Abstract
The exact mechanisms of prion misfolding and factors that predispose an individual to prion diseases are largely unknown. Our approach to identifying candidate factors in-silico relies on contrasting the C-terminal domain of PrPC sequences from two groups of vertebrate species: those that have been found to suffer from prion diseases, and those that have not. We propose that any significant differences between the two groups are candidate factors that may predispose individuals to develop prion disease, which should be further analyzed by wet-lab investigations. Using an array of computational methods we identified possible point mutations that could predispose PrPC to misfold into PrPSc. Our results include confirmatory findings such as the V210I mutation, and new findings including P137M, G142D, G142N, D144P, K185T, V189I, H187Y and T191P mutations, which could impact structural stability. We also propose new hypotheses that give insights into the stability of helix-2 and -3. These include destabilizing effects of Histidine and T188-T193 segment in helix-2 in the disease-prone prions, and a stabilizing effect of Leucine on helix-3 in the disease-resistant prions.
Keywords: prions, prion misfolding, point mutations, sequence alignment, exchange groups, conformational transition
Introduction
Misfolding of the prion protein (PrP) is believed to be responsible for the Transmissible Spongiform Encephalopathy (TSE) diseases (Prusiner, 1998). Experimental investigations suggest that the pathogenesis of TSE is characterized by the unfolding of the normal Prion protein (PrPC) followed by misfolding into an infectious “scrapie” isoform (PrPSc) (Pan et al. 1993). According to the protein-only hypothesis, PrPSc promotes structural conversion of the cellular PrPC into the pathogenic conformation (Prusiner, 1998; Prusiner et al. 1998). The pathogenesis presumably involves the initial formation of PrPSc, which is a result of a point mutation(s) or some exogenous factors, and which subsequently interacts with and converts PrPC molecules into PrPSc molecules. The last decade of research has provided a significant amount of evidence that supports this hypothesis (Mead, 2006).
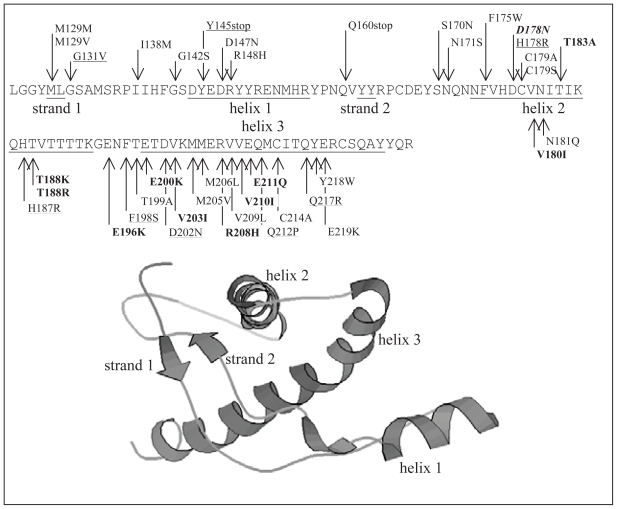
Known PrPC structures reveal that the C-terminal domain (positions 125 to 230) is structured and contains three α-helices and a short β-sheet that includes two strands (see Fig. 1), whereas the N-terminal domain (positions 23 to 126) is highly flexible and cannot be assigned a particular conformation (Riek et al. 1997; Riek et al. 1998; Lopez-Garcia et al. 2000). At the same time, the structure of the PrPSc isoform is currently still unknown.
Figure 1.
Sequence and mutations in the C-terminal domain of huPrP together with the ribbon drawing of the corresponding 3D structure (positions 125 to 228 of 1HJM). The secondary structure segments are denoted by underscores. Bold indicates pathogenic mutations associated with the CJD phenotype, underline indicates GSS, and italic indicates FFI.
Spectroscopic studies have shown that PrPC is composed of about 42% α-helices and 3% β-sheets, whereas PrPSc is composed of only 30% α-helices and 43% β-sheets (Pan et al. 1993). Thus, the conformational transition of PrPC into PrPSc has to involve unfolding of some α-helices and formation of new β-sheets. Helix-1 is the most conserved in PrP sequences and forms only a few interactions with the rest of the C-terminal domain. These facts led to a model in which helix-1 was considered as a starting point for conformational transition and forms a β-like aggregate, whereas helix-2 and helix-3 retain their conformation (Huang et al. 1995; Morrisey et al. 1999; Wille et al. 2002). Some recent models of the pathologically misfolded form of PrP also show that the helix-1 region is unstable and has to unfold during the conformational transition (Eghiaian et al. 2004). At the same time, recent results provide strong evidence that helix-1 is not converted into a β-sheet during the aggregation of PrPC to PrPSc (Watzlawik et al. 2006). This conclusion is also supported by experimental data obtained using low-resolution electron crystallography which suggest that helix-1 in PrPSc refolds into a left-handed β-helix (Wille et al. 2002), while subsequent work shows that helix-1 is not included in the β-helix but forms an unstructured loop (Govaerts et al. 2004). These discrepancies motivate this work, in which we use sequence based analysis to find factors that could impact the stability of particular secondary structure segments.
A number of point mutations in the human prion have been identified. A significant proportion of all mutations are found within the structured C-terminal domain; 27 out of total of 30 as reported in (Kovacs et al. 2002) and 37 out of 55 as reported in PrionDB at http://www.receptors.org/Prion/ (Horn et al. 2001). Thus, we focus our attention on the C-terminal domain (see Fig. 1). Pathogenic mutations are classified based on their association with prion diseases that include Gerstmann-Straussler-Scheinker disease (GSS), Creutzfeld-Jakob disease (CJD), and Fatal Familial Insomnia (FFI). The number of possible single-point mutations in the C-terminal domain is relatively large (109 positions * 19 = 2071), and thus it is not feasible to physically check every one of them using wet-lab techniques. Well-designed computational experiments (such as the design we propose) can reveal promising candidate factors, which serve as new hypotheses for wet-lab investigation. To this end, another of our goals is to use sequence based analysis to find point mutations that could predispose PrPC to misfold into PrPSc.
In contrast to other sequence analysis based approaches that contrast prion proteins with structurally similar proteins such as Doppel (Kuznetsov and Rackovsky, 2004), we present a novel in-silico approach based on the assumption that some species are susceptible and others are resistant to prion disease (PD). We divide the available prion sequences from vertebrate animals into those that are prone to PD, and those that are apparently resistant, i.e. there are no reports of any known PD in that species and research suggests that they do not develop PD. We then compare the PrP sequences from these two groups (hereafter “the contrasts”), with a focus on the C-terminal domain. To the best of the authors’ knowledge, only two prior sequence-analysis-based contributions perform similar contrasting analysis, but they focused on identification of β-aggregating stretches (Tartagia et al. 2005) or contrasted just four prion proteins (Pappalardo et al. 2007). We used an array of computational techniques including multiple sequence alignment, exchange group similarities, and feature selection methods to identify possible factors that distinguish the contrasts for a larger set of 11 proteins. We suggest that such discriminating factors are potentially important in the conformational change from PrPC to PrPSC. The results of this analysis are best viewed as either evidence confirming known factors associated with prion misfolding, or newly hypothesized factors that predispose the misfolding.
Materials and Methods
Dataset
We extracted the sequences of all prions that were deposited in Protein Data Bank (PDB) (Berman et al. 2000) as of September 2007. This database is expert-curated, which assures high quality of the data, and includes structural information, which allows us to identify secondary structure regions and perform structural analysis. The 70 prion sequences stored in PDB belong to 15 species: chicken (1 sequence), ovine (4 sequences), human (29), elk (1), rabbit (1), canine (1), frog (1), turtle (1), bovine (5), mouse (4), cat (1), pig (1), syrian hamster (2), sheep (5), and yeast (13). Yeast prions were removed since they have no homology with the remaining vertebrate prions, and are shown to have substantially different properties (Bousset and Melki, 2002). We filtered out redundant sequences, i.e. we selected the newest deposition for each species (except for sheep prions, for which there are two depositions from 2004; we selected the slightly older 1UW3 that does not include polymorphisms), and eliminated sequences that did not cover the C-terminal domain. We note that among the C-terminal domain sequences the four bovine sequences and the two mouse sequences are identical, while the only differences between the two sheep sequences are C148R and Q168H mutations, and among ten human prions nine sequences are identical and one differs from them by two mutations M166C and E221C. The positions associated with these mutations do not show any consistent pattern vs. our contrasts (i.e. they do not serve to differentiate PD-prone from PD-resistant species), and so the duplicate sequences are redundant and could be safely removed. It is in fact necessary to remove them; data-mining techniques such as feature selection assume that there is no redundancy in a dataset (deletion of redundant data items is a standard preprocessing step in data mining), and so the presence of redundant sequences would undermine our results.
Next, for the remaining 14 species we searched for evidence in the literature that supports existence of PD, or which suggests that they are PD resistant. Eight mammalian species (human, bovine, sheep, elk, cat, mouse, syrian hamster, and ovine) are shown to develop PD (Prusiner, 1997; Prusiner, 1998; Benkel et al. 2007; Murayama et al. 2007). At the same time, prion diseases were never confirmed for the non-mammalian species turtle, chicken and frog, and several studies suggest that they do not develop prion diseases (De Simone et al. 2006; Ji et al. 2007). For the remaining 3 species, i.e. pig, canine, and rabbit, we could not find sufficient evidence to categorize them to either class (Wells et al. 2003; Vorberg et al. 2003; Lysek et al. 2005). We note that canine shares high sequence similarity with PD-prone species, i.e. between 88% for human and 98% for cat, and moderate similarity with PD-resistant species, i.e. between 30% for frog and 41% for turtle. Similarly for rabbit and pig the sequence similarity to PD-prone species ranges between 90% (for human) and 96% (for sheep and ovine), and between 86% (for hamster) and 93% (for elk), respectively, while for PD-resistant the similarity ranges between 31% (for frog) and 42% (for turtle), and between 28% (for frog) and 41% (for turtle), respectively. It is of course possible to simply add the three uncategorized species to the “PD-resistant” class, since no evidence has been produced that they do experience prion disease. However, this would, in our view, be a serious methodological error. Our analysis contrasts species that are known to develop PDs against those that clearly do not, and this distinction directly affects all of the computational techniques (discussed below) that will be employed in our work. The inclusion of pig, canine and rabbit prions would undermine the contrasts, because we could not positively assert that these are truly PD-resistant species. Our methods are fundamentally intended to identify only those differences that perfectly distinguish between the two classes; if the classes themselves become uncertain, our entire methodology becomes merely a “shotgun correlation.” The eleven species we have selected already represent the maximal set of species that we can confidently differentiate into our two classes at the present time. It would be highly desirable to include more species in each class; data-mining techniques such as feature selection are generally intended to operate over thousands or tens of thousands of examples. Obtaining a firm determination of susceptibility to prion disease in canines, rabbits and pigs would be an excellent start.
Point mutations
We performed multiple sequence alignment of the 11 PrP C-terminal domain sequences using ClustalW version 1.83 (Chenna et al. 2003). ClustalW produces biologically meaningful alignments that allow finding identities, similarities and differences between a set of protein sequences. Next, we searched for significant mutations based on positions that are conserved within PD-prone and PD-resistant species. Each position was categorized as follows:
Each position that includes a conserved (the same) amino acid (AA) in the PD-prone species and a conserved (the same) AA (different from the AA conserved for the PD-prone) in the PD-resistant species is categorized as significant. Such a position shows conservation within each group while at the same time it differentiates the contrasts.
Each position that has different AAs over different PD-prone species and/or PD-resistant species is categorized as insignificant. These positions show no significant conservation pattern.
Each position that has conserved (the same) AA over all PD-prone and resistant species is considered insignificant. Although these positions show significant conservation, these residues do not differentiate the contrasts.
Working from the hypothesis that TSE mutations are exclusive to PD-prone species, each significant position is a candidate factor that predisposes PrPC to misfold into PrPSc.
We repeated the same procedure using exchange groups, which represent conservative replacements of AAs through evolution (Dayhoff et al. 1978). They relax the constraint of conservation by defining equivalence classes of AAs, as derived from the BLOSUM AA substitution matrix (Henikoff and Henikoff, 1992), which in turn was derived based on the BLOCKS database (Henikoff and Henikoff, 1991). This reduces the alphabet of 20 AAs to the following six exchange groups: E1 = {H,R,K}, E2 = {D,E,N,Q}, E3 = {C}, E4 = {S,T,P,A,G}, E5 = {M,I,L,V}, and E6 = {F,Y,W}, and we consider a position to be conserved if all corresponding AAs belong to the same exchange group. We then label each position according to the three rules above, using exchange groups instead of individual AAs. Again, any position with conserved (but different) exchange groups in PD-prone and resistant species is another candidate factor that predisposes PrPC to misfold into PrPSc.
Stability of secondary structure
Each prion sequence was converted into a feature-based vector, and the features that differentiate the contrasts were identified using a combination of feature selection methods and correlation analysis. The features represent physicochemical properties of protein sequences that were previously used to characterize and predict certain properties related to the secondary structure of protein sequences, including structural class (Feng et al. 2005; Cao et al. 2006; Kedarisaetti et al. 2006; Kurgan and Chen, 2007) and secondary structure content (Zhang et al. 2001; Ruan et al. 2005; Homaeian et al. 2007). As such, features that discriminate between the contrasts are candidate factors that predispose β-sheet poor PrPC to misfold into β-sheet rich PrPSc. As the conformational change from PrPC to PrPSc will ultimately be driven by physiochemical properties, these features are a promising source of candidate factors. The features we analyze include:
-
Molecular weight, MolW (Kedarisaetti et al. 2006; Homaeian et al. 2007), of a protein sequence is the result of adding up the average molecular weight MolWi values of its residues (see Table. 1) plus the mass of a water molecule (MolWH2O) that is approximately 18 daltons
where N denotes the total number of residues in the sequence.
-
Average isoelectric point, pI (Kedarisaetti et al. 2006; Kurgan and Chen, 2007; Homaeian et al. 2007), of a protein sequence is computed based on the average isoelectric point pIi values of its residues (see Table. 1)
-
Composition vector, CV, and composition moment vector, CMV (Zhang et al. 2001; Feng et al. 2005; Ruan et al. 2005; Cao et al. 2006; Kedarisaetti et al. 2006; Kurgan and Chen, 2007; Homaeian et al. 2007) are defined as the composition percentage of each residue in the sequence that incorporates information about the position of residues
where nij represents the jth position of the ith amino acid, ni is the frequency of ith amino acid in the sequence, and k is the order of the CMV. We apply CMVs for k = 0, 1, 2. Note that CMVi0 reduces to CVi.
-
Order n hydrophobicity auto-correlation function, Ana (Zhang et al. 2001; Kedarisaetti et al. 2006; Homaeian et al. 2007; Kurgan and Chen, 2007), is computed by summing up the products of amino acid indices ai (see Table 1) of every pair of residues separated by n residues.
where a denotes the following hydrophobicity indices: Fauchere-Pliska’s (FH) index (Fauchere and Pliska, 1983) with n = 1,2, …,10 and Eisenberg’s (EH) index (Eisenberg et al. 1984) with n = 1,2, …,6.
-
sum, Hsuma, average, Havga, and 3-point running average, Hsum3a, of the above hydrophobicity indices, (Kedarisaetti et al. 2006; Homaeian et al. 2007; Kurgan and Chen, 2007)
where a = {FH, EH}.
Composition of property groups, PGi, where i denotes a given property (Cao et al. 2006; Kedarisaetti et al. 2006; Homaeian et al. 2007; Kurgan and Chen 2007). AAs are clustered based on their properties (see Table 2) and composition is computed for each of the groups and subgroups. The hydrophobicity group includes hydrophilic and hydrophobic AAs. R group classification is based on molecular weight, hydropathy and isoelectric point. Exchange groups cluster AAs based on accepted point mutations to represent conservative replacements through revolution. Electronic group classification is based on the tendency of AAs to accept or donate electrons. Other groups are defined based on molecular weights, polarity, aromaticity and charge. Finally, chemical groups are based on the composition of chemical groups that constitute the side chains, see Table 1.
Table 1.
List of physicochemical amino acid indices and chemical groups used to derive features.
| Amino acid | Code | Index | Physicochemical index/chemical groups
|
||||
|---|---|---|---|---|---|---|---|
| MolW | pI | FH | EH | Associated chemical groups | |||
| Alanine | A | 1 | 71.0791 | 6.01 | 0.42 | 0.62 | CH CO NH CH3 |
| Cysteine | C | 2 | 103.1437 | 5.07 | 1.34 | 0.29 | CH CO NH CH2 SH |
| Aspartate | D | 3 | 115.0887 | 2.77 | −1.05 | −0.9 | CH CO NH CH2 CO COO− |
| Glutamate | E | 4 | 129.1157 | 3.22 | −0.87 | −0.74 | CH CO NH CH2 CH2 CO COO− |
| Phenylalanine | F | 5 | 147.1772 | 5.48 | 2.44 | 1.19 | CH CO NH CH2 CAROM
CHAROM CHAROM CHAROM CHAROM CHAROM |
| Glycine | G | 6 | 57.0521 | 5.97 | 0 | 0.48 | CH2 CO NH |
| Histidine | H | 7 | 137.1414 | 7.59 | 0.18 | −0.4 | CH CO NH CH2 CAROM
CHAROM N CHAROM NH |
| Isoleucine | I | 8 | 113.16 | 6.02 | 2.46 | 1.38 | CH CO NH CH2 CH CH3 CH3 |
| Lysine | K | 9 | 128.1792 | 9.74 | −1.35 | −1.5 | CH CO NH CH2 CH2 CH2 CH2 NH3+ |
| Leucine | L | 10 | 113.16 | 5.98 | 2.32 | 1.06 | CH CO NH CH2 CH CH3 CH3 |
| Methionine | M | 11 | 131.1977 | 5.47 | 1.68 | 0.64 | CH CO NH CH2 CH2 S CH3 |
| Asparagine | N | 12 | 114.104 | 5.41 | −0.82 | −0.78 | CH CO NH CH2 CO C NH2 |
| Proline | P | 13 | 97.1171 | 6.48 | 0.98 | 0.12 | CHRING CO NHRING CH2RING
CH2RING CH2RING |
| Glutamine | Q | 14 | 128.131 | 5.65 | −0.3 | −0.85 | CH CO NH CH2 CH2 CO C NH2 |
| Arginine | R | 15 | 156.188 | 10.76 | −1.37 | −2.53 | CH CO NH CH2 CH2 CH2 NH C
NH2 NH2+ |
| Serine | S | 16 | 87.0784 | 5.68 | −0.05 | −0.18 | CH CO NH CH2 OH |
| Threonine | T | 17 | 101.1054 | 5.87 | 0.35 | −0.05 | CH CO NH CH CH3 OH |
| Valine | V | 18 | 99.133 | 5.97 | 1.66 | 1.08 | CH CO NH CH CH3 CH3 |
| Tryptophan | W | 19 | 186.2139 | 5.89 | 3.07 | 0.81 | CH CO NH CH2 CAROM
CAROM CAROM NH CHAROM CHAROM CHAROM CHAROM CHAROM |
| Tyrosine | Y | 20 | 163.1756 | 5.67 | 1.31 | 0.26 | CH CO NH CH2 CAROM
CHAROM CHAROM CHAROM CHAROM CAROM OH |
Table 2.
Property groups of amino acids used to derive features.
| Groups | Subgroups | AAs | Groups | Subgroups | AAs |
|---|---|---|---|---|---|
| R groups | Nonpolar aliphatic | AVLIMG | Hydrophobicity groups | Hydrophobic | VLIMAFPWYCG |
| Polar uncharged | SPTCNQ | Hydrophilic basic | KHR | ||
| Positively charged | KHR | Hydrophilic acidic | DE | ||
| Negative | DE | Hydrophilic polar with uncharged side chain | STNQ | ||
| Aromatic | FYW | ||||
| Exchange groups | E1 | KHR | Electronic groups | Electron donor | DEPA |
| E2 | DENQ | Weak electron donor | VLI | ||
| E3 | C | Electron acceptor | KNR | ||
| E4 | AGPST | Weak electron acceptor | FYMTQ | ||
| E5 | ILMV | Neutral | GHWS | ||
| E6 | FYW | Special AA | C | ||
| Other groups | Charged | DEKHRVLI | Other groups | Tiny | AG |
| Polar | DEKHRNTQSYW | Bulky | FHWYR | ||
| Aromatic | FHWY | Polar-uncharged | NQ | ||
| Small | AGST |
We employed three feature selection techniques to minimize bias in our results. These are the ReliefF (Robnik-Sikonja and Kononenko, 2003), information gain (Quinlan, 1993), and the χ2 statistics, taken between a given attribute and the binary class (PD-prone/PD-resistant). The ReliefF algorithm estimates the ability of features to separate classes. This algorithm examines nearest-neighbors of a feature vector that belong to the same or a different class as the vector under consideration. Features that categorize these nearest neighbors correctly receive a high score, and the process is repeated for each feature vector. The second selection technique is based on the concept of minimization of information entropy, while the chi-square statistic measures deviation from an assumed (normal) distribution of values for independent variables. All three feature selection algorithms are implemented in the WEKA data-mining software package (Witten and Frank, 2005). As a cross-check on the three selection algorithms, we also compute the bi-serial correlation between each feature and the binary class variable.
Results and Discussion
Point mutations
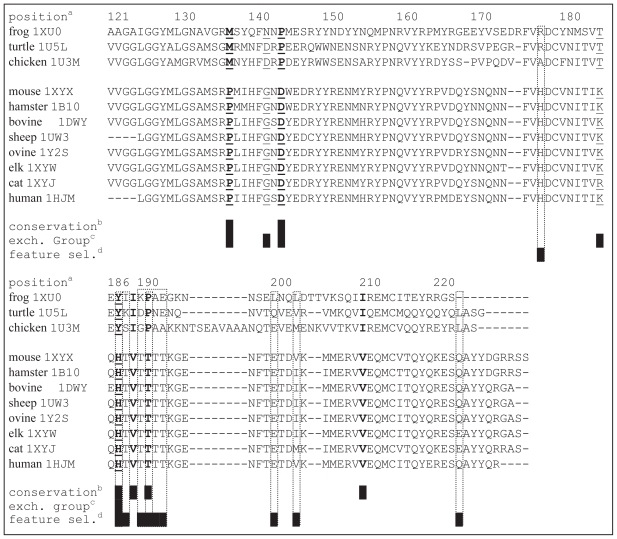
The aligned prion sequences are shown in Figure 2. Our analysis shows the following significant positions: 137, 144, 187, 189, 191, and 210, which are associated with the following point mutations with respect to huPrP: P137M, D144P, H187Y, V189I, T191P, V210I (see Fig. 2). Similarly, when considering conservation at the level of exchange groups, the following positions were found significant: 137, 142, 144, 185, and 187. The positions 137, 144, and 187 overlap with the results of residue conservation, while the remaining two positions are associated with G142D, G142N, and K185T point mutations. One mutation is a confirmatory result, while the remaining eight are new findings:
Figure 2.
Results of sequence alignment between the three PD-resistant prions (top) and the eight PD-prone prions (identified by the PDB ID for the protein).
aPositions are encoded with respect to the huPrP.
b“Conservation” line shows positions (black squares with the corresponding residues shown in bold) that were significant based on the conservation of amino acids.
c“Exch. group” line shows positions (black squares with the corresponding residues underlined) that were significant based on the conservation of amino acids grouped in exchange groups.
d“Feature sel.” line shows positions (black squares with the corresponding residues denoted by dotted line boxes) that were significant based on the feature selection.
P137M (new finding). Residues that compose helix-1 are not involved in hydrogen bonds with the rest of the C-terminal domain. This is true except for Y149 and Y150 which belong to helix- 1 and whose side chain hydroxyls donate to the carboxyl groups of D202 and the CO of P137 (Riek et al. 1998). Therefore, a mutation at P137 could further weaken interaction between helix-1 and the rest of the C-terminal domain. At the same time, several studies report importance of weakened interactions between helix-1 and other segments in the C-terminal domain on the folding into a stable native structure (Hirschberger et al. 2006; Schwarzinger et al. 2006; Eghiaian et al. 2007)
G142D and G142N (new findings). A mutation at the same position, i.e. G142S, was previously classified as having a CJD-like phenotype (Gambetti et al. 2003). For this mutation, Glycine at position 142 was substituted with a polar, hydrophilic Serine. Using our approach, we identified mutations at that position involving Aspartate and Asparagine, which are very similar to each other and both also polar and hydrophilic, similar to the known mutation.
D144P (new finding). Previous research shows that D144 forms a salt bridge with H140, R148 and R208 (Zuegg and Gready, 1999). The salt bridge between D144 and R208 links helix-1 and helix-3, while the R208H mutation is associated with CJD (Riek et al. 1998). Since salt bridges are suggested to increase the stability of proteins, mutation at this position could potentially lead to destabilization of the prion’s structure. Recent results also show that a point mutation leading to the disruption of a single salt bridge in p53 increases propensity to form amyloid fibrils (Galea et al. 2005).
H187Y (new finding). This position is associated with a known H187R mutation that results in GSS (Cervenakova et al. 1999). At the same time, both Tyrosine and Arginine are polar and similar in size, i.e. their van der Waals volumes are 141 and 148, respectively.
V210I (confirmatory finding). This mutation is well-known and is associated with CJD in humans (Riek et al. 1998).
We have shown that several of the new mutations we have found are closely related to known mutations involved in TSE diseases, while others may impact structural stability of the prion protein. While we were unable to find established research that would directly corroborate the remaining new mutations (K185T, V189I, and T191P), existing research indicates that mutations in this segment (which contains helix-2) may have β-sheet promoting effects. Helix-2 is characterized by a strong propensity for the extended conformation, and a single AA replacement in the vicinity of this helix is shown to significantly affect the conformational preference of the entire helix-2–helix-3 segment and to further increase the propensity for the extended conformation, facilitating conformational rearrangement in this region (Knaus et al. 2001; Kuznetsov and Rackovsky, 2004). These findings also correlate well with the high number of disease-promoting mutations in helices-2 and -3, which also points to the particular importance of these helices for conformational transition (only one disease-promoting mutation is found in helix-1 while seven and eight such mutations are found in helix-2 and helix-3, respectively).
Stability of secondary structures
Our feature selection was performed using tenfold cross-validation to assure statistical validity for our results. Features are evaluated in each fold, and then they are ranked on their performance across all ten folds. Higher-ranked features have greater discriminatory power for the contrasts than lower-ranked ones. We average the ranks reported for each feature across our three feature selection methods. We report the top five features, ordered by average rank, which have biserial correlation coefficient values >0.9 in Table 3. The biserial correlation coefficient measures correlations between ratio-scale and binary variables, and is interpreted in the usual manner (values >0.8 indicate strong correlations).
Table 3.
Top five features that differentiate between PD-prone and resistant prions.
| Feature | Avg. rank | Bi-serial correlation coefficient |
|---|---|---|
| CMVP1 | 7.6 | 0.97 |
| Chemical N group | 9.3 | 0.94 |
| CMVH1 | 11.1 | 0.97 |
| CMVT1 | 12.2 | 0.96 |
| CMVL2 | 12.8 | 0.99 |
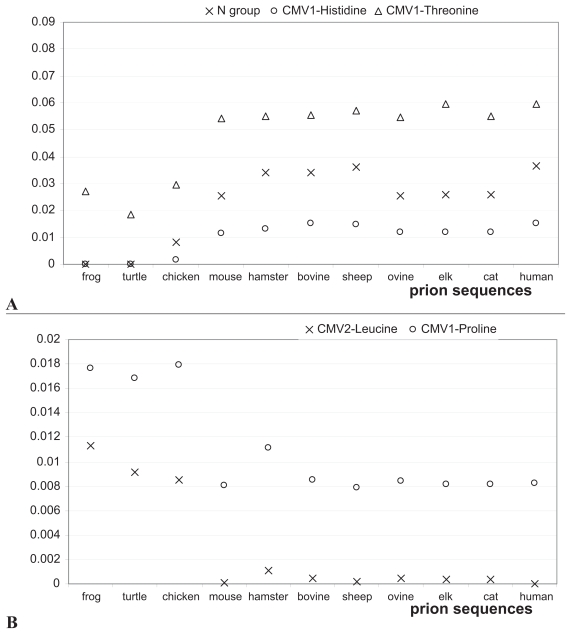
The five features in Table 3 fall into two groups: those that show higher values for PD-prone species than PD-resistant species, and those that show higher values for PD-resistant species than PD-prone species. We begin our discussion with the former group. The second feature in Table 3 is related to the composition of the N group in the AA side chains. Since N group occurs only in Histidine, this feature indicates that presence of this AA is specific to one group of prion proteins. This finding is also supported by the third feature, CMVH1, which reveals additional details. Values of these two correlated features for the ten prion sequences are shown in Figure 3(a). The plots shows higher values of the composition moment vector for Histidine for the PD-prone species when compared with the PD-resistant species. Since the composition moment values are proportional to the distance of the corresponding residue from the N-terminal, high values indicate the presence of Histidine near the C-terminal in the PD-prone prions. Figure 2 shows two highly conserved Histidine positions in helix-2, i.e. 177 and 187, that are specific to PD-prone prions, while the only position in the PD-resistant chicken prion that contains Histidine is 140. This finding is supported by prior research which shows that charged Histidine side chains in the middle of α-helices have a destabilizing effect on the structure because of the unfavorable interaction with the helix macrodipole (Armstrong and Baldwin, 1993). This destabilizing effect in the context of protonation of H187 (Langella et al. 2004) provides some explanation for the weak stability of helix-2. We note that this finding can also be related to the H187R mutation, associated with GSS.
Figure 3.
Values of top five features for the 11 prion sequences: features that indicate abundance of the associated amino acids in A) PD-prone prions, and B) PD-resistant prions. The first three sequences correspond to the PD-resistant prions, and the remaining sequences to the PD-prone prions.
The CMVT1 feature, which again is characterized by higher values for PD-prone species (see Fig. 3A) reveals that Threonine is significantly more abundant in this group of species. Figure 2 reveals that a highly conserved TVTTTT segment in helix-2 is specific to these prions. This segment is surface exposed and located between two glycosylation sites and most likely “covered” by the glycan side chains. It was previously found to be significant in the context of a potential molecular mechanism leading to the destabilization of the helix-2 segment, which postulates formation of a hydrogen bond between residues T188 and T193 that drives the unwinding of the α-helix (Pappalardoa et al. 2004). Another study that looked into the TTTT sub-segment (positions 190–193) concluded that this sub-segment is usually found in a strand and/or loop conformation and that the second half of helix-2 would be better accommodated in non-helical conformations (Dima and Thirumalai, 2004).
In contrast, the remaining two features have higher values for the PD-resistant prions; see Figure 3B. Analysis of the aligned sequences shown in Figure 2 reveals that although Leucine is present at positions 125, 130, and 138 in both types of prions, this AA is only present in the vicinity of the C-terminal in the PD-resistant prions. As a result, positions 200, 203, and 223 (located within helix-3) were identified as significant locations based on the position-sensitive CMVL2 feature (see Fig. 2). Recent computational analysis of local interactions that promote formation of secondary structures shows that Alanine, Glutamine, Glutamate, and Leucine are strongly associated with formation of helices (Chen et al. 2006). We also note that positions 200 and 203 are associated with known mutations. Position 203 is associated with the V203I mutation that causes CJD (Peoc’h et al. 2000). E200K, which results in CJD, is one of the most common worldwide prion mutations (Mead, 2006). This mutation results in loss of a salt-bridge interaction between the side chains of E200 and K204 (Zhang et al. 2000). In the native huPrP these side chains are intimately juxtaposed (within 5 Å) and therefore they could be involved in a salt bridge. In the E200K mutant protein, the nearest negatively charged side chain to E200 is that of D196 which is 13 Å from E200 (Zhang et al. 2000). Therefore, mutation on this position could result in destabilization of the structure.
Finally, the CMVP1 feature indicates that location 191, which contains a highly conserved Pro-line residue, is specific to the PD-resistant prions (see Fig. 2). We were unable to find existing research that would corroborate the significance of this position, due to the limited amount of work on non-mammalian prions.
Conclusions
We present a novel, in-silico approach to identify factors related to misfolding of prion proteins. We contrasted PrPC sequences of the C-terminal domains of PD-prone and PD-resistant species. The analysis focused on finding significant point mutations and investigating structural stability of secondary structures that comprise the C-terminal domain. We confirmed the V210I mutation, which is associated with CJD, and present several new findings that include P137M, G142D, G142N, D144P, K185T, V189I, H187Y and T191P mutations; destabilizing effects of Histidine and the T188-T193 segment on stability of helix-2 in the PD-prone prions; and stabilizing effects of Leucine on helix-3 in the PD-resistant species. All of these new findings are possible candidate factors that could influence conformational change from PrPC to PrPSc. They are a new set of hypotheses that should be investigated via wet-lab experimentation or (at a minimum) molecular dynamics simulations. In addition, if and when additional species can be definitively classified as PD-prone or PD-resistant, it would be quite interesting to repeat our experiments with these additional species included in the contrasts. Finally, we note that the resistance to prion diseases of the PD-resistant species could be a result of other factors besides the differences in their sequences, which should be addressed in future studies.
Acknowledgements
This research was partially supported by NSERC Canada, the Province of Alberta’s Queen Elizabeth II graduate scholarship, and the Alberta Ingenuity Fund.
References
- Armstrong KM, Baldwin RL. Charged histidine affects a-helix stability at all positions in the helix by interacting with the backbone charges. Proc. Natl. Acad. Sci., U.S.A. 1993;90:11337–40. doi: 10.1073/pnas.90.23.11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkel BF, Valle E, Bissonnette N, et al. Simultaneous detection of eight single nucleotide polymorphisms in the ovine prion protein gene. Mol. Cell. Probes. 2007;21(5–6):363–7. doi: 10.1016/j.mcp.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset L, Melki R. Similar and divergent features in mammalian and yeast prions. Microbes Infect. 2002;4(4):461–9. doi: 10.1016/s1286-4579(02)01561-7. [DOI] [PubMed] [Google Scholar]
- Cao Y, Liu S, Zhang L, et al. Prediction of protein structural class with rough sets. BMC Bioinformatics. 2006;7:20. doi: 10.1186/1471-2105-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervenakova L, Butefisch C, Lee HS, et al. Novel PRNP sequence variant associated with familial encephalopathy. Am. J. Med. Genet. 1999;88:653–656. doi: 10.1002/(sici)1096-8628(19991215)88:6<653::aid-ajmg14>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Chen K, Kurgan L, Ruan J. Optimization of the Sliding Window Size for Protein Structure Prediction. 2006 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology; Toronto. 2006. pp. 366–372. [Google Scholar]
- Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. A Model of Evolutionary Change in Proteins. Atlas of Protein Sequence and Structure. 1978;15(Suppl 3):345–58. [Google Scholar]
- De Simone A, Dodson GG, Fraternali F, Zagari A. Water molecules as structural determinants among prions of low sequence identity. FEBS Lett. 2006;580:2488–94. doi: 10.1016/j.febslet.2006.02.083. [DOI] [PubMed] [Google Scholar]
- Dima RI, Thirumalai D. Probing the instabilities in the dynamics of helical fragments from mouse PrPC. Proc. Natl. Acad. Sci. U.S.A. 2004;101(43):15335–40. doi: 10.1073/pnas.0404235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghiaian F, Grosclaude J, Lesceu S, et al. Insight into the PrPC –> PrPSc conversion from the structures of antibody-bound bovine prion scrapie-susceptibility variants. Proc. Natl. Acad. Sci., U.S.A. 2004;101:10254–9. doi: 10.1073/pnas.0400014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghiaian F, Daubenfeld T, Quenet Y, et al. Diversity in prion protein oligomerization pathways results from domain expansion as revealed by hydrogen/deuterium exchange and disulfide linkage. Proc. Natl. Acad. Sci., U.S.A. 2007;104(18):7414–9. doi: 10.1073/pnas.0607745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Weiss RM, Trewilliger TC. The Hydrophobic Moment Detects Periodicity in Protein Hydrophobicity. Proc. Natl. Acad. Sci., U.S.A. 1984;81:140–4. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchere JL, Pliska V. Hydrophobic parameters p of amino-acid side chains from the partitioning of N.-acetyl-amino-acid amides. European J. of Medicinal Chemistry. 1983;18:369–75. [Google Scholar]
- Feng KY, Cai YD, Chou KC. Boosting classifier for predicting protein domain structural class. Biochem. Biophys. Res. Commun. 2005;334(1):213–7. doi: 10.1016/j.bbrc.2005.06.075. [DOI] [PubMed] [Google Scholar]
- Galea C, Bowman P, Kriwacki RW. Disruption of an inter-monomer salt bridge in the p53 tetramerization domain results in an increased propensity to form amyloid fibrils. Protein Sci. 2005;14(12):2993–3003. doi: 10.1110/ps.051622005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetti P, Kong Q, Zou W, et al. Sporadic and familial CJD: classification and characterisation. British Medical Bulletin. 2003;66:213–39. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- Govaerts C, Wille H, Prusiner SB, et al. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci., U.S.A. 2004;90:8342–7. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino Acid Substitution Matrices from Protein Blocks. Proc. Natl. Acad. Sci., U.S.A. 1992;89:10915–19. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Automated Assembly of Protein Blocks for Database Searching. Nucleic Acids Res. 1991;19:6565–72. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberger T, Stork M, Schropp B, et al. Structural instability of the prion protein upon M205S/R. mutations revealed by molecular dynamics simulations. Biophys. J. 2006;90(11):3908–18. doi: 10.1529/biophysj.105.075341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn F, Vriend G, Cohen FE. Collecting and Harvesting Biological Data: The GPCRDB and NucleaRDB Databases. Nucleic Acids Res. 2001;29:346–9. doi: 10.1093/nar/29.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homaeian L, Kurgan L, Cios KJ, et al. Prediction of Protein Secondary Structure Content for the Twilight Zone Sequences. Proteins. 2007;69(3):486–98. doi: 10.1002/prot.21527. [DOI] [PubMed] [Google Scholar]
- Huang Z, Prusiner S, Cohen FE. Scrapie prions: A three-dimensional model of an infectious fragment. Fold. Des. 1995;1:13–19. [PubMed] [Google Scholar]
- Ji HF, Zhang HY, Chen LL. Why are prion diseases precluded by non-mammals? . Trends Biochem. Sci. 2007;32(5):206–8. doi: 10.1016/j.tibs.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Kedarisetti KD, Kurgan L, Dick S. Classifier Ensembles for Protein Structural Class Prediction with Varying Homology. Biochem. Biophys. Res. Commun. 2006;348(3):981–8. doi: 10.1016/j.bbrc.2006.07.141. [DOI] [PubMed] [Google Scholar]
- Knaus KJ, Morillas M, Swietnicki W, et al. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001;8:770–4. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- Kovacs GG, Trabattoni G, Hainfellner JA, et al. Mutations of the prion protein gene phenotypic spectrum. J. Neurol. 2002;249:1567–82. doi: 10.1007/s00415-002-0896-9. [DOI] [PubMed] [Google Scholar]
- Kurgan L, Chen K. Prediction of Protein Structural Class for the Twilight Zone Sequences. Biochem. Biophys. Res. Commun. 2007;357(2):453–60. doi: 10.1016/j.bbrc.2007.03.164. [DOI] [PubMed] [Google Scholar]
- Kuznetsov IB, Rackovsky S. Comparative computational analysis of prion proteins reveals two fragments with unusual structural properties and a pattern of increase in hydrophobicity associated with disease-promoting mutations. Protein Science. 2004;13:3230–44. doi: 10.1110/ps.04833404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langella E, Improta R, Barone V. Checking the pH-induced conformational transition of prion protein by molecular dynamics simulations: effect of protonation of histidine residues. Biophys. J. 2004;87:3623–32. doi: 10.1529/biophysj.104.043448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia F, Zahn R, Riek R, et al. NMR. structure of the bovine prion protein. Proc. Natl. Acad. Sci., U.S.A. 2000;97:8334–9. doi: 10.1073/pnas.97.15.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysek DA, Schorn C, Nivon LG, et al. Prion protein NMR. structures of cats, dogs, pigs, and sheep. Proc. Natl. Acad. Sci., U.S.A. 2005;102(3):640–5. doi: 10.1073/pnas.0408937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S. Prion disease genetics. Eur. J. Hum. Gen. 2006;14:273–81. doi: 10.1038/sj.ejhg.5201544. [DOI] [PubMed] [Google Scholar]
- Morrisey MP, Shakhnovich EI. Evidence for the role of PrPC helix in the hydrophilic seeding of prion aggregates. Proc. Natl. Acad. Sci., U.S.A. 1999;96:11293–8. doi: 10.1073/pnas.96.20.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Yoshioka M, Okada H, et al. Urinary excretion and blood level of prions in scrapie-infected hamsters. J. Gen. Virol. 2007;88:2890–8. doi: 10.1099/vir.0.82786-0. [DOI] [PubMed] [Google Scholar]
- Pan KM, Baldwin M, Nguyen J, et al. Conversion of (-helices into (-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci., U.S.A. 1993;90:10962–6. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardoa M, Milardib D, La Rosaa C, et al. A molecular dynamics study on the conformational stability of PrP 180–193 helix II prion fragment. Chem. Phys. Lett. 2004;390(4–6):511–6. [Google Scholar]
- Peoc’h K, Manivet P, Beaudry P, et al. Identification of three novel mutations (E196K, V203I, E211Q.) in the prion protein gene (PRNP) in inherited prion diseases with Creutzfeldt-Jakob disease phenotype. Hum. Mutat. 2000;15(5):482. doi: 10.1002/(SICI)1098-1004(200005)15:5<482::AID-HUMU16>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Pappalardo M, Milardi D, Grasso D, et al. Steered molecular dynamics studies reveal different unfolding pathways of prions from mammalian and non-mammalian species. New J. Chem. 2007;31:901–5. [Google Scholar]
- Prusiner SB. Prions. Proc. Natl. Acad. Sci., U.S.A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prion Diseases and the BSE Crisis. Science. 1997;278:245. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Scott MR, DeArmond SJ, et al. Prion protein biology. Cell. 1998;93:337–8. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- Quinlan JR. C45: Programs for Machine Learning. San Mateo, CA: Morgan Kaufmann; 1993. [Google Scholar]
- Riek R, Hornemann S, Wider G, et al. NMR. characterization of the full-length recombinant murine prion protein, mPrP(23–231) FEBS Lett. 1997;413:282–8. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- Riek R, Wider G, Billiter M, et al. Prion protein NMR. structure and familial human spongiform encephalopathies. Proc. Natl. Acad. Sci., U.S.A. 1998;95:11667–72. doi: 10.1073/pnas.95.20.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robnik-Sikonja M, Kononenko I. Theoretical and Empirical Analysis of ReliefF and RReliefF. Machine Learning J. 2003;53:23–69. [Google Scholar]
- Ruan J, Wang K, Yang J, et al. Highly accurate and consistent method for prediction of helix and strand content from primary protein sequences. Artificial Intelligence in Medicine. 2005;35(1–2):9–35. doi: 10.1016/j.artmed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Schwarzinger S, Horn AH, Ziegler J, et al. Rare large scale subdomain motions in prion protein can initiate aggregation. J. Biomol Struct. Dyn. 2006;23(6):581–90. doi: 10.1080/07391102.2006.10507083. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Cavalli A, Pellarin R, et al. Prediction of aggregation rate and aggregation-prone segments in polypeptide sequences. Protein Science. 2005;14:2723–34. doi: 10.1110/ps.051471205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watzlawik J, Skora L, Frense D, et al. Prion protein helix1 promotes aggregation but is not converted into β (-sheet. J. Biol. Chem. 2006;281(40):30242–50. doi: 10.1074/jbc.m605141200. [DOI] [PubMed] [Google Scholar]
- Wells G, Hawkins S, Austin A, et al. Studies of the transmissibility of the agent of bovine spongiform encephalopathy to pigs. J. Gen. Virol. 2003;84:1021–31. doi: 10.1099/vir.0.18788-0. [DOI] [PubMed] [Google Scholar]
- Wille H, Michelitsch MD, Guenebaut V, et al. Structural studies of the scrapie prion protein by electron crystallography. Proc. Natl. Acad. Sci., U.S.A. 2002;99:3563–8. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten I, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd edition. Morgan Kaufmann; San Francisco: 2005. [Google Scholar]
- Vorberg I, Groschup MH, Pfaff E, et al. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J. Virol. 2003;77:2003–9. doi: 10.1128/JVI.77.3.2003-2009.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Sun ZR, Zhang CT. A New Approach to Predict the Helix/Strand Content of Globular Proteins. J. Theor. Biol. 2001;208:65–78. doi: 10.1006/jtbi.2000.2201. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Swietnicki W, Zagorski MG, et al. Solution structure of the E200K variant of human prion protein: Implications for the mechanism of pathogenesis in familial prion diseases. J. Biol. Chem. 2000;275(43):33650–4. doi: 10.1074/jbc.C000483200. [DOI] [PubMed] [Google Scholar]
- Zuegg J, Gready JE. Molecular Dynamics Simulations of Human Prion Protein: Importance of Correct Treatment of Electrostatic Interactions. Biochemistry. 1999;38(42):13862–76. doi: 10.1021/bi991469d. [DOI] [PubMed] [Google Scholar]