Abstract
The food-borne pathogen Campylobacter jejuni is one of the leading causes of bacterial gastroenteritis worldwide and the most frequent antecedent in neuropathies such as the Guillain-Barré and Miller Fisher syndromes. C. jejuni was demonstrated to possess an N-linked protein glycosylation pathway that adds a conserved heptasaccharide to >40 periplasmic and membrane proteins. Recently, we showed that C. jejuni also produces free heptasaccharides derived from the N-glycan pathway reminiscent of the free oligosaccharides (fOS) produced by eukaryotes. Herein, we demonstrate that C. jejuni fOS are produced in response to changes in the osmolarity of the environment and bacterial growth phase. We provide evidence showing the conserved WWDYG motif of the oligosaccharyltransferase, PglB, is necessary for fOS release into the periplasm. This report demonstrates that fOS from an N-glycosylation pathway in bacteria are potentially equivalent to osmoregulated periplasmic glucans in other Gram-negative organisms.
Keywords: Campylobacter jejuni, N-linked protein glycosylation, periplasmic glucans, osmolarity
Free sugars are widely distributed in nature and have been found in all three domains of life. Periplasmic glucans are a common feature in Gram-negative Proteobacterial species (1) and they can represent up to 10% of the cellular dry weight when their synthesis is maximum in response to low osmotic environments (2). While these glycans are comprised exclusively of glucose, they can be structurally diverse, and four distinct families were defined based on variations in chain length, glycosyl linkage, branching pattern, and presence of modifications (1). In addition, periplasmic glucans were reported to be important for biofilm development (3), antibiotic resistance (4), host-pathogen and plant-microbe interactions (5–9), as well as intracellular multiplication (10). Free oligosaccharides (fOS) have also been described in mammalian cells, yeast, plants, and fish (11). In these cases, fOS are released from N-linked glycoproteins by peptide-N-glycanase (PNGase) within the endoplasmic reticulum associated protein degradation pathway (12) or from dolichol-pyrophosphate-linked oligosaccharides (Dol-PP-OS) by a pyrophosphatase that generates phosphorylated free oligosaccharides (fOS-P) (13, 14). Other studies have suggested that in the absence of a sufficient acceptor, OS are transferred from their Dol-PP carrier to water within the ER (15, 16). Although it is well established that several eukaryotic species produce large quantities of diverse fOS, their biological roles and physiological relevance are unknown.
The food-borne pathogen Campylobacter jejuni is one of the leading causes of bacterial gastroenteritis worldwide (17) and most frequently associated with Guillain-Barré syndrome, the most common cause of paralysis since the eradication of polio (18). C. jejuni is a commensal in the avian gut and is typically associated with poultry meat and products, but can also contaminate other food and water systems. The mechanisms that allow Campylobacter to adapt and survive in different environments are poorly understood. Recently, we identified fOS in C. jejuni 11168 and provided preliminary data showing that these sugars were the same as the heptasaccharide attached to proteins modified by the N-linked protein glycosylation (pgl) pathway (19). In this pathway, the central oligosaccharyltransferase, PglB, transfers the assembled heptasaccharides consisting of GalNAc-GalNAc-(Glc)-GalNAc-GalNAc-GalNAc-diNAcBac (where diNAcBac is 2,4-di-acetamido-2,4,6-trideoxy-D-Glc) (20, 21) from an undecaprenyl-pyrophosphate (Und-PP) carrier to specific asparagine residues within acceptor proteins (20, 22) (Fig. S1). In vitro and in vivo studies have shown that pgl mutants display reduced ability to invade intestinal epithelial cells (23), reduced levels of colonization in animals (23–26), and reduced DNA uptake (27), establishing a critical role for the N-glycosylation pathway in the pathogenicity of C. jejuni.
We investigated the biological role, structure, biosynthesis, and regulation of the recently identified fOS in C. jejuni. We show that fOS production changes in the presence of salts and sucrose and with bacterial growth phase. In addition, C. jejuni growth is sensitive to changes in sodium chloride concentrations, although a direct link between these two phenotypes still needs to be established. Release of fOS into the periplasm is directly dependent on PglB and is regulated at the level of protein activity; moreover the WWDYG motif that is highly conserved among oligosaccharyltransferase orthologs (21) is critical for this function. Thus, C. jejuni heptasaccharides play a dual role in the periplasm being involved in the posttranslational modification of proteins as well as existing in the free form (i.e., fOS) as a function of osmolarity.
Results
Periplasmic fOS Is Similar to the N-Linked Heptasaccharide.
In a previous study we demonstrated that fOS derived from the N-glycosylation pathway were present in whole cell lysates of wild-type C. jejuni 11168, but were absent in the isogenic oligosaccharyltransferase mutant, pglB (19). To determine the cellular distribution of fOS, semiquantitative mass spectrometry (sqMS) was used to examine the amount of fOS in various cell fractions. Over 90% of the total cellular fOS was found in the periplasm, ≈10% in the cytoplasmic fraction, and no fOS was detected in membrane preparations (Fig. S2). This suggests that production of fOS is similar to the process of N-glycosylation and is restricted to the periplasm. We attribute the low levels of fOS in the cytoplasmic fraction were due to the cell fractionation method as complete fractionation of C. Jejuni is difficult to achieve (28).
fOS and N-Glycans in Select pgl Mutants.
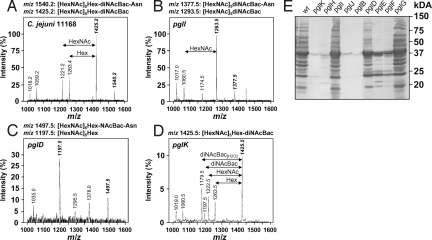
To investigate fOS biosynthesis in more detail, sqMS was used to determine fOS and N-linked glycan levels within whole cell lysates of select pgl pathway mutants (Fig. 1 and Table S1). Ions at m/z 1425.0 and m/z of 1540.2 were previously shown to correspond to full-length fOS and full-length N-linked glycan and are thus unique markers that can be used to quickly quantify the levels of these structures. Contrary to the wild-type strain, which was shown to have a fOS:N-glycan ratio of 9:1 (19) (Fig. 1A), fOS and N-glycans were not detected in pglB, in the GalNAc transferase mutants pglH and pglJ and in the pglE (aminotransferase) and pglF (dehydratase) mutants deficient in diNAcBac biosynthesis (Table S1). In contrast, wild-type levels of fOS and N-glycan were present in pglG (previously shown not to affect N-glycosylation) (26) and in pglI (Glc transferase, lacking Hex with the mass of 162 Da) (Fig. 1B). The pglK (flippase) mutant contained a 10-fold lower fOS amount and no detectable N-linked heptasaccharide (Fig. 1D). The pglD (acetyltransferase for diNAcBac synthesis) mutant lacked full-length fOS (Fig. 1C), however, MS2 analysis identified the predominant ions at m/z 1197.5 as [HexNAc]5Hex and m/z 1497.5 as [HexNAc]5Hex-NAcBac-Asn (Fig. S3) that were present at levels of 15–25% and 45–55% relative to the wild-type, respectively. These findings, corroborated by our NMR results establish that N-linked glycan biosynthesis genes are implicated in fOS biosynthesis concluding that fOS is derived from the N-glycan pathway (26).
Fig. 1.
Extracted MS spectra of underivatized C. jejuni glycans. (A) C. jejuni 11168 (wild-type), (B) pglI, (C) pglD, and (D) pglK mutants. Precursor ions (m/z) corresponding to fOS and N-linked glycans are in bold and italics. Intensities scales for pglD and pglK were shown as an expansion of the wild-type scale. (E) C. jejuni 11168 whole-cell lysate (50 μg/lane) reactivity of the wild-type (WT) and various pgl mutant strains after immunodetection with N-glycan-specific hR6 antiserum. The masses (in kilodaltons) of standard proteins are indicated on the right.
To further support the results obtained by sqMS, we investigated whole-cell lysates with an N-glycan-specific (hR6) antiserum that exclusively recognizes N-glycosylated C. jejuni proteins (Fig. 1E). Immunodetection confirmed the absence of N-glycoproteins in the pglB, pglH, pglJ, and pglK mutants, while comparable N-glycoprotein profiles were present in the wild-type, pglG and pglI mutants. For the pglD mutant, the reduced N-glycoprotein reactive profile after signal quantification at 50–60% wild-type level is comparable to our sqMS data and to what we previously described (26). Also, low amounts of N-linked glycoproteins were detected in pglE and pglF most probably due to crosstalk between the diNAcBac and the pseudaminic-acid (Pse) biosynthetic pathway needed to glycosylate flagellin (26, 29). This indicates that hR6-based immunodetection not only allows quantification and visual N-glycoprotein profiling but also is a more sensitive method for the analyses of N-linked protein glycosylation compared to our mass spectrometry approach. Therefore, we used immunodetection in the subsequent experiments to detect and compare protein N-glycosylation patterns.
fOS Levels Decrease in High Osmolarity Media.
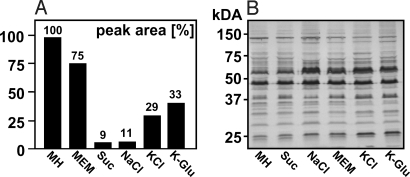
SqMS analyses revealed substantially lower fOS levels in wild-type cells grown on Mueller Hinton (MH) medium supplemented with 300 mM sucrose, 100 mM NaCl, 100 mM KCl, or 100 mM K-glutamate compared to cells grown on MH or MEM (Fig. 2A). In contrast, cells grown on MH at acidic (pH 4.0–5.0) or basic (pH 9.0–10.0) pH, in the presence of excess iron (Fe, 50 mM) or exposure to variable atmospheric oxygen levels did not affect the levels of fOS (Fig. S4A) or the N-glycosylation profile (Fig. S4B). These findings show that the osmolarity of the media is an important factor that influences the amount of fOS produced in C. jejuni.
Fig. 2.
fOS and N-linked glycoprotein production under different growth conditions. (A) Relative fOS amounts were determined by sqMS from wild-type cells grown on MH, and on MH supplemented with 300 mM sucrose (Suc), 100 mM NaCl, KCl, K-glutamate (K-Glu), or on MEM, and normalized relative to the level detected in MH-grown wild-type cells. (B) N-linked glycoprotein profiles of wild-type whole cell lysates (25 μg/lane) were immunodetected with hR6 antiserum. The masses (in kilodaltons) of standard proteins are indicated on the left. Values represent the mean of at least two independent experiments.
In contrast, the N-glycoprotein profiles in the corresponding whole cell lysates, investigated using N-glycan-specific (hR6) antiserum, showed no differences compared to MH-grown cells indicating that none of the conditions examined affected the N-glycosylation process (Fig. 2B and Fig. S4B).
PglB-Dependent fOS Release Is Sensitive to Changes in Sodium Chloride Concentrations.
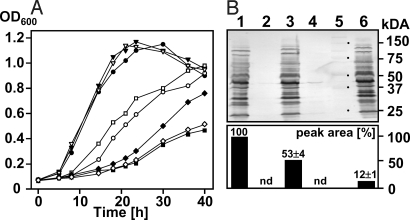
Growth rates among wild-type, pglB, pglD, pglE, and pglF cells were similar on MH supplemented with sucrose, KCl, and K-glutamate (Fig. S5), but were markedly different on MH supplemented with NaCl (Fig. 3A). While growth rates for wild-type and pglD cells were reduced but similar in MH supplemented with 100 mM NaCl, the pglB mutant showed almost no growth under these conditions indicating the requirement of a functional oligosaccharyltransferase for growth at high NaCl concentrations. Growth of pglE and pglF in MH supplemented with 100 mM NaCl was also reduced compared to wild-type and pglD, but less affected compared to the pglB mutant most likely due to the partial complementation of these enzymes by the Pse pathway (Fig. S6D). This indicates that minor amounts of fOS, nondetectable by mass spectrometry, might still be present in these strains, or that N-glycosylation somehow affects C. jejuni growth at high salt concentrations. The high salt pglB growth phenotype could be partially restored by complementing pglB in trans (pCj-pglB) but could not be restored by expressing a pglB allele with point mutations in the conserved WWDYG motif (457WWDYG461 to 457WAAYG461; pCj-pglBmut) (Fig. 3A). In MH, grown pglB(pCj-pglB) fOS amounts reached 50 ± 6% of the wild-type and were found to be decreased to 12 ± 1% when pglB(pCj-pglB) was grown on MH with 100 mM NaCl (Fig. 3B Lower), whereas the N-glycoprotein profile was completely restored, independent of the salt concentration (Fig. 3B Upper). This clearly demonstrates that the conserved amino acid sequence previously shown to be indispensable for PglB-dependent protein N-glycosylation (21) is also necessary for fOS release. It is worth mentioning that growth differences among wild-type and pglB mutant were less distinct at lower NaCl concentrations (Fig. S6 A–C) with similar growth rates below 25 mM further indicating that the N-glycan pathway is needed to cope with high salt concentrations.
Fig. 3.
The 457WWDYG461 motif of PglB is necessary for fOS release. (A) Growth of the wild-type (WT), the pglB mutant, and the mutant expressing either a native (Cj-pglB) or a mutated pglB allele [Cj-pglBmut(458WD459 to 458AA459)] in trans on MH and MH supplemented with 100 mM NaCl. Wild-type MH (●); wild-type MH NaCl (○); pglB MH(▾); pglB MH NaCl (■); pglD MH(▿); pglD MH NaCl (□); pglB (pCj-pglB) MH NaCl (▴); pglB (pCj-pglBmut) MH NaCl (◇). (B) N-linked glycoprotein profiles in select whole-cell lysates (25 μg/lane) of cells (as indicated below) were immunodetected with hR6 antiserum (Upper), standard proteins were marked in lane 5, the corresponding masses in kilodaltons are indicated on the right; lanes: 1, wild-type (MH); 2, pglB (MH); 3, pglB (pCj-pglB) (MH); 4, pglB (pCj-pglBmut) (MH); 6, pglB (pCj-pglB) (MH, 100 mM NaCl); corresponding fOS amounts (Lower) were determined by sqMS. Relative peak areas were normalized relative to levels detected in MH-grown wild-type cells. Values represent the mean of three independent experiments; nd, below detection limit.
fOS Production Peaks During the Logarithmic Growth Phase.
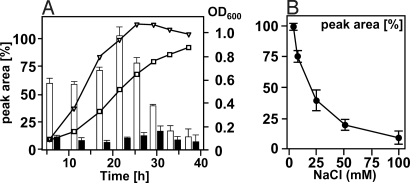
To investigate if the observed difference in fOS levels resulted from slower growth rates in high salt medium, we determined fOS production during growth of the wild-type using sqMS. fOS concentrations were found to be highest in the late exponential stage (OD600 = 0.8–0.9) in MH-grown cells and dropped when cells entered stationary phase (Fig. 4A). In MH supplemented with 100 mM NaCl, fOS levels were substantially lower, but also showed a slight increase at an OD600 of 0.8 ± 0.05. The corresponding whole cell lysates, probed with N-glycan-specific hR6 antiserum (Fig. S7), showed no significant changes in the N-glycoprotein profile. To investigate the correlation between fOS release and the salt concentration in the medium, wild-type cells were grown in MH with increasing amounts of NaCl. sqMS-based quantification of fOS in cells grown to an identical OD600 of 0.8 clearly indicated that the amount of fOS is inversely proportional to the salt concentration in the medium (Fig. 4B).
Fig. 4.
Effects of NaCl and growth phase on fOS production. (A) Relative fOS amounts (bars) were determined by sqMS and plotted against the growth curve of C. jejuni wild-type in MH (fOS, open bars; OD600, open triangles) and MH 100 mM NaCl (fOS, filled bars; OD600, open squares). (B) Relative amounts of fOS were determined in whole cell lysates of wild-type cells grown to an OD600 of 0.80 ± 0.05 on MH or MH supplemented with increasing NaCl concentrations (in mM). Values represent the mean of three independent experiments.
fOS production in response to changes in salt concentration was further investigated (Fig. S8). fOS levels slowly but markedly decreased when logarithmic phase wild-type cells (OD600 of 0.83) were transferred to MH containing 100 mM NaCl, while fOS amounts slowly increased when cells were transferred from high to low salt medium. In addition, no detectable fOS was present in the culture supernatants obtained from cells grown for 36 h in low salt medium or from cells after the transition from low to high salt medium (Fig. S8). Only the full-length heptasaccharide was detected in these cells, indicating that fOS is diluted out, while cells are dividing rather than being degraded or secreted into the medium.
fOS Release Is Regulated Through PglB Activity Rather than pglB Transcription.
To investigate the underlying regulatory mechanism of fOS release, pglB transcription was analyzed throughout the bacterial growth cycle. PglB-specific RT-PCR products were highest at the end of the logarithmic growth phase, regardless of the salt concentration (Fig. S9A), indicating that pglB transcription itself is primarily dependent on the growth phase. Growth phase dependent pglB transcription was lost in pglB(pCj-pglB) (Fig. S9C), probably due to the sigma28 controlled flaA promoter that drives gene expression on plasmid pCj-pglB. The difference in the expression profile when compared to the wild-type might explain the lack of complete plasmid-derived PglB complementation in Fig. 3B. Equal amounts of the 16sRNA-specific control PCR product were observed for all preparations (Fig. S9 B and D).
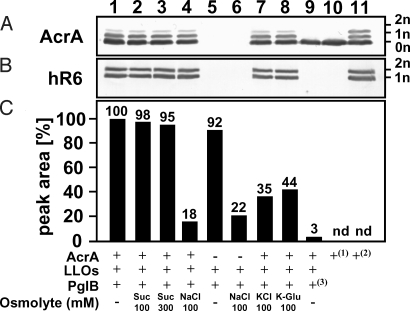
In vitro PglB activity assays were carried out to investigate if PglB-dependent fOS release is regulated at the protein activity level (Fig. 5). fOS release from lipid-linked oligosaccharides (LLOs), followed by sqMS, showed that PglB-dependent fOS release was 5.9 times lower in the presence of NaCl (Fig. 5, lane 4), 2.7 times lower in the presence of KCl (Fig. 5, lane 6), and 2.3 times lower in the presence of K-glutamate (Fig. 5, lane 7), but was not affected in the presence of sucrose (Fig. 5, lanes 2 and 3) when compared to standard low ionic-strength assay conditions that were used to normalize the results (Fig. 5, lane 1). This ionic strength-dependent decrease in fOS formation was independent of the presence of the N-glycan acceptor protein AcrA (Fig. 5, lanes 5 and 6). Low amounts of fOS were detected with Escherichia coli membranes containing PglBmut (457WAAYG461) (Fig. 5, lane 9) and with membranes lacking PglB, whereas fOS was not detectable in the absence of LLOs indicating the presence of minor amounts of fOS in the LLO preparation.
Fig. 5.
In vitro PglB-dependent fOS release decreases under high salt conditions. Western blots from 10% SDS/PAGE show immunoreactive signals obtained with (A) AcrA-specific antiserum that correspond to none (0n), mono (1n), and double (2n) glycosylated AcrA-His6 proteins, and (B) with hR6 antiserum that correspond to mono (1n) and double (2n) glycosylated AcrA-His6 proteins. Purified AcrA-His6 from E. coli BL21(pglmut)1 or E. coli BL21(pgl)2 were used as references for nonglycosylated and glycosylated AcrA-His6. (C) In vitro fOS release followed by sqMS. fOS amounts (represented by bars), obtained in the presence of the indicated osmolyte (concentration in mM) or using membranes of E. coli expressing PglBmut3, were depicted as a fraction of the amount obtained in the absence of osmolytes that was set to 100% (nd, below detection limit). Note that lanes in the immunoblots (A and B) correspond to the in vitro conditions described below (C).
In parallel, the protein N-glycosylation activity of PglB was evaluated by detecting the N-glycosylation state of AcrA with hR6 antiserum. PglB-dependent AcrA N-glycosylation activity resulted in comparable signal intensities for mono and diglycosylated AcrA that were independent on the ionic strength (Fig. 5, lanes 1–4, 7, and 8). As expected, only nonglycosylated AcrA was found in the presence of E. coli membranes containing PglBmut or by omitting LLOs from the reaction mixture. Based on these results, we conclude that the mechanism of fOS release is solely regulated by altering PglB activity in response to high salt concentrations, whereas PglB-dependent protein N-glycosylation is not affected.
Discussion
We characterized periplasmic fOS in C. jejuni 11168 derived from the N-glycosylation pathway. Wild-type fOS was structurally identical to the N-linked heptasaccharide that is added to over 40 periplasmic and membrane proteins (20, 22). In vivo, fOS production was inversely proportional to solute concentration in the growth media with decreased levels of fOS produced in the presence of high salt and sucrose concentrations. Thus, the underlying regulatory mechanism of C. jejuni fOS production is similar to osmoregulated periplasmic glucans (OPGs) in other Proteobacteria (1, 5). Furthermore, we demonstrated that C. jejuni fOS release into the periplasmic space requires the PglB OTase and that the conserved sequence motif 457WWDYG461, shown to be necessary for PglB-dependent protein N-glycosylation (21), is also required for fOS formation. The large drop in in vitro PglB-fOS release activity in response to ionic solutes suggests a comparable protein activity inhibition mechanism described for the cyclic β-1,2-glucan glucosytransferase of Agrobacterium tumefaciens and Rhizobium meliloti (30, 31). Also, consistent with our findings, in vitro assays with the R. meliloti glucosyltransferase showed inhibition of enzymatic activity in the presence of ionic solutes (KCl and NaCl) under the same salt concentrations, but very little change in activity when the neutral solute, sucrose, was used in the 0–200 mM range (30). Similarly, the addition of ionic solutes to the C. jejuni in vitro PglB assay demonstrated the same trends as the in vivo assay (i.e., inhibition of fOS production), but addition of sucrose in vitro showed no change. Our data suggest that NaCl has a direct affect on PglB activity at the protein level while sucrose may exert its effects indirectly on the whole cell.
It is also worth pointing out that although the C. jejuni lipid-linked heptasaccharide only represents a minor fraction of the purified LLOs used in the in vitro assay (32), only fOS containing the full-length heptasaccharide were detected in vitro. This indicates that PglB selectively releases the full length wild-type glycan which is in contrast to its relaxed N-glycosylation substrate specificity shown in the heterologous E. coli system (33, 34). This in vitro PglB activity might also explain the low levels of fOS in the pglK mutant, potentially released from the lipid during the cell extract preparation process.
Although we cannot draw a conclusion about the exact release mechanism, the linear nature of C. jejuni fOS most likely excludes the mechanism known from cyclic glucan synthesis in other Proteobacteria that was suggested to be coupled to cyclization (10, 35). The absence of a eukaryotic PNGaseF-like protein that would release fOS in C. jejuni, and the absence of a gene encoding a pyrophosphatase homolog protein (36) favors a mechanism proposed for the eukaryotic OTase that, in the absence of sufficient acceptor sequences, the OTase exhibits hydrolytic activity and transfers OSs from their lipid-linked intermediates to water (15, 16).
Questions about additional functions for the C. jejuni fOS are evident: Mutation in the biosynthetic periplasmic glucan pathways in other organisms are reported to result in defects in microbe-host attachment (8), impede normal intracellular multiplication (in Brucella sp.) (37, 38), reduce flagella assembly, and increase sensitivity to antibiotics and detergents (in Rhizobium and Agrobacterium sp and P. aeruginosa) (4, 39, 40), and to have pleiotropic effects on polysaccharide biosynthesis, protein folding and degradation, and carbohydrate catabolism (in Erwinia chrysanthemi) (41). Loss of periplasmic glucan production in R. meliloti and Bradyrhizobium japonicum affected growth in various hypoosmotic environments (42, 43), whereas the opposite effect, decreased growth rates in a hyperosmotic high sodium chloride environment was observed for the C. jejuni pglB, pglE, and pglF mutants where no detectable fOS was present. In contrast, loss of periplasmic glucan production in Brucella spp., Azorhizobium and some S. meliloti strains has no osmo-dependent growth phenotype (10, 44), and it was suggested that osmoprotection is not likely to be the primary function of their OPGs (1).
In C. jejuni, pgl pathway mutant phenotypes that resulted in impaired adherence, invasion, and colonization were always thought to be a result of impaired protein N-glycosylation leading to loss of protein complex formation and/or protein instability e.g., VirB10 of the type IV secretion system (T4SS) in C. jejuni 81–176 (27). Interestingly, reduced stability of the T4SS complex in A. tumefaciens (39) was attributed to loss of periplasmic cyclic β-1,2-glucan production, thus raising the question whether a similar mechanism exists in C. jejuni. If fOS would be directly involved in the infection/colonization process, this would imply fOS to be released in the environment; however, we could not detect fOS in the culture supernatant in our experimental setup. Although periplasmic glucans from E. chrysanthemi and A. tumefaciens were found in certain infected plant tissues, external OPGs could not complement an opg mutant phenotype (9, 45). Therefore, it was suggested that OPGs need to be inside the periplasmic space for expression of virulence (37), which would resemble the C. jejuni-fOS situation. Another hint that favors a role of fOS in pathogenicity is provided by the fact that the C. jejuni pglD mutant N-glycosylates its proteins at ≈50% wild-type levels but exhibits the same loss of chicken colonization as the pglB, pglE, and pglF mutants (26), which completely lack fOS. Therefore, free glycan levels and/or the diNAcBac residue at the reducing end that is absent in the aberrant pglD-fOS, might play critical roles and require further characterization. On the other hand, mutations in STT3 orthologous proteins as well as defects in maturation of N-glycosylated proteins in Arabidopsis thaliana resulted in reduced salt tolerance and reduced growth when compared to low salt conditions (46, 47). However, no information is available on potential changes in fOS levels under different osmolarities in this species although A. thaliana was reported to possess a cytoplasmic PNGase protein for fOS release (11). Since inactivation of C. jejuni fOS production accompanies loss of N-glycosylation, it remains to be determined whether the growth defect in high salt is a direct consequence of the absence of fOS or due to loss of N-glycosylation of certain proteins/protein complexes or both. We are currently attempting to separate the C. jejuni fOS and N-linked heptasaccharide activities of PglB to determine if the reported pgl mutant phenotypes can be directly related back to one of these two forms of the C. jejuni heptasaccharide. Because OPGs are unable to diffuse across the outer or cytoplasmic membranes, they increase the solute concentration in the periplasm during low osmotic shifts. The increase in OPG concentration helps to increase the cell volume of the periplasm and reduce macromolecular crowding (48). Shifts in C. jejuni fOS concentrations are anticipated to also play this general role and may also benefit the N-glycan pathway by providing the appropriate cellular density for the protein glycosylation machinery.
The discovery of fOS adds another player to the complex glycome of C. jejuni. It will be interesting to determine whether all Proteobacteria with orthologous N-glycan pathways also produce fOS and whether this unique mechanism to cope with salt stress has provided evolutionary pressure to maintain the N-glycosylation pathway in these organisms. Also, comparisons are warranted between the N-glycosylation pathway and OPG biosynthesis in some Proteobacteria that use a remarkable mechanism of cyclic glucan production in which one polyfunctional protein of ≈320 kDa is responsible for covalent attachment of nucleotide-activated Glc onto itself, elongation, polymerization control, flipping, cyclization, and release (49).
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacterial strains, plasmids and oligonucleotides are listed in Table S2 and Table S3. C. jejuni was routinely grown using MH medium (Difco) under microaerobic conditions (10% CO2, 5% O2, 85% N2) at 37 °C or as indicated. Growth curves were recorded as described (23). Change of growth media was performed with cells washed twice in 50 mL MH medium before inoculation. E. coli was grown on 2×YT agar (50) at 37 °C. If needed, antibiotics were added to the following final concentrations: 30 μg/mL kanamycin, 150 μg/mL ampicillin, 15 μg/mL tetracycline, 30 μg/mL chloramphenicol, and 20 μg/mL trimethoprim.
Preparation of Protein Extracts.
Whole cell lysates of C. jejuni were prepared as described previously (19). Periplasmic fractions were prepared according to the method of Neu and Heppel (51). Outer and inner membrane proteins were prepared by centrifugation (60 min at 135,000 × g at 4 °C) of the aqueous, and the supernatant of the pellet/spheroplasts fraction that was obtained after sonication (3 × 1 min) with a Sonicator Ultrasonic Processor XL 2020 (4-s pulse, 1 s off; Mandel Scientific) and low-spin centrifugation (30 min at 5,400 × g at 4 °C). Pellets containing membrane fractions were resuspended in 100 mM Tris-HCl, pH 7.2, to 1/100 of the original culture volume. Protein extracts were stored at −80 °C until further use.
sqMS-Based fOS Analyses.
Free glycans were purified using the same method as for N-linked glycans (19) but without pronase E digest. For comparison of relative free glycan amounts, whole-cell lysates obtained from 108–1011 C. jejuni cells were adjusted to a final concentration of 20 mg/mL with 100 mM Tris-HCl, pH 7.2. FOS secretion was investigated using freeze-dried culture supernatants after low-spin centrifugation of one-fourth of the original culture volume and resuspension in 1/200 of the original culture volume with 100 mM Tris-HCl, pH 7.2. Mass spectra were acquired using a 4000 Q-Trap mass spectrometer (Applied Biosystems/MDS Sciex) with a capillary electrophoresis interface as described previously (19). The precursor scans for m/z 407.3 were performed with m/z step size 0.5 Th, dwell time 2.0 ms, and a collision energy of 50 eV. The relative amount of each glycoform, including free and N-linked, was compared based on the peak area of the extracted electropherogram for each corresponding ion.
Immunodetection by Western Blotting.
Immunodetection of proteins by Western blotting was performed as described (26). N-glycan-specific (hR6) (S. Amber and M. Aebi, manuscript in preparation) or AcrA-specific (21) polyclonal antiserum served as primary antibodies applied at 1:100,000 and 1:40,000 dilutions, respectively. Alkaline phosphatase conjugated anti-mouse or anti-rabbit IgG (Santa Cruz Biotechnology) served as the secondary antibodies at 1:500 dilutions. Signal intensities of digitized images were quantified by densitometric analysis of the gray scales with the AlphaImager software (AlphaInnotech).
Expression of Recombinant C. jejuni Proteins in E. coli.
Expression of recombinant PglB proteins from plasmids pWA1, pMAF10 (34) in E. coli TOP10, and preparation of PglB-containing membranes was performed as described (52) with the exception that 50 mM HEPES, pH 7.5, was used throughout the membrane preparation procedure. Soluble periplasmic Hexa-Histidine-tagged AcrA (AcrA-His6) constitutively expressed on plasmid pWA2 in E. coli SCM7 in the presence of pACYC(pgl) or pACYC(pglmut) was purified as described (34), dialyzed twice against 10 mM HEPES, pH 7.2, and stored at 4 °C until further use.
Preparation of LLOs.
LLOs were obtained from E. coli DH5α pACYC(pglmut) as described (32).
Genetic Manipulations of C. jejuni 11168.
Shuttle plasmids pCj-pglB, pCj-pglBmut, and pRY111(wlaA-wlaF)pglB::Kan (Table S2) were mobilized from E. coli pRK211.2 to C. jejuni 81–176 as described (53). Plasmids with the Campylobacter methylation signature were isolated from 1–2 × 108 C. jejuni 81–176 cells grown on MH for 18–20 h using the PureLink Quick Plasmid Miniprep Kit (Invitrogen). Then 1 μg C. jejuni 81–176 derived plasmid DNA was used to electroporate C. jejuni 11168 (54). Plasmid stability in C. jejuni 11168 was confirmed after plasmid isolation and transformation of E. coli DH5α followed by restriction analyses. Integration of the Kan-Cassette into the pglB locus by double homologous recombination of KanR and CmS candidates was verified by PCR using pglB and Kan-specific oligonucleotides. One positive KanR and CmS clone was used for further studies.
Semi-Quantitative RT-PCR (sqRT-PCR).
sqRT-PCR experiments were carried out with 50-ng total RNA prepared as described in ref. 55 using the One-Step RT-PCR kit (Qiagen) according to the manufacturer's instructions. Specific PCR products obtained with oligonucleotides PglBGsp2 and CS8 designed to amplify nt 23 to nt 345 of pglB were obtained after the 42nd cycle of the PCR and analyzed on 0.7% agarose gels. Signal intensities were quantified as described above. Control experiments were performed with heat-inactivated reverse transcriptase, RT-PCRs with oligonucleotides 16S-1 and 16S-2 specific to amplify a 518-bp 16sRNA fragment were obtained after the 30th PCR cycle and served as an invariant internal standard.
In Vitro PglB Activity.
In vitro PglB activity was assayed following a modified protocol by Glover and coworkers (52). First, 50 pmol purified AcrA-His6 proteins, 25 μg LLOs, and 100 μg membranes containing either PglB or PglBmut were combined in a total volume of 200 μL assay buffer [50 mM HEPES, pH 7.5, 1.2% (vol/vol) Triton X-100, 20 mM MnCl2] in the absence or presence of sucrose (100 mM, 300 mM) or ionic solutes (100 mM). After incubation for 15 min at 30 °C, AcrA-His6 proteins were immediately separated from the reaction mixture by Ni-NTA agarose pull-down (using 100 μL Ni-NTA agarose, washed twice with 500 μL 100 mM Tris-HCl, pH 7.2). After two washes with 500 μL 100 mM Tris-HCl, pH 7.2, followed by centrifugation for 30 s at 5,000 rpm, bound proteins were eluted with 200 μL 250 mM imidazole in 0.1 M Tris-HCl, pH 7.2. Recovered AcrA-His6 proteins were dialyzed against 10 mM Tris-HCl, pH 7.5, lyophilized, resuspended in one-tenth the original assay volume in 1× SDS Sample Buffer, and applied to 10% PAGE. AcrA-His6 proteins were detected by western blotting with AcrA- and N-glycan-specific (hR6) antisera. The flow-through and washing fractions of the Ni-NTA AcrA-His6 purification were combined and incubated with Soybean-agglutinin-agarose (50 μL SBA-agarose; EY-Labs) to enrich for fOS. After two washing steps with 500 μL 100 mM Tris-HCl, pH 7.2, bound glycans were eluted twice with 50 μL 250 mM galactose in 0.1 M Tris-HCl, pH 7.2. Elution fractions were combined and analyzed by sqMS.
Supplementary Material
Acknowledgments.
The authors acknowledge Dr. Susan Jensen (University of Alberta, Edmonton, Alberta, Canada) for helpful discussions. We also thank Dr. Patricia Guerry (Naval Medical Research Center, Silver Spring, MD) for the pCE plasmids, and Dr. Markus Aebi (Institute of Microbiology, Department of Biology, Swiss Federal Institute of Technology Zurich, Switzerland) for providing hR6 and AcrA antisera. This work was funded by the National Research Council (NRC) Genomics and Health Initiative and the Alberta Ingenuity Centre for Carbohydrate Research. C.M.S. holds an Alberta Ingenuity Scholar Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903078106/DCSupplemental.
References
- 1.Bohin JP. Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol Lett. 2000;186:11–19. doi: 10.1111/j.1574-6968.2000.tb09075.x. [DOI] [PubMed] [Google Scholar]
- 2.Bohin JP, Lacroix JM. In: The Periplasm. Ehrmann M, editor. Washington, DC: American Society for Microbiology; 2006. pp. 325–341. [Google Scholar]
- 3.Lequette Y, et al. Linear osmoregulated periplasmic glucans are encoded by the opgGH locus of Pseudomonas aeruginosa. Microbiology. 2007;153:3255–3263. doi: 10.1099/mic.0.2007/008953-0. [DOI] [PubMed] [Google Scholar]
- 4.Mah TF, et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 5.Breedveld MW, Miller KJ. Cyclic beta-glucans of members of the family Rhizobiaceae. Microbiol Rev. 1994;58:145–161. doi: 10.1128/mr.58.2.145-161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhagwat AA, et al. Further studies of the role of cyclic beta-glucans in symbiosis. An NdvC mutant of Bradyrhizobium japonicum synthesizes cyclodecakis-(1→3)-beta-glucosyl. Plant Physiol. 1999;119:1057–1064. doi: 10.1104/pp.119.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geremia RA, et al. A Rhizobium meliloti mutant that forms ineffective pseudonodules in alfalfa produces exopolysaccharide but fails to form beta-(1→2) glucan. J Bacteriol. 1987;169:880–884. doi: 10.1128/jb.169.2.880-884.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puvanesarajah V, et al. Role for 2-linked-beta-D-glucan in the virulence of Agrobacterium tumefaciens. J Bacteriol. 1985;164:102–106. doi: 10.1128/jb.164.1.102-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page F, et al. Osmoregulated periplasmic glucan synthesis is required for Erwinia chrysanthemi pathogenicity. J Bacteriol. 2001;183:3134–3141. doi: 10.1128/JB.183.10.3134-3141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briones G, et al. Brucella abortus cyclic beta-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect Immun. 2001;69:4528–4535. doi: 10.1128/IAI.69.7.4528-4535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Funakoshi Y. Free N-linked oligosaccharide chains: Formation and degradation. Glycoconj J. 2006;23:291–302. doi: 10.1007/s10719-006-6975-x. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Park H, Kitajima K, Lennarz WJ. Peptides glycosylated in the endoplasmic reticulum of yeast are subsequently deglycosylated by a soluble peptide: N-glycanase activity. J Biol Chem. 1998;273:21526–21530. doi: 10.1074/jbc.273.34.21526. [DOI] [PubMed] [Google Scholar]
- 13.Belard M, Cacan R, Verbert A. Characterization of an oligosaccharide-pyrophosphodolichol pyrophosphatase activity in yeast. Biochem J. 1988;255:235–242. [PMC free article] [PubMed] [Google Scholar]
- 14.Kmiecik D, et al. Catabolism of glycan moieties of lipid intermediates leads to a single Man5GlcNAc oligosaccharide isomer: A study with permeabilized CHO cells. Glycobiology. 1995;5:483–494. doi: 10.1093/glycob/5.5.483. [DOI] [PubMed] [Google Scholar]
- 15.Spiro MJ, Spiro RG. Potential regulation of N-glycosylation precursor through oligosaccharide-lipid hydrolase action and glucosyltransferase-glucosidase shuttle. J Biol Chem. 1991;266:5311–5317. [PubMed] [Google Scholar]
- 16.Villers C, et al. Release of oligomannoside-type glycans as a marker of the degradation of newly synthesized glycoproteins. Biochem J. 1994;298:135–142. doi: 10.1042/bj2980135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allos BM. Campylobacter jejuni infections: Update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 18.Nachamkin I. Chronic effects of Campylobacter infection. Microbes Infect. 2002;4:399–403. doi: 10.1016/s1286-4579(02)01553-8. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, et al. Mass spectrometry-based glycomics strategy for exploring N-linked glycosylation in eukaryotes and bacteria. Anal Chem. 2006;78:6081–6087. doi: 10.1021/ac060516m. [DOI] [PubMed] [Google Scholar]
- 20.Young NM, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 21.Wacker M, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 22.Kowarik M, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szymanski CM, Burr DH, Guerry P. Campylobacter protein glycosylation affects host cell interactions. Infect Immun. 2002;70:2242–2244. doi: 10.1128/IAI.70.4.2242-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlyshev AV, et al. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology. 2004;150:1957–1964. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- 25.Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 26.Kelly J, et al. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J Bacteriol. 2006;188:2427–2434. doi: 10.1128/JB.188.7.2427-2434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsen JC, Szymanski CM, Guerry P. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81–176. J Bacteriol. 2004;186:6508–6514. doi: 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers JD, Kelly DJ. A sulphite respiration system in the chemoheterotrophic human pathogen Campylobacter jejuni. Microbiology. 2005;151:233–242. doi: 10.1099/mic.0.27573-0. [DOI] [PubMed] [Google Scholar]
- 29.Guerry P, Ewing CP, Schoenhofen IC, Logan SM. Protein glycosylation in Campylobacter jejuni: Partial suppression of pglF by mutation of pseC. J Bacteriol. 2007;189:6731–6733. doi: 10.1128/JB.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingram-Smith C, Miller KJ. Effects of ionic and osmotic strength on the glucosyltransferase of Rhizobium meliloti responsible for cyclic beta-(1,2)-glucan biosynthesis. Appl Environ Microbiol. 1998;64:1290–1297. doi: 10.1128/aem.64.4.1290-1297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.IIñón de Iannino N, Briones G, Iannino F, Ugalde RA. Osmotic regulation of cyclic 1,2-β-glucan synthesis. Microbiology. 2000;146:1735–1742. doi: 10.1099/00221287-146-7-1735. [DOI] [PubMed] [Google Scholar]
- 32.Reid CW, et al. Affinity-capture tandem mass spectrometric characterization of polyprenyl-linked oligosaccharides: Tool to study protein N-glycosylation pathways. Anal Chem. 2008;80:5468–5475. doi: 10.1021/ac800079r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wacker M, et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci USA. 2006;103:7088–7093. doi: 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman MF, et al. Engineering N-linked protein glycosylation with diverse O-antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci USA. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.IIñón de Iannino N, Briones G, Tolmasky M, Ugalde RA. Molecular cloning and characterization of cgs, the Brucella abortus cyclic beta(1–2) glucan synthetase gene: Genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J Bacteriol. 1998;180:4392–4400. doi: 10.1128/jb.180.17.4392-4400.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhill J, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 37.Roset MS, Ciocchini AE, Ugalde RA, IIñón de Iannino N. Molecular cloning and characterization of cgt, the Brucella abortus cyclic beta-1,2-glucan transporter gene, and its role in virulence. Infect Immun. 2004;72:2263–2271. doi: 10.1128/IAI.72.4.2263-2271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arellano-Reynoso B, et al. Cyclic beta-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat Immunol. 2005;6:618–625. doi: 10.1038/ni1202. [DOI] [PubMed] [Google Scholar]
- 39.Banta LM, Bohne J, Lovejoy SD, Dostal K. Stability of the Agrobacterium tumefaciens VirB10 protein is modulated by growth temperature and periplasmic osmoadaption. J Bacteriol. 1998;180:6597–6606. doi: 10.1128/jb.180.24.6597-6606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 41.Bouchart F, et al. Proteomic analysis of a non-virulent mutant of the phytopathogenic bacterium Erwinia chrysanthemi deficient in osmoregulated periplasmic glucans: Change in protein expression is not restricted to the envelope, but affects general metabolism. Microbiology. 2007;153:760–767. doi: 10.1099/mic.0.2006/000372-0. [DOI] [PubMed] [Google Scholar]
- 42.Dylan T, Helinski DR, Ditta GS. Hypoosmotic adaptation in Rhizobium meliloti requires beta-(1→2)-glucan. J Bacteriol. 1990;172:1400–1408. doi: 10.1128/jb.172.3.1400-1408.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhagwat AA, Keister DL. Site-directed mutagenesis of the cyclic β-(1→3)(1→6)-glucan synthesis locus of Bradyrhizobium japonicum. Mol Plant Microbe Interact. 1995;8:366–370. [Google Scholar]
- 44.Komaniecka I, Choma A. Isolation and characterization of periplasmic cyclic beta-glucans of Azorhizobium caulinodans. FEMS Microbiol Lett. 2003;227:263–269. doi: 10.1016/S0378-1097(03)00690-6. [DOI] [PubMed] [Google Scholar]
- 45.Cangelosi GA, et al. Role for Agrobacterium tumefaciens ChvA protein in export of beta-1,2-glucan. J Bacteriol. 1989;171:1609–1615. doi: 10.1128/jb.171.3.1609-1615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang JS, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA. 2008;105:5933–5938. doi: 10.1073/pnas.0800237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koiwa H, et al. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell. 2003;15:2273–2284. doi: 10.1105/tpc.013862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood JM. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guidolin LS, Ciocchini AE, Inon de Iannino N, Ugalde RA. Functional mapping of Brucella abortus cyclic beta-1,2-glucan synthase: Identification of the protein domain required for cyclization. J Bacteriol. 2009;191:1230–1238. doi: 10.1128/JB.01108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 51.Neu HC, Heppel LA. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965;240:3685–3692. [PubMed] [Google Scholar]
- 52.Glover KJ, Weerapana E, Numao S, Imperiali B. Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem Biol. 2005;12:1311–1315. doi: 10.1016/j.chembiol.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Labigne-Roussel A, Harel J, Tompkins L. Gene transfer from Escherichia coli to Campylobacter species: Development of shuttle vectors for genetic analysis of Campylobacter jejuni. J Bacteriol. 1987;169:5320–5323. doi: 10.1128/jb.169.11.5320-5323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerry P, et al. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 1994;235:474–481. doi: 10.1016/0076-6879(94)35163-5. [DOI] [PubMed] [Google Scholar]
- 55.Carrillo CD, et al. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.