Abstract
The integument forms a number of different types of mineralized element, including dermal denticles, scutes, ganoid scales, elasmoid scales, fin rays and osteoderms found in certain fish, reptiles, amphibians and xenarthran mammals. To this list can be added teeth, which are far more widely represented and studied than any of the other mineralized elements mentioned above, and as such can be thought of as a model mineralized system. In recent years the focus for studies on tooth development has been the mouse, with a wealth of genetic information accrued and the availability of cutting edge techniques. It is the mouse dentition that this review will concentrate on. The development of the tooth will be followed, looking at what controls the shape of the tooth and how signals from the mesenchyme and epithelium interact to lead to formation of a molar or incisor. The number of teeth generated will then be investigated, looking at how tooth germ number can be reduced or increased by apoptosis, fusion of tooth germs, creation of new tooth germs, and the generation of additional teeth from existing tooth germs. The development of mineralized tissue will then be detailed, looking at how the asymmetrical deposition of enamel is controlled in the mouse incisor. The continued importance of epithelial–mesenchymal interactions at these later stages of tooth development will also be discussed. Tooth anomalies and human disorders have been well covered by recent reviews, therefore in this paper we wish to present a classical review of current knowledge of tooth development, fitting together data from a large number of recent research papers to draw general conclusions about tooth development.
Keywords: cusp, enamel knot, homeobox code, incisor, mineralization, molar, pattern, tooth development
The mouse has a highly reduced dentition, with one incisor, separated by a diastema region to three molars, in each quadrant. The mouse only has one generation of teeth, and the incisors continuously grow throughout the animal's life. The mouse can therefore tell us much about patterning of teeth (molar vs. incisor), control of tooth number, and the role of stem cells in tooth development. Although the exact tooth germ morphology may vary in different-shaped teeth, the stages of tooth development are well conserved throughout toothed vertebrates, and data from the mouse should provide clues to tooth development in diverse groups (Streelman et al. 2003). In keeping with this, expression patterns of key molecules, such as Shh, have been shown to be largely conserved in the dentition in mouse, fish and snakes (Fraser et al. 2006; Buchtova et al. 2008). Information about replacement generations of teeth and formation of other tooth types, such as canines, are difficult to assess using the mouse model, and other model organisms are required.
Tooth development
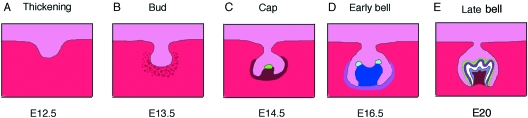
Tooth development progresses through a series of well-defined stages – epithelial thickening, bud, cap and bell (Fig. 1). In the mouse a thickening of the oral epithelium is first visible at around E11.5 (embryonic day 11.5). This thickening expresses key signalling molecules such as Shh that act to increase cell proliferation at the sites of tooth development (Hardcastle et al. 1998). The proliferating epithelium invaginates further into the underlying neural crest-derived mesenchyme and forms a bud. Before E12.0 the instructive information for initiation of a tooth resides in the epithelium, but at E12.0 the mesenchyme that starts to condense around the forming bud takes over this instructive role. Thus early oral epithelium and any source of neural crest-derived mesenchyme can form a tooth (Mina & Kollar, 1987; Lumsden, 1988; Tucker et al. 1999) and later dental mesenchyme and a source of non-oral epithelium, can also form a tooth (Ruch et al. 1984; Mina & Kollar, 1987). The bud is clearly formed at E13.5, and surrounded by condensing mesenchyme that expresses a host of signalling molecules and transcription factors such as Bmp4, Msx1 and Pax9. Loss of Msx1 and Pax9 leads to a downregulation of Bmp4 and an arrest of tooth development at the bud stage in mice (Satokata & Maas, 1994; Chen et al. 1996; Peters et al. 1998). Interestingly, mutations in MSX1 and PAX9 are also associated with tooth agenesis in humans (Fleischmannova et al. 2008). Msx1and Bmp4 are closely associated during tooth development, acting as part of a positive feedback loop, and addition of Bmp4 can partially rescue the tooth defect in Msx1 mutant mice (Chen et al. 1996; Bei et al. 2000; Zhao et al. 2000). A loss of mesenchymal Bmp4 and an arrest of tooth development at early stages are also observed after loss of Wnt signalling, placing the Wnt pathway upstream of mesenchymal Bmp4(Liu et al. 2008). The dental mesenchyme then signals back to the tooth and induces the formation of a structure known as the enamel knot at the tip of the bud. The enamel knot is visible histologically as a bulge in the centre of the inner enamel epithelium at the cap stage and was first described almost 100 years ago (Ahrens, 1913). This knot of cells expresses a host of signalling molecules, such as Shh, Fgf4, Bmp4 and Wnt10b, and as such has been classed as an important signalling centre for tooth development (Vaahtokari et al. 1996; Sarkar & Sharpe, 1999).
Fig. 1.
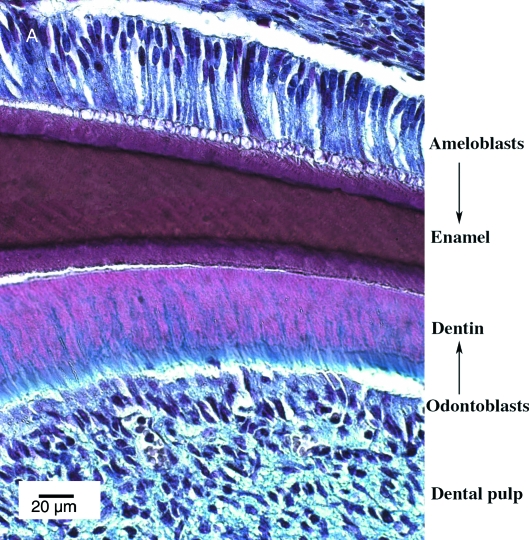
Molar tooth development schematic. (A) At E12.5 an obvious invagination of the dental epithelium is visible. Light pink (epithelium). Dark pink (mesenchyme). (B) By E13.5 the invagination has formed a bud and the underlying neural crest-derived mesenchyme starts to condense (brown spots). (C) By E14.5 the epithelium starts to fold, forming the cap stage tooth germ, with the primary enamel knot (green) visible as a bulge in the dental epithelium, surrounded by the condensing mesenchyme (brown). (D) By E16.5 the epithelium has extended further into the mesenchyme, forming a bell. The inner enamel epithelium (IEE) encloses the dental papilla (blue), and the dental follicle (purple) surrounds the outer dental epithelium. The primary enamel knot has disappeared, to be replaced by the secondary enamel knots (light blue). (E) In the newborn mouse (E20) the adjacent ameloblasts and odontoblasts have differentiated and start producing enamel and dentin. Purple (Enamel). White (Dentin). Grey (Odontoblasts). Green (Ameloblasts).
The role of the enamel knot
Bmp4 signalling from the condensing mesenchyme is thought to play a critical role in induction of the enamel knot. Addition of Bmp4 to the oral epithelium leads to an upregulation of enamel knot markers such as p21 (Jernvall et al. 1998). The range of Bmp action is restricted by the expression of a Bmp antagonist, known as ectodin or wise. Bmp4 induces ectodin expression, which then acts back on Bmp4 as part of a negative feedback loop, leading to restricted induction of markers such as p21 (Laurikkala et al. 2003). The ectodin knockout mouse is characterized by overexpression of p21, enlarged enamel knots and cuspal defects (Kassai et al. 2005). Loss of Bmp4 signalling in the dental epithelium by conditional knockout of the receptor Bmpr1a leads to arrest of tooth development at the bud stage, confirming the signalling role of Bmp4 from the mesenchyme to the epithelium at this stage (Andl et al. 2004). Although the enamel knot expresses a number of signalling molecules, the cells of the enamel knot do not proliferate and thus act like an anchor, constricting the movement of cells in the tooth (Jernvall et al. 1994; Vaahtokari et al. 1996). High proliferation outside the enamel knot and low proliferation within the knot therefore act to fold the epithelium of the tooth germ, forming a cap-shaped structure at E14.5. The folding divides the epithelium into the inner and outer enamel epithelium. The mesenchymal cells adjacent to the inner enamel epithelium will form the dental papilla; those on the outside form the dental follicle.
The enamel knots are transient structures and disappear by apoptosis, driven by Bmp4, by the end of the cap stage (Lesot et al. 1996; Vaahtokari et al. 1996; Jernvall et al. 1998). Incisor tooth germs only have a single enamel knot, whereas molars go on to form secondary enamel knots (Kettunen & Thesleff, 1998). Whether the primary enamel knot contributes to the secondary enamel knots is unclear, with fate-mapping experiments and proliferation studies providing both positive and negative evidence (Coin et al. 1999; Matalova et al. 2005; Cho et al. 2007). The secondary enamel knots again lead to folding of the inner enamel epithelium at the bell stage (E16.5), resulting in the formation of a complex multi-cuspid tooth. The hard tissues, dentin and enamel, are then laid down following the contours of the epithelium, defining the tooth shape. Molars go on to form tertiary enamel knots, which appear as Slit1-expressing epithelial clusters next to the enamel-free areas at the cusp tips (Luukko et al. 2003). The number of enamel knots is thought to determine the cuspal pattern of the resulting tooth (Vaahtokari et al. 1996), hence simple conical teeth, like incisors, only have one knot.
The size and shape of the primary enamel knot is key to the production of the exact degree of curvature of the oral epithelium. In molars if the enamel knot is too small, as observed in mice with mutations in Eda or Edaradd, this affects the folding of the tooth and the positioning of the secondary enamel knots and leads to a molar tooth with few flattened cusps (Pispa et al. 1999; Ohazama et al. 2004). The incisors, however, appear largely unaffected. If the receptor for Eda, Edar, is mutated, the enamel knot is not reduced in size but fails to form the correct shape (Tucker et al. 2000). The effect on the final tooth morphology, however, is identical to that of Eda and Edaradd. Compromising signalling from the primary enamel knot, by changing its size or shape, therefore leads to changes in the arrangement of the secondary enamel knots in molars and to cusp defects. Edaexpression is regulated by Wnt (Laurikkala et al. 2001). If Wnt signalling is blocked at the early bell stage, when the secondary enamel knots are forming, the expression of Eda is reduced and the molars form with flattened cusps, similar to those observed in the Eda (Tabby) mutant (Liu et al. 2008). In humans, mutations in EDA, EDAR and EDARADD lead to hypohidrotic ectodermal dysplasia, which is characterized by the presence of molars with reduced cusps and peg-like teeth (Kere et al. 1996; Monreal et al. 1999; Headon et al. 2001).
The homeobox code
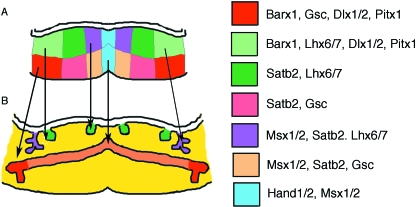
The shape of the tooth crown that develops is therefore driven by the number of enamel knots that form. This patterning information appears to reside in the mesenchyme into which the tooth germs invaginates, and as such is determined very early on in tooth development. The mandible and maxilla do not express Hox genes, the anterior border of which sits between the first and second pharyngeal arch. However, they express a number of homeobox-containing genes that are expressed in nested patterns across the jaw. This overlapping pattern of homeobox genes has been termed the ‘homeobox code’ (Sharpe, 1995) (Fig. 2). The mandible is divided into oral (Lhx6,7), aboral (Gsc), distal (presumptive incisor) (Msx1.2) and proximal (presumptive molar) (Dlx1,2,Barx1, Pitx1) domains (Thomas et al. 1997; Tucker et al. 1998b, 1999; Mitsiadis & Drouin, 2008). Evidence that this code is instructing patterning information to the developing teeth comes from misexpression and knockout studies. If a presumptive molar gene, such as Barx1, is misexpressed in the presumptive incisor region (either directly or by changing the expression of factors that control its expression domain), the tooth germs that develop in the presumptive incisor region will develop as molars, with the formation of multiple cusps (Tucker et al. 1998b; Miletich et al. 2005). In contrast, if presumptive molar genes, such as Dlx1and 2, are knocked out in the mouse, then molars fail to develop in the maxilla, whereas incisor development is unaffected (Thomas et al. 1997). In this case, the mandibular molars still form, probably due to compensation by other members of the Dlx family (such as Dlx5,6) that are expressed in the mandible but not the maxilla.
Fig. 2.
Division of the mandible mesenchyme into modules by the nested expression of transcription factors. (A) Schematic of E9.5–10.0 mouse mandible, showing areas of mesenchymal gene expression. For simplicity, expression in the epithelium is not shown. (B) Schematic of E13.5 mouse mandible (flat mounted). Arrows link the earlier expression patterns to the developing structures: teeth (green), salivary glands (submandibular/sublingual) (purple), Meckel's cartilage (orange), and the developing middle ear (red).
The expression patterns of the homeobox genes are defined by positive and negative signals from the oral epithelium. Bmp4, for example, is initially expressed in the distal epithelium prior to any signs of tooth development and induces expression of Msx1 in the underlying presumptive incisor mesenchyme, while at the same time negatively regulating the expression of Barx1, so as to restrict Barx1 expression to the presumptive molar region (Tucker et al. 1998b). Fgf8, meanwhile, is expressed adjacent to Bmp4 in the proximal oral epithelium and positively induces Barx1expression in the underlying presumptive molar epithelium (Tucker et al. 1998b). The expression of Fgf8 and Bmp4 in the oral epithelium is initially induced by Shh signalling from the pharyngeal endoderm during pharyngeal arch formation (Haworth et al. 2007; Brito et al. 2008). Bmp4 and Fgf8 negatively regulate each other so that loss of Bmp4 signalling leads to an expansion of Fgf8 expression into the distal epithelium (Wilson & Tucker, 2004). This mutual antagonism acts to further delineate the boundary between the presumptive molar- and presumptive incisor-forming regions. The expression of Fgf8 is positively controlled by the paired-related homeobox gene Pitx2 (Otlx2), which is co-expressed in the oral epithelium (Mucchielli et al. 1997; Lu et al. 1999). In the Pitx2knockout, Fgf8expression is reduced, whereas Bmp4 expression is upregulated (Lu et al. 1999). In a similar positive feedback loop, epithelial Bmp4 signalling induces and is induced by Islet-1, a member of the LIM homeodomain family. Overexpression of Islet-1 in the epithelium overlying the molar region leads to ectopic expression of Bmp4, and loss of molar markers in the underlying mesenchyme, resulting in a failure in molar development (Mitsiadis et al. 2003). This does not, however, lead to a corresponding upregulation of incisor markers, such as Msx1, and no transformation of molars to incisors occurs.
The expression levels of the genes are also important. Pitx1(orPtx1) is a member of the paired family of homeobox genes. Like Barx1, Pitx1 is positively regulated by Fgf8 and negatively regulated by Bmp4(St Amand et al. 2000). Pitx1is initially expressed in the presumptive mandibular molar mesenchyme overlapping with Barx1 and the oral epithelium, but is excluded from the presumptive maxillary molar mesenchyme (Mitsiadis & Drouin, 2008). In the Pitx1knockout the level of Barx1 is reduced in the mandible, but the expression of Barx1 in the maxilla is unaffected (Mitsiadis & Drouin, 2008). Loss of Pitx1 leads to a reduction of cusps in the mandibular molars, whereas the maxillary molars appear normal. Indirectly reducing the level of Barx1, by loss of Pitx1, thus changes the shape of the tooth, with molars taking on a more premolar appearance. Low levels of Barx1 may therefore play an important role in the specification of premolars in species with such teeth (Fig. 3B).
Fig. 3.
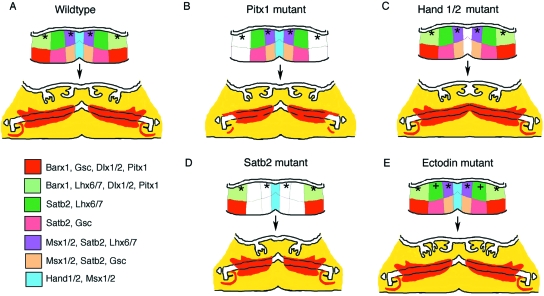
Loss of gene expression disrupts different regions of the developing mandible. (A) Schematic showing wild-type expression of transcription factors at E10 in the mouse mandible and the structures that form from these regions at E16.5. *Position of presumptive tooth development. At E16.5 the bones of the mandible have formed around Meckel's cartilage and the middle ear is distinct at the end of Meckel's cartilage (malleus, incus and tympanic ring). Bones are shown in red. (B–E) Mutant phenotypes. Area in white indicates region affected by loss of gene. (B) Pitx1knockout. Loss of Pitx1 leads to a reduction of the cusps in the mandibular molars. The proximal mandible and tympanic ring of the middle ear are also affected. (C) Hand1/2 double knockout. Loss of Hand1/2 expression in the midline leads to loss of the midline structures, such as the rostral symphysis, and fusion of the incisors. (D) Satb2 knockout. Loss of Satb2 leads to loss of intermediate regions of the mandible, resulting in loss of the incisors. (E) Ectodinknockout. Additional teeth develop in the diastema region (+). The forming teeth develop a shape analogous to that of a premolar.
Patterning of the mandible is not restricted to homeobox genes; many other transcription factors play key roles. At the distal end of the developing mandible, the bHLH transcription factors Hand 1and 2 are expressed and define the midline. When these genes are knocked out together, the midline region is lost, as shown by loss of the rostral process of Meckel's cartilage and fusion of the incisor tooth germs (Barbosa et al. 2007) (Fig. 3C). These midline regions remain intact after loss of Satb2, a member of a novel transcription factor gene family, the members of which bind to nuclear matrix attachment regions and regulate tissue-specific organization of chromatin (Britanova et al. 2006). Satb2 appears to play a role in setting up the intermediate region between the midline and more proximal areas of the mandible (Britanova et al. 2006; Depew and Compagnucci, 2008). In the knockout the incisors and associated bone are missing, but the more proximal areas are unaffected (Fig. 3D). Patterning information is thus generated very early on during a key window of time before the tooth germ is visible.
If such a homeobox code is responsible for patterning the type of tooth that forms, it might be expected that such a code would not be in place in species with a homodont dentition. A similar nested pattern of homeobox genes, however, is found in the developing jaws of birds, which lost their homodont dentition approximately 100 ma (mega annum) (Barlow & Francis-West, 1997; Barlow et al. 1999; Wilson & Tucker, 2004). The presence of such a code, therefore, does not indicate the presence of teeth of different shape. This is perhaps unsurprising as the homeobox code plays an important role in patterning, not only mammalian teeth but also the skeletal elements that hold the teeth and form part of the jaws and middle ear (Figs 2 and 3). For example, loss of the Dlx genes as well as affecting molar development leads to defects in the formation of the proximal parts of the dentary and squamosal bones of the jaw (Qiu et al. 1995; Depew et al. 2005). Such links are not restricted to the skeletal tissues of the jaw, with soft tissues such as the tongue and salivary glands being affected in many homeobox gene knockouts (Lanctot et al. 1999; Szeto et al. 1999). It would thus appear that the homeobox code was in place to pattern the jaw and was then later co-opted for patterning of the dentition (Stock, 2001). This may explain why teeth occupying the same relative position within the jaw of two different species often exhibit a similar morphology, regardless of their numerical position in the tooth row (Butler, 1995).
The pattern of the homeobox genes is very dynamic within the jaw. Thus, early on before any signs of tooth development Msx1 can be thought of as a presumptive incisor marker, expressed in the distal part of the jaw. A few days later in mouse development, however, when the tooth germs have reached the bud stage, Msx1 is associated with the mesenchyme surrounding all the tooth germs (Tucker et al. 1998a). This change in expression pattern, and role, explains why all teeth are arrested at the bud stage in the Msx1 knockout (Satokata & Maas, 1994). This dynamic pattern appears to be driven by the changing expression pattern of signalling molecules. For example, Bmp4is first expressed in the epithelium of the presumptive incisor region but later shifts to the mesenchyme and is expressed around the tooth germs, in partnership with Msx1 (Tucker et al. 1998a). It has been proposed that epithelial Bmp4 is in fact expressed in a series of ancestral signalling centres that successively appear at three distinct positions along the mandible, corresponding to the anterior and posterior antemolar rudiment, and the first molar enamel knot at E12, E13 and E14, respectively (Peterkova et al. 2000). This alternative interpretation of the expression of Bmp4 allows for an interesting re-evaluation of the role of Bmp4 in the tooth.
Although the mouse has a reduced dentition, consisting solely of molars and incisors, the ancestor of the lineage including Glires (rodents and lagomorphs) and primates would have also possessed premolars and canines, similar to the dentition observed in primates, carnivores and insectivores, such as the shrew (Yamanaka et al. 2007). Some rodents, such as squirrels and guinea pigs, indeed still have premolar teeth. In the mouse, tooth germs do start to develop in the diastema region but these quickly abort and undergo apoptosis (Peterkova et al. 2003). Although these teeth do not normally develop past the bud stage in the mouse, it is predicted that the homeobox code specifying premolar and canine tooth shapes would still be in place. This can be tested by investigating the phenotype of mutant mice that form additional teeth in the diastema region. Such diastema teeth are observed in a number of knockout/transgenic mice, where the action of signalling molecules is heightened by removal of an inhibitor (ectodin in the case of Bmp and Wnt signalling, and sprouty in the case of Fgf signalling) (Kassai et al. 2005; Klein et al. 2006) and when the Eda pathway is disrupted, such as loss of Eda in the Tabby mutant mouse or when Eda and Edar are overexpressed (Mustonen et al. 2004; Tucker et al. 2004; Peterkova et al. 2005). In such mice an extra tooth forms directly in front of the first molar and develops a shape reminiscent of a premolar, a tooth type last seen in the putative murine lineage around 50 ma as evidenced in the primitive rodent Tribosphenomys. Tribospenomys dates back to between 60 and 52 Ma and has a P4 premolar tooth, whereas more recent primitive rodents, such as Pappocricetodon, from about 45 Ma, lack premolars (Viriot et al. 2002). Thus although the mouse does not form a premolar tooth, the information required for patterning a premolar is still present, locked in the homeobox code of the jaw (Fig. 3E).
Reducing tooth number
In the mouse, the number of tooth germs that form has been reduced by the death of tooth germs that start to develop in the diastema region. The number of teeth that form can also be reduced by fusion of existing tooth germs to form a single compound tooth. This process of fusion occurs normally for many teeth, as exemplified by the maxillary incisors. These have been shown to form in both rat and mouse by a fusion of tooth germs on the medial nasal and maxillary processes (Peterkova et al. 1993; Kriangkrai et al. 2006a,b). In humans the deciduous lateral incisors of the maxilla form in this way (Hovorakova et al. 2006). The compound origin of these teeth has been linked to the high incidence of defects associated with this tooth, in particular the formation of supernumerary teeth. In a normal population of 3–3.5-year-olds, half of the supernumerary teeth in the primary dentition were maxillary lateral incisors, with the majority of other supernumeraries also being located in the maxilla (Ravn, 1971). In the rat, disruption of the fusion process, as occurs in cases of cleft lip or by insertion of a mechanical barrier, leads to the formation of supernumerary incisors (Kriangkrai et al. 2006a).
Fusion of the maxillary central incisors is observed in patients with SMMCI (single median maxillary central incisor) syndrome. This is thought to be caused by a failure in growth at the midline, stimulated by a defect in the SHH signalling pathway (Nanni et al. 2001). In keeping with this, a single fused incisor is observed in mice with mutations in the Shhsignalling pathway (Hardcastle et al. 1998). In the ectodin knockout, in addition to the formation of an ectopic tooth, the molar teeth fuse together and form one large tooth (Kassai et al. 2005). Thus the normal separation of the molar primordium may involve Bmp signalling. Fusion is therefore observed in both normal and abnormal situations.
In the mouse, although the majority of diastema tooth buds undergo apoptosis and regress, the residual tooth germ immediately mesial (distal) to the first molar (known as R2) is partly incorporated into the developing first molar (M1) (Viriot et al. 2000). R2 is believed to form the mesial region of the developing first molar, thereby increasing the size and complexity of M1. In keeping with this, in some mouse mutants with supernumerary diastema teeth, the R2 fails to fuse with the M1, leading to a decrease in the size and complexity of the first molar (Peterkova et al. 2005).
Fused teeth are also seen in many other mammals, such as in the premolar region in many talpids (Kawada et al. 2006). Here the number of premolars varies subtly throughout the population, with many individuals showing fusion of two premolars into a larger, more complex tooth. Tooth fusion thus represents a method not only for reducing the number of teeth but also for increasing the size and complexity of a given tooth.
Increasing tooth number
In the wild-type mouse mandible, one incisor forms from each incisor placode, with three molars (M1, M2 and M3) forming from the molar placode. If the molar region is dissected out and cultured at E12, the complete molar dentition forms in normal sequence and with normal crown shapes and proportions, indicating that at this early stage all the patterning information is already present (Lumsden, 1979). One way to increase the number of teeth is to increase the number of placodes initially laid down. This is perhaps more difficult to study in the mouse where extra teeth that appear to develop from new placodes are often in fact a rescue of the aborted teeth in the diastema, as in the case of the ectodin and sprouty mutants, or are due to a failure of fusion in those teeth that develop initially from two separate sites, as in the case of the pax6 mutant (Kassai et al. 2005; Klein et al. 2006; Kriangkrai et al. 2006a). Ectopic application of Shh has been shown to lead to formation of ectopic tooth germs, but the development of these has not been followed past the bud stage of development and it is therefore unclear whether they would have formed into distinct teeth (Hardcastle et al. 1998). Additional teeth may also form by the splitting of a tooth germ. For example, when rabbit molars were halved, two miniature molars formed (Glasstone, 1952). At E14.5, splitting of the first molar tooth germ led to two molar teeth, with the anterior molar forming two to four cusps, whereas the posterior tooth developed four to seven cusps. Regeneration of the normal cusp pattern was therefore limited, and concentrated in the posterior halves (Fisher, 1971; Coin et al. 2000).
Multiple teeth have been shown to arise from the molar field in mice where β-catenin has been overexpressed (Jarvinen et al. 2006; Liu et al. 2008). The number of teeth that can develop from the molar field, therefore, would appear to be restricted by Wnt signalling. In wild-type mice the molar field will form up to three molars when explanted and grafted to a kidney capsule, whereas up to 40 teeth have been shown to develop from the same region in the β-catenin mouse (Jarvinen et al. 2006). The initial tooth buds form normally, but the dental epithelium then starts to undergo further budding and invaginations, leading to the formation of additional enamel knots and additional teeth. It is unclear whether these extra teeth represent the formation of successional teeth, which normally do not form in the mouse, or they result from the formation of an odontoma. A similar overproduction of teeth has been recently described in the Epfn mutant mouse (Nakamura et al. 2008). Epfn is a zinc-finger transcription factor, homologous to Sp6. In this case the mice survive up to 2 years old and develop up to 50 incisors by the age of 1 year, with eight molars on each side of the mandible. Loss of Epfn leads to an upregulation of Lef-1, a target of Wnt signalling; thus again the formation of supernumerary teeth is linked to stimulation of Wnt signalling. Overexpression of Lef-1 in a transgenic mouse has also been associated with the formation of ectopic teeth (Zhou et al. 1995). Apc(adenomatous polyposis coli) is a Wnt modifier that organizes the complex that degrades β-catenin. In human patients with a mutation in APC, supernumerary teeth and odontomas are observed (Fader et al. 1962; Wolf et al. 1986; Wang et al. 1998). In mice where Apc has been conditionally knocked out in the oral epithelium, multiple tooth buds form during embryonic development (Kuraguchi et al. 2006). This thus fits with the theory that enhancing Wnt signalling, by preventing degradation of β-catenin, leads to supernumerary teeth. In contrast, in patients with a mutation in AXIN2 (axis inhibitor 2) severe tooth loss is observed in permanent molars, premolars, lower incisors and maxillary lateral incisors (Lammi et al. 2004). Axin 2, like Apc, is a Wnt signal modifier acting to organize the β-catenin degradation complex. Mutations in Axin2 would therefore be assumed to lead to increased tooth number rather than a reduction. This could be explained by Axin2 acting in a negative feedback loop with Wnt signalling, and thus loss of this gene might lead to a drop in Wnt signalling (Lammi et al. 2004).
In both the β-catenin overexpression and Epfn knockout mice, the molar teeth that form have a much simpler shape than those associated with the wild-type molars, many forming only a single cusp. There appears to be a trade-off, therefore, between the number of teeth and the complexity of the tooth generated. This concurs with the general trend of mammals, with more complex tooth shapes, restricted to a single set of replacement teeth, or in many cases no replacement teeth at all.
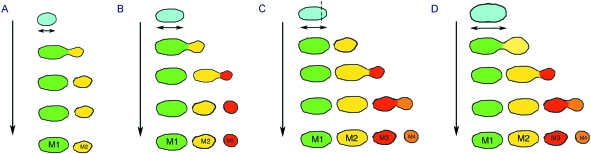
The size and number of molar teeth that normally develop from the molar field appear to be regulated by a system of positive signals from the mesenchyme and negative signals from the intermolar region (Kavanagh et al. 2007). In this way M1 inhibits development of M2, and M2 inhibits M3. If the primordia for M2 are removed from M1 in culture, the inhibition is removed and M2 develops earlier than normally observed in culture and reaches a greater size (Kavanagh et al. 2007). In extreme cases, removal of M1 results in the formation of a fourth molar M4, not normally seen in the mouse. This occurred in cases where the M3 was able to reach the size of the M2. In contrast, when the M2 failed to reach half the size of the M1, the M3 failed to form (Kavanagh et al. 2007) (Fig. 4). This links in with a recent study recombining molar tissue that suggested that the number of mesenchymal cells regulates the tooth number, not the final tooth size (Cai et al. 2007). Thus once a tooth is a given size it will lose its inhibition of the next tooth in the series, which will then develop, this occurring more readily when the original primordium is large. Thus if the number of mesenchymal cells in the tooth germ is increased, four molars are able to form from this region in culture, with the largest tooth that forms showing a similar size to a cultured M1 (Cai et al. 2007) (Fig. 4). If the number of mesenchymal cells is reduced, however, as in the case of dissociated molar mesenchyme from a single tooth germ recombined with a single tooth epithelium, the tooth is indeed smaller in size (Hu et al. 2006) and it would be predicted that an M2 would fail to form in many cases (Fig. 4). This model is supported by the phenotype of mouse mutants; for example, in Eda(Tabby) mutants the original molar field is smaller in size (from histology and as indicated by Shh expression) and the M3 often fails to form (Pispa et al. 1999; Kangas et al. 2004).
Fig. 4.
Changing molar tooth number. The size of the molar field affects the number of teeth that form. (A) A small molar field at E13.5, as generated by recombination or in an Edamutant, leads to the formation of a reduced number of teeth. (B) A wild-type molar field at E13.5. Inhibitory signals from the intermolar region lead to the formation of three molars of diminishing size. (C) If the anterior part of the molar field is cut off at E13.5, the posterior part is released from inhibition by M1 and up to four molars can form, with M2 reaching the normal size of M1. M2 and M3 when isolated from M1 have an accelerated initiation compared to that of whole cultured explants or in vivo (compare to B). (D) If the molar field is large in size, as after recombination with large numbers of mesenchymal cells, four molars can form, with M2 reaching the normal size of M1. Downward arrows indicate development of the molar field from E13.5 to formation of distinct teeth.
Changing tooth complexity
The complexity of the tooth can be altered by the addition or removal of cusps. As has been mentioned earlier, this alteration can be created by the fusion of tooth germs, but it can also be caused by the formation of additional cusps in an existing tooth germ. Changes in tooth complexity appear to be driven by diet, with diet-dependent changes of complexity of dental patterns being observed in both carnivores and rodents (Evans et al. 2007). The generation of cusps is hypothesized to be driven by the formation of additional enamel knots. Additional cusps can be formed by overstimulation of the Eda signalling pathway. If the receptor Edar is constitutively activated in oral epithelium, a large number of spiky cusps develop; whether this is due to an increased number of enamel knots, however, is unclear (Tucker et al. 2004). The number of cusps appears to be a factor of both cusp size and tooth size. Thus a large tooth can incorporate more cusps by physically having enough space to generate additional enamel knots, which are spaced far enough apart to allow folding of the epithelium. In addition, if the cusps are small, more can be fitted in (Cai et al. 2007). If a small molar is artificially created by recombining a small number of mesenchymal cells with tooth epithelium, then the number of cusps generated is reduced, with a single cusp forming in the majority of cases (Hu et al. 2006). Increasing the number of mesenchymal cells increases the tooth size towards that of the normal M1 and increases the number of cusps that form.
It is not only the number of enamel knots that is key but also the distribution of the knots within the developing tooth. This can be clearly seen when the molar teeth of mice and voles are compared (Keranen et al. 1998). In mice the molar cusps sit in parallel and in keeping with this, the secondary enamel knots are induced in parallel within the developing tooth germ. In the vole, in contrast, the secondary enamel knots develop at an angle to each other, producing the zig-zag pattern of cusps in the final molar tooth.
Mineralization: epithelial–mesenchymal interactions
After the disappearance of the secondary enamel knot signalling centres, cells constituting the tooth organ terminally differentiate, with the inner enamel epithelial cells differentiating into ameloblasts and the dental pulp mesenchymal cells into odontoblasts (Fig. 5). The differentiation of these cells is regulated, as in the previous stages, by epithelial–mesenchymal interactions. Signals from the inner enamel epithelium (IEE) induce the formation of odontoblasts in the dental mesenchyme (Ruch et al. 1982; Begue-Kirn et al. 1994). In vitro studies have shown that odontoblasts can be induced by Tgfb1, Fgfs, and Bmp2, whereas Igf1 (insulin-like growth factor 1) promotes polarization of these cells (Begue-Kirn et al. 1994; Martin et al. 1998). Interestingly, the basement membrane is essential for odontoblast differentiation, implicating cell–matrix interactions in the signalling between the two tissues (Thesleff & Hurmerinta, 1981). Odontoblast differentiation starts from the tips of the developing cusps and proceeds in a cervical or intercuspal direction (Thesleff et al. 2001). This differentiation pattern has been recently linked to the changing pattern of Wnt10a, which moves from the secondary enamel knots to the underlying preodontoblasts (Yamashiro et al. 2007). Overexpression of Wnt 10a in culture leads to induction of Dspp, one of the key markers of odontoblasts, indicating a role for Wnt signalling in the early stages of odontoblast formation (Yamashiro et al. 2007). Signals from the odontoblasts then pass to the overlying epithelium, triggering the terminal differentiation of ameloblasts. Ameloblast differentiation has been shown to require the presence of functional odontoblasts or predentin matrix (Karcher-Djuricic et al. 1985; Zeichner-David et al. 1995).
Fig. 5.
Cross-section of a mineralizing tooth. (A) Incisor P21. Ameloblasts are columnar epithelium polarized cells secreting to the extracellular matrix and forming the enamel. The enamel lies next to the dentin secreted by the underlying odontoblast layer of mesenchyme-derived cells.
Continuously growing teeth and asymmetrical enamel formation in incisors
In mice, replacement teeth are unnecessary in the incisors, as these are able to grow continuously. In other rodents, such as the guinea pig, the molars are also able to grow continuously throughout the animal's life. This is possible due to the presence of a stem cell niche located within the cervical loops, at the apical part of the tooth (Smith, 1980; Harada et al. 1999; Ohshima et al. 2005). In the case of mouse incisors only the labial cervical loop functions to generate ameloblasts that produce enamel on the labial tooth surface. Cells within the labial cervical loop are capable of differentiating into enamel-producing ameloblasts, stratum intermedium, stellate reticulum and outer enamel epithelium (Kawano et al. 2004). On the lingual side the cervical loop is much smaller and its progeny do not give rise to ameloblasts. Instead, the lingual cervical loop functions as a root analogue, forming epithelial cell rests of Malassez (ERM) and anchoring the incisor in the jaw (Tummers & Thesleff, 2008). The asymmetric deposition of enamel results in the sharpening of the mouse incisor by single face erosion.
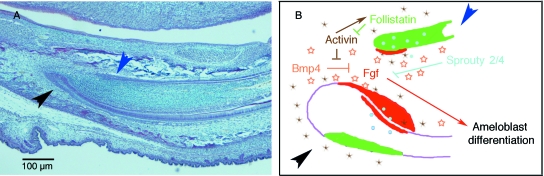
How this asymmetry is set up is not clear, but recent studies suggest that it is a very tightly regulated process. Recombination studies have shown that both labial and lingual incisor dental mesenchyme can induce ameloblast differentiation in labial dental epithelium, but neither can induce it in lingual epithelium, indicating a failure of the dental epithelium to respond (Amar et al. 1986, 1989). Follistatin is expressed in the dental epithelium of the lingual cervical loop and overexpression in the dental epithelium has been found to inhibit ameloblast differentiation in vivo, suggesting that follistatin is responsible for the lack of enamel on the lingual side of the mouse incisor (Wang et al. 2004). Follistatin is also expressed in the enamel-free area of molars, reinforcing this idea. In keeping with this, follistatin null mutant mice have polarized epithelial cells on the lingual side of the incisors resembling labial ameloblasts (Wang et al. 2004). Follistatin is an inhibitor of the Tgfβ superfamily including Bmps and Activin, suggesting that ameloblast differentiation on the lingual side is triggered by a loss of repression of a member of this family. Culture experiments have shown that Bmp4 can induce ameloblast differentiation, making this the most likely candidate for the signalling molecule from mesenchymal odontoblasts (Wang et al. 2004).
Follistatin may also be involved indirectly in the regulation of mesenchymal Fgfs, which are normally expressed in the dental papilla adjacent to the cervical loops. Fgf3 is only expressed on the labial side and plays an important role, interacting with Fgf10 to maintain ameloblast precursor cells (Harada et al. 1999, 2002). Follistatin-overexpressing transgenic mice fail to express Fgf3 in the dental papilla next to the labial cervical loop, whereas Fgf3is ectopically expressed on the lingual cervical loop in the follistatin null mouse (Wang et al. 2007). Bmp4 represses Fgf3, whereas Activin, which is preferentially expressed in labial mesenchyme, inhibits the effect of Bmp4 and stimulates Fgf3 expression in the labial mesenchyme. The presence of Fgf3 then results in the formation of a large cervical loop on this side. In this case, Follistatin appears to inhibit the proliferation of ameloblast precursor cells by inhibiting Activin, allowing Bmp4 to block the expression of Fgf3 on the lingual side (Thesleff et al. 2007). Recent studies carried out on the Sprouty genes show the effect of these receptor tyrosine kinase antagonists on the maintenance of ameloblasts precursor cells in the incisors. The inactivation of Sprouty 4 in conjunction with the inactivation of one allele of Sprouty 2 creates incisors with enamel on both lingual and labial sides. These ‘tusk’ mutant mice have excessively long incisors due to the inability of these incisors to erode (Klein et al. 2008). This phenotype is achieved by the disruption of the inhibitory effect Sprouty has on the Fgf signalling loop on the lingual side of the incisors. Fgf9 is expressed in the inner enamel epithelium on the labial side, overlying the expression domain of Fgf3 and Fgf10 in the mesenchyme. In the Sprouty 2/4 mutant, Fgf9, 3 and 10 are all upregulated on the lingual side, and enamel is formed (Klein et al. 2008). Again this stresses the importance of Fgf3 being asymmetrically expressed, producing a greater dose of Fgf signalling on the labial side that allows the differentiation of ameloblasts (Fig. 6).
Fig. 6.
Mouse incisor cervical loops: regulation of ameloblast differentiation. (A) Sagittal section of a P0 mouse incisor showing the cervical loops, stained with Haematoxylin and Eosin. Black arrowhead points to the labial cervical loop and blue arrowhead to the lingual cervical loop. These correspond to the same regions arrowed in (B). (B) Schematic representation of the regulation of ameloblast differentiation in the incisor leading to the asymmetric deposition of enamel. Follistatin (green), induced by activin (brown stars), is expressed mainly in the lingual cervical loop. Follistatin inhibits activin in the dental papilla. The absence of activin in this area allows for the inhibition of Fgf (red) by Bmp4 (orange stars). At the same time, Sprouty 2 and 4 (blue dots) inhibit an Fgf regulatory loop between epithelial and mesenchymally expressed Fgfs. This decreases the concentration of Fgfs, inhibiting ameloblast differentiation in the labial cervical loop.
Tooth hard tissue
The later stages of tooth development are characterized by the formation of the mineralized tissue: dentin, cementum and enamel. Dentin and cementum have significant similarities with bone (Linde & Goldberg, 1993), but enamel is the only epithelially derived calcified tissue in mammals and is unique in its structure.
Enamel
Differentiated ameloblasts express tissue-specific genes whose extracellular matrix products result in the formation of mineralized enamel matrix. Ameloblasts secrete two major classes of proteins, glycosylated and nonglycosylated. The nonglycosylated are the hydrophobic amelogenins, which are the most abundant proteins in the enamel matrix, constituting around 90%. These proteins are believed to function as the principal enamel deposition organizers, but lately they have been suspected to be involved in root formation, periodontium regeneration and to function as growth factors (Zeichner-David, 2001). Amelogenin expression has also been found in soft tissues suggesting functions other than as a nucleator of mineralized tissue (Deutsch et al. 2006). Leucine-rich amelogenin peptide(LRAP), an alternatively spliced amelogenin, has been shown to have a role in the upregulation of osteopontin in cementogenesis via the MAPK pathway (Boabaid et al. 2004). In addition, Fanchon and colleagues hypothesized that the deficiency of dentin mineralization in tooth organs treated with matrix metalloproteinase inhibitors could be the result of the infiltration and accumulation of amelogenin in the matrix adjacent to the odontoblasts (Fanchon et al. 2004). The glycosylated or non-amelogenin enamel proteins include tuftelin, ameloblastin and enamelin. Tuftelin was the first non-amelogenin protein characterized, but its function on tooth development is still not well understood (Deutsch et al. 1991; Zeichner-David et al. 1997). Tuftelin has also been found to be expressed in other organs such as kidney, lung, liver and testis (MacDougall et al. 1998; Leiser et al. 2007). Ameloblastin represents about 5% of the enamel matrix and has been immuno-localized in the secretory ameloblast (Uchida et al. 1998; Brookes et al. 2001). Studies carried out on the interaction between ameloblastin and amelogenin suggest a co-operative function in the scaffolding needed for the formation of enamel (Ravindranath et al. 2004). Mouse recombinant ameloblastin acts as a growth factor increasing cell attachment and proliferation of periodontal ligament cells in vitro (Zeichner-David et al. 2006). Ameloblastin has also been found in pre-odontoblast, pulpal mesenchymal cells and Hertwig's epithelial root sheath (HERS) cells, but its function in these tissues as well as in ameloblasts is still not fully understood (Zeichner-David et al. 2003). Enamelin is a large enamel protein that has also been immuno-localized to the secretory ameloblast (Hu et al. 1997). ENAMELIN is thought to be the main candidate gene responsible for the autosomal-inherited form of amelogenesis imperfecta (AI), and mutations in AMELOGENINlead to X-linked AI (Kim et al. 2004; Kim et al. 2005). In organ cultures the addition of insulin, or insulin-like growth factor-I and -II (Igf-I and -II), results in the induction of ameloblastin, amelogenin, and enamelin and may represent one of the signalling pathways involved in ameloblast induction (Takahashi et al. 1998; Caton et al. 2005).
Dentin
Odontoblasts produce a dentin layer that has many phenotypic similarities to bone produced by osteoblasts. Dentin is mainly composed of Collagen type I, but its histological appearance is derived from the presence of non-collagenous proteins (NCPs). These include dentin sialoprotein (Dsp) and dentin phosphoprotein (Dpp), two proteins encoded by a single gene dentin sialophosphoprotein (Dspp) that is differentially spliced to form the two proteins (MacDougall et al. 1997). This gene was believed to be expressed exclusively in odontoblasts, but it has also been detected in pre-ameloblast, alveolar bone osteoblasts, cementoblasts and periodontal fibroblasts (Qin et al. 2003; Baba et al. 2004) Other NCPs include osteocalcin (Bglap), bone sialoprotein (Bsp/Ibsp), osteopontin (Spp1) and dentin matrix protein-1 (Dmp-1). This last protein has been suggested to play a role in the induction of mesenchyme to terminally differentiate into odontoblasts (Narayanan et al. 2001). Recent studies have shown that Dmp-1 binds specifically to the Dspp promoter and activates transcription (Narayanan et al. 2006). All of these proteins are also present in bone. The uniqueness of dentin is thought to be due to the difference in the concentrations of some of these proteins (Butler et al. 2003). This similarity with bone can be mimicked by the addition of Igfs to odontoblast-like cell lines, which leads to inhibition of Dspp and Dmp1 while inducing Collagen type I, suggestive of a transformation from an odontoblast-like to a bone-like cell (Caton et al. 2007).
Conclusions
The tooth provides an excellent model for studying how an organ develops. In particular the large variety of tooth shapes, sizes and numbers allows multiple important developmental questions to be addressed. As a mineralized element it also provides an opportunity to study how different mineral layers are formed and interact with each other. The ability of the mouse incisor to grow continuously also provides an entrance into the world of stem cells, stem cell niches and tissue engineering.
The need for models other than the mouse, however, is apparent for our understanding of other aspects of the dentition, most notably that of replacement teeth. The shrew, for example, offers a good opportunity to study a mammal where the deciduous tooth development is suppressed, indicating that the early activation of replacement teeth may lead to suppression of the deciduous tooth (Jarvinen et al. 2008). The ferret provides a more standard model for tooth replacement, with 28–30 deciduous teeth being replaced by approximately 34 permanent teeth (Berkovitz, 1973; He et al. 2002; Jarvinen, 2008). The ferret, shrew and opossum retain a full complement of tooth types (molars, premolars, canines and incisors), allowing for a more complex study of tooth patterning (Torres et al. 2008). Reptiles such as lizards and snakes offer a wonderful opportunity to study successive tooth replacement patterns (Delgado et al. 2003; Buchtova et al. 2008), along with fish species such as the trout and Atlantic salmon (Fraser et al. 2006; Huysseune & Witten, 2008). From the point of view of tooth shape, cichlids show an amazing range of tooth morphologies (Streelman et al. 2003), and we can also learn from unusual tooth morphologies, such as the hollow fangs of poisonous snakes (Zahradnicek et al. 2008). Our understanding of tooth development in these species is still in its infancy, and with the help of genetic information identified in the mouse such exciting new avenues will quickly provide fascinating insights into tooth development and mineralization.
References
- Ahrens K. Die Entwicklung der menschlichen Zahne. Arb Anat Inst Wiesbaden. 1913;48:169–266. [Google Scholar]
- Amar S, Karcher-Djuricic V, Meyer JM, Ruch JV. The lingual (root analogue) and the labial (crown analogue) mouse incisor dentin promotes ameloblast differentiation. Arch Anat Microsc Morphol Exp. 1986;75:229–239. [PubMed] [Google Scholar]
- Amar S, Luo W, Snead ML, Ruch JV. Amelogenin gene expression in mouse incisor heterotopic recombinations. Differentiation. 1989;41:56–61. doi: 10.1111/j.1432-0436.1989.tb00732.x. [DOI] [PubMed] [Google Scholar]
- Andl T, Ahn K, Kairo A, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Baba O, Qin C, Brunn JC, et al. Detection of dentin sialoprotein in rat periodontium. Eur J Oral Sci. 2004;112:163–170. doi: 10.1111/j.0909-8836.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, et al. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AJ, Francis-West PH. Ectopic application of recombinant BMP-2 and BMP-4 can change patterning of developing chick facial primordia. Development. 1997;124:391–398. doi: 10.1242/dev.124.2.391. [DOI] [PubMed] [Google Scholar]
- Barlow AJ, Bogardi JP, Ladher R, Francis-West PH. Expression of chick Barx-1 and its differential regulation by FGF-8 and BMP signaling in the maxillary primordia. Dev Dyn. 1999;214:291–302. doi: 10.1002/(SICI)1097-0177(199904)214:4<291::AID-AJA2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Begue-Kirn C, Smith AJ, Loriot M, Kupferle C, Ruch JV, Lesot H. Comparative analysis of TGF beta s, BMPs, IGF1, msxs, fibronectin, osteonectin and bone sialoprotein gene expression during normal and in vitro-induced odontoblast differentiation. Int J Dev Biol. 1994;38:405–420. [PubMed] [Google Scholar]
- Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- Berkovitz BK. Tooth development in the albino ferret (Mustela putorius) with special reference to the permanent carnassial. Arch Oral Biol. 1973;18:465–471. doi: 10.1016/0003-9969(73)90066-6. [DOI] [PubMed] [Google Scholar]
- Boabaid F, Gibson CW, Kuehl MA, et al. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–1136. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- Britanova O, Depew MJ, Schwark M, et al. Satb2 haploinsufficiency phenocopies 2q32–q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am J Hum Genet. 2006;79:668–678. doi: 10.1086/508214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito JM, Teillet MA, Le Douarin NM. Induction of mirror-image supernumerary jaws in chicken mandibular mesenchyme by Sonic Hedgehog-producing cells. Development. 2008;135:2311–2329. doi: 10.1242/dev.019125. [DOI] [PubMed] [Google Scholar]
- Brookes SJ, Kirkham J, Shore RC, Wood SR, Slaby I, Robinson C. Amelin extracellular processing and aggregation during rat incisor amelogenesis. Arch Oral Biol. 2001;46:201–208. doi: 10.1016/s0003-9969(00)00121-7. [DOI] [PubMed] [Google Scholar]
- Buchtova M, Handrigan GR, Tucker AS, et al. Initiation and patterning of the snake dentition are dependent on Sonic hedgehog signaling. Dev Biol. 2008;319:132–145. doi: 10.1016/j.ydbio.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Butler PM. Ontogenetic aspects of dental evolution. Int J Dev Biol. 1995;39:25–34. [PubMed] [Google Scholar]
- Butler WT, Brunn JC, Qin C. Dentin extracellular matrix (ECM) proteins: comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect Tissue Res. 2003;44(Suppl. 1):171–178. [PubMed] [Google Scholar]
- Cai J, Cho SW, Kim JY, Lee MJ, Cha YG, Jung HS. Patterning the size and number of tooth and its cusps. Dev Biol. 2007;304:499–507. doi: 10.1016/j.ydbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Caton J, Bringas P, Jr, Zeichner-David M. IGFs increase enamel formation by inducing expression of enamel mineralizing specific genes. Arch Oral Biol. 2005;50:123–129. doi: 10.1016/j.archoralbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Caton J, Bringas P, Jr, Zeichner-David M. Establishment and characterization of an immortomouse-derived odontoblast-like cell line to evaluate the effect of insulin-like growth factors on odontoblast differentiation. J Cell Biochem. 2007;100:450–463. doi: 10.1002/jcb.21053. [DOI] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Cho SW, Lee HA, Cai J, et al. The primary enamel knot determines the position of the first buccal cusp in developing mice molars. Differentiation. 2007;75:441–451. doi: 10.1111/j.1432-0436.2006.00153.x. [DOI] [PubMed] [Google Scholar]
- Coin R, Lesot H, Vonesch JL, Haikel Y, Ruch JV. Aspects of cell proliferation kinetics of the inner dental epithelium during mouse molar and incisor morphogenesis: a reappraisal of the role of the enamel knot area. Int J Dev Biol. 1999;43:261–267. [PubMed] [Google Scholar]
- Coin R, Schmitt R, Lesot H, Vonesch JL, Ruch JV. Regeneration of halved embryonic lower first mouse molars: correlation with the distribution pattern of non dividing IDE cells, the putative organizers of morphogenetic units, the cusps. Int J Dev Biol. 2000;44:289–295. [PubMed] [Google Scholar]
- Delgado S, Davit-Beal T, Sire JY. Dentition and tooth replacement pattern in Chalcides (Squamata; Scincidae) J Morphol. 2003;256:146–159. doi: 10.1002/jmor.10080. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Compagnucci C. Tweaking the hinge and caps: testing a model of the organization of jaws. J Exp Zoolg B Mol Dev Evol. 2008;310:315–335. doi: 10.1002/jez.b.21205. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA, Morasso M, Rubenstein JL. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J Anat. 2005;207:501–561. doi: 10.1111/j.1469-7580.2005.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch D, Palmon A, Fisher LW, Kolodny N, Termine JD, Young MF. Sequencing of bovine enamelin (‘tuftelin’) a novel acidic enamel protein. J Biol Chem. 1991;266:16021–16028. [PubMed] [Google Scholar]
- Deutsch D, Haze-Filderman A, Blumenfeld A, et al. Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: brain and cells of the hematopoietic system. Eur J Oral Sci. 2006;114(Suppl. 1):183–189. doi: 10.1111/j.1600-0722.2006.00301.x. discussion 201–202, 381. [DOI] [PubMed] [Google Scholar]
- Evans AR, Wilson GP, Fortelius M, Jernvall J. High-level similarity of dentitions in carnivorans and rodents. Nature. 2007;445:78–81. doi: 10.1038/nature05433. [DOI] [PubMed] [Google Scholar]
- Fader M, Kline SN, Spatz SS, Zubrow HJ. Gardner's syndrome (intestinal polyposis, osteomas, sebaceous cysts) and a new dental discovery. Oral Surg Oral Med Oral Pathol. 1962;15:153–172. doi: 10.1016/0030-4220(62)90004-x. [DOI] [PubMed] [Google Scholar]
- Fanchon S, Bourd K, Septier D, et al. Involvement of matrix metalloproteinases in the onset of dentin mineralization. Eur J Oral Sci. 2004;112:171–176. doi: 10.1111/j.1600-0722.2004.00120.x. [DOI] [PubMed] [Google Scholar]
- Fisher AR. Morphological development in vitro of the whole and halved lower molar tooth germ of the mouse. Arch Oral Biol. 1971;16:1481–1496. doi: 10.1016/0003-9969(71)90084-7. [DOI] [PubMed] [Google Scholar]
- Fleischmannova J, Matalova E, Tucker AS, Sharpe PT. Mouse models of tooth abnormalities. Eur J Oral Sci. 2008;116:1–10. doi: 10.1111/j.1600-0722.2007.00504.x. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Berkovitz BK, Graham A, Smith MM. Gene deployment for tooth replacement in the rainbow trout (Oncorhynchus mykiss): a developmental model for evolution of the osteichthyan dentition. Evol Dev. 2006;8:446–457. doi: 10.1111/j.1525-142X.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- Glasstone S. The development of halved tooth germs; a study in experimental embryology. J Anat. 1952;86:12–15. [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Toyono T, Toyoshima K, et al. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z, Mo R, Hui CC, Sharpe PT. The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development. 1998;125:2803–2811. doi: 10.1242/dev.125.15.2803. [DOI] [PubMed] [Google Scholar]
- Haworth KE, Wilson JM, Grevellec A, et al. Sonic hedgehog in the pharyngeal endoderm controls arch pattern via regulation of Fgf8 in head ectoderm. Dev Biol. 2007;303:244–258. doi: 10.1016/j.ydbio.2006.11.009. [DOI] [PubMed] [Google Scholar]
- He T, Friede H, Kiliaridis S. Dental eruption and exfoliation chronology in the ferret (Mustela putorius furo) Arch Oral Biol. 2002;47:619–623. doi: 10.1016/s0003-9969(02)00043-2. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, et al. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;414:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- Hovorakova M, Lesot H, Peterkova R, Peterka M. Origin of the deciduous upper lateral incisor and its clinical aspects. J Dent Res. 2006;85:167–171. doi: 10.1177/154405910608500210. [DOI] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, et al. Cloning and characterization of porcine enamelin mRNAs. J Dent Res. 1997;76:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- Hu B, Nadiri A, Kuchler-Bopp S, Perrin-Schmitt F, Peters H, Lesot H. Tissue engineering of tooth crown, root, and periodontium. Tissue Eng. 2006;12:2069–2075. doi: 10.1089/ten.2006.12.2069. [DOI] [PubMed] [Google Scholar]
- Huysseune A, Witten PE. An evolutionary view on tooth development and replacement in wild Atlantic salmon (Salmo salar L.) Evol Dev. 2008;10:6–14. doi: 10.1111/j.1525-142X.2007.00209.x. [DOI] [PubMed] [Google Scholar]
- Jarvinen E. Mechanisms and molecular regulation of mammalian tooth replacement. Dissertationes Bioscientiarum Molecularium Universitatis Helsingiensis in Viikki.
- Jarvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen E, Valimaki K, Pummila M, Thesleff I, Jernvall J. The taming of the shrew milk teeth. Evol Dev. 2008;10:477–486. doi: 10.1111/j.1525-142X.2008.00258.x. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- Kangas AT, Evans AR, Thesleff I, Jernvall J. Nonindependence of mammalian dental characters. Nature. 2004;432:211–214. doi: 10.1038/nature02927. [DOI] [PubMed] [Google Scholar]
- Karcher-Djuricic V, Staubli A, Meyer JM, Ruch JV. Acellular dental matrices promote functional differentiation of ameloblasts. Differentiation. 1985;29:169–175. doi: 10.1111/j.1432-0436.1985.tb00311.x. [DOI] [PubMed] [Google Scholar]
- Kassai Y, Munne P, Hotta Y, et al. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- Kavanagh KD, Evans AR, Jernvall J. Predicting evolutionary patterns of mammalian teeth from development. Nature. 2007;449:427–432. doi: 10.1038/nature06153. [DOI] [PubMed] [Google Scholar]
- Kawada S, Koyasu K, Zholnerovskaya EI, Oda S. Analysis of dental anomalies in the Siberian mole, Talpa altaica (Insectivora, Talpidae) Arch Oral Biol. 2006;51:1029–1039. doi: 10.1016/j.archoralbio.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Kawano S, Saito M, Handa K, et al. Characterization of dental epithelial progenitor cells derived from cervical-loop epithelium in a rat lower incisor. J Dent Res. 2004;83:129–133. doi: 10.1177/154405910408300209. [DOI] [PubMed] [Google Scholar]
- Keranen SV, Aberg T, Kettunen P, Thesleff I, Jernvall J. Association of developmental regulatory genes with the development of different molar tooth shapes in two species of rodents. Dev Genes Evol. 1998;208:477–486. doi: 10.1007/s004270050206. [DOI] [PubMed] [Google Scholar]
- Kere J, Srivastava AK, Montonen O, et al. X-linked anhydrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Thesleff I. Expression and function of FGFs-4, -8, and -9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256–268. doi: 10.1002/(SICI)1097-0177(199803)211:3<256::AID-AJA7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hu YY, et al. Amelogenin p.M1T and p.W4S mutations underlying hypoplastic X-linked amelogenesis imperfecta. J Dent Res. 2004;83:378–383. doi: 10.1177/154405910408300505. [DOI] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lin BP, et al. ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res. 2005;84:278–282. doi: 10.1177/154405910508400314. [DOI] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, et al. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial–mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriangkrai R, Chareonvit S, Yahagi K, Fujiwara M, Eto K, Iseki S. Study of Pax6 mutant rat revealed the association between upper incisor formation and midface formation. Dev Dyn. 2006a;235:2134–2143. doi: 10.1002/dvdy.20875. [DOI] [PubMed] [Google Scholar]
- Kriangkrai R, Iseki S, Eto K, Chareonvit S. Dual odontogenic origins develop at the early stage of rat maxillary incisor development. Anat Embryol (Berl) 2006b;211:101–108. doi: 10.1007/s00429-005-0068-7. [DOI] [PubMed] [Google Scholar]
- Kuraguchi M, Wang XP, Bronson RT, et al. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2006;2:e146. doi: 10.1371/journal.pgen.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C, Moreau A, Chamberland M, Tremblay ML, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola M, Mustonen T, et al. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev Biol. 2001;229:443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Kassai Y, Pakkasjarvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Leiser Y, Blumenfeld A, Haze A, et al. Localization, quantification, and characterization of tuftelin in soft tissues. Anat Rec (Hoboken) 2007;290:449–454. doi: 10.1002/ar.20512. [DOI] [PubMed] [Google Scholar]
- Lesot H, Vonesch JL, Peterka M, Tureckova J, Peterkova R, Ruch JV. Mouse molar morphogenesis revisited by three-dimensional reconstruction. II. Spatial distribution of mitoses and apoptosis in cap to bell staged first and second upper molar teeth. Int J Dev Biol. 1996;40:1017–1031. [PubMed] [Google Scholar]
- Linde A, Goldberg M. Dentinogenesis. Crit Rev Oral Biol Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- Liu F, Chu EY, Watt B, et al. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–268. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Lumsden AG. Pattern formation in the molar dentition of the mouse. J Biol Buccale. 1979;7:77–103. [PubMed] [Google Scholar]
- Lumsden AG. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103(Suppl):155–169. doi: 10.1242/dev.103.Supplement.155. [DOI] [PubMed] [Google Scholar]
- Luukko K, Loes S, Furmanek T, Fjeld K, Kvinnsland IH, Kettunen P. Identification of a novel putative signaling center, the tertiary enamel knot in the postnatal mouse molar tooth. Mech Dev. 2003;120:270–276. doi: 10.1016/s0925-4773(02)00458-6. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J Biol Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Simmons D, Dodds A, et al. Cloning, characterization, and tissue expression pattern of mouse tuftelin cDNA. J Dent Res. 1998;77:1970–1978. doi: 10.1177/00220345980770120401. [DOI] [PubMed] [Google Scholar]
- Martin A, Unda FJ, Begue-Kirn C, Ruch JV, Arechaga J. Effects of aFGF, bFGF, TGFbeta1 and IGF-I on odontoblast differentiation in vitro. Eur J Oral Sci. 1998;106(Suppl. 1):117–121. doi: 10.1111/j.1600-0722.1998.tb02162.x. [DOI] [PubMed] [Google Scholar]
- Matalova E, Antonarakis GS, Sharpe PT, Tucker AS. Cell lineage of primary and secondary enamel knots. Dev Dyn. 2005;233:754–759. doi: 10.1002/dvdy.20396. [DOI] [PubMed] [Google Scholar]
- Miletich I, Buchner G, Sharpe PT. Barx1 and evolutionary changes in feeding. J Anat. 2005;207:619–622. doi: 10.1111/j.1469-7580.2005.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Drouin J. Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev Biol. 2008;313:887–896. doi: 10.1016/j.ydbio.2007.10.055. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Angeli I, James C, Lendahl U, Sharpe PT. Role of Islet1 in the patterning of murine dentition. Development. 2003;130:4451–4460. doi: 10.1242/dev.00631. [DOI] [PubMed] [Google Scholar]
- Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 1999;22:366–369. doi: 10.1038/11937. [DOI] [PubMed] [Google Scholar]
- Mucchielli ML, Mitsiadis TA, Raffo S, Brunet JF, Proust JP, Goridis C. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev Biol. 1997;189:275–284. doi: 10.1006/dbio.1997.8672. [DOI] [PubMed] [Google Scholar]
- Mustonen T, Ilmonen M, Pummila M, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131:4907–4919. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- Nakamura T, de Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y. Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J Biol Chem. 2008;283:4825–4833. doi: 10.1074/jbc.M708388200. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Du Y, et al. SHH mutation is associated with solitary median maxillary central incisor: a study of 13 patients and review of the literature. Am J Med Genet. 2001;102:1–10. doi: 10.1002/1096-8628(20010722)102:1<1::aid-ajmg1336>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Narayanan K, Srinivas R, Ramachandran A, Hao J, Quinn B, George A. Differentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1. Proc Natl Acad Sci U S A. 2001;98:4516–4521. doi: 10.1073/pnas.081075198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K, Gajjeraman S, Ramachandran A, Hao J, George A. Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J Biol Chem. 2006;281:19064–19071. doi: 10.1074/jbc.M600714200. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Courtney JM, Tucker AS, et al. Traf6 is essential for murine tooth cusp morphogenesis. Dev Dyn. 2004;229:131–135. doi: 10.1002/dvdy.10400. [DOI] [PubMed] [Google Scholar]
- Ohshima H, Nakasone N, Hashimoto E, Sakai H, Nakakura-Ohshima K, Harada H. The eternal tooth germ is formed at the apical end of continuously growing teeth. Arch Oral Biol. 2005;50:153–157. doi: 10.1016/j.archoralbio.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Peterkova R, Peterka M, Vonesch JL, Ruch JV. Multiple developmental origin of the upper incisor in mouse: histological and computer assisted 3-D-reconstruction studies. Int J Dev Biol. 1993;37:581–588. [PubMed] [Google Scholar]
- Peterkova R, Peterka M, Viriot L, Lesot H. Dentition development and budding morphogenesis. J Craniofac Genet Dev Biol. 2000;20:158–172. [PubMed] [Google Scholar]
- Peterkova R, Peterka M, Lesot H. The developing mouse dentition: a new tool for apoptosis study. Ann N Y Acad Sci. 2003;1010:453–466. doi: 10.1196/annals.1299.083. [DOI] [PubMed] [Google Scholar]
- Peterkova R, Lesot H, Viriot L, Peterka M. The supernumerary cheek tooth in tabby/EDA mice – a reminiscence of the premolar in mouse ancestors. Arch Oral Biol. 2005;50:219–225. doi: 10.1016/j.archoralbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J, Jung HS, Jernvall J, et al. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev Biol. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- Qin C, Brunn JC, Cadena E, Ridall A, Butler WT. Dentin sialoprotein in bone and dentin sialophosphoprotein gene expressed by osteoblasts. Connect Tissue Res. 2003;44(Suppl. 1):179–183. [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, et al. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Ravindranath HH, Chen LS, Zeichner-David M, Ishima R, Ravindranath RM. Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem Biophys Res Commun. 2004;323:1075–1083. doi: 10.1016/j.bbrc.2004.08.207. [DOI] [PubMed] [Google Scholar]
- Ravn JJ. Aplasia, supernumerary teeth and fused teeth in the primary dentition. An epidemiologic study. Scand J Dent Res. 1971;79:1–6. doi: 10.1111/j.1600-0722.1971.tb01986.x. [DOI] [PubMed] [Google Scholar]
- Ruch JV, Lesot H, Karcher-Djuricic V, Meyer JM, Olive M. Facts and hypotheses concerning the control of odontoblast differentiation. Differentiation. 1982;21:7–12. doi: 10.1111/j.1432-0436.1982.tb01187.x. [DOI] [PubMed] [Google Scholar]
- Ruch JV, Lesot H, Karcher-Djuricic V, Meyer JM. Extracellular matrix-mediated interactions during odontogenesis. Prog Clin Biol Res. 1984;151:103–114. [PubMed] [Google Scholar]
- Sarkar L, Sharpe PT. Expression of Wnt signalling pathway genes during tooth development. Mech Dev. 1999;85:197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–536. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Sharpe PT. Homeobox genes and orofacial development. Connect Tissue Res. 1995;32:17–25. doi: 10.3109/03008209509013701. [DOI] [PubMed] [Google Scholar]
- Smith CE. Cell turnover in the odontogenic organ of the rat incisor as visualized by graphic reconstructions following a single injection of 3H-thymidine. Am J Anat. 1980;158:321–343. doi: 10.1002/aja.1001580307. [DOI] [PubMed] [Google Scholar]
- St Amand TR, Zhang Y, Semina EV, et al. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev Biol. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- Stock DW. The genetic basis of modularity in the development and evolution of the vertebrate dentition. Philos Trans R Soc Lond B Biol Sci. 2001;356:1633–1653. doi: 10.1098/rstb.2001.0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streelman JT, Webb JF, Albertson RC, Kocher TD. The cusp of evolution and development: a model of cichlid tooth shape diversity. Evol Dev. 2003;5:600–608. doi: 10.1046/j.1525-142x.2003.03065.x. [DOI] [PubMed] [Google Scholar]
- Szeto DP, Rodriguez-Esteban C, Ryan AK, et al. Role of the Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamane A, Bringas P, Caton J, Slavkin HC, Zeichner-David M. Induction of amelogenin and ameloblastin by insulin and insulin-like growth factors (IGF-I and IGF-II) during embryonic mouse tooth development in vitro. Connect Tissue Res. 1998;38:269–278. doi: 10.3109/03008209809017047. discussion 295–303. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Hurmerinta K. Tissue interactions in tooth develop-ment. Differentiation. 1981;18:75–88. doi: 10.1111/j.1432-0436.1981.tb01107.x. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Keranen S, Jernvall J. Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv Dent Res. 2001;15:14–18. doi: 10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Wang XP, Suomalainen M. Regulation of epithelial stem cells in tooth regeneration. C R Biol. 2007;330:561–564. doi: 10.1016/j.crvi.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Tucker AS, Qui M, et al. Role of Dlx-1 and Dlx-2 genes in patterning of the murine dentition. Development. 1997;124:4811–4818. doi: 10.1242/dev.124.23.4811. [DOI] [PubMed] [Google Scholar]
- Torres CB, Alves JB, Silva GA, Goes VS, Nakao LY, Goes AM. Role of BMP-4 during tooth development in a model with complete dentition. Arch Oral Biol. 2008;53:2–8. doi: 10.1016/j.archoralbio.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Sharpe PT. Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev Dyn. 1998a;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT. The activation level of the TNF family receptor, Edar, determines cusp number and tooth number during tooth development. Dev Biol. 2004;268:185–194. doi: 10.1016/j.ydbio.2003.12.019. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Headon DJ, Schneider P, et al. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998b;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126:51–61. doi: 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- Tummers M, Thesleff I. Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evol Dev. 2008;10:187–195. doi: 10.1111/j.1525-142X.2008.00226.x. [DOI] [PubMed] [Google Scholar]
- Uchida T, Murakami C, Wakida K, et al. Sheath proteins: synthesis, secretion, degradation and fate in forming enamel. Eur J Oral Sci. 1998;106(Suppl. 1):308–314. doi: 10.1111/j.1600-0722.1998.tb02191.x. [DOI] [PubMed] [Google Scholar]
- Vaahtokari A, Aberg T, Jernvall J, Keranen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Viriot L, Lesot H, Vonesch JL, Ruch JV, Peterka M, Peterkova R. The presence of rudimentary odontogenic structures in the mouse embryonic mandible requires reinterpretation of developmental control of first lower molar histomorphogenesis. Int J Dev Biol. 2000;44:233–240. [PubMed] [Google Scholar]
- Viriot L, Peterkova R, Peterka M, Lesot H. Evolutionary implications of the occurrence of two vestigial tooth germs during early odontogenesis in the mouse lower jaw. Connect Tissue Res. 2002;43:129–133. doi: 10.1080/03008200290001168. [DOI] [PubMed] [Google Scholar]
- Wang M, Dobeck JM, Sorkin BC, Skobe Z. Adenomatous polyposis coli protein is expressed in alternate stages of the ameloblast life cycle. J Dent Res. 1998;77:1979–1982. doi: 10.1177/00220345980770120501. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, et al. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J, Tucker AS. Fgf and Bmp signals repress the expression of Bapx1 in the mandibular mesenchyme and control the position of the developing jaw joint. Dev Biol. 2004;266:138–150. doi: 10.1016/j.ydbio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Wolf J, Jarvinen HJ, Hietanen J. Gardner's dento-maxillary stigmas in patients with familial adenomatosis coli. Br J Oral Maxillofac Surg. 1986;24:410–416. doi: 10.1016/0266-4356(86)90054-9. [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Yasui K, Sonomura T, Uemura M. Development of heterodont dentition in house shrew (Suncus murinus) Eur J Oral Sci. 2007;115:4334–4340. doi: 10.1111/j.1600-0722.2007.00499.x. [DOI] [PubMed] [Google Scholar]
- Yamashiro T, Zheng L, Shitaku Y, et al. Wnt10a regulates dentin sialophosphoprotein mRNA expression and possibly links odontoblast differentiation and tooth morphogenesis. Differentiation. 2007;75:452–462. doi: 10.1111/j.1432-0436.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- Zahradnicek O, Horacek I, Tucker AS. Viperous fangs: development and evolution of the venom canal. Mech Dev. 2008;125:786–796. doi: 10.1016/j.mod.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Zeichner-David M. Is there more to enamel matrix proteins than biomineralization? Matrix Biol. 2001;20:307–316. doi: 10.1016/s0945-053x(01)00155-x. [DOI] [PubMed] [Google Scholar]
- Zeichner-David M, Diekwisch T, Fincham A, et al. Control of ameloblast differentiation. Int J Dev Biol. 1995;39:69–92. [PubMed] [Google Scholar]
- Zeichner-David M, Vo H, Tan H, et al. Timing of the expression of enamel gene products during mouse tooth development. Int J Dev Biol. 1997;41:27–38. [PubMed] [Google Scholar]
- Zeichner-David M, Oishi K, Su Z, et al. Role of Hertwig's epithelial root sheath cells in tooth root development. Dev Dyn. 2003;228:651–663. doi: 10.1002/dvdy.10404. [DOI] [PubMed] [Google Scholar]
- Zeichner-David M, Chen LS, Hsu Z, Reyna J, Caton J, Bringas P. Amelogenin and ameloblastin show growth-factor like activity in periodontal ligament cells. Eur J Oral Sci. 2006;114(Suppl. 1):244–53. doi: 10.1111/j.1600-0722.2006.00322.x. discussion 254–6, 381–2. [DOI] [PubMed] [Google Scholar]
- Zhao X, Zhang Z, Song Y, et al. Transgenically ectopic expression of Bmp4 to the Msx1 mutant dental mesenchyme restores downstream gene expression but represses Shh and Bmp2 in the enamel knot of wild type tooth germ. Mech Dev. 2000;99:29–38. doi: 10.1016/s0925-4773(00)00467-6. [DOI] [PubMed] [Google Scholar]
- Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]