Abstract
Paleogenomics seeks to reconstruct ancestral genomes from the genes of today's species. The characterization of paleo-duplications represented by 11,737 orthologs and 4,382 paralogs identified in five species belonging to three of the agronomically most important subfamilies of grasses, that is, Ehrhartoideae (rice) Panicoideae (sorghum, maize), and Pooideae (wheat, barley), permitted us to propose a model for an ancestral genome with a minimal size of 33.6 Mb structured in five proto-chromosomes containing at least 9,138 predicted proto-genes. It appears that only four major evolutionary shuffling events (α, β, γ, and δ) explain the divergence of these five cereal genomes during their evolution from a common paleo-ancestor. Comparative analysis of ancestral gene function with rice as a reference indicated that five categories of genes were preferentially modified during evolution. Furthermore, alignments between the five grass proto-chromosomes and the recently identified seven eudicot proto-chromosomes indicated that additional very active episodes of genome rearrangements and gene mobility occurred during angiosperm evolution. If one compares the pace of primate evolution of 90 million years (233 species) to 60 million years of the Poaceae (10,000 species), change in chromosome structure through speciation has accelerated significantly in plants.
Keywords: grasses, paleogenomics
Paleogenomics, the study of ancestral genome structures, allows the identification and characterization of mechanisms (e.g., duplications, translocations, and inversions) that have shaped genome species during their evolution and provides a framework to better integrate results from genetics, genomics, and comparative analyses. Studies of fossils and lower taxa organisms [Neanderthal (1), Echinoderms (2), Mammoth (3), Sponge (4), and Moss (5)] have yielded unprecedented information on the evolution of animal species and the relationships between them. When fossil DNA is not available, paleogenomics can be performed through large-scale comparative analyses of actual species and through ancestor modeling.
In silico colinearity studies and ancestral genome reconstruction in mammals have been facilitated by a generally moderate reshuffling of chromosomal segments since their divergence from a common ancestor ≈130 million years ago (mya) (6–9). Recently, Nakatani et al. (10) provided an integrated view of vertebrate paleogenomics with an ancestor of 10 to 13 proto-chromosomes. In contrast to mammals, paleogenomics has been poorly investigated in plants as angiosperm species have undergone serial whole genome or segmental duplications, diploidization, small-scale rearrangements (translocations, gene conversions), and gene copying events that make comparative studies between and within the monocotyledon (mainly grasses) and eudicot families very challenging. For the eudicots, two scenarios based on comparisons between the grape, Arabidopsis thaliana, and poplar genome sequences have been proposed recently. In the first one, the eudicots were proposed to descend from a paleo-hexaploid ancestor with seven proto-chromosomes (11) whereas, in the second, they originated from a paleo-tetraploid ancestor with seven proto-chromosomes (12). Comparative genomics studies in the monocots and most particularly in grasses has been the subject of intense research in the past decade (13, 14). Recently, we published an original and robust method for the identification of orthologous regions between genomes as well as for the detection of duplications within genomes based on integrative sequence alignment criteria combined with a statistical validation (15). This approach has been applied to identify paleo-duplications between the rice, wheat, sorghum, and maize genomes and to propose a common ancestor for the grasses with five proto-chromosomes (15). However, sequence alignments were performed only between rice and wheat, and the relationships with the maize and sorghum genomes were established using lower resolution, marker-based macrocolinearity studies. Here, we were able to use a much higher resolution to delineate synteny blocks from sequences of the maize, rice, and sorghum genomes (16, 17), as well as from large sets of genetically mapped genes in wheat and barley. This difference in resolution was critical to estimate the size and gene content of the grass ancestral genome as well as identify classes of genes that were particularly affected by rearrangements during the evolution of these species. Finally, comparison of the five monocot proto-chromosomes with the seven eudicot proto-chromosomes demonstrated the faster pace of changes in chromosomal structure in the plant versus the animal kingdom, particularly in respect to conserved gene order and mobility.
Results
Cereal Genome Synteny and Duplication Pattern.
By using alignment parameters and statistical tests described in ref. 15 we analyzed the syntenic relationships between the rice, maize, sorghum, wheat, and barley genomes using various resources as described in SI Appendix. Using rice as a reference genome with 41,046 gene models, we identified 4,454 maize orthologs (defining 30 syntenic blocks), 6,147 sorghum orthologs (12 syntenic blocks), 827 wheat orthologs (13 syntenic blocks), and 309 barley orthologs (13 syntenic blocks) (Fig. 1A and Fig. S1 in SI Appendix). Thus, similar numbers of syntenic blocks were identified in genomes for which a complete sequence is available and in those with limited sets of sequence data (e.g., barley and wheat) thereby demonstrating the efficiency of the method. In maize, the higher number of blocks reflects the recent tetraploidization of the genome. In total, 11,737 orthologous pairs (Table S1 in SI Appendix) and 68 synteny blocks that covered 99%, 82%, 99%, 91%, and 84% of the rice, maize, sorghum, wheat, and barley genomes, respectively, were observed. A recent comparison of the sorghum genome with different numbers of gene models for rice and maize identified a set of 11,502 ancestral gene families (17), a number that is very close to ours, indicating the robustness of synteny alignments. This result complements and greatly refines previous marker-based and low resolution, sequence-based macrocolinearity studies [for review Salse and Feuillet (13)], thereby allowing us to better characterize the duplication patterns in the different genomes.
Fig. 1.
Identification of 11,737 orthologs and 4,382 paralogs in five cereal genomes. (A) Schematic representation of the 11,737 orthologs identified between the rice chromosomes (r1 to r12) used as a reference, and the barley (b1 to b7), wheat (w1 to w7), sorghum (s1 to s10), and maize (m1 to m10) chromosomes. Each line represents an orthologous gene. The five different colors used to represent the blocks reflect the origin from the five ancestral proto-chromosomes (15). (B) Schematic representation of the 4,382 paralogous pairs identified within the rice (r1 to r12), barley (b1 to b7), wheat (w1 to w7), sorghum (s1 to s10), and maize (m1 to m10) genomes. Each line represents a duplicated gene. The different colors reflect the origin from the five ancestral proto-chromosomes. Black lines represent lineage specific duplicated paralogs. (C) Distribution of the average CIP/CALP values observed for the orthologous (colored bars) and paralogous (colored curves) genes in the five cereal genomes. The number of genes in each category (paralogous vs. orthologous) is displayed within five classes (from 70 to 100%) of average CIP/CALP values. The asterisks indicate lineage specific events that affect the distribution, that is, the w1-w5-w7 translocation in wheat, the recent r11-r12 duplication in rice and the tetraploidisation (WGD) in maize. A time scale is provided on the left side of the diagram.
Two methods can be used for the in silico identification and characterization of genome duplications. The most robust and direct approach, called “intragenome duplication” (ID), consists in aligning a genome sequence against itself with stringent alignment criteria and statistical validation. We have used it recently to initiate paleogenomics studies in grasses (15). The second indirect approach, called “double synteny” (DS) or “doubly conserved synteny” (DCS), is based on the identification of chromosomal duplications through the detection of regions showing a high proportion of gene matches on two different chromosomes within a genome and corresponding to two syntenic regions in another genome (for review, see ref. 13). In this study, we reassessed duplications in the rice, maize, sorghum, wheat, and barley genomes through a combination of ID and DS approaches with the stringent alignment parameters defined in Salse et al. (15) (SI Appendix). Ten (383 paralogs), 17 (3,469 paralogs), 8 (390 paralogs), 10 (102 paralogs), and 9 (38 paralogs) intragenomic duplications were identified and characterized in the rice, maize, sorghum, wheat, and barley genomes, respectively (Fig. 1B and Fig. S2 in SI Appendix). In total, 54 interchromosomal duplications were characterized individually in the five cereal genomes, compared with the 31 previously identified in the rice (18), sorghum (19), barley (20), maize (16), and wheat (21) genomes using the DS approach, illustrating the advantage of combining the two methods. They represent 76%, 83%, 82%, 73%, and 75% of the rice, maize, sorghum, wheat, and barley genomes, respectively. Thus, in total, 4,382 paralogous genes were identified for the five cereal genomes providing the largest set of conserved duplicated genes in cereals to date (13, 15).
Integration of the independent analyses of the duplications within and synteny between the 5 major cereal genomes [hereafter referred to r for rice, m for maize, s for sorghum, and t for the Triticeae (wheat and barley)] allowed us to the characterize more precisely the seven shared duplications identified recently among the ancestral grass chromosomal groups (15, 22). These paleo-duplications were found on the following chromosome pair combinations: t4-t5/r11-r12/s5-s8/m2-m4-m1-m3-m10, t1-t3/r5-r1/s9-s3/m6-m8-m3, t1-t4/r10-r3/s1/m1-m5-m9, t2-t4/r7-r3/s2-s1/m2-m7-m1-m9-m5, t2-t6/r4-r2/s6-s4/m2-m10-m4-m5, t5-t7/r9-r8/s2-s7/m2-m7-m1-m4-m10-m6, and t6-t7/r2-r6/s4-s10/m4-m5-m6-m9 (Table S1 in SI Appendix). Sequence similarity comparisons (SI Appendix) of the 11,737 orthologous and 4,382 paralogous gene pairs identified in the five species clearly confirmed the coexistence within each genome of ancestral shared duplications and recent lineage-specific duplications (Fig. 1B). Analysis of the distribution of sequence similarity between all orthologous gene pairs (Fig. 1C, bars) showed a peak for average CIP/CALP values of 85–80% which reflects the speciation of the five genomes from a common ancestor 50–70 mya. When the distribution of sequence similarity between paralogous genes is compared, two peaks are observed (Fig. 1C, curves). The first one (average CIP/CALP value of 85–80%) overlaps with the speciation of the 5 genomes from a common ancestor 50–70 mya thereby reflecting the ancestral shared duplications whereas, the second peak (average CIP/CALP value of 100–95%) is a result of lineage-specific and recent duplications such as the r11-r12 duplication in rice and the maize tetraploidisation (Fig. 1C).
To support the use of the comparative analyses in genetic mapping, we developed a user-friendly online Web tool called “Narcisse-Cereals” based on the public “Narcisse” platform (23) that allows us to visualize the 11,737 orthologs and the 4,382 paralogs characterized in the five cereal genomes (www.clermont.inra.fr/umr1095/narcisse_cereals) as well as gain access to the raw data (gene name, sequence, position, and alignment criteria) obtained from the analysis of the synteny and duplication of the rice, maize, sorghum, wheat, and barley genomes.
Cereal Genome Ancestor Structure and Function.
The integration of rice, maize, and sorghum whole genome sequences in the comparative analysis allowed us to further characterize the ancestral genome structure in terms of size and gene content. Reconstruction of the five proto-chromosomes structural content was performed following the approach described by Murphy et al. (6) and Nakatani et al. (10) in the mammalian and early vertebrate paleogenomics studies, respectively, in which orthologous chromosomal segments are compared systematically in dot plots (Fig. 2). Here, the dot plot graph theory approach was applied by taking into account the syntenic as well as the shared and lineage specific duplications described above (SI Appendix). A set of 12 dot plots was first created for each orthologous chromosomal segment, and these were then assembled by the combination of two or three blocks into five nonredundant sets of duplicates that correspond to the five proto-chromosomes (Fig. 2 and Fig. S3 in SI Appendix). Despite numerous re arrange ments (Fig. 2), a sufficient number of gene pairs remained that allowed us to identify large syntenic blocks, shared between the three genomes and derived (diagonal dot plots) from the common ancestor with five proto-chromosomes. We then estimated the ancestral or minimum gene number by calculating the number of genes that are conserved between at least two genomes. Moreover, the size of the ancestral genome was estimated on the basis of the longest shared sequence conserved between at least two genomes. In both cases, knowledge about the regions corresponding to the seven ancestral shared duplications (gray areas in Fig. 2 and Table S1 in SI Appendix) allowed us to eliminate redundancy and provide a robust ancestor model (14). For example, the first proto-chromosome (ancestral chromosome A5) exemplifies the synteny between r5-s9-m6/8 (first dot plot) and r1-s3-m3/8 (second dot plot) as well as the paleo-duplication identified between r5-r1, s9-s3, and m6/8-m3/8 (represented with a gray block) (15). For this chromosome, the dot plot analysis revealed seven regions including the centromere (Fig. 2) that are delimited by the conserved duplication boundaries. These blocks were then used to estimate the number of conserved genes between rice and maize (blue and green) and, rice and sorghum (red). This resulted in a final set of 2,145 nonredundant ancestral genes located within seven chromosomal regions on proto-chromosome A5 (Fig. 2). If one considers the sum of the length for the 2,145 conserved CDSs to calculate the minimal size for this chromosome (with 3.8 kb per gene on average), then proto-chromosome A5 was at least 6.3 Mb in size. Using the same approach, we analyzed the four other proto-chromosomes and identified 563 genes/2.8 Mb, 1,083 genes/4.1 Mb, 2,754 genes/9.3 Mb, and 2,639 genes/11.1 Mb for proto-chromosomes A11, A8, A4, and A7, respectively (Fig. 2). Thus, we conclude that the ancestral genome with five proto-chromosomes contained at least 9,138 protogenes (located on 22 proto-chromosome blocks) representing a minimum size of 33.6 Mb. Here, the ancestor genome size is considered as a minimal size corresponding to the cumulative “gene space” free of transposable elements (TEs) or repeated elements. Of course, we cannot exclude the presence of TEs in the cereal ancestor. However, it is not possible to estimate their amount based on comparative analyses because of high transposition activity and rapid turnover of TEs in the cereal genomes (14) which only allow the detection of less than 4 million years old insertions through intraspecific comparisons (24).
Fig. 2.
Cereal ancestor proto-chromosomes structure. The synteny between rice (r), considered as the reference sequence (vertical), and maize (m) or sorghum (s) (horizontal) is shown as 12 dot-plots. The rice/sorghum synteny is depicted with 12 red dot-plots. The synteny between rice and maize is displayed as 12 blue and green dot-plots (reflecting the tetraploid nature of the maize genome). The seven paleo-duplications are indicated by gray blocks within the dot plots. Twenty-two ancestral proto-chromosome blocks (from A5S3 to A7L2) were identified with respect to paleo-duplication boundaries shown with gray blocks on A5, A11, A8, A4, and A7, harboring different colored blocks and reflecting the origin from the five ancestral proto-chromosomes. The number of conserved genes (cumulative diagonal dot-plots) and the physical size (cumulative coding sequence length) of each proto-chromosome block is shown in parenthesis on the right end side of the figure.
To assign potential function to the 9,138 ancestral genes and identify those that were modified preferentially during evolution, we selected a dataset consisting of 65 gene ontology (GO) classes associated with the 41,046 rice gene models (http://gnn.tigr.org/tdb/e2k1/osa1/GO.retrieval.shtml) and compared it with the relative distribution of gene ontology in the 9,138 ancestral gene set. A two-sample χ2 test of proportions was performed with the number of genes observed in each of the 65 GO classes between the rice and the ancestor gene contents. A P value >10e−5 was found for 28 GO classes and was considered to reflect the under-representation of these categories between the rice and ancestor genomes. Among them, five classes (corresponding to transcription factor activity, transcription, biological process, DNA binding, and structural molecule activity) were underrepresented in the ancestor compared with the rice genome gene content (Fig. S4 in SI Appendix). This suggests that genes in these five classes were affected particularly by ancestral and lineage specific rearrangements (mainly duplications) that resulted in additional copies potentially providing a selective advantage during evolution and adaptation.
Accelerated Evolution of Flowering Plants.
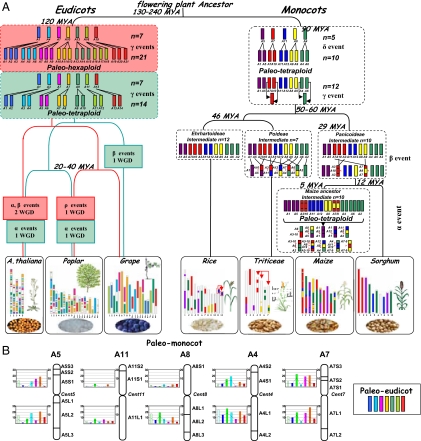
To reconstruct the rice, maize, sorghum, wheat, and barley genomes from the five ancestral proto-chromosomes containing 9,138 genes, we propose an evolutionary model that involves 4 major (α, β, δ, and γ) events named after the nomenclature defined in studies of the A. thaliana genome evolution (25). Before the divergence of the five grass genomes, the ancestor with n = 5 proto-chromosomes underwent a whole genome duplication (WGD) that resulted in an n = 10 chromosomes intermediate (δ event, Fig. 3A). After this tetraploidization, 2 interchromosomal translocations and fusions (γ event) resulted in two new chromosomes and an n = 12 intermediate ancestor (Fig. 3A). The Panicoideae have evolved from this ancestral 12 chromosomes genome after two chromosomal fusions (β event) that resulted in an intermediate ancestor with n = 10 chromosomes (Fig. 3A). Then, maize and sorghum evolved independently from this ancestor. Whereas the sorghum genome structure remained similar to the ancestral genome, maize underwent a lineage-specific whole genome duplication (α event) that produced an intermediate with n = 20 chromosomes. Rapidly, 17 translocations and chromosome fusions led to a final number of 10 chromosomes (Fig. 3A). The α event corresponds to the tetraploidization described in previous studies (15). In this model, the Ehrhartoideae have retained the original 12 chromosomes and rice underwent lineage specific rearrangements with recent duplications between the r11 and r12 chromosomes (as α events in Fig. 3A). Finally, from the intermediate ancestral genome with 12 chromosomes, the Pooideae underwent 5 chromosomal fusions that resulted in an ancestral Triticeae genome with n = 7 chromosomes (β event in the Fig. 3A). The Triticeae (wheat, barley, and rye), represented as a single genome in Fig. 3A, have retained the seven chromosomes as a basic chromosome number and underwent additional minor polyploidization events, segmental duplications, and translocations. For example, our analysis clearly established that a translocation between chromosomes 4 and 5 is common to wheat and barley whereas the previously reported translocation between chromosomes 4–7 is shared between the wheat and the rye genome only (15) (Fig. 3A).
Fig. 3.
Angiosperm evolutionary models. (A) Schematic representation of the monocot (Right) and eudicot (Left) evolutionary scenarios. The monocot chromosomes are represented with a five-color code to illuminate the evolution of segments from a common ancestor with five proto-chromosomes (named according to the rice nomenclature). The four shuffling events that have shaped the structure of the different grass genomes during their evolution from the common ancestor are indicated as δ (whole genome duplication), γ (ancestral chromosome translocations and fusions), β (family specific shuffling), and α (lineage specific shuffling). The seven eudicots proto-chromosomes are represented with different colors. The two scenarios proposed by Jaillon et al. (10) and Velasco et al. (11) are shown in red and green boxes, respectively. The different shuffling events that have shaped the structure of the A. thaliana, poplar and grape genomes during their evolution from a common ancestor are indicated as γ (WGD), β, and α; the cereal ancestor intermediate or lineage-specific shuffling events depend on the model. The current structure of the genomes with the representation of the remaining ancestral duplicated blocks is represented at the bottom. (B) Monocot/Eudicot ancestral genomes comparison. The five monocot proto-chromosomes (with the associated 22 chromosome blocks) are depicted as vertical bars. The number (bars) and origin (seven colors) of orthologs identified between the 9,138 monocot and 9,731 eudicot genes is indicated for each of the 10 monocot proto-chromosome arms.
Recent paleogenomics analyses within the eudicot family (11, 12, 26) led to two models illustrated in Fig. 3A. In the first one, the grape, Arabidopsis, and poplar genomes derive from a hexaploid ancestor with seven proto-chromosomes that underwent one and two specific whole genome duplications in poplar and Arabidopsis, respectively, whereas the grape remained unduplicated (11, 26, 27). In the second scenario, the eudicot genomes derive from a tetraploid ancestor with 7 proto-chromosomes that underwent specific whole genome duplications in the poplar, grape, and Arabidopsis lineages (12). Here, we wanted to exploit knowledge about the eudicots and monocots proto-chromosomes structures to see whether we can increase our understanding of the events that led to the divergence between these two main plant lineages. The five monocots proto-chromosomes (with 9,138 genes) were aligned with the seven eudicots proto-chromosomes [with 9,731 genes; Jaillon et al. (11)] using the approach describe previously for the identification of orthologs between the five cereal genomes. The results (Fig. 3B) show no orthologous chromosomal relationships between the five monocots proto-chromosomes (or chromosome arms or even chromosome blocks) and the seven eudicot proto-chromosomes. This indicates clearly that macrocolinearity has been largely eroded since the two major groups of angiosperms diverged from a common ancestor 150–300 mya and that the lack of colinearity observed previously between the monocot and dicot genomes was not due to a limited dataset or statistical methods but really reflects an active history of rearrangements during the evolution of the plant genomes.
Discussion
Paleology of the Monocot Chromosome Structural Evolution.
As the number of sequenced genomes grows, paleogenomics is becoming an increasingly important research area that provides insights into plant and animal genome evolution. By combining new alignment criteria and statistical validation that take into account the gene position information, we improved the characterization of paralogous or orthologous gene pairs previously identified in cereals with less stringent methods such as orthoMCL (28), INPARANOID (29) and MCscan (26). The conserved genes constitute syntenic blocks (characterized by the distance between two genes and the number of genes within a block) that then can be efficiently used to infer ancestral relationships even in the absence of complete contiguous genome sequences, such as barley and wheat. When taking this analysis into the perspective of parallel evolution of the plant and animal kingdoms, our data suggest that plants genomes were affected by more rapid changes in chromosomal architecture and frequency of manifesting these changes in speciation than mammalian genomes. We would attribute these features to the evolution of DNA replication and repair mechanisms in plants that have to account for the immobility of plants versus animals and their vulnerability to environmental changes.
We show that overall, among 73% of homologous genes identified between the five cereal genomes (i.e., 27% of species-specific genes, so called orphans, or possibly artefacts of genome annotation), only 12.8% are still conserved at orthologous positions (i.e., 87.2% of gene transposition) after 50–70 my of evolution, demonstrating a high rate of gene translocation in these genomes. This is a much higher rate than previously reported (<50%) with smaller datasets (30), which is likely due to the conservative approach we have used in this study or inflated gene counts in previous whole genome annotation projects. In any case, polyploidization (either as part of whole genome duplications or genome hybridizations) and the degree of gene copying events appear to be major factors involved in the deterioration of syntenic relationships in plant genomes (31). Freeling et al. (32) reported recently that up to 75% of the genes in Arabidopsis transposed after the origin of the Brassicale, compared with 87.2% estimated in the current analysis. The authors suggest that negative (i.e., purifying) selection may remove transposed copies of members of some gene families preferentially, whereas positive selection favors transposition of copies from other gene families. Our data confirm these observations and support the hypothesis that genome plasticity resulting from high gene transposition frequency offers the opportunity to positively select useful physical gene interaction considered selectively advantageous and to remove any other combinations considered selectively deleterious.
The characterization of the largest number of orthologs (11,737) and paralogs (4,382) within a single analysis across five cereal genomes and the identification of shared (seven paleo-duplications) and lineage specific duplications, allowed us to describe precisely four successive evolutionary events, δ (whole genome duplication event), γ (cereal ancestor shuffling events), β (cereal ancestor intermediate shuffling events), and α (lineage-specific shuffling events) that have shaped the grass genomes during the 50–70 my of evolution from a five proto-chromosomes ancestor. Such fundamental evolutionary mechanisms have been revealed also through recent studies in vertebrates (10 to 13 proto-chromosomes more than 400 mya) (10) and in the dicotyledons (seven proto-chromosomes 120 mya) (11). Interestingly, both in animal and plants, similar evolutionary mechanisms have been described with a reduced number of proto-chromosomes and several rounds of WGD followed by lineage-specific rearrangements leading to different chromosome numbers in today's species. Although there are many similarities among the eukaryotic kingdoms with respect to the characteristics of such chromosomal rearrangements, there are also significant differences. Polyploidization, a dominant force in the evolution of plant and fungi, is a rare event in most vertebrate lineages indicating differences in the capacity to adapt to genome duplications.
Here, we used the set of genes that were conserved at orthologous positions between five grass genomes and corrected from gene redundancy resulting from ancestral- and lineage-specific duplications to estimate the minimal ancestral genome gene number. We proposed that the cereals derive from a 33.6 Mb ancestor structured in five proto-chromosomes containing 9,138 proto-genes, a similar estimate to the 9,731 ancestral eudicot (11) and 11,502 ancestral angiosperm (17) gene repertories. The 33.6-Mb ancestral genome size is also consistent with the estimate of Bennett and Leitch (33), who suggested a minimal angiosperm genome without duplicated gene copies and repeated elements of maximum 50 Mb, based on a 1C-value of 157 Mb for A. thaliana as a reference.
The pace of gene mobility in plants becomes obvious if one draws from chromosomal alignments of species separated by 300 my of evolution between monocots and eudicots ancestors. Here, gene order has deteriorated to a degree that no synteny blocks can be identified anymore. These results shed light on previous analyses of synteny between Arabidopsis and rice where synteny was detected only at the micro level for a few (<100) loci (34–36). The results obtained through comparisons between the eudicot and monocot proto-chromosomes demonstrate that, in contrast to animal genomes, it is not possible to identify synteny at the genome or chromosome levels across plant classes.
Evolutionary Fitness of the Protogene Battery.
Comparison of ancestral gene contents with those of current genomes permits the identification of “duplication-sensitive” gene families (for which 1 paralogous copy is lost in 1 genome compared with the others) and “duplication-resistant” gene families (for which paralogous copies are maintained leading to copy number amplification) (26). From the established monocot ancestor structure and using the rice gene ontology (GO) as a reference, analysis of the evolution of the 9,138 ancestral grass genome gene set showed that 5 major GO classes are duplication-resistant as they have been subjected particularly to duplications resulting in additional copies that potentially provided a selective advantage during evolution and adaptation. Interestingly, these duplication-resistant genes have been conserved in different genomes since the ancestral whole genome duplication δ event.
Our results for the monocot ancestor are consistent with the results obtained by Paterson and colleagues for the eudicots who showed that duplication-resistant gene families correspond to transcriptional regulators that are retained more significantly after WGD events (26, 32, 37). We recently demonstrated that within the 10 major paleo-duplications that cover 47.8% of the rice genome, only 12.6% of paralogous gene pairs are still conserved within sister blocks, leading to the conclusion that pseudogenization (loss of one copy) occurred for the vast majority of the paralogous pairs (87.4%) during 50–70 my of evolution (38). In contrast, genes that may not provide a selective advantage when duplicated would be restored rapidly to a singleton status. Thus, our data support previous findings in the eudicots and suggest preferential retention of duplicated genes involved in signal transduction, and transcription, in response to rapidly changing biotic and abiotic extrinsic factors compared with genes encoding products involved in relatively stable processes.
Additional sequences from other grass (i.e., Brachypodium) and non-cereal monocot genomes such as Musa acuminata (banana) or Ananas comosus (pineapple), along with sequences of basal eudicots such as Eschscholzia california (california poppy) or Papaver somnifera (opium poppy) and Aquile giaformosa (columbine), and basal angiosperms such as Amborella trichopoda will further improve the accuracy of the paleogenomics studies in the major angiosperm clades and help to refine our model of plant genome evolution.
Materials and Methods
Details about the materials and methods used for the analysis regarding (i) nucleic acid sequence alignments, (ii) genome sequence databases, (iii) identification of duplicated and syntenic regions, (iv) graphical display, and (v) ancestor genome reconstruction can be found in SI Appendix. Enlarged resolution format of the figures is also available in SI Appendix.
Supplementary Material
Acknowledgments.
We thank Prasanna R. Bhat, Stefano Lonardi, Yonghui Wu, Steve Wanamaker, Nils Rostoks, Luke Ramsay, Nils Stein, Jan T. Svensson, Serdar Bozdag, Matthew Moscou, Rajeev Varshney, Kazuhiro Sato, and David Marshall for their contributions of generating the barley resources. We also gratefully acknowledge Olivier Jaillon and Jérôme Gouzy for fruitful discussions. This work was supported by Agence Nationale de la Recherche Grant ANR-05-BLANC-0258–01 from Institut National de la Recherche Agronomique (to C.F.), the USDA Barley CAP, U.K.-LINK AGOUEB, National Science Foundation DBI Grant 0321756, USDA-CSREES-NRI Grant 2006–55606-16722 (to T.J.C.), and the Selman A. Waksman Chair in Molecular Genetics (to J.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902350106/DCSupplemental.
References
- 1.Green RE, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bottjer DJ, et al. Paleogenomics of echinoderms. Science. 2006;314:956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 3.Poinar HN, et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 4.Jackson DJ, et al. Sponge paleogenomics reveals an ancient role for carbonic anhydrase in skeletogenesis. Science. 2007;316:1893–1895. doi: 10.1126/science.1141560. [DOI] [PubMed] [Google Scholar]
- 5.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 6.Murphy WJ, et al. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- 8.Lopez Rascol V, et al. Ancestral animal genomes reconstruction. Curr Op Immunology. 2007;19:542–546. doi: 10.1016/j.coi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 10.Nakatani Y, et al. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 12.Velasco R, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salse J, Feuillet C. In: Comparative Genomics of Cereals. Varshney RK, Tuberosa R, editors. Amsterdam: Springer; 2007. pp. 177–205. [Google Scholar]
- 14.Messing J, Bennetzen J. Grass genome structure and evolution. Genome Dynamics. 2008;4:41–56. doi: 10.1159/000126005. [DOI] [PubMed] [Google Scholar]
- 15.Salse J, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei F, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paterson AH, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005;3:e38. doi: 10.1371/journal.pbio.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson AH, et al. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein N, et al. A 1,000-loci transcript map of the barley genome: New anchoring points for integrative grass genomics. Theor Appl Genet. 2007;114:823–839. doi: 10.1007/s00122-006-0480-2. [DOI] [PubMed] [Google Scholar]
- 21.Singh NK, et al. Single-copy genes define a conserved order between rice and wheat for understanding differences caused by duplication, deletion, and transposition of genes. Funct Integr Genomics. 2007;7:17–35. doi: 10.1007/s10142-006-0033-4. [DOI] [PubMed] [Google Scholar]
- 22.Bolot S, et al. The ‘inner circle’ of the cereal genomes. Curr Opin Plant Biol. 2008;12:1–7. doi: 10.1016/j.pbi.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Courcelle E, et al. Narcisse: A mirror view of conserved syntenies. Nucleic Acids Res. 2008;36:485–490. doi: 10.1093/nar/gkm805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chantret N, et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops) Plant Cell. 2005;17:1033–1045. doi: 10.1105/tpc.104.029181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang H, et al. Unraveling ancient hexaploidy through multiply aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ming R, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien KP, et al. Inparanoid: A comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:476–480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song R, et al. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 2002;12:1549–1555. doi: 10.1101/gr.268302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu JH, Messing J. Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proc Natl Acad Sci USA. 2008;105:14330–14335. doi: 10.1073/pnas.0807026105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeling M, et al. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: Progress, problems and prospects. Ann Bot. 2005;95:45–90. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salse J, et al. Synteny between Arabidopsis thaliana and rice at the genome level: A tool to identify conservation in the ongoing rice genome sequencing project. Nucleic Acids Res. 2002;30:2316–2328. doi: 10.1093/nar/30.11.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandepoele K, et al. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 2002;12:1792–1801. doi: 10.1101/gr.400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. Statistical inference of chromosomal homology based on gene colinearity and applications to Arabidopsis and rice. BMC Genomics. 2006;12:447. doi: 10.1186/1471-2105-7-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seoighe C, Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Throude M, et al. Structure and expression analysis of rice paleo-duplications. Nucleic Acid Res. 2009;37:1248–1259. doi: 10.1093/nar/gkn1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.