Summary
Background
The let-7 and lin-4 microRNAs belong to a class of temporally expressed, noncoding regulatory RNAs that function as heterochronic switch genes in the nematode C. elegans. Heterochronic genes control the relative timing of events during development and are considered a major force in the rapid evolution of new morphologies. let-7 is highly conserved and in Drosophila is temporally coregulated with the lin-4 homolog, miR-125. Little is known, however, about their requirement outside the nematode or whether they universally control the timing of developmental processes.
Results
We report the generation of a Drosophila mutant that lacks let-7 and miR-125 activities and that leads to a pleiotropic phenotype arising during metamorphosis. We focus on two defects and demonstrate that loss of let-7 and miR-125 results in temporal delays in two distinct metamorphic processes: the terminal cell-cycle exit in the wing and maturation of neuromuscular junctions (NMJs) at adult abdominal muscles. We identify the abrupt (ab) gene, encoding a nuclear protein, as a bona fide let-7 target and provide evidence that let-7 governs the maturation rate of abdominal NMJs during metamorphosis by regulating ab expression.
Conclusions
Drosophila let-7 and miR-125 mutants exhibit temporal misregulation of specific metamorphic processes. As in C. elegans, Drosophila let-7 is both necessary and sufficient for the appropriate timing of a specific cell-cycle exit, indicating that its function as a heterochronic microRNA is conserved. The ab gene is a target of let-7, and its repression in muscle is essential for the timing of NMJ maturation during metamorphosis. Our results suggest that let-7 and miR-125 serve as conserved regulators of events necessary for the transition from juvenile to adult life stages.
Introduction
The heterochronic genes of C. elegans form a regulatory network that temporally regulates specific cell fates [1]. Mutations in heterochronic genes alter the timing of stage-specific processes, resulting in reiteration of larval patterns or execution of precocious adult fates. Two of the best-studied heterochronic genes in C. elegans are lin-4 and let-7, which serve as early and late developmental timers, respectively [2, 3]. Both let-7 and lin-4 are microRNAs, small noncoding RNAs that posttranscriptionally regulate gene expression by binding to semicomplementary sequences on mRNA targets and thus cause their destruction or prevent their translation into protein [4, 5]. In the worm, only a few targets of these microRNAs have been identified, and each target participates in the heterochronic gene network [1, 6].
In C. elegans, lin-4 is required to downregulate target genes in early larval stages, whereas let-7 acts in the last larval stage, just prior to the transition to adulthood, and loss of either microRNA alters the timing of decisions affecting cell fates [1]. Notably, the defects of heterochronic mutants are limited to specific tissues and specific developmental stages. Loss of let-7, for example, prevents the programmed cell-cycle exit of hypodermal blast cells at the L4-adult transition, causing them to reiterate an earlier program of cell division and preventing their differentiation at the appropriate time; however, cell division is unaffected in other tissues or at earlier stages of development [1, 3].
The sequence of the mature 21–22 nucleotide (nt) let-7 microRNA is 100% conserved among bilaterians [7]. Temporal regulation of let-7 is also conserved, and its expression is largely limited to developmental stages corresponding to the transitional period between the juvenile and adult in the nematode, fly, zebrafish, and mouse [7, 8]. Drosophila let-7 is coexpressed with the lin-4 homolog, miR-125, in numerous tissues beginning late in the third larval instar (L3) and peaking in pupae during metamorphosis [7, 9, 10]. Their coregulation arises from a common genomic locus on the left arm of the second chromosome that also includes another microRNA, miR-100 [11, 12]. All three microRNAs are expressed together in one primary transcript. Interestingly, paralogs of let-7 and miR-125 are also clustered in the mouse genome and are expressed with temporal overlap [8, 13], and in C. elegans, certain let-7 and lin-4 family members are expressed at the same time [14]. The overlapping expression of let-7 and miR-125 and their genomic clustering in many species suggests that these microRNAs share a subset of functions. Computational analyses of microRNAs have predicted hundreds of targets for let-7 and for miR-125, although none have been functionally validated within any organism other than C. elegans.
We sought to determine whether the heterochronic functions of let-7 and miR-125 are conserved in Drosophila and here analyze a mutation that eliminates both microRNAs. Loss of let-7 and miR-125 leads to widespread defects during metamorphosis. The most severe of these, impaired eclosion of newly formed adult flies, results in 70% lethality. We map this defect to a delay in maturation of abdominal neuromuscular junctions (NMJs) and establish as its cause the persistence of a let-7 target, Abrupt, in mutant muscle cells. In addition, we demonstrate that let-7 is critical for the appropriate timing of cell-cycle exit of developing wing cells; this role is remarkably similar to a let-7 function in C. elegans.
Results
Loss of let-7 and miR-125 Leads to Defects Specifically during Metamorphosis
In Drosophila, let-7 and the lin-4 homolog, miR-125, are coregulated along with miR-100 (Figure 1A). All three miRNAs are expressed as one primary, poly-adenylated RNA (Figure S1 available online) that is processed by the enzymes Dicer and Drosha to yield the functional miRNAs [9, 15]. Expression of the three miRNAs is temporally regulated, and the 21–22 nt RNAs are observed in larvae beginning at the L3-pupal transition and peaking in pupae during metamorphosis [7, 9].
Figure 1. Generation of a Drosophila let-7, miR-125 Mutant.
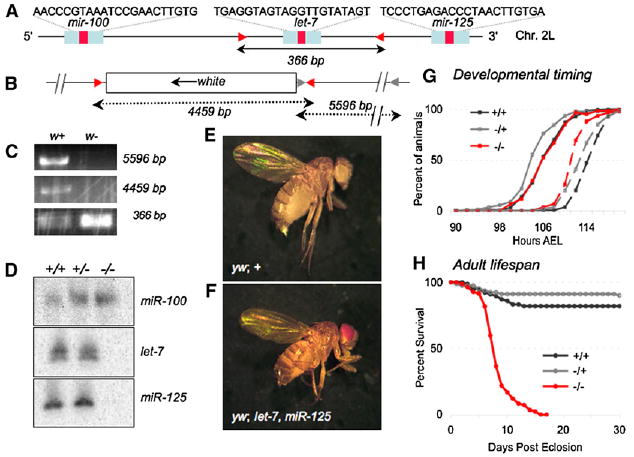
(A) The wild-type let-7, miR-125 locus. Red arrowheads indicate positions of primers (set 1, see Experimental Procedures) that amplify a 366 bp product in both wild-type and heterozygous mutants (C).
(B) The mutated let-7, miR-125 locus, primer set 1 (red arrow heads), now amplifies an additional 4459 bp product in heterozygous mutants; gray arrowheads indicate the positions of primer set 2, which amplifies a 5596 bp product in heterozygous mutants (C).
(C) PCR analysis (using the primer sets described in [A] and [B]) of sibling w+ (heterozygous mutant) and w− (non mutant) flies confirms the deletion of let-7 (loss of the 366 bp band; one copy is left in the heterozygous mutant) and insertion of the mini-w gene (gain of the 4459 bp and 5596 bp bands).
(D) Northern analysis of total pupal RNA from animals 42 hr APF. Probes for miR-100, let-7, and miR-125 confirm the absence of let-7 and miR-125.
(E and F) Wild-type female fly (E) and female let-7, miR-125 homozygous mutant fly (F).
(G) Developmental timing of larval wandering (solid lines) and pupariation (dashed lines) of let-7, miR-125 (−/−, n = 61) is unchanged relative to wild-type (+/+, n = 244) and heterozygous mutant (−/+, n = 92) larvae.
(H) Lifespan of let-7, miR-125 (−/−, n = 82) adult female flies is severely shortened relative to the wild-type (+/+, n = 77) and the heterozygous mutants (−/+, n = 79).
We initiated studies of Drosophila let-7 and miR-125 to determine whether their role in timing developmental processes was conserved. Using the Gal 4-UAS expression system [16], we found that extensive misexpression of let-7 or miR-125 at early stages arrested development at the L1 or L2 stage and led to lethality (Table S1), confirming the importance of temporal restriction of their expression to late in development. To examine the physiological role of these microRNAs, we generated a mutant by using homologous recombination [17]. We replaced a 36 bp region of the let-7 precursor hairpin with the white eye-color gene and verified the deletion by PCR (Figures 1A–1C). Northern blot analysis indicated that expression of both let-7 and miR-125 was lost as a result of the white insertion (Figure 1D).
Consistent with the timing of their expression, loss of let-7 and miR-125 did not alter embryonic or larval development. All mutant animals entered the pupal stage with normal timing and viability and developed as superficially normal pharate adults (Figures 1E–1G). Strikingly, however, the majority of mutant pharates were unable to eclose, suggesting underlying metamorphic defects (Tables 1 and 2; see below). Moreover, animals successful at eclosion had severely shortened life-spans; in longevity assays, 50% of homozygous mutant females died just 8 days after eclosion (Figure 1H). In addition, we observed many locomoter and behavioral defects in the mutants prior to their death (Table 1 and Figure S4). Close examination of eclosed mutant animals also revealed that their wings were significantly smaller than normal (Figure 2D).
Table 1.
Defects in let-7, miR-125 Mutants
| Defect | Description |
|---|---|
| Eclosion | Low eclosion frequency (Table 2) |
| NMJ size | Fewer, smaller boutons in dorsal abdominal (DAb) muscles (Figure 5) |
| Lifespana | Reduced longevity (Figure 1H) |
| Fertilitya | Sterile, morphologically normal gonads, no oviposition |
| Locomotion | No flight, uncoordinated movements, tremblingb |
| Behavior | Aberrant grooming, aborted courtship, male-male courtship |
| Wing and cell size | Small wings, small cells (Figure 2) |
Both males and females.
Some defects increase in severity with age.
Table 2.
Genetic Suppression of Eclosion Defect
| Genotypea | % eclosedb | nc |
|---|---|---|
| yw; + | 97 | 259 |
| yw; let-7 miR-125 | 31 | 185 |
| w; ab1/ab1D | 87 | 124 |
| w; ab1/abclu1 | 100 | 126 |
| w; let-7 miR-125 ab1D/+ | 60 | 133 |
| w; let-7 miR-125 ab1/ab1D | 53 | 227 |
| w; let-7 miR-125 ab1/abclu1 | 59 | 227 |
Includes males and females for each genotype.
Percentages are of expected genotype from each cross.
n = number of flies.
Figure 2. let-7 Is Required to Time the Terminal Cell-Cycle Exit in the Wing.
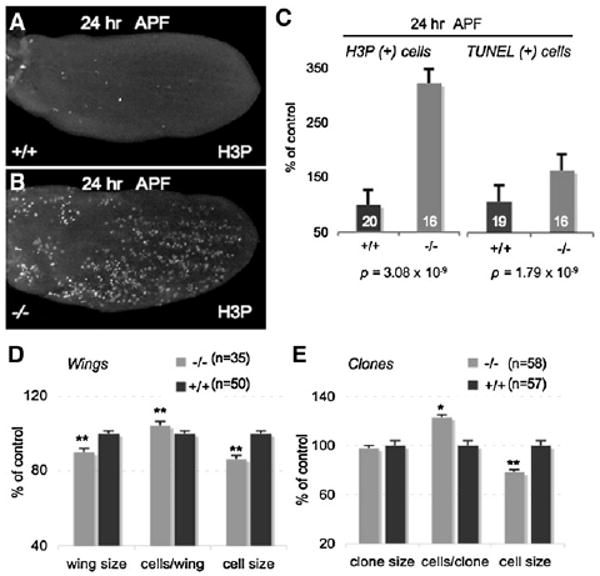
(A and B) Most cell division has ceased by 24 hr APF in wild-type wing discs (A), whereas many cells continue to divide in let-7, miR-125 mutant wing discs at 24 hr APF (B). Discs in (A) and (B) are oriented with anterior up and are stained for phospho-Histone 3 (H3P), a marker of mitosis. Wing discs are magnified 1000×.
(C) Mutant wing discs at 24 hr APF contain more mitotic or dying cells than wild-type controls. Mitotic cells, marked by H3P (left), and dying cells, marked by TUNEL assay (right), in mutant wings at 24 hr APF, expressed as a percentage of wild-type controls. Error bars represent standard error.
(D and E) Wings from eclosed let-7, miR-125 mutants (−/−) are significantly smaller and contain more, smaller cells than wild-type (+/+) wings (D). Clones mutant for let-7, miR-125 contain 25% more and smaller cells than wild-type clones (E). Number of wings or clones examined is indicated next to the graph legend. * p < 0.05; ** p < 0.001, compared to wild-type; error bars represent standard error.
let-7 Is Required for Terminal Exit from the Cell Cycle in Wing Imaginal Discs
In C. elegans, loss of let-7 prevents the programmed cell-cycle exit of the hypodermal blast cells at the L4-adult transition, causing them to reiterate an earlier program of cell division [3]. Expression of let-7 at earlier stages is sufficient to force precocious exit from the cell cycle and differentiation of these cells [3, 18]. The wing phenotype in our Drosophila mutant suggested a functional parallel with C. elegans let-7 in cell-division control, and we therefore examined wings from let-7, miR-125 mutants for proliferation defects. Prior to wing differentiation, cell division in the primordial wing imaginal disc ceases approximately 24 hr after puparium formation (APF), concomitant with the expression of the miRNAs (Figure 2A) [19, 20]. Although control wing discs had few mitotic cells at 24 hr APF, age-matched wing discs from let-7, miR-125 mutant pupae contained numerous dividing cells (Figures 2A–2C). This led to significantly more cells in adult mutant wings (Figure 2D). In mosaic wings, clones of let-7, miR-125 mutant cells contained 25% more cells than wild-type clones in control wings (Figure 2E). These data indicate that most let-7, miR-125 mutant cells continued to divide when wild-type cells had exited the cell cycle in preparation for differentiation. Strikingly, the mutant cells were significantly smaller than wild-type controls (Figures 2D and 2E), suggesting that the extra divisions were uncoupled from cellular growth. We also found that cell death was increased in pupal wings of mutant animals compared to controls (Figure 2C), and early events of wing metamorphosis such as the proximal-distal extension that separates the anterior and posterior crossveins also appeared to be delayed Figure S2A). These data suggest that the small wings of mutant animals resulted from a combination of small cell size and increased cell death during pupal development.
Given the precedent from C. elegans, the failure of the double-mutant cells to exit the cell cycle at the appropriate time suggested that loss of let-7 was responsible for this defect. We tested this idea by examining whether misexpression of let-7 was sufficient to induce cell-cycle withdrawal in larval-stage wing discs. Indeed, clonal expression of UAS-let-7 via a “flp-out” Gal 4 cassette forced cells in the wing disc to arrest after only 30 hr of growth (Figure 3A). Moreover, broad misexpression of let-7 in larval wing discs significantly reduced their size relative to controls (Figure 3B) and led to markedly undersized adult wings (Figure 3C). The effect of let-7 was specific; expression of UAS-miR-125 did not alter wing disc or adult wing size (Figures 3B and 3C). The correspondence between the misexpression and loss-of-function experiments suggests that loss of let-7 is responsible for the cell division defect in let-7, miR-125 mutant wings. Taken together, these data suggest that let-7 is required for enforcing the terminal cell-cycle arrest in pupal-stage wing cells and that Drosophila let-7 functions in a conserved heterochronic role as a stage-specific timer of cell-cycle exit.
Figure 3. let-7 Misexpression Causes Premature Cell-Cycle Exit in the Wing.
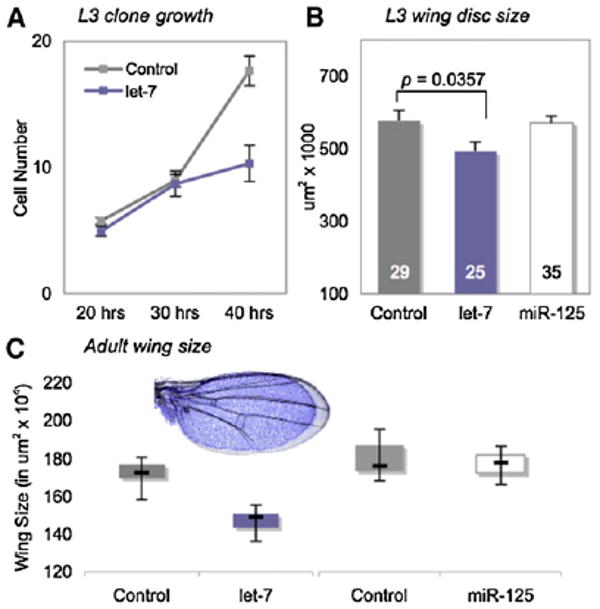
(A) Cell number in “flp-out” clones of cells misexpressing UAS-let-7 and UAS-GFP; control clones express UAS-GFP alone (see Experimental Procedures). Most let-7-expressing cells stop dividing after 30 hr of clonal growth. Controls: n = 52 at 20 hr, n = 41 at 30 hr, and n = 34 at 40 hr; UAS-let-7: n = 34 at 20 hr, n = 31 at 30 hr, and n = 31 at 40 hr. Error bars represent standard error.
(B and C) The wing-specific driver, Vestigial (Vg) Gal4, was used to misexpress UAS-GFP alone (control), or in combination with UAS-let-7 or UAS-miR-125 in larval wing discs. UAS-let-7 expression significantly reduced disc size, whereas UAS-miR-125 had no effect. Numbers in each bar represent discs measured. (C) Misexpression of UAS-let-7 under Vg Gal4 control significantly reduces adult wing size compared to controls that express UAS-GFP alone (p < 0.001; control: n = 50; UAS-let-7: n = 50). In contrast, misexpression of UAS-miR-125 does not alter wing size (p > 0.05; control: n = 20; UAS-miR-125: n = 20). Boxes represent second- and third-quartile values; whiskers extend to the maximum and minimum values. Inset shows the size difference of overlaid UAS-let-7-misexpressing (blue) and control (gray) wings.
Neuromuscular Junctions Mature Slowly in let-7, miR-125 Mutants
To determine whether other abnormalities in the let-7, miR-125 mutants were temporal in nature, we focused on the eclosion defect. We found that 100% of let-7, miR-125 mutant animals initiated eclosion behavior with the same timing as age-matched controls (wild-type, n = 16; mutant, n = 15), indicating that the circadian and endocrine triggers for eclosion were intact [21, 22]. However, although let-7, miR-125 mutants carried out the typical pattern of abdominal and thoracic contractions [22, 23], the contractions were largely unproductive. Up to 70% of let-7, miR-125 mutants were unable to generate the force required to emerge from the pupal case (Table 2), suggesting the flies suffered from severe muscle weakness.
Muscle weakness can result from reduced synaptic strength, and we therefore examined the NMJs of wild-type and mutant abdominal muscles at different times during pupal development. Midway through the pupal stage, at 54 hr APF, the extent of nascent NMJs on dorsal abdominal muscles of mutant and wild-type animals was similar (Figures 4A and 4B). However, although small boutons formed over the next 42 hr, mutant NMJs grew at a significantly slower rate than controls and were 50% smaller than NMJs from aged-matched controls at each time point (Figure 4C). The immaturity of mutant dorsal abdominal NMJs was such that at 96 hr APF their size approximated that of NMJs in wild-type animals that were 18 hr younger (Figure 4C). Staining with antibodies against the synaptic bouton marker, cysteine string protein (CSP) [24, 25], in animals just prior to eclosion revealed that the NMJs in abdominal muscles of mutant flies contained significantly fewer and visibly smaller boutons (Figures 5F–5H).
Figure 4. NMJ Maturation Is Delayed in let-7, miR-125 Mutants.
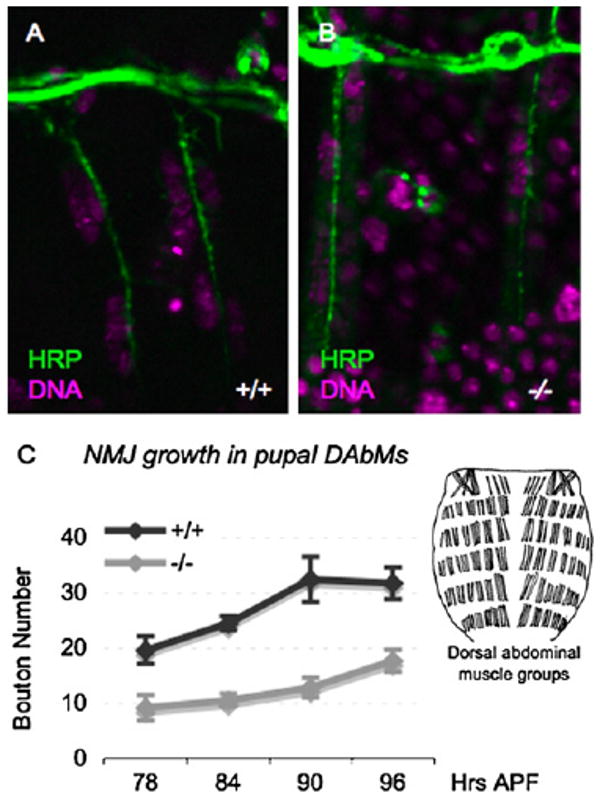
(A and B) NMJ growth begins with normal kinetics in let-7, miR-125 mutants. Nascent NMJs containing a similar extent of boutons are present in both wild-type (A) and let-7, miR-125 (B) dorsal abdominal muscles at 54 hr APF. Images are magnified 4000×.
(C) NMJ growth (measured by increase in bouton number during pupal development) is significantly delayed in let-7, miR-125 mutants (−/−) relative to wild-type (+/+). Ninety-six hour APF corresponds to the onset of eclosion. Error bars represent standard error. Inset: representation of dorsal abdominal muscles of the adult fly (modified with permission from [23]).
Figure 5. The Eclosion Defect and Delayed NMJ Maturation in let-7, miR-125 Mutants Are Due to Persistent Expression of Abrupt.
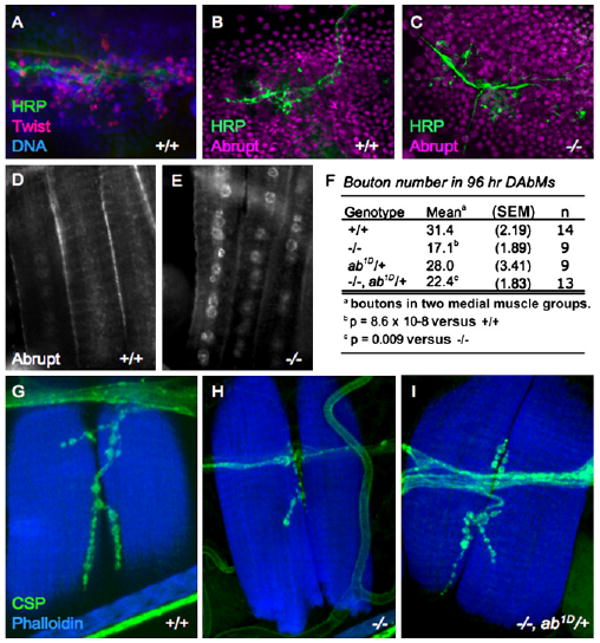
(A) Wild-type pupal epidermis at 24 hr APF, stained with Twist antibodies to show the myoblasts, which reside within the histoblast nests (marked in blue with Hoechst stain of DNA) and migrate along Horseradish peroxidase (HRP)-positive neurons (green). Images are magnified 4000×.
(B and C) Ab is expressed in myoblasts and histoblasts at 26 hr APF in both wild-type (B) and let-7, miR-125 mutants (C). HRP marks the growing neurons (green). Images are magnified 4000×.
(D and E) Ab expression is downregulated in the nuclei of freshly eclosed wild-type adult dorsal abdominal muscles (D), but nuclear Ab expression persists in dorsal muscles from let-7, miR-125 mutant adults (E). Images are magnified 2000×.
(F–I) NMJs of let-7, miR-125 mutants at 96 hr APF have significantly fewer boutons than do controls (F–H). Reducing the dose of ab with the strong ab1D allele (which alone has no effect on NMJs) significantly rescues bouton number in the mutants; SEM denotes standard error of the mean.
(G–I) Dorsal abdominal muscles (blue, Phalloidin) and synaptic boutons (green, CSP) from wild-type (G), let-7, miR-125 mutant (H), and let-7, miR-125, ab1D/+ double mutant (I) adult females. Images are magnified 4000×.
The abrupt Gene Is an In Vivo let-7 Target that Persists Aberrantly in let-7, miR-125 Mutants
miRNAs function by binding to and targeting specific mRNAs for translational repression or destruction [26], and both miRNAs are predicted by several databases to individually target hundreds of mRNAs [27, 28], although none of these have been verified in vivo. To get at the mechanism by which loss of let-7 and miR-125 could reduce the size and elaboration of abdominal NMJs, we searched for candidate targets of let-7 and miR-125 activity that might bring about the NMJ defect. The abrupt (ab) gene, which encodes a nuclear, BTB-POZ, Zn-finger containing protein, is a highly ranked candidate whose 3′ untranslated region (UTR) contains several predicted let-7 binding sites (Figure 6A) and is regulated by let-7 expression in cell-culture assays [29]. ab (also called clueless (clu [30]) is widely expressed during fly development and is required for diverse functions, including neuromuscular targeting and dendrite arborization in embryos [31–33], and the patterning of the fifth longitudinal vein (LV5) of the wing [34].
Figure 6. Abrupt Is a Bona Fide let-7 Target.
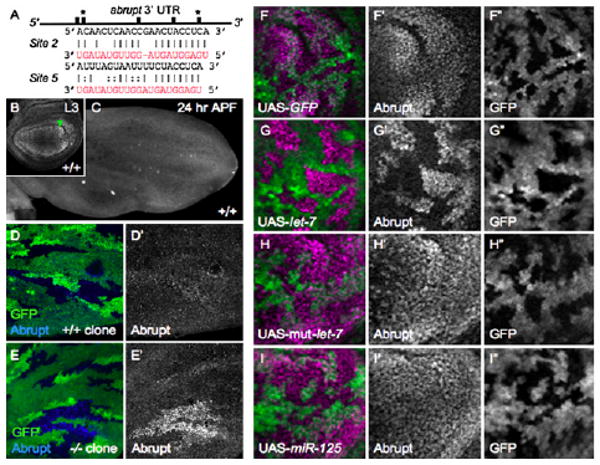
(A) Representation of the 3′ UTR of the abrupt mRNA, depicting the positions of five predicted let-7 binding sites as black bars. The sequences of two sites (*) are shown. The let-7 sequence is shown in red; lines indicate complementarity, and dots indicate G:U wobble basepairs.
(B and C) Ab is expressed broadly in mature wild-type (+/+) L3 wing discs, with highest expression in the region of LV5 ([B], arrowhead; n = 25); its expression is lost from the discs by 24 hr APF ([C], n = 10).
(D and E) Loss of let-7, miR-125 prevents downregulation of Ab in 24 hr APF wing discs. (D and D′) Control wing-disc clones generated by mitotic recombination, showing merged (D) and single-channel (D′) images. Clones are marked by the absence of GFP (lack of green). One hundred percent of control clones show downregulation of Ab (n = 10 discs), whereas in let-7, miR-125 mutant clones (E and E′), Ab expression (blue) persists, most strongly in the LV5 region (92%, n = 13 discs).
(F–I) Misexpression of let-7, but not miR-125, is sufficient to repress Ab expression. L3 wing discs with “flp-out” clones expressing UAS-GFP (green) alone (F–F″ n = 13), or in combination with UAS-let-7 (G–G″, n = 17), a mutated form of UAS-let-7 (H–H″, n = 9), or UAS-miR-125 (I–I″; n = 14), are stained for Ab (purple). Images are magnified 2000× in (B), (D), and (E) and 4000× in (F)–(I). Image in (C) is magnified 1000×.
Ab protein is expressed throughout the wing disc in wild-type late L3 animals (the highest levels are in cells that will form LV5) (Figure 6B) [34], but its expression is reduced to barely detectable levels early in the pupal stage (Figure 6C). Strikingly, however, Ab protein continued to be expressed specifically in let-7, miR-125 mutant cells, particularly in the LV5 region, in mosaic pupal-stage wing discs (Figures 6D–6E). We took advantage of the wing expression of Ab to test for microRNA specificity by using Gal 4-mediated misexpression. Expression of UAS-let-7 in L3 wing disc cells was sufficient to repress Ab expression prematurely (Figures 6F–6G). In contrast, Ab expression was not altered by expression of UAS-mut-let-7, a mutated form of let-7 (Figure 6H), or by expression of UAS-miR-125 (Figure 6I). Thus, the persistent expression of Ab in mutant pupal wing-disc cells was likely due to loss of let-7. The data indicate that let-7 is both necessary and sufficient for the appropriate temporal repression of Ab in wing discs and establishes ab as a bona fide, in vivo target of Drosophila let-7.
We then examined whether Ab was involved in the abdominal NMJ defects in let-7, miR-125 mutants. Ab is expressed in the nuclei of both wild-type and mutant adult myoblasts within the first 26 hr APF (Figures 5A–5C). However, Ab was rarely observed in freshly eclosed wild-type adults, indicating that its expression is lost from abdominal muscles as pupae mature (Figure 5D). In contrast, many nuclei from abdominal muscles of freshly eclosed let-7, miR-125 mutants contained high levels of Ab protein (Figure 5E). Taken with our observation that expression of UAS-let-7, but not UAS-miR-125, is sufficient to repress Ab expression (Figures 6G and 6I), these data suggest that let-7 functions during pupal development to repress Ab expression in muscle nuclei.
let-7-Mediated Repression of Abrupt Expression in Muscle Nuclei Is Required for NMJ Growth and for Eclosion
Retrograde signaling from muscle to motoneuron controls NMJ development in the Drosophila adult [35]. We reasoned that the persistence of Ab expression in mutant muscles could interfere with this signaling, cause the delay in NMJ maturation, and possibly, form the basis of the eclosion defect. Although Ab was present in the pupal brain of wild-type and let-7, miR-125 mutants (Figures S3C and S3D), we did not detect Ab in abdominal ganglia or motoneurons from either genotype (Figures S3A and S3B), supporting this idea. We attempted to phenocopy the eclosion defect by expressing a UAS-ab transgene in muscle or neuronal lineages with Gal 4 drivers specific to these cell types. Expression of ab in muscle cells was lethal at the L1 or L2 stage, as was expression in cholinergic neurons. In contrast, a pan-neuronal or a motoneuron driver allowed animals to eclose (Table S2).
As a more definitive test of whether persistent Ab expression in let-7, miR-125 mutant muscles reduced their ability to form mature NMJs, we carried out genetic suppression experiments by using mutant alleles of ab. Bouton numbers were significantly increased in abdominal NMJs of let-7, miR-125, ab1D/+ adults (Figures 5F–5I). Moreover, reduction of ab gene dosage significantly suppressed the eclosion defect. let-7, miR-125 mutants carrying one copy of the strong ab1D allele eclosed at a frequency nearly double that of let-7, miR-125 alone (Table 2). These data provide compelling evidence that the persistence of Ab in the abdominal muscles of let-7, miR-125 mutants underlies the NMJ growth defect and leads to muscle weakness that impairs adult eclosion.
We also tested for a causal role for Ab in other defects in the let-7, miR-125 mutants. Ab was expressed in cells of the larval wing disc and was downregulated early in the pupal stage (Figures 6B and 6C). Interestingly, in a wild-type backgound, one copy of the strong ab1D or abclu1 allele produced a dominant increase in wild-type wing size (Figure S4A and data not shown). Thus, although reducing the genetic dose of ab led to an increase in let-7, miR-125 mutant wing size, this probably does not reflect an epistatic genetic relationship. In support of this interpretation, cell size was not altered in let-7, miR-125, ab1D/+ wings (Figure S4A; or abclu1, data not shown). Reducing the dose of ab also did not suppress the defects in locomotion (Figure S4B), the sterility of either female or males (Figure S4C), or the reduced lifespan (Figure S4D) of let-7, miR-125 mutants. Therefore, these defects must be due to other let-7 targets, to loss of miR-125, or to contributions from a combination of the microRNAs.
Discussion
Our results indicate that in Drosophila, loss of the let-7 and miR-125 microRNAs leads to numerous defects that alter the morphology and behavior of adult flies. The expression of these microRNAs is restricted to the metamorphic stage of development, and we show that both loss and premature expression of let-7 and miR-125 are detrimental to animal survival, indicating that temporal restriction of their expression is essential. Many defects in the mutants affect sensory and motor behaviors, and our results indicate that neuromuscular connectivity in abdominal muscles is severely hampered and that there are direct consequences on the rate of adult eclosion. Both the NMJ and eclosion defects result from deregulated expression of a single let-7 target in abdominal muscles. Control of NMJ growth represents a new regulatory role for microRNAs in general, and given the extensive neurological problems of let-7, miR-125 mutants, it is possible that synapse development is broadly regulated by these particular micoRNAs.
let-7-Regulated Repression of abrupt Is Required for Timing NMJ Maturation During Metamorphosis
Our data clearly implicate regulation of ab by let-7 as a critical factor in the rate of NMJ development during Drosophila metamorphosis. The aberrant persistence of Ab in muscle nuclei in the mutant significantly slows NMJ development, perhaps by interfering with synapse-strengthening signals exchanged between muscle and motoneurons [35]. Because Ab expression is widespread during Drosophila development, the need for its downregulation late in development is in striking contrast to the requirement for Ab at earlier stages: in muscle for neuromuscular targeting and for epidermal attachment of specific muscle groups [31] and in a class of embryonic and larval sensory neurons to limit dendritic branching [32, 33]. Based on its dose sensitivity, Ab has been proposed to function in a concentration-dependent manner [31]. By examining the expression of Ab protein directly, we find that it is downregulated to nearly undetectable levels in most abdominal-muscle nuclei by the pharate adult stage (Figure 5D and data not shown). Its decline in the myoblasts may begin as early as 26 hr APF (Figure 5B), a time when let-7 expression is rising [10, 11]. However, our experiments indicate that deregulation of Ab cannot account for all of the mutant defects, indicating that other targets of let-7 and/or miR-125 must function as effectors for these processes (Figure S4). To date, Ab is the only target of Drosophila let-7 to be verified in vivo, and no miR-125 targets have been authenticated.
Temporal Control of Cell Division by Drosophila let-7
In addition to its role in NMJ maturation, our results implicate let-7 in a wing defect that causes cells to continue to divide after wild-type wing cells have exited the cell cycle. This phenotype is strikingly similar to the reiterated divisions of hypodermal blast cells in C. elegans let-7 mutants [3]. In both cases, the extra divisions do not continue indefinitely, nor do they occur in all tissues (data not shown) [1]. Little is known about the mechanism of cell-cycle regulation by let-7 in either organism. In C. elegans, persistent expression of lin-41, encoding a RBCC (ring B-box coiled-coil) protein, in let-7 mutants accounts for much of the mutant phenotype [36]. Drosophila homologs of lin-41 include dappled (shown to be a let-7 target in cell-culture assays [37]) and brat (brain tumor), a translational inhibitor [38]. brat encodes a potent tumor suppressor whose absence causes metastatic brain tumors [39] and that is predicted by one computational program to contain let-7 binding sites [13]. Interestingly, overexpression of brat suppresses wing growth [38]. Thus, a plausible hypothesis for the wing defect in let-7, miR-125 mutants is that loss of let-7 simultaneously deregulates a cell-cycle regulator(s) and brat and that the former drives additional cell divisions in the wing while the latter suppresses growth.
Practical and Evolutionary Implications of let-7 and miR-125 Function during Metamorphosis
The expression of many genes required during larval stages is downregulated during metamorphosis, in part because their presence during pupal development hinders the ordered progression of events such as the primary and secondary responses to ecdysone, the hormone that controls insect metamorphosis [40, 41]. Thus, a plausible role for let-7 and miR-125 is to rid cells of unnecessary larval mRNAs quickly at the transition to pupal development; such a role might be similar to the clearing of maternal mRNAs at the maternal–zygotic transition as recently demonstrated in zebrafish and in Drosophila [42, 43]. Heterochrony facilitates rapid evolution of new morphologies through changes in the timing of developmental processes [44, 45]. The pleiotropic phenotype associated with let-7, mir-125 mutants suggests that each microRNA contributes to multiple processes during metamorphosis, and the identification of additional targets of both is therefore an important future quest that should clarify their contribution to developmental timing and morphological evolution.
Conclusions
We have generated a mutant of Drosophila let-7 and miR-125 that exhibits temporal misregulation of specific metamorphic processes. Drosophila let-7 is both necessary and sufficient for the appropriate timing of a specific cell-cycle exit and thus functions as a conserved heterochronic microRNA. The abrupt gene is a target of let-7, and its repression in muscle is essential for the timing of NMJ maturation during metamorphosis. let-7 and miR-125 could serve as conserved regulators of processes necessary for the transition from juvenile to adult life stages.
Experimental Procedures
Fly Strains
yw;+;hsflp hsp70-I-SceI/TM6B, yw eyflp, w; EnGal 4, w; DppGal 4, w; TubGal 4, w; VgGal 4, yw; Act>y+>Gal 4, w; MEF2Gal 4, w; 24BGal 4, w;CHAGal 4, w; UAS-GFP, and w; UAS-P35 [46] were obtained from the Bloomington Stock Center. w;ab1, w;ab1D, and w;clu1 were gifts of S. Crews. C155Gal 4 and OK6Gal 4 were gifts of B. McCabe. yw hsflp; FRT40A, Ubi-GFP, yw hsflp; FRT40A let-7, and yw hsflp; FRT40A let-7 ck were generated in this work. Our control strain (designated +/+) was yw; +; +, which was used to isogenize all other strains.
Fly Husbandry
Eggs from appropriate crosses were collected on yeasted grape plates for 2 hr periods. Larvae were staged from egg deposition and after hatching were raised in uncrowded vials containing yeasted molasses and cornmeal food at 25°C for defined periods of time. Pupae were staged from white pre-pupa formation as in [47] and were raised on glass slides at 25°C for defined periods of time. Adult viability was measured daily from vials of ten flies each. Fertility was assessed by observation of progeny production from single-pair crosses with wild-type partners.
Homologous Recombination
Genomic fragments (5.1 kb) flanking let-7 were PCR amplified and cloned into an ends-out homologous-recombination vector kindly provided by S. Chen and G. Struhl. Standard procedures were used for injecting the plasmid into w1118 embryos and isolating transformants. Virgin female flies carrying the targeting construct on the X chromosome were crossed to yw hsflp hsp70-I-SceI-expressing males. Progeny were collected for 24 hr periods and heat shocked for 1 hr at 37°C each day for three consecutive days. Eclosing progeny were crossed to yw eyflp-expressing males, and female progeny that retained eye color were PCR screened for the insertion of the mini-white gene into the let-7 locus. Primer set 1: forward, CAAACCACCT AGCAAAAAGGA; reverse, TAGGGTCTCAGGGAATCAGC. Primer set 2: forward, TTCCGCAAAAATGGGTTTTA; reverse, AGGCTTCCCCTGCTATCATT.
Molecular Biology
The UAS-let-7 and UAS-miR-125 lines were generated by PCR amplification of 366 bp and 188 bp fragments, respectively, and cloning into pUAST [16]. The UAS-mut-let-7 line was generated by site-directed mutagenesis of the UAS-let-7 construct, with G-C transitions at nucleotides 4 and 5 of the mature let-7 miRNA and complementary mutations on the opposing side of the hairpin. Standard procedures were used for injecting the plasmids into w− embryos and isolating transformants. Northern analysis confirmed the production of processed let-7 and miR-125 RNAs from Gal 4 activation (data not shown).
Reverse-Transcription PCR
Poly (A) + RNA was generated from total RNA isolated from whole 42 hr APF pupae. Primers flanking all three miRNAs were then used for amplification of a 1400 bp product. Primer set 3: forward, AAAAGGGGTTGCTTTTCGAT; reverse, CCTTCCTTTGCAGTCAAAATG.
Northern Blotting
Total RNA was isolated from 42 hr APF pupae, separated on a 15% denaturing polyacrylamide gel, and transferred to Genescreen Plus membrane (Perkin Elmer). Membranes were probed with either Starfire (Integrated DNA technologies) or LNA (Exiqon) probes, radioactively labeled according to the manufacturers' instructions.
Immunocytochemistry
Fixation and immunocytochemistry of imaginal discs and epidermal preparations was carried out as described [48]. The following primary antibodies and dilutions were used: rabbit anti-Abrupt, 1:500 (gift of E. Bier) [34], mouse anti-abrupt 1:100 (DSHB), rabbit anti-H3P, 1:1000 (Upstate Biotechnology), mouse anti-CSP, 1:200 (DSHB); rabbit anti-Twist, 1:5000 (gift of M. Baylies). Secondary antibodies were as follows: goat anti-rabbit Alexa 488 (1:2000), donkey anti-mouse Cy-5 (1:600), and donkey anti-rabbit Cy3 (1:600) (Jackson ImmunoResearch). Alexa 647-Phalloidin (Molecular Probes) was used at 1:100 for labeling F-actin, and Hoechst 33258 (Sigma) was used for marking DNA. Cy3-HRP (1:200, Molecular Probes) was used for marking neurons. TUNEL assays (a specific protocol is available upon request) were carried out with Apoptag Red (Chemicon International). Images were captured on a Zeiss Axiophot equipped with Apotome or Bio-Rad 1000 or Leica confocal microscopes and processed in Adobe Photoshop or Image J.
Measurements of Wing and Cell Size
The Gal 4-UAS system [16] was used for misexpression of let-7 and miR-125. For measurement of wing and disc size, images were captured on a Zeiss Axiophot; size was measured in microns with Axiovision software (Zeiss). Cell size was calculated from a count of trichome number in two defined areas per wing and subsequent calculation of an average cell size (each trichome corresponds to one cell [49]). Cell number per wing was extrapolated from these measurements. For mitotic clones in adult wings, trichomes (marked with crinkled (ck) in both control and mutant wings) were counted as above. Clone size was determined from the inclusive area of ck mutant cells; conditions were optimized so that between 2 and 6 clones per wing were obtained. Cell size in clones was calculated from the number of cells per clone/clone size. All measurements were done with female wings.
Generation of Clones and Cell-Proliferation Measurements
For the cell-number assay, Gal 4 “flp-out” clones expressing UAS-GFP ± UAS-let-7 were generated by heat-shock-induced Flp recombinase at 60 hr AEL and dissected 20, 30, or 40 hr later [50]. Clone cell number was determined from a count of GFP-positive nuclei. For analysis of Ab expression, clones expressing UAS-GFP ± UAS-let-7, UAS-mut-let-7, or UAS-miR-125 were generated at 70 hr AEL and dissected at 100 hr AEL. For mutant clones, mitotic recombination was induced at 72 hr AEL; the ck mutation was used for marking control and let-7, miR-125 mutant clones in the adult wing.
Bouton Measurements
Abdomens of female flies were dissected down the ventral midline, pinned, fixed with 4% paraformaldehyde, and stained with anti-CSP, Cy3-HRP, and Alexa-647-conjugated Phalloidin. CSP-positive boutons were examined for size and counted in two medial muscle groups of one hemitergite of abdominal segment IV for each fly.
Locomotor Assay
Groups of ten 2-day-old adult flies were tapped to the bottom of an empty vial. The percentage of flies that successfully climbed back to the top of the vial in 15 s was recorded and averaged from ten consecutive trials.
Statistical Analysis
Two-tailed Student's t tests with unequal variance were used for significance assessments.
Supplementary Material
Acknowledgments
We are grateful to E. Yoshida for fly injections, R. Hartog and S. K. Brady for technical assistance, M. Schubiger and O. Hobert for advice, J. Parker for confocal assistance, and J. Truman, B. McCabe, W. Grueber, J.-P. Kruse, and members of the Johnston lab for advice and comments on the manuscript. This work was supported by grants from the National Institutes of Health and the Irma T. Hirschl Charitable Trust to L.A.J.
Footnotes
Supplemental Data: Four figures and two tables are available at http://www.current-biology.com/cgi/content/full/18/13/■■■/DC1/.
References
- 1.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Rougvie AE. Control of developmental timing in animals. Nat Rev Genet. 2001;2:690–701. doi: 10.1038/35088566. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 8.Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 10.Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- 11.Bashirullah A, Pasquinelli AE, Kiger AA, Perrimon N, Ruvkun G, Thummel CS. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev Biol. 2003;259:1–8. doi: 10.1016/s0012-1606(03)00063-0. [DOI] [PubMed] [Google Scholar]
- 12.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 16.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 17.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes GD, Ruvkun G. Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb Symp Quant Biol. 2006;71:21–27. doi: 10.1101/sqb.2006.71.018. [DOI] [PubMed] [Google Scholar]
- 19.Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- 20.Schubiger M, Palka J. Changing spatial patterns of DNA replication in developing wing of Drosophila. Dev Biol. 1987;123:145–153. doi: 10.1016/0012-1606(87)90436-2. [DOI] [PubMed] [Google Scholar]
- 21.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNabb SL, Baker JD, Agapite J, Steller H, Riddiford LM, Truman JW. Disruption of a behavioral sequence by targeted death of peptidergic neurons in Drosophila. Neuron. 1997;19:813–823. doi: 10.1016/s0896-6273(00)80963-0. [DOI] [PubMed] [Google Scholar]
- 23.Kimura KI, Truman JW. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci. 1990;10:403–411. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bronk P, Nie Z, Klose MK, Dawson-Scully K, Zhang J, Robertson RM, Atwood HL, Zinsmaier KE. The multiple functions of cysteine-string protein analyzed at Drosophila nerve terminals. J Neurosci. 2005;25:2204–2214. doi: 10.1523/JNEUROSCI.3610-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohan SA, Pescatori M, Brecha NC, Mastrogiacomo A, Umbach JA, Gundersen CB. Cysteine string protein immunoreactivity in the nervous system and adrenal gland of rat. J Neurosci. 1995;15:6230–6238. doi: 10.1523/JNEUROSCI.15-09-06230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 27.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgler C, Macdonald PM. Prediction and verification of microRNA targets by MovingTargets, a highly adaptable prediction method. BMC Genomics. 2005;6:88. doi: 10.1186/1471-2164-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vactor DV, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- 31.Hu S, Fambrough D, Atashi JR, Goodman CS, Crews ST. The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev. 1995;9:2936–2948. doi: 10.1101/gad.9.23.2936. [DOI] [PubMed] [Google Scholar]
- 32.Sugimura K, Satoh D, Estes P, Crews S, Uemura T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron. 2004;43:809–822. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 33.Li W, Wang F, Menut L, Gao FB. BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron. 2004;43:823–834. doi: 10.1016/j.neuron.2004.08.040. [DOI] [PubMed] [Google Scholar]
- 34.Cook O, Biehs B, Bier E. brinker and optomotor-blind act coordinately to initiate development of the L5 wing vein primordium in Drosophila. Development. 2004;131:2113–2124. doi: 10.1242/dev.01100. [DOI] [PubMed] [Google Scholar]
- 35.Marques G, Zhang B. Retrograde signaling that regulates synaptic development and function at the Drosophila neuromuscular junction. Int Rev Neurobiol. 2006;75:267–285. doi: 10.1016/S0074-7742(06)75012-7. [DOI] [PubMed] [Google Scholar]
- 36.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 37.O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
- 38.Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- 40.Fletcher JC, Thummel CS. The Drosophila E74 gene is required for the proper stage- and tissue-specific transcription of ecdysone-regulated genes at the onset of metamorphosis. Development. 1995;121:1411–1421. doi: 10.1242/dev.121.5.1411. [DOI] [PubMed] [Google Scholar]
- 41.Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- 42.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 43.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 44.Gould SJ. Ontogeny and Phylogeny. Cambridge: Harvard University Press; 1977. [Google Scholar]
- 45.Klingenberg CP. Heterochrony and allometry: the analysis of evolutionary change in ontogeny. Biol Rev Camb Philos Soc. 1998;73:79–123. doi: 10.1017/s000632319800512x. [DOI] [PubMed] [Google Scholar]
- 46.Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 47.Bainbridge SP, Bownes M. Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- 48.Johnston LA, Edgar BA. Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature. 1998;394:82–84. doi: 10.1038/27925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobzhansky T. The influence of the quantity and quality of chromosomal material on the size of the cells in Drosophila melanogaster. Arch Entwicklungsmech Organismen. 1929;115:363–379. doi: 10.1007/BF02078996. [DOI] [PubMed] [Google Scholar]
- 50.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


