Abstract
Covalent attachment of ubiquitin to substrates is crucial to protein degradation, transcription regulation and cell signalling. Highly specific interactions between ubiquitin-conjugating enzymes (E2) and ubiquitin protein E3 ligases fulfil essential roles in this process. We performed a global yeast-two hybrid screen to study the specificity of interactions between catalytic domains of the 35 human E2s with 250 RING-type E3s. Our analysis showed over 300 high-quality interactions, uncovering a large fraction of new E2–E3 pairs. Both within the E2 and the E3 cohorts, several members were identified that are more versatile in their interaction behaviour than others. We also found that the physical interactions of our screen compare well with reported functional E2–E3 pairs in in vitro ubiquitination experiments. For validation we confirmed the interaction of several versatile E2s with E3s in in vitro protein interaction assays and we used mutagenesis to alter the E3 interactions of the E2 specific for K63 linkages, UBE2N(Ubc13), towards the K48-specific UBE2D2(UbcH5B). Our data provide a detailed, genome-wide overview of binary E2–E3 interactions of the human ubiquitination system.
Keywords: protein network, protein–protein interaction networks, ubiquitin-conjugating enzymes, ubiquitin–protein ligases, yeast two-hybrid
Introduction
Modification of proteins with ubiquitin (ubiquitination) regulates the degradation of proteins by the 26S proteasome, but also serves a wide variety of other cellular processes, ranging from endocytosis to cell death (Pickart, 2001). Complex and tightly regulated protein–protein interactions (PPIs) of three key enzymes, ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin protein ligases (E3s), allow that ubiquitin becomes activated and covalently linked to substrates (Pickart, 2001; Dye and Schulman, 2007). These E1, E2 and E3 enzymes are operating in a hierarchical system. In the human genome, two E1s have been identified, responsible for the activation and transfer of ubiquitin (Ub) to the E2s (Haas et al, 1982; Jin et al, 2007). In contrast, over 30 E2s have been identified, and these have in common a highly conserved 150–200 amino-acid catalytic domain (Pickart, 2001). This UBC domain consists of several α-helices, β-sheets and variable loop regions, which are surrounding an active-site cysteine residue (Burroughs et al, 2008). Flanking this conserved region, additional N- and/or C-terminal extensions have been found that are involved in substrate selection, dimerization and additional processes (Silver et al, 1992; Haldeman et al, 1997; Summers et al, 2008). E2s are transferring the activated ubiquitin to E3s, characterized by the presence of either an HECT (Homologous to E6-AP Carboxy-Terminus) (Hatakeyama and Nakayama, 2003; Bernassola et al, 2008), U-box (Hatakeyama and Nakayama, 2003) or Really Interesting New Genes (RING) domain (Ardley and Robinson, 2005). The RING-finger domain is a highly conserved pattern of cysteine and/or histidine residues, chelating two atoms of zinc in a highly typical cross-brace structure. RING-type E3s can bind E2s through their RING-finger, but in contrast with HECT-type E3s, they catalyse the direct transfer of ubiquitin from the E2 to a substrate (Ardley and Robinson, 2005). Interactions between E2s and E3s are thought to be important because the interacting E2 dictates the type of inter-ubiquitin linkages and thereby determines the fate of the substrate. For example, ubiquitin chains linked through lysine 48 (i.e. by UBE2D2(UbcH5B)) are targeted by the 26S proteasome for degradation (Finley et al, 1994), whereas UBE2N(Ubc13)-mediated K63-linked ubiquitin chains are involved in signalling (Deng et al, 2000).
A limited number of clues, obtained by experimental structures, showed that E3 interactions are occurring in the UBC-fold (Huang et al, 1999; Schulman et al, 2000; Zheng et al, 2000; Christensen et al, 2007; Poyurovsky et al, 2007; Xu et al, 2008). Large-scale, genome-wide PPI studies can make an important contribution in the understanding of the specificity of E2–E3 interactions. Aiming to map E2–E3 interactions on a genome-wide scale and to study the underlying selectivity of these binary complexes, we have used a yeast two-hybrid (Y2H) screen, including 35 E2 UBC-folds and 250 E3 RING-finger domains. Our analysis showed more than 300 binary E2–E3 interactions, of which a large proportion was not known earlier. As expected we found that multiple E2 and E3 enzymes could interact with multiple E3s and E2s, respectively. Some of the E2s and E3s seemed to be more versatile than others and showed more interactions than the average number one would expect, thereby conferring to the concept of hubs in protein interaction networks. Comparing the physical E2–E3 interactions with reported E2–E3 pairs assayed in in vitro ubiquitination reactions showed a high level of agreement. One of the highly interacting E2s, UBE2U, an earlier unknown C-terminal extended, class III E2, showed the highest number of interactions. We validated the interaction between UBE2U and the E3 ligase MDM2 and also several interactions between hub E2s and E3s by independent GST-pull-down analysis, indicating a high level of confidence of the found E2–E3 interactions. On the basis of the interaction network we generated mutants of UBE2N(Ubc13) to mimic the E3 interaction pattern of UBE2D2(UbcH5B). This resulted in new E3 interactions, which allows manipulation of ubiquitination pathways in living cells. The experimental interactions found in this study provide a global, high-resolution framework for structural and functional studies towards the organization and selectivity between E2 and E3 enzymes.
Results
Central in the process of ubiquitination are the interactions between ubiquitin-conjugating enzymes (E2) and ubiquitin protein ligases (E3). To gain more insight into the selectivity of binary E2–E3 complexes, we devised a systematic and genome-wide Y2H screen. Earlier work by others and by our group showed that these interactions are highly selective, and that Y2H assays are powerful tools to study these specificities (Winkler et al, 2004; Christensen et al, 2007). This screen was performed to generate a framework of binary E2–E3 interactions. Interactions between the individual UBC-folds and the RING-finger domains are the major contributors to E2–E3 interactions (Lorick et al, 1999). Therefore, in a first approach we limited ourselves to screen for the interactions of human UBC-folds with RING-finger domains.
Annotating the human ubiquitin-conjugating enzymes
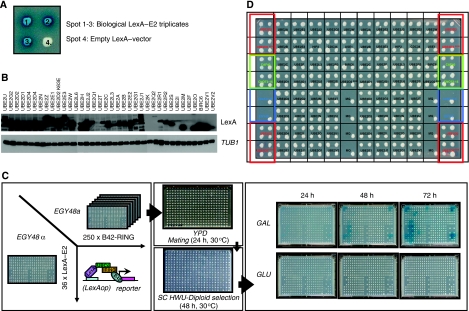
It has been estimated earlier that within the human genome, 30–40 E2s are present (Pickart, 2001). To precisely annotate the complete human E2 superfamily, we searched human cDNA databases for candidates. Our search showed 52 E2 family members that contain the UBC-fold (Supplementary Table I). Of these 52 E2s, we identified 16 pseudogenes for which no expressed protein-coding mRNAs could be found. We also identified an additional E2-like enzyme, UBE2N-like, lacking the central ubiquitin-accepting cysteine. Among the remaining 35 enzymes, three E2s were annotated as putative E2s and no ubiquitin(-like)-conjugating activity has been reported for these (UBE2D4, UBE2W and UBE2U) so far. Recently, it has been shown that UBE2F has the ability to act as a second NEDD8 E2 and prefers interaction with the Rbx2 RING protein, adding divergence to the NEDDylation system (Huang et al, 2009). So at this moment, 31 E2 enzymes are described to be involved in the process of ubiquitin or ubiquitin-like protein conjugation. Among these E2s, there are well-known examples with biochemically active roles in protein ubiquitination, such as UBE2D2(UbcH5B) and UBE2L3(UbcH7) (Shimura et al, 2000; Ozkan et al, 2005). Apart from the wild-type (WT) E2s, we also included the UbcH5B(K63E) mutant, which has been shown earlier to fail to interact with the WT CNOT4 RING-finger, but showed an altered-specificity interaction with the D48K/E49K double mutated CNOT4 (Winkler et al, 2004). Using multiple sequence alignments, we determined the boundaries of the UBC-fold. These 36 UBC-folds were fused to the C-terminus of LexA, DNA sequence verified, transformed in MATα cells and arranged in a standardized array (BD–E2 array) (Figure 1B and D).
Figure 1.
Outline array based, mating yeast two-hybrid matrix screen. (A) Organization of LexA–E2 fusions as three independent bacterial triplicate clones and an empty LexA vector. (B) LexA–E2 protein expression levels. Total protein lysates of yeast cells transformed with LexA–E2 fusions were resolved on SDS–PAGE, transferred to membranes and probed with either LexA antibody or yeast tubulin (TUB1). (C) Overview of experimental yeast two-hybrid screening procedure. EGY48α cells were transformed with 36 LexA–E2 fusions and EGY48a cells were transformed with 250 B42–RING-fusions. Screening for pair-wise E2–E3 interactions was carried out by standardized array-based mating between EGY48α and EGY48a cells on YPD for 24 h at 30°C, followed by diploid selection on SC HWU− for 48 h at 30°C. Interactions were visualized by transferring diploids on SC HWU− X-Gal (colorimetric selection) or on SC HWUL− (auxotrophic selection) under both B42-inducing (galactose as main carbon source) and repressive (glucose) conditions. Digital images of the interactions were taken at 3, 24-h interval time points. (D) Overview of standardized LexA–E2 array. Each spot represents EGY48α cells transformed with the indicated LexA–E2 fusion construct. Spots with a red square contained LexA–UBE2D2 and LexA–UBE2D2 K63E constructs and were mated throughout the entire procedure with EGY48a cells transformed with B42–CNOT4 N63 as positive and negative interaction controls, respectively. Spots with a green square were transformed with LexA–Caf40 and LexA–CNOT2-15 and served as auto-activating controls. Blue squared spots represent either EGY48a cells transformed with LexA-empty vector and an untransformed EGY48α strain, as mating/growing controls.
Selection of human RING-type E3s
Recent genome-wide annotations of human E3 ubiquitin protein ligases have indicated that there are 300 RING-type and 9 U-box-type E3s present within the human genome (Li et al, 2008). To select a representative collection of these domains, genome-wide database searches were initiated. First, we performed a SMART search using the curator-defined RING signature (SM00184) and this yielded 520 hits of which 314 of them were unique. Second, we performed a ScanProSite using ProSite pattern PS50089 against the human sequences deposited in SwissProt+Trembl (UniProt) generating a multiple sequence alignment of 627 hits in 611 UniProt sequences. Third, we performed a PSI-BLAST search of the human CNOT4 sequence against SwissProt, sequences with ‘RING' in their name were retained during the process, with ‘ubiquitin ligase' in some steps retained and without these names removed in some steps. Finally, all sequences with an expectation of 1.1 and lower were selected and this yielded 217 hits. Finally, we constructed a list of 308 Ensembl entries containing InterPro domain IPR001841. Next, these entries were cleared of redundancies and converted to UniProt codes and full-length protein sequences, which were retrieved from SwissProt. To locate the RING signature, a RING-finger matching tool was written and from the 295 sequences, we retrieved 313 RING-finger sequences. All RING-fingers were aligned in a multiple sequence alignment using Clustal W. The resulting alignment was carefully corrected by hand, especially with respect to zinc-binding regions. Regions that could not be aligned with any degree of reliability, for example sequences containing more than four C/HXXC/H, were removed from the alignment. In total, 313 RINGs were successfully aligned, 161 of them being classified as proper RING (matching the CXXC[X](10–51)C[X](1–3)H[X](2–5)CXXC[X](4–60)CXXC signature), 16 as PHD (matching CXXC[X](8–31)C[X](2–4)C[X](2–5)HXXC[X](12–60)CXXC) and 11 of them as FYVE (matching CXXC[X](12–40)CXXXXCXXC[X](13–80)CXXC). We identified 111 E3s being uncommon, as they match the following signature [C/H]XX[C/H][X](0–86)[C/H][X](0–5)[C/H][X](0–29)[C/H]XX[C/H][X](0–86)[C/H/D], wherein X could be any amino acid, except a C or an H. We also found 14 E3s being incomplete, belonging to none of the signatures. Together, 177 sequences are expected to act like E3 ligases, for 25 this is not expected, because they lack a complete RING signature and for the remaining 111 this remains unclear. Of these RING sequences we obtained clones for 250 human RING-finger/U-box E3 ligases (80.9% coverage of total (250/309)), based on cDNA availability. The RING-finger/U-box domains were subcloned as C-terminal fusions with the B42 activation domain (AD), under control of a galactose-inducible promoter. All B42–E3 constructs were verified by DNA sequencing and arrayed as a library of 250 B42–RING fusions in MATa yeast cells (AD–E3 array) (Supplementary Table II). Correct expression of the B42 fusions was evaluated using immunoblotting (data not shown).
Genome-wide E2–E3 yeast-two hybrid screen
Systematic mating crosses between the BD–RING and AD–E2 arrays allowed scoring for putative interactions between the arrays of E2 domain and E3 domain. Each of the 36 LexA–E2 fusions was arrayed in three independent yeast transformants. These three spots were placed in a small square together with LexA-only, measuring E2–E3 interactions in triplicate parallel to the empty vector control (Figure 1A). EGY48a cells transformed with B42–RING fusions were spotted on top of the BD–E2 arrays. After mating, diploids were selected and screened for interactions based on colorimetric (LacZ-set) or auxotrophic (LEU2-set) selection. A semi-automated pipeline of digitalized plate photographs followed by spot size/colour intensity quantification on three time points, allowed us to quantify interactions in an unbiased and sensitive manner (Figure 1C).
Selection of interactors
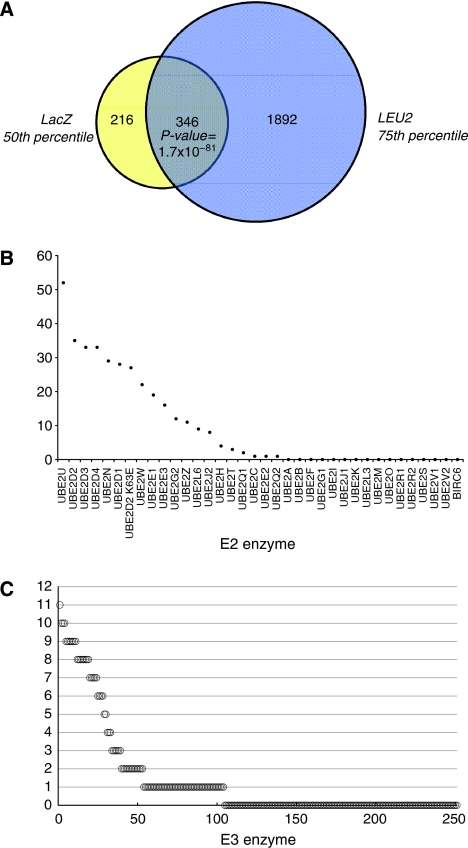
Close to 10 000 interactions within the 36 × 250 E2–E3 matrix were assessed independently on LacZ and LEU2 reporter read-outs. It has been shown earlier that, in general, interactions that activate both reporters are of higher confidence and more reproducible than interactions that activate only one reporter (Golemis et al, 2001). Therefore, our definition of an E2–E3 interaction is a galactose-dependent increase in both reporter activities. To select the shared interactions, we determined percentile values for both the LacZ- and the LEU2-set. As any selection strategy has the potential risk of discarding true positive and including false-positive interactions, selection of the most suitable percentile cut-off was based on all E3 interactions for a given E2. Given the variability of spot sizes in the LEU2 dataset, we concluded that interaction values from 50–200 pixels are the consequence of either noise (false positives), absence of LEU2-reporter activation (true-negative interactions) or weak-activating interactions. To avoid the risk of discarding true interactions, all interactors in the LEU2-set above the 75th percentile were selected (2238 signals). From the LacZ-set with a total of 577 blue signals we selected all interactors above the 50th percentile (562 interactors, 94.4% of total). Combining these two sets resulted in 346 high-confidence interactors (15.5% of LEU2-set and 61.6% of the LacZ-set), which we named the LL-set (Figure 2A; Supplementary Table III). An overview of all the identified E2–E3 interactions scored by both LacZ staining and LEU2 selection is given in Supplementary Tables IV and V, respectively.
Figure 2.
Topological characterization of the E2–E3 network. (A) Venn diagram showing the shared E2–E3 interactions in the 75th percentile selected LEU2-set and the 50th percentile selected LacZ-set and the overlap between them. (B) Distribution of the E3 connectivity for each individual E2. Depicted are the numbers of E3 interactions observed in the shared LacZ- and LEU2-set among all interactions of a single E2. (C) Distribution of the E2 connectivity for each individual E3, idem as panel B.
Human E2–E3 domain–domain interactions
Inspecting the connectivity, that is the numbers of E3 interactions for each E2 in the LL-set, we identified 20 E2s that were connected to at least one E3 and 16 E2s that did not show any E3 interaction (Figure 2B). Screening the E3 domains, we found 104 E3s that were connected to at least one E2 and 147 E3s that did not show any E2 interactions. An overview of the E3 connectivity is shown in Figure 2C.
The E2s showing the highest number of interactions were UBE2U (52 interactors), followed by UBE2D2 (35), UBE2D3 and UBE2D4 (33), UBE2N (29), UBE2D1 (28) and the K63E mutant of UBE2D2 (27). Next, UBE2W, UBE2E1, UBE2E3, UBE2G2, UBE2Z, UBE2L6 and UBE2J2 showed 8–22 E2 interactions. Smaller number of interactions was found for UBE2H (three), UBE2Q1 (two), UBE2C (one), UBE2E2 (one) and UBE2Q2 (one) (Figure 2A). To investigate the possibility that differences in E3 interaction patterns were because of differences in LexA protein expression, immunoblot analysis was performed. Figure 1B shows that the majority of LexA–E2 fusions are expressed at similar levels and that only some E2s are expressed at low levels (UBE2Q2 and UBE2I(Ubc9)). Please note that we find one E3 interaction (PCGF2) for UBE2Q2 and, as expected, no interactions for the SUMO-specific UBE2I(Ubc9). Expression of UBE2F and UBE2K could not be detected, which correlates with the absence of E3 interactions. Of the other 13 non-interactors, five E2s (UBE2M, BIRC6, UBE2O, UBE2V1 and UBE2V2) have not been linked to ubiquitin conjugation, but in some cases to conjugation of ubiquitin-like proteins. It is important to note that we do not observe an over-representation of a particular class of E2s, indicating that N- or C-terminal extensions to the UBC-fold are not confounding the results (data not shown). We also analysed a potential effect of the position of the RING within the full-length E3 on interaction potential (Supplementary Table II). This indicated that N-terminal RING domains score slightly lower in E2 interactions as compared C-terminal RINGs (Supplementary Figure 1). Together, these analyses indicate that the interactions are distributed broadly over the different classes and types of E2 and E3 enzymes.
Topological characteristics of the E2–E3 interaction network
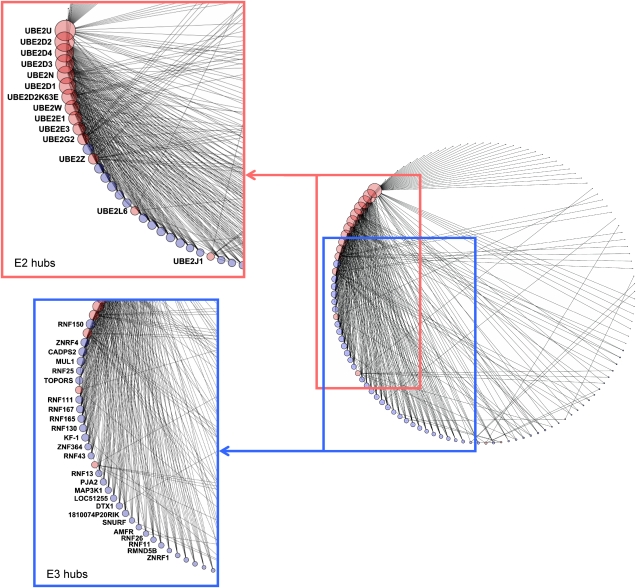
The E2–E3 interaction map can be regarded as a network, in which individual E2s and E3s are considered as nodes (Figure 3), which are connected to each other through interactions. In total, we found 346 interactions between 20 E2 and 104 E3 domains, with an average of 2.8 interactions per domain (Figure 3). The connectivity of both the E2s and E3s interactions indicate the presence of several high and lesser connected E2 and E3 domains. Our analysis showed that UBE2U, the UbcH5 family of highly homologous E2s (UBE2D1-4), UBE2N and UBE2E1 and UBE2E3 showed a high number of E3 interactions (Figures 2B and 3). UBE2U is an E2 enzyme, which carries a C-terminal extension attached to its UBC-fold and thus belongs to the class III group of E2 enzymes. At present no interaction data for UBE2U or an ubiquitin(-like) conjugation activity have been described in the literature, which may be related it its restricted expression pattern in the urogenital tract (based on mRNA expression profile, data not shown). Members of the UbcH5 family are highly conserved E2 enzymes and homologous to yeast UBC4/5. These E2s are involved in the degradation of misfolded and short-lived proteins and have been shown to be active in ubiquitination (Seufert and Jentsch, 1990). UBE2N(Ubc13) forms heterodimers with UBE2V1 and UBE2V2 and is involved in K63 ubiquitin chain assembly (Deng et al, 2000). UBE2G2 is the human homolog of the yeast UBC7 and resides in the endoplasmatic reticulum (ER). UBE2G2 has been reported to interact with the E3 ligase gp78 (AMFR) and this E2–E3 pair has a role in ER-associated degradation (ERAD) (Chen et al, 2006). Another characteristic of hub proteins is that they connect to proteins with only a few interactions (Maslov and Sneppen, 2002). This is clearly illustrated by the E3 interaction patterns of UBE2U and UBE2G2. On the other hand, UBE2D1-4 interact with well-connected E3s.
Figure 3.
E2–E3 network. A network graph of binary E2–E3 domain interactions involving 124 E2 and E3 domains linked through 346 interactions. Nodes (domains) are shown as circles and interactions between them as black lines. E2 domains are shown as red circles and E3 domains as blue circles. The size of each node is linear related to the number of links of that node. Red square depicts E2 hub proteins, blue square hub E3s. Figure was generated using Cytoscape v.2.6.1.
The hierarchical nature of the ubiquitin-conjugation machinery allows a flux of activated ubiquitin from the activation by the E1 through E3s to downstream targets. The role of E2s in this process is to maintain the robustness of the system as a whole and to connect the activation of ubiquitin with its conjugation to substrates. Therefore, deletion of the E2s with high numbers of E3 interactions could have severe consequences for the complete ubiquitination network. To investigate this, we systematically removed (hub) E2s from the E2–E3 network and quantified the number of E3s that become unconnected (Supplementary Figure 2). By removing UBE2U, 32 E3s become unconnected, whereas 20 remain connected to another E2. In contrast, removing UBE2D2 has only very mild effects, because many E3s interact with the homologous UBE2D1, -D3 and -D4 and to a lesser extent with UBE2E1 and UBE2E3. Interestingly, removal of UBE2G2 from the network leaves seven E3s unbound. As this E2 is localized in the ER and is involved in ERAD, it fulfils a central role in the network. Finally, deletion of UBE2Z from the network disconnects two RING domains from the recently identified second E1 for ubiquitin, Uba6.
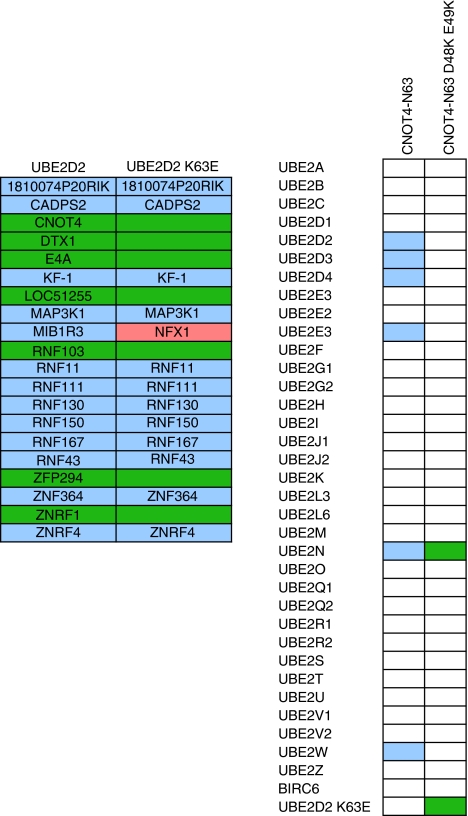
Apart from the WT E2s tested in this screen, the UBE2D2(UbcH5B) loop 1 mutant (K63E) was also included. As shown earlier, contrary to WT UBE2D2, the K63E mutant is not able to bind to WT CNOT4 (Winkler et al, 2004). However, when mutating the UBE2D2-interacting residues in the CNOT4 RING-finger such that they swap their charges (D48K/E49K), complete rescue of interaction could be achieved. Interestingly, UBE2D2 K63E does not show a dramatic change in its E3 interaction pattern compared with WT UBE2D2 and several E3s are interacting with both WT and the mutant UBE2D2. Strikingly, UBE2D2 K63E gained interactions with LOC51136, NFX1 and RNF8 as compared with the WT enzyme. Inclusion of the D48K/E49K mutant of the CNOT4 RING-finger shows loss of binding with the UbcH5 family, UBE2E1, UBE2E3 and UBE2W and, as expected it displays specific binding with UBE2D2 K63E. The mutant CNOT4 also showed a stronger interaction with UBE2N(Ubc13) as compared with the WT E3 (Figure 4).
Figure 4.
Mutant UbcH5B and mutant CNOT4–N63 interactions. (A) Overview of RING E3 interaction patterns between wild type and UBE2D2(UbcH5B) (K63E). E3s in purple interacted with both wild type and mutant UBE2D2(UbcH5B), in green only with wild-type UBE2D2(UbcH5B) and in pink only with the K63E mutant. (B) E2 interaction pattern of wild type and mutant CNOT-N63. White indicates no interaction, purple or green an interaction with CNOT4-N63 or CNOT4-N63 D49K/E49K, respectively.
The presence of highly connected E2s and E3s provides the network with robustness and resistance against small perturbations. This implies the presence of selective structural features involved in E2–E3 interaction specificity. To study the underlying molecular determinants, E2s were selected that exhibit more than one E3 interaction. First, the degree of overlap in E3 interaction between these E2s was quantified as the ‘hub overlap index' (Supplementary Figure 3). These numbers indicate the extent by which a certain E3 is used for E2 binding. The hub overlap index indicates that the homologous E2s, like UBE2D2, -D3, -D4 and to a lesser extent UBE2D1, cluster together in E3 interaction pattern, indicating that enzyme function is directly coupled to structural similarities. The same is true for the homologous UBE2E1 and UBE2E3 proteins, which share interactions with many E3s. Surprisingly, the highly similar UBE2E2 displayed an E3 interaction pattern distinct from the profiles of UBE2E1 and UBE2E3. The E2s UBE2N and UBE2W are highly connected, but they exhibit lower amounts of overlap. Unexpectedly, the highly connected UBE2U does not cluster together with the other highly interacting E2s, but it interacts with many E3s, suggesting a general role in ubiquitination in the urogenital tract.
To compare the hub overlap index with structural similarities between E2s, we calculated the percentages of sequence identity either for the complete UBC-fold or only for the predicted E3-interacting interface (Supplementary Figure 4). The distribution of sequence identity between E2 enzymes roughly equals the pattern of the hub overlap index. For most pairs, the sequence conservation clearly relates to the hub overlap, as indicated for the sub-clusters UBE2D1–D4, UBE2E1 and -E3 and UBE2N and UBE2W. This underscores a structural basis among these E2 enzymes for E2 enzymes. In contrast, UBE2E2 is highly similar to UBE2E1 and UBE2E3, but it displays little interaction overlap. UBE2T is interesting in this respect as it is equally similar to UBE2E1, UBE2E2 and UBE2E3, but the UBE2T interaction pattern only overlaps with that of UBE2E2. The similarity in the E3 interaction surfaces of UBE2L6 and the UBE2D family is limited, but their E3 interaction profiles display a significant degree of overlap.
Finally, the screen identified 11 E3 interactions for the UBE2Z(Use1) E2 enzyme. Recently, it has been shown that this E2 enzyme can become ubiquitin loaded by the newly identified E1 enzyme, Uba6, in a manner discriminating against other E2 enzymes (Jin et al, 2007). The observed UBE2Z interactions could provide a link between Uba6 and RING E3 ligases.
Among the screened E3 domains, RNF150, ZNRF4, CADPS2 and MUL1 showed the highest number of E2 interactions (Figure 3). For RNF150, no E3 ligase activity has been described, but the protein belongs to the human Goliath family (Anandasabapathy et al, 2003). Strikingly, two other Goliath family member E3s, RNF130 and RNF167 were also found to act as hub proteins, with only 52% sequence identity between the RING-fingers. The RNF130 protein (hGoliath) contains a protease-associated domain, a transmembrane domain and a RING-finger domain and is the human homolog of Drosophila's g1, a zinc-finger protein, involved in embryonic development (Guais et al, 2006). RNF130 displayed auto-ubiquitination activity when interacting with UBE2D1(UbcH5A) and UBE2D3(UbcH5C), but not with a number of other E2s (Guais et al, 2006). It would be interesting to test whether RNF167 and RNF130 are also functional E3s. A second example of multiple, highly homologous family members acting as hub E3s come from the E3 ligases ZNRF4, ZNRF1 and ZNRF2, having 51% sequence identity between their RING-finger domains. These show 10, 6 and 5 E2 interactions, respectively. The ZNRF proteins are implicated in spermatogenesis and the establishment and maintenance of neuronal transmission and plasticity mediated by their E3 ligase activity (Fujii et al, 1999; Araki and Milbrandt, 2003). CAPDS2 (calcium-dependent secretion activator 2) is involved in the exocytosis of neurotransmitter vesicles and cerebellar development (Sadakata et al, 2007). No E3 domain annotations could be found for this protein, and information about interacting E2s or ubiquitin-ligating activity is absent. MUL1 (MULAN) has been implicated recently in the establishment of mitochondrial dynamics and mitochondria-to-nucleus signalling, in a manner dependent on its RING-finger domain. MUL1 displayed auto-ubiquitination activity when tested with Ubc4, the yeast homolog of the human UbcH5 family (Li et al, 2008).
Quality of the physical E2–E3 interaction network
Comparing the E2–E3 interactions obtained in this screen with E2–E3 literature-curated interactions evaluates the quality of the interactions in the E2–E3 network. Physical interactions between E2 and E3 enzymes are required for efficient ubiquitination in vivo and in vitro and therefore for their catalytic function. Besides this, comparing the novel E2–E3 pairs generated by our screen with annotated functional E2–E3 pairs provides insight into the functionality of the E2–E3 pairs. We first selected the top 10% interacting E3s and compared their physical E2 interaction patterns with information obtained by low-throughput E2–E3 experiments. As shown in Figure 5, 25 hub E3s showed a total number of 209 E2 interactions covering 60% of the total number of E2–E3 two-hybrid interactions. For the published literature we recovered 64 E2–E3 pairs for the 25 hub E3s; 35 of the 64 pairs were positive for in vitro or in vivo ubiquitination activity and 29 were tested negative. Of the 35 positive literature interactions we confirmed 33 in our screen, which we regard as true positives (Figure 5). We found only two pairs that were biochemically reported positive, but did not occur in the Y2H screen (false negatives). The observed literature interactions were mostly concerning the members of the UbcH5 family and UBE2E1 and UBE2E3. To investigate the possibility that these E2s are more versatile in ubiquitination reactions, we compared reported E2–E3 combinations that were negative in biochemical reactions. Of the 29 E2–E3 interactions reported negative we recovered 24 in our dataset (true negatives). We also found five pairs that were reported negative in ubiquitination assays, but showed an interaction in the Y2H screen (false positives). In conclusion, this indicates a specificity of 83% (24/(5+24)) and a sensitivity of 94% (33/(2+33)) between biochemically active E2–E3 pairs and pairs found interacting in our screen. In the above analysis we focussed on the top 10% of interactors. Analysis of sensitivity and specificity of the complete data set is confounded by uncertainty of proper folding and/or localization of the Y2H proteins (Supplementary Table VI), which are potential pitfalls of Y2H screening. Therefore, we focussed on the E2 and E3s, which display at least one interaction. This criterion retains 20 E2s and 104 E3s from our screen. Literature inspection of this set showed 118 E2–E3 interactions of which 83 scored positive and 35 negative. Of the 83 positive interactions we recovered 48 interactions in our dataset. Of the 35 negative interactions 30 were also scored in Y2H interaction. This yields a sensitivity of 86% and a specificity of 58%. The verification rate of our screen (true positives plus true negatives/total interactions) is 66%, which compares well to global Y2H screens reported earlier (Li et al, 2004; Rual et al, 2005). On the basis of these data, we conclude that the detected E2 interactions for the hub E3s are in agreement with functional E2–E3 pairs reported in the literature, emphasizing the quality of interactions found in our screen.
Figure 5.
Quality of the E2–E3 network. Physical E2–E3 interactions in relation to literature-curated, functional E2–E3 pairs. Hub node E3s were selected and interactions with E2s were scored when biochemically tested in in vitro ubiquitination assays as reported in literature.
Validation of E2–E3 interactions by in vitro binding
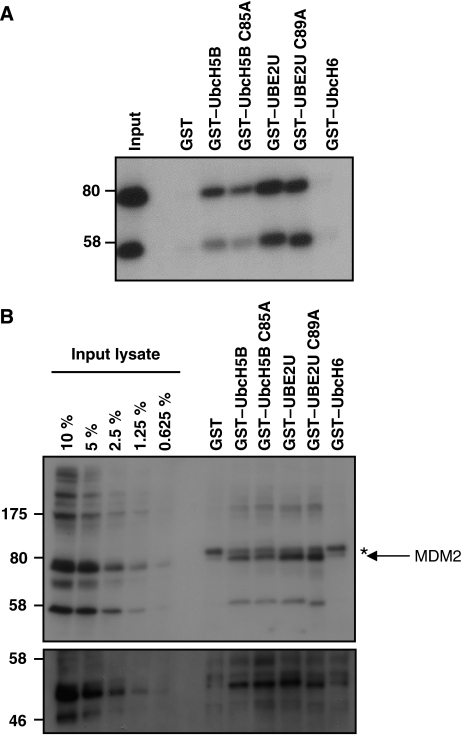
Interestingly, we observed a high number of E3 interactions with an uncharacterized E2 annotated as UBE2U. The UBE2U protein (321 amino acids) is composed of a typical UBC-domain, with a C-terminal tail attached. No biological function has been described for this E2 yet, but microarray experiments showed that UBE2U mRNA transcripts have been identified mainly in tissues belonging to the urogenital tract. The UBE2U gene is only present in mammalian genomes. We looked in detail, which E3 enzymes were able to interact with this E2. One of the interacting E3s was MDM2. MDM2 has been identified as a major regulator of the cellular level and activity of the transcriptional regulator p53 (Momand et al, 1992). The significance of MDM2 in controlling normal cellular behaviour has become clear by the observation that in more than 50% of human tumours alterations in the p53 has been observed and in at least 7% of these cases, the MDM2 protein is affected (Momand et al, 1998). MDM2 has been identified as an E3 ligase, capable of ubiquitinating both itself and p53, targeting them for proteasomal degradation (Nakamura et al, 2000). For this action, MDM2 has a C-terminal Cys3–His2–Cys3 RING-finger, which was shown earlier to be able to interact with UBE2D2(UbcH5B) (Saville et al, 2004). To investigate whether other E2s identified in the two-hybrid screen can bind to MDM2, we focussed on the MDM2–UBE2U interaction that seemed quite strong from the Y2H. To investigate the MDM2–UBE2U interaction in more detail, we performed GST-pull-down assays with immobilized WT and catalytic mutant (C85A or C89A) GST–E2 UBC domains. As a negative control, GST–UBE2E1(UbcH6) was included. As shown in Figure 6A, MDM2 is captured by UBE2U and as expected also by UBE2D2(UbcH5B). Interestingly, UBE2U binds more efficiently to MDM2 than UBE2D2(UbcH5B). No binding could be observed with GST alone or GST–UBE2E1(UbcH6), reflecting the specific nature of MDM2–E2 interactions. As expected, mutation of the catalytic cysteine did not affect the interactions. Next, the GST–E2 enzymes were applied to lysates of human osteosarcoma cells (U2OS), which contain higher levels of MDM2 because of amplification or over-expression (Florenes et al, 1994). Figure 6B shows an efficient pull-down of endogenous MDM2 with both GST–UBE2U and GST–UBE2U C89A enzymes. Binding could also be observed when GST–UBE2D2(UbcH5B) WT and C85A where used, but no binding could be seen with GST alone or GST–UBE2E1(UbcH6). The GST–E2 bound MDM2 fractions were also tested by antibodies against p53. Figure 6B shows that p53 could be detected when MDM2 was captured on GST–E2 enzymes. These results indicate that UBE2U can bind to MDM2, confirming the Y2H results, and that the p53 substrate is part of the captured complex.
Figure 6.
UBE2U physically interacts with MDM2. (A) HEK293T cells were transiently transfected with myc-tagged MDM2, lysed and incubated with GST–E2s immobilized on gluthathion-agarose beads. Bound material was resolved on 7.5% SDS–PAGE and proteins were visualized after immunoblotting with antibodies against myc. (B) Untransfected U2OS cells were lysed and combined with GST–E2s as in panel A. Immunoblotting was carried out using MDM2 antibodies (upper panel) and using p53 antibodies after reprobing (lower panel). Arrow indicates MDM2 signal, asterisk indicates background band.
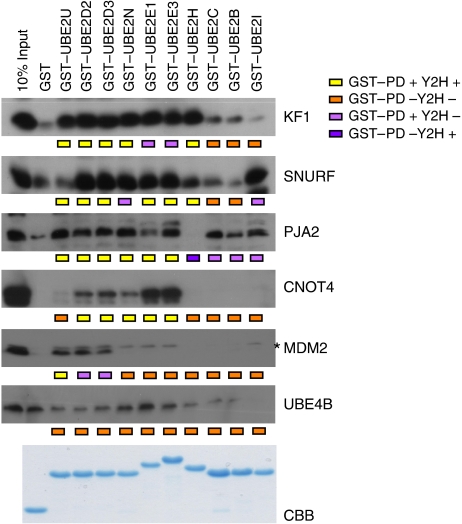
To validate the Y2H interactions for other E2 proteins we used the GST-pull-down approach for several E3 nodes expressed in crude bacterial lysates as 6xhistidine-tagged proteins. Sixty binary interactions were tested, which cover the spectrum of different distributions between hub and non-hub proteins. Figure 7 shows that 50 interactions were in agreement with the Y2H analysis: 23 interactions were positive in both assays (true positives) and 27 scored negative in both assays. Of the other 10 interactions 9 were only positive in the GST-pull-down approach, whereas one interaction (PJA2-UBE2H) only scored positive in the Y2H screen. Comparison of the data sets indicates 96% specificity (TN/(FP+TN)=27/(1+27)) and 72% sensitivity (TP/(TP+FN)=23/(23+9)) (Figure 5) and with a verification rate of 83%, which is in agreement with earlier reported verification rates of global Y2H screens (Li et al, 2004; Rual et al, 2005). Together, this underscores the high quality of our E2–E3 interaction dataset.
Figure 7.
GST-pull-down analysis between (hub) E2 and E3 enzymes. To validate yeast two-hybrid interactions, GST-pull-down analysis was performed between ten E2 enzymes and six E3 ligases. Approximately 10 μg GST–E2 enzyme were immobilized on G/A-beads and incubated with crude lysates expressing 6xhis–RING constructs. Bound proteins were resolved on SDS–PAGE, immono blotted and proteins were visualized using anti-6xhis antibodies. GST–E2 levels were visualized using Coomassie brilliant blue staining. An asterisk indicates a specific signal.
Altering the E3-interaction specificity of the K63-specific UBE2N(Ubc 13)
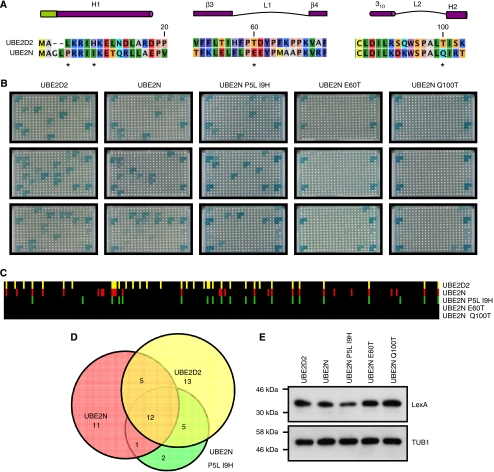
Elucidation of the 3D structures of several E2–E3 complexes showed that the E3 interaction surface of E2s comprises the N-terminal helix 1, loop 1 between β-strands 3 and 4 and loop 2 between helices 310 and 2 (Figure 8A) (Huang et al, 1999; Zheng et al, 2000; Dominguez et al, 2004; Christensen et al, 2007; Xu et al, 2008). We noted that the E3 interaction patterns of UBE2D2(UbcH5B) and of UBE2N(Ubc13) display a ∼60% overlap (Figure 8D). This E2 pair is particularly interesting as the primary sequences are rather divergent and UBE2D2 directs poly-ubiquitin conjugation through K48 linkages (Brzovic et al, 2006), whereas UBE2N forms K63-linked ubiquitin chains as a heterodimer with the catalytically inactive E2s, UBE2V1 and UBE2V2(MMS2) (VanDemark et al, 2001).
Figure 8.
Design of a UBE2N derivative with a new E3 interaction specificity. (A) Sequence alignment of UBE2D2(UbcH5B) and UBE2N(Ubc13). Secondary structure elements (indicated on top in purple and green for the UBE2D2-specific extension of helix 1) and regions involved in E3 RING interactions were derived from the E2 crystal structures (PDB codes 2ESK and 1JBB for UBE2D2 and UBE2N, respectively). Amino acids are coloured according to their physico-chemical properties. (B) E3 interaction patterns of UBE2D2, UBE2N and UBE2N mutants. The library of 251 yeast clones expressing B42–RING fusions were re-arrayed on three 96-well plates and mated with LexA–E2 expressing yeast clones. Diploids were selected and interactions were visualized as described in Figure 1. Pictures were taken after 72 h incubation at 30°C. Upper and lower three squares in the outer two columns of each plate represent plate controls. (C) Quantification of wild type and mutant E2–E3 interactions. Spot staining (LacZ) and spot sizes (LEU2) of each E2–E3 interaction were quantified and corrected for background signals. Individual E2–E3 interactions are depicted as bars; black bars indicate no interaction and coloured bars indicate E2–E3 interactions. (D) Venn diagram showing the overlap in E3 interactions between the different E2 enzymes. (E) Expression of wild type and mutant E2 enzymes. Total cell lysates of yeast cells expressing the indicated LexA–E2 fusions were separated on 12.5% SDS–PAGE and analysed by immunoblotting using antibodies against LexA or tubulin (TUB1) as loading control.
To obtain more insight into the E3 interaction specificity of UBE2N we decided to mutate residues in helix 1, loop 1 or loop 2. We reasoned that mutation of such residues into their UBE2D2 counterparts could allow interactions, which would otherwise be specific to UBE2D2. Comparing residues between UBE2D2 and UBE2N pointed to P5 and I9 in helix 1, E60 of loop 1 and Q100 in loop 2 as specificity candidates in UBE2N (Figure 8A). We changed these positions into the UBE2D2 residues and re-arrayed our library of 251 B42–E3(RING) clones to analyse the E3 interaction profiles of the UBE2N mutants. First, we noted that loop 1 and loop 2 of UBE2N are particularly sensitive to mutation as the E60T and Q100T mutants fail to interact with any E3 despite normal expression levels (Figure 8B and D). Interestingly, the E3 interaction pattern of P5L/I9H differs from both UBE2N and UBE2D2 (Figure 8B and C). This mutant still shares 13/29 interactions with WT UBE2N and gains seven new E3 interactions. Of these, five E3s (RMND5B, 1810074P20RIK, RNF150, MAP3K1 and MUL1) are shared with UBE2D2 and two (RNF183 and CHFR) are unique to the P5L/I9H mutant (Figure 8C; Supplementary Table III). MAP3K1 and MUL1 (or MULAN) are particularly interesting as MAP3K1 has been identified as the E3 enzyme responsible for degradation of the ERK1/2 kinases involved in signal transduction pathways (Lu et al, 2002) and MUL1 has a role in mitochondrial dynamics (Li et al, 2008). The P5L/I9H-specific interactor CHFR has been identified as the E3 ligase responsible for degradation of HDAC1, leading to upregulation of the Cdk inhibitor p21 and the metastasis markers KAI1 and E-cadherin (Oh et al, 2009). These observations indicate that exogenous expression of P5L/I9H UBE2N may result in switching from K48-linked to K63-linked polyubiquitin chains conjugated to substrates of the above-mentioned E3 ligases. In this respect it is important to note that the UBE2N surface responsible for heterodimerization with UBE2V1 and UBE2V2 does not overlap with the E3 interaction surface that comprises helix 1, loop 1 and loop 2 (VanDemark et al, 2001).
In conclusion, our mutational approach exploits the high-quality E2–E3 interaction dataset to design E2 enzymes with new E3 interaction specificities, which can be applied to alter ubiquitination pathways to manipulate protein function in cells.
Discussion
The selectivity of interactions between E2 enzymes and RING-type E3 ligases represent a central and crucial part of the ubiquitin-conjugation pathways in organisms. To explore the genome-wide landscape of E2–E3 complexes, we performed a comprehensive Y2H screen to study the selective behaviour of these binary interactions. Specific interactions mediated by the E2 UBC-fold and the E3 RING-finger domain were tested between arrayed collections of 35 UBC-folds and 250 RING-finger domains. RING-finger E3 ligases are binding ubiquitin-loaded E2s on their RING domain on one end and substrates on the other acting like a scaffold (Ardley and Robinson, 2005). The few structures known for E2–E3 complexes indicate that the major determinants of E2–E3 binding and selectivity reside within these domains. Our screen focussed on the molecular determinants of E2–E3 selectivity as the primary characteristic of specificity that does not take the secondary parameters such as spatial and temporal interactions into account.
Interactions between human E2 and E3 enzymes
Our database searches retrieved 35 human E2 enzymes that contain the highly conserved UBC-fold. Besides well-known E2 enzymes such as UBE2C(UbcH10) (Summers et al, 2008), members of the UbcH5 subfamily (Seufert and Jentsch, 1990) and UBE2N(Ubc13) (Deng et al, 2000) all having clear roles in ubiquitination; more divergent E2 enzymes have been identified. Among these E2s are UBE2F, UBE2W, BIRC6 (Apollon), UBE2O(E2-230K) and UBE2V1 and UBE2V2. All functional E2s contain the UBC-fold, providing them with a molecular platform for RING-domain interaction and rendering them as catalysts in the process of ubiquitin (-like) conjugation (Burroughs et al, 2008). We also included the UBE2D2 K63E mutant in the E2 arrays, evaluating the effects of this E2–E3 interaction mutant. The selection of RING-type E3 domains included in our screen covers the majority of the human E3 enzymes.
Our results showed that multiple E2s are able to interact with more than one E3 and vice versa. E2 enzymes that interact with many E3s were UBE2U, members of the UbcH5 family and UBE2N. UBE2U showed the highest levels of E3 interactions. The restricted expression pattern of UBE2U could explain why it has not been identified earlier as an E3(RING)-interactor. E2s such as UBE2V1 and UBE2V2 did not show E3 interactions. Although they contain a UBC-fold that could potentially interact with E3s, they lack the catalytic cysteine residue required for ubiquitin acceptance (Deng et al, 2000). Some E2s are involved in the conjugation of ubiquitin-like proteins, such as UBE2M(UbcH12) and UBE2F(NCE2) for NEDD8 and UBEI(Ubc9) for SUMO (Dye and Schulman, 2007); (Kerscher et al, 2006; Huang et al, 2009). In addition, UBE2O(E2-230K) and BIRC6(apollon) did not interact with E3s. Both BIRC6 and UBE2O are thought to contain a chimeric E2/E3 domain, which would obviate the need for an exogenous E3 ligase (Berleth and Pickart, 1996). Other E2s failed to interact with E3s in our screen, like the human homologs of the yeast Rad6p, UBE2A(hHr6A) and UBE2B(hHr6B). A possible explanation is that the Rad6p is phosphorylated at serine 120 by CDK-1 and -2 (Sarcevic et al, 2002) and this modification increases the E2 activity. Residue Ser120 is conserved in Rad6p homologs, but it is unclear whether the modification increases the binding between the E2 and the E3 or whether it influences the activity. Another E2, UBE2C(UbcH10), is required for degradation of mitotic cyclins and cell-cycle progression. Recently, the N-terminal extension of UBE2C, which is missing in our construct, has been implicated in regulating substrate ubiquitination and the number of lysines ubiquitinated in these substrates through interaction with the APC E3 ligase (Summers et al, 2008).
Some E3 enzymes were more connected than others. Strikingly, CADPS2 showed the highest number of interactions, but it has not been annotated as an E3 enzyme yet. This observation could indicate the identification of a new or deviant domain that could interact with E2 enzymes and potentially be involved in ubiquitination. Within the E3s with the highest number of E2 interactions, members of two closely related proteins families were found. The RNF130, RNF167 and RNF150 proteins belong to the Goliath family and are involved in apoptosis and embryonic development (Bouchard and Cote, 1993; Anandasabapathy et al, 2003; Guais et al, 2006). These E3s showed a pattern of overlapping E2 interactions. In addition, ZNRF1, ZNRF2 and ZNRF4 were found among the highest interactors. The ZNRF proteins shared many interactions with the same E2s. These two protein families indicate that conserved E3s are clustered together to interact with the same panel of many E2s.
Heterodimeric complexes, such as BRCA1–BARD1, MDM2–MDMX and BMI1–Ring1B, represent a special group of E3 enzymes. In those cases, one RING provides the complex with E3 activity, but only in association with the partnering RING. The active RING serves as a binding subunit for E2 enzymes, but the function of the other RING is often unclear. This situation is not represented in our two-hybrid set-up. In a recent Y2H study, the BRCA1–BARD1 RING domains were fused and tested for E2 interactions (Christensen et al, 2007). In this set-up, the E2 proteins UBE2D1, -D2, -D3, -E1, -E2, -E3, -I, -K, -L3, -N and W scored positive. In contrast, we only observe interactions with UBE2J2, -U and -W for the isolated BRCA1 RING domain in the LacZ read-out. This clearly indicates that screening with fused RING dimers yields different results from screening with isolated RING domains. It remains, however, unclear what the underlying molecular determinants of these differences are and a potential role for the inactive dimerization partners needs to be established.
We have shown earlier that the CNOT4 D48K E49K mutant has lost its interactions with UBE2D1-4(UbcH5A-D), UBE2E1(UbcH6) and UBE2E3(UbcH9) (Winkler et al, 2004). We show here that the RING-finger of CNOT4 is able to interact with UBE2W and that the mutant also loses this interaction. These observations indicate that the residues within UBE2D1-4, UBE2E1, UBE2E3 and UBE2W required for RING interactions are coupled with respect to their involvement in determining RING specificity. The CNOT4 double mutant, however, gained a stronger signal with UBE2N(Ubc13) when compared with WT CNOT4. From this point of view, the RING-interacting residues in UBE2N are likely to mediate binding in an opposite manner, gaining a stronger RING association. Unexpectedly, residues at position 63 are not highly conserved between the CNOT4-interacting E2 domains, indicating a more complex recognition mode. The mutant RING-finger displays a specific interaction with the UBE2D2 K63E mutant, but not with the WT UBE2D2, altering the specificity of interaction. When testing the UBE2D2 K63E mutant, it gained an interaction with NFX1. In addition, the change in E3 interaction pattern of UBE2D2 indicates that, although the E2–E3 interface is conserved, individual residues in this interface have different contributions in achieving interaction specificity among different pairs of E2s and E3s. Molecular modelling and docking approaches of E2–E3 pairs may provide more insight into the specific nature of these interactions. It is generally regarded that the UBC-fold of the E2 and the RING-finger domain of the E3s represent the interaction surfaces for E2–E3(RING) interactions (Joazeiro et al, 1999; Dye and Schulman, 2007). However, in some cases residues outside these domains contribute as exemplified by the UBE2L3(UbcH7)-c-Cbl structure. The RING domain of c-Cbl packs onto the tyrosine kinase-binding (TKB) domain and a connecting linker harbours several E2-interacting residues (Zheng et al, 2000). The absence of the TKB domain and linker can explain why no c-Cbl interactions were recovered in our interaction screen. Another example is the Hsp70-interacting protein (CHIP), which exclusively interacts with the UBC-fold of UBE2D1(UbcH5A) through its RING-type U-box domain (Xu et al, 2008). No CHIP interactions were recovered in our screen. The false negatives of our screen as exemplified by c-Cbl and CHIP can be related to the absence of interaction residues outside the E3(RING) domain, misfolding and/or mislocalization of the E3s in our screen. Nevertheless, we describe 295 novel E2–E3 interactions and we identify 66 E3(RING) proteins as novel E2 interactors. Together, this represents an important step forward in our knowledge of the E2 and E3 proteins of the human ubiquitin system.
In a first attempt to understand E3 specificities in directing different types of ubiquitin linkages we focussed on the UBE2D2(UbcH5b)–UBE2N(Ubc13) E2 pair. Substitution of residues in the E3 interaction surfaces of the K63-specific UBE2N yielded the P5L/I9H mutant (Figure 8). This mutant in the N-terminal helix of UBE2N gained five E3 interactions shared with UBE2D2 (specific for K48 linkages directing proteosomal degradation) and two unique interactions. UBE2N is catalytically active as a heterodimeric E2 with UBE2V1(Mms2) or UBE2V2. Importantly, the crystal structure of yeast Ubc13/MMS2 indicates that the P5L/I9H mutations will not interfere with heterodimer formation of UBE2N (VanDemark et al, 2001). Thus, the P5L/I9H mutant of UBE2N may redirect K63- to K48-linked poly-ubiquitin chains on a selected set of UBE2D2 substrates on expression in cells. We expect that the UBE2N–UBE2D2 pair represents only the first example and that the E2–E3 interaction data harbour many more possibilities for re-designing ubiquitin pathways.
Topology of the E2–E3 network
It is clear that E2–E3 interactions are not following a normal distribution. Both E2s and E3s showed a small number of highly connected proteins, followed by a larger group of enzymes with lesser interactions and so on. The identified highly interacting E2 and E3 proteins share many structural features within their complete UBC-fold and the E3-interacting interface residues. These high levels of sequence identity allow these hubs to interact with certain clusters of RINGs. For some of these hubs, their biological function is expected to involve redundancy. Evolutionary duplication of, for example, members of the UBE2D(UbcH5)-subgroup allows these E2s to interact with the same set of RING-fingers and, possibly, to ubiquitinate the same substrates. E2 enzymes that are more divergent in their interface sequence, such as UBE2N, have a different E3 interaction profile and are interacting with a different subcluster of RING E3s.
Proteins with many connections are holding large sub-networks together, forming complete, interconnected networks (Jeong et al, 2001). Deletion of these nodes has been shown to be associated with network robustness, pointing out that many hub proteins are essential for remaining the integrity of the network and the complete system (Jeong et al, 2001). In the E2–E3 network, UBE2U, members of the UBE2D1-4(UbcH5A-B) family, UBE2E1(UbcH6) and UBE2E3(UbcH9), UBE2N and UBE2G2 behave as hub proteins. Inactivation of these E2 proteins could potentially have dramatic contributions to large regions of the E2–E3 network and to cellular ubiquitination. As only a few substrates have been identified yet, we evaluated the in silico effect of deletion of hub E2s, by counting the numbers of E2s that become unconnected. Deletion of UBE2U, UBE2G2 or UBE2N has dramatic effects on the number of unconnected E3s, indicating that these hubs are indeed holding the E2–E3 network together. Strikingly, two of these three hubs have specific biological roles, UBE2N is involved in K63-chain synthesis and UBE2G2 is an ER-resident E2 involved in ERAD. In contrast, deletion of any of the UBE2Ds or UBE2E1 has little effect on the network; all E3s that remain connected are already bound to a closely related E2 enzyme. The observation that members of the UBE2D1–4 family, are biologically implicated in the degradation of abnormally folded and short-lived proteins, could be related to the expansion of the UBE2D family. Indeed, both the UBE2D1–4 orthologs UBC4 and UBC5 in Saccharomyces cerevisiae are functionally redundant and deletion impairs growth, temperature tolerance, protein misfolding and stress responses (Seufert and Jentsch, 1990). Interestingly, deletion of E2s known to conjugate UBLs, like UBE2I (for SUMO) and UBE2M or UBE2F (for NEDD8), does not give rise to any unconnected E3, indicating that these E2s are less important for maintaining the integrity of the ubiquitin network.
Physical E2–E3 interactions form the basis of ubiquitination activity and are required for an E2–E3 pair to become functional. By comparing the Y2H data with E2–E3 pairs that are reported to be functional in ubiquitination, we showed that the physical interactions among hub proteins for the majority of E2–E3 pairs correlate very well with reported biochemically active E2–E3 pairs. In addition, comparing all detected Y2H interactions or those that are having only one or more interactions was in good agreement with literature-curated E2–E3 pairs. These comparisons indicate that the ability of an E2–E3 pair to be functional in ubiquitination reactions is based on determinants of physical interaction. In addition, physical interactions could be used to predict the functionality of an E2–E3 interaction. The independent validation of a subset of E2–E3 in GST-pull-down-binding experiments provides a good coverage of the found Y2H interactions and, combined with the above-mentioned, indicates a high quality of the E2–E3 interactions determined by Y2H.
The E2–E3 network can be fitted into the overall organization of the ubiquitination process. The two known ubiquitin E1s are activating ubiquitin and interact with the E2s to transfer ubiquitin in a strictly hierarchical organization (Haas et al, 1982; Jin et al, 2007). Then the E2–E3 interactions take place in their specific manner and finally the ubiquitin is transferred to the E3-bound substrate (Dye and Schulman, 2007). Although no global information about E3-substrate specificity is available, some well-characterized E3s have been shown to mediate ubiquitination of multiple substrates (Tanaka et al, 2004; Boulton, 2006; Lin and Mak, 2007). This stresses the need for genome-wide analysis of interactions between E3 enzymes and their substrates. However, this may be hampered by the fact that for many E3s and substrates the binding sites are poorly characterized, a small amount of conservation among these sites is observed and the potential need for the post-translational modifications before E3 recognition of the substrate. Besides this, the E1–E2–E3-substrate ubiquitination super network can be regarded as a system that combines both hierarchical and the highly interconnected properties of E2–E3 interactions.
Taken all together, we report a high-quality framework centred on the functional domains of E2 and E3 enzymes in the human ubiquitination system. Exhaustive screening of E2–E3 interactions provides insight into the arrangement of proteins in this complex biological system. These binary interactions can be used to initiate follow-up studies concerning the structural aspects of E2–E3 pairs, such as modelling and docking of E2–E3 interactions to gain more insight into the underlying specificity. Although not all of these interactions will occur physiologically, for example because there are barriers in space and time preventing the coming together of E2s and E3s, these results provide a highly detailed, global network of E2–E3 interactions and their role in ubiquitination as a whole system. The interactions obtained with the mutant E2 could initiate new lines of synthetic ubiquitin research aimed at designed modulation of the system. Together, these experimental interactions provide a rich source of information and can be used to guide future research in ubiquitination and E2–E3 selectivity.
Materials and methods
Plasmid procedures
Full length or EST cDNAs of human ubiquitin-conjugating enzymes and human RING-type E3 ubiquitin protein ligases were obtained from the Deutsches Ressourcenzentrum fur Genomforschung GmbH (RZPD, Berlin, Germany). PCR primers (Biolegio, Nijmegen, the Netherlands) were used to amplify the E2 UBC-fold and the E3 RING-finger catalytic domains (including 10 N-terminal and 10 C-terminal residues) and these domains were sub-cloned either in pEG202-NLS or in the pJG4-5 plasmid to generate E2 or RING constructs fused to the C-terminus of the Escherichia coli LexA (DNA-binding domain) and B42 (AD) proteins, respectively. cDNAs encoding E2 enzymes were sub-cloned in pGEX-4T-1 to generate GST fusions. Mutagenesis was performed using QuickChange II Site-Directed mutagenesis (Stratagene), according to the manufacturer's instructions. The mammalian expression plasmid myc-Mdm2 has been described earlier (van der Horst et al, 2006). All constructs were completely verified by DNA sequencing. Additional information concerning cDNAs, amplification primers and cloning procedures is available on request.
Antibodies
Monoclonal antibodies recognizing LexA(2–12), GST(B14), MDM2(SMP-14) and p53(DO1) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and an HRP-coupled anti-penta-his antibody was purchased from Qiagen (Qiagen, Venlo, the Netherlands). Monoclonal antibodies against HA(12CA5) and CNOT4(19A20) were purified from hybridomas as reported elsewhere (Winkler et al, 2004).
Yeast manipulation and array generation
Growth and manipulation of the parental yeast strain EGY48 (MAT α, his3, trp1, ura3, 6LexAop–LEU2) was carried out as described before (Winkler et al, 2004). Mating type changing was carried out by using HO-endonuclease as described earlier (Burke et al, 2000). To generate a standardized array of yeast cells expressing all LexA–E2 fusions, three individual verified DB–E2 bacterial shuffle plasmids were co-transformed together with the LacZ reporter plasmid (LexAop)8 pSH18-34 in EGY48α, whereas AD-RING fusions were transformed in EGY48a by using a LiOAc/DMSO transformation in 96-well plates (Gietz and Woods, 2002). Transformants were selected on solid synthetic complete (SC) medium without histidine (H) and uracil (U) in the case of EGY48α (LexA–UBC/pSH18-34) or without tryptophan (W) for EGY48a (B42–RING) by incubation at 30°C for 2–3 days. Correct transformation with the appropriate fusion protein was verified using colony PCR as described before (Burke et al, 2000). In the E2 array, transformants of the EGY48α strains were arranged in a square of four spots, LexA–E2s were spotted in three independent biological transformants, combined with a transformant expressing LexA alone.
Semi-high-throughput yeast two-hybrid analysis
Individual EGY48a transformants bearing B42–RING fusions were inoculated in liquid SC W− medium, grown overnight at 30°C shaken at 230 r.p.m. Liquid cultures were transferred on solid SC W− by manual pinning in 384 format using manual pinning tools (VP384F), library copiers (VP381) and colony copiers (VP380) from V & P Scientific, Inc. (San Diego, CA, USA) and grown overnight to increase the amount of cells used for mating. In parallel, LexA–E2-transformed EGY48α cells were pinned on solid SC HU−. Mating was performed by pinning the LexA–E2 and the B42–RING transformants at exactly the same position on solid, non-selective YPD medium for 24 h at 30°C. Diploids were selected at 30°C for 48 h on SC HWU− plates. To visualize putative E2–E3 interactions, pre-selected diploids were transferred to read-out plates. For colorimetric selection, diploids were pinned on SC HWU− plates supplemented with X-Gal, and for auxotrophic selection, diploids were transferred to SC HWUL− plates. Both types of read-out were assessed under both inducing conditions, with galactose as the sole carbon source as well as under repressing conditions (with glucose as carbon source). Plates were incubated for a period of 24–96 h and pictures of the yeast colonies were taken at fixed intervals.
Spot quantification analysis
Digital images of yeast plates to quantify yeast spot size and blue staining intensity were carried out systematically at fixed intervals by using a standardized digital camera set-up. Spot size and blue staining intensity were quantified using a earlier reported Java-based spot-size measuring software algorithm (Collins et al, 2006), according to the developer's instructions and using an in-house written grid-based blue intensity measure algorithm, respectively.
Western blot verification of protein expression
To confirm correct protein expression, total protein lysates were prepared (Kushnirov, 2000). Briefly, individual, freshly pinned yeast colonies were grown in appropriate medium, overnight in 96-well plates at 30°C under continuous shaking. Cells were centrifuged, re-suspended in 0.1 M NaOH and incubated for 5 min at room temperature. Next, cells were centrifuged and resuspended in 1 × sample buffer, boiled for 5 min and centrifuged. Standard, proteins were separated on a 17.5% SDS–PAGE gel, transferred to nitrocellulose membranes and probed with mouse anti-HA or anti-LexA antibodies and visualization was carried out with ECL according to the supplier's instructions (Pierce).
Recombinant proteins
Full length or regions encoding the UBC-fold of individual E2s were subcloned in pGEX-4T-1 and expressed as GST–E2 fusion proteins in E. coli BL21(DE3) as described earlier (Winkler et al, 2004). Briefly, freshly transformed colonies were grown to mid-log phase in LB-medium, induced with 0.4 mM isopropyl-D-thiogalactopyranoside (IPTG) for 3 h at 30°C, and lysed in 25 ml ice-cold lysis buffer (300 mM KCl, 50 mM Tris–HCl pH 8.0, 2 mM EDTA 0.1% Triton X-100, 20% sucrose) containing 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), protease inhibitor cocktail (Roche) and 25 mg/ml lysozyme. After freeze thawing and sonification, lysates were centrifuged at 50 000 r.p.m. for 1 h at 4°C. Crude lysates were stored at −80°C. Constructs expressing recombinant human his–thioredoxin–RING fusions in the pLICHIS vector backbone were expressed in E. coli BL21(DE3) and protein expression except that IPTG induction was overnight at 24°C in LB-medium supplemented with 100 μM ZnCl2.
Mammalian cell culture
Human embryonic kidney cells (HEK293T) and human osteosarcoma cells (U2OS) were maintained in DMEM supplemented with glutamine, penicillin/streptomycin and 10% FBS under 5% CO2 at 37°C. 293T cells were grown to 60–70% confluency and transfections were carried out using FuGENE6 according to the suppliers' instructions with myc-tagged MDM2; 24 h after transfection, cells were washed twice with ice-cold PBS, lysed in RIPA (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM DTT, 0.5 mM PMSF and 1 μg/ml aprotonin, leupeptin and pepstatin), cleared by centrifugation at 14 kr.p.m. at 4°C. U2OS cells were treated with or without 20 μM MG132 for 4 h before lysis in RIPA-buffer.
GST-pull-down assays
GST–E2 proteins were immobilized on gluthathione-agarose beads (Sigma) and incubated with 5 μg of His6-CNOT4-N78/His6-CNOT4-N227 or U2OS or 293T lysates for 2 h at 4°C in G/A-buffer (50 mM potassium phosphate pH 6.6, 50 mM KCl, 0.1% NP-40, 10 μM ZnCl2, 0.5 mM PMSF, 1 mM DTT, 1 μg/ml aprotonin, leupeptin and pepstatin (All Sigma)). GST–E2–RING pull-down analysis was performed by incubation of GST–E2 proteins on beads with his–RING bacterial lysate (approximately 5 μg of RING-fusion protein) for 3 h at 4°C. Beads were washed three times with G/A-buffer, bound proteins resolved by SDS–PAGE and visualized with immunoblotting.
Data set
The protein interactions from this publication have been submitted to the IMEx (http://imex.sf.net) consortium through IntAct (Kerrien et al, 2007) and assigned the identifier IM-9597.
Note added in proof
Although the revised version of this manuscript was under review, Markson et al published a global screen of human E2–E3 interactions using full-length cDNAs in the Gal4-based Y2H system (Markson et al, Genome Res. 2009, doi:10.1101/gr.093963.109).
Supplementary Material
Overview of human ubiquitin-conjugating enzymes
Overview of human RING-type E3 ligases
Overview of binary human E2-E3 interactions
Overview of all background-corrected E2-E3 interactions detected on LacZstaining
Overview of all background-corrected E2-E3 interactions detected on LEU2-auxotrophy
Overview of observed E2-E3 interactions compared with literature-curated, biochemically functional interactions
Acknowledgments
The authors thank Adrien SL Melquiond and members of the Timmers laboratory for helpful discussions. This work was funded by support from the Netherlands Proteomic Center (NPC) and the Netherlands Society for Scientific Research (700.50.034).
Footnotes
The authors declare that they have no conflict of interest.
References
- Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, Skrenta H, Hollenhorst M, Fathman CG, Soares L (2003) GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity 18: 535–547 [DOI] [PubMed] [Google Scholar]
- Araki T, Milbrandt J (2003) ZNRF proteins constitute a family of presynaptic E3 ubiquitin ligases. J Neurosci 23: 9385–9394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley HC, Robinson PA (2005) E3 ubiquitin ligases. Essays Biochem 41: 15–30 [DOI] [PubMed] [Google Scholar]
- Berleth ES, Pickart CM (1996) Mechanism of ubiquitin conjugating enzyme E2-230K: catalysis involving a thiol relay? Biochemistry 35: 1664–1671 [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14: 10–21 [DOI] [PubMed] [Google Scholar]
- Bouchard ML, Cote S (1993) The Drosophila melanogaster developmental gene g1 encodes a variant zinc-finger-motif protein. Gene 125: 205–209 [DOI] [PubMed] [Google Scholar]
- Boulton SJ (2006) BRCA1-mediated ubiquitylation. Cell Cycle 5: 1481–1486 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 21: 873–880 [DOI] [PubMed] [Google Scholar]
- Burke D, Dawson D, Stearns T (2000) Methods in yeast genetics, a cold spring harbor laboratory course manual
- Burroughs AM, Jaffee M, Iyer LM, Aravind L (2008) Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J Struct Biol 162: 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM (2006) The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA 103: 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol 14: 941–948 [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS (2006) A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol 7: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R (2004) Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure 12: 633–644 [DOI] [PubMed] [Google Scholar]
- Dye BT, Schulman BA (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol 14: 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florenes VA, Maelandsmo GM, Forus A, Andreassen A, Myklebost O, Fodstad O (1994) MDM2 gene amplification and transcript levels in human sarcomas: relationship to TP53 gene status. J Natl Cancer Inst 86: 1297–1302 [DOI] [PubMed] [Google Scholar]
- Fujii T, Tamura K, Copeland NG, Gilbert DJ, Jenkins NA, Yomogida K, Tanaka H, Nishimune Y, Nojima H, Abiko Y (1999) Sperizin is a murine RING zinc-finger protein specifically expressed in Haploid germ cells. Genomics 57: 94–101 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Golemis EA, Serebriiski I, Finley RL Jr, Kolonin MG, Gyuris J, Brent R (2001) Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Mol Biol Chapter 20: Unit 20.1. [DOI] [PubMed] [Google Scholar]
- Guais A, Siegrist S, Solhonne B, Jouault H, Guellaen G, Bulle F (2006) h-Goliath, paralog of GRAIL, is a new E3 ligase protein, expressed in human leukocytes. Gene 374: 112–120 [DOI] [PubMed] [Google Scholar]
- Haas AL, Warms JV, Hershko A, Rose IA (1982) Ubiquitin-activating enzyme. mechanism and role in protein-ubiquitin conjugation. J Biol Chem 257: 2543–2548 [PubMed] [Google Scholar]
- Haldeman MT, Xia G, Kasperek EM, Pickart CM (1997) Structure and function of ubiquitin conjugating enzyme E2-25K: the tail is a core-dependent activity element. Biochemistry 36: 10526–10537 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Nakayama KI (2003) U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun 302: 635–645 [DOI] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA (2009) E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell 33: 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (1999) Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science 286: 1321–1326 [DOI] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN (2001) Lethality and centrality in protein networks. Nature 411: 41–42 [DOI] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW (2007) Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447: 1135–1138 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC (1999) The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase.[see comment]. Science 286: 309–312 [DOI] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B et al. (2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kushnirov VV (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF et al. (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3: e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Mak TW (2007) The role of E3 ligases in autoimmunity and the regulation of autoreactive T cells. Curr Opin Immunol 19: 665–673 [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell 9: 945–956 [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K (2002) Specificity and stability in topology of protein networks. Science 296: 910–913 [DOI] [PubMed] [Google Scholar]
- Momand J, Jung D, Wilczynski S, Niland J (1998) The MDM2 gene amplification database. Nucleic Acids Res 26: 3453–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Roth JA, Mukhopadhyay T (2000) Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol Cell Biol 20: 9391–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh YM, Kwon YE, Kim JM, Bae SJ, Lee BK, Yoo CH, Deshaies RJ, Seol JH (2009) Chfr is linked to tumour metastasis through the downregulation of HDAC1. Nat Cell Biol 11: 295–302 [DOI] [PubMed] [Google Scholar]
- Ozkan E, Yu H, Deisenhofer J (2005) Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA 102: 18890–18895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C (2007) The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 26: 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sadakata T, Kakegawa W, Mizoguchi A, Washida M, Katoh-Semba R, Shutoh F, Okamoto T, Nakashima H, Kimura K, Tanaka M, Sekine Y, Itohara S, Yuzaki M, Nagao S, Furuichi T (2007) Impaired cerebellar development and function in mice lacking CAPS2, a protein involved in neurotrophin release. J Neurosci 27: 2472–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarcevic B, Mawson A, Baker RT, Sutherland RL (2002) Regulation of the ubiquitin-conjugating enzyme hHR6A by CDK-mediated phosphorylation. EMBO J 21: 2009–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP (2004) Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem 279: 42169–42181 [DOI] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408: 381–386 [DOI] [PubMed] [Google Scholar]
- Seufert W, Jentsch S (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, Suzuki T (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25: 302–305 [DOI] [PubMed] [Google Scholar]
- Silver ET, Gwozd TJ, Ptak C, Goebl M, Ellison MJ (1992) A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J 11: 3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers MK, Pan B, Mukhyala K, Jackson PK (2008) The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell 31: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Suzuki T, Hattori N, Mizuno Y (2004) Ubiquitin, proteasome and parkin. Biochim Biophys Acta 1695: 235–247 [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073 [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C (2001) Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105: 711–720 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Albert TK, Dominguez C, Legtenberg YI, Boelens R, Timmers HT (2004) An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J Mol Biol 337: 157–165 [DOI] [PubMed] [Google Scholar]
- Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S (2008) Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of human ubiquitin-conjugating enzymes
Overview of human RING-type E3 ligases
Overview of binary human E2-E3 interactions
Overview of all background-corrected E2-E3 interactions detected on LacZstaining
Overview of all background-corrected E2-E3 interactions detected on LEU2-auxotrophy
Overview of observed E2-E3 interactions compared with literature-curated, biochemically functional interactions