Abstract
Asparagine-linked protein glycosylation is essential for the virulence of the human gut mucosal pathogen Campylobacter jejuni. The heptasaccharide that is transferred to proteins is biosynthesized via the glycosyltransferase-catalyzed addition of sugar units to an undecaprenyl diphosphate-linked carrier. Genetic studies on the heptasaccharide assembly enzymes have shown that PglH, which transfers three terminal N-acetyl-galactosamine (GalNAc) residues to the carrier polyisoprene, is essential for chick colonization by C. jejuni. While it is now clear that PglH catalyzes multiple transfer reactions, the mechanism whereby the reactions cease after the addition of just three GalNAc residues has yet to be understood. To address this issue, a series of mechanistic biochemical studies was conducted with purified native PglH. This enzyme was found to follow a processive mechanism under initial rate conditions, however, product inhibition and product accumulation led to PglH release of intermediate products prior to complete conversion to the native ultimate product. Point mutations of an essential EX7E sequence motif were used to demonstrate that a single active site was responsible for all three transferase reactions, and a homology model with the mannosyltransferase PimA, from Mycobacteria smegmatis, establish the requirement of the EX7E motif in catalysis. Finally, increased binding affinity with increasing glycan size is proposed to provide PglH with a counting mechanism that does not allow the transfer of more than three GalNAc residues. These results provide important mechanistic insights into the function of the glycosyl transfer polymerase that is related to the virulence of C. jejuni.
Keywords: Campylobacter jejuni, asparagine-linked glycosylation, N-linked glycosylation PglH, glycosyl transferase
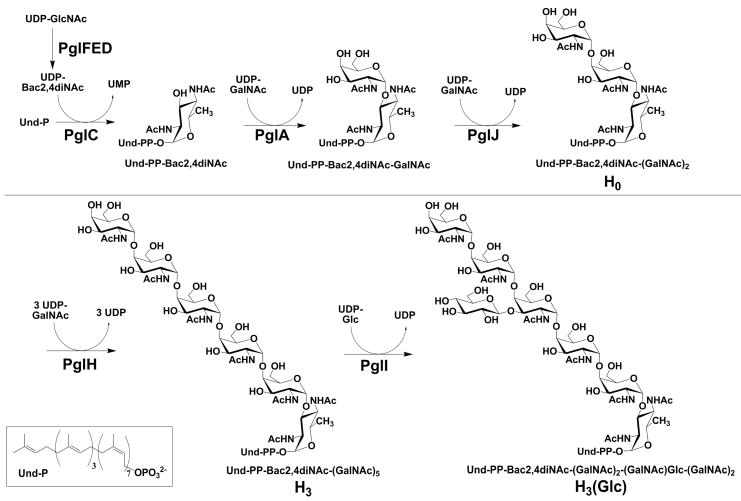
Asparagine-linked glycosylation (N-linked glycosylation) is an essential protein modification that entails the formation of a glycosylamide bond between a glycan and a target asparagine in protein substrates. The addition of an N-linked glycan is required for the function of numerous proteins, including those that are related to virulence in the gram-negative enteropathogen Campylobacter jejuni (C. jejuni) (1–9). C. jejuni, a human-gut mucosal pathogen, is implicated in gastroenteritis and is the leading cause of food-borne illness in North America (10, 11). The enzymes PglC, A, J, H and I (Fig. 1) are involved in oligosaccharide donor assembly for C. jejuni N-linked glycosylation in strain 11168. Deletion of the genes encoding several of the Pgl proteins lead to a loss in chick colonization efficiency, and therefore a decrease in the virulence of the organism (12–15). Importantly, C. jejuni strains with a mutated PglH gene have a significantly reduced ability to adhere to and invade human epithelial Caco-2 cells in vitro as well as chicks (14). Therefore, understanding the activity of the enzymes that build the glycan may lead to new therapeutic strategies for treating C. jejuni infections, or methods for mitigating C. jejuni contamination of food stocks (14, 16).
Figure 1.
Campylobacter jejuni polyisoprene-linked oligosaccharide biosynthesis. It is important to note that the stereochemistry of the bacterial undecaprenol double bonds are 2-trans to 8-cis rather than the 3-trans to 7-cis shown for the plant derived undecaprenol used here.
The C. jejuni N-linked glycan structure is formed through a series of sequential glycosyl transfer reactions from uridine diphosphate (UDP)-activated sugars to an undecaprenyl diphosphate carrier (Fig. 1) (13, 18, 19). The structure of the N-linked glycan including the anomeric stereochemistry of the glycan was solved by Young and co-workers utilizing NMR spectroscopy (17). The enzymes responsible for building the polyisoprene-linked heptasaccharide have previously been heterologously expressed, isolated, and functionally characterized (18–22). The first step in the polyisoprene-linked glycan assembly is catalyzed by PglC, which transfers N,N′-diacetylbacillosamine (Bac2,4diNAc, 2,4-diacetamido-2,4,6-trideoxyglucopyranose) phosphate from UDP-Bac2,4diNAc to undecaprenyl phosphate (Und-P) (20). UDP-Bac2,4diNAc is biosynthesized from UDP-N-acetyl-glucosamine (UDP-GlcNAc) by the enzymes PglF, E and D (19, 20, 23). Once undecaprenyl-diphospho-Bac2,4diNAc (Und-PP-Bac2,4diNAc) is formed, sequential N-acetyl-galactosamine (GalNAc) transfer reactions are catalyzed by PglA, PglJ and PglH to provide Und-PP-Bac2,4diNAc-GalNAc, Und-PP-Bac2,4diNAc-(GalNAc)2 and Und-PP-Bac2,4diNAc-(GalNAc)5, respectively (20, 22). Interestingly, PglH, the focus of this study, acts as a polymerase adding three 1,4-linked GalNAc residues to the growing glycan (18–20, 22, 24). The last step in the biosynthesis of the polyisoprene-linked heptasaccharide is addition of a branching β-1,3-glucose (Glc) residue to the third GalNAc by PglI (22). Each of the polyiosprenoid-linked glycan products of the PglA, PglJ, PglH and PglI enzymes have been produced in vitro then characterized by 2-aminobenzamide fluorescent labeling followed by MALDI-MS characterization (18). After completion of glycan assembly, PglK (WlaB), a putative flippase ABC transporter, transfers the Und-PP-linked glycan from the cytosolic to the periplasmic face of the bacterial inner membrane (25). Once the glycan is delivered to the periplasm, it is available as a substrate for the oligosaccharyl transferase, PglB (25). PglB catalyzes the N-linked glycosylation reaction, transferring Bac2,4diNAc-(GalNAc)2-GalNAc(Glc)-(GalNAc)2 from the Und-PP carrier to selected asparagine residues in target proteins (26).
Numerous glycosyltransferase proteins contain conserved EX7E sequence motifs including PglH and the recently crystallized and structurally characterized PimA, from Mycobacteria smegmatis. PimA catalyzes the transfer of mannose to the 2′-OH of phosphatidyl inositide. The crystal structure of PimA with bound UDP-mannose clearly shows that the E274, in the EX7E motif (residues 274–282), binds the 4′-OH of the mannosyl donor and the E282 binds the 2′- and 3′- OH of the ribose in the UDP moiety. In addition, the membrane-bound proteins, Alg2 and Alg11, that are involved in N-linked glycosylation in Saccharomyces cerevisiae catalyze multiple mannose transfer reactions and contain the conserved EX7E sequence motif (27). The eukaryotic and bacterial N-linked glycosylation pathways are similar in that they both assemble a glycan onto a membrane-bound isoprenoid carrier. In contrast to the Alg proteins, PglH does not require membranes for function and does not include a predicted and well-defined transmembrane domain (18).
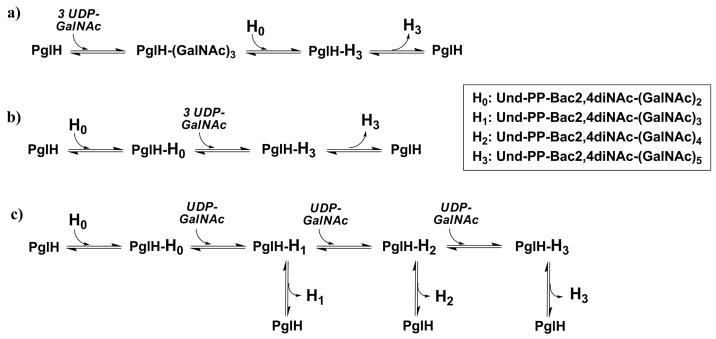
The mechanism by which PglH catalyzes specifically three glycosyl transfer reactions may involve three limiting models, which utilize either a single or multiple active sites. As illustrated in Fig. 2, these models are: a) a block transfer of a tri-GalNAc moiety to Und-PP-Bac2,4diNAc-(GalNAc)2, b) a processive mechanism in which the polyisoprene-linked substrate binds and is not released until the final product is formed or c) a three-step process in which each intermediate can readily dissociate and then rebind to the enzyme for subsequent reactions. In this report, we provide evidence for a sequential dissociative and processive reaction catalyzed by PglH utilizing a single enzyme active site. The results presented also suggest a mechanism of transfer that utilizes product inhibition to stop the enzyme from carrying out more than three GalNAc transfer reactions.
Figure 2.
Potential mechanisms for the PglH reaction. a) Block transfer of a tri-GalNAc moiety to H0, b) Processive mechanism in which intermediates do not dissociate from the enzyme, c) Dissociative mechanism in which intermediates freely dissociate and re-associate as substrates.
Materials and Methods
Common materials
The radioactive substrates and undecaprenol were purchased from American Radiolabeled Chemicals, Inc. The UDP-N,N′-diacetylbacillosamine was prepared as previously described (28) and the UDP-GalNAc was purchased from Sigma–Aldrich. Polyisoprenyl phosphates were prepared as described previously (29). The pure solvent upper phase (PSUP), modified from previous studies for optimal extraction of the hexasaccharide polyisoprene-linked product, was prepared by mixing 240 ml of H2O, 20 g of KCl and 240 ml methanol. The isoprenyl derivatives were separated on a normal-phase Varian Microsorb HPLC column using the following gradient at 1 mL/min: 0→3 min: 0%B; 3→5min: 0→20%B; 5→35min: 20→30%B; 35→60min: 30→45%B; 60→65min: 45→100%B; 65→70min: 100%B, where A was 4:1 chloroform/methanol and B was 10:10:3 chloroform/methanol/2M ammonium acetate. A LS6500 Beckman Scintillation Counter was used to determine the radioactivity present in the assay samples with Formula 989 (Beckman-Coulter) as the scintillation fluid. All reactions were performed in 100 μL unless noted otherwise. UDP-GalNAc was [3H] labeled on C-6 and [14C] labeled on the N-acetyl group.
Native PglH expression
The PglH gene containing a stop codon was ligated into a modified pET32 (Novagen) vector that encodes an N-terminal octahistidine-tag followed by a tobacco etch virus protease (TEV) cleavage sequence (30). BL-21 RIL cells (Stratagene) were transformed with the PglH encoding vector, and protein was expressed using a modified auto-induction method described by Studier (31). In this method, 1 mL of an overnight cell culture was added to expression media containing 30 μg/mL kanamycin in 1L of auto-induction media (0.1% w/v tryptone, 0.05% w/v yeast extract, 2 mM MgSO4, 0.05% v/v glycerol, 0.005% w/v glucose, 0.02% w/v α-lactose, 2.5 mM Na2HPO4, 2.5 mM KH2PO4, 5 mM NH4Cl, 0.5 mM Na2SO4). Cells were allowed to grow with shaking for three hours at 37°C. After three hours, the temperature was decreased to 16°C and the cells were left shaking overnight. Cells were harvested by centrifugation, washed with 0.9% NaCl and lysed in 50 mL of Buffer A [50 mM TrisOAc (pH 8), 200 mM NaCl, 1% Triton X-100, 20 mM imidazole]. Cell debris was removed by ultracentrifugation at 142,000 × g and the supernatant was mixed with 6 mL of 50% Ni-NTA (Qiagen) pre-equilibrated in buffer B [50 mM TrisOAc (pH 8), 200 mM NaCl, 20 mM imidazole]. The sample was left on a rotator for one hour at 4°C then poured into a column. The resin was washed (4 × 5 mL) with Buffer C [50 mM TrisOAc (pH 8), 200 mM NaCl, 50 mM imidazole]. His-TEV-PglH was then eluted with Buffer D [50mM TrisOAc (pH 8), 500 mM imidazole]. The 10 mL elution was dialyzed overnight in 4L of Buffer E [50 mM bicine (pH 8.5), 100 mM NaCl], then again for one hour in a fresh 4 L of Buffer E. Dialyzed protein was mixed with His-tagged TEV protease (prepared in house) in Buffer F [50 mM bicine (pH 8.5), 100 mM NaCl, 1 mM DTT, 0.5 mM EDTA]. The reaction was left for three hours at room temperature then overnight at 4°C. The resulting product was mixed with 2 mL of 50% Ni-NTA pre-equilibrated in Buffer B for 1 hour at 4°C to remove His-tagged TEV. The product was poured into a column and the flow through was collected. Three mL of Buffer B was added to the column and an additional 3 mL of flow through was collected. Product was confirmed through activity assays, SDS-PAGE, and Western-blot analysis in which the histidine-tag can be observed prior to cleavage but not after. The near native PglH protein was approximately 90% pure based on SDS-PAGE GelCode Blue (Pierce) stain analysis of the purified TEV cleavage product. PglJ and PglA were prepared in a T7-Pgl-His6 construct as described previously (18) except cells were grown using the above auto-induction method (18). PglH mutants were prepared using the primers shown in supporting figure 1 in a T7-Pgl-His or His-TEV-Pgl construct. E41A, E49A, E171A, E179A, E265A, E273A, E308A, E346A, E354A, D170A, D306A, D307A or R191A and wild type were prepared in the T7-PglH-His6 construct for comparison and the E316A mutant was prepared in the His8-TEV-PglH construct. All mutants were expressed using the auto-induction method above and purified as described above depending on the construct.
PglA product biosynthesis
Und-PP-Bac2,4diNAc-[3H]GalNAc was produced in a single pot reaction containing the following material in 200 μL (supporting Scheme 1): 50 mM TrisOAc (pH 8), 1% Triton X-100, 10 mM MgCl2, 10 mM ATP, 10% DMSO, 100 μM undecaprenol, 400 μM UDP-Bac2,4diNAc, 320 μM UDP-[3H]GalNAc (0.016 Ci/mmol), 1 μM PglA and 5 μL of Streptococcus mutans (S. mutans) kinase (32) and PglC prepared as cell envelope fractions (20). After two hours rocking at room temperature 400 μL of 2:1 chloroform/methanol was added to the reaction mixture. The organic layer was removed and the aqueous layer was washed again with 400 μL of 2:1 chloroform/methanol. Organic layers were combined and washed with 2 × 200 μL of PSUP. The organic layer was then dried with a stream of nitrogen and product was purified by normal phase HPLC. One mL HPLC fractions were collected and then 100 μL aliquots of each were dried under a stream of nitrogen. DMSO (200 μL) was added to the samples, vortexed, and mixed with 4.8 mL scintillation fluid. The retention time of radioactive PglA product was 30 minutes under these conditions. This retention time was identical to that of the product from a coupled PglC/PglA reaction with chemically synthesized undecaprenyl phosphate as described previously (29). For quantification of the polyisoprene-linked material, product was initially diluted into 100 μL of DMSO and sonicated, then the radioactivity of a 10 μL aliquot was measured. The final yield of pure product was approximately 60% based on the undecaprenyl-linked substrate.
PglJ product biosynthesis
Und-PP-Bac2,4diNAc-[3H]GalNAc-GalNAc (H0, Fig. 1) was prepared in a reaction mixture including the following components: 50mM bicine (pH 8.5), 2.5 mM DTT, 10 mM MnCl2, 0.04% Triton, and 10% DMSO, 320 μM UDP-GalNAc, Und-PP-Bac2,4diNAc-[3H]GalNAc (0.016 Ci/mmol, final concentration varied from 10–50 μM), and 1 μM PglJ. Product was extracted and purified as described for the PglA product. The retention time of H0 was 37 min under the conditions described above. Alternatively, PglJ was added to the PglC/A reaction mixture to obtain Und-PP-Bac2,4diNAc-([3H]GalNAc)2 (0.032 Ci/mmol). The final yield was 80% of the input H0.
PglH intermediate and product biosynthesis
Und-PP-Bac2,4diNAc-[3H]GalNAc-(GalNAc)2–4 (0.016 Ci/mmol H1, H2, and H3) was prepared using a reaction mixture identical to the PglJ product biosynthesis reaction except Und-PP-Bac2,4diNAc-[3H]GalNAc-GalNAc (H0; 0.016 Ci/mmol) was the substrate and 0.5 equivalents of UDP-GalNAc relative to Und-PP-Bac2,4diNAc-[3H]GalNAc-GalNAc (H0) was used to enhance formation of the intermediate products. PglH was added to provide a concentration of 100 nM. Product was isolated as described for the PglA and PglJ reactions and had the following retention times: H1 Und-PP-Bac2,4diNAc-[3H]GalNAc-(GalNAc)2: 47 min; H2 Und-PP-Bac2,4diNAc-[3H]GalNAc-(GalNAc)3: 53 min; H3 Und-PP-Bac2,4diNAc-[3H]GalNAc-(GalNAc)3: 59 min. Alternatively, PglJ and PglH were added to the PglA product synthesis reaction mixtures to give H1, H2 and H3 with 0.048, 0.064, and 0.08 Ci/mmol specific activity, respectively. For the synthesis of Und-PP-Bac2,4diNAc-([14C]GalNAc)3–4, 1 mM UDP-[14C] GalNAc (0.05 Ci/mmol) replaced UDP-[3H]GalNAc in the PglA reaction mixture. In addition, 1 μM PglJ and 0.1 μM PglH were also included in the mixture. The product was purified by NP-HPLC.
PglH Mutant assays
Assays were prepared with 78 nM Und-PP-Bac2,4diNAc-[3H]GalNAc-GalNAc and 250 nM UDP-[3H]GalNAc then initiated with 20 nM PglH wild type and mutants E41A, E49A, E171A, E179A, E265A, E273A, E308A, E346A, E354A, D170A, D306A, D307A or R191A in the T7-PglH-His6 construct or wild type and E316A in the His8-TEV-PglH construct. A 10 μL aliquot was removed after an overnight incubation at room temperature to determine total product using the organic/aqueous extraction described above.
PglH radiolabel kinetic and product inhibition assays
PglH reactions were prepared with the following components: 50mM bicine (pH 8.5), 2.5 mM DTT, 10 mM MnCl2, 0.04% Triton, and 10% DMSO, 250 nM UDP-[3H]GalNAc (20 Ci/mmol), unlabeled UDP-GalNAc (variable up to 50 μM), Und-PP-Bac2,4diNAc-[3H]GalNAc-(GalNAc)2–4 (variable from 5 nM to 800 nM, 0.016 Ci/mmol), PglH (variable concentration at a maximum of 1/3 the concentration of limiting polyisoprene-linked substrate). Aliquots of 10 μL were removed at various time points, and then partitioned between 200 μL of PSUP and 800 μL of chloroform/methanol (2:1). Reactions did not exceed 10% substrate turnover over the time courses used. The organic layer was dried and counted. Alternatively, PglH kinetic assays coupled with NP-HPLC were performed as described above, except 10 μL aliquots were removed, extracted and analyzed by NP-HPLC. PglH reaction intermediates were isolated by using the Pgl product extraction method described above with chloroform/methanol and PSUP. The organic layer was dried, dissolved in 4:1 chloroform/methanol and injected onto the NP-HPLC column. The product was eluted using the gradient described above. Fractions were collected (1mL) then dried using a stream of nitrogen. Residue from each fraction was counted and retention times were determined. The radioactivity from each fraction was counted and correlated to the known intermediate retention times. Product inhibition assays were performed in an identical manner to the kinetic assays with either 10 μM UDP or 94 nM Und-PP-Bac2,4diNAc-([3H]GalNAc)5 H3 (0.08 Ci.mmol).
Interference Assay
Und-PP-Bac2,4diNAc-([3H]GalNAc)2 (40 Ci/mmol) and Und-PP-Bac2,4diNAc-([14C]GalNAc)3–4(0.15 and 0.2 Ci/mmol) were prepared using the methods described above for PglA product preparation, except the UDP-[3H]GalNAc concentration was 5 μM (20 Ci/mmol) and PglJ (1 μM) was added to the mixture for the formation of Und-PP-Bac2,4diNAc-([3H]GalNAc)2. After NP-HPLC purification, 100 μL of each product and intermediate was separated into aliquots, then dried and stored at −20°C until needed. The Und-PP-Bac2,4diNAc-([3H]GalNAc)2 was dissolved in 100 μL of DMSO, sonicated and then a 10 μL aliquot was counted to determine the concentration using the known specific activity. Und-PP-Bac2,4diNAc-([14C]GalNAc)3–4 was dissolved in 20 μL of DMSO, sonicated, and then 10 μL was counted to determine the concentration. Und-PP-Bac2,4diNAc-([3H]GalNAc)2 (40 Ci/mmol), at a final reaction concentration of 75 nM, was added to 75 nM Und-PP-Bac2,4diNAc-([14C]GalNAc)3 (0.15 Ci/mmol) or Und-PP-Bac-([14C]GalNAc)4 (0.2 Ci/mmol). UDP-GalNAc was added at a final concentration of 32 μM. The reaction was initiated by the addition of PglH at a final concentration of 20 nM. After 2.5 (H1) or 20 (H2) minutes, 400 μL of 2:1 chloroform/methanol was added to stop the reaction. The organic layer was then immediately analyzed by NP-HPLC to determine the distribution of [14C] and [3H] label in the reaction product as described above for other polyisoprene-linked products.
Results
PglH utilizes a single active site for multiple GalNAc transfers
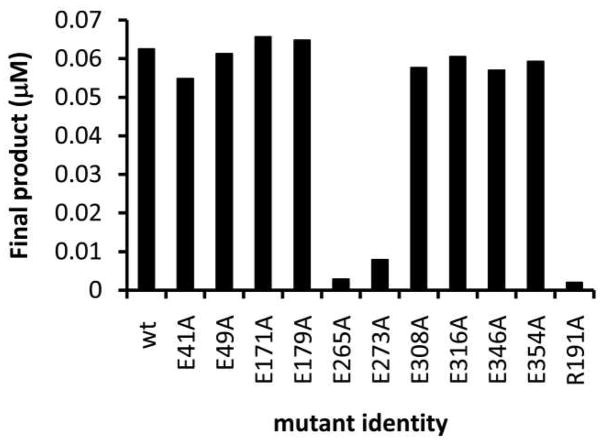
Sequence analysis of the Pgl proteins indicates that PglC, A, and J all contain two EX7E sequence motifs, while surprisingly, PglH contains five such motifs (Supporting Fig. 2) which were initially proposed to be required for multiple transfer reactions. The EX7E motif flanked by E265 and E273, is the most conserved among the Pgl proteins. PimA contains two EX7E motifs, one of which plays a role in GDP-Mannose binding (residues 274–282) and the other which has no known role in PimA catalysis (residues 157–165). The EX7E sequence motif that plays a role in GDP-Mannose binding aligns with E265 and E273 of PglH, therefore this sequence motif was anticipated to be critical for PglH function. The other motif (residues 157–165) does not align with any of the PglH EX7E motifs. To test the role of the EX7E sequences in PglH catalysis, alanine mutants were prepared in a T7-PglH-His6 or His8-TEV-PglH construct at each of the glutamate residues that flank the PglH EX7E motifs (E41, E49, E171, E179, E265, E273, E308, E316, E346 and E354), as described in material and methods. All of the mutant proteins expressed at levels typical of the wild type enzyme. The activity of the purified PglH mutants and wild type enzyme were assayed by measuring the incorporation of tritium from UDP-[3H]GalNAc into the Und-PP-Bac2,4diNAc-(GalNAc)2 (H0) substrate by organic/aqueous extraction followed by scintillation counting (Fig. 3; H0 biosynthesized as shown in the supporting Scheme 1 and materials and methods). Only the mutations at residues E265 and E273 had an effect on the total product formed by PglH. In overnight reactions the E265A and E273A mutants catalyzed the transfer of only trace levels of GalNAc to H0 (Fig. 3). In addition, when Und-PP-Bac2,4diNAc-(GalNAc)3 (H1), or Und-PP-Bac2,4diNAc-(GalNAc)4 (H2) isolated as described below were used as substrates, no product was observed with the E265A or E273A mutant (data not shown). However, total product turnover was identical to wild type under the same conditions with all the other EX7E alanine mutants (E41, E49, E171, E179, E308, E316, E346 and E354) whether H1, H2 (data not shown) or the native substrate H0 was used (Fig. 3). Normal phase (NP) HPLC analysis confirmed that Und-PP-Bac2,4diNAc-(GalNAc)5 (H3) was formed with all of the active mutants of PglH (data not shown).
Figure 3.
PglH utilizes the highly conserved EX7E motif flanked by residues E265 and E273 for catalysis of all reactions. Reactions containing the indicated PglH mutants (20 nM) were prepared and the amount of product determined for an overnight reaction with H0 (68 nM) and UDP-[3H]GalNAc (20 Ci/mmol; 250 nM).
PimA was found to have 14.5% identity and 29.3% similarity to PglH. The ESyPred3D homology modeling software (33) was able to thread the sequence of PglH into the structure of PimA (34), showing several interesting relationships between the enzymes (supporting Fig. 3). First, the EX7E motif flanked by E265 and E273 lies at the hypothesized active site of PglH and may fulfill the same role as in PimA. None of the other EX7E motifs appear at or near the active site in the model protein, consistent with near wild-type activity of these EX7E mutants. Additional fully-active alanine mutations of D170, D306 and D307 were spatially remote from the proposed active site. In PimA there are a number of arginine residues in the active site that are critical for function. In the threaded PglH model, R191 is predicted to lie in the same region as the E265 and E273 residues at the face of the proposed active site. To test the active site model, an alanine mutant of R191 was prepared and the activity of the expressed protein was analyzed. As predicted by the model, activity in the R191A mutant was abolished (Fig. 3). These results correlate directly with the PimA studies, and strongly suggest that the same site of the protein is required for catalyzing the addition of all three GalNAc residues. These results are also consistent with studies of the bi-functional enzyme Alg11 in the eukaryotic Asn-linked oligosaccharide biosynthesis pathway in which a single EX7E motif is involved in multiple glycosyl transfers (27).
PglH does not follow a block transfer mechanism
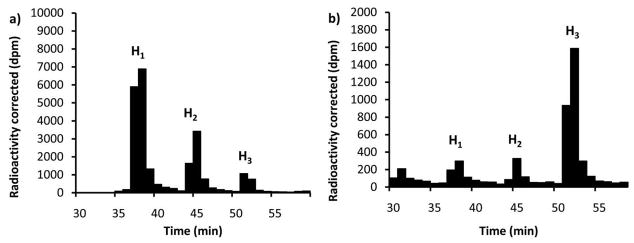
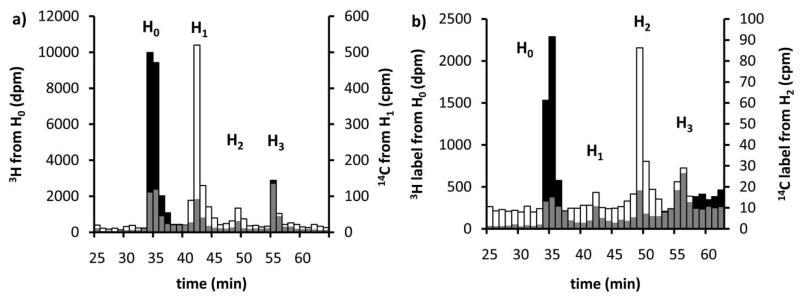
Of the three potential mechanisms illustrated in Fig. 2 only the block transfer would not produce intermediate tetra- and penta- saccharide-linked polysioprenes. In order to determine whether intermediate glycans are formed and released in the PglH reaction, native PglH was prepared and mixed with H0 and UDP-[3H]GalNAc. The total reaction product was then analyzed by NP-HPLC followed by scintillation counting. As shown in Fig. 4, when 17:1 excess H0 (4.3 μM) to UDP-GalNAc (0.25 μM) was allowed to react overnight, the first polyisoprene-linked intermediate H1 was more than two times the concentration of the second intermediate H2 and nine times the concentration of the final product H3 (Fig. 4a). This result ruled out the block transfer mechanism shown in Fig. 2 leaving only the processive and dissociative mechanisms as a possibility. Interestingly, when the concentration of polyisoprene-linked substrate was decreased, to provide an equimolar ratio of H0 (0.26 μM) to UDP-GalNAc (0.25 μM), only 20% of the product was H1 and H2 while the remaining product was H3 (Fig. 4b). Surprisingly, even with a high concentration of H0 (4.3 μM) and seven-fold excess UDP-GalNAc (28 μM) the major product was the H3 (45%) form of the glycan while 31% was H1 and 25% was H2 (data not shown). To further demonstrate that the tritium-labeled NP-HPLC purified intermediates, which had retention times consistent with the addition of one and two GalNAc units to H0, were indeed the PglH intermediates, each was converted to H3 quantitatively by further incubation with PglH and fresh UDP-GalNAc (data not shown). These results indicated that the concentration of substrates had a major influence on the total amount of intermediate and product formed in a PglH reaction, and that the reaction does not utilize a trisaccharide block transfer mechanism.
Figure 4.
The concentration of substrate determines the distribution of products and intermediates in a PglH reaction. PglH reactions were prepared with either a) 4.3 μM Und-PP-Bac2,4diNAc-[3H]GalNAc-GalNAc (H0) and 0.25 μM UDP-[3H]GalNAc or b) 0.26 μM Und-PP-Bac2,4diNAc-[3H]GalNAc-GalNAc (H0) and 0.25 μM UDP-[3H]GalNAc. Each column represents the amount of radioactivity in a 1 mL fraction at the corresponding time corrected for the number of [3H]GalNAc units incorporated into each. Note that with high polyisoprene-linked substrate relative to UDP-[3H]GalNAc the majority of the product is the first intermediate (H1), and when the substrates are approximately equimolar the major product is the final product (H3). The identity of H1and H2 were based on consistent normal phase HPLC retention times of the tritium-labeled PglH products and quantitative conversion of the isolated intermediate to H3 upon further incubation with PglH.
PglH follows a processive and dissociative mechanism
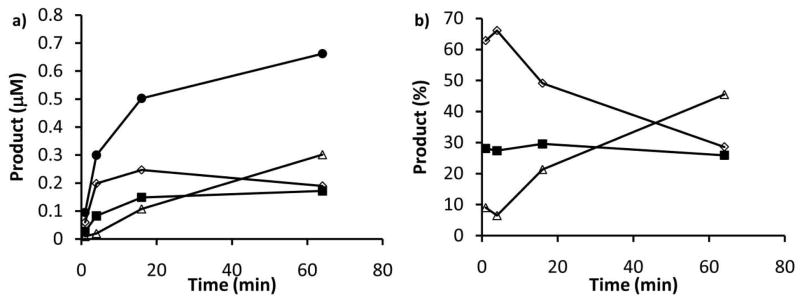
The PglH initial reaction rate with H0 is likely to be represented by a complex combination of H0, H1 and H2 reaction rates. To test this possibility we prepared PglH reactions, then removed aliquots at specific times and separated the products by NP-HPLC. The separated products were then counted and the amount of each polyisoprene-linked intermediate present in the initial aliquot was calculated based on the amount of radioactivity associated with each HPLC peak (Fig. 5a). Interestingly, as shown in Fig. 5b, the percentage of H2 was approximately 30% throughout the reaction, suggesting that a steady-state was reached with the formation of this material, and the rate limiting step of the reaction likely involves either conversion of H2 to H3 or release of the H3 product. It is important to note that within the first minute very little H3 was detected; therefore, the initial rate would include all of the steps in the reaction, but would be dominated by the first reaction. Additionally, under these conditions the H1 and H2 levels reach a maximum then do not fluctuate appreciably over the full hour of the experiment. However, H3 does continue to form throughout the time course. These data suggest that H1 and H2 can be formed early in the process and dissociate from the enzyme, yet the major product is H3.
Figure 5.
A distribution of products and intermediates are formed in the PglH reaction. PglH reactions were prepared with 28 μM UDP-GalNAc and 4 μM Und-PP-Bac2,4diNAc-(GalNAc)2 (H0) and aliquots were extracted at the given time points then analyzed by NP-HPLC. a) The quantity of each intermediate was calculated based on their distribution as analyzed by NP-HPLC and the total counts in the reaction. Product concentration is provided at the indicated time points for total (closed circles), H1 (open diamond), H2 (closed squares) and H3 (open triangle). b) The percentage of the total product was calculated for each time point and plotted H1 (open diamond), H2 (closed squares) and H3 (open triangle). Note that H2 stays at the same percentage throughout the reaction and that H1 and H2 levels sharply increase initially, but then stabilize as H3 is the major product. Also, note that the lines are drawn for illustrative purposes only.
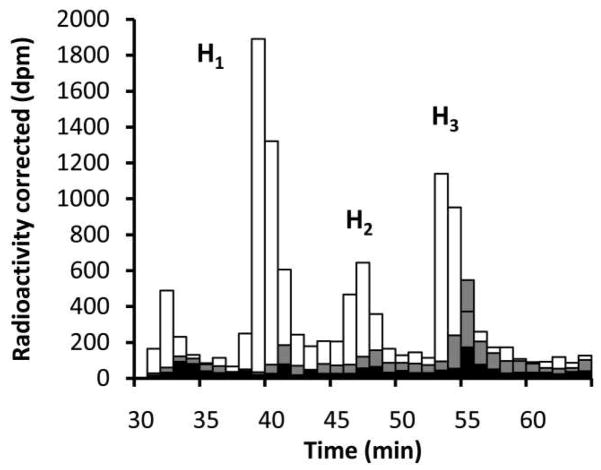
As outlined in Fig. 2, PglH was expected to follow one of the three potential mechanisms. While H3 was the major product under the conditions of the above experiment, it was not clear when H1 was formed in the large relative quantities as shown in Fig. 4a. In the above reaction the UDP-GalNAc concentration was four-times that of the polyisoprene and would not be expected to produce significant amounts of H1 relative to H3. To determine when the H1 product was predominantly formed, a reaction was prepared with 3 μM H0 and 0.25 μM UDP-GalNAc. Aliquots were extracted at several time points, including overnight, and the resulting intermediate distribution was analyzed. As shown in Fig. 6, at 8 and 32 minutes the final product H3 made up approximately 52% of the total product formed, while H1 made up less than 22%. However, after the reaction was allowed to continue overnight the major product was the H1 intermediate, which was 48% of the total final product, while H3 only made up 32%. Importantly, like the reaction shown in Fig. 5 the total H2 formed in the first hour never went above 30% of the total product. These results suggest that as more product was formed, the ability of the enzyme to carry the intermediates through to completion decreased. These results also show that in the initial stages, PglH is more processive and does not significantly release intermediates. Taken together, PglH follows a more processive mechanism under initial rate conditions, but as intermediates and products accumulate, the enzyme begins to release the intermediates prior to complete conversion.
Figure 6.
PglH is initially a processive enzyme that builds up product to a limit then intermediates form and dissociate. PglH reactions were prepared with 3 μM Und-PP-Bac2,4diNAc-(GalNAc)2 (H0) and 0.25 μM UDP-GalNAc. Reactions were quenched and extracted after 8 minutes (black), 32 minutes (gray) and overnight (white) then analyzed by normal phase HPLC. Each bar represents the total radioactivity associated with 1 mL fractions at the given time points. Radioactivity was corrected for the number of GalNAc units incorporated into each molecule. Note that the quantity of final product is high relative to the others initially, but at the end of the reaction the product is mostly the first intermediate.
PglH catalyzes a sequential transfer of GalNAc to the polyisoprene-linked oligosaccharide
The ability of PglH to utilize H0, H1, and H2 as substrates was measured using steady-state kinetics. The enzyme followed a Michaelis-Menten type of relationship between substrate concentrations and initial reaction rate with H0 and H1 even though the individual kinetic parameters should include more than one reaction. This is consistent with the results shown in figure 5b suggesting that in the initial phase of the reaction only a single reaction provides the major product when H0 was the substrate. At higher concentrations of H2 (>250 nM) the initial rate was reduced by more than an order of magnitude relative to the maximum rate at lower concentrations (100 nM). Therefore, at high concentrations H2 inhibits the reaction. As shown in Table 1, when the UDP-GalNAc concentration was held constant, the apparent kcatpolyisoprene and apparent Kmpolyisoprene decreased as much as four-fold with the H1 and H2 intermediates. Assuming that the Kmpolyisoprene represents the relative binding efficiency of the intermediates, the slow turnover with increasing glycan size may be due to a slow release of tightly bound product and intermediates. When the polyisoprene-linked substrate concentration was held constant and the UDP-GalNAc concentration was varied, as shown in Table 2, the KmUDP-GalNAc was the same with H0, H1 and H2. These results suggest that the binding of UDP-GalNAc was identical for all three glycans, which is consistent with a single UDP-GalNAc binding site model for the reaction.
Table 1.
PglH intermediate and substrate polyisoprene kinetics
| glycan | kcat (min−1)a |
Kmpolyisoprene (μM)a |
kcat/Kmpolyisoprene (μM−1*min−1)a |
|---|---|---|---|
| H0 | 3.4 ± 0.6 | 0.28 ± 0.08 | 12 ± 4 |
| H1 | 0.84 ± 0.08 | 0.09 ± 0.02 | 9 ± 2 |
| H2 | 0.7 ± 0.1 | 0.06 ± 0.02 | 12 ± 4 |
Steady-state kinetic measurements were performed with 1.05 μM UDP-GalNAc as described in the experimental section. H2 kinetics were measured at concentrations below 100 nM H2. The values reported are from a minimum of eight substrate concentrations, and fit using the Michaelis-Menten equation. Kinetic parameters are apparent. The identity of H1and H2 were based on consistent normal phase HPLC retention times of the tritium-labeled PglH products and quantitative conversion of the isolated intermeidate to H3 upon further incubation with PglH.
Table 2.
PglH intermediate and substrate UDP-GalNAc kinetics
| glycan | kcat (min−1)a |
KmUDP-GalNAc (μM)a |
kcat/KmUDP-GalNAc (μM−1*min−1)a |
|---|---|---|---|
| H0 | 4.4 ± 0.2 | 2.6 ± 0.3 | 1.7 ± 0.2 |
| H1 | 0.9 ± 0.1 | 2.3 ± 0.7 | 0.4 ± 0.1 |
| H2 | 1.74 ± 0.04 | 2.6 ± 0.2 | 0.67 ± 0.05 |
Steady-state kinetic measurements were performed with 0.13 μM polyisoprene-linked substrates as described in the experimental section. The values reported are from a minimum of eight substrate concentrations, and fit using the Michaelis-Menten equation. Kinetic parameters are apparent. The identity of H1and H2 were based on consistent normal phase HPLC retention times of the tritium-labeled PglH products and quantitative conversion of the isolated intermediate to H3 upon further incubation with PglH.
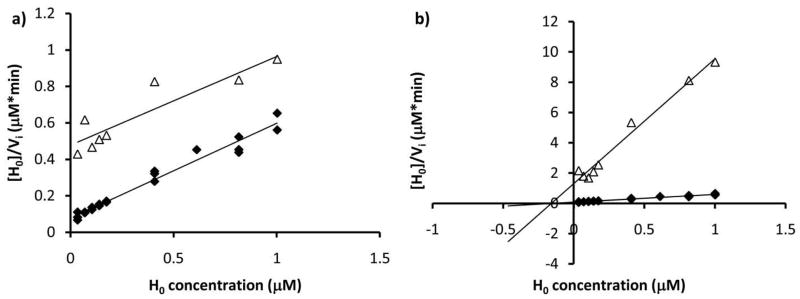
To determine the order of substrate binding and inhibition activity of the PglH reaction intermediates and products, the ability of UDP and H3 to inhibit PglH was measured. Based on the diagnostic parallel and intersecting lines shown in the Fig. 7 Hanes-Woolf plots, H3 inhibition was clearly competitive with H0 (Fig. 7a), and UDP was a noncompetitive inhibitor with respect to H0 (Fig. 7b). These results suggest that H0 binds first to the enzyme followed by UDP-GalNAc. UDP is then released from the active site and H1 remains bound to the enzyme or dissociates and re-associates for the next step. Importantly, in the presence of 250 nM UDP-GalNAc the Ki measured for H3 against H0 was 20 nM, which was 4-fold lower than the Kmpolyisoprene of H0 under identical conditions (Kmpolyisoprene = 80 nM at 250 nM UDP-GalNAc). In addition, UDP was a much weaker inhibitor as the apparent competitive and noncompetitive components of the Ki were in the μM range. These results are consistent with the kinetic analysis that suggests that the binding affinity is enhanced with increasing GalNAc units.
Figure 7.
PglH follows a sequential ordered path of substrate addition. PglH reactions were prepared with variable H0 alone (closed diamonds) or with a) 94 nM H3, (open triangles) or b) 10 μM UDP (open triangles) and the initial rates were determined then analyzed by Hanes-Woolf plots. Note that the H3 product inhibition and H0 lines are parallel indicating competitive inhibition and that the UDP product inhibition lines intersect at the x-axis suggesting noncompetitive inhibition.
PglH intermediates are competitive with the initial substrate
From the above experiments it was not clear whether intermediates that dissociate from PglH can re-associate in the presence of H0. The catalytic efficiency (kcat/Kmpolyisoprene) shown in Table 1 is an indicator of how well the intermediates compete as substrates for the enzyme. However, in situ formed intermediates and exogenously added intermediates may not have similar kinetics. To determine whether the intermediates were competitive with H0, uniquely labeled H1 or H2 intermediates were introduced in H0 reactions and the ability of the intermediates and substrates to compete were assayed. In order to determine if the intermediates could compete with H0, [14C]-labeled H1 and H2 were enzymatically synthesized and isolated. The [14C]-labeled H1 and H2 were then mixed with [3H]-labeled H0, UDP-GalNAc, and PglH (supporting Scheme 2). NP-HPLC analysis of the reaction products after several minutes (Fig. 8) showed clearly that the [14C]-labeled H1 and H2 were able to compete with H0. In addition, the amount of total product when [14C]-labeled H1 or H2 were present with H0 was considerably lower than when only the H0 substrate was present (data not shown). Reactions that required only five minutes for complete consumption of H0 were not complete even after 30 minutes when equimolar H1 or H2 were present. The decrease in total product formation suggested that the intermediates acted as inhibitors of the overall reaction with H0. Therefore, H1 and H2 were likely competing for the same active site.
Figure 8.
PglH intermediates can compete with the natural substrate of the enzyme. Tritium-labeled Und-PP-Bac2,4diNAc-(GalNAc)2 (H0) and [14C]-labeled Und-PP-Bac2,4diNAc-(GalNAc)3 (H1) and Und-PP-Bac2,4diNAc-(GalNAc)4 (H2) were enzymatically synthesized then mixed with PglH. H1 or H2 were equimolar with H0 (75 nM). The reactions were stopped after 2.5 minutes with H1 or 20 minutes with H2 and the distribution of products was analyzed by NP-HPLC. Each bar represents the number of counts detected in 1 mL fractions at the given time and the [14C] label is associated with the right-hand y-axis while tritium is associated with the left-hand y-axis. Overlap between the [3H] and [14C] are shown in gray. a) [3H]-labeled H0 (black) and [14C]-labeled H1 (white) b) [3H] labeled H0 (black) and [14C] labeled H2 (white) NP-HPLC traces. Note that the distribution of products is the same with the [14C] and [3H]-labeled substrates.
Discussion
The results presented in this report suggest a mechanism in which PglH is able to stop catalysis after the formation of the hexasaccharide product, rather than continuing to catalyze the transfer of more GalNAc units. The mutagenesis, modeling, UDP-GalNAc kinetics and competition results all suggest that only one active site is involved in catalysis by PglH. With only a single active site, the substrate must slip through this site as the same amino acid residues catalyze the transfer of additional GalNAc units to the elongating glycan. The steady-state polyisoprene kinetics, product inhibition and product distribution analyses all suggest that as the glycan size increases the binding affinity increases. An increase in binding affinity of the intermediates and eventually the final product would lead to a decrease in the rate of transfer as was evident with the decrease in kcat with larger glycans. This decrease in the reaction rate could be due to an increasing difficulty for the glycan to slip through the active site for additional transfer reactions. This culminates in the formation of H3, which based on the product inhibition studies is an extremely potent inhibitor of the PglH reaction, with a low Ki relative to the Kmpolyisoprene of the initial substrate. Therefore the increasing binding affinity with increasing glycan size leads to a maximum number of GalNAc residues incorporated into the PglH product, and serves as a counting mechanism for the enzyme.
The initial rate of the H1 and H2 reaction was considerably slower than that of the H0 reaction (Table 1). The H1 intermediate was the major product of reactions with high H0 concentrations relative to UDP-GalNAc (fig 4a and 6). Since the H0 reaction is fast relative to the H1 reaction this provides more time for the H1 intermediate to dissociate from the enzyme. Under conditions of high H0 concentration, H0 would then be able to rapidly displace this intermediate allowing for another round of H1 formation. Under conditions of increasing H3 concentration similar results would occur displacing primarily the H1 intermediate due to how quickly it is formed relative to H2 and H3. These kinetic results provide a mechanistic explanation for the surprising intermediate distribution observed with high concentrations of the H0 substrate (fig 4a and 6).
Previous chick colonization studies with C. jejuni demonstrated that the minimal structural requirement for efficient N-linked glycosylation and C. jejuni colonization was the hexasaccharide backbone of the N-linked glycan (13). Deletion of the next enzyme in the pathway, PglI, leads to a decrease in the efficiency of N-linked glycosylation and cell colonization, but does not ablate the process (13, 17). The potent product inhibition demonstrated by H3 may be relieved in the presence of PglI. Since a flippase (Fig. 1) is proposed to function downstream of the PglI modification, it is likely that H3 inhibition is effectively removed by glucose branching followed by translocation of the PglI product into the periplasm. PglB (the oligosaccharyl transferase) and PglK (the putative flippase) have relaxed specificities for the glycan (26, 35, 36). PglH product inhibition, PglI removal and sequestration by PglK may provide a mechanism to avoid incorporation of incomplete glycans into target proteins. Therefore, PglH acts as a gatekeeper for N-linked glycosylation, which is essential for native efficient chick colonization (14, 37). While the importance of PglH has previously been demonstrated using genetic studies (14, 38), this report provides a biochemical rationale for the effects of PglH and possibly also PglI deletion from C. jejuni.
It is important to note that the PglH assays performed in this report utilize triton X-100 concentrations that are just above the critical micelle concentration. This concentration of detergent was required for optimal activity of PglH. Therefore it is possible that PglH acts at the interface of mixed-micelles containing the polyisoprenoid-linked substrate. While this does complicate the interpretation of the kinetic results shown in tables 1 and 2, it does not change the overall conclusions reported. It was clear from the experiments performed that the final product of the PglH reaction was a potent competitive inhibitor of PglH. It was also clear that mixtures of the intermediate polyisoprenoid-linked glycans were competitive with one another in a manner consistent with the kinetic results. In addition, the distribution of products in PglH reactions appeared to be dependent on relative ratios of substrates rather than the concentrations of substrate relative to detergent (which was the same in all assays). While we have not ruled out a role for interfacial interactions for understating the PglH reaction, the fact that catalysis occurs three sugar residues away from the polyisoprene may remove the dependence of these interactions on the activity of the enzyme. The PglH reaction may be promoted by interactions with other Pgl proteins as well as the interactions of those proteins with membranes or membrane mimics. Studies designed to understand the role of membranes and the other Pgl proteins including PglI are currently underway in our laboratory.
Supplementary Material
Acknowledgments
We thank Meredith Hartley, Angelyn Larkin, Dr. James Morrison and Dr. Nelson Olivier for critical reading and editing of this manuscript. We thank, Meredith Hartley for preparation of the S. mutans kinase. We also thank Mark Chen and Meredith Hartley for help in the biosynthesis of UDP-Bac2,4diNAc.
Abbreviations
- C. jejuni
campylobacter jejuni
- UDP
uridine diphosphate
- Pgl
protein glycosylation, N-linked asparagine-linked
- Und
undecaprenyl
- P
phosphate
- PP
diphosphate
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetyl-glucosamine
- Glc
glucose
- Bac2
4diNAc, 2,4-diacetamido-2,4,6-trideoxyglucopyranose
- Alg
Asparagine-linked glycosylation (eukaryotic)
- TEV
Tobacco Etch Virus
- PSUP
pure solvent upper phase
- NP-HPLC
normal phase high performance liquid chromatography
Footnotes
This work was supported by NIH grants GM039334 (B. I.) and GM080794 (to J.M.T.)
Supporting Information Available Biosynthetic Schemes for the interference assay and biosynthesis of the PglH substrate and intermediates, PglC, A, J and H amino acid sequences with highlighted EX7E motifs, Homology model of PglH threaded through PimA structure, and primers for PglH mutagenesis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sun J, Duffy KE, Ranjith-Kumar CT, Xiong J, Lamb RJ, Santos J, Masarapu H, Cunningham M, Holzenburg A, Sarisky RT, Mbow ML, Kao C. Structural and functional analyses of the human Toll-like receptor 3. Role of glycosylation. J Biol Chem. 2006;281:11144–11151. doi: 10.1074/jbc.M510442200. [DOI] [PubMed] [Google Scholar]
- 2.Clevestig P, Pramanik L, Leitner T, Ehrnst A. CCR5 use by human immunodeficiency virus type 1 is associated closely with the gp120 V3 loop N-linked glycosylation site. J Gen Virol. 2006;87:607–612. doi: 10.1099/vir.0.81510-0. [DOI] [PubMed] [Google Scholar]
- 3.Standley S, Baudry M. The role of glycosylation in ionotropic glutamate receptor ligand binding, function, and trafficking. Cell Mol Life Sci. 2000;57:1508–1516. doi: 10.1007/PL00000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrecca K, Atanasiu R, Akhavan A, Shrier A. N-linked glycosylation sites determine HERG channel surface membrane expression. J Physiol. 1999;515(Pt 1):41–48. doi: 10.1111/j.1469-7793.1999.041ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres GE, Egan TM, Voigt MM. N-Linked glycosylation is essential for the functional expression of the recombinant P2X2 receptor. Biochemistry. 1998;37:14845–14851. doi: 10.1021/bi981209g. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Dang H, Patrick JW. Contributions of N-linked glycosylation to the expression of a functional alpha7-nicotinic receptor in Xenopus oocytes. J Neurochem. 1998;70:349–357. doi: 10.1046/j.1471-4159.1998.70010349.x. [DOI] [PubMed] [Google Scholar]
- 7.Warner JB, Thalhauser C, Tao K, Sahagian GG. Role of N-linked oligosaccharide flexibility in mannose phosphorylation of lysosomal enzyme cathepsin L. J Biol Chem. 2002;277:41897–41905. doi: 10.1074/jbc.M203097200. [DOI] [PubMed] [Google Scholar]
- 8.Chandra NC, Spiro MJ, Spiro RG. Identification of a glycoprotein from rat liver mitochondrial inner membrane and demonstration of its origin in the endoplasmic reticulum. J Biol Chem. 1998;273:19715–19721. doi: 10.1074/jbc.273.31.19715. [DOI] [PubMed] [Google Scholar]
- 9.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 10.Wooldridge KG, Ketley JM. Campylobacter-host cell interactions. Trends Microbiol. 1997;5:96–102. doi: 10.1016/S0966-842X(97)01004-4. [DOI] [PubMed] [Google Scholar]
- 11.Ketley JM. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143(Pt 1):5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 12.Szymanski CM, Michael FS, Jarrell HC, Li J, Gilbert M, Larocque S, Vinogradov E, Brisson JR. Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J Biol Chem. 2003;278:24509–24520. doi: 10.1074/jbc.M301273200. [DOI] [PubMed] [Google Scholar]
- 13.Kelly J, Jarrell H, Millar L, Tessier L, Fiori LM, Lau PC, Allan B, Szymanski CM. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J Bacteriol. 2006;188:2427–2434. doi: 10.1128/JB.188.7.2427-2434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlyshev AV, Everest P, Linton D, Cawthraw S, Newell DG, Wren BW. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology. 2004;150:1957–1964. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- 15.Wood AC, Oldfield NJ, O’Dwyer CA, Ketley JM. Cloning, mutation and distribution of a putative lipopolysaccharide biosynthesis locus in Campylobacter jejuni. Microbiology. 1999;145(Pt 2):379–388. doi: 10.1099/13500872-145-2-379. [DOI] [PubMed] [Google Scholar]
- 16.Szymanski CM, Burr DH, Guerry P. Campylobacter protein glycosylation affects host cell interactions. Infect Immun. 2002;70:2242–2244. doi: 10.1128/IAI.70.4.2242-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young NM, Brisson JR, Kelly J, Watson DC, Tessier L, Lanthier PH, Jarrell HC, Cadotte N, St Michael F, Aberg E, Szymanski CM. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 18.Glover KJ, Weerapana E, Imperiali B. In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc Natl Acad Sci U S A. 2005;102:14255–14259. doi: 10.1073/pnas.0507311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, Aebi M. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 20.Glover KJ, Weerapana E, Chen MM, Imperiali B. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2006;45:5343–5350. doi: 10.1021/bi0602056. [DOI] [PubMed] [Google Scholar]
- 21.Glover KJ, Weerapana E, Numao S, Imperiali B. Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem Biol. 2005;12:1311–1315. doi: 10.1016/j.chembiol.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weerapana E, Glover KJ, Chen MM, Imperiali B. Investigating bacterial N-linked glycosylation: synthesis and glycosyl acceptor activity of the undecaprenyl pyrophosphate-linked bacillosamine. J Am Chem Soc. 2005;127:13766–13767. doi: 10.1021/ja054265v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenhofen IC, McNally DJ, Vinogradov E, Whitfield D, Young NM, Dick S, Wakarchuk WW, Brisson JR, Logan SM. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J Biol Chem. 2006;281:723–732. doi: 10.1074/jbc.M511021200. [DOI] [PubMed] [Google Scholar]
- 24.Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- 25.Linton D, Allan E, Karlyshev AV, Cronshaw AD, Wren BW. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol Microbiol. 2002;43:497–508. doi: 10.1046/j.1365-2958.2002.02762.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen MM, Glover KJ, Imperiali B. From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry. 2007;46:5579–5585. doi: 10.1021/bi602633n. [DOI] [PubMed] [Google Scholar]
- 27.O’Reilly MK, Zhang G, Imperiali B. In vitro evidence for the dual function of Alg2 and Alg11: essential mannosyltransferases in N-linked glycoprotein biosynthesis. Biochemistry. 2006;45:9593–9603. doi: 10.1021/bi060878o. [DOI] [PubMed] [Google Scholar]
- 28.Olivier NB, Chen MM, Behr JR, Imperiali B. In Vitro Biosynthesis of UDP-N,N′-Diacetylbacillosamine by Enzymes of the Campylobacter jejuni General Protein Glycosylation System. Biochemistry. 2006;45:13659–13669. doi: 10.1021/bi061456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B. Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2007;46:14342–14348. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 31.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Hartley MD, Larkin A, Imperiali B. Chemoenzymatic synthesis of polyprenyl phosphates. Bioorg Med Chem. 2008;16:5149–5156. doi: 10.1016/j.bmc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 34.Guerin ME, Kordulakova J, Schaeffer F, Svetlikova Z, Buschiazzo A, Giganti D, Gicquel B, Mikusova K, Jackson M, Alzari PM. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J Biol Chem. 2007;282:20705–20714. doi: 10.1074/jbc.M702087200. [DOI] [PubMed] [Google Scholar]
- 35.Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alaimo C, Catrein I, Morf L, Marolda CL, Callewaert N, Valvano MA, Feldman MF, Aebi M. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 2006;25:967–976. doi: 10.1038/sj.emboj.7601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect Immun. 2004;72:3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen JC, Szymanski C, Guerry P. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81–176. J Bacteriol. 2004;186:6508–6514. doi: 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.