Abstract
A significant fraction of the Saccharomyces cerevisiae genome is transcribed periodically during the cell division cycle1, 2, suggesting that properly timed gene expression is important for regulating cell cycle events. Genomic analyses of transcription factor localization and expression dynamics suggest that a network of sequentially expressed transcription factors could control the temporal program of transcription during the cell cycle3. However, directed studies interrogating small numbers of genes indicate that their periodic transcription is governed by the activity of cyclin-dependent kinases (CDKs)4. To determine the extent to which the global cell cycle transcription program is controlled by cyclin/CDK complexes, we examined genome-wide transcription dynamics in budding yeast mutant cells that do not express S-phase and mitotic cyclins. Here we show that a significant fraction of periodic genes were aberrantly expressed in the cyclin mutant. Surprisingly, although cells lacking cyclins are blocked at the G1/S border, nearly 70% of periodic genes continued to be expressed periodically and on schedule. Our findings reveal that while CDKs play a role in the regulation of cell cycle transcription, they are not solely responsible for establishing the global periodic transcription program. We propose that periodic transcription is an emergent property of a transcription factor network that can function as a cell cycle oscillator independent of, and in tandem with, the CDK oscillator.
The biochemical oscillator controlling periodic events during the cell cycle is centered on the activity of cyclin-dependent kinases (CDKs) (reviewed in ref. 5). The cyclin/CDK oscillator governs the major events of the cell cycle, and in embryonic systems this oscillator functions in the absence of transcription, relying only on maternal stockpiles of mRNAs and proteins. CDKs are also thought to act as the central oscillator in somatic cells and yeast, and directed studies suggest that they play an essential role in controlling the temporally ordered program of transcription (reviewed in refs. 4, 6). However, systems-level analyses using high throughput technologies1, 2, 7, 8 have suggested alternative models for regulation of periodic transcription during the yeast cell cycle1, 3, 9. By correlating genome-wide transcription data with global transcription factor binding data, models have been constructed in which periodic transcription is an emergent property of a transcription network1, 3, 9. In these networks, transcription factors expressed in one cell cycle phase bind to the promoters of genes encoding transcription factors that function in a subsequent phase. Thus, the temporal program of transcription could be controlled by sequential waves of transcription factor expression, even in the absence of extrinsic control by cyclin/CDK complexes.
The validity and relevance of the intrinsically oscillatory transcription factor network hypotheses remain uncertain, because for the limited number of periodic genes that have been dissected in detail, periodic transcription was found to be governed by CDKs (reviewed in ref. 4). Thus, we sought to determine to what extent CDKs and transcription factor networks contribute to global regulation of the cell cycle transcription program. To this end, we investigated the dynamics of genome-wide transcription in budding yeast cells disrupted for all S-phase and mitotic cyclins (clb1,2,3,4,5,6). These cyclin mutant cells are unable to replicate DNA, separate SPBs, undergo isotropic bud growth, or complete nuclear division, indicating that they are devoid of functional Clb/CDK complexes10-12. So by conventional cell cycle measures, clb1,2,3,4,5,6 cells arrest at the G1/S border. We have previously shown that clb1,2,3,4,5,6 cells cyclically trigger G1 events10, including the activation of G1-specific transcription and bud emergence. Nevertheless, if Clb/CDK activities are essential for triggering the transcriptional program, then periodic expression of S-phase and G2/M-specific genes should not be observed.
We examined global transcription dynamics in synchronized populations of both wild-type cells and cyclin mutant cells. Synchronous populations of early G1 cells were collected by centrifugal elutriation. Cell aliquots were then harvested at 16 min intervals for 270 min (equivalent to ~2 cell cycles in wild-type and ~1.5 in the cyclin mutant). Transcript levels were measured genome-wide for each time point using Yeast 2.0 oligonucleotide arrays (Affymetrix, Santa Clara, CA). Results from two independent experiments each for both wild-type and cyclin mutant cells were highly reproducible, with adjusted r2 values of 0.995 and 0.989, respectively (Supplementary Fig. 1). All statistical analyses were performed using replicate data sets, but to facilitate illustration, single data sets were used for all graphical representations.
To identify periodically transcribed genes, we applied a modification of the method developed by de Lichtenberg et al.13 to data acquired from our wild-type cells. We established a set of 1271 genes that were transcribed periodically (Fig. 1a, and Supplementary Table 1). This set of periodic genes shares 510 and 577 genes with those sets previously identified as periodic by Spellman et al.2 and Pramila et al.1, respectively (Supplementary Fig. 2), with 440 consensus periodic genes identified by all three studies (Supplementary Table 2). We then examined the transcriptional dynamics of our set of 1271 periodic genes in the cyclin mutant (Fig. 1b). The behavior of many genes changed significantly in the cyclin mutant, supporting previous findings. However, despite the fact that cyclin mutant cells arrest at the G1/S border, a large fraction of periodic genes in all cell cycle phases continued to be expressed on schedule (Fig. 1b). Similar cyclin-dependent and -independent behaviors are also observed in the set of 440 consensus periodic genes (Supplementary Fig. 3).
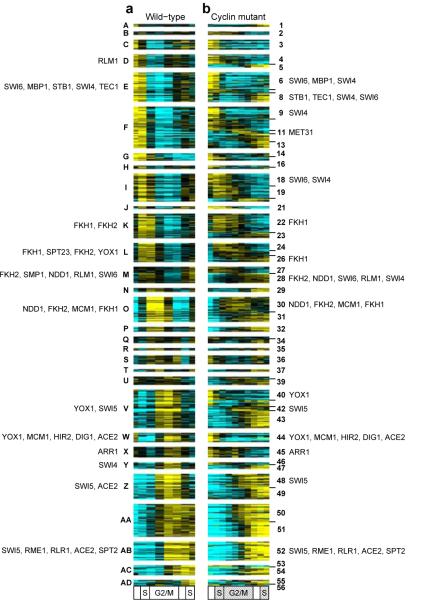
Figure 1.

Dynamics of periodic transcripts in wild-type and cyclin mutant cells. Heat maps depicting mRNA levels of periodic genes are shown for a, wild-type and b, cyclin mutant cells. Each row in a and b represents data for the same gene (Supplementary Table 1). Transcript levels are expressed as log2-fold change vs. mean expression. Transcript levels at each point in the time series were mapped onto a cell cycle timeline31 (see Methods). The S and G2/M phases of the cyclin mutant timeline are shaded, indicating that by conventional definitions, cyclin mutant cells arrest at the G1/S-phase transition.
Using absolute change and Pearson correlation analyses (see Supplementary Information), we determined that 833 of the periodic genes exhibited changes in expression behavior in the cyclin mutant and thus are likely to be directly or indirectly regulated by B-cyclin/CDK.
Our genome-level experiments accurately reproduced previous findings regarding several well-studied B-cyclin/CDK-regulated genes (Fig. 2). We observed that a subset of late G1 transcripts (SBF-regulated genes like CLN2 but not MBF-regulated genes like RNR1) were not fully repressed (Fig. 2a and b) as expected in mitotic cyclin mutant cells 14, 15. A subset of M/G1 transcripts (including SIC1 and NIS1), are targets of the transcription factors Swi5 and Ace2, which are normally excluded from the nucleus by CDK phosphorylation until late mitosis16-19. SIC1 and NIS1 were expressed earlier in the cyclin mutant (Fig. 2c and d) presumably because nuclear exclusion of Swi5 and Ace2 is lost in cyclin mutant cells. The modest degree of shift in the timing of SIC1 and NIS1 transcription likely reflects the fact that SWI5 and ACE2 transcripts do not accumulate to maximal levels in cyclin mutant cells as expected for Clb2 cluster genes (including CDC20) (Fig. 2e and f) 14, 20, 21. Although a significant fraction of periodic genes exhibited changes in the amplitude of expression (increased or decreased), a statistical analysis of the dynamic range of expression across all periodic genes revealed that the majority of genes in cyclin mutant cells exhibit only modest changes, if any, with respect to wild-type cells (Supplementary Fig. 4).
Figure 2.
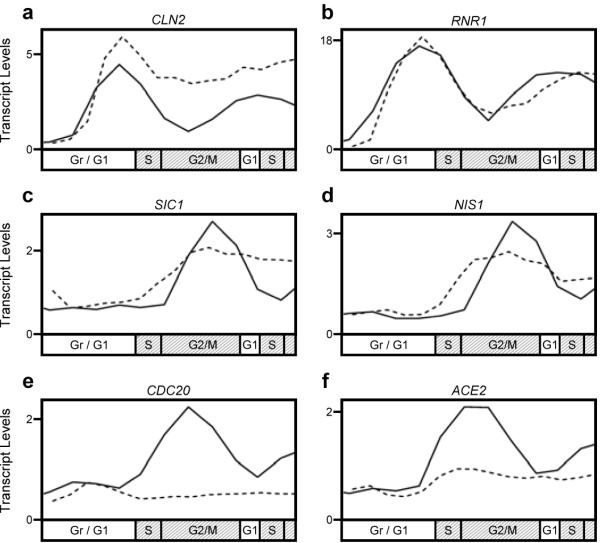
Transcription dynamics of established cyclin/CDK-regulated genes. Absolute transcript levels (dChip-normalized Affymetrix intensity units/1000) are shown for the SBF- and MBF-regulated genes a, CLN2 and b, RNR1; the Ace2/Swi5-regulated genes c, SIC1 and d, NIS1; and the Clb2 cluster genes e, CDC20 and f, ACE2. Wild-type cells (solid line) and cyclin mutant cells (dashed line).
To identify novel subsets of co-regulated genes based on transcriptional behaviors observed in both wild-type and cyclin mutant cells, we employed the affinity propagation algorithm22 to first cluster genes based on expression in wild-type cells, and then subcluster genes based on their behavior in cyclin mutant cells (Fig. 3 and Supplementary Fig. 5). Of the 833 cyclin-regulated genes, 513 were assigned to 30 discrete clusters exhibiting similar behaviors in wild-type cells (Fig. 3a, and Supplementary Fig. 6), and were then subclustered into 56 novel clusters based on their transcription profiles in cyclin mutant cells (Fig. 3b and Supplementary Table 3). Using data from global transcription factor localization studies23, we identified subsets of transcription factors that may regulate these subclusters using an over-representation analyses (Fig. 3 and Supplementary Table 4). Based on their association with the promoters of genes in cyclin-regulated subclusters, these factors are likely to be directly or indirectly regulated by cyclins. Consistent with this hypothesis, several of these factors have already been shown to be CDK targets14, 15, 18, 19, 24-29. These findings lay the groundwork for elucidating the full range of mechanisms by which cyclin/CDKs regulate transcription during the cell cycle.
Figure 3.
Genes exhibiting altered behaviors in cyclin mutant cells. a, Clusters of genes with similar expression patterns in wild-type cells. b, Subclusters of genes with similarly altered expression patterns in cyclin mutant cells. Each row in a and b represents data for the same gene (Supplementary Table 1). Transcript levels are depicted as shown in Fig. 1. Up to five over-represented transcription factors for each cluster are shown (see Methods and Supplementary Table 4 for complete lists).
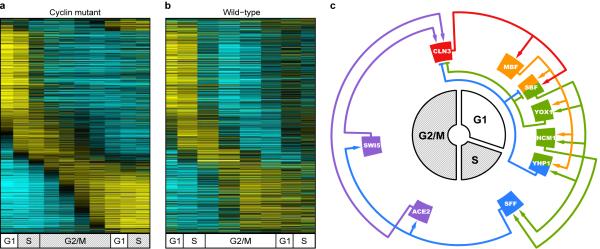
Strikingly, 882 of the genes identified as periodic in wild-type cells, continued to be expressed on schedule in cyclin mutant cells despite cell cycle “arrest” at the G1/S border (Fig. 4a and b). Some of these genes (450 in total) exhibited minor changes in transcript behavior but continued to be expressed at the proper time, as shown above for ACE2. Thus, some genes that were cyclin-regulated are also included in the set of genes that maintain periodicity. Nevertheless, a statistical analysis of the dynamic range of expression of these genes in wild-type and cyclin mutant cells indicates that the amplitude changes for most of these genes is quite modest (see Supplementary Figs. 7, 8 and 9). The finding that nearly 70% of the genes identified as periodic in wild-type cells are still expressed on schedule in cyclin mutant cells demonstrates the existence of a cyclin/CDK-independent mechanism that regulates temporal transcription dynamics during the cell cycle. This observation is supported by the analysis of the set of 440 consensus periodic genes, the bulk of which maintain periodicity in the cyclin mutant (Supplementary Fig. 10).
Figure 4.
The periodic transcription program is largely intact in cyclin mutant cells that arrest at the G1/S border. a, Genes maintaining periodic expression in cyclin mutant cells exhibit similar dynamics in b, wild-type cells. Each row in a and b represents the same gene (Supplementary Table 1). Transcript levels are depicted as shown in Fig. 1. c, Synchronously updating Boolean network model. Transcription factors are arranged based on the time of peak transcript levels in cyclin mutant cells. Arrows indicate transcription factor/promoter interaction. Activating interactions, outer rings; repressive interactions, inner rings. Coloring indicates activity in one of five successive states; SBF and YHP1 are active in two states (Supplementary Table 6).
In principle, a transcription network defined by sequential waves of transcription factor expression1, 3, 9 might function independent of any extrinsic control by CDKs. To determine if a transcription network could account for cyclin/CDK-independent periodic transcription, we constructed a synchronously updating Boolean network model and determined that such a model can indeed explain the periodic expression patterns we observed in cyclin mutant cells (Fig. 4c). Transcription factors that maintained periodicity in the cyclin mutant were placed on a circularized cell cycle time line based on their peak time of transcription in the cyclin mutant. Connections were drawn based on documented physical interactions23, 30 (Supplementary Table 5) between a transcription factor and the promoter region of a gene encoding a transcription factor expressed subsequently (see Supplementary Information). The architecture of the network in cyclin mutant cells is virtually identical to that in wild-type cells (Supplementary Fig. 11), and is also remarkably similar to models based on wild-type expression data from previous studies1, 3, 9.
When the network is endowed with Boolean logic functions (Supplementary Table 6a), synchronous updating of the model leads to a cycle that produces successive waves of transcription by progressing through five distinct states before returning to the initial state (Supplementary Fig. 12a and b). Thus, the model functions as an oscillator and produces a correctly-sequenced temporal program of transcription.
To examine the robustness of the network oscillator, we evaluated outcomes when initializing the network from all possible starting states. Over 80% of the 512 starting states entered the oscillatory cycle depicted in Fig. 4c, with the remainder terminating in a steady state where all genes were transcriptionally inactive (Supplementary Table 6b and c). We also examined whether the oscillations were sensitive to the choice of the Boolean logic functions assigned to nodes with multiple inputs, specifically, the activating inputs to Cln3 and SFF, and the repressors of SBF and Cln3. For most of the logic functions, the predominant outcome was again the oscillatory cycle depicted in Fig. 4c, but in some cases, the model enters two qualitatively similar cycles (Supplementary Fig. 12c and d, and Supplementary Table 6), with the remainder again terminating in a transcriptionally inactive steady state. Several Boolean logic functions produce the same cycles (Supplementary Table 6b), so the model cannot precisely determine the true logic of the network connections. Nevertheless, the fact that the model can produce qualitatively similar cycles, and that these cycles can be reached from many initial states, suggests that robust oscillation is an emergent property of the network architecture.
Previous studies proposed that a cyclin/CDK-independent oscillator could trigger some periodic events, including bud emergence10. The robust oscillating character of our model suggests that a transcription factor network may function as this cyclin/CDK-independent oscillator. Because cyclin genes are themselves among the periodic genes targeted by this network, and because cyclin/CDKs can, in turn, influence the behavior of transcription factors in the network, precise cell cycle control could be achieved by coupling a transcription factor network oscillator with the cyclin/CDK oscillator. The existence of coupled oscillators could explain why the cell cycle is so robust to significant perturbations in gene expression or cyclin/CDK activity.
Our findings also suggest that the properly scheduled expression of genes required for cell cycle regulated processes such as DNA synthesis and mitosis is not sufficient for triggering these events. The execution of cell cycle events in wild-type cells is likely to require both properly timed transcription and post-transcriptional modifications mediated by CDKs.
Methods Summary
Strains and cell synchronization
Wild-type and cyclin mutant strains of S. cerevisiae are derivatives of BF264-15Dau, and were constructed by standard yeast methods. The clb1,2,3,4,5,6 GAL1-CLB1 mutant strain along with growth conditions and synchrony procedures was described previously10, 11.
RNA isolation and microarray analysis
Total RNA was isolated time points (every 16 min for a total of 15 time points) as described previously10. mRNA was amplified and fluorescently labeled using GeneChip One-Cycle Target Labeling (Affymetrix, Santa Clara, CA) . Hybridization to Yeast 2.0 oligonucleotide arrays (Affymetrix, Santa Clara, CA) and image collection were performed at the Duke Microarray Core Facility (http://microarray.genome.duke.edu/) according to standard Affymetrix protocols.
Data analysis
A workflow diagram for data analysis is depicted in Supplementary Fig. 13.
Methods
Strains and cell synchronization
Yeast strains were grown in rich YEP medium (1% yeast extract, 2% peptone, 0.012% adenine, 0.006% uracil) containing 2% galactose. Forty-five minutes prior to elutriation, dextrose was added to YEP 2% galactose medium to terminate CLB1 expression from the GAL1 promoter. After elutriation, wild-type and clb1,2,3,4,5,6 GAL1-CLB1 cells were grown in rich YEP 2% dextrose,1M sorbitol at 30°C at a density of 107/ml. Sorbitol was added to stabilize cells with elongated buds. Aliquots of 50ml (cell density = 107/ml) were harvested every 8 min for 4hr. Budding index was determined microscopically by counting ≥ 200 cells for each time point.
Data analysis
CEL files from all 60 oligonucleotide arrays were normalized, and summarized using the dChip method32 as implemented in the affy package (v.1.8.1) within Bioconductor using default parameters. The output of this package is a measure of absolute expression levels for each probe in arbitrary expression units (Fig. 2). Data presented in heat maps and centroid line graphs (Figs. 1, 3, and 4, and Supplementary Figs. 3, 5, 8, 11,12, 15-18) are expressed as log2-fold change for each gene relative to its mean expression over the interval from the first G1 to the second S phase.
The CLOCCS population synchrony model31 was used to temporally align expression data from our two wild-type and two cyclin mutant experiments. Briefly, the CLOCCS model allows the alignment of data from multiple synchrony/time series experiments to a common cell cycle time line using budding as a parameter measured on single cells at each time point31. Although cyclin mutant cells arrest at the G1/S border by conventional measures, G1 events, such as bud emergence, are activated periodically with a cycle time similar to wild-type cells10. Thus, the CLOCCS model can be used to temporally align cycles in wild-type and cyclin mutant cells. Because the kinetics of synchrony/release experiments can vary, and wild-type and cyclin mutant cells have marginally different cycle times, alignment is imperative for meaningful comparison of data. We used the CLOCCS parameter estimates to align all four data sets such that the population level measurements were mapped onto a common cell cycle timeline. The timeline utilizes standard cell cycle phases (as determined by measured parameters) and an additional phase (Gr) corresponding to a period of recovery from the initial synchrony procedure that overlaps with early G131. The recovery period (Gr) was eliminated from most of the data displayed, as the genes expressed in this period tend to be specific to this period and are not expressed again in the next cell cycle in either wild-type or cyclin mutant cells. The CLOCCS model was designed for wild-type yeast populations but can accommodate data from the cyclin mutant with minor modifications (see Supplementary Information). CLOCCS model fits for both the wild-type and cyclin mutant datasets are shown in Supplementary Fig. 14, and the corresponding parameter estimates are shown in Supplementary Table 7.
A modification of the method described by de Lichtenberg13 was used to determine the subset of genes exhibiting periodic transcription (see Supplementary Information for details). The methods used to identify genes with altered transcriptional profiles (Fig. 3), and similar profiles (Figs. 4a and b) in cyclin mutant cells with respect to wild-type cells are also described in detail in Supplementary Information. Methods for determining over-represented transcription factors in the clusters (Fig. 3) and details regarding the construction of the synchronously updating Boolean network model (Fig. 4c) can also be found in Supplementary Information. Two additional analyses similar to that performed in Fig. 3 were performed on consensus periodic genes (Supplemental Table 2) identified as changing expression as well as those identified as maintaining periodicity. Details and results of those analyses can be found in Supplementary Information (Supplementary Figs. 15-19 and Supplementary Tables 8 and 9)
Supplementary Material
Acknowledgments
We would like to thank D. Lew and L. Simmons Kovacs for helpful discussions and critical reading of the manuscript, and P. Benfey for helpful discussions and support. We gratefully acknowledge financial support from the American Cancer Society (to S.B.H.), Alfred P. Sloan Foundation (to A.J.H.), National Science Foundation (to A.J.H. and J.E.S.S.), and National Institutes of Health (to S.B.H., A.J.H., and J.E.S.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE8799. Reprints and permissions information is available at www.nature.com/reprints. Correspondence and requests for materials should be addressed to S.B.H (shaase@duke.edu).
References
- 1.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–78. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon I, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 4.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–55. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 5.Murray AW. Recycling the cell cycle: Cyclins revisited. Cell. 2004;116:221–34. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 6.Zwicker J, Muller R. Cell cycle-regulated transcription in mammalian cells. Prog Cell Cycle Res. 1995;1:91–9. doi: 10.1007/978-1-4615-1809-9_7. [DOI] [PubMed] [Google Scholar]
- 7.Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 8.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 9.Lee TI, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 10.Haase SB, Reed SI. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature. 1999;401:394–7. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- 11.Haase SB, Winey M, Reed SI. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nature Cell Biology. 2001;3(1):38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- 12.Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: Regulation by Cdc28 and cyclins. J. Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lichtenberg U, et al. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2005;21:1164–71. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 14.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 15.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–41. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 16.Toyn JH, Johnson AL, Donovan JD, Toone WM, Johnston LH. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the CDK-inhibitor Sic1 in telophase. Genetics. 1997;145:85–96. doi: 10.1093/genetics/145.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knapp D, Bhoite L, Stillman DJ, Nasmyth K. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol Cell Biol. 1996;16:5701–7. doi: 10.1128/mcb.16.10.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor, SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 19.O’Conallain C, Doolin MT, Taggart C, Thornton F, Butler G. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol Gen Genet. 1999;262:275–82. doi: 10.1007/s004380051084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu G, et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–4. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 21.Kumar R, et al. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- 22.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–6. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 23.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costanzo M, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 25.de Bruin RA, et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23:483–96. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 26.Ho Y, Costanzo M, Moore L, Kobayashi R, Andrews BJ. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol Cell Biol. 1999;19:5267–78. doi: 10.1128/mcb.19.8.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pic-Taylor A, Darieva Z, Morgan BA, Sharrocks AD. Regulation of cell cycle-specific gene expression through cyclin-dependent kinase-mediated phosphorylation of the forkhead transcription factor Fkh2p. Mol Cell Biol. 2004;24:10036–46. doi: 10.1128/MCB.24.22.10036-10046.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidorova JM, Mikesell GE, Breeden LL. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995;6:1641–58. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 30.Teixeira MC, et al. The YEASTRACT database: A tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–51. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlando DA, et al. A probabilistic model for cell cycle distributions in synchrony experiments. Cell Cycle. 2007;6:478–88. doi: 10.4161/cc.6.4.3859. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:0032.1–0032.11. doi: 10.1186/gb-2001-2-8-research0032. res. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.