infections, PVL is often assumed to be the driving force behind the current CA-MRSA epidemic in the United States. However, PVL-encoding genes are found in less than 5% of all S. aureus clinical isolates (124–127), and studies of S. aureus nasal carriage isolates reveal a relatively low prevalence of those that are PVL-positive (0.6% in one study) (50, 128). If PVL were critically important for the success of S. aureus as a human pathogen, including the success of CA-MRSA, it should be found in higher abundance overall. Moreover, PVL has been reported in S. aureus since at least the early 1900s, yet CA-MRSA emerged only recently, suggesting that emergence of PVL is not driving the CA-MRSA epidemic. In support of this notion, comparative genome sequencing studies suggest that association of PVL with USA300 is not sufficient to account for the current epidemic (80), findings consistent with the coexistence of PVL-positive and PVL-negative USA400 strains in Canada (129). As mentioned above, two recent studies found no difference in clinical outcomes or mortality between patients with CA-MRSA and CA-MSSA skin and soft tissue or bloodstream infections (92, 93) (although an independent report found otherwise; ref. 91), and Kaplan and colleagues reported that CA-MSSA was associated with more invasive disease compared with CA-MRSA (37). These observations are important because PVL is infrequently associated with CA-MSSA strains other than USA300 (95).
To directly address the role of PVL in CA-MRSA pathogenesis, Voyich et al. compared USA300 and USA400 wild-type and isogenic PVL deletion (Δpvl) strains in mouse models of abscess and sepsis (79). PVL did not contribute to disease in these models, and survival of wild-type and Δpvl strains was equivalent following phagocytosis by human neutrophils (79). Moreover, PVL did not contribute to neutrophil lysis after phagocytosis, despite the observation that there is substantial upregulation of lukS-PV and lukF-PV transcripts under those conditions (79). USA300 and USA400 encode several two-component toxins that have up to 82% amino acid identity with PVL subunits and therefore possess redundant mechanisms to kill leukocytes.
Labandeira-Rey et al. studied laboratory S. aureus strains expressing PVL in a mouse model of pneumonia (130). An intriguing idea suggested by this study was that PVL functions as a regulator of global gene expression, promoting not only lung injury but also colonization and transmission (130). Unfortunately, this proved to be incorrect, as microarray- and proteomics-based studies demonstrated that PVL has no impact on either USA300 or USA400 gene or protein expression (131). Subsequently, Bubeck Wardenburg et al. evaluated USA300 and USA400 wild-type and isogenic Δpvl strains in both C57BL/6J and BALB/c models of S. aureus–induced pneumonia (82, 105) and skin infection (105). In contrast to the observations of Labandeira-Rey et al., but consistent with the findings of Voyich et al., wild-type and isogenic CA-MRSA Δpvl strains were equally virulent in these mouse models (82, 105). Recent follow-up studies by Bowden and colleagues used the same USA300 wild-type and Δpvl mutant strains in similar mouse infection models and reported that the wild-type strain was more virulent than the mutant (132). The reason for the discrepancy between studies is unclear, but collectively the observations indicate that PVL is not a major virulence determinant in mouse models of CA-MRSA infection. Consistent with this notion, Daum and Montgomery found comparable virulence of USA300 wild-type and Δpvl mutant strains in a rat pneumonia model (133). As a next step toward addressing the role of PVL in CA-MRSA virulence, Diep et al. used two rabbit bacteremia models to compare hematogenous dissemination of wild-type and Δpvl CA-MRSA strains to major organs (131). PVL did not promote seeding of lungs, spleen, or blood by USA300; there was a modest and transient effect of PVL in the kidney (131).
Data therefore suggest that if PVL contributes to CA-MRSA pathogenesis in humans, its overall role is relatively minor. For the rare cases of severe CA-MRSA disease, such as necrotizing pneumonia, there is likely a host susceptibility component, which could involve PVL, but this topic is not discussed here due to space limitations.
ACME.
ACME was discovered by whole genome sequencing of USA300. It is juxtaposed to SCCmecIV and can be mobilized by recombinases encoded within it (94). ACME contains genes encoding a complete arginine deiminase pathway; these genes are within an arc cluster and an oligopeptide permease operon (Opp-3) (70). Arginine deiminase is a virulence factor of Streptococcus pyogenes (134), and other chromosomally encoded oligopeptide permease operons appear to be important for S. aureus virulence (135); thus, ACME, as a newly described component of USA300, might contribute to fitness and epidemic spread. This hypothesis was tested in follow-up studies, which demonstrated that deletion of ACME but not SCCmec decreased the fitness of USA300 (94). Specifically, ACME promoted hematogenous dissemination to major organs in a rabbit bacteremia model of USA300 infection. Studies by Diep and colleagues also demonstrated a strong epidemiological link between ACME and the epidemic USA300 clone (94). Collectively, these findings indicate that ACME has contributed to the success (and perhaps to the emergence) of the epidemic USA300 clone.
α-Type phenol-soluble modulins.
Wang et al. discovered a group of peptides in S. aureus that have similarity with the phenol-soluble modulins (PSMs) of Staphylococcus epidermidis (81). These PSMs were produced at much higher levels in vitro by prominent CA-MRSA strains, including USA300 and USA400 (81), than by health care–associated strains, leading to the hypothesis that PSMs of USA300 and USA400 might contribute to the enhanced virulence of CA-MRSA. Wild-type and isogenic PSM deletion strains of USA300 and USA400, when tested in mouse models of skin infection and sepsis, demonstrated that four of the S. aureus PSMs, which are named α-type PSMs (PSMα), contribute significantly to USA300 and USA400 virulence (81). PSMα peptides were shown to recruit, activate, and ultimately lyse human neutrophils, thus promoting S. aureus pathogenesis. This study was the first to identify molecules encoded by CA-MRSA that account at least in part for the enhanced virulence of USA300 and USA400.
α-Hemolysin.
α-hemolysin (Hla; also known as α-toxin), which probably is the most widely studied S. aureus toxin, is lethal in animal models of S. aureus bacteremia (136, 137). This pore-forming toxin destroys a wide range of host cells, including epithelial cells, erythrocytes, fibroblasts, and monocytes. Hla is not unique to CA-MRSA strains and for years has been known to play a major role in S. aureus pathogenesis (137). However, Bubeck Wardenburg et al. demonstrated recently that Hla was essential for the ability of USA300 and USA400 to cause pneumonia in a mouse model of infection (82). Moreover, the level of Hla produced by these strains in vitro correlates with severity of lung disease in mice and rats (82, 89, 138). It is likely that enhanced in vivo expression of selected virulence factors, many of which are regulated by the accessory gene regulator (Agr) system of S. aureus, is a critical component of the success of CA-MRSA, as proposed by Wang et al. (81) and Daum and colleagues (89).
Concluding remarks
Several questions must ultimately be addressed in order to make significant advances toward reducing disease caused by antibiotic-resistant strains of S. aureus, including those that cause CA-MRSA infections. For example, what is the driving force behind the recent emergence of CA-MRSA? Antibiotics, although highly effective, have been overused and misused for decades, inevitably contributing to the growing problem of resistance (13) (Figure 1). A better understanding of the basis of pathogen emergence is essential to developing alternative approaches for controlling the current CA-MRSA epidemic. New technologies, especially high-throughput whole genome sequencing, will be a major asset in our quest to fully understand, control, and prevent emergence and reemergence of drug-resistant S. aureus. Notably, the question of how new, more virulent strains emerge would be more readily addressed if we had available several hundred S. aureus genome sequences that span the current CA-MRSA epidemic.
Acknowledgments
This work was supported by US Public Health Service Grant AI070289 (H.F. Chambers) and the Intramural Research Program of the NIH, National Institutes of Allergy and Infectious Diseases (NIAID; F.R. DeLeo). Electron microscopy was performed by David W. Dorward (NIAID), and images were colorized by Anita Mora (NIAID).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:2464–2474 (2009). doi:10.1172/JCI38226
References
- 1.Diekema D.J., et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 2001;32(Suppl. 2):S114–S132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 2.Kirby W.M. Extraction of a highly potent penicillin inactivator from penicillin resistant Staphylococci. Science. 1944;99:452–453. doi: 10.1126/science.99.2579.452. [DOI] [PubMed] [Google Scholar]
- 3.Barber M., Rozwadowska-Dowzenko M. Infection by penicillin-resistant staphylococci. Lancet. 1948;252:641–644. doi: 10.1016/s0140-6736(48)92166-7. [DOI] [PubMed] [Google Scholar]
- 4.Roundtree P.M., Freeman B.M. Infections caused by a particular phage type of Staphylococcus aureus. . Med. J. Aust. 1956;42:157–161. [PubMed] [Google Scholar]
- 5.Jevons M.P., Parker M.T. The evolution of new hospital strains of Staphylococcus aureus. . J. Clin. Pathol. 1964;17:243–250. doi: 10.1136/jcp.17.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jevons M.P.1961“Celbenin”-resistant Staphylococci. Br. Med. J. 1124–125.760984 [Google Scholar]
- 7.Stewart G.T., Holt R.J. Evolution of natural resistance to the newer penicillins. Br. Med. J. 1963;1:308–311. doi: 10.1136/bmj.1.5326.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klevens R.M., et al. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. . Clin. Infect. Dis. 2006;42:389–391. doi: 10.1086/499367. [DOI] [PubMed] [Google Scholar]
- 9.Klein E., Smith D.L., Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. . Emerg. Infect. Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis W.R., Schlosser J., Chinn R.Y., Tweeten S., Jackson M. National prevalence of methicillin-resistant Staphylococcus aureus in inpatients at US health care facilities, 2006. . Am. J. Infect. Control. 2007;35:631–637. doi: 10.1016/j.ajic.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Klevens R.M., et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. . JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 12.Heron M.P., Hoyert D.L., Xu J., Scott C., Tejada-Vera B.2008Deaths: preliminary data for 2006. National vital statistics reports. 561–52. http://www.cdc.gov/nchs/data/nvsr/nvsr56/nvsr56_16.pdf . [PubMed] [Google Scholar]
- 13.Chambers H.F. The changing epidemiology of Staphylococcus aureus? . Emerg. Infect. Dis. 2001;7:178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold B.C., et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. . JAMA. 1998;279:593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. . Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus--Minnesota and North Dakota, 1997-1999. . JAMA. 1999;282:1123–1125. doi: 10.1001/jama.282.12.1123. [DOI] [PubMed] [Google Scholar]
- 16.Fridkin S.K., et al. Methicillin-resistant Staphylococcus aureus disease in three communities. . N. Engl. J. Med. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 17.Moran G.J., et al. Methicillin-resistant S. aureus infections among patients in the emergency department. . N. Engl. J. Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 18.Udo E.E., Pearman J.W., Grubb W.B. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. . J. Hosp. Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-E. [DOI] [PubMed] [Google Scholar]
- 19.Vandenesch F., et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. . Emerg. Infect. Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright M.C., et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). . Proc. Natl. Acad. Sci. U. S. A. 2002;99:7687–7692. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil E.J., et al. How clonal is Staphylococcus aureus? . J. Bacteriol. 2003;185:3307–3316. doi: 10.1128/JB.185.11.3307-3316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsay J.A., et al. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. . J. Bacteriol. 2006;188:669–676. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDougal L.K., et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. . J. Clin. Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuma K., et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. . J. Clin. Microbiol. 2002;40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lencastre H., Oliveira D., Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. . Curr. Opin. Microbiol. 2007;10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien F.G., et al. Diversity among community isolates of methicillin-resistant Staphylococcus aureus in Australia. . J. Clin. Microbiol. 2004;42:3185–3190. doi: 10.1128/JCM.42.7.3185-3190.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baggett H.C., et al. Community-onset methicillin-resistant Staphylococcus aureus associated with antibiotic use and the cytotoxin Panton-Valentine leukocidin during a furunculosis outbreak in rural Alaska. . J. Infect. Dis. 2004;189:1565–1573. doi: 10.1086/383247. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. Community-associated methicillin-resistant Staphylococcus aureus infections in Pacific Islanders Hawaii, 2001–2003. . MMWR Morb. Mortal. Wkly. Rep. 2004;53:767–770. [PubMed] [Google Scholar]
- 29.Begier E.M., et al. A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. . Clin. Infect. Dis. 2004;39:1446–1453. doi: 10.1086/425313. [DOI] [PubMed] [Google Scholar]
- 30.Aiello A.E., Lowy F.D., Wright L.N., Larson E.L. Meticillin-resistant Staphylococcus aureus among US prisoners and military personnel: review and recommendations for future studies. . Lancet Infect. Dis. 2006;6:335–341. doi: 10.1016/S1473-3099(06)70491-1. [DOI] [PubMed] [Google Scholar]
- 31.Diep B.A., et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. . Ann. Intern. Med. 2008;148:249–257. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- 32.Adcock P.M., Pastor P., Medley F., Patterson J.E., Murphy T.V. Methicillin-resistant Staphylococcus aureus in two child care centers. . J. Infect. Dis. 1998;178:577–580. doi: 10.1086/517478. [DOI] [PubMed] [Google Scholar]
- 33.Miller L.G., Diep B.A. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. . Clin. Infect. Dis. 2008;46:752–760. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- 34.Liu C., et al. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004-2005. . Clin. Infect. Dis. 2008;46:1637–1646. doi: 10.1086/587893. [DOI] [PubMed] [Google Scholar]
- 35.Seybold U., et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. . Clin. Infect. Dis. 2006;42:647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 36.Faria N.A., et al. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. . J. Clin. Microbiol. 2005;43:1836–1842. doi: 10.1128/JCM.43.4.1836-1842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan S.L., et al. Three-year surveillance of community-acquired Staphylococcus aureus infections in children. . Clin. Infect. Dis. 2005;40:1785–1791. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 38.Laupland K.B., Ross T., Gregson D.B. Staphylococcus aureus bloodstream infections: risk factors, outcomes, and the influence of methicillin resistance in Calgary, Canada, 2000-2006. . J. Infect. Dis. 2008;198:336–343. doi: 10.1086/589717. [DOI] [PubMed] [Google Scholar]
- 39.Chi C.Y., Wang S.M., Lin H.C., Liu C.C. A clinical and microbiological comparison of Staphylococcus aureus toxic shock and scalded skin syndromes in children. . Clin. Infect. Dis. 2006;42:181–185. doi: 10.1086/498901. [DOI] [PubMed] [Google Scholar]
- 40.Adem P.V., et al. Staphylococcus aureus sepsis and the Waterhouse-Friderichsen syndrome in children. . N. Engl. J. Med. 2005;353:1245–1251. doi: 10.1056/NEJMoa044194. [DOI] [PubMed] [Google Scholar]
- 41.Kravitz G.R., Dries D.J., Peterson M.L., Schlievert P.M. Purpura fulminans due to Staphylococcus aureus. . Clin. Infect. Dis. 2005;40:941–947. doi: 10.1086/428573. [DOI] [PubMed] [Google Scholar]
- 42.Miller L.G., et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. . N. Engl. J. Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 43.Naimi T.S., et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. . JAMA. 2003;290:2976–2984. doi: 10.1001/jama.290.22.2976. [DOI] [PubMed] [Google Scholar]
- 44.Francis J.S., et al. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. . Clin. Infect. Dis. 2005;40:100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 45.Gonzalez B.E., et al. Pulmonary manifestations in children with invasive community-acquired Staphylococcus aureus infection. . Clin. Infect. Dis. 2005;41:583–590. doi: 10.1086/432475. [DOI] [PubMed] [Google Scholar]
- 46.Hageman J.C., et al. Severe community-acquired pneumonia due to Staphylococcus aureus, 2003-04 influenza season. . Emerg. Infect. Dis. 2006;12:894–899. doi: 10.3201/eid1206.051141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King M.D., et al. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. . Ann. Intern. Med. 2006;144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez B.E., et al. Severe staphylococcal sepsis in adolescents in the era of community-acquired methicillin-resistant Staphylococcus aureus. . Pediatrics. 2005;115:642–648. doi: 10.1542/peds.2004-2300. [DOI] [PubMed] [Google Scholar]
- 49.Graham P.L., III, Lin S.X., Larson E.L. A U.S. population-based survey of Staphylococcus aureus colonization. . Ann. Intern. Med. 2006;144:318–325. doi: 10.7326/0003-4819-144-5-200603070-00006. [DOI] [PubMed] [Google Scholar]
- 50.Gorwitz R.J., et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. . J. Infect. Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 51.Crum N.F., et al. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. . Am. J. Med. 2006;119:943–951. doi: 10.1016/j.amjmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 52.Centers for Disease Control and Prevention (CDC). . Methicillin-resistant Staphylococcus aureus infections among competitive sports participants--Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-2003. . MMWR Morb. Mortal. Wkly. Rep. 2003;52:793–795. [PubMed] [Google Scholar]
- 53.Kazakova S.V., et al. A clone of methicillin-resistant Staphylococcus aureus among professional football players. . N. Engl. J. Med. 2005;352:468–475. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- 54.Charlebois E.D., et al. Origins of community strains of methicillin-resistant Staphylococcus aureus. . Clin. Infect. Dis. 2004;39:47–54. doi: 10.1086/421090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H., et al. Comparisons of community-associated methicillin-resistant Staphylococcus aureus (MRSA) and hospital-associated MSRA infections in Sacramento, California. . J. Clin. Microbiol. 2006;44:2423–2427. doi: 10.1128/JCM.00254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller L.G., et al. Clinical and epidemiologic characteristics cannot distinguish community-associated methicillin-resistant Staphylococcus aureus infection from methicillin-susceptible S. aureus infection: a prospective investigation. . Clin. Infect. Dis. 2007;44:471–482. doi: 10.1086/511033. [DOI] [PubMed] [Google Scholar]
- 57.Sattler C.A., Mason E.O., Jr., Kaplan S.L. Prospective comparison of risk factors and demographic and clinical characteristics of community-acquired, methicillin-resistant versus methicillin-susceptible Staphylococcus aureus infection in children. . Pediatr. Infect. Dis. J. 2002;21:910–917. doi: 10.1097/00006454-200210000-00005. [DOI] [PubMed] [Google Scholar]
- 58.Young D.M., et al. An epidemic of methicillin-resistant Staphylococcus aureus soft tissue infections among medically underserved patients. . Arch. Surg. 2004;139:947–951. doi: 10.1001/archsurg.139.9.947. [DOI] [PubMed] [Google Scholar]
- 59.Cookson B.D., et al. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. . J. Clin. Microbiol. 2007;45:1830–1837. doi: 10.1128/JCM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. . J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shopsin B., et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. . J. Clin. Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma X.X., et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. . Antimicrob. Agents Chemother. 2002;46:1147–1152. doi: 10.1128/AAC.46.4.1147-1152.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hawkins C., et al. Persistent Staphylococcus aureus bacteremia: an analysis of risk factors and outcomes. . Arch. Intern. Med. 2007;167:1861–1867. doi: 10.1001/archinte.167.17.1861. [DOI] [PubMed] [Google Scholar]
- 64.Dombrowski J.C., Winston L.G. Clinical failures of appropriately-treated methicillin-resistant Staphylococcus aureus infections. . J. Infect. 2008;57:110–115. doi: 10.1016/j.jinf.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lodise T.P., Lomaestro B., Graves J., Drusano G.L. Larger vancomycin doses (at least four grams per day) are associated with an increased incidence of nephrotoxicity. Antimicrob. Agents Chemother. 2008;52:1330–1336. doi: 10.1128/AAC.01602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howe R.A., Monk A., Wootton M., Walsh T.R., Enright M.C. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. . Emerg. Infect. Dis. 2004;10:855–857. doi: 10.3201/eid1005.030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuroda M., et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. . Lancet. 2001;357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 68.Baba T., et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 69.Holden M.T., et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. . Proc. Natl. Acad. Sci. U. S. A. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diep B.A., et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. . Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 71.Sievert D.M., et al. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. . Clin. Infect. Dis. 2008;46:668–674. doi: 10.1086/527392. [DOI] [PubMed] [Google Scholar]
- 72.Mwangi M.M., et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. . Proc. Natl. Acad. Sci. U. S. A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voyich J.M., et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. . J. Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 74.Tenover F.C., et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. . J. Clin. Microbiol. 2006;44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cassat J.E., et al. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. . J. Bacteriol. 2005;187:576–592. doi: 10.1128/JB.187.2.576-592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fitzgerald J.R., Sturdevant D.E., Mackie S.M., Gill S.R., Musser J.M. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. . Proc. Natl. Acad. Sci. U. S. A. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson D.A., Enright M.C. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. . Antimicrob. Agents Chemother. 2003;47:3926–3934. doi: 10.1128/AAC.47.12.3926-3934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Highlander S.K., et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. . BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voyich J.M., et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? . J. Infect. Dis. 2006;194:1761–1770. doi: 10.1086/509506. [DOI] [PubMed] [Google Scholar]
- 80.Kennedy A.D., et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. . Proc. Natl. Acad. Sci. U. S. A. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang R., et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 82.Bubeck Wardenburg J., Bae T., Otto M., DeLeo F.R., Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. . Nat. Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
- 83.Van Wamel W.J., Rooijakkers S.H., Ruyken M., Van Kessel K.P., Van Strijp J.A. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. . J. Bacteriol. 2006;188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yarwood J.M., et al. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. . J. Biol. Chem. 2002;277:13138–13147. doi: 10.1074/jbc.M111661200. [DOI] [PubMed] [Google Scholar]
- 85.Diep B.A., Carleton H.A., Chang R.F., Sensabaugh G.F., Perdreau-Remington F. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. . J. Infect. Dis. 2006;193:1495–1503. doi: 10.1086/503777. [DOI] [PubMed] [Google Scholar]
- 86.Fowler V.G., Jr., et al. Potential associations between hematogenous complications and bacterial genotype in Staphylococcus aureus infection. . J. Infect. Dis. 2007;196:738–747. doi: 10.1086/520088. [DOI] [PubMed] [Google Scholar]
- 87.Gonzalez B.E., et al. Community-associated strains of methicillin-resistant Staphylococcus aureus as the cause of healthcare-associated infection. . Infect. Control Hosp. Epidemiol. 2006;27:1051–1056. doi: 10.1086/507923. [DOI] [PubMed] [Google Scholar]
- 88.Maree C.L., Daum R.S., Boyle-Vavra S., Matayoshi K., Miller L.G. Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. . Emerg. Infect. Dis. 2007;13:236–242. doi: 10.3201/eid1302.060781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Montgomery C.P., et al. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. . J. Infect. Dis. 2008;198:561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 90.Rogers D.E., Tompsett R. The survival of staphylococci within human leukocytes. J. Exp. Med. 1952;95:209–230. doi: 10.1084/jem.95.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davis S.L., et al. Epidemiology and outcomes of community-associated methicillin-resistant Staphylococcus aureus infection. . J. Clin. Microbiol. 2007;45:1705–1711. doi: 10.1128/JCM.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller L.G., et al. A prospective investigation of outcomes after hospital discharge for endemic, community-acquired methicillin-resistant and -susceptible Staphylococcus aureus skin infection. . Clin. Infect. Dis. 2007;44:483–492. doi: 10.1086/511041. [DOI] [PubMed] [Google Scholar]
- 93.Wang J.L., et al. Comparison of both clinical features and mortality risk associated with bacteremia due to community-acquired methicillin-resistant Staphylococcus aureus and methicillin-susceptible S. aureus. . Clin. Infect. Dis. 2008;46:799–806. doi: 10.1086/527389. [DOI] [PubMed] [Google Scholar]
- 94.Diep B.A., et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. . J. Infect. Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 95.McCaskill M.L., et al. Increase of the USA300 clone among community-acquired methicillin-susceptible Staphylococcus aureus causing invasive infections. . Pediatr. Infect. Dis. J. 2007;26:1122–1127. doi: 10.1097/INF.0b013e31814536e0. [DOI] [PubMed] [Google Scholar]
- 96.Foster T.J. Immune evasion by staphylococci. Nat. Rev. Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 97.Rooijakkers S.H., Van Strijp J.A. Bacterial complement evasion. Mol. Immunol. 2007;44:23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 98.Collins L.V., et al. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. . J. Infect. Dis. 2002;186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 99.Peschel A., et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. . J. Exp. Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peschel A., et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. . J. Biol. Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 101.Li M., et al. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. . Mol. Microbiol. 2007;66:1136–1147. doi: 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 102.Valeva A., et al. Transmembrane beta-barrel of staphylococcal alpha-toxin forms in sensitive but not in resistant cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11607–11611. doi: 10.1073/pnas.94.21.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Panton P.N., Valentine F.C.O.1932Staphylococcal toxin. Lancet. 1506–508.2563883 [Google Scholar]
- 104.Prevost G., et al. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. . Infect. Immun. 1995;63:4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bubeck Wardenburg J., Palazzolo-Ballance A.M., Otto M., Schneewind O., DeLeo F.R. Panton-Valentine leukocidin is not a virulence determinant in murine models of community-associated methicillin-resistant Staphylococcus aureus disease. . J. Infect. Dis. 2008;198:1166–1170. doi: 10.1086/592053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bubeck Wardenburg J., Schneewind O. Vaccine protection against Staphylococcus aureus pneumonia. . J. Exp. Med. 2008;205:287–294. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van de Velde H. Etude sur le mécanisme de la virulence du Staphylocoque pyogène. . La Cellule. 1894;10:401–460. [Google Scholar]
- 108.Woodin A.M. Purification of the two components of leucocidin from Staphylococcus aureus. . Biochem. J. 1960;75:158–165. doi: 10.1042/bj0750158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaneko J., Kimura T., Narita S., Tomita T., Kamio Y. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene. 1998;215:57–67. doi: 10.1016/S0378-1119(98)00278-9. [DOI] [PubMed] [Google Scholar]
- 110.Jayasinghe L., Bayley H. The leukocidin pore: evidence for an octamer with four LukF subunits and four LukS subunits alternating around a central axis. Protein Sci. 2005;14:2550–2561. doi: 10.1110/ps.051648505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Colin D.A., Mazurier I., Sire S., Finck-Barbancon V. Interaction of the two components of leukocidin from Staphylococcus aureus with human polymorphonuclear leukocyte membranes: sequential binding and subsequent activation. . Infect. Immun. 1994;62:3184–3188. doi: 10.1128/iai.62.8.3184-3188.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Szmigielski S., Prevost G., Monteil H., Colin D.A., Jeljaszewicz J. Leukocidal toxins of staphylococci. Zentralbl. Bakteriol. 1999;289:185–201. doi: 10.1016/s0934-8840(99)80105-4. [DOI] [PubMed] [Google Scholar]
- 113.Genestier A.L., et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. . J. Clin. Invest. 2005;115:3117–3127. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Woodin A.M., Wieneke A.A. The participation of calcium, adenosine triphosphate and adenosine triphosphatase in the extrusion of the granule proteins from the polymorphonuclear leucocyte. Biochem. J. 1964;90:498–509. doi: 10.1042/bj0900498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Konig B., Prevost G., Piemont Y., Konig W. Effects of Staphylococcus aureus leukocidins on inflammatory mediator release from human granulocytes. . J. Infect. Dis. 1995;171:607–613. doi: 10.1093/infdis/171.3.607. [DOI] [PubMed] [Google Scholar]
- 116.Colin D.A., Monteil H. Control of the oxidative burst of human neutrophils by staphylococcal leukotoxins. Infect. Immun. 2003;71:3724–3729. doi: 10.1128/IAI.71.7.3724-3729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Szmigielski S., Jeljaszewicz J., Wilczynski J., Korbecki M. Reaction of rabbit leucocytes to staphylococcal (Panton-Valentine) leucocidin in vivo. J. Pathol. Bacteriol. 1966;91:599–604. doi: 10.1002/path.1700910237. [DOI] [PubMed] [Google Scholar]
- 118.Grojec L.P., Jelijaszewicz J. Effect of staphylococcal leukocidin on mouse leukocyte system. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 1981;250:446–455. [PubMed] [Google Scholar]
- 119.Ward P.D., Turner W.H. Identification of staphylococcal Panton-Valentine leukocidin as a potent dermonecrotic toxin. Infect. Immun. 1980;28:393–397. doi: 10.1128/iai.28.2.393-397.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cribier B., et al. Staphylococcus aureus leukocidin: a new virulence factor in cutaneous infections? An epidemiological and experimental study. . Dermatology. 1992;185:175–180. doi: 10.1159/000247443. [DOI] [PubMed] [Google Scholar]
- 121.Finck-Barbancon V., Prevost G., Piemont Y. Improved purification of leukocidin from Staphylococcus aureus and toxin distribution among hospital strains. . Res. Microbiol. 1991;142:75–85. doi: 10.1016/0923-2508(91)90099-V. [DOI] [PubMed] [Google Scholar]
- 122.Lina G., et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. . Clin. Infect. Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 123.Gillet Y., et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. . Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 124.Ellington M.J., et al. Is Panton-Valentine leucocidin associated with the pathogenesis of Staphylococcus aureus bacteraemia in the UK? . J. Antimicrob. Chemother. 2007;60:402–405. doi: 10.1093/jac/dkm206. [DOI] [PubMed] [Google Scholar]
- 125.Couppie P., Cribier B., Prevost G. Leukocidin from Staphylococcus aureus and cutaneous infections: an epidemiologic study. . Arch. Dermatol. 1994;130:1208–1209. doi: 10.1001/archderm.130.9.1208. [DOI] [PubMed] [Google Scholar]
- 126.Holmes A., et al. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. . J. Clin. Microbiol. 2005;43:2384–2390. doi: 10.1128/JCM.43.5.2384-2390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Robert J., Etienne J., Bertrand X. Methicillin-resistant Staphylococcus aureus producing Panton-Valentine leukocidin in a retrospective case series from 12 French hospital laboratories, 2000-2003. . Clin. Microbiol. Infect. 2005;11:585–587. doi: 10.1111/j.1469-0691.2005.01173.x. [DOI] [PubMed] [Google Scholar]
- 128.Melles D.C., et al. Panton-Valentine leukocidin genes in Staphylococcus aureus. . Emerg. Infect. Dis. 2006;12:1174–1175. doi: 10.3201/eid1207.050865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang K., McClure J.A., Elsayed S., Tan J., Conly J.M. Coexistence of Panton-Valentine leukocidin-positive and -negative community-associated methicillin-resistant Staphylococcus aureus USA400 sibling strains in a large Canadian health-care region. . J. Infect. Dis. 2008;197:195–204. doi: 10.1086/523763. [DOI] [PubMed] [Google Scholar]
- 130.Labandeira-Rey M., et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. . Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 131.Diep B.A., et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. . PLoS ONE. 2008;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brown E.L., et al. The Panton-Valentine leukocidin vaccine protects mice against lung and skin infections caused by Staphylococcus aureus USA300. . Clin. Microbiol. Infect. 2009;15:156–154. doi: 10.1111/j.1469-0691.2008.02648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Montgomery C.P., Daum R.S. Transcription of inflammatory genes in the lung after infection with community-associated methicillin-resistant Staphylococcus aureus: a role for Panton-Valentine Leukocidin? . Infect. Immun. 2009;77:2159–2169. doi: 10.1128/IAI.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Degnan B.A., et al. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. . Infect. Immun. 1998;66:3050–3058. doi: 10.1128/iai.66.7.3050-3058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Coulter S.N., et al. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. . Mol. Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 136.Bhakdi S., Tranum-Jensen J. Alpha-toxin of Staphylococcus aureus. . Microbiol. Rev. 1991;55:733–751. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Menzies B.E., Kernodle D.S. Site-directed mutagenesis of the alpha-toxin gene of Staphylococcus aureus: role of histidines in toxin activity in vitro and in a murine model. . Infect. Immun. 1994;62:1843–1847. doi: 10.1128/iai.62.5.1843-1847.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burlak C., et al. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. . Cell. Microbiol. 2007;9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Purcell K., Fergie J. Epidemic of community-acquired methicillin-resistant Staphylococcus aureus infections: a 14-year study at Driscoll Children’s Hospital. . Arch. Pediatr. Adolesc. Med. 2005;159:980–985. doi: 10.1001/archpedi.159.10.980. [DOI] [PubMed] [Google Scholar]
- 140.Ito T., Kuwahara K., Hiramatsu K. Staphylococcal cassette chromosome mec (SCC mec) analysis of MRSA. Methods Mol. Biol. 2007;391:87–102. doi: 10.1007/978-1-59745-468-1_7. [DOI] [PubMed] [Google Scholar]