Abstract
A new member of the AP2/ERF transcription factor family, GmERF3, was isolated from soybean. Sequence analysis showed that GmERF3 contained an AP2/ERF domain of 58 amino acids and two putative nuclear localization signal (NLS) domains. It belonged to a group IV protein in the ERF (ethylene response factor) subfamily as typified by a conserved N-terminal motif [MCGGAI(I/L)]. Expression of GmERF3 was induced by treatments with high salinity, drought, abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and soybean mosaic virus (SMV), whereas there was no significant GmERF3 mRNA accumulation under cold stress treatment. GmERF3 could bind to the GCC box and DRE/CRT element, and was targeted to the nucleus when transiently expressed in onion epidermal cells. The GmERF3 protein fused to the GAL4 DNA-binding domain to activate transcription of reporter genes in yeast. Ectopic expression of the GmERF3 gene in transgenic tobacco plants induced the expression of some PR genes and enhanced resistance against infection by Ralstonia solanacearum, Alternaria alternata, and tobacco mosaic virus (TMV), and gave tolerance to high salinity and dehydration stresses. Furthermore, overexpression of GmERF3 in transgenic tobacco led to higher levels of free proline and soluble carbohydrates compared to wild-type plants under drought conditions. The overall results suggested that GmERF3 as an AP2/ERF transcription factor may play dual roles in response to biotic and abiotic stresses in plants.
Keywords: Abiotic stress, biotic stress, ethylene response factor, pathogen
Introduction
AP2/ERF transcription factors belong to one of the largest plant transcription factor families, and are characterized by conserved AP2/ERF DNA binding domains of 57–66 amino acids in size (Okamuro et al., 1997). The AP2/ERF genes constitute a large multigene family divided into four subfamilies named AP2, CBF/DREB, ERF, and RAV based on their sequence similarities and numbers of AP2/ERF domains (Sakuma et al., 2002). AP2 subfamily proteins contain two AP2/ERF domains, and genes in this subfamily participate in the regulation of developmental processes (Elliott et al., 1996; Chuck et al., 1998; Boutilier et al., 2002). The RAV subfamily proteins contain one AP2/ERF domain and a B3 domain, which differ in biological functions and are involved in distinct types of the transcription. Recently, the involvement of RAV subfamily members in ethylene response (Alonso et al., 2003), brassinosteroid response (Hu et al., 2004), and biotic and abiotic stress responses (Sohn et al., 2006) was reported. In contrast to the AP2 and RAV subfamily members, the CBF/DREB and ERF subfamily proteins contain single AP2/ERF domains. The genes in the CBF/DREB subfamily play a crucial role in the resistance of plants to abiotic stresses by recognizing the dehydration responsive or cold-repeat element (DRE/CRT) with a core motif of A/GCCGAC (Yamaguchi-Shinozaki and Shinozaki, 1994; Thomashow, 1999). The ERF subfamily is mainly involved in the response to biotic stresses like pathogenesis by recognizing the cis-acting element AGCCGCC, known as the GCC box (Hao et al., 1998). Many of the ERF subfamily members also bind DRE/CRT elements (Lee et al., 2004; Xu et al., 2007).
ERF transcription factors have been identified in various plant species, such as Arabidopsis thaliana (Oñate-Sánchez and Singh, 2002), tobacco (Nicotiana tabacum) (Fischer and Dröge-Laser, 2004), and tomato (Lycopersicon esculentum) (Tournier et al., 2003). The ERF proteins involved in defence responses against pathogen infection have also been extensively documented (Park et al., 2001; Shin et al., 2002; Gutterson and Reuber, 2004), and overexpression of ERF genes in transgenic tobacco or Arabidopsis plants induces expression of several PR genes, resulting in enhanced resistance to bacterial, fungal, or viral pathogen (Yi et al., 2004; Zuo et al., 2007; Fischer and Dröge-Laser, 2004). Recent studies revealed the role of some ERF proteins during hormone and abiotic stress responses in plants (Shinozaki et al., 2003). The pepper ERF gene, CaPF1, was induced by ET, JA, and cold stress, and overexpression of CaPF1 in transgenic Arabidopsis plants gave tolerance to freezing temperatures and enhanced resistance to Pseudomonas syringae pv. tomato DC3000 (Yi et al., 2004). Moreover, overexpression of CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) conferred tolerance to heavy metals (cadmium, copper, and zinc), heat, and to pathogens (Bacillus thuringiensis and Staphylococcus epidermidis) (Tang et al., 2005). Thus, ERF proteins play an important role, not only in pathogen defence responses but also in tolerance to abiotic stresses.
Although ERF subfamily transcription factors have been identified in different plant species, only a few have been characterized (Oñate-Sánchez and Singh, 2002; Sakuma et al., 2002). In fact, in soybean, only the role of GmEREBP1 has been reported (Mazarei et al., 2002, 2007). In this study, a new member of the soybean ERF subfamily was isolated, designated GmERF3, and its expression patterns in response to different stress conditions were analysed. According to Tournier et al. (2003), GmERF3 belongs to a novel ERF class IV, which is typified by a conserved N-terminal signature sequence (MCGGAII/L). Interestingly, full-length GmERF3 protein was able to bind with both the GCC box and DRE/CRT motifs with different binding affinities. GmERF3 as a transcriptional activator can activate expression of reporter genes in yeast. Previous research has demonstrated that overexpression of ERF genes enhances resistance to biotic and abiotic stresses. Further evidence is provided here that resistance to bacterial, fungal, and viral pathogens (biotic stress), and also to high salinity and drought stresses (abiotic stress), can be engineered by overexpression of the GmERF3 transcription factor.
Materials and methods
Plant materials and stress treatments
Soybean cultivar ‘Tiefeng 8’, a genetic resource for salinity tolerance and ‘Zhongpin 95-5383’, a soybean mosaic virus-resistant variety, were used throughout the study. Plant treatments were performed at the two true-leaf stage. ‘Tiefeng 8’ was used for salt, dehydration, cold, and ABA stress treatments. ‘Zhongpin 95-5383’ was used for SA, ET, and JA treatments and SMV inoculation. Stress treatments were performed as follows: For salt stress, the roots of seedlings were dipped into solutions of 200 mM NaCl. For chilling treatment, seedlings were put into a 4 °C growth chamber. For dehydration, the root systems of whole plants were washed gently with water to remove soil and then the plants were put on filter paper for drought induction. For ABA, SA, and JA treatments, soybean seedlings were sprayed with 200 μM ABA dissolved in 0.01% ethanol, 2 mM SA in water, and 100 μM JA in 0.01% ethanol, respectively. Ethylene treatment was performed in a gas-tight plexiglass chamber by dissolving 2 ml 40% ethephon and 1 g NaHCO3 in 200 ml H2O (under these conditions ethephon will liberate ethylene gas). For SMV treatment, mechanical inoculation was carried out by rubbing leaves with a brush dipped in a mixture of carborundum and an extract of infected leaves ground in phosphate buffer (pH 7.2). After inoculation, the leaves were rinsed with tap water. To test the expression pattern of the GmERF3 gene during the early phase of the stress response (0–24 h) and specifically from 0 h to 5 h after treatment, the discontinuous time points were selected to sample at 0 h, 1 h, 2 h, 5 h, 10 h, and 24 h, respectively, and it was then frozen in liquid nitrogen and kept at –80 °C for further analysis.
Isolation of the GmERF3 gene
The soybean unigene set and expressed sequence tags (ESTs) used as a primary sequence data set are available on the DFCI soybean gene index database (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=soybean), from which 148 tentative consensus (TC) sequences/EST sequences containing the AP2/ERF domain were acquired. Therein, 98 unigenes were predicted to encode a complete AP2/ERF domain; 62 of them encoding ERF-like proteins were assigned to the ERF subfamily (Zhang et al., 2008), of which, TC225486, that could be induced to different degrees by biotic and abiotic stresses, was subjected to further functional analysis. The gene was cloned by RT-PCR from cDNA of soybean ‘Tiefeng 8’ using the primer pair 5′-AGGGACGCTTTTGTTATTCTTCG-3′ (forward) and 5′-GCTTTATTTACACACAGGGAGACCA-3′ (reverse), and designated GmERF3. PCR was performed as follows: 35 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 90 s.
Preparation of recombinant proteins and electrophoretic mobility shift assays
A GmERF3 cDNA fragment containing the entire AP2/ERF domain was prepared by PCR and cloned into the EcoRI/XhoI site of the pGEX-4T-1 vector (Pharmacia, Piscataway, NJ). The primer set used for amplification was: 5′-CGGAATTCGCCGCCTCTTCTCGTCTGTCT-3′ (forward) and 5′-CCGCTCGAGATTGTTGCCAGCACTGAACTTGT-3′ (reverse). The resulting in-frame fusion plasmid was transformed into Escherichia coli strain BL21 (DE3).
Overexpression of GST:
GmERF3 was induced by 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 37 °C for 4 h. Purification of the fusion protein was conducted by affinity chromatography using a Glutathione Sepharose 4B Microspin column according to the manufacturer's instructions (Amersham, Arlington Heights, IL, USA). EMSA was performed as described by Choi et al. (2005). Gels were exposed to Imaging Screening-K and visualized with a Molecular Imager FX (Bio-Rad, Richmond, CA, USA).
Transcriptional activation assay of GmERF3 protein
The GmERF3 gene was amplified by PCR and the EcoRI site and PstI sites were added at the 5′ and 3′ ends, respectively. The amplified fragments were digested with EcoRI and PstI, and inserted downstream of the GAL4 DNA-binding domain of vector pGBKT7 linearized with EcoRI and PstI. The primers used for the PCR amplification were: 5′-CGGAATTCATGTGTGGAGGAGCTATCATCTCTG-3′ (forward) and 5′-AACTGCAGTGGAAGGAATGTCATCAAGGCAC-3′ (reverse). The transcriptional activation activity of GmERF3 was identified by yeast two-hybrid analysis using Saccharomyces cerevisiae strain AH109. pGBKT7, a vector containing the TRP1 nutritional marker for selection in yeast, and GAL4, a DNA-binding domain under the control of the ADH1 promoter, were used to transform the yeast as described in the manufacturer's instructions (Clontech, Palo Alto, CA). After selection of the yeast transformants carrying the GmERF3 gene on SD (–Trp, –Leu) medium, they were transferred to SD (–Trp, –His, –Leu, –Ade) medium to identify the transcriptional activation.
Subcellular localization of GmERF3
The full-length GmERF3 coding region was fused to the N-terminus of the hGFP gene under the control of the CaMV 35S promoter. Transient expression of the GmERF3:hGFP fusion construct and the hGFP control vector were performed by introducing the resultant DNAs into onion epidermal cells using the particle bombardment method, according to the manufacturer's protocol (Bio-Rad, Richmond, CA, USA). Transformed cells were cultured on MS medium for 16–24 h and observed under a confocal microscope (Leica Microsystem, Heidelberg, Germany).
Plant transformation
GmERF3 full-length cDNA was inserted into a polylinker site of binary vector pBI121 in the sense orientation. The constructs were introduced into tobacco W38 using Agrobacterium-mediated transformation as described by Hoekema et al. (1983). Transgenic plants were selected on MS medium containing 200 μg ml−1 kanamycin and 500 μg ml−1 carbenicillin.
Salt- and drought-stress tolerance analysis of transgenic plants and determination of chlorophyll content
Shoot tips excised from aseptic seedlings of both wild-type and GmERF3 transgenic tobacco were submerged into half-strength MS medium containing 200 mM NaCl or 2% PEG for salt and drought tolerance assays, respectively, and were incubated for 1 month. Changes in leaf morphology and degrees of root elongation were measured. For the determination of chlorophyll content, healthy and fully expanded leaves from wild-type and transgenic plants were detached. Leaf discs of 1 cm diameter were cut and floated in MS medium containing 400 mM NaCl for 6 d. The treatment was performed in continuous white light at 25 °C. The chlorophyll content was measured as described by Aono et al. (1993).
Pathogen response assays of transgenic plants
For bacterial infection analysis, Ralstonia solanacearum strain BJ1057 was grown in LB broth, and bacterial cells were collected, washed, and resuspended in 10 mM MgCl2. The density of bacterial cells was determined by measuring absorbance at OD600. Bacterial cells in suspension (1×107 cfu ml−1) were infiltrated into fully expanded 6-week-old tobacco leaves using a 10 ml plastic syringe without a needle, using the method of Thilmony et al. (1995). The bacterial population in leaves was measured by grinding four leaf discs in 10 mM MgCl2, plating serial dilutions on LB plates, and counting colony-forming units.
For fungal resistance analysis, mycelia of Alternaria alternata were cultured on potato dextrose agar at 28 °C. When the mycelia reached the edge of the plate, 0.5 cm diameter agar discs were excised from the edges of growing colonies using a cork borer and inverted onto detached leaves from transgenic and wild-type control plants. All leaves were placed on wet filter paper in Petri dishes and incubated at 28 °C to permit normal disease development under high humidity. After 7 d, lesion diameters were measured, and the average lesion area for each independent transgenic line was calculated and compared with that of wild-type tobacco.
For TMV infection, fully expanded leaves of 6–8-week-old soil-grown tobacco plants were used. Inoculation with TMV strain U1 was performed in 50 mM potassium phosphate buffer, pH 7.5, by gently rubbing the leaves with carborundum. Mock-inoculations were done with carborundum and buffer only.
Expression analysis of putative downstream genes of GmERF3
Total RNA was isolated from wild-type and transgenic plants with Trizol (Tiangen Biotech., Beijing) according to the manufacturer's instructions. Poly (A)+ RNA was used as template for synthesis of the first-strand cDNA. Complementary DNA was generated with reverse transcriptase (TaKaRa, Dalian). Gene-specific primers were designed from the flanking sequence of the AP2/ERF domain as follows: 5′-CTTGGACGTTGACTTCGAGGCTGAT-3′ (forward) and 5′-AGAGTTAGGCTGCTGCTGGTTGGC-3′ (reverse). The specific primer pair (5′-GTCTGGTGATGGTGTTAGC-3′ and 5′-CCTATCAGCAATTCCAGGAAAC-3′) for the soybean Actin gene was used as an internal control to normalize the amount of template added. For expression analysis of stress-responsive genes, RT-PCR amplifications were performed using the following specific primers: 5′-GGTGTAGAACCTTTGACCTGGGACG-3′ and 5′-GAACCCTAGCACATCCAACACGAAC-3′ for PR1 (X12737); 5′-GCAAACAATTTACCATCAGACCA-3′ and 5′-GAGTCCAAAGTGTTTCTCTGTGATA-3′ for PR2 (M60460); 5′-CAGAACATTAACTGGGATTTGAGAG-3′ and 5′-CTCCATTGCTGCATTGATCTACT-3′ for PR4 (X58546); 5′-AAGTTGATGCAAGGGAGATGTCT-3′ and 5′-ATGACATTTAGGACATTTGCTGC-3′ for SAR8.2 (M97194); 5′-TTCCTCCTTGCCTTGGTGACTT-3′ and 5′-ATTCGCCGTTTATAGCCGTACAT-3′ for Osmotin (M29279).
Measurement of volumetric soil water content, free proline, and soluble carbohydrates contents
GmERF3 transgenic and control tobacco plants were withheld from watering for 4 weeks, and were then rewatered. Volumetric soil water contents were measured using TDR-3 soil moisture sensors, and leaves were collected from transgenic and control plants at weekly intervals for measurement of free proline and soluble carbohydrates contents.
Proline analysis was carried out as described by Shan et al. (2007). Fresh leaf material (0.5 g) was extracted with 5 ml of 3% sulphosalicylic acid at 100 °C for 10 min with shaking. The extracts were filtered through glass wool and analysed for proline content using the acid ninhydrin method. Briefly, 2 ml of aqueous extract was mixed with 2 ml of glacial acetic acid and 2 ml of acid ninhydrin reagent (1.25 g of ninhydrin, 30 ml of glacial acetic acid, and 20 ml of 6 M orthophosphoric acid) and heated at 100 °C for 30 min. After cooling, the reaction mixture was partitioned against toluene (4 ml) and the absorbance of the organic phase was determined at 520 nm. The resulting values were compared with a standard curve constructed using known amounts of proline (Sigma, St Louis, MO, USA).
Soluble carbohydrate contents were determined following a method of Xiang et al. (2007). 0.1 g tobacco leaf was cut into small pieces and put into a tube; 10 ml of water was added and the tube was then put into a boiling water bath to allow a 30 min extraction. After cooling to room temperature, 0.5 ml of extract solution was sampled and placed in another tube, to which 1.5 ml of water was added, followed by 1 ml of 90% phenol and 5 ml of concentrated sulphuric acid. The tube was then well shaken. After standing for 30 min, the aqueous extract was assayed for soluble carbohydrates content at the wavelength of 485 nm.
Statistical analysis
The mean values of proline and soluble sugar content were taken from the measurements of four replicates and the ‘Standard Error’ of the means was calculated. Data were analysed by SAS (Statistical Analysis System) software (SAS Corporation, Cory, NC, USA) by the t test to assess significant differences among means.
Results
Isolation and structure analysis of GmERF3
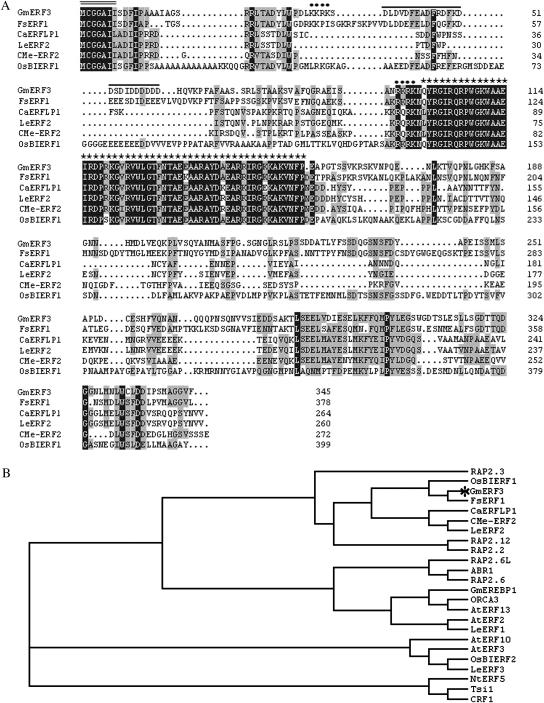
The full-length cDNA sequence of an ERF homologue, GmERF3 gene (GenBank Accession EU681278) was isolated from soybean total RNA by reverse transcription polymerase chain reaction (RT-PCR). Sequence analysis revealed that it contained a 1038 bp open reading frame encoding a polypeptide of 345 amino acids with predicted molecular mass of 38 kDa (pI 4.78). The predicted GmERF3 protein contained a conserved DNA-binding domain (AP2/ERF domain) of 58 amino acids, two putative basic amino acid regions (K32KRK and R94KRK) that potentially acted as NLS, an acidic amino acid-rich region (D37LDVDFEADFRDFKDDSDIDDDDDD) that might act as a transcriptional activation domain in the N-terminal region, and a conserved N-terminal motif of unknown function (MCGGAII) (Fig. 1A). Alignment and phylogenetic tree analysis revealed that GmERF3 is most similar to previously described ERF class B-2 subgroup members (RAP2.3, RAP2.2, and RAP2.12; Sakuma et al., 2002) (Fig. 1B). GmERF3 has 38–59% identity for overall amino acid sequence and 94–99% for the AP2/ERF domain to Fagus sylvatica FsERF1, rice OsBIERF1, Capsicum annuum CaERFLP1, Cucumis melo CMe-ERF2, and tomato LeERF2. Tournier et al. (2003) identified tomato LeERF2, a novel class IV ERF, characterized by an N-terminal signature sequence, MCGGAII/L. This motif was also present in GmERF3 and members of the recently identified Class IV (Tournier et al., 2003; Lee et al., 2004; Cao et al., 2006; Mizuno et al., 2006).
Fig. 1.
Deduced amino acid sequences of AP2/ERF-related proteins and phylogenic relationships of selected AP2/ERF domains from AP2/ERF-related proteins. (A) Comparison of deduced amino acid sequences of AP2/ERF-related proteins that have high sequence similarity with GmERF3. Amino acid residues that are conserved in at least three of the six sequences are shaded, whereas amino acids identical in all six proteins are shown in dark grey. The black bar above the sequences represents the putative acidic domain. Double overline represents the highly conserved N-terminal MCGGAII/L motif of unknown function. Dots (·) represent putative nuclear localization signals. An asterisk (*) indicates a conserved DNA-binding domain (AP2/ERF domain). Dashes show gaps in the amino acid sequences introduced to optimize alignment. (B) Phylogenic comparison of the GmERF3 protein and some AP2/ERF-related protein sequences, based on the selected AP2/ERF domain amino acid sequences of those proteins. Alignments were made in Clustal X using the default parameters. Accession numbers for the AP2/ERF proteins used are as follows: RAP2.3, NP188299; OsBIERF1, AAV98700; FsERF1, CAE54591; CaERFLP1, AAS20427; CMeERF2, BAD01556; LeERF2, AAO34704; RAP2.12, NP001077718; RAP2.2, NP850583; RAP2.6L, NP196837; ABR1, NP201280; RAP2.6, NP175008; AtERF10, NP171876; AtERF3, NP175479; OsBIERF2, AAV98701; LeERF3, AAO34705; NtERF5, AAU81956; GmEREBP1, AAM45475; ORCA3, CAB96899; ATERF13, NP182011; ATERF2, NP199533; LeERF1, AAO34703; Tsi1, AAC14323; CRF1, NP192852.
Expression pattern of the GmERF3 gene under various stress conditions
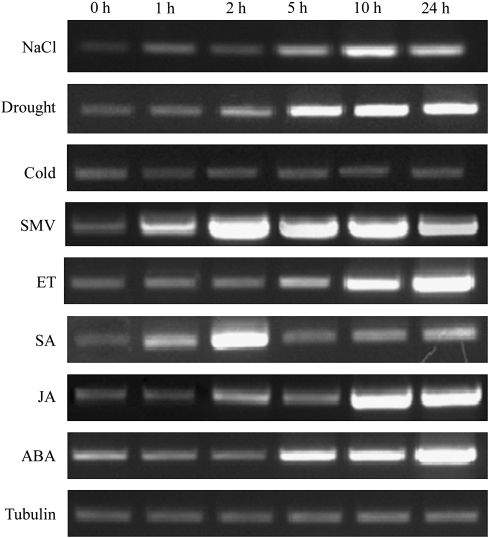
In plants, ERFs play a direct regulatory role in response to multiple signal stimulation. In order to clarify the potential function of GmERF3 in response to different stimuli, the expression pattern of the GmERF3 gene was analysed under various abiotic and biotic stress conditions. As shown in Fig. 2, GmERF3 expression was induced after salt, drought, SMV, SA, ET, ABA, and JA treatments. However, there was no significant GmERF3 mRNA accumulation under cold stress treatment. GmERF3 had a low expression level under normal conditions. Under salt treatment, GmERF3 mRNA accumulated at 5 h after initiation of the treatment and reached a maximum at 10 h. At 24 h, the mRNA levels declined to the level of 5 h. For drought and JA treatments, transcripts of GmERF3 were induced at 2 h, and remained at a very high level at 5–24 h and at 10–24 h, respectively. For SMV and SA treatments, GmERF3 mRNA accumulated within 1 h and reached maxima at 2 h; thereafter, mRNA levels declined. For ET and ABA treatments, GmERF3 mRNA accumulated within 5 h, and thereafter, expression levels increased gradually for at least 24 h.
Fig. 2.
Expression of the GmERF3 gene in response to biotic and abiotic stresses. Time-course of GmERF3 induction after treatment with 200 mM NaCl, drought, cold, SMV, 2 mM SA, 1 mM ET, 100 mM ABA, and 100 mM JA for the indicated times. Total RNAs were purified from 2-week-old seedlings of wild-type soybean after the above treatments and reverse-transcribed. The resultant cDNAs were used as templates for RT-PCR and the tubulin gene was amplified as a template control. The tubulin gene amplification results for NaCl-treated samples are presented. The amplifications of the tubulin gene for other treatments proved similar and were omitted for simplicity.
GmERF3 protein specifically binds to both the GCC box and DRE/CRT element in vitro
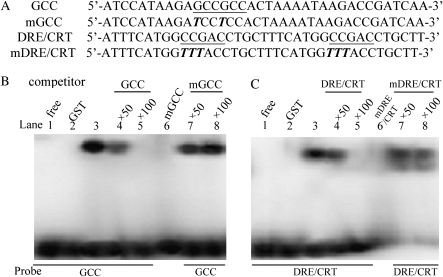
Some proteins in the ERF subfamily have dual binding activities with the GCC box and DRE/CRT sequence (Park et al., 2001; Hao et al., 2002; Zhang et al., 2004; Lee et al., 2005; Tang et al., 2007). To test whether GmERF3 binds to the cis-acting elements of the GCC box and DRE/CRT element in vitro, gel retardation assays were performed. Purified proteins from E. coli cultures harbouring the appropriate vector without the GmERF3 gene were isolated as a control (GST control). The sequences of GCC, mutated GCC (mGCC), DRE/CRT, and mutated DRE/CRT (mDRE/CRT) are shown in Fig. 3A. When the GCC box was used as a probe, the soybean GmERF3 protein caused a mobility shift in the radioactively labelled GCC probe (Fig. 3B, lane 3), which migrated more slowly than the free probe (Fig. 3B, lane 1). This shift was specific to the soybean protein and did not occur in the GST control (Fig. 3B, lane 2). Furthermore, when a mutated version of the GCC box (mGCC) was used in the assay, the mobility shift was not observed (Fig. 3B, lane 6). Competition experiments were conducted to examine the specificity of the mobility shift. The purified soybean GmERF3 protein was mixed with the radiolabelled GCC probe, and an unlabelled competitor (either the GCC probe or the mGCC probe) was then added to the mixture in varying amounts. As the levels of unlabelled GCC increased, the amount of bound labelled probe decreased. A 50-fold excess of a cold GCC sequence was sufficient to displace the labelled probe (Fig. 3B, lane 4). When the ratio of unlabelled to labelled GCC probe reached approximately 100:1, virtually no labelled probe was bound (Fig. 3B, lane 5). When the unlabelled mGCC probe was used as the competitor, no binding competition was observed, even at a ratio of 100:1 unlabelled mGCC to labelled GCC (Fig. 3B, lanes 7 and 8). Similar experiments were carried out with DRE/CRT and its mutated sequence. Recombinant GmERF3 was also able to bind the DRE/CRT sequence in a specific manner (Fig. 3C), although this binding capacity seemed to be lower than to the GCC box.
Fig. 3.
Sequence-specific binding activity of GmERF3 to the GCC box and DRE/CRT element. (A) Nucleotide sequences of GCC, mutated GCC (mGCC), DRE/CRT, and mutated DRE/CRT (mDRE/CRT) probes. The core sequences of GCC and DRE/CRT elements are underlined and mutated nucleotides in mGCC and mDRE/CRT probes are expressed in bold italic. (B) Gel retardation assay showing sequence-specific binding to the GCC box of the recombinant GmERF3 protein. 0.2 μg GST-GmERF3 protein was added to each reaction mixture. Lane 1 contained only the free GCC probes, and lane 2 contained free GCC probe and GST protein. Lane 3, radiolabelled GCC probe; lanes 4 and 5, titration with a cold GCC sequence as a competitor; lane 6, radiolabelled mutated GCC (mGCC) probe; lanes 7 and 8, titration with cold mGCC sequence as a competitor. (C) Gel retardation assay showing sequence-specific binding to the DRE/CRT element of the recombinant GmERF3 protein. 0.2 μg GST-GmERF3 protein was added to each reaction mixture. Lane 1 contained only the free DRE/CRT probe, and lane 2 contained free DRE/CRT probe; GST protein; lane 3, radiolabelled DRE/CRT probe; lanes 4 and 5, titration with a cold DRE/CRT sequence as a competitor; lane 6, radiolabelled mutated DRE/CRT (mDRE/CRT) probe; lanes 7 and 8, titration with a cold mDRE/CRT sequence as a competitor.
Transcriptional activation activity of GmERF3
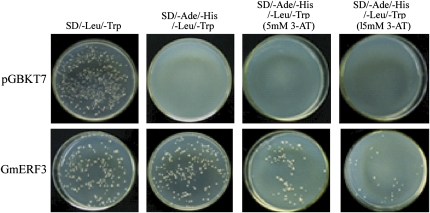
GmERF3 has an acidic N-terminal region (aa 37–61, pI=4.78), which could act as a transcriptional activation domain (Liu et al., 1999). To determine whether GmERF3 could act as a transcriptional activator in yeast, the yeast two-hybrid analysis was used. The full-length GmERF3 gene was fused to the DNA-binding domain of GAL4 (Clontech, Palo Alto, CA, USA) to identify the transcriptional activation activity by growing the yeast cells on SD/–Leu/–Trp and SD/–Ade/–His/–Leu/–Trp media. Yeast cells carrying the pGBKT7 plasmid, which contains only the GAL4 DNA-binding domain, were used as the negative control. It grows on SD/–Leu/–Trp, but not on the SD/–Ade/–His/–Leu/–Trp medium. Together with the GAL4 activation domain, yeast cells carrying full-length GmERF3 fused to the GAL4 DNA binding domain, activated the transcription of downstream reporter genes and grew on SD/–Ade/–His/–Leu/–Trp medium (Fig. 4). Further analysis suggested yeast cells carrying full-length GmERF3 can also grow on SD/–Ade/–His/–Leu/–Trp medium with 5 mM 3-amino-1, 2, 4-triazole (3-AT), and even on 15 mM 3-AT (Fig. 4).
Fig. 4.
Transactivation activity of GmERF3 in yeast cells. The GmERF3 gene was fused in-frame to the GAL4 DNA-binding domain (DB) expression vector and then transformed into yeast strain AH109. The transformants were selected by growth on SD media (–Trp, –Leu), (–Trp, –Leu, –Ade, –His), without 3-AT or with 5 mM and 15 mM 3-AT, respectively. Plates were incubated at 30 °C for 4 d, and then photographed. pGBKT7 indicates the vector used in this experiment, it expressed GAL4 DB alone. (This figure is available in colour at JXB online.)
Targeting of GmERF3 to the nucleus
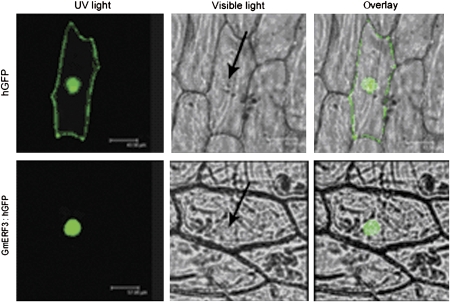
The N-terminal region of the putative GmERF3 protein contained two putative basic amino acid regions (K32KRK and R94KRK) that potentially act as nuclear localization signals, and indicate that the protein is likely to localize in the nucleus. To confirm this, an in vivo targeting experiment was performed whereby the GmERF3 coding region was fused to the N-terminus of the hGFP gene under the control of the CaMV 35S promoter and transferred into onion epidermal cells using the particle bombardment method. Localization of the fusion protein was then determined by visualization with a fluorescence confocal microscope. As shown in Fig. 5, the control hGFP was uniformly distributed throughout the cell, whereas the GmERF3:hGFP fusion protein was exclusively localized to the nucleus.
Fig. 5.
Nuclear localization of the GmERF3 protein in onion epidermal cells. Onion epidermal cells were transiently transformed with constructs containing either control hGFP or GmERF3:hGFP under the control of the CaMV35S promoter by the particle bombardment transformation method. Subcellular localization of the GmERF3-hGFP fusion protein or control hGFP alone was viewed with a fluorescent confocal microscope 16 h after bombardment. Fluorescence images (left), bright-field images (middle), and the corresponding overlay images (right) of representative cells expressing hGFP or GmERF3:hGFP fusion protein are shown. Arrows indicate the location of nuclei. (This figure is available in colour at JXB online.)
Overexpression of GmERF3 in transgenic tobacco enhances expression of PR genes and confers pathogen tolerance
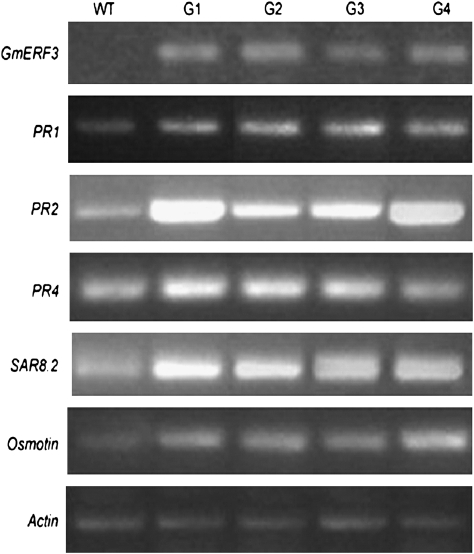
To investigate the functions of GmERF3 further and to determine whether it activates expression of downstream PR genes, transgenic tobacco plants that constitutively express the GmERF3 gene under the control of the 35S promoter were developed. The expression of GmERF3 and its possible target genes was analysed in 35S::GmERF3 tobacco plants and compared with that of wild-type plants. Transcripts of PR1, Osmotin, SAR8.2, and PR2 were increased in all the transgenic lines, and the transcript of PR4 also increased, except in line G4 (Fig. 6). It has been well demonstrated that PR1, PR2, PR4, Osmotin, and SAR8.2 are GCC-box containing genes and their expression can be activated or repressed by different ERF transcription factors (Park et al., 2001; Gu et al., 2002; Fischer and Dröge-Laser, 2004), and the expression of these genes is related to the onset of systemic acquired resistance in tobacco (Ward et al., 1991).
Fig. 6.
Expression analysis of GmERF3 putative downstream genes in wild-type and 35S::GmERF3 transgenic plants under normal conditions. Total RNA was isolated from the plants and analysed by RT-PCR using Actin as a template control, G1 to G4 indicate GmERF3-transformed independent tobacco lines.
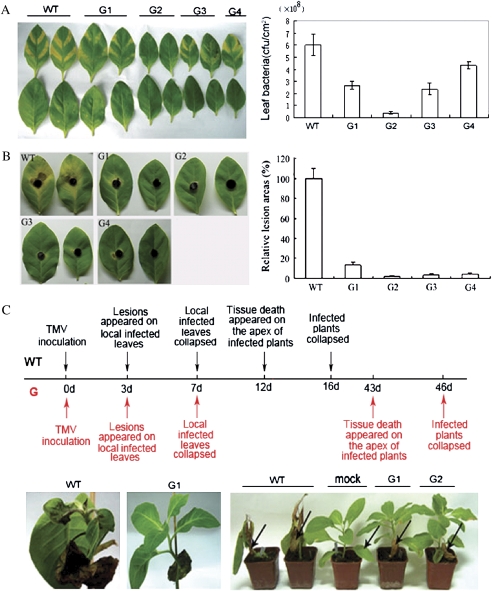
To determine whether overexpression of GmERF3 enhanced resistance to pathogens in transgenic tobacco plants, the bacterial pathogen Ralstonia solanacearum was injected into leaves of wild-type and transgenic plants, and symptom development was subsequently monitored for 7 d. As shown in Fig. 7A, all transgenic lines exhibited significantly reduced disease lesions and leaf bacterial numbers compared to wild-type plants. Approximately 50% inhibition of bacterial growth was detected in 35S::GmERF3 tobacco plants during the 7-d infection period (P <0.01). Line G2 showed stronger resistance than other transgenic lines tested.
Fig. 7.
Resistance analysis of GmERF3 transgenic tobacco plants. (A) Resistance of transgenic tobacco to the bacterial pathogen Ralstonia solanacearum. Fully expanded leaves of 7-week-old tobacco plants were syringe-infiltrated with 107 cfu ml−1 solution of R. solanacearum. Disease symptoms in wild-type and GmERF3 transgenic plants are shown in the upper panel; leaves in the upper line were inoculated with bacteria, and leaves below the lower line were mock-inoculated. The photograph was taken 7 d after inoculation. Infected leaves were collected and bacterial populations are shown in the lower panel. Values are means of three different experiments. Error bars indicate the SE. (B) Responses of transgenic tobacco to the fungal pathogen Alternaria alternata. Detached leaves were challenged with mycelia of A. alternata. The photograph was taken 7 d following inoculation. Disease symptoms are shown on the right. The average lesion area of each independent transgenic line (n=4) was calculated and their relative lesion areas are shown in columns after comparison with the average lesion area on wild-type tobacco. (C) Time-course for the systemic spread of TMV in wild-type and GmERF3 transgenic tobacco plants. The line in the middle of the upper panel indicates time. Notes above and below the line describe symptoms of wild-type and GmERF3 transgenic tobacco plants caused by TMV infection and spread, respectively. WT and G represent the wild-type and GmERF3 transgenic plants, respectively. Photographs were taken 12 d (right and middle of the lower panel) and 16 d (left of the lower panel) after inoculation. Arrows indicate TMV-inoculated leaves or mock-inoculated leaves. (This figure is available in colour at JXB online.)
Resistance to the fungal pathogen Alternaria alternata was also identified in GmERF3 transgenic tobacco plants by a detached leaf inoculation test. Forty-eight tobacco plants from 12 transgenic lines were assayed, and 7 d after inoculation, tobacco controls showed typical necrosis symptoms surrounded by chlorotic halos and extensive pathogen sporulation whereas symptoms on transgenics, if present, were faint chloroses or hypersensitive-like necroses with little sporulation (Fig. 7B). Symptoms of infection appeared at 83% of the inoculation sites on the control plants, whereas the comparative value for all transgenics was 21%. As shown in Fig. 7B, four transgenic lines, G1–G4, had significantly (P <0.05) smaller lesion sizes than wild-type plants at 7 d after inoculation. The levels of resistance varied among transgenic lines. Approximately 30% of the transgenic lines were completely resistant to A. alternata and failed to develop symptoms even when exposed to the pathogen repeatedly and for long periods.
To quantify resistance responses to the virus pathogen, wild-type and transgenic tobacco plants were inoculated with TMV. All infected transgenic lines had no significantly reduced lesion numbers and sizes on inoculated leaves relative to wild-type plants. Infected leaves of both wild-type and transgenic plants were collapsed within 7 d after inoculation (Fig. 7C). The virus continued to spread, and leaves at the apices of wild-type plants were collapsed 16 d after inoculation (Fig. 7C). Comparatively, apices of GmERF3 overexpressing plants showed no symptoms, indicating that the virus had not translocated to that tissue. Forty-six days after inoculation, leaves at the apices of GmERF3 overexpressing plants were collapsed. Mock-inoculated wild-type tobacco plants produced no symptoms.
Overexpression of GmERF3 enhances salt and drought tolerance in transgenic tobacco plants
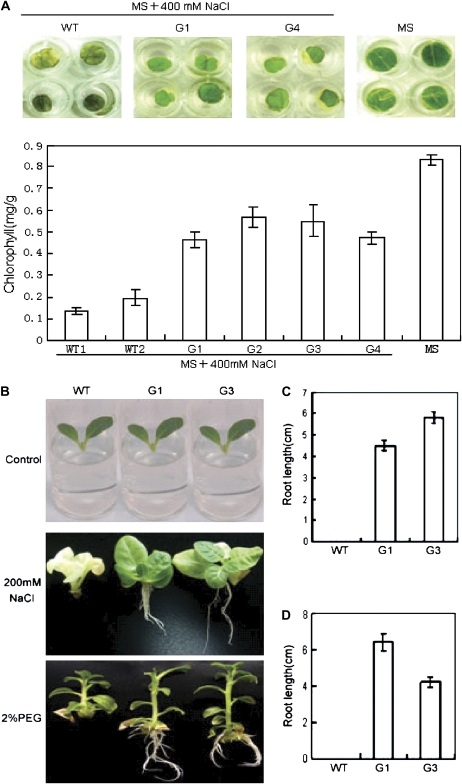
In order to test whether GmERF3 overexpression changes the response of plants to abiotic stresses, leaf discs of transgenic and wild-type tobacco plants were first floated on MS solution containing 400 mM NaCl for 5 d and plant salt tolerances were examined by comparing phenotypes and chlorophyll contents. After 5 d of salt treatment, leaf discs from the wild type were bleached, whereas leaf discs from transgenic GmERF3 plants remained green (upper panel, Fig. 8A). Chlorophyll content measurements in these plants confirmed the observed phenotypic differences (Fig. 8A, lower panel). Although there were differences between transgenic lines, their abilities to tolerate salt were higher than those of wild-type plants. Shoot tips excised from aseptic seedlings of both wild-type and GmERF3 transgenic tobacco were transferred to half-strength MS medium containing 200 mM NaCl. Significant phenotypic differences between wild-type plants and transgenic lines were observed after 30 d (Fig. 8B, middle panel). During that period, leaves of wild-type plants gradually lost greenness and root elongation was severely retarded, whereas leaves of the transgenic plants remained green and the roots displayed tolerance against salt stress (Fig. 8C). Root formation on wild-type plants was zero.
Fig. 8.
GmERF3 enhances tolerance to salt and drought in tobacco. (A) Chlorophyll contents in transgenic tobacco leaf tissues after salt treatment. Leaf discs from transgenic plants carrying the GmERF3 gene and wild-type plants were floated on half-strength MS liquid medium containing 400 mM NaCl for 5 d. As a control, wild-type leaf discs were floated on half-strength MS liquid medium. Phenotypic differences were observed and chlorophyll contents (mg g−1 fresh weight) were measured from NaCl-treated leaf discs of 35S::GmERF3 transgenic and wild-type tobacco plants. The experiments were repeated twice, each time with 4–8 leaf discs. (B) Phenotypes on half-strength MS medium containing 200 mM NaCl or 2% PEG were photographed 30 d after treatment. Control indicates seedlings before treatment. (C) Root lengths of seedlings after salt treatment; the data shown are relative to the control plus SD. (D) Root lengths of seedlings after drought treatment; data are relative to the control plus SD. (This figure is available in colour at JXB online.)
To analyse the effect of overexpression of GmERF3 on drought tolerance, shoot tips excised from aseptic seedlings of both wild-type and GmERF3 transgenic tobacco were submerged in half-strength MS medium containing 2% PEG6000 and rooted for 30 d. As shown in Fig. 8B (lower panel), leaves of the transgenic plants remained green and the roots displayed tolerance against drought stress. Although the leaves of wild-type plants remained green, root elongation was severely retarded (Fig. 8D).
These results suggested that overexpression of GmERF3 enhanced salt and drought tolerance in transgenic tobacco plants. On the contrary, 35S::GmERF3 transgenic tobacco plants did not exhibit detectable tolerance against cold stress (data not shown).
Overexpression of GmERF3 improved accumulation of free proline and soluble carbohydrate contents
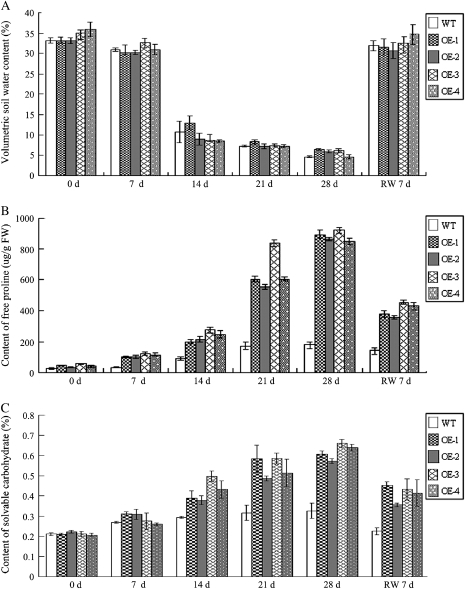
To evaluate physiological changes in transgenic plants, the contents of free proline and soluble carbohydrates as osmotic regulators in the wild-type and 35S::GmERF3 transgenic tobaccos were measured following drought stress. Under normal growth conditions, free proline concentrations in GmERF3-overexpressing plants were approximately 2-fold higher than those in the control plants (P <0.01) (Fig. 9B). During drought treatment, the contents of proline in both GmERF3 transgenic plants and the control plants rose continuously. This effect was especially significant after 14 d. After 4 weeks, transgenic line G3 had the highest proline content, 5-fold that in the control (P <0.01). The content of proline in other transgenic plants was also higher than the control (Fig. 9B). The level of proline accumulation decreased significantly during the recovery period. Furthermore, volumetric soil water content of the transgenic lines at different time points under drought stress were about the same as those of the control plants. Both decreased gradually and returned to the normal level 7 d after rewatering (Fig. 9A). Comparison of soluble carbohydrates in wild-type and GmERF3 transgenic plants showed remarkably higher levels of soluble carbohydrates in transgenic plants than wild-type plants. A low but significant amount of soluble sugars was detected in wild-type plants; these levels increased significantly under drought stress. Transgenic plants grown under control conditions exhibited levels of soluble carbohydrates comparable with wild-type plants (Fig. 9C). After drought stress, the transgenic lines (G1–G4) showed 1.5–2 times higher levels of soluble sugars compared to wild type plants (P <0.05) (Fig. 9C).
Fig. 9.
Effects of water stress on free proline and soluble carbohydrate contents of wild-type and 35S::GmERF3 transgenic tobacco plants. Wild-type and 35S::GmERF3 transgenic tobacco plants were grown under normal conditions for 7 weeks and then exposed to water deprivation stress for 28 d, before resumption of normal conditions. Volumetric soil water content was measured and samples for total soluble carbohydrates and proline contents measurements were taken at different times as indicated. (A) Comparison of volumetric soil water contents of wild-type and 35S::GmERF3 transgenic tobacco plants. (B) Free proline contents of wild-type and 35S::GmERF3 transgenic tobacco plants. (C) Soluble carbohydrate contents of wild-type and 35S::GmERF3 transgenic tobacco plants. WT, wild-type control; G1–G4, different transgenic lines.
Discussion
In this study, a new member of the AP2/ERF transcription factor family, GmERF3, was identified in soybean. According to Sakuma et al. (2002), the DREB and ERF subfamilies differ by two conserved amino acid residues in the AP2/ERF domain. That is, the 14th valine and the 19th glutamic acid are conserved in the DREB proteins, whereas alanine and aspartic acid residues are conserved at the corresponding positions of ERF proteins. In agreement with other ERF proteins, GmERF3 has the 14th alanine and 19th aspartic acid in the 58 amino acid AP2/ERF domain, suggesting that GmERF3 is a member of the ERF subfamily. ERFs have been shown to act as activators or repressors of transcription. NtERF3, AtERF3/4, and AtERF7-12 characterized by the well-defined EAR repressor domain, were shown to be active repressors (Fujimoto et al., 2000; Ohta et al., 2000; Yang et al., 2005). Tobacco NtERF2/4, Arabidopsis AtERF1/2/5, periwinkle ORCA2/3, and tomato Pti4 function as activators of transcription (Zhou et al., 1997; Menke et al., 1999; Fujimoto et al., 2000; Ohta et al., 2000; van der Fits and Memelink, 2000). In this study, stretches of acidic amino acids in GmERF3 were located at the N termini. These are capable of activating transcription of reporter genes in yeast cells, suggesting that GmERF3 is a transcriptional activator. In addition, GmERF3 contains two basic amino acid stretches (K32KRK and R94KRK) and a conserved N-terminal MCGGAII/L sequence. Recombinant GmERF3 protein localized to the nucleus and bound specifically to the GCC box and DRE/CRT sequences in vitro. The function of the N-terminal MCGGAII/L motif is unknown, but is unlikely to be required for nuclear localization or for binding to the GCC box (Tournier et al., 2003). Tournier et al. (2003) isolated four new members of the ERF subfamily from tomato (LeERF1–4), and phylogenetic analysis indicated that LeERF2 belonged to a new ERF class characterized by a conserved N-terminal MCGGAII/L sequence. This N-terminal motif is found only in ERF genes. In addition to GmERF3, CaERFLP1 (Lee et al., 2004), TaERF1 (Xu et al., 2007), Tsi1 (Park et al., 2001), and CaPF1 (Yi et al., 2004) also contain the N-terminal signature sequence and belong to the class IV ERF subfamily, which specifically binds both the GCC box and DRE/CRT element. It is worth noting that the AP2/EREBP domain fragment of Arabidopsis CBF1 can interact in vitro with the sequence containing not only the DRE/CRT motif, but also the GCC element (Hao et al., 2002). This dual-binding activity was probably due to the common core sequence –CCGNC–, which occurs in two different upstream elements. This, in conjunction with our data, may be indicative of possible cross-functionality between the ERF and DREB transcription factors.
In plants, SA, JA, and ethylene are important inducers of defence-related genes. By contrast, the plant hormone ABA regulates interacting signalling pathways involved in plant responses to several abiotic stresses (Shinozaki et al., 2003), such as drought, salt, and cold, as well as plant growth and development. Recent research showed that ABA is implicated in increasing the resistance of plants towards pathogens via its positive effect on callose deposition (Ton et al., 2004, 2005). In the present study, the expression of GmERF3 was induced not only by SA, JA, ET, and SMV, but also by ABA, high salinity, and drought. It was proposed that biotic and abiotic stresses induce overlapping sets of genes in plants (Fujita et al., 2006). Biotic stress (SMV infection), abiotic stress (high salinity and drought) or plant hormones (JA, SA, ET, and ABA) induce GmERF3 expression in soybean. This has led to the hypothesis that GmERF3 might act as a connector linking different signalling pathways that mediate biotic and abiotic stress responses.
ERF subfamily genes play various roles in plant growth, development, and response to different environmental stress factors (Okamuro et al., 1997). It is a common observation that certain transcription factors serve as targets for different signalling pathways. In this study, transgenic tobacco plants overexpressing the GmERF3 gene showed increased resistance to Ralstonia solanacearum, Alternaria alternata, and TMV, and enhanced tolerance to high salinity and drought stresses. This may be due to the increased expression of stress-inducible genes induced by the overexpression of GmERF3. GmERF3 binds specifically to the GCC box sequence in vitro; this motif is present in the promoter regions of many ethylene-inducible genes, including PR genes (Ohme-Takagi and Shinshi, 1995), and also binds to the DRE/CRT element (Park et al., 2001), present in the promoter regions of drought-, salt-, and freezing-related genes (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999). In this study, overexpression of GmERF3 in transgenic tobacco activated the expression of several PR genes, such as PR1, PR2, PR4, Osmotin, and SAR8.2 under normal unstressed conditions. Analysis of these PR promoters revealed that GCC cis-elements were crucial for pathogen-induced gene activation. Therefore, it is possible that biotic and abiotic stresses regulate GmERF3 expression; the abundance of GmERF3 transcriptional activator then most likely up-regulates the transcription of several stress-inducible genes, and their protein products contribute to the increased levels of endurance under the stress conditions. In addition, soluble sugars and free proline accumulation may play a highly protective role in plants that are exposed to biotic or abiotic stresses. In wheat, many studies showed that wheat cultivars with higher K+, Pro, and soluble sugar, and lower MDA at different growth stages had better stress resistances (Hongbo et al., 2006). The involvement of proline in response to water deficit was demonstrated in transgenic tobacco overexpressing proline biosynthesis enzymes (Roosens et al., 2002). Transgenic Arabidopsis plants overexpressing CBF3/DREB1A were shown to accumulate proline even under unstressed control conditions (Gilmour et al., 2000). In this study, 35S::GmERF3 transgenic tobacco also accumulated higher levels of free proline and soluble sugars than wild-type plants under unstressed and drought stress conditions (Fig. 9). Those results together prove that the function of ERF genes is conservative in different plant species. However, more evidence is needed to support the conclusion that the accumulation of sugars or proline is a direct reason for more stress tolerance in transgenic plants, such as Gene Chip or MicroArray analysis.
Transgenic tobacco plants overexpressing the GmERF3 gene showed increased resistance to the bacterial pathogen Ralstonia solanacearum and the fungal pathogen Alternaria alternata, which exhibited significantly reduced disease lesions compared with wild-type controls. What kind of mechanism(s) will inhibit or kill these pathogens? The high antioxidant capacity of plants may play an important role in resistance to necrotic symptoms caused by abiotic or biotic stresses (Mittler et al., 1999). Tobacco plants exhibiting high antioxidant capacity develop multi-resistance to several stresses (Barna et al., 1993). Breeding plants for high antioxidant capacity is one effective way to create multi-resistant crops (Kira'ly, 2000). In addition, an increase in the activity of chitinases and some other cell-wall-degrading enzymes or the accumulation of antimicrobial compounds may have a role in arresting the spread of the pathogen (Kira'ly et al., 2007). Increased activity of chitinases degrades chitin in fungal cell walls (van Loon et al., 2006). Interestingly, the activity of chitinases is characteristic of plants resistant to viral or bacterial attack. In the case of bacterial infections, a likely explanation is that most chitinases exhibit lysozyme activity (Kira'ly et al., 2007). Using Gene Chip or MicroArray analysis tools in further studies should indicate whether or not elevated antioxidant capacity, induced expression of chitinases, or antimicrobial compounds in GmERF3 transgenic plants cause the enhanced resistance to bacterial and fungal pathogens. In contrast to NtERF5, which when overexpressed, resulted in enhanced resistance to TMV by suppressing viral replication (Fischer and Dröge-Laser, 2004), resistance to TMV in GmERF3 overexpressing tobacco plant might be due to significantly suppressed viral spread. GmERF3 transgenic lines failed to exhibit significantly reduced lesion size and leaf lesion numbers compared with wild-type plants in the infected leaves. However, the rate of collapse of the apex tissues of the infected transgenic plants was clearly delayed relative to infected wild-type plants. The possibility cannot be ruled out that the suppression of viral spread was a consequence of the reduced TMV propagation. The mechanism of this effect remains unknown. In order to demonstrate whether GmERF3 is part of a naturally-occurring resistance pathway, RNAi plants will need to be analysed. In conclusion, the data presented in this report suggest that GmERF3 as a positive transcription factor might have a role in cross-talk between the biotic and abiotic stress signal pathways in plants.
Acknowledgments
The authors are grateful to Dr RA McIntosh (Plant Breeding Institute, University of Sydney, Australia) for reviewing this manuscript. This work was funded by the National HITECH Research and Development Program of China (‘863’ program, No. 2008AA10Z124 and No. 2006AA10A111), the National Natural Science Foundation of China (No. 30700508), the National Key Project for Researches on Transgenic Plant (No. 2008ZX08002-002). We thank Dr Lijuan Qiu (Soybean Molecular Breeding Group, Institute of Crop Sciences, CAAS) for the gift of soybean cultivars.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Aono M, Kubo A, Saji H, Tanaka K, Kondo N. Enhanced tolerance to photooxidative stress of transgenic Nicotiana tabacum with high chloroplastic glutathione reductase activity. Plant and Cell Physiology. 1993;34:129–136. [Google Scholar]
- Barna B, Ádám L, Király Z. Juvenility and resistance of a superoxide-tolerant plant to diseases and other stresses. Naturwissenschaften. 1993;80:420–422. [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. Journal of Plant Physiology. 2006;163:1167–1178. doi: 10.1016/j.jplph.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Choi MS, Kim MC, Yoo JH, et al. Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.) Journal of Biological Chemistry. 2005;280:40820–40831. doi: 10.1074/jbc.M504616200. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S. The control of maize spikelet meristem fate by the APETALA/12-like gene indeterminate spikelet l. Genes and Development. 1998;12:1145–1154. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Dröge-Laser W. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Molecular Plant–Microbe Interactions. 2004;17:1162–1171. doi: 10.1094/MPMI.2004.17.10.1162. [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. The Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signalling networks. Current Opinion in Plant Biology. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB. Tomato transcription factors Pti4, Pti5, and Pti6 activate defence responses when expressed in Arabidopsis. The Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology. 2004;7:465–471. doi: 10.1016/j.pbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Hao D, Yamasaki K, Sarai A, Ohme-Takagi M. Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry. 2002;41:4202–4208. doi: 10.1021/bi015979v. [DOI] [PubMed] [Google Scholar]
- Hao DY, Ohme-Takagi M, Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. Journal of Biological Chemistry. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Hirsch P, Hooykaas PJJ, Schilperoort R. A binary plant vector strategy based on separation of virand T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature. 1983;303:179–180. [Google Scholar]
- Hongbo S, Zongsuo L, Mingan S. Osmotic regulation of 10 wheat (Triticum aestivum L.) genotypes at soil water deficits. Colloids Surf B Biointerfaces. 2006;47:132–139. doi: 10.1016/j.colsurfb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAVI is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Research. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenbergerm O, Thomashow MF. Arabidopsis CPF1 overexpression induces cor genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nature Biotechnology. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kira'ly Z. New aspects of breeding crops for disease resistance: the role of antioxidants. In: Hrazdina G, editor. Use of agriculturally important genes in biotechnology. Amsterdam, The Netherlands: IOS Press; 2000. pp. 124–130. [Google Scholar]
- Kira'ly L, Barna B, Kira'ly Z. Plant resistance to pathogen infection: forms and mechanisms of innate and acquired resistance. Journal of Phytopathology. 2007;155:385–396. [Google Scholar]
- Lee JH, Hong JP, Oh SK, Lee S, Choi D, Kim WT. The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Molecular Biology. 2004;55:61–81. doi: 10.1007/s11103-004-0417-6. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim DM, Lee JH, Kim J, Bang JW, Kim WT, Pai HS. Functional characterization of NtCEF1, an AP2/EREBP-type transcriptional activator highly expressed in tobacco callus. Planta. 2005;222:211–224. doi: 10.1007/s00425-005-1525-5. [DOI] [PubMed] [Google Scholar]
- Liu L, White MJ, MacRae TH. Transcription factors and their genes in higher plants: functional domains, evolution and regulation. European Journal of Biochemistry. 1999;262:247–257. doi: 10.1046/j.1432-1327.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- Mazarei M, Elling AA, Maier TR, Puthoff DP, Baum TJ. GmEREBP1 is a transcription factor activating defence genes in soybean and Arabidopsis. Molecular Plant–Microbe Interactions. 2007;20:107–119. doi: 10.1094/MPMI-20-2-0107. [DOI] [PubMed] [Google Scholar]
- Mazarei M, Puthoff DP, Hart JK, Rodermel SR, Baum TJ. Identification and characterization of a soybean ethylene-responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Molecular Plant–Microbe Interactions. 2002;15:577–586. doi: 10.1094/MPMI.2002.15.6.577. [DOI] [PubMed] [Google Scholar]
- Menke FL, Champion A, Kijne JW, Memelink J. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO Journal. 1999;16:4455–4463. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Herr EH, Orvar BL, Van Camp W, Inzé D, Ellis E. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proceedings of the National Academy of Sciences, USA. 1999;96:14165–14170. doi: 10.1073/pnas.96.24.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno S, Hirasawa Y, Sonoda M, Nakagawa H, Sato T. Isolation and characterization of three DREB/ERF-transcription factors from melon (Cucumis melo) Plant Science. 2006;170:1156–1163. [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H. Three ethylene responsive- transcription factors in tobacco with distinct transactivation functions. The Plant Journal. 2000;22:29–38. doi: 10.1046/j.1365-313x.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van-Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Singh KB. Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiology. 2002;128:1313–1322. doi: 10.1104/pp.010862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. The Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosens NH, Al Bitar F, Loenders K, Angenon G, Jacobs M. Overexpression of ornithine-δ-aminotransferase increases Pro biosynthesis and confers osmotolerance in transgenic plants. Molecular Breeding. 2002;9:73–80. [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Shan DP, Huang JG, Yang YT, Guo YH, Wu CA, Yang GD, Gao Z, Zheng CC. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytologist. 2007;176:70–81. doi: 10.1111/j.1469-8137.2007.02160.x. [DOI] [PubMed] [Google Scholar]
- Shin R, Park JM, An JM, Paek KH. Ectopic expression of Tsi1 in transgenic hot pepper plants enhances host resistance to viral, bacterial, and oomycete pathogens. Molecular Plant–Microbe Interactions. 2002;15:983–989. doi: 10.1094/MPMI.2002.15.10.983. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Current Opinion in Plant Biology. 2003;6:410–417. doi: 10.1016/s1369-5266(03)00092-x. [DOI] [PubMed] [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Molecular Biology. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- Tang M, Sun J, Liu Y, Chen F, Shen S. Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Molecular Biology. 2007;63:419–428. doi: 10.1007/s11103-006-9098-7. [DOI] [PubMed] [Google Scholar]
- Tang W, Charles TM, Newton RJ. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Molecular Biology. 2005;59:603–617. doi: 10.1007/s11103-005-0451-z. [DOI] [PubMed] [Google Scholar]
- Thilmony RL, Chen Z, Bressan RA, Martin GB. Expression of the tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv. tabaci expressing avrPto. The Plant Cell. 1995;7:1529–1536. doi: 10.1105/tpc.7.10.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanism. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Ton J, Jakab G, Toquin V, Flors V, Iavicoli A, Maeder MN, Métraux JP, Mauch-Mani B. Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. The Plant Cell. 2005;17:987–999. doi: 10.1105/tpc.104.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. The Plant Journal. 2004;38:119–130. doi: 10.1111/j.1365-313X.2004.02028.x. [DOI] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech JC, Bouzayen M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Letters. 2003;550:149–154. doi: 10.1016/s0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Rep M, Pieterse CM. Significance of inducible defence-related proteins in infected plants. Annual Review of Phytopathology. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux JP, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. The Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Huang Y, Xiong L. Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiology. 2007;144:1416–1428. doi: 10.1104/pp.107.101295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZS, Xia LQ, Chen M, et al. Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Molecular Biology. 2007;65:719–732. doi: 10.1007/s11103-007-9237-9. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. The Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D. The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiology. 2004;136:2862–2874. doi: 10.1104/pp.104.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, Xu Z, Guan S, Li LC, Li A, Guo J, Mao L, Ma Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) Journal of Experimental Botany. 2008;59:4095–4107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Huang Z, Xie B, Chen Q, Tian X, Zhang X, Zhang H, Lu X, Huang D, Huang R. The ethylene-, jasmonate-, abscisic acid-, and NaCl-responsive tomato transcription factor JERF1 modulates expression of GCC box-containing genes and salt tolerance in tobacco. Planta. 2004;220:262–270. doi: 10.1007/s00425-004-1347-x. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO Journal. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo KJ, Qin J, Zhao JY, Ling H, Zhang LD, Cao YF, Tang KX. Over-expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes. Gene. 2007;391:80–90. doi: 10.1016/j.gene.2006.12.019. [DOI] [PubMed] [Google Scholar]