Abstract
The single-celled trichome of Arabidopsis thaliana is a widely used model system for studying cell development. While the pathways that control the later stages of trichome development are well characterized, the early signalling events that co-ordinate these pathways are less well understood. Hormones such as gibberellic acid, salicylic acid, cytokinins, and ethylene are known to affect trichome initiation and development. To understand the role of the plant hormone ethylene in trichome development, an Arabidopsis loss-of-function ethylene receptor mutant, etr2-3, which has completely unbranched trichomes, is analysed in this study. It was hypothesized that ETR2 might affect the assembly of the microtubule cytoskeleton based on analysis of the cytoskeleton in developing trichomes, and exposures to paclitaxol and oryzalin, which respectively act either to stabilize or depolymerize the cytoskeleton. Through epistatic and gene expression analyses it is shown that ETR2 is positioned upstream of CHROMATIN ASSEMBLY FACTOR1 and TRYPTICHON and is independent of the GLABRA2 and GLABRA3 pathways. These results help extend understanding of the early events that control trichome development and identify a signalling pathway through which ethylene affects trichome branching.
Keywords: Cytoskeleton, endoreduplication, epigenetic, hormone, signal transduction, tubulin
Introduction
Trichome development in Arabidopsis thaliana has six distinct steps (Syzmanski et al., 1998) that are controlled by over 30 genes, the majority of which signal to affect trichome branching (reviewed in Schellmann and Hülskamp, 2005). Endoreduplication during early trichome development requires the activity of the transcription factor GLABRA3 (GL3), which is thought to facilitate rapid growth of the cell (Hülskamp et al., 1994). Once the trichome has attained a designated size, the microtubule cytoskeleton re-orients to cause two or more branching events controlled by several independent pathways. GL3 is thought to act upstream of FURCA4 (FRC4) to regulate trichome branching positively (Luo and Oppenheimer, 1999), while in another pathway, ANGUSTIFOLIA (AN) is negatively regulated by the MYB transcription factor NOEK (NOK) to restrict branching (Folkers et al., 1997; Jakoby et al., 2008). TRYPTICHON (TRY) has been found, through epistatic analysis, to act upstream of FRC4, ZWICHEL (ZWI), and STICHEL (STI) to restrict branching and has also been proposed to affect GL3 negatively (Esch et al., 2003). STI, ZWI, and AN are implicated in the assembly of microtubules with consequences for branching pattern and number (Oppenheimer et al., 1997; Mathur and Chua, 2000; Folkers et al., 2002). STI is considered to be one of the most important contributors to trichome branching, as mutations to this gene yield predominantly unbranched trichomes, while mutations to the other genes mentioned above yield predominantly two-branched trichomes (Hülskamp et al., 1994). The epigenetic state of the cell also appears to play a role in the final shape of the trichome based on loss-of-function mutations to the trimeric protein CHROMATIN ASSEMBLY FACTOR1 (CAF1) (Exner et al., 2006, 2008; Ono et al., 2006). Mutations either to the FASCIATA1 (FAS1) or to the FASCIATA2 (FAS2) subunits of CAF1 show increased trichome branching and are thought to act through regulation of STI (Exner et al., 2008).
Hormones can also affect trichome development. For example, the gibberellic acid (GA) mutant spy-5 has increased trichome branching (Perazza et al., 1998). Exogenous application of ethylene meanwhile, has been found to increase branch number in cucumber trichomes (Kazama et al., 2004) and increased ethylene synthesis has been correlated with branch extension in cotton trichomes (Shi et al., 2006; Qin et al., 2007). Ethylene is a gaseous plant hormone that, in Arabidopsis, is sensed by five receptors (ERS1, ERS2, ETR1, ETR2, and EIN4) (Hua and Meyerowitz, 1998). These receptors, which localize to the endoplasmic reticulum (ER), form homo- and heterodimers with each other and associate with CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), which represses activation of downstream pathways in the absence of ethylene (Grefen et al., 2008). Upon binding of ethylene through a copper co-factor, repression of the ethylene response pathways is relieved through inactivation of CTR1 (Rodríguez et al., 1999). It has also been shown that ethylene can affect downstream responses in a CTR1 independent pathway (Hass et al., 2004). There are two classes of ethylene receptor mutants in Arabidopsis. Gain-Of-Function (GOF) mutations disrupt ethylene binding (Bleeker et al., 1988; Hua et al., 1995, 1998; Sakai et al., 1998; Hall et al., 1999), while Loss-Of-Function (LOF) mutations result in a protein that is unable to associate with CTR1 (Hua and Meyerowitz, 1998). LOF mutations cause truncations to the signalling portion of the ethylene receptor such that CTR1 is no longer activated (thus a loss of function). Loss of CTR1 activity alleviates the repression on the ethylene signalling pathway. Therefore LOF mutants have a constitutively active ethylene response pathway. GOF mutants, conversely, can never bind ethylene and thus CTR1 is constantly activated by the mutated ethylene receptor (thus a gain of function). These mutants, therefore, have a complete inhibition of the ethylene signalling pathway. Single loss-of-function mutants generally result in no obvious phenotype, although etr1-7 has been shown to have increased sensitivity to ethylene (Cancel and Larsen, 2002). In the past, only triple LOF mutants, such as etr1-7etr2-3ein4-4, exhibit developmental characteristics consistent with a constitutive ethylene response (Hua and Meyerowitz, 1998). Study of both GOF and LOF ethylene receptor mutants help determine the impact of receptor function in ethylene responsive developmental pathways.
To better understand the role of ethylene in trichome morphogenesis, a screen of GOF and LOF ethylene receptor mutants for alterations in trichome branching was undertaken. Unlike all the other mutants, the etr2-3 mutant has only unbranched trichomes, suggesting that this mutation impacts the early stages of trichome development. Here it is demonstrated that signalling through ETR2 participates in the control of microtubule dynamics, and that it is an upstream regulator of the TRY-mediated trichome branching pathway.
Materials and methods
Plant growth conditions
Arabidopsis thaliana seeds (source: Arabidopsis Biological Resource Center) were stratified for 4 d at 4 °C in the dark and grown in soil under a long photoperiod (16/8 h light/dark) at a light intensity of 180 μE m−2 s−1 at the rosette level at 22 °C. For paclitaxol and oryzalin experiments, plants were grown on Murashige and Skoog basal salt mixture agar plates (Sigma, Oakville, ON, Canada) pH 5.7–5.8, 0.6% (w/v) agar for one week. Paclitaxol (MP Biomedicals, Solon, OH, USA) tests were performed according to Mathur and Chua (2000) (n=60 plants). Plant roots were soaked in an oryzalin (Riedel-de Haën, Pestanal) solution (0, 0.01, 0.1 or 1 μM) for 72 h and then root hairs were counted (n=900 root hairs per concentration). For epistatic analysis, all double mutants were followed to homozygous F3 plants and were back-crossed to the wild type (Col-0) to ascertain the parental genotype.
Microscopy
The degree of trichome branching was analysed on leaf 5 of 30-d-old soil-grown Arabidopsis plants using a Carl Zeiss Stemi 2000-C dissecting microscope (Carl Zeiss, Germany). Representative images of leaves for the wild type (Col-0) and all single and double mutants were taken using a cryo-Scanning Electron Microscope as per Harrison et al. (2007). For analysis of trichome nuclear ploidy, trichomes were fixed and removed as per Zhang and Oppenheimer (2004) and stained with 4′6-diamino-2-phenylindole (DAPI) as per Folkers et al. (1997) and pictured using a Carl Zeiss Axioplan Fluorescent microscope (Carl Zeiss, Germany). Images were captured using an AxioCam HRc CCD camera and fluorescence levels of nuclei for the wild type and etr2-3 were analysed using AlphaEase FC Imaging System software (Alpha Innotech; San Leandro, CA). Levels of endoreduplication were determined by comparing the fluorescence of stomatal guard cell nuclei, which are known to be 2C (Melaragno et al., 1993), to the fluorescence of wild-type and etr2-3 trichome nuclei. Levels of endoreduplication were normalized to the nearest multiple of 2C.
To determine the effect of the etr2-3 mutation on the microtubule cytoskeleton, double mutants were constructed between etr2-3 and 35S:MAP4-GFP and the GFP-labelled cytoskeleton in the trichomes was viewed as per Mathur and Chua (2000).
Histochemical GUS assay
To analyse the cellular expression of ETR2, the entire 5′ upstream region of the gene was cloned using the primers: ETR2pro fw: 5′-GTCGACAGAAGAACGCATGAGAGCC-3′; ETR2pro rv: 5′-CCATGGCACCACCATTGATAGTATC-3′ into the pCAMBIA 1305.1 vector (Centre for the Application of Molecular Biology to International Agriculture, Canberra, Australia; http://www.cambia.org) and transformed into wild-type Arabidopsis, Columbia ecotype, as per Clough and Bent (1998). Leaves from young and mature homozygous T3 plants were GUS stained according to Regan et al. (1999) and photographed using either a Carl Zeiss Axioplan microscope or Carl Zeiss Stemi 2000-C dissecting microscope with an AxioCam HRc CCD camera.
Quantitative PCR
Quantitative PCR was used to determine the expression differences of several genes. Plants of the wild type (Col-0 and Enk-2), etr2-1, etr2-3, and fas1-1 were grown as outlined above for 10 d at which point they were harvested and frozen in N2. Total RNA was extracted using the Qiagen RNeasy Plant Kit (Mississauga, ON) according to the manufacturer's instructions and 1 μg of total RNA was used for the synthesis of cDNA using AMV reverse transcriptase (Promega; Madison, WI). At least three independent biological replicates were used for the wild type and each mutant with at least two technical replicates. Quantitative PCR was performed using the Cepheid OmniMix HS system following the manufacturer's instructions (Sunnyvale, CA) on the SmartCycler system (Cepheid; Sunnyvale, CA). Ubiquitin-10 was used as an internal control. Primers used for the analyses were as follows: ANGUSTIFOLIA (AN) fw: 5′-TCGCATACAGAAACAAGGACAC-3′, AN rv: 5′-ACACGTCAAAACTATGG CTAGC-3′; STICHEL (STI) fw: 5′-GCTTTAGTAAACGAGCTAGTTGG-3′, STI rv: 5′-CTAGCTCGCTTAACAGTCTCTG-3′; TRIPTYCHON (TRY) fw: 5′-TCGCCCTCCAT GACTCTGAAGAAG-3′, TRY rv: 5′-CTCTTCCTGCTATCAAATCCCACC-3′; ZWICHEL (ZWI) fw: 5′-CCACAGTGTCTGATGCTGTTGAGGAG-3′; ZWI rv: 5′-CTGGAGGAGATCTCCAATATACTTGTTATC-3′; FASCIATA1 (FAS1) fw: 5′-TTCTGAATCTGTCTTTGGTGCTGGGAGACG-3′; FAS1 rv: 5′-CCATGAATGATCGAATATCCACCTCACTCAGT-3′; UBIQUITIN-10 (UBQ10) fw: 5′-GTCCTCAGGCTCCGTGGTG-3′; UBQ10 rv: 5′-GCCATCCTCCAACTGCTTTC-3′.
Results
Loss-of-function ETR2 mutants display altered trichome branching
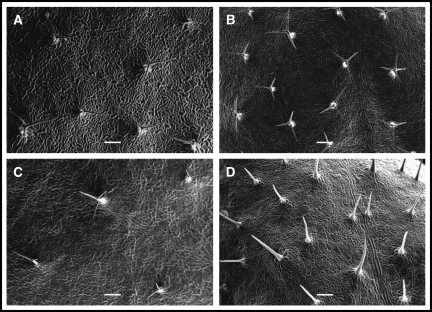
To determine if any of the Arabidopsis ethylene receptor GOF or LOF mutants had altered trichome development, as compared to the wild type (Columbia), mutant lines of all five Arabidopsis receptors were screened. Only LOF mutations to the ETR2 receptor caused trichome branching abnormalities among the ethylene receptor mutants tested (etr1-1, etr1-5, etr1-6, etr1-7, ers1-1, ers1-3, ers2-1, ers2-3, etr2-1, etr2-2, etr2-3, ein4-1, ein4-4, ein4-7). Wild-type rosette leaves in Arabidopsis have 1% two-branched trichomes, 97% three-branched trichomes, and 2% four-branched trichomes (as measured on leaf 5; n=798 trichomes). The LOF mutants of ETR2 differed from the wild type (Fig. 1). While the GOF mutant, etr2-1 (Fig. 1B), had trichomes similar to the wild type (Fig. 1A), with 2% two-branched trichomes, 93% three-branched trichomes, and 5% four-branched trichomes (n=950 trichomes), etr2-2 (a LOF mutant; Fig. 1C) lacked four-branched trichomes and instead had 17% two-branched trichomes, and 83% three-branched trichomes (n=1349 trichomes). The most altered phenotype was observed in the LOF mutant etr2-3 (Fig. 1D), which had 100% unbranched trichomes (n=1253 trichomes). This was true on rosette leaves, flowering stalks, and on sepals, as compared to the wild type which has both two-branched and unbranched trichomes on the flowering stalks and sepals. Representative images of the wild type, etr2-1, etr2-2, and etr2-3 are shown in Fig. 1.
Fig. 1.
Representative trichome branching in wild type and three etr2 mutants. Trichomes of the wild type are predominantly three-branched (A) as are trichomes of etr2-1 (B). The etr2-2 mutant has a higher number of two-branched trichomes (C) while etr2-3 has only unbranched trichomes (D). Scale bars=200 μm.
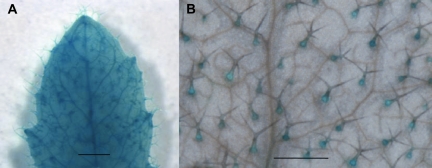
It has previously been shown that ETR2 is expressed in mature leaves (Sakai et al., 1998; Grefen et al., 2008). Transgenic Arabidopsis expressing an ETR2pro:GUS construct were created to determine if there is a cellular specific expression related to trichomes. It was found in all 13 lines recovered that ETR2 is expressed in juvenile leaves, and that GUS expression was observed in young trichomes in half of these lines (Fig. 2A). During leaf expansion, ETR2 expression became restricted to trichomes (Fig. 2B), while in fully expanded leaves no GUS staining was found in either the leaf epidermal cells or trichomes as has also been shown by Jakoby et al. (2008). These data demonstrate that ETR2 is expressed in trichomes early in development during which the initiation of trichome branching occurs.
Fig. 2.
ETR2 is expressed in young leaves and mature trichomes. (A) ETR2pro:GUS is expressed in very young leaves and trichomes (Scale bar=1 mm). (B) ETR2pro:GUS is restricted to fully branched trichomes only (Scale bar=1 mm).
Microtubule cytoskeleton organization is altered in etr2-3
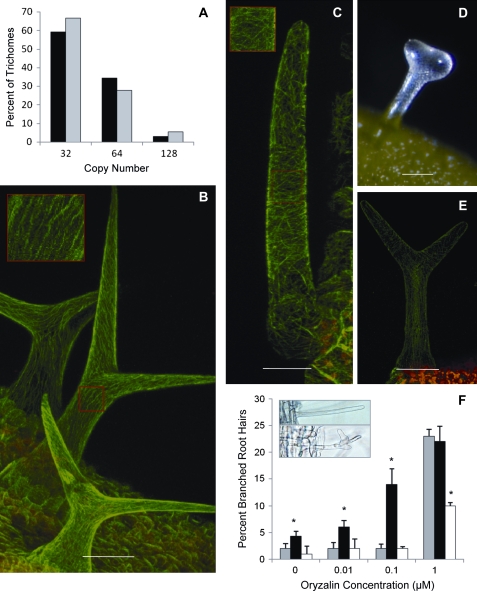
To date, mutations that cause a reduction in trichome branching either affect the level of endoreduplication within the cell or the organization of the microtubule cytoskeleton (Mathur and Chua, 2000). Trichomes were stained with 4′,6-diamidino-2-phenylindole (DAPI) to determine the DNA content of etr2-3 trichomes as compared to the wild type. Epidermal pavement cells generally have between 2C and 16C of DNA, while trichomes have between 4C to 64C, with the majority containing an average of 32C (Melaragno et al., 1993). It was found that average DNA content between etr2-3 and wild-type trichomes was not significantly different (Fig. 3A; P <0.05). This indicates that ETR2 affects trichome branching in an endoreduplication-independent manner.
Fig. 3.
ETR2 affects microtubule assembly. (A) Both wild-type (black bar) and etr2-3 (grey bar) nuclei show the same level of endoreduplication (P <0.05, t test). (B) Wild-type trichomes expressing 35S:MAP4-GFP show a longitudinal fine mesh of microtubules (scale bar=50 μm). Inset is a magnification of longitudinally oriented microtubules in the stalk of the trichome. (C) Some etr2-3×35S:MAP4-GFP trichomes nearing maturity show disorganized microtubule cytoskeleton and early signs of depolymerization at the base of the trichome (scale bar=30 μm). Inset is a magnification of disorganized microtubule assembly discussed in the text. (D) Branching is induced in etr2-3 when treated with 20 μM paclitaxol for 2 h and then grown on MS medium until new leaves are produced (scale bar=100 μm). (E) etr2-3×35S:MAP4-GFP lines which express the highest level of MAP4-GFP show trichome branching and normal microtubule assembly (scale bar=70 μm). (F) Oryzalin causes depolymerization of microtubules and induces unbranched root hairs to become branched (inset). Bar chart of dose-response of the wild type (grey bars), etr2-3 (black bars), and etr2-1 (white bars) to different oryzalin concentrations. Asterisk indicates statistically significant difference from the wild type (P <0.05, t test).
Assembly and structural arrangement of microtubules are maintained in part by MICROTUBULE-ASSOCIATED PROTEINs (MAPs) (Igarashi et al., 2000; Smertenko et al., 2000). This association has been exploited through the use of a GFP-labelled MAP4 protein to image the microtubule cytoskeleton in a number of living plant cell types (Olson et al., 1994; Marc et al., 1998). To image the microtubule cytoskeleton in developing trichomes, etr2-3 was crossed with transgenic Arabidopsis expressing 35S:MICROTUBULE-ASSOCIATED PROTEIN4-GFP (35S:MAP4-GFP). Overexpression of the MAP4 has also been shown to cause stabilization of microtubule assembly (Marc et al., 1998). In 35S:MAP4-GFP lines this can lead to artefacts such as bulging hypocotyl cells, thickened trichomes or induced trichome branching. Despite this drawback, with careful screening these lines have been widely used throughout the literature (Olson et al., 1995; Marc et al., 1998; Kragler et al., 2003; Shevchenko et al., 2008). The etr2-3 35S:MAP4-GFP double mutants were therefore carefully screened for lines which did not display these artefacts. It was found that in the early stages of wild-type trichome outgrowth, the cortical microtubule cytoskeleton appears to criss-cross the stalk of the trichome. A similar pattern was observed in very young trichomes of etr2-3. Following trichome branching, the microtubule cytoskeleton re-aligns into a longitudinal orientation (Fig. 3B, inset). It is at this stage that differences between wild-type and etr2-3 trichomes were observed. While some of the etr2-3 trichomes showed normal longitudinal microtubules in nearly mature trichomes, a subset of trichomes exhibited a random and disorganized microtubule network that showed evidence of depolymerization (Fig. 3C, inset). Trichomes of etr2-3 also lacked the dense knots of microtubules typically present in branching trichomes. There was no visible GFP signal in fully mature etr2-3 trichomes. Thus, microtubule assembly in etr2-3 trichomes nearing maturity do not consistently show the same organization as in the wild type.
In previously characterized mutants that show altered microtubule assembly, it has been shown that pharmacological stabilization of microtubules can induce branching in trichomes (Mathur and Chua, 2000). When etr2-3 was treated with high concentrations of paclitaxol, a known stabilizer of microtubule assembly (Schiff et al., 1979; De Brabander et al., 1981; Verde et al., 1991; Hyman and Karsenti, 1998), approximately 50% of the trichomes developed branch initials or unbranched stalks with incipient branch bulges (Fig. 3D). The cytoskeleton of 2 etr2-3, 35S:MAP4-GFP double mutant lines that were identified as having a highly stabilized microtubule cytoskeleton (evidenced by bulging hypocotyl cells, intense GFP signal, as well as a mixture of unbranched and branched trichomes) were also analysed. The two-branched trichomes in these lines had blunt-ended branch tips, a phenotype that has been associated with stabilized microtubules in sti (Mathur and Chua, 2000) and the microtubule cytoskeleton appeared normal (Fig. 3E). Therefore stabilization of microtubule assembly by either overexpression of MAP4 or application of paclitaxol can partially rescue the etr2-3 mutant phenotype.
If the etr2-3 mutant is affected in the assembly of the microtubule cytoskeleton, it should be possible to disrupt cytoskeletal assembly more easily in the etr2-3 mutant compared to the wild type. This was studied by treating etr2-1, etr2-3, and the wild type with increasing concentrations of oryzalin. Oryzalin has been shown to depolymerize microtubules, leading to root hair branching, an increase in the girth of trichomes, and alteration of the shape of the root tip (Baskin et al., 1994). While analysis of root hair branching is not directly associated with trichome branching, branching of the root hairs can be accurately quantified and used to support our findings that cytoskeletal assembly is impaired in these mutants. In MS media, etr2-3 had a higher number of branched root hairs than the wild type (Fig. 3F). Three days after the addition of 0.1 μM oryzalin to the media, the number of branched root hairs in etr2-3 increased significantly as compared to the wild type or etr2-1 (P <0.05). The wild type required a concentration of 1.0 μM oryzalin before a significant increase in root hair branching was detected. By contrast, the GOF mutant etr2-1, had a significantly lower number of branched root hairs at 1.0 μM oryzalin compared to the wild type (P <0.05). Therefore, LOF and GOF mutations to ETR2 have opposite effects on the sensitivity to oryzalin.
Epistatic crosses and expression analysis place ETR2 in the trichome development pathway
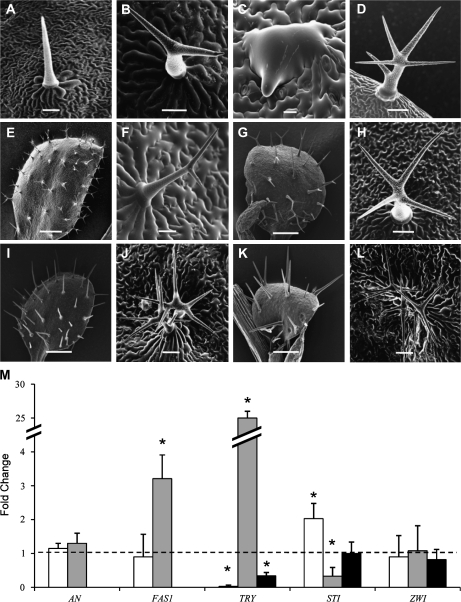
The etr2-3 mutant was crossed with several trichome branching mutants to gain insight into the position of ETR2 in the known trichome morphogenesis signalling pathway. Representative pictures of the original mutants (Fig. 4B, D, F, H, J) and the resulting double mutants (Fig. 4C, E, G, I, K, L) are shown and percentages of branch types are shown in Table 1. When crossed with glabra3 (gl3), which has two-branched trichomes (Fig. 4B), the double mutant had puddle-like, unbranched trichomes that grew parallel to the leaf surface (Fig. 4C). When crossed with spy-5 (Fig. 4D), a mutant in the gibberellin pathway with increased trichome branching, the resulting double mutant had a mixture of two- and three-branched trichomes (Fig. 4E). Crossing of etr2-3 with glabra2 (gl2) (Fig. 4F) resulted in leaves with a mixture of two branched and unbranched trichomes (Fig. 4G). The glabra2 and etr2-3 glabra2 double mutants have approximately the same proportion of unbranched and two branched trichomes (Table 1). There were, however, distinct differences in the morphology of the trichomes between gl2 and the double mutant. Trichomes of the double mutant were fully extended and sharp tipped. Conversely, trichomes on the first leaves of gl2 either did not fully extend (for unbranched trichomes) or showed a high proportion of branched trichomes with very short branches. When etr2-3 was crossed to fasciata1 (fas1), a loss-of-function mutation to CAF1 that has trichomes with increased branching (Fig. 4H), the resulting double mutant had 100% unbranched trichomes (Fig. 4I). Based on the epistatic relationship between FAS1 and ETR2, etr2-3 was crossed to try, a mutant with clumped trichomes that exhibits supernumerary branches. The try etr2-3 double mutant had unbranched trichomes that, in a few cases appeared clumped (Fig. 4K). A cross was also performed between fas1-1 and try and the double mutant had the trichome branching and clustering phenotype of try (Fig. 4L). Taken together, these results suggest that ETR2 signals in a pathway that is parallel to GL2, GL3, and gibberellic acid, but that ETR2 operates in the same pathway as CAF1 and TRY.
Fig. 4.
Epistatic analysis places ETR2 in the TRY and CAF1 controlled pathway. Representative cryo-SEM images of (A) etr2-3 (scale bar=100 μm), (B) gl3 (scale bar=100 μm) (C) etr2-3 gl3 (scale bar=10 μm), (D) spy-5 (scale bar=100 μm), (E) etr2-3 spy-5 (scale bar=1 mm), (F) gl2 (scale bar=100 μm), (G) etr2-3 gl2 (scale bar=1 mm), (H) fas1 (scale bar=100 μm), (I) etr2-3 fas1 (scale bar=100 μm). (J) try (scale bar=100 μm), (K) try etr2-3 (scale bar=100 μm), (L) try fas1-1 (scale bar=100 μm), (M) quantitative PCR analysis of ANGUSTIFOLIA, TRYPTICHON, and STICHEL expression in etr2-1 (white bars), etr2-3 (light grey bars), and fas1-1 (black bars) (n=3–5) as compared to wild-type levels (dashed line). asterisk indicates significant differences from Col-0 (for etr2-1 and etr2-3) and Enk-2 (for fas1-1) (P <0.05).
Table 1.
Trichome branching phenotypes of single and double mutants in the trichome branching pathway
| Unbranched | 2 Branches | 3 Branches | 4 Branches | 5 Branches | |
| Wild type | 1 | 97 | 2 | ||
| etr2-3 | 100 | ||||
| gl2 | 59 | 41 | |||
| etr2-3 gl2 | 57 | 41 | 2 | ||
| gl3 | 78 | 22 | |||
| etr2-3 gl3 | 97 | 3 | |||
| spy5 | 37 | 47 | 16 | ||
| etr2-3 spy5 | 6 | 81 | 13 | ||
| fas1-1 | 52 | 27 | 21 | ||
| etr2-3 fas1-1 | 100 | ||||
| trya | 34 | 54 | 12 | ||
| etr2-3 try | 100 |
Trichomes on leaf 2 or 3 of five plants were counted, and the relative percentages of each type of trichome recorded.
Values from Perazza et al. (1999).
To characterize further how ETR2 affects the expression of genes involved in the regulation of trichome branching, the expression levels of five key trichome branching regulators were analysed (Fig. 4M). Expression was performed in etr2-1, etr2-3, and fas1-1 and compared to their respective wild type (Col-0 for the first two mutants and Enk-2 for fas1-1). Because epistatic analysis suggested that ETR2 signals in the same pathway as CAF1, the expression level of FAS1 in the etr2-3 and etr2-1 mutants was also tested. It was found that expression levels of FAS1 in etr2-3 were significantly increased, while they were not significantly affected in etr2-1 (P <0.05). This would support the epistatic results which suggest that ETR2 acts upstream of CAF1. It was found that TRY expression was significantly increased in etr2-3, while it was significantly repressed in etr2-1 (Fig. 4M). In the fas1-1 mutant TRY expression is also significantly repressed. As TRY is a known regulator of STI and ZWI, the expression patterns of these two genes were considered. It was determined that STI was significantly down-regulated in etr2-3 and increased in etr2-1 but was unaffected in fas1-1. There was no significant difference in expression of ZWI in any of the mutants tested. As a control, the level of expression of AN was tested as it is a gene known to be in a separate trichome branching pathway from TRY, and presumably unaffected by ETR2. The expression levels in both ethylene mutants was not significantly different from the wild type (Fig. 4M). These data confirm that ETR2 is in the same signalling pathway as CAF1 and that downstream components of this regulatory network are TRY and STI.
Discussion
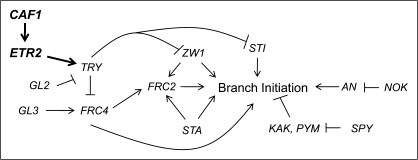
Re-orientation of cellular growth to produce branching in Arabidopsis trichomes is a highly orchestrated process that requires input from several independent pathways (Hülskamp et al., 1994, 1999; Folkers et al., 1997; Krishnakumar and Oppenheimer, 1999; Luo and Oppenheimer, 1999). There is some evidence that the plant hormone ethylene is implicated in regulating the development of trichomes in cucumber (Kazama et al., 2004) and cotton (Shi et al., 2006; Qin et al., 2007). This study has demonstrated that LOF mutations to the Arabidopsis ethylene receptor ETR2 yield leaf trichomes with fewer branches. This reduction in branching is most likely due to the altered stability of microtubule assembly, while the mutation does not affect the level of endoreduplication. Based on the epistatic and gene expression analyses presented here, a model whereby ETR2 affects CAF1, in the TRY branching pathway has been proposed (Fig. 5).
Fig. 5.
Placement of ETR2 and CAF1 within the trichome branching pathway. Known network of trichome branching with ETR2 and CAF1 (in bold) as negative regulators of trichome branching through TRY (adapted from Luo and Oppenheimer (1999) with kind permission from The Company of Biologists).
Multiple ethylene receptors have been conserved throughout the plant kingdom. While these receptors show a high degree of homology in the ethylene binding domain, they show sequence variability in the C-terminal signalling domain. Recent work has shown that this variability may allow for differential transduction of the ethylene signal (Cancel and Larsen, 2002; Grefen et al., 2008; Gao et al., 2008). The data presented here has shown that truncations to the signalling domain of the Arabidopsis ETR2 receptor affect trichome development and that the extent of these truncations is correlated with the severity of the trichome branching phenotype. In the etr2-3 mutant, only 45% of the protein is made (Hua and Meyerowitz, 1998) and only unbranched trichomes are observed on the leaf surface. While in the etr2-2 mutant, 73% of the protein is produced and this correlates with the appearance of a significantly higher number of two-branched trichomes, compared to the wild type. A similar dependence of phenotype on protein length has been previously described in LOF mutations to ETR1 with regard to plant growth when exposed to ethylene (Cancel and Larsen, 2002). This trichome phenotype is unique to the ETR2 receptor, as loss of part, or all, of the signalling domains in any of the other four Arabidopsis ethylene receptors did not affect trichome morphology.
Previous research suggests that two major pathways which contribute to alterations in trichome branch number is a result of different levels of endoreduplication in the cell or due to altered microtubule assembly (Hülskamp et al., 1994, 1999; Oppenheimer et al., 1997; Folkers et al., 2002; Downes et al., 2003; Castelano et al., 2004; El Refy et al., 2004; Desvoyes et al., 2006; Ramirez-Parra et al., 2007; Larson-Rabin et al., 2009). Using analysis of ploidy level in both wild-type and etr2-3 trichomes, it was demonstrated that average endoreduplication levels were not significantly different between the two plant lines. This led to the hypothesis that mutations to ETR2 were affecting the microtubule cytoskeleton. There is a growing body of evidence that ties ethylene to the control of microtubule orientation. Applications of 1-aminocyclopropane-1-carboxylic acid (ACC) to lettuce roots have been shown to induce randomization of the microtubule array and aid the induction of root hairs (Takahashi et al., 2003). ACC and ethylene are also known to alter the re-orientation of microtubules into a longitudinal orientation (Apelbaum and Burg, 1971; Roberts et al., 1984; Yuan et al., 1994; Le et al., 2004). Analysis of the microtubule cytoskeleton in developing etr2-3 trichomes indicated that there was a higher degree of microtubule disorganization, compared to the wild type, and a complete lack of microtubule knots in the cytoskeleton that are normally associated with branch initiation. By stabilization of microtubule assembly through either pharmacological means (paclitaxol administration) or through transgenic strategies (overexpression of MAP4) induction of either branch initials or full trichome branching was achieved. Conversely, destabilization of the cytoskeleton was more easily accomplished at lower concentrations of oryzalin in etr2-3, compared to the wild type. These data would indicate that branch formation in etr2-3 is impeded by the lack of proper microtubule organization, and that the LOF mutant still retains the ability to branch if microtubule assembly is artificially stabilized.
Two other proteins, STI and ZWI, have been proposed to have a similar stabilizing effect on microtubule assembly as their loss can also be partially replaced by paclitaxol (Ilgenfritz et al., 2003). Based on epigenetic and gene expression analysis in the etr2-3 mutant, a model is proposed concerning ETR2's role in trichome branching (Fig. 5). As etr2-3 is a LOF mutant, ethylene responses controlled by ETR2 are constitutively active in this background. Therefore, in the model proposed, as CAF1 expression was found to be increased in etr2-3, it can be hypothesized that ethylene bound to ETR2 would likewise cause an increase in CAF1 expression. Through epigenetic silencing, CAF1 would then initiate a signal cascade that culminates in the repression of STI through increased activity of TRY. Exner and colleagues (2008) have implicated CAF1 in the control of STI and ZWI activity, although it was not known through what intermediate. Ethylene has previously been proposed to affect transcriptional control through the recruitment of histone deacetylases which would alter chromatin structure and block cis-elements (Pazin and Kadonaga, 1997; Fujimoto et al., 2000; Zhou et al., 2005). The demonstration that ethylene controls the epigenetic state of a cell through transcriptional control of CAF1 opens a new avenue through which ethylene signal transduction can control a suite of genes through the regulation of one intermediate. Interestingly, it was found that there were changes to STI expression in etr2-3 and etr2-1 but not in fas1-1 (the CAF1 mutant), even though epigenetic analysis places ETR2 upstream of CAF1 (Fig. 5). This may indicate that CAF1 is not the only regulator through which ETR2 signals affecting STI. The results of this study, together with those of Exner and colleagues (2008), also indicate that CAF1 and ETR2 affect trichome branching independent of the GL2 and GL3 endoreduplication pathways (Fig. 5). This is striking as TRY, also implicated in the same pathway, is known to affect the level of endoreduplication in cells. However, as TRY can affect multiple cell developmental pathways independently (Hülskamp et al., 1994; Schnittger et al., 1999; Schellmann et al., 2002), this may be a separate mechanism whereby TRY affects trichome branching independent of its role in the cell cycle.
It has been shown in this study that ETR2 is an important regulator of trichome development through the modulation of CAF1, TRY, and STI expression. The data from imaging of the microtubule cytoskeleton and through drug treatment also suggest that signalling through ETR2 is responsible for maintaining microtubule stability. As mutations to the other ethylene receptors do not affect trichome morphology in a similar manner, this suggests that the ETR2 receptor has a unique role in mediating ethylene's involvement in the establishment of cell shape. This study is an important step forward in the understanding of how ethylene affects cellular development.
Acknowledgments
We thank the Arabidopsis Biological Resource Center, Caren Chang, and Elliot Meyerowitz for providing the Arabidopsis thaliana seed stocks. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canada Foundation for Innovation grants to SR and JM, a Canada Research Chair to SR, and an NSERC graduate scholarship to JP.
References
- Apelbaum A, Burg SP. Altered cell microfibrillar orientation in ethylene-treated Pisum sativum stems. Plant Physiology. 1971;48:648–652. doi: 10.1104/pp.48.5.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Wilson JE, Cork A, Williamson RE. Morphology and microtubule organization in Arabidopsis roots exposed to oryzalin or taxol. Plant and Cell Physiology. 1994;35:935–942. [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB. Loss-of-function mutations in the ethylene receptor ETR1 cause enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiology. 2002;129:1557–1567. doi: 10.1104/pp.003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano MM, Boniotti MB, Caro E, Schnittger A, Gutierrez C. DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. The Plant Cell. 2004;16:2380–2393. doi: 10.1105/tpc.104.022400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- De Brabander M, Geuens G, Nuydens R, Willebrords R, Mey JD. Taxol induces the assembly of free microtubules in living cells and blocks the organizing capacity of the centrosomes and kinetochores. Proceedings of the National Academy of Sciences, USA. 1981;78:5608–5612. doi: 10.1073/pnas.78.9.5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiology. 2006;140:67–80. doi: 10.1104/pp.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. The Plant Journal. 2003;35:729–742. doi: 10.1046/j.1365-313x.2003.01844.x. [DOI] [PubMed] [Google Scholar]
- El Refy AE, Perazza D, Zekraoui L, Valay J, Bechtold N, Brown S, Hülskamp M, Herzog M, Bonneville JM. The Arabidopsis KAKTUS gene encodes a HECT protein and controls the number of endoreduplication cycles. Molecular Genetics and Genomics. 2004;270:403–414. doi: 10.1007/s00438-003-0932-1. [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development. 2003;130:5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- Exner V, Gruissem W, Hennig L. Control of trichome branching by Chromatin Assembly Factor-I. BMC Plant Biology. 2008;8:54–66. doi: 10.1186/1471-2229-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V, Taranto P, Schonrock N, Gruissem W, Hennig L. Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development. 2006;133:4163–4172. doi: 10.1242/dev.02599. [DOI] [PubMed] [Google Scholar]
- Folkers U, Berger J, Hülskamp M. Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development. 1997;124:3779–3786. doi: 10.1242/dev.124.19.3779. [DOI] [PubMed] [Google Scholar]
- Folkers U, Kirik V, Schöbinger U, et al. The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO Journal. 2002;21:1280–1288. doi: 10.1093/emboj/21.6.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. The Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Wen C, Binder BM, Chen Y, Chang J, Chiang Y, Kerris RJ, III, Chang C, Schaller GE. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. Journal of Biological Chemistry. 2008;283:23801–23810. doi: 10.1074/jbc.M800641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Stadele K, Ruzicka K, Obrdlik P, Harter K, Horak J. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Molecular Plant. 2008;1:308–320. doi: 10.1093/mp/ssm015. [DOI] [PubMed] [Google Scholar]
- Harrison EJ, Bush M, Plett JM, et al. Diverse developmental mutants revealed in an activation-tagged population of poplar. Canadian Journal of Botany. 2007;85:1071–1081. [Google Scholar]
- Hall AE, Chen QG, Findell JL, Schaller GE, Bleecker AB. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiology. 1999;121:291–299. doi: 10.1104/pp.121.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Lohrmann J, Albrecht V, et al. The response regulator 2 mediates ethylene signaling and hormone signal integration in Arabidopsis. EMBO Journal. 2004;23:3290–3302. doi: 10.1038/sj.emboj.7600337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Misera S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–566. doi: 10.1016/0092-8674(94)90118-x. [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A, Folkers U. Pattern formation and cell differentiation: trichomes in Arabidopsis as a genetic model system. International Review of Cytology. 1999;186:147–178. doi: 10.1016/s0074-7696(08)61053-0. [DOI] [PubMed] [Google Scholar]
- Hyman A, Karsenti E. The role of nucleation in patterning microtubule networks. Journal of Cell Science. 1998;111:2077–2083. doi: 10.1242/jcs.111.15.2077. [DOI] [PubMed] [Google Scholar]
- Igarashi H, Orii H, Mori H, Shimmen T, Sonobe S. Isolation of a novel 190 kDa protein from tobacco BY-2 cells: possible involvement in the interaction between actin filaments and microtubules. Plant and Cell Physiology. 2000;41:920–931. doi: 10.1093/pcp/pcd015. [DOI] [PubMed] [Google Scholar]
- Ilgenfritz H, Bouyer D, Schnittger A, Mathur J, Kirik V, Schwab B, Chua N, Jürgens G, Hülskamp M. The Arabidopsis STICHEL gene is a regulator of trichome branch number and encodes a novel protein. Plant Physiology. 2003;131:643–655. doi: 10.1104/pp.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiology. 2008;148:1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama H, Dan H, Imaseki H, Wasteneys GO. Transient exposure to ethylene stimulates cell division and alters the fate and polarity of hypocotyl epidermal cells. Plant Physiology. 2004;134:1614–1623. doi: 10.1104/pp.103.031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F, Curin M, Trutnyeva K, Gansch A, Waigmann E. MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiology. 2003;132:1870–1883. doi: 10.1104/pp.103.022269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, Oppenheimer D. Extragenic suppressors of the Arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth. Development. 1999;126:3079–3088. doi: 10.1242/dev.126.14.3079. [DOI] [PubMed] [Google Scholar]
- Larson-Rabin Z, Li Z, Masson PH, Day CD. FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiology. 2009;149:874–884. doi: 10.1104/pp.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J, Vandenbussche F, Van Der Straeten D, Verbelen J. Position and cell type-dependent microtubule reorientation characterizes the early response of the Arabidopsis root epidermis to ethylene. Physiologia Plantarum. 2004;121:513–519. [Google Scholar]
- Luo D, Oppenheimer D. Genetic control of trichome branch number in Arabidopsis: the roles of the FURCA loci. Development. 1999;126:5547–5557. doi: 10.1242/dev.126.24.5547. [DOI] [PubMed] [Google Scholar]
- Marc J, Granger C, Brincat J, Fisher D, Kao T, McCubbin A, Cyr R. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangement in living epidermal cells. The Plant Cell. 1998;10:1927–1939. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Chua N. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. The Plant Cell. 2000;12:465–478. doi: 10.1105/tpc.12.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. The Plant Cell. 1993;5:1661–1668. doi: 10.1105/tpc.5.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KR, McIntosh JR, Olmsted JB. Analysis of MAP4 function in living cells using green fluorescent protein (GFP) chimeras. Journal of Cell Biology. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Kaya H, Takeda S, Abe M, Ogawa Y, Kato M, Kakutani T, Scheid OM, Araki T, Shibahara K. CHROMATIN ASSEMBLY FACTOR1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes to Cells. 2006;11:153–162. doi: 10.1111/j.1365-2443.2006.00928.x. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Pollock MA, Vacik J, Szymanski DB, Ericson B, Feldmann K, Marks MD. Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proceedings of the National Academy of Sciences, USA. 1997;94:6261–6266. doi: 10.1073/pnas.94.12.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne A-M, Bonneville J-M. Trichome cell growth in Arabidopsis thaliana can be depressed by mutations in at least five genes. Genetics. 1999;152:461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M. Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiology. 1998;117:375–383. doi: 10.1104/pp.117.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Hu C, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu Y. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. The Plant Cell. 2007;19:3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Parra E, Gutierrez C. E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiology. 2007;144:105–120. doi: 10.1104/pp.106.094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan S, Bourquin V, Tuominen H, Sundberg B. Accurate and high resolution in situ hybridization analysis of gene expression in secondary stem tissues. The Plant Journal. 1999;19:363–369. doi: 10.1046/j.1365-313x.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- Roberts IN, Lloyd CW, Roberts K. Ethylene-induced microtubule reorientations: mediation by helical arrays. Planta. 1984;164:439–447. doi: 10.1007/BF00395959. [DOI] [PubMed] [Google Scholar]
- Rodríguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO Journal. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M. Epidermal differentiation: trichomes in Arabidopsis as a model system. International Journal of Development Biology. 2005;49:579–584. doi: 10.1387/ijdb.051983ss. [DOI] [PubMed] [Google Scholar]
- Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jürgens G, Hülskamp M. Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. The Plant Cell. 1999;11:1105–1116. doi: 10.1105/tpc.11.6.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko G, Kalinina Y, Kordyum E. Role of cytoskeleton in gravisening of the root elongation zone in Arabidopsis thaliana plants. Cell Biology International. 2008;32:560–562. doi: 10.1016/j.cellbi.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Shi Y, Zhu S, Mao X, Feng J, Qin Y, Zhang L, Cheng J, Wei L, Wang Z, Zhu Y. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. The Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A, Saleh N, Igarashi H, Mori H, Hauser-Hahn I, Jiang C, Sonobe S, Lloyd CW, Hussey PJ. A new class of microtubule-associated proteins in plants. Nature Cell Biology. 2000;2:750–753. doi: 10.1038/35036390. [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD. Control of GL2 expression in Arabidopsis leaves and trichomes. Development. 1998;125:1161–1171. doi: 10.1242/dev.125.7.1161. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kawahara A, Inoue Y. Ethylene promotes the induction by auxin of the cortical microtubule randomization required for low-pH-induced root hair initiation in lettuce (Lactuca sativa L.) seedlings. Plant and Cell Physiology. 2003;44:932–940. doi: 10.1093/pcp/pcg119. [DOI] [PubMed] [Google Scholar]
- Verde F, Berrez J, Antony C, Karsenti E. Taxol-induced microtubule asters in mitotic extracts of Xenopus eggs: requirement for phosphorylated factors and cytoplasmic dynein. Journal of Cell Biology. 1991;112:1177–1187. doi: 10.1083/jcb.112.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Shaw PJ, Warn RM, Lloyd CW. Dynamic reorientation of cortical microtubules from transverse to longitudinal in living plant cells. Proceedings of the National Academy of Sciences, USA. 1994;91:6050–6053. doi: 10.1073/pnas.91.13.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Oppenheimer DG. A simple and efficient method for isolating trichomes for downstream analyses. Plant and Cell Physiology. 2004;45:221–224. doi: 10.1093/pcp/pch016. [DOI] [PubMed] [Google Scholar]
- Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. The Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]