Abstract
Extrarenal viral infections commonly trigger glomerulonephritis, usually in association with immune complex disease. The Ig component of immune complexes can activate glomerular cell Fc receptors, but whether complexed viral nucleic acids contribute to glomerular inflammation remains unknown. Because of the types of Toll-like receptors (Tlrs) expressed by glomerular mesangial cells, we hypothesized that viral single-stranded RNA and DNA would activate mesangial cells via Tlr-independent pathways and trigger overlapping antiviral immune responses. Consistent with this hypothesis, 5′-triphosphate RNA (3P-RNA) and non-CpG DNA activated murine primary glomerular mesangial cells to secrete Cxcl10 and Il-6 even in cells derived from mice deficient in the Tlr adaptor proteins Myd88 and Trif. Transcriptome analysis revealed that 3P-RNA and non-CpG-DNA triggered almost identical gene expression programs, especially the proinflammatory cytokine Il-6, several chemokines, and genes related to type I IFN. We observed similar findings in glomerular preparations after injecting 3P-RNA and non-CpG-DNA in vivo. These effects depended on the formation of complexes with cationic lipids, which enhanced nucleic acid uptake into the cytosol of mesangial cells. Small interfering RNA studies revealed that 3P-RNA recognition involves Rig-1, whereas non-CpG-DNA did not require Rig-1 or Dai to activate glomerular mesangial cells. We conclude that 3P-RNA and double-stranded DNA trigger a common, TLR-independent, antiviral response in glomerular mesangial cells, which may promote glomerulonephritis in the setting of viral infection.
Viral infections can induce de novo immune complex glomerulonephritis (e.g., hepatitis-C-virus-associated glomerulonephritis). Even more frequently, acute viral infections trigger disease activity of preexisting glomerular diseases, such as IgA nephropathy, lupus nephritis, or renal vasculitis.1 Infection of glomerular cells (i.e., viral glomerulitis) does not appear to account for most of such cases.1 So what are the molecular mechanisms that link viral infection to glomerular pathology?
Viral infection activates systemic antiviral immune responses that can contribute to glomerular disease by enhancing autoantibody production, immune complex deposition, or systemic interferon (IFN) production. Type I IFNs inhibit viral replication in infected cells and have pleiotropic immunomodulatory effects on macrophages, T cells, and natural killer cells.2 In the intravascular compartment, plasmacytoid dendritic cells are the main source of type I IFNs. Viral proteins activate plasmacytoid dendritic cells via Toll-like receptor (Tlr) 2 and 4 signaling from the cell surface.3 In addition, various shapes of viral nucleic acids activate dendritic cells in intracellular compartments. Viral double-stranded RNA (dsRNA) and U-rich single-stranded RNA activate Tlr3 and Tlr7 in intracellular endosomes, respectively.4–6 Viral CpG DNA ligates Tlr9 in the same compartment.7 More recently, viral RNA and DNA also were discovered to activate dendritic cells via TLR-independent recognition and signaling pathways that locate to the intracellular cytosol.3,8 Viral dsRNA and 5′-triphosphate RNA (3P-RNA) interact with retinoic-acid-inducible protein-1 (Rig-1) and melanoma-differentiation-associated gene-5 (Mda-5).9–12 Viral non-CpG-DNA recognition was shown to trigger type I IFN via the DNA-dependent activator of IFN regulatory factors (Dai), previously named Dlm-1 or Z-DNA-binding protein-1 (Zbp1), and Tank-binding kinase-1.13,14
Immune recognition of viral nucleic acids is not limited to antigen-presenting cells. For example, Tlr3-mediated recognition of viral dsRNA in pancreatic islet cells can trigger autoimmune pancreatic islet destruction via local production of Ifn-α.15 Viral dsRNA and double-stranded (dsDNA) both induce multiple antiviral genes in human hepatoma cells, both involving Rig-1 and mitochondrial antiviral signaling protein (Mavs).16 But whether local recognition of viral nucleic acids contributes to glomerular disease remains largely unknown.
We have recently shown that glomerular mesangial cells (MCs) express functional Tlr3 but lack Tlr7 or Tlr9 expression, which supports the endosomal recognition of viral dsRNA but not of viral 3P-RNA or CpG DNA.17,18 However, we previously have shown that transient systemic exposure to 3P-RNA or non-CpG DNA aggravates lupus nephritis in autoimmune MRL(fas)lpr mice via TLR-independent mechanisms.19 This phenomenon was associated with distinct effects of RNA and DNA on systemic autoimmunity (e.g., enhanced proliferation of autoreactive B and T cells by non-CpG-DNA and enhanced type I IFN signaling by 3P-RNA). We also could show that both nucleic acid formats shuttled to the glomerulus and colocalized with MCs.19 But whether MCs express the respective cytosolic RNA and DNA sensors and trigger antiviral immune responses upon exposure to 3P-RNA or non-CpG DNA remains unknown. We hypothesized that viral 3P-RNA and non-CpG DNA both trigger innate antiviral responses in glomerular MCs, including the release of type I IFN, and that this effect is mediated by different Tlr-independent recognition machineries in the intracellular cytosol.
Results
3P-RNA and Non-CpG-DNA Activate Mesangial Cells in an Intracellular Compartment
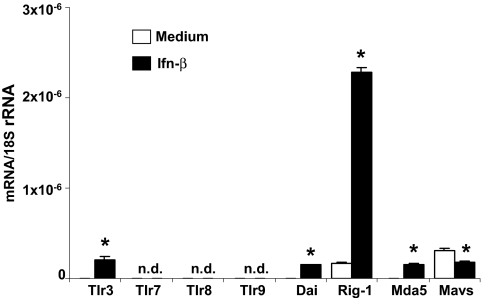
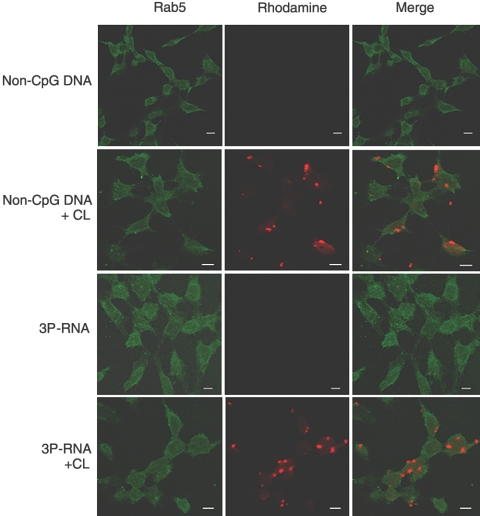
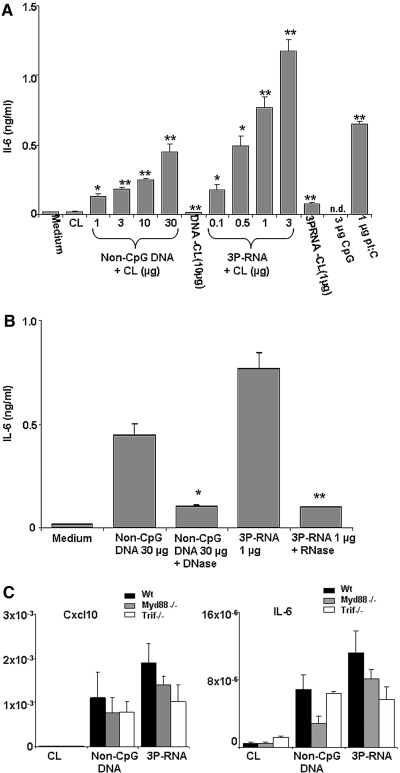
We prepared primary MCs (pMCs) from C57BL/6 mice and first determined the mRNA expression of the nucleic-acid-specific pattern recognition molecules Tlr3, Tlr7, Tlr8, Tlr9, Rig-1, Mda5, Dai, and Mavs. Under basal culture conditions pMCs expressed Rig-1 and Mavs but not Tlr3, Tlr7, Tlr8, Tlr9, Dai, or Mda5 mRNA (Figure 1). Prestimulation with Ifn-β induced Tlr3, Dai, Rig-1, and Mda5 mRNA but suppressed Mavs mRNA (Figure 1). In dendritic cells and embryonic fibroblasts 3P-RNA and non-CpG-DNA need to reach the intracellular cytosol (e.g., in cationic lipid (CL) complexes), before they can interact with their putative receptors.9,10,13,14,20 Consistently, rhodamine-labeled 3P-RNA or non-CpG DNA was barely detectable in the intracellular cytosol of pMCs unless complexed with CL as assessed by confocal microscopy (Figure 2). In the cytosol, 3P-RNA and non-CpG-DNA should be able to access their respective recognition receptors and activate pMCs. In fact, 3P-RNA and non-CpG-DNA both induced pMCs to produce Il-6 in a dose-dependent manner only when complexed with CL (Figure 3A). Pretreatment with RNAse or DNAse drastically reduced this effect, indicating that the immunostimulatory effect of these complexes derives from their RNA or DNA content, respectively (Figure 3, A and B).
Figure 1.
Ifn-β induces Tlr3, Dai, Rig-1, and Mda5 mRNA in pMCs. Total RNA was isolated from pMCs after 6 h of exposure to medium or 2000 U of Ifn-β. Real-time RT-PCR data for Tlr3, Tlr7, Tlr8, Tlr9, Dai, Rig-1, Mda5, and Mavs mRNA are expressed relative to 18S rRNA expression. Data are means ± SD from three experiments, each analyzed in duplicate. n.d., not detectable. *P < 0.05 versus medium.
Figure 2.
Cationic lipid enhances the uptake of non-CpG DNA and 3P-RNA in MCs. Primary MCs were exposed to either 1 μg of rhodamine-labeled 3P-RNA or 5 μg of rhodamine-labeled non-CpG-DNA in the presence or absence of CL for 2 h. Intracellular uptake was detected by confocal microscopy and appears as red staining inside of the MCs. Fluorescein-isothiocyanate-labeled anti-Rab5 was used to mark early endosomes of MCs and appears as green staining. Note that the intracellular uptake strongly increased when 3P-RNA or non-CpG-DNA were complexed with CL. Images are representative of three independent experiments. Original magnification, ×400; scale bar, 10 μm.
Figure 3.
3P-RNA and non-CpG-DNA complexed with CL induce Il-6 release in MCs. (A) Primary MCs were stimulated with increasing doses of 3P-RNA/CL and non-CpG-DNA/CL complexes. The highest doses also were tested in the absence of CL (−CL). PolyI:polyC RNA and CpG oligodeoxynucleotide 1668 were used as positive and negative controls, respectively. (B) In similar experiments, 3P-RNA/CL and non-CpG-DNA/CL were preincubated with either RNAse or DNAse. Supernatants were harvested after 24 h and analyzed by Il-6 ELISA. Data are means ± SD. *P < 0.05 versus medium; **P < 0.01 versus medium. (C) Myd88−/−, Trif mutant, and wild-type primary MC mRNA were isolated after 6 h of exposure to 3P-RNA/CL or non-CpG-DNA/CL. Real-time RT-PCR was performed for the indicated targets as described in Concise Methods. Data are expressed relative to 18S rRNA expression and are means ± SD from three experiments, each analyzed in duplicate.
Rig-1 Mediates 3P-RNA- but Not Non-CpG-DNA-Induced Activation of Mesangial Cells
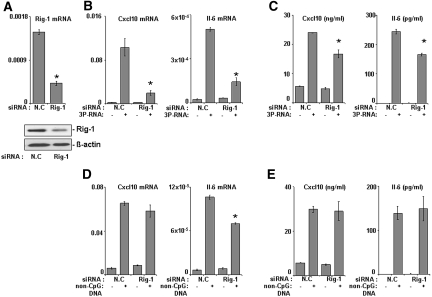
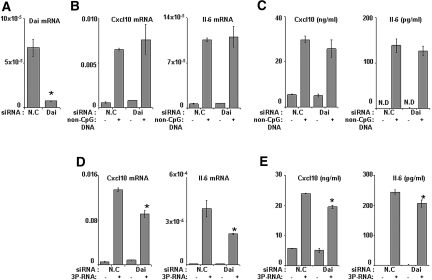
Consistent with multiple previous studies in other cell types 3P-RNA and non-CpG-DNA activated MCs via TLR-independent pathways because pMCs prepared from either Myd88-deficient (myeloid differentiation primary response gene 88), Trif-mutant (TIR-domain-containing adapter-inducing IFN-β), or wild-type mice showed comparable Cxcl10, Il-6, and Ccl5 induction upon 3P-RNA and non-CpG-DNA stimulation (Figure 3C and data not shown). The recognition of 3P-RNA was reported to involve Rig-1 in dendritic cells9,10; hence, Rig-1-specific small interfering RNA (siRNA) studies were carried out to confirm the role of Rig-1 in 3P-RNA recognition in MCs. Rig-1-specific siRNA significantly suppressed Rig-1 mRNA levels and protein levels (Figure 4A) and largely prevented Il-6 or Cxcl10 mRNA expression and protein secretion in MCs upon exposure to 3P-RNA (Figure 4, B and C). The 3P-RNA-induced cytokine release was not completely prevented by Rig-1 siRNA, which may relate to either other contributing signaling pathways or the technically incomplete knockdown of Rig-1. By contrast, knockdown of Rig-1 did not significantly impair the non-CpG-DNA-induced expression of Cxcl10 mRNA in MCs and had no effect on Cxcl10 release of MCs (Figure 4, D and E). Suppression of Rig-1 somewhat reduced the non-CpG-DNA-induced expression of Il-6 mRNA at 6 h, but this did not translate to the protein level during 24 h of stimulation (Figure 4, D and E). These data show that Rig-1 mediates 3P-RNA- but not non-CpG-DNA-induced activation of MCs.
Figure 4.
Rig-1 is responsible for 3P-RNA recognition in MCs. (A) Mouse MCs were transfected with Rig-1-specific siRNA or nonspecific control siRNA and subjected to real-time RT-PCR and Western blotting to evaluate the expression of Rig-1. (B) The mRNA expression of Cxcl10 and Il-6 in siRNA-treated MMCs was measured by real-time RT-PCR after 6 h of treatment with 3P-RNA. (C) Cxcl10 and Il-6 protein levels were measured by ELISA in siRNA-treated MMCs after 24 h of treatment with 3P-RNA. (D and E) The mRNA expression and protein levels of Cxcl10 and Il-6 were measured by real-time RT-PCR and ELISA, respectively, after treatment with non-CpG-DNA. All real-time RT-PCR data are expressed relative to 18S rRNA expression. Data are means ± SD from three experiments, each analyzed in duplicate. *P < 0.05 Rig-1 siRNA versus nonspecific control RNA.
Dai Contributes to 3P-RNA- but Not Non-CpG-DNA-Induced Activation of Mesangial Cells
Dai has been suggested to mediate the recognition of double-stranded B-DNA in vitro.13 But a recent study reported that mouse embryonic fibroblasts and bone-marrow-derived dendritic cells from Dai knockdown mice showed a considerable response to B-DNA compared with that of wild-type cells.14 To test the role of Dai in MCs, we used a Dai-specific siRNA that effectively suppressed Dai mRNA levels in MCs (Figure 5A). However, non-CpG-DNA-induced Cxcl10 or Il-6 mRNA expression and protein release were independent of Dai (Figure 5, B and C). Interestingly, suppression of Dai significantly reduced the 3P-RNA-induced expression of Cxcl10 and Il-6 mRNA and protein release in MCs (Figure 5, D and E). Thus, Dai contributes to 3P-RNA- but not non-CpG-DNA-induced activation of MCs.
Figure 5.
Dai is dispensable for non-CpG-DNA recognition in MCs. (A) Mouse MCs were transfected with Dai (Zbp1)-specific siRNA or nonspecific control siRNA and subjected real-time RT-PCR to evaluate the expression of Dai. (B) The mRNA expression of Cxcl10 and Il-6 in siRNA-treated MCs was measured by real-time RT-PCR after 6 h of treatment with non-CpG-DNA. (C) The Cxcl10 and Il-6 protein levels were measured by ELISA in supernatants of siRNA-treated MC after 24 h of treatment with non-CpG-DNA. (D and E) The mRNA expression and protein levels of Cxcl10 and Il-6 were measured by real-time RT-PCR and ELISA, respectively, after treatment with 3P-RNA. All real-time RT-PCR data are expressed as relative to 18S rRNA expression. Data are means ± SD from three experiments, each analyzed in duplicate. *P < 0.05 Dai siRNA versus nonspecific control RNA.
3P-RNA and Non-CpG-DNA Both Activate IFN-Regulated Factor-3 in Mesangial Cells
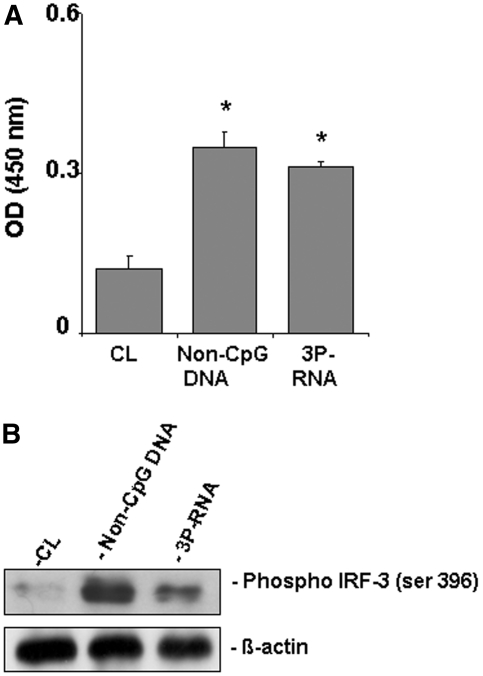
Innate RNA and DNA recognition receptors can specifically activate a group of transcription factors called the IFN regulatory factors (Irf).21 We isolated nuclear extracts from MCs after stimulation with 3P-RNA or non-CpG-DNA for 2 h. The 3P-RNA and non-CpG-DNA both increased the phosphorylation of Irf3, suggesting that despite different recognition machineries 3P-RNA and non-CpG-DNA both share Irf3 as a transcription factor to induce gene expression in glomerular MCs (Figure 6, A and B).
Figure 6.
3P-RNA and non-CpG-DNA both activated Irf3 in MCs. Mouse MCs were stimulated with 3P-RNA/CL and non-CpG-DNA/CL complexes. Nuclear extracts were harvested after 2 h and analyzed for phosphorylated Irf3 by ELISA and Western blotting. Data are means ± SD. *P < 0.05 versus CL.
3P-RNA and Non-CpG-DNA Trigger Proinflammatory Cytokines in Mesangial Cells
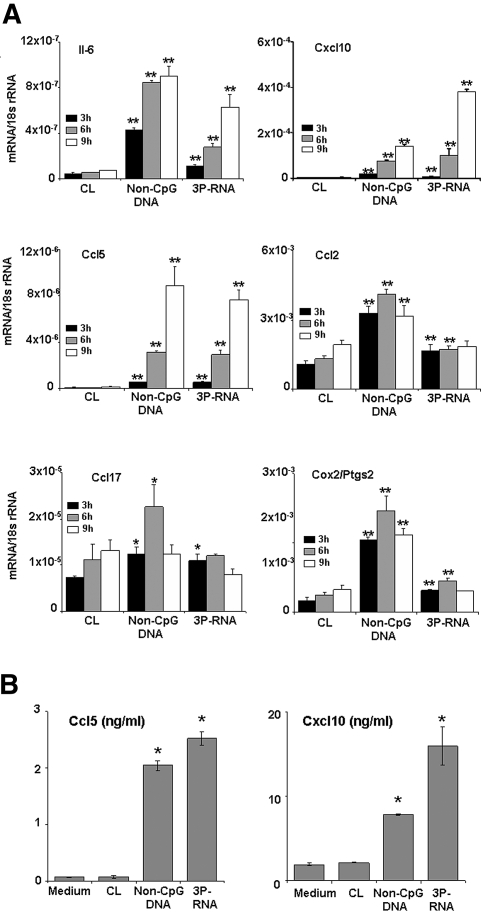
Shared Irf3 phosphorylation would propose that 3P-RNA and non-CpG DNA induce similar rather than different gene expression patterns in MCs. We used the Affymetrix mouse genome 430 2.0 array to characterize the pMC mRNA expression profiles upon stimulation with complexes of 0.5 μg/ml 3P-RNA/CL and 30 μg/ml non-CpG-DNA/CL, two doses that stimulated pMCs to secrete comparable levels of Il-6 within 24 h of stimulation (Figure 3A). A large number of genes were found to be coinduced by 3P-RNA/CL and non-CpG-DNA/CL; the 30 most coinduced genes are listed in Table 1. The Affymetrix gene array analysis revealed that the genes strongly coinduced by 3P-RNA/CL and non-CpG DNA/CL included Il-6 and multiple proinflammatory CC and CXC chemokines (i.e., Cxcl10/Ip10, Ccl5/Rantes, Ccl2/Mcp-1, Ccl7/Mcp-3, and Ccl17/Tarc) (Table 1). We confirmed the induction of these factors and that of Cox2/Ptgs2 in pMCs at 3, 6, and 9 h of 3P-RNA/CL or non-CpG-DNA/CL stimulation by real-time reverse transcription PCR (RT-PCR) (Figure 7A). Real-time RT-PCR also revealed that 3P-RNA and non-CpG-DNA induced these genes to a similar extent, as suggested by the microarray analysis. We also used real-time RT-PCR to confirm that 3P-RNA/CL and non-CpG DNA/CL did not induce Il-1-β, Il-2, Tnf-α, Tgf-β, and Hif-1α in pMCs as suggested by the Affymetrix gene array (data not shown). Do these mRNA expression data translate to the protein level? 3P-RNA/CL and non-CpG DNA/CL both induced pMCs to secrete Il-6, Ccl5/Rantes, and Cxcl10/Ip10 within 24 h (Figures 3A and 7B). Together, these data show that 3P-RNA and non-CpG DNA stimulate MCs to produce multiple proinflammatory cytokines and chemokines.
Table 1.
Thirty genes most induced by 3P-RNA and Non-CpG-DNA in MCs
| Gene Name | Affymetrix ID | Fold Change Non-CpG-DNA | Fold Change 3P-RNA |
|---|---|---|---|
| Ifit1 | 1450783_at | 199 | 14.5 |
| Cxcl10 | 1418930_at | 61.7 | 4.4 |
| Amphiregulin | 1421134_at | 52.7 | 11.6 |
| Usp18 | 1418191_at | 52.4 | 2.2 |
| Isg15 | 1431591_s_at | 34.6 | 3.2 |
| Il-6 | 1450297_at | 25.2 | 14.1 |
| Ccl5 | 1418126_at | 19.9 | 4.6 |
| Epiregulin | 1419431_at | 14.9 | 5.3 |
| Ptgs2 | 1417263_at | 7.0 | 4.2 |
| Hmox1 | 1448239_at | 6.8 | 6.3 |
| Stanniocalcin 1 | 1450448_at | 5.2 | 4.1 |
| Nr4a2 | 1447863_s_at | 5.1 | 5.2 |
| Ccl2 | 1420380_at | 4.4 | 3.0 |
| Ccl7 | 1421228_at | 4.2 | 2.7 |
| Zfp36 | 1452519_a_at | 3.6 | 1.6 |
| Poliovirus receptor | 1423905_at | 3.3 | 2.0 |
| Dio2 | 1418937_at | 3.1 | 7.5 |
| Mmp13 | 1417256_at | 3.1 | 3.9 |
| Csf2 | 1427429_at | 3.4 | 1.8 |
| Mcpt8 | 1449965_at | 2.6 | 1.8 |
| Hmga2 | 1450781_at | 2.5 | 1.7 |
| Il-1rl1 | 1425145_at | 2.5 | 2.1 |
| Cd44 antigen | 1452483_a_at | 2.3 | 2.1 |
| Cebpb | 1427844_a_at | 2.5 | 2.4 |
| Ccl17 | 1419413_at | 2.2 | 1.5 |
| IFrd1 | 1416067_at | 2.2 | 1.5 |
| Bcl6 | 1421818_at | 2.0 | 1.6 |
Figure 7.
3P-RNA and non-CpG-DNA trigger proinflammatory cytokines and chemokines in MCs. (A) Primary MC mRNA was isolated 3, 6, and 9 h after exposure to 3P-RNA/CL or non-CpG-DNA/CL. Real-time RT-PCR was performed for the indicated targets as described in Concise Methods. Data are expressed relative to 18S rRNA expression and are means ± SD from three experiments, each analyzed in duplicate. *P < 0.05 versus CL; **P < 0.01 versus CL. (B) In similar experiments, cell supernatants were harvested from stimulated MCs after 24 h of stimulation and analyzed by ELISA for Ccl5 and Cxcl10. Data are means ± SD from three experiments, each analyzed in duplicate.*P < 0.05 versus medium.
3P-RNA and Non-CpG-DNA Trigger Type I IFN and IFN-Related Mediators in Mesangial Cells
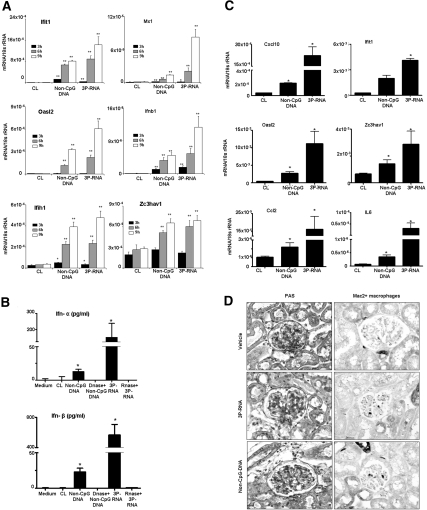
The gene array analysis suggested that many of the genes induced by both 3P-RNA/CL and non-CpG-DNA/CL relate to the type I IFN signaling cascade (Table 1). We used real-time RT-PCR to confirm the induction for several of these antiviral molecules at 3, 6, and 9 h of stimulation in pMCs. For example, the translation regulator IFN-induced protein with tetratricopeptide repeats-1 (Ifit-1), the nuclear GTPase myxovirus resistance-1 (Mx-1), and the nuclear RNAse L activator 2′,5′-oligoadenylate synthetase-like 2 (Oasl2) were induced like Ifn-β mRNA (Figure 8A). We confirmed the production of IFN-α and IFN-β by ELISA in pMC supernatants 24 h after stimulation with 3P-RNA/CL or non-CpG-DNA/CL (Figure 8B). Together, these data show that 3P-RNA and non-CpG DNA stimulate MCs to produce Ifn-α and Ifn-β and multiple other IFN-induced genes with specific antiviral functions, such as Mx-1, Oasl2, and Ifit1 proteins.
Figure 8.
3P-RNA and non-CpG-DNA trigger IFN-related genes in mesangial cells. (A) Primary MC mRNA was isolated 3, 6, and 9 h after exposure to 3P-RNA/CL or non-CpG-DNA/CL. Real-time RT-PCR was performed for the indicated targets as described in Concise Methods. Data are expressed relative to 18S rRNA expression and are means ± SD from three experiments, each analyzed in duplicate. *P < 0.05 versus CL; **P < 0.01 versus CL. (B) In similar experiments, cell supernatants were harvested from stimulated MCs after 24 h of stimulation and analyzed by ELISA for Ifn-α and Ifn-β. Data are means ± SD from three experiments, each analyzed in duplicate. *P < 0.05 versus medium. (C) The C57BL/6 mice were injected three times on alternate days with CL, non-CpG DNA dissolved in CL, or 3P-RNA in CL (n = 6), and glomerular RNA was isolated 12 h after the last injection, as described in Concise Methods. Data are means ± SD. *P < 0.05 versus CL. (D) Renal sections were stained with periodic acid–Schiff (left panel) or a Mac2-specific antibody (right panel) to identify intrarenal macrophages. Original magnification, ×400.
3P-RNA and Non-CpG-DNA Injections Induce IFN-Related Mediators in Glomeruli of C57BL/6 Mice
Do these in vitro effects translate in vivo? To answer this question, we intravenously injected either CL, 30 μg of 3P-RNA, or 100 μg of non-CpG-DNA into C57BL/6 mice three times on alternate days. 3P-RNA and non-CpG-DNA were dissolved in CL before injecting into the mice. Glomeruli were isolated from kidney, and mRNA expression profiles were determined by real-time RT-PCR. The 3P-RNA and non-CpG DNA both induced the glomerular expression of Cxcl10/Ip10, Ifit1, Oasl2, Zc3hav1, Ccl2, and Il-6 (Figure 8C). This effect was associated with a significant increase of glomerular Mac2+ macrophages (vehicle, 0.2 ± 0.1; 3P-RNA, 1.0 ± 0.1; non-CpG DNA, 1.1 ± 0.1; P < 0.01 for both versus vehicle by t test) but not with major glomerular pathology (Figure 8D). Thus, complexed 3P-RNA and non-CpG-DNA induce multiple IFN-related mediators, proinflammatory cytokines, and macrophage recruitment in glomeruli of C57BL/6 mice.
3P-RNA and Non-CpG-DNA Both Trigger Mesangial Cell Apoptosis
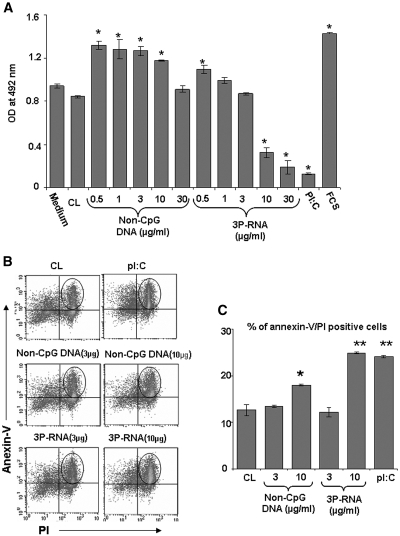
Viral recognition commonly triggers apoptotic cell death, which is thought to contribute to the control of viral replication and spreading. The gene array analysis suggested that many of the genes induced by both 3P-RNA/CL and non-CpG-DNA/CL relate to cell cycle regulation (Table 1). We therefore analyzed the impact of 3P-RNA/CL and non-CpG-DNA/CL on pMC proliferation. At lower concentrations, 3P-RNA/CL and non-CpG-DNA/CL increased the number of pMCs over a period of 72 h (Figure 9A). At higher concentrations, the number of pMCs declined with both nucleic acids in a dose-dependent manner (Figure 9A). Flow cytometry revealed that 3P-RNA/CL and non-CpG-DNA/CL stimulation mainly increased the numbers of propidium iodine and annexin V double-positive MCs, indicating late apoptotic MCs (Figure 9, B and C). Thus, in addition to cytokine and chemokine release and type I IFN signaling, 3P-RNA and non-CpG-DNA trigger MC apoptosis, especially at higher concentrations.
Figure 9.
3P-RNA and non-CpG-DNA trigger apoptosis in MCs. (A) Primary MCs were stimulated with increasing doses of 3P-RNA/CL or non-CpG-DNA/CL as indicated. poly I:polyC RNA 50 μg and 30% FCS were used as controls. After 72 h, the number of proliferating cells was determined by a proliferation assay, as described in Concise Methods. (B) In similar experiments, stimulated MCs were stained with anti-annexin V and PI for 24 h. Flow cytometry analyzed annexin V and PI double-positive cells (encircled). (C) Quantitative analysis of the flow cytometry data shows the percentage of annexin V and PI double-positive cells as means ± SD. *P < 0.05 versus CL. **P < 0.01 versus CL.
Discussion
The glomerular sieving process exposes MCs to all types of circulating micro- and macromolecules, including viral particles during extrarenal viral infections. Viral nucleic acids remain partially protected from nuclease digestion when complexed to immunoglobulins, nucleoproteins, or lipid particles, a process that also supports the uptake of such particles into intracellular compartments.22–24 Hence, glomerular immune complex deposits usually are taken up and processed by glomerular MCs.25 Viral dsRNA activates human and murine MCs to produce Il-6 and Ccl2 in vitro and in vivo, which is attributed to Tlr3 (i.e., the only nucleic-acid-specific Tlr expressed by MCs).17,18 But viral nucleic acids occur in formats other than dsRNA that do not ligate Tlr3, such as the 5′-triphosphate and homopolymeric ribonucleotide motifs of hepatitis C virus RNA.11 Here, we demonstrate that 3P-RNA and unmethylated non-CpG-DNA are potent activators of MCs. Consistent with the published literature, we confirmed that 3P-RNA and non-CpG-DNA activate MCs via Tlr-independent recognition pathways.3,26 Rig-1 and Dai were shown to be involved in the recognition of 3P-RNA and viral non-CpG-DNA, respectively.9,10,13 The role of Rig-1 for the recognition of 3P-RNA has been confirmed with Rig-1-deficient mice by several groups.9,10 The role of Dai in the recognition of unmethylated double-stranded B-DNA is in doubt because cells derived from Dai-deficient mice still respond to B-DNA.14 In a follow-up study, Wang et al. admitted that other cytosolic DNA sensors must exist.27 In fact, our data from Dai knockdown experiments demonstrate that Dai (and Rig-1) are dispensable for non-CpG-DNA recognition in MCs. Still, the recognition of non-CpG-DNA seems to take place inside MCs, because complex formation with CL was required for dsDNA-induced MC activation. To our surprise, transfection with Dai-specific siRNA reduced Il-6 and Cxcl10 mRNA expression in MCs stimulated with 3P-RNA. The significance of this finding remains doubtful because ELISA-based quantification of the respective proteins in MC supernatants showed significant but only slight reduction with the Dai knockdown. To our best knowledge, previous studies have not addressed a potential role for Dai in 3P-RNA-induced cell activation. In view of a recent study that reported the role of Rig-1 in the recognition of dsDNA in human hepatoma cells,16 the role of cytosolic RNA- or DNA-binding proteins in innate pathogen recognition appears to vary between different cell types and species.
Although 3P-RNA and dsDNA recognition in MCs involves different pathways, both activate the transcription factor Irf3.20 This may explain why 3P-RNA and non-CpG-DNA induce almost identical gene expression programs in MCs. Our transcriptome analysis revealed that 3P-RNA and non-CpG-DNA predominantly induce the expression of four groups of genes [i.e., proinflammatory cytokines (Il-6, Ifn-α, and Ifn-β), chemokines (Ccl2, Ccl5, Ccl7, Ccl17, and Cxcl10), a type I IFN-related gene signature (Ifit1, Mx1, Oasl2, Ifnb1, Ifih1, and Zc3hav1), and cell-growth-related genes]. The local expression of cytokines and chemokines is important for antiviral defense, because they create a local inflammatory environment and promote the recruitment of antigen-presenting cells and immune effector cells. Among the proinflammatory chemokines, Cxcl10 is of particular interest, because it is specifically induced by IFN and preferentially recruits cytotoxic T cells via the chemokine receptor Cxcr3.28 Type I IFNs have multiple other immunoregulatory functions and specifically trigger antiviral effectors.2,29 For example, MCs expressed the nuclear GTPase Mx1 that can bind to viral nucleocapsids or other viral components and degrade them.29 Mesangial cells also induced Oasl2, which activates the antiviral endoribonuclease RNAse L, a mechanism that initiates the cleavage of viral RNA26 and that can produce small RNA self-cleavage products that enhance type I signaling via Rig-1, Mda5, and Mavs in a positive amplification loop.30 Note that type I IFN release has not been documented previously in MCs.
The 3P-RNA and non-CpG-DNA also induced MC apoptosis, which is in line with the concept that apoptotic suicide is the ultimate mechanism to prevent viral replication and spreading. Our analysis defines 3P-RNA- and non-CpG-DNA-induced antiviral effector mechanisms of MCs. However, the response spectrum remains limited compared with that of professional antigen-presenting cells. In antigen-presenting cells, the recognition of viral RNA and DNA also reduces antigen uptake and fosters cell maturation, antigen-presentation, and costimulation. Nonimmune cells, such as MCs, lack many of these functions, because they are functionally and structurally differentiated to serve specialized functions in their microenvironments. Mesangial cells are considered to represent specialized pericytes that maintain the structure of the glomerular capillary loop, control glomerular hemodynamics, and clear macromolecules that cannot pass the glomerular filter membrane.31 Our data suggest a novel and previously unrecognized function of MCs to trigger specific antiviral immune responses upon immune recognition of viral 3P-RNA and non-CpG-DNA. These data implicate a novel mechanism for the pathogenesis of viral-infection-associated glomerulonephritis. In fact, the same set of antiviral genes was found to be induced in glomeruli after systemic exposure to 3P-RNA and non-CpG DNA in C57BL/6 mice. In these experiments, the mRNA expression also may originate from other glomerular cells than MCs. In fact, podocytes and glomerular endothelial cells have a similar capacity to that MCs to trigger antiviral genes upon immune recognition of viral RNA and DNA (H-JA, unpublished observations). Direct viral infection and replication in glomerular cells rarely has been documented.32,33 However, de novo glomerulonephritis or flares of preexisting chronic glomerulonephritis are common during extrarenal viral infections. The activation of antiviral immune responses in glomerular cells, as described here, will always be unable to control the extrarenal viral infection but can induce or aggravate glomerular pathology.
Together, 3P-RNA but not non-CpG-DNA recognition involves Rig-1 and Dai in glomerular MCs. Together with recent reports, these data suggest that the roles of Rig-1 and Dai in innate pathogen recognition can vary between cell types and species. Nevertheless, complexed 3P-RNA and dsDNA trigger a common antiviral response program in MCs and in the intrarenal glomerular compartment, which may explain how extrarenal viral infections can trigger glomerulonephritis. This mechanism seems to represent another example how functionally inappropriate immune responses trigger immunity-related tissue damage.
Concise Methods
Mice and Cell Lines
The 6-wk-old female C57BL/6 mice were obtained from Charles River (Sulzfeld, Germany). Myd88-deficient C57BL/6 mice (F8) were obtained from S. Akira (Department of Host Defense, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan). Trif-mutant C57BL/6 mice (F10) were obtained from B. Beutler (Department of Genetics, The Scripps Research Institute, LA Jolla, CA). All experimental procedures were approved by the local government authorities. All mice were housed in filter-top cages with a 12 h dark/light cycle and unlimited access to food and water. For the preparation of pMCs, capsule and medulla of the kidney were removed, and the renal cortices were diced in cold PBS and sequentially passed through a series of stainless steel sieves (150, 103, 63, 50, and 45 μm) and treated with a 1 mg/ml solution of type IV collagenase (Worthington, Lakewood, NY) for 15 min at 37°C. Finally, the digested glomeruli were seeded into 6-well plates with RPMI 1640 containing 20% FCS, 1% insulin, transferrine, and selenium (Roche, Mannheim, Germany). After five passages, >99% of pMCs were positive for smooth muscle actin, and >99% were negative for cytokeratin 18. Mesangial cells were stimulated with different concentrations of phosphodiester 3P-RNA (5′PPP-GAAAAGGGGACACACACACACACACACACAC-3′) complexed with the CL lipofectamine 2000 (Invitrogen, Karlsruhe, Germany). Mesangial cells also were stimulated with phosphodiester non-CpG dsDNA, (sense, 5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′) complexed with CL. Non-CpG-DNA was generated by annealing complementary single strands of DNA as described previously.20 For siRNA studies and Irf3 phosphorylation, a murine MC (MMC) line was used.34
Microarray Studies
pMCs were stimulated with 0.5 μg of 3P-RNA/CL and 30 μg of non-CpG-DNA/CL. Medium/CL-treated cells were used as a control. After 6 h, total RNA was prepared using RNeasy Mini Kit (Qiagen, Hilden, Germany). Total RNA (6 μg) from three independent preparations in each group was used for biotin-labeled complementary RNA probe synthesis and hybridization of MOE 430Av2 arrays according to the Affymetrix Expression Analysis Technical Manual. Triplicate arrays were scanned and analyzed using the Affymetrix GeneChip Operating Software (GCOS1.0). Each array was checked for general assay quality (3′:5′ ratios for glyceraldehydes 3-phosphate dehydrogenase and β-actin <1.1, average background <75 fluorescence units, and scaling factors within a two-fold range). The complete data set was deposited into the GEO database (http://www.ncbi.nlm.nih.gov/geo/; submission no. GSE11898). All nine Affymetrix Microarray CEL files were normalized together using RMA Express, version 1.0 beta 2 (http://rmaexpress.bmbolstad.com/). As the probe sets definition, the default Mouse 430_2.cdf (included in the library files from the Affymetrix Support Page http://www.affymetrix.com/support/technical/byproduct.affx?product=moe430–20) was used, which defines 45,101 probe sets. Probe data were background-adjusted, quantile-normalized, and summarized to probe set signals using median polish. Probe set signals were logarithmized to base 2. To exclude probe sets in the lower end of the signal range that have large signal variation, a background filter cutoff value was defined as the maximal signal value obtained from nonhuman Affymetrix control probe sets multiplied by a factor of 1.2.35 Probe sets with a signal below cutoff in every array of the corresponding comparison and Affymetrix control probe sets were excluded from the analysis. The cutoff value in controls versus 3P-RNA/CL was 3.24, and 22,855 probe sets were retained; in controls versus non-CpG-DNA/CL the cutoff value was 3.10, and 24,097 probe sets were retained. Differentially expressed probe sets between controls and 3P-RNA/CL or non-CpG-DNA/CL were computed using the Microsoft Excel plugin of SAM, version 1.21.36 Parameters were unpaired tests for significance analysis and 100 permutations for false discovery rate estimation. Accepting a false discovery rate of 1%, 4575 (controls versus 3P-RNA/CL) and 7331 probe sets (controls versus non-CpG-DNA/CL) were considered differentially expressed. Of these, the number of probe sets induced or reduced more than 1.5-fold were 855 in controls versus 3P-RNA/CL and 2576 in controls versus non-CpG-DNA/CL.
Cytokine ELISA
Cell culture supernatant cytokine levels were determined using commercial ELISA kits: Il-6 (OptEiA; BD Biosciences, Heidelberg, Germany), Ifn-α and Ifn-β (PBL Biomedical Labs, NJ.), Cxcl10 and Ccl5 (R&D Systems, Minneapolis, MN) following the manufacturer's protocols.
Immunofluorescence Microscopy
Mesangial cells cells were stimulated with 5′-rhodamine-labeled 3P-RNA and non-CpG-DNA complexed with or without CL. After 2 h, cells were fixed with 2% paraformaldehyde, 10 mM Pipes, and 15% saturated picric acid at pH 6.0. Fixed MCs were incubated overnight with mouse anti-Rab5 (BD, Biosciences, Heidelberg, Germany) to mark early endosomes. Fluorescein-isothiocyanate-labeled goat anti-mouse IgG was used for detection. Mesangial cells were scanned using a LSM510 laser scanning microscope (Carl Zeiss, Jena, Germany).
Proliferation Assay
Cell proliferation was determined using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI). Briefly, pMCs were grown in 96-well plates for 24 h before stimulation with various doses of 3P-RNA/CL and non-CpG-DNA/CL. After 72 h, the solution was added, and the cells were incubated at 37°C in 5% CO2 for 2 h before absorbance was determined at a wavelength of 492 nm.
Flow Cytometry for Annexin V–FITC and Propidium Iodide
The 3P-RNA/CL- or non-CpG-DNA/CL-stimulated pMCs were washed with PBS and incubated with annexin V binding buffer containing FITC–anti-annexin V and propidium iodide (PI) for 15 min at room temperature. The cells were analyzed by flow cytometry (FACSCalibur, BD Biosciences, Mannheim, Germany) with acquisition of 30,000 events per sample.
RNA Silencing Studies
The DDX58 (Rig-1) siRNA ON-TARGETplus SMART pool oligonucleotides (Dharmacon), Dai siRNA, and negative control siRNA (Ambion/Applied Biosystems, Darmstadt, Germany) sequences were as follows: DDX58 (Rig-1), 5′-CAAGAAGAGUACCACUUAAUU-3′, 5′-GUUAGAGGAACACAGAUUAUU-3′, 5′-GUUCGAGAUUCCAGUCAUAUU-3′, 5′-GAAGAGCACGAGAUAGCAAUU-3′; Dai, 5′-ACAGUCCAGACAGUCCACAUCAAAU-3′, 5′-GGCAACAAGAUGACCAUCCACCUUA-3′, 5′-GGAAGACACAGGUACAAGCUCUGAA-3′. Mouse MCs (1 × 105) were plated in 12-well plates in antibiotic-free 2% FCS–Dulbecco modified Eagle medium. Small interfering RNA (40 nM) was transfected twice with CL as mentioned above. Twenty-four hours after the second transfection, the cells were stimulated with 1 μg of 3P-RNA and 5 μg of non-CpG-DNA for 6 h. Knockdown efficacy of Rig-1 and Dai as well as Cxcl10 and Il-6 mRNA expression were determined by real-time RT-PCR after 6 h.
Real-Time RT-PCR
Complementary DNA was generated from total RNA using random priming and Moloney murine leukemia virus reverse transcriptase (Invitrogen). Real-time RT-PCR was performed using SYBR Green PCR Master Mix or TaqMan (Applied Biosystems, Weiterstadt, Germany) and analyzed on an AB Prism 7000 analyzer (Applied Biosystems). All values were normalized to 18S ribosomal RNA (rRNA).
The SYBR Green forward (f) and reverse (r) oligonucleotide sequences used in the study were:
18S rRNA, 5′-GCAATTATTCCCCATGAACG-3′ (f), 5′-AGGGCCTCACTAAACCATCC-3′ (r); Ifit1, 5′-CAAGGCAGGTTTCTGAGGAG-3′ (f), 5′-GACTGGTCACCATCAGCAT-3′ (r); Oasl2, 5′-TCTGTTGCACGACTGTAGGC-3′ (f), 5′-GTGTCCAATCCCTGTTCCC-3′ (r); Zc3hav1, 5′-TTGCAAGCTTAATCTGCTCG-3′ (f), 5′-ACCTGGAAGTTCTGTTCCGA-3′ (r); Ifih1, 5′-GCCTGGAACGTAGACGACAT-3′ (f), 5′-TCATCGAAGCAGCTGACACT-3′ (r); Mx1, 5′-TCTGAGGAGAGCCAGACGAT-3′ (f), 5′-CCAGGTCCTGCTCCACAC-3′ (r); Ifnb1, 5′-TCCCTATGGAGATGACGGAG-3′ (f), 5′-ACCCATGCTGGAGAAATTG-3′ (r); Ccl5, 5′-GTGCCCACGTCAAGGAGTAT-3′ (f), 5′-CACTTCTTCTCTGGGTTGG-3′ (r); Ccl17, 5′-TGCTTCTGGGGACTTTTCTG-3′ (f), 5′-ATAGGAATGGCCCCTTTGAA-3′ (r), Hif-1α, 5′-CGGCGAGAACGAGAAGAA-3′ (f), 5′-AAACTTCAGACTCTTTGCTTCG-3′ (r); Il-1-β, 5′-TTCCTTGTGCAAGTGTCTGAAG-3′ (f), 5′-CACTGTCAAAAGGTGGCATTT-3′.
The TaqMan probes used in the study were: Il-6, ID Mm00446190_m1, FAM 5′-AAATGAGAAAAGAGTTGTGCAATGG-3′; Ddx58/Rig-1, ID Mm00554529_m1, FAM 5′-CCAAACCAGAGGCCGAGGAAGAGCA-3′; Ifih1/Mda5, ID Mm00459183_m1, FAM 5′-GACACCAGAGAAAATCCATTTAAAG-3′; Cxcl10, Mm00445235_ml, FAM 5′-GACTCAAGGATCCCTCTCGCAAGG-3′; Tlr3, Mm01207403_ml, FAM 5′ CTTTCAAAAACCAGAAGAATCTAAT-3′; Tlr7, Mm00446590_ml, FAM 5′ AAAATGGTGTTTTCGATGTGGACAC-3′; Mavs, ID Mm00523168_m1, FAM 5′-AGTGACCAGGATCGACTGCGGGCTT-3′; Tnf, Mm00443258_m1, FAM 5′-GTCCCCAAAGGGATGAGAAGTTCCC-3′; Tlr9, 5′-CAATCTGACCTCCCTTCGAGTACTT-3′ (f), 5-GCCACATTCTATACAGGGATTGG-3′(r), core sequence 5′-ATTGCCGTCGCTGCGACCATG-3′; Dai, 5′-CAGGGAAGCACCCCTCTTAT-3′ (f), 5′-GAATGAAGCTCCTGGGTCAG-3′ (r), core sequence FAM 5′-CCCCCAGAAGTGTCAACCACCACT-3′; Cox2/Ptgs2, 5′-GGACTGGATTCTATGGTGAAAACTG-3′ (f), 5′-GGCTTCAGCAGTAATTTGATTCTTG-3′ (r), core sequence FAM 5′-ACTACACCTGAATTTC-3′; Tgf-β, 5′-CACAGTACAGCAAGGTCCTTGC-3′ (f), 5′-AGTAGACGATGGGCAGTGGCT-3′ (r), core sequence FAM 5′-C GCTTCGGCGTCACCGTGCT-3′; Il-2, 5′-GACTGGTTCTTCTGGTGGAAGCT-3′ (f), 5′-TGGGATGCTTGGCCATATG-3′ (r), core sequence FAM 5′-TGGGAGTCCAGCCACCAACATTACTTCT-3′.
Irf3 Phosphorylation Assay
Nuclear extracts were prepared after stimulating MMCs with 1 μg of 3P-RNA and 30 μg of non-CpG-DNA for 2 h. Irf3 DNA-binding activity in nuclear extracts was measured by TransAM Irf3 assay (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. In brief, 20 μg of nuclear extract was incubated in 96-well plates coated with consensus Ifn-stimulated response elements. Plates were washed, and anti-Irf3 antibodies were added to the wells. Antibody binding was detected with a secondary horseradish-peroxidase-conjugated antibody, developed with tetramethyl benzidine substrate, and read at a wavelength of 450 nm.
Western Blotting
Cells were lysed in radioimmunoprecipitation assay buffer [50 mM Tris-HCl (pH 7.4), 0.25% Na-deoxycholate, 1% Nonidet P-40, 150 mM NaCl, and 1 mM EDTA] in the presence of protease and phosphatase inhibitors (Sigma-Aldrich). Western blotting was performed for Phospho-IRF3 (Ser396), Rig-1, and β-actin according to the protocol described in the manual (Cell Signaling Technology, Beverly, MA). ECL Plus Western blotting detection reagents were used for the detection of proteins, as described in the manual (GE Healthcare, Buckinghamshire, UK).
In Vivo Experiments
The C57BL/6 mice received three intravenous injections every other day with either CL alone, 30 μg of phosphodiester 3P-RNA dissolved in CL, or 100 μg of phosphodiester non-CpG-DNA in CL. Glomeruli were purified 12 h after the latest injection by applying a paramagnetic isolation method following perfusion of mice with magnetic 4.5-μm Dynabeads (Invitrogen), as described elsewhere.37 Renal paraffin-embedded sections were stained with periodic acid–Schiff or a Mac2-specific antibody, as described previously.13 Glomerular macrophages were quantified in 15 cortical glomeruli by a blinded observer.
Statistical Analysis
Data were expressed as mean ± SD. Comparison between two groups was performed by two-tailed t test or one-way ANOVA. A value of P < 0.05 was considered to be statistically significant.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank S. Endres, H. Poek, and V. Hornung, Munich, for their help in preparing 3P-RNA and S. Pfeiffer for technical support. We thank R. Hoffmann, J. Mages, and M.T. Lindenmeyer, Munich, for supporting the microarray studies and N. Ramalingam, Munich, for his help conducting confocal microscopy. We thank B. Beutler, La Jolla, CA, and S. Akira, Osaka, Japan, for providing Trif and Myd88 mutant mice, respectively. Parts of this project were prepared as a doctoral thesis at the Faculty of Medicine, University of Munich, by R.A. This work was supported by grants from the Deutsche Forschungsgemeinschaft (AN372/9-1, GRK 1202). A.T. and V.V. were supported by the Deutsche Forschungsgemeinschaft (Grant VI231/2-1).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Lai AS, Lai KN:Viral nephropathy. Nat Clin Pract Nephrol 2: 254–262, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theofilopoulos AN, Baccala R, Beutler B, Kono DH:Type I interferons (α/β) in immunity and autoimmunity. Annu Rev Immunol 23: 307–336, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Pichlmair A, Reis e Sousa C:Innate recognition of viruses. Immunity 27: 370–383, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA:Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413: 732–738, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S:Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303: 1526–1529, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C:Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303: 1529–1531, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Krieg AM, Yi AK, Matson S, Waldschmidt TJ, Bishop GA, Teasdale R, Koretzky GA, Klinman DM:CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374: 546–549, 1995 [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi O, Akira S:Recognition of viruses by innate immunity. Immunol Rev 220: 214–224, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G:5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C:RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M:Innate immunity induced by composition-dependent RIG-1 recognition of hepatitis C virus RNA. Nature 454: 523–527, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T:The RNA helicase RIG-1 has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T:DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuch O, Takeshita F, Coban C, Akira S:TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451: 725–729, 2008 [DOI] [PubMed] [Google Scholar]
- 15.Lang KS, Recher M, Junt T, Navarini AA, Harris NL, Freigang S, Odermatt B, Conrad C, Ittner LM, Bauer S, Luther SA, Uematsu S, Akira S, Hengartner H, Zinkernagel RM:Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med 11: 138–145, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Cheng G, Zhong J, Chung J, Chisari FV:Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci USA 104: 9035–9040, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wörnle M, Schmid H, Banas B, Merkle M, Henger A, Roeder M, Blattner S, Bock E, Kretzler M, Grone HJ, Schlondorff D:Novel role of toll-like receptor 3 in hepatitis C-associated glomerulonephritis. Am J Pathol 168: 370–385, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patole PS, Grone HJ, Segerer S, Ciubar R, Belemezova E, Henger A, Kretzler M, Schlondorff D, Anders HJ:Viral double-stranded RNA aggravates lupus nephritis through Toll-like receptor 3 on glomerular mesangial cells and antigen-presenting cells. J Am Soc Nephrol 16: 1326–1338, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Allam R, Pawar RD, Kulkarni OP, Hornung V, Hartman G, Segerer S, Akira S, Endres S, Anders HJ:Viral 5′-triphosphate RNA and non-CpG-DNA aggravate systemic autoimmunity and lupus nephritis by distinct TLR-independent immune responses. Eur J Immunol 38: 3487–3498, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Stetson DB, Medzhitov R:Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24: 93–103, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Tamura T, Yanai H, Savitsky D, Taniguchi T:The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26: 535–584, 2008 [DOI] [PubMed] [Google Scholar]
- 22.Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen SR, Shlomchik MJ, Viglianti GA, Rifkin IR, Marshak-Rothstein A:RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med 202: 1171–1177, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savarese E, Chae OW, Trowitzsch S, Weber G, Kastner B, Akira S, Wagner H, Schmid RM, Bauer S, Krug A:U1 small nuclear ribonucleo protein immune complexes induce type I interferon in plasmacytoid dendritic cells through TLR7. Blood 107: 3229–3234, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Yasuda K, Richez C, Maciaszek JW, Agrawal N, Akira S, Marshak-Rothstein A, Rifkin IR:Murine dendritic cell type I IFN production induced by human IgG-RNA immune complexes is IFN regulatory factor (IRF) 5 and IRF7 dependent and is required for IL-6 production. J Immunol 178: 6876–6885, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Gomez-Guerrero C, Lopez-Armada MJ, Gonzalez E, Egido J:Soluble IgA and IgG aggregates are catabolized by cultured rat mesangial cells and induce production of TNF-α and IL-6, and proliferation. J Immunol 153: 5247–5255, 1994 [PubMed] [Google Scholar]
- 26.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN:TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med 13: 543–551, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T:Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA 105: 5477–5482, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh MF, Lay SL, Chen JP, Sung JM, Lin YL, Wu-Hsieh BA, Gerard C, Luster A, Liao F:Both CXCR3 and CXCL10/IFN-inducible protein 10 are required for resistance to primary infection by dengue virus. J Immunol 177: 1855–1863, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Sadler AJ, Williams BR:Interferon-inducible antiviral effectors Nat Rev Immunol 8: 559–568, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malathi K, Dong B, Gale M, Jr, Silvermann RH:Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448: 816–819, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlöndorff D:The glomerular mesangial cell: an expanding role for a specialized pericyte. FASEB J 1: 272–281, 1987 [DOI] [PubMed] [Google Scholar]
- 32.Bruggeman LA, Ross MD, Tanji N, Cara A, Dikman S, Gordon RE, Burns GC, D'Agati VD, Winston JA, Klotman ME, Klotman PE:Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol 11: 2079–2087, 2000 [DOI] [PubMed] [Google Scholar]
- 33.Winston JA, Bruggeman LA, Ross MD, Jacobson J, Ross L, D'Agati VD, Klotman PE, Klotman ME:Nephropathy and establishment of a renal reservoir of HIV type 1 during primary infection. N Engl J Med 344: 1979–1984, 2001 [DOI] [PubMed] [Google Scholar]
- 34.Satriano JA, Banas B, Luckow B, Nelson P, Schlondorff D:Regulation of RANTES and ICAM-1 expression in murine mesangial cells. J Am Soc Nephrol 8: 596–603, 1997 [DOI] [PubMed] [Google Scholar]
- 35.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Grone HJ, Nelson PJ, Schlondorff D, Cohen CD, Kretzler M:Modular activation of nuclear factor-κB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Tusher VG, Tibshirani R, Chu G:Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takemoto M, Asker N, Gerhardt H, Lundkvist A, Johansson BR, Saito Y, Betsholtz C:A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol 161: 799–805, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.