Abstract
The explosive 2,4,6-trinitrotoluene (TNT) is a significant environmental pollutant that is both toxic and recalcitrant to degradation. Phytoremediation is being increasingly proposed as a viable alternative to conventional remediation technologies to clean up explosives-contaminated sites. Despite the potential of this technology, relatively little is known about the innate enzymology of TNT detoxification in plants. To further elucidate this, we used microarray analysis to identify Arabidopsis (Arabidopsis thaliana) genes up-regulated by exposure to TNT and found that the expression of oxophytodienoate reductases (OPRs) increased in response to TNT. The OPRs share similarity with the Old Yellow Enzyme family, bacterial members of which have been shown to transform explosives. The three predominantly expressed forms, OPR1, OPR2, and OPR3, were recombinantly expressed and affinity purified. Subsequent biochemical characterization revealed that all three OPRs are able to transform TNT to yield nitro-reduced TNT derivatives, with OPR1 additionally producing the aromatic ring-reduced products hydride and dihydride Meisenheimer complexes. Arabidopsis plants overexpressing OPR1 removed TNT more quickly from liquid culture, produced increased levels of transformation products, and maintained higher fresh weight biomasses than wild-type plants. In contrast, OPR1,2 RNA interference lines removed less TNT, produced fewer transformation products, and had lower biomasses. When grown on solid medium, two of the three OPR1 lines and all of the OPR2-overexpressing lines exhibited significantly enhanced tolerance to TNT. These data suggest that, in concert with other detoxification mechanisms, OPRs play a physiological role in xenobiotic detoxification.
Large amounts of land and water are heavily contaminated by explosives, mainly as a result of the manufacture and military use of munitions. The high financial cost associated with cleaning up these contaminated sites largely precludes the use of many existing remediation technologies, such as soil excavation and incineration or disposal to landfill. There is a great deal of work documenting the global contamination, general toxicity, and microbial metabolism of 2,4,6-trinitrotoluene (TNT) in the environment; however, relatively little is known about the enzymes mediating the detoxification of TNT in plants (for review, see Rylott and Bruce, 2009).
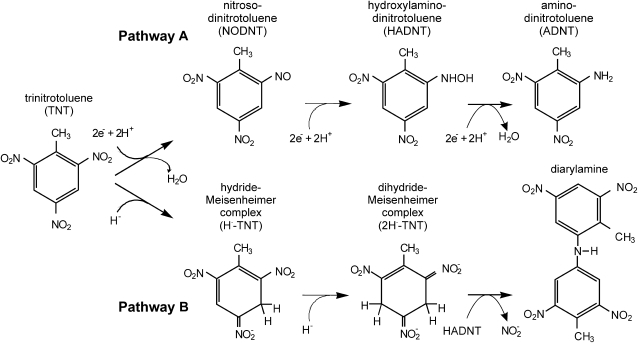
Phytoremediation, the use of plants to remove environmental pollutants, offers a low-cost, sustainable alternative to conventional remediation technologies and is attracting considerable attention as a means to clean up sites contaminated with explosives. While TNT is a potent phytotoxin, plants are able to detoxify low levels of TNT. In an effort to determine how plant tolerance could be further improved, we are investigating the biochemistry and enzymology underlying the innate ability of plants to detoxify TNT. The detoxification of xenobiotics has been loosely categorized into three phases (Sandermann, 1992): activation, conjugation, and compartmentation. The proposed route of TNT detoxification follows these phases. The electron-withdrawing properties of the nitro groups of TNT make the aromatic ring electron deficient. This favors reductive transformation reactions in plants, and the TNT molecule is most commonly activated by the reduction of a nitro group to give hydroxylamino and then amino derivatives (Fig. 1, pathway A). Following the introduction of a functional group, more hydrophilic molecules such as Glc are conjugated to the activated TNT molecule (Gandia-Herrero et al., 2008), facilitating transport and subsequent compartmentation or sequestration.
Figure 1.
The transformation of TNT by pentaerythritol tetranitrate reductase. Pathway A shows the transformation of the nitro group to nitroso-dinitrotoluene (NODNT), HADNT, and then ADNT products. Pathway B shows the reduction of the aromatic ring to form hydride and dihydride Meisenheimer complexes, then chemical condensation with HADNT to form diarylamines.
Data from both our microarray experiments (Gandia-Herrero et al., 2008) and other expression studies (Ekman et al., 2003; Mezzari et al., 2005) have found that members of the small gene family of oxophytodienoate reductases (OPRs) in Arabidopsis (Arabidopsis thaliana) are up-regulated following exposure to TNT. The Arabidopsis genome contains three characterized OPRs: OPR1, OPR2, and OPR3. In addition, there are three as yet uncharacterized putative OPRs, named here as OPR4, OPR5, and OPR6, with OPR4 and OPR5 being identical. The physiological functions of the OPRs remain obscure, with the exception of OPR3, which is involved in jasmonic acid biosynthesis, converting (9S,13S)-12-oxophytodienoic acid to 3-2(2′(Z)-pentyl)cyclopentane-1-octanoic acid in the peroxisome (Sanders et al., 2000; Stintzi and Browse, 2000; for review, see Wasternack, 2007). OPR3 is located on chromosome II within the Arabidopsis genome and contains a C-terminal Ser-Arg-Leu type 1 peroxisome-targeting sequence. The remaining OPRs are all located on chromosome I and do not possess any known organelle-targeting sequences. The spatial expression of OPR1 and OPR2 across root cells where TNT accumulates (Biesgen and Weiler, 1999; Baerenfaller et al., 2008) favors a role in detoxification.
The OPRs share similarity with the Old Yellow Enzyme family, a group of flavoenzymes that has been repeatedly associated with the transformation of explosives (Binks et al., 1996; Schaller and Weiler, 1997; Snape et al., 1997; Basran et al., 1998; French et al., 1998; Blehert et al., 1999; Pak et al., 2000; Fitzpatrick et al., 2003; Williams et al., 2004). Studies also indicate that Old Yellow Enzyme homologs function as antioxidants, detoxifying the breakdown products of lipid peroxidation and other toxic electrophilic compounds (Kohli and Massey, 1998; Williams and Bruce, 2002; Fitzpatrick et al., 2003; Trotter et al., 2006). This oxidative stress could result from exposure to xenobiotics including TNT, wounding, or pathogen attack.
Pentaerythritol tetranitrate reductase, an Old Yellow Enzyme homolog isolated from Enterobacter cloacae (Binks et al., 1996), possesses two catalytic activities toward TNT (Fig. 1): nitroreduction of TNT to form hydroxylamino-dinitrotoluene (HADNT) and then amino-dinitrotoluene (ADNT), and aromatic ring reduction of TNT to yield hydride and dihydride (2H−-TNT) Meisenheimer TNT adducts (French et al., 1998; Williams et al., 2004). The TNT ring-reduced compounds condense via a nonenzymatic reaction with HADNTs to form diarylamines, with the liberation of nitrite (Wittich et al., 2008). Expression of pentaerythritol tetranitrate reductase in tobacco (Nicotiana tabacum) confers both resistance to, and the ability to transform, TNT (French et al., 1999). OPR1, OPR2, and OPR3 share 43%, 44%, and 36% identity, respectively, with pentaerythritol tetranitrate reductase, and all possess the conserved active site amino acids crucial for TNT transformation by pentaerythritol tetranitrate reductase and other members of the Old Yellow Enzyme family (Snape et al., 1997; French et al., 1998; Khan et al., 2004), suggesting that they are capable of transforming TNT.
The OPR4/5 protein is predicted to have reduced activity toward TNT, compared with the other OPRs, owing to a C-terminal truncation that removes residues thought to be important in binding the cofactor NADH, Thus, we investigated OPR1, -2, and -3 as likely candidates for the TNT nitroreduction activity in Arabidopsis.
RESULTS
Expression of OPRs in Arabidopsis in Response to TNT
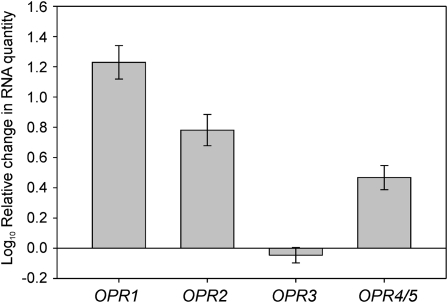
To identify genes up-regulated in response to TNT treatment, we performed a microarray experiment (Gandia-Herrero et al., 2008). Liquid culture-grown 14-d-old seedlings were treated with 60 μm TNT for 6 h, and then gene expression was analyzed. The microarray data revealed 633 genes that were significantly up-regulated between 2- and 204-fold, with a median value of 3.0. Many were genes involved in the activation and conjugation phases of xenobiotic detoxification, including glycosyltransferases, glutathione reductases, and OPRs. The microarray probes used did not distinguish between OPR1 and OPR2; thus, the 14-fold up-regulation observed represented a pooled value for the expression of both genes. For OPR4/5, a 3-fold increase was seen; OPR3 was not up-regulated, and OPR6 expression was not detected in the microarray data. Searches of publicly available microarray expression data sets revealed that OPR6 was rarely up-regulated, and no OPR6 expression was found in liquid culture samples exposed to TNT using reverse transcription (RT)-PCR (data not shown). To better quantify the expression levels of the OPRs in response to the TNT treatment, quantitative RT-PCR was performed using gene-specific probes. Analysis showed that OPR1 expression increased by approximately 17-fold, OPR2 expression by 6-fold, and OPR4/5 by 3-fold, while no specific increase in OPR3 expression was seen (Fig. 2). Given that expression levels of OPR4/5 and OPR6 in response to TNT were low; that expression in the roots, the site of TNT accumulation, was predicted to be low relative to OPR1, OPR2, and OPR3; and that OPR4/5 has a C-terminal truncation predicted to reduce activity, we concluded that OPR4/5 and OPR6 were unlikely to play a significant role in TNT detoxification in planta, and further characterization was focused on OPR1, OPR2, and OPR3.
Figure 2.
Quantitative RT-PCR of OPR expression in response to TNT treatment. Fourteen-day-old, liquid culture-grown Arabidopsis plants were treated with 60 μm TNT for 6 h, and the level of OPR expression was determined by quantitative RT-PCR. ACT2 mRNA was used to normalize cDNA levels. Results are means of three independent RNA isolations measured in triplicate ± se.
Transformation of TNT by Recombinant OPRs
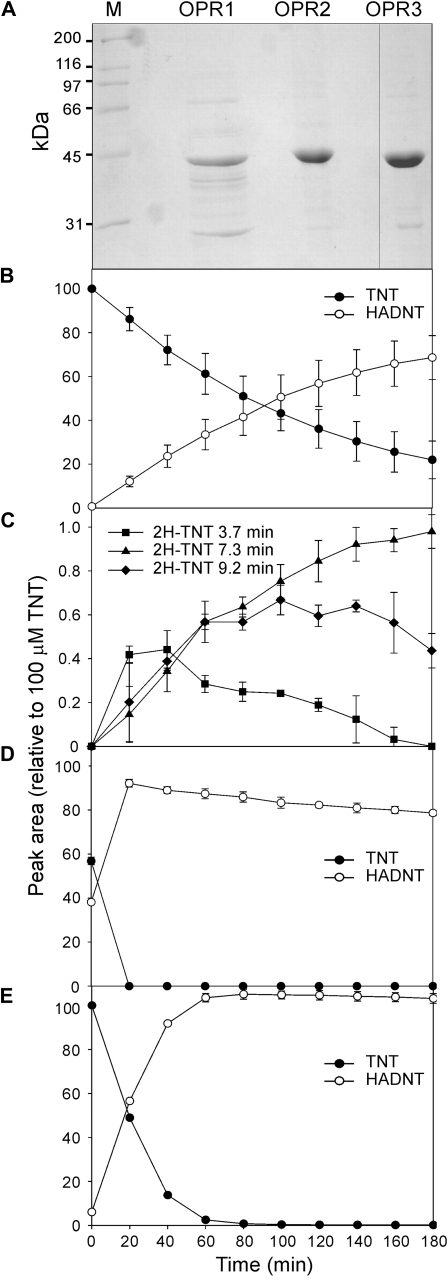
To determine whether OPR1, OPR2, and OPR3 were capable of producing reduced TNT products and hydride Meisenheimer complexes, OPR1, -2, and -3 were cloned into pET-15b expression vectors and transformed into Escherichia coli. Following expression optimization steps, soluble, active recombinant protein was obtained for OPR1, OPR2, and OPR3. The recombinant proteins were purified by affinity chromatography (Fig. 3A), and the identities of these proteins were confirmed using trypsin digestion and matrix-assisted laser-desorption ionization time of flight/time of flight (MALDI-TOF/TOF) tandem mass spectrometry.
Figure 3.
Purification and characterization of Arabidopsis OPRs. A, SDS-PAGE analysis of affinity-purified Arabidopsis OPR1, OPR2, and OPR3 from E. coli. Lane M contains protein markers. B to E, Time course of TNT disappearance and product formation from purified in vitro assays analyzed by HPLC. B and C, OPR1. D, OPR2. E, OPR3. 2H-TNT, Hydride Meisenheimer complexes and elution times. Results are means of three replicates ± sd.
The NADH-dependent transformation of TNT by the purified OPRs was followed by HPLC over a period of 3 h. An NADH-dependent alcohol dehydrogenase was employed to keep the levels of NADH constant during the course of the assay. Figure 3 shows that all three OPRs were able to transform TNT to HADNT intermediates. No ADNTs were observed in the time scale monitored, although the decrease in HADNT concentration in the OPR2 and OPR3 assays (Fig. 3, D and E) suggests further transformation. The accumulation of an additional peak, more polar than HADNT, was observed in both OPR2 and -3 assays, which potentially represents dihydroxylamino-nitrotoluene (data not shown). Reduction of TNT by OPR1 resulted in the formation of products with retention times of 3.7, 7.3, and 9.2 min and spectra with absorbance maxima at approximately 500 nm (Fig. 3C). These were not seen in reaction mixtures catalyzed by OPR2 and OPR3 and were identified as dihydride Meisenheimer TNT complexes by reference to characterized standards generated by pentaerythritol tetranitrate reductase (Williams et al., 2004). 2H−-TNT adducts have been shown to yield four isomers in dynamic equilibrium. These are accounted for as the 1,3- and 3,5-hydride additions to the TNT molecule, with the respective tautomers forming protonated nitroalkanes (Williams et al., 2004). Figure 3C shows that levels of 2H−-TNT 3.7 and 9.2 decreased after 40 and 140 min, respectively, but no corresponding increases in any detected compounds were seen. The purified preparation of OPR1 showed the presence of a significant level of a contaminating protein (Fig. 3A). Trypsin digestion and MALDI-TOF/TOF tandem mass spectrometry analysis revealed this to be an E. coli 2-deoxy-d-gluconate 3-dehydrogenase. Although deemed unlikely to catalyze the observed reaction with TNT, we assayed cell extracts of NovaBlue (DE3) E. coli, containing pET-15b with and without OPR1. Only cell extract containing OPR1 was able to transform TNT to yield the dihydride-Meisenheimer TNT adducts, confirming that the observed activity with TNT was due to OPR1.
Characterization of Knockout and Overexpression OPR Lines
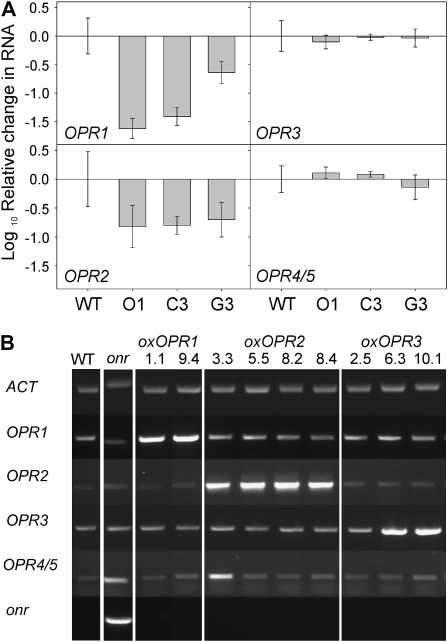
To dissect the individual contributions of the OPRs toward TNT detoxification in Arabidopsis, T-DNA insertional mutants opr1 and opr2 (Alonso et al., 2003) and opr3 (Stintzi and Browse, 2000) were obtained. Expression analysis on opr1 and opr2 demonstrated that OPR1 transcript was absent from opr1 and OPR2 transcript was absent from opr2 (Supplemental Fig. S1); opr3 had been characterized previously (Stintzi and Browse, 2000). OPR1 and OPR2, the two OPRs most strongly up-regulated in response to TNT, are closely linked on chromosome I, making the likelihood of generating opr1opr2 mutants by genetic crossing extremely unlikely. Thus, RNA interference (RNAi) was used to investigate the effect of knockdown expression of both OPR1 and OPR2. The RNAi gene construct included 155 bp of OPR1 and 451 bp of OPR2 (construction details are presented in Supplemental Fig. S2). Although the OPR1 sequence was shorter than the optimum 300- to 600-bp length for efficient RNAi, much of the selected OPR2 sequence was identical to OPR1. The full knockdown OPR1,2 sequence shared 96% and 92% identity with OPR1 and OPR2, respectively, but only 59% and 73% identity with OPR3 and OPR4/5, respectively. The knockdown OPR1,2 sequence was under the control of the 35S promoter, resulting in near constitutive expression, thus providing expression in the root tissues. Independently transformed knockdown OPR1,2 lines were generated with homozygous T3 lines C3, G3, and O1 used for further analysis. There were no phenotypic differences observed in the T3 knockdown OPR1,2 plants compared with the untransformed wild-type Arabidopsis when grown on plant medium or soil. Quantitative RT-PCR analysis (Fig. 4A) showed that OPR1 transcript levels were significantly reduced in all three knockdown OPR1,2 lines, with line O1 showing the greatest reduction in transcript. The levels of OPR2 were reduced by an equal amount in all three knockdown OPR1,2 lines. The levels of transcript for OPR3 and OPR4/5 were not significantly different from the levels in untransformed wild-type plants, indicating that the knockdown OPR1,2 transcript was specific to OPR1 and OPR2.
Figure 4.
Transcript levels of knockdown and overexpressing OPR lines. A, Quantitative RT-PCR expression analysis on 14-d-old wild-type (WT) plants and knockdown OPR1,2 lines. Results are means of three independent RNA isolations measured in triplicate ± se. B, Representative semiquantitative RT-PCR on 14-d-old wild-type plants, overexpressing (oxOPR) lines, and pentaerythritol tetranitrate reductase-expressing (onr) lines. The experiment was repeated twice with similar results.
The OPR1, OPR2, and OPR3 genes were overexpressed using the 35S promoter, with the C-terminal Ser-Arg-Leu type 1 peroxisome-targeting sequence present in OPR3 conserved in the overexpressing OPR3 construct. As a control to monitor the detection of ADNT and dihydride Meisenheimer TNT complexes in our experiments, the onr gene for the bacterial Old Yellow Enzyme homolog, pentaerythritol tetranitrate reductase, was transformed into Arabidopsis. Transformed lines were generated, and four independent lines were characterized for each gene. As predicted from our earlier studies on tobacco transformed with the same construct, all four 35S-onr lines exhibited enhanced tolerance to TNT. The expression of line 35S-onr1 is shown in Figure 4B, and this line was used as a control in our liquid culture and solid medium experiments.
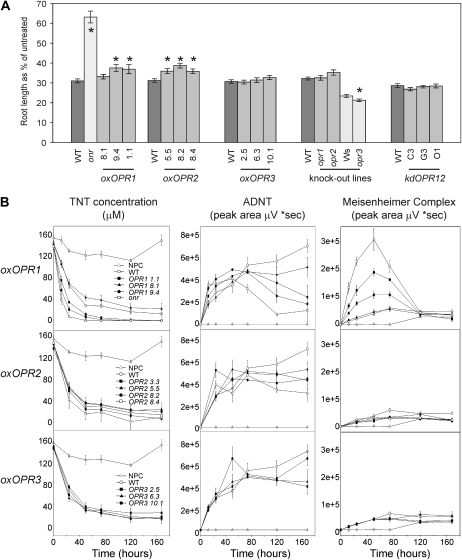
To investigate the tolerance of the knockout, knockdown, and overexpressing lines to TNT, these lines were grown on solid medium containing 2 μm TNT. Arabidopsis seedlings expressing onr had root lengths 2-fold longer than wild-type plants when grown on 2 μm TNT (Fig. 5A). This enhanced tolerance to TNT agrees with that previously observed in transgenic tobacco (French et al., 1999). Two of the OPR1-overexpressing lines (1.1 and 9.4) and all three of the OPR2-overexpressing lines had seedling root lengths significantly longer than those of wild-type plants (P < 0.05), while root lengths of OPR3-overexpressing seedlings were similar to those in the wild type. Decreased root length was observed in the opr3 mutant seedlings, but root lengths of opr1, opr2, or knockdown OPR1,2 lines were not significantly different from those of the wild type. No significant differences were observed, compared with the wild type, in the normal growth and development of the opr1 or opr2 knockout or overexpressing lines throughout the life cycle of the plants, with the exception of opr2, which exhibited reduced fertility (data not shown), and the previously reported male-sterile phenotype of opr3 (Stintzi and Browse, 2000). The opr2 phenotype links with the specific expression of the OPR2 gene during late pollen development (Biesgen and Weiler, 1999).
Figure 5.
Root length and liquid culture analysis of Arabidopsis lines with altered levels of OPR transcript. A, Root length of 7-d-old seedlings grown on medium containing 2 μm TNT. Values are expressed as percentage of the root length of seedlings grown on medium without TNT ± se. Results are from 40 to 60 seedlings from three independent seed batches. * Significantly different from the wild type (P < 0.05). B, TNT removal and ADNT and Meisenheimer complex formation detected in the medium from 14-d-old, liquid culture-grown Arabidopsis dosed with 200 μm TNT. Results are means of four to five biological replicates ± se. kd, Knockdown; NPC, no plant control; onr, pentaerythritol tetranitrate reductase-expressing lines; ox, overexpressing; Ws, Wassilewskija, background ecotype of opr3; WT, Col-0 wild type.
TNT Transformation in Liquid Culture
As expected from earlier results (French et al., 1999), the 35S-onr Arabidopsis plants exhibited enhanced uptake of TNT from liquid culture when compared with the wild type. Of the overexpressing OPR lines examined, the OPR1-overexpressing lines 1.1 and 9.4 showed enhanced TNT uptake (Fig. 5B), with OPR2-overexpressing and OPR3-overexpressing lines both behaving like the wild type. After 3 d, ADNT in the medium from OPR1-overexpressing lines 1.1 and 9.4 and 35S-onr lines started to decline, whereas ADNT levels in the medium from the remaining overexpressing OPR lines reached a plateau and ADNT levels in medium from wild-type samples continued to rise. No HADNTs were detected in the medium. After 8 h, medium in flasks containing overexpressing OPR1 1.1 and 9.4 and 35S-onr lines turned a translucent red/brown color. Absorbance in this 400- to 500-nm waveband region is characteristic of Meisenheimer adducts, which we observed in recombinant OPR1 assays (Fig. 3C). The spectra of these species shared similarities to characterized Meisenheimer adducts (Williams et al., 2004; data not shown), although the spectra of the wild type, OPR1 overexpressing, and 35S-onr were different from each other, suggesting, as in vitro data have shown (Williams et al., 2004), that the profile of Meisenheimer complexes produced is dependent on the Old Yellow Enzyme homolog.
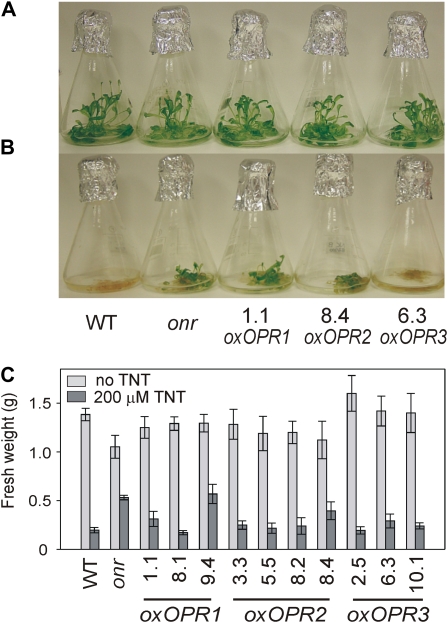
After 7 d of growth in medium containing 200 μm TNT, seedlings of the 35S-onr line, OPR1-overexpressing lines 1.1, and 9.4, and OPR2-overexpressing line 8.4 were less chlorotic and had greater biomasses than wild-type seedlings (Fig. 6). Biomass was not significantly altered from the wild type in the control flasks without TNT; however, in the absence of TNT, it was noted that the OPR-overexpressing lines showed enhanced survival under hydroponic conditions (data not shown).
Figure 6.
Morphology and biomass of overexpressing OPR (oxOPR) lines treated with TNT. A, Three-week-old, liquid culture-grown Arabidopsis growing without TNT. B, Appearance 7 d after dosing with 200 μm TNT. C, Fresh weight biomass after 2 weeks. Results are means of four to five biological replicates ± se. onr, Pentaerythritol tetranitrate reductase-expressing lines; WT, wild type.
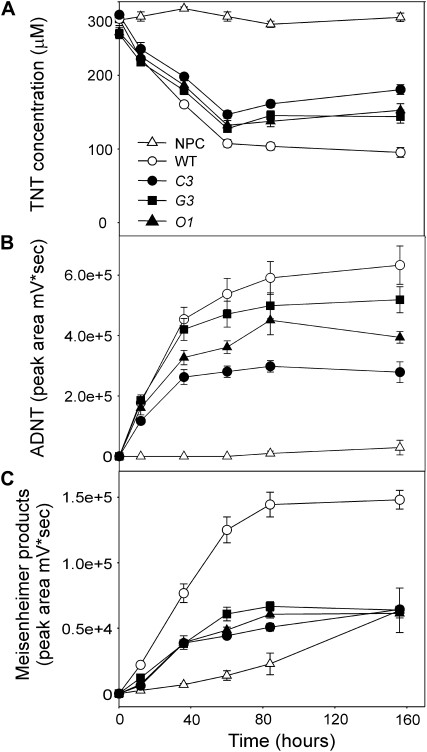
In the flasks containing the knockdown OPR1,2 lines grown in liquid culture with 300 μm TNT, TNT was taken up more slowly than in the wild type by all three knockdown lines in the first 2 d. Thereafter, no more TNT was removed from the medium by the knockdown OPR1,2 lines, and significantly less ADNT and Meisenheimer complex compounds were released into the medium than from the wild type (Fig. 7).
Figure 7.
Uptake and product formation of TNT-treated Arabidopsis OPR knockdown lines. A, TNT removal. B, ADNT. C, Meisenheimer complex formation in the medium of 14-d-old, liquid culture-grown Arabidopsis dosed with 300 μm TNT. Results are means of four to five biological replicates ± se. NPC, No plant control; WT, wild type.
DISCUSSION
We have cloned and purified three recombinant Arabidopsis OPRs, OPR1, OPR2, and OPR3, which exhibit significant (43%, 44%, and 36%, respectively) identity with bacterial pentaerythritol tetranitrate reductase, a member of the Old Yellow Enzyme family that is able to transform TNT. The greatest degree of similarity, both structurally and in regulation, exists between OPR1 and OPR2, and both are up-regulated in the root on exposure to TNT (Biesgen and Weiler, 1999). However, OPR1 had the greatest increase in expression in response to TNT treatment, suggesting a more dominant role in detoxification. The mechanism of TNT uptake and the intracellular site of accumulation are not known; however, while OPR1 and OPR2 are predicted to be active in the cytosol, OPR3 is targeted to the peroxisome, and this localization is likely to reduce accessibility toward TNT in planta.
The in vitro activity data clearly show that OPR1, OPR2, and OPR3 all have activity toward TNT. In liquid culture, the overexpressing OPR1 lines 1.1 and 9.4 had increased TNT uptake and produced more TNT transformation products than the wild type, suggesting increased TNT metabolism. Conversely, the knockdown OPR1,2 lines removed less TNT from liquid culture and produced fewer TNT transformation products. When grown on solid medium, two of the three OPR1-overexpressing lines and all of the OPR2-overexpressing lines exhibited significantly enhanced tolerance to TNT (as measured by root growth). This agrees with previous data in tobacco expressing onr, which showed that enhanced TNT transformation directly increased TNT tolerance (French et al., 1999). These results suggest that OPR1 and OPR2 are involved in TNT detoxification in planta. However, the tolerance to TNT of the opr1 and opr2 mutants and knockdown OPR1,2 seedlings was not significantly less than in wild-type seedlings, suggesting that there is some redundancy between the OPR activities, and other detoxification mechanisms are also likely to be involved (Gandia-Herrero et al., 2008; Rylott and Bruce, 2009). OPR1 is the only OPR in which both nitroreduction and aromatic ring reduction were observed in vitro, and both overexpressing OPR1 and 35S-onr plants released measurable levels of Meisenheimer complexes into the medium.
Expression analysis (Fig. 4) showed that in both the OPR-overexpressing and knockdown OPR1,2 transgenic lines, only the targeted OPR expression levels were altered; the expression levels of the remaining OPR homologs remained unchanged. While this result shows that OPR transcript levels are regulated independently from each other, it is possible that posttranscriptional regulation inhibited the generation of higher levels of active protein in the overexpressing lines. Previous studies using promoter β-glucuronidase fusions with Arabidopsis OPR1 and OPR2 showed that although transcript levels transiently increased in response to local and systemic wounding, UV-C illumination, and coldness, no subsequent changes in protein levels were detected (Biesgen and Weiler, 1999). Attempts to determine OPR activity in plant extracts using TNT as substrate were unsuccessful. However, the plant lines with altered levels of OPR transcripts did produce significantly different TNT metabolite profiles from those of untransformed plants when grown in liquid culture. Furthermore, seedlings of the OPR1- and OPR2-overexpressing lines had significantly enhanced seedling root growth in the presence of TNT, and these changes were observed in replicate, independently transformed lines. These findings indicate that the alterations seen in transcript levels reflect changes in OPR activity levels.
A wide range of α/β-unsaturated ketones and aldehydes, including vinyl ketones, smaller molecules such as acrolein, and those with more extended αβ-carbonyl chains such as traumatin, are produced in response to stress-related lipid oxygenation as a result of interaction with xenobiotics, pathogen attack, or wounding. Many of these compounds are substrates for Old Yellow Enzyme homologs, and it has been proposed that they could reduce reactive oxygen species in vivo (Vaz et al., 1995). These alternative, physiological substrates could compete with TNT for OPR-mediated catalysis. We have shown here that OPR1, OPR2, and OPR3 all have direct activity toward TNT, and their overexpression confers significantly enhanced tolerance to TNT. However, members of the Old Yellow Enzyme family are better known for their role as antioxidants (Kohli and Massey, 1998; Williams and Bruce, 2002; Fitzpatrick et al., 2003; Trotter et al., 2006), and OPRs may also have activity toward reactive oxygen species and an additional role protecting the plant from the oxidative stress resulting from TNT exposure.
CONCLUSION
In summary, we demonstrate that, in agreement with characterized bacterial Old Yellow Enzyme homologs, OPR1, -2, and -3 display activities toward TNT and OPRs contribute to the detoxification of TNT in the roots of Arabidopsis. A more detailed understanding of the biochemical mechanism of TNT detoxification in plants will allow opportunities to select or breed robust plant phenotypes in a rational way for field applications. This work further supports the view that members of the Old Yellow Enzyme family of flavoproteins play a role in the general detoxification of xenobiotic compounds in plants as well as microorganisms.
MATERIALS AND METHODS
Biochemicals
TNT was kindly provided by the Defence Science and Technology Laboratory at Fort Halstead, Kent, United Kingdom.
Recombinant OPRs
OPR1, OPR2, and OPR3 were cloned from Arabidopsis (Arabidopsis thaliana ecotype Columbia-0 [Col-0]) cDNA into pET-15b (Novagen). OPR1 and OPR3 were expressed in NovaBlue (DE3) Escherichia coli (Novagen), and OPR2 was expressed in Rosetta-gami B (DE3) E. coli (Novagen). Cells were grown to mid-log phase at 37°C and induced with 0.4 mm isopropylthio-β-galactoside. After 20 h, cells were spun at 12,000g for 10 min at 4°C, washed in buffer A (50 mm potassium phosphate buffer, pH 7), sonicated, and centrifuged at 4°C and 12,000g for 30 min. Soluble extract of OPR1 was applied to a Mimetic Orange 1 affinity chromatography column, and OPR2 and -3 were applied to a Mimetic Blue 1 column (Prometic Biosciences). Columns were washed with buffer A, then protein was eluted using a sodium chloride gradient, concentrated on 10-kD spin filters (Perbio Science), and dialyzed into buffer A. Mimetic Orange 2 matrix was added to the OPR1 fraction to bind and remove contaminants. Protein identities were confirmed following SDS-PAGE and trypsin digest, by MALDI-TOF peptide mass fingerprinting, immediately followed by more detailed protein characterization using MALDI-TOF/TOF tandem mass spectrometry on the same prepared sample.
Transformation of TNT by OPR1, -2, and -3
Assays and HPLC analysis were performed as described previously (Williams et al., 2004). The identities of ADNT and HADNT were confirmed using standards (Accustandard). Hydride Meisenheimer adduct standards were generated using purified pentaerythritol tetranitrate reductase (Williams et al., 2004).
Plant Material and Growth Conditions
Wild-type Arabidopsis ecotypes Col-0 and Wassilewskija were obtained from the Nottingham Arabidopsis Stock Centre at the University of Nottingham. The opr1 (SALK_145355) and opr2 (SALK_014855) mutants were obtained from T-DNA express (Alonso et al., 2003). The opr3 mutant (Stintzi and Browse, 2000), in the Wassilewskija background, was kindly provided by John Browse (Institute of Biological Chemistry, Washington State University). Seeds were imbibed for 4 d at 4°C on agar plates containing 20 mm Suc and half-strength Murashige and Skoog salts (Sigma-Aldrich) with TNT dissolved in dimethylformamide when present (equivalent volumes of dimethylformamide were added to TNT-free flasks). Seedlings were grown in 16 h of light with 125 μmol m−2 s−1 white light. For liquid culture experiments, eight 1-d-old seedlings were transferred to 100-mL conical flasks containing 20 mL of half-strength Murashige and Skoog medium and 20 mm Suc. Plants were grown under 20 μmol m−2 s−1 light on a rotary shaker at 130 rpm.
Generation of OPR-Overexpression and Knockdown Constructs and Identification of opr1 and opr2
The 35S overexpression lines were generated using the pART27 binary cassette system (Gleave, 1992). The OPRs were amplified by PCR using overexpressing OPR primers and Arabidopsis Col-0 cDNA. Sequences were cloned into pART7 KpnI and BamHI sites for OPR1, KpnI and XbaI sites for OPR2, and XhoI and ClaI sites for OPR3. The resulting 3SS-OPR-ocs cassettes were cloned into pART27 via NotI sites. The pART27-onr construct used was as reported previously (French et al., 1999).
The knockdown OPR1,2 chimera was constructed as follows. OPR1 and OPR2 sequences were amplified using kdOPR1 and kdOPR2 primers, and the fragments were digested with BfaI and ligated together. The OPR1-OPR2 ligated sequence was then reamplified using kdOPR1F and kdOPR2R primers, sequenced, and cloned into pHellsgate 8 (Wesley et al., 2001). Construction details are presented in Supplemental Figure S2.
All constructs was transformed into Arabidopsis Col-0 using Agrobacterium tumefaciens strain GV3101. Transformants and homozygous lines were selected using kanamycin resistance.
The T-DNA border regions of opr1 and opr2 were amplified for sequencing using T-DNA left border primer LBa1 in combination with OPR1F for opr1 and OPR2-seq for opr2. Insertion sequences are shown in Supplemental Figure S1. Endogenous OPR1 and OPR2 were amplified using OPR1F/OPR1R and OPR2F/OPR2R primers. All primer sequences are described in Supplemental Table S1.
RNA Isolation and RT-PCR
The DNase-treated total RNA was isolated from 14-d-old liquid culture-grown plants using the RNeasy kit (Qiagen). The synthesis of cDNA was primed with oligo(dT)12-18 using SuperScript II reverse transcriptase (Invitrogen) containing RNasin (Promega) at 42°C for 90 min, before inactivation at 70°C for 15 min.
For quantitative RT-PCR, cDNA was purified using a Promega Wizard SV gel and PCR cleanup system and quantified. The rtOPR primers and probe sets (Supplemental Table S1), specific for each OPR gene, were tested for primer efficiency, then TaqMan quantitative PCR was performed using 50 ng of cDNA in an Applied Biosystems 7300 real-time PCR machine with a cycle of 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s, 60°C for 1 min using FAM (5′) and TAMRA (3′) fluorescent dyes. The level of expression was normalized against ACT2 (GenBank accession no. NM_180280). Data were analyzed using ABI software according to the manufacturer's recommendations.
Semiquantitative RT-PCR was performed on the overexpressing OPR lines using serial dilutions of cDNA and sqOPR primers. Reactions were denatured at 94°C for 3 min, cycled 25 times at 94°C for 15 s, 55°C for 30 s, 1 min for 72°C, and then held at 72°C for 10 min. For reactions with OPR2 primers, the annealing step was 60°C. ACT2 gene expression using sqACT primers was used as a reference. Primer sequences are shown Supplemental Table S1.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: OPR1 (NM_202428), OPR2 (NM_106319), OPR3 (NM_126619), OPR4 (NM_101664), OPR5 (NM_179352), and OPR6 (NM_100810).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic diagram of OPR1 and OPR2 loci and transcript analysis for opr1 and opr2 mutations.
Supplemental Figure S2. Construction of the RNAi OPR1,2 vector.
Supplemental Table S1. DNA primer sequences used for cloning OPRs.
Supplementary Material
Acknowledgments
We are grateful to Prof. John Browse for kindly providing the opr3 mutant. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and Nottingham Arabidopsis Stock Centre for distributing plant lines.
This work was supported by the Strategic Environmental Research and Development Program of the U.S. Department of Defense, by a Burgess studentship to E.R.B., and by a Biotechnology and Biological Sciences Research Council studentship to Z.C.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Neil C. Bruce (ncb5@york.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320 938–941 [DOI] [PubMed] [Google Scholar]
- Basran A, French CE, Williams RE, Nicklin S, Bruce NC (1998) Degradation of nitrate ester and nitroaromatic explosives by Enterobacter cloacae PB2. Biochem Soc Trans 26 680–685 [DOI] [PubMed] [Google Scholar]
- Biesgen C, Weiler EW (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208 155–165 [DOI] [PubMed] [Google Scholar]
- Binks PR, French CE, Nicklin S, Bruce NC (1996) Degradation of pentaerythritol tetranitrate by Enterobacter cloacae PB2. Appl Environ Microbiol 62 1214–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blehert DS, Fox BG, Chambliss GH (1999) Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J Bacteriol 181 6254–6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman DR, Lorenz WW, Przybyla AE, Wolfe NL, Dean JF (2003) SAGE analysis of transcriptome responses in Arabidopsis roots exposed to 2,4,6-trinitrotoluene. Plant Physiol 133 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick TB, Amrhein N, Macheroux P (2003) Characterization of YqjM, an Old Yellow Enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J Biol Chem 278 19891–19897 [DOI] [PubMed] [Google Scholar]
- French CE, Nicklin S, Bruce NC (1998) Aerobic degradation of 2,4,6-trinitrotoluene by Enterobacter cloacae PB2 and by pentaerythritol tetranitrate reductase. Appl Environ Microbiol 64 2864–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CE, Rosser SJ, Davies GJ, Nicklin S, Bruce NC (1999) Biodegradation of explosives by transgenic plants expressing pentaerythritol tetranitrate reductase. Nat Biotechnol 17 491–494 [DOI] [PubMed] [Google Scholar]
- Gandia-Herrero F, Lorenz A, Larson T, Graham IA, Bowles DJ, Rylott EL, Bruce NC (2008) Detoxification of the explosive 2,4,6-trinitrotoluene in Arabidopsis: discovery of bifunctional O- and C-glucosyltransferases. Plant J 56 963–974 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Khan H, Barna T, Harris RJ, Bruce NC, Barsukov I, Munro AW, Moody PC, Scrutton NS (2004) Atomic resolution structures and solution behavior of enzyme-substrate complexes of Enterobacter cloacae PB2 pentaerythritol tetranitrate reductase: multiple conformational states and implications for the mechanism of nitroaromatic explosive degradation. J Biol Chem 279 30563–30572 [DOI] [PubMed] [Google Scholar]
- Kohli RM, Massey V (1998) The oxidative half-reaction of Old Yellow Enzyme: the role of tyrosine 196. J Biol Chem 273 32763–32770 [DOI] [PubMed] [Google Scholar]
- Mezzari MP, Walters K, Jelinkova M, Shih MC, Just CL, Schnoor JL (2005) Gene expression and microscopic analysis of Arabidopsis exposed to chloroacetanilide herbicides and explosive compounds: a phytoremediation approach. Plant Physiol 138 858–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JW, Knoke KL, Noguera DR, Fox BG, Chambliss GH (2000) Transformation of 2,4,6-trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C. Appl Environ Microbiol 66 4742–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Bruce NC (2009) Plants disarm soil: engineering plants for the phytoremediation of explosives. Trends Biotechnol 27 73–81 [DOI] [PubMed] [Google Scholar]
- Sandermann H (1992) Plant metabolism of xenobiotics. Trends Biochem Sci 17 82–84 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Weiler EW (1997) Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana: structural and functional relationship to yeast Old Yellow Enzyme. J Biol Chem 272 28066–28072 [DOI] [PubMed] [Google Scholar]
- Snape JR, Walkley NA, Morby AP, Nicklin S, White GF (1997) Purification, properties, and sequence of glycerol trinitrate reductase from Agrobacterium radiobacter. J Bacteriol 179 7796–7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter EW, Collinson EJ, Dawes IW, Grant CM (2006) Old Yellow Enzymes protect against acrolein toxicity in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 72 4885–4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz AD, Chakraborty S, Massey V (1995) Old Yellow Enzyme: aromatization of cyclic enones and the mechanism of a novel dismutation reaction. Biochemistry 34 4246–4256 [DOI] [PubMed] [Google Scholar]
- Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Williams RE, Bruce NC (2002) ‘New uses for an old enzyme’: the Old Yellow Enzyme family of flavoenzymes. Microbiology 148 1607–1614 [DOI] [PubMed] [Google Scholar]
- Williams RE, Rathbone DA, Scrutton NS, Bruce NC (2004) Biotransformation of explosives by the Old Yellow Enzyme family of flavoproteins. Appl Environ Microbiol 70 3566–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittich RM, Haidour A, Van Dillewijn P, Ramos JL (2008) OYE flavoprotein reductases initiate the condensation of TNT-derived intermediates to secondary diarylamines and nitrite. Environ Sci Technol 42 734–739 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.