Abstract
The correction of exogenous and endogenous environmental insult to DNA involves a series of DNA repair mechanisms that reduce the likelihood of mutation accumulation and hence an increased probability of tumour development. The mechanisms underlying the process of base excision repair are relatively well understood and are placed in context with how deterioration of this process is associated with an increased risk of malignancy.
Keywords: base excision repair, MUTYH, DNA repair, mutations
Base excision repair
Base excision repair (BER) is one of the most important DNA repair pathways, which ameliorates environmentally induced DNA damage, including that which arises spontaneously as a result of alkylation, oxidation, and deamination events during normal metabolic processes [1]. BER is also responsible for repairing small, non-helix distorting lesions that may be induced by chemical carcinogens [2]. In addition, it is also responsible for the repair of abasic sites, which may arise spontaneously as a function of temperature fluctuation, or it could arise as intermediates in the DNA repair process [3]. Compared with other repair machinery, such as nucleotide excision repair, the core components of BER machinery have been well conserved from bacteria to humans both structurally and functionally during evolution [4-7], underscoring the vital role BER plays in maintaining genome integrity. BER is believed to be the simplest and most defined of all DNA repair processes. The molecular mechanism of BER has been resolved to the tertiary structure for all core components [8-10].
The BER pathway functions by a series of well-coordinated enzymatic events which can overall be divided into two steps. The first step of BER is the recognition and excision of a damaged base or an abasic site by a series of specific DNA glycosylases. The next step involves the sequential action of different proteins which correct DNA by template-directed insertion of one or a few nucleotides, starting at the damaged site.
The first step of BER relies on glycosylases that recognize and remove the damaged base through N-glycosylic bond hydrolysis to generate abasic or apurinic/apyrimidinic (AP) sites. These AP sites are identical to spontaneous DNA depurination or depyrimidation. Each of the glycosylases has specificity to a relatively narrow, partially overlapping spectrum of lesions, and may function as a monofunctional enzyme (exclusively removes damaged base) or a bifunctional enzyme (removes damaged base and incises DNA backbone) [11]. Bifunctional glycosylases possess AP-lyase activity that hydrolyses the 3'-phosphodiester bond of the AP site by a β- or β-d-elimination mechanism generating 3' a,β-unsaturated aldehyde and 5'-phosphate products at the termini [12]. This terminus is then cleaved by a pivotal enzyme APE1 (also called HAP1 or Ref) which has lyase and phosphodiesterase activities [13]. Monofunctional DNA glycosylases need the assistance of APE1 to hydrolyse the phosphodiester bond at the 5' end of the AP site via its lyase activity, producing a single strand break (SSB) with a normal 3'-hydroxyl group and an abnormal 5'-deoxyribose 5-phosphate (dRP) residue [14].
Numerous prokaryotic and eukaryotic DNA glycosylases have been isolated and purified. Their substrate specificities have been determined using various types of substrates. Moreover the crystal structure of numerous DNA glycosylases has been solved. Data obtained from structure determination have allowed glycosylases to be grouped into several major structural families by architectural folds [7], including helix-hairpin-helix [15], helix-two-turn-helix (H2TH) [16], and uracil DNA glycosylases (UDGs) [17].
In humans about 11 mono-functional and bifunctional glycosylases have been identified [12,18] (Table 1). The human mono-functional glycosylases include methyl-CpG-binding domain protein 4 (MBD4), N-methylpurine-DNA glycosylase (MPG), single-strand selective mono-functional uracil DNA glycosylase (SMUG1), thymine/uracil mismatch glycosylase (TDG), uracil-DNA glycosylase 2 (UDG2), uracil DNA N-glycosylase (UNG1), and UNG2. The bifunctional glycosylases include A/G-specific adenine DNA glycosylase (MUTYH), endonuclease VIII-like 1 (NEIL1), endonuclease III-like protein 1 (NTH1), and N-glyco-sylase/DNA lyase (OGG1).
Table 1.
| Name | Lyase activity | Chromosome location | Cellular localization | Known substrates | References |
| MBD4 (also called MED1) | No | 3q21–22 | Nuclei | G:T mismatches within methylated and unmethylated CpG sites. Can also remove uracil or 5-fluorouracil in G:U mismatches | [19-22] |
| MPG (also called AAG, ANPG, or MID1) | No | 16p13.3 | Nuclei | 3-methyladenine, 7-methyl-guanine, hypo-xanthine, ethenoA | [23-26] |
| SMUG1 | No | 12q13.3 | Nuclei | Uracil (U) in single or double strand DNA, with a preference for single strand DNA | [27,28] |
| TDG | No | 12q24.1 | Nuclei | G/T, C/T, and T/T mispairs. Also uracil and 5-bromouracil mispairs with guanine | [29-31] |
| UDG2 | No | 5 | Nuclei | U:A mismatch | [32,33] |
| UNG1 (also called DGU, UNG15, UDG1 or UDG1M) | No | 12q24.1 | Mitochondria | Uracil from misincorporation of dUMP residues by DNA polymerase or due to deamination of cytosine | [34-36] |
| UNG2 (also called UDG1A or UDG1N) | No | 12q24.1 | Nuclei | Uracil from misincorporation of dUMP residues by DNA polymerase or due to deamination of cytosine | [35,36] |
| MUTYH (also called MYH) | Yes (weak) | 1p32.1–34.3 | Nuclei and mitochondria | A:8-oxoG and 2-OH-A:G | [37,38] |
| NEIL1 (also called NEH1or FPG1) | Yes | 15q22–24 | Nuclei | Oxidized pyrimidines, such as thymine glycol, formamidopyrimidine (Fapy) and 5-hydroxyuracil. Also has a marginal activity towards 8-oxoguanine | [39,40] |
| NTH1 (also called NTHL1 or OCTS3) | Yes | 16p13.2–13.13 | Nuclei and mitochondria | Damaged DNA at cytosines, thymines and guanines | [41,42] |
| OGG1 (also called MMH, MUTM, or OGH1) | Yes | 3p26.2 | Nuclei and mitochondria | 8-oxoG:C, 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylforma-midopyrimidine (FAPY) | [43,44] |
Determination of the crystal structure of glycosylases has revealed that there exist differences in the folds and specific residues used to recognize damaged bases. This information, coupled by experimental characterization of DNA base repair processes, has allowed enzymatic mechanisms of cleavage by DNA glycosylases. Based on specific mechanisms for recognition of damage, DNA glycosylases can generally be grouped into those that remove oxidative damage, deamination products, and alkylation damage.
As presented in Table 1, in human cells, the DNA glycosylases that are involved in the removal of oxidized bases include MUTYH, NEIL1, NTH1, OGG1, and SMUG1. NEIL1, NTH1 and SMUG1 catalyze excision of oxidized pyrimidines, such as 5-OHC, whereas MUTYH and OGG1 repair oxidized purines, such as 8-oxoG. The human glycosylases that remove uracil are uracil-DNA glycosylase (UNG/UDG), thymine-DNA glycosylase (TDG), SMUG1, and MBD4. The only glycosylase known to be involved in repair of alkylation damage is MPG.
To complete repair after glycosylase action BER can proceed through two different sub-pathways, short-patch and long-patch pathway. These pathways are differentiated by the enzymes involved and the number of nucleotides removed. When BER is initiated by bifunctional glycosylases, short patch is the main pathway, whereas when BER is initiated by monofunctional glycosylases it might proceed through either pathway [45].
In the short-patch pathway, DNA polymerase β (Pol β), which is recruited upon direct interaction of damaged DNA with APE1, extends the 3'-OH terminus by inserting 1 nucleotide and at the same time removes 5' terminal deoxyribose phosphate (5'-dRP) by its AP lyase activity [46]. Finally, the single strand nick is sealed by either DNA ligase I or DNA ligase III in a complex with the scaffolding protein XRCC1 [47,48].
Long-patch BER is initiated in a manner similar to the short-patch pathway. Initially 2 to 12 nucleotides are incorporated by a sequential action of three different DNA polymerases (Pol β, δ, or ε) by elongating the 3' end by a few nucleotides and moving aside a DNA fragment which contains 5' deoxyribophosphate [49]. Next, this flap structure is cleaved out by specific flap endonuclease, FEN1 [50,51]. In addition to FEN1, DNA synthesis and strand displacement is stimulated by the combined presence of proliferating cell nuclear antigen (PCNA) [52], replication factor C (RFC) [53], and poly(ADP-ribose)polymerase 1 (PARP1) [54,55]. Finally, the intact DNA strand can be restored by DNA ligase I or III. However, since this pathway is stimulated by PCNA, it has been suggested that ligase I is the predominant enzyme due to its interaction with PCNA [56].
MutY homologue (MUTYH)
Structure of the MUTYH gene
The first human homologue of the E. coli MutY gene was cloned by Slupska et al. [37]. By screening a human cDNA sequence database they identified an expressed sequence tag (EST) which was then used to probe a human bacterial artificial chromosome (BAC) library to isolate genomic human mutY, which was referred to as MUTYH, also called MYH. A full-length MUTYH cDNA clone from a human brain tissue cDNA library was also isolated which localized to the short arm of chromosome 1 between p32.1 and p34.3. MUTYH encompasses 7.1 kb and has 16 exons encoding a 535 amino acid protein displaying 41% identity with E. coli protein [37]. Ohtsubo and co-workers [38] identified 10 forms of MUTYH transcripts, each with a different 5' sequence or first exon and each transcript being alternatively spliced, that were sub-grouped into 3 types, isoform MUTYHα (splice variants α1, 2, 3, and 4), isoform MUTYHβ (splice variants β1, 3, and 5), and isoform MUTYHγ (splice variants γ2, 3, and 4). The authors also showed that MUTYH protein encoded by type α mRNA possesses a mitochondrial targeting sequence (MTS), consisting of the amino terminal 14 residues which are required for its localization in the mitochondria [57], while those encoded by type β and γ mRNAs lack the MTS, and are localized in the nuclei. MUTYHα3 is the major isoform expressed in most cells and corresponds to the cDNA sequence isolated and characterized by Slupska et al. [37]. In addition to human, homologues of MutY have been cloned from other mammals, including cow [58], mouse [59] and rat [60].
The protein structure and function of MUTYH
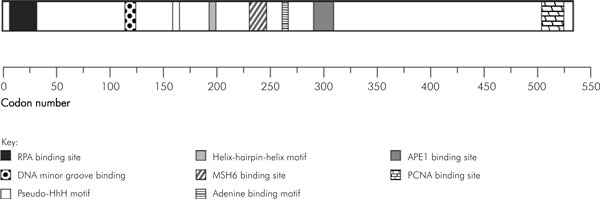
The open reading frame (ORF) of the full-length major isoform of MUTYH (MUTYHα3) encodes a 535 amino acid protein [38]. This protein displays very high identity with MutY homologues from other mammals, 78 and 74.1% identity with mouse and rat MutY, respectively [61]. Structurally, MUTYH reveals extensive homology and conservation with established structural domains found in E. coli MutY and many prokaryotic and eukaryotic BER enzymes, in addition to domains unique to MUTYH (Fig. 1).
Figure 1.
Diagram of the functional domains of MUTYH (adopted from [107]).
MutY and its homologues contain the catalytic domain which shares several motifs with other glycosylases, including the helix-hairpin-helix (HhH), pseudo-HhH and an [4Fe-4S] iron sulphur cluster in the N terminus [62,63]. The latter has a high overall similarity to endonuclease III that excises oxidized pyrimidines [64,65]. HhH (aa 114–273 in MUTYH) functions to detect, recognize and remove adenines opposite to 8-oxoG by binding the phosphate backbone of the substrate. It includes a highly conserved aspartic acid residue (Asp222 in MUTYH), which is required for nucleophilic attack of the adenine base (reviewed in Ref. [61]). MutY has a special carboxy-terminal domain that is not found in other BER glycosylases, with sequence and structural homology to MutT (an 8-oxoGTPase), and this domain has an important role in the recognition of 8-oxoG because the truncation of the domain results in loss of discrimination between 8-oxoG:A and G:A mispairs [66-68]. In addition, ORF of MUTYH also contains other domains that are shown to be the binding sites of other proteins. By using HeLa nuclear extracts, Parker and co-workers [69] demonstrated that MUTYH contains an APE1 binding site (aa 300), a replication protein A (RPA) binding site (aa 6–32), and a PCNA binding site (aa 505–527).
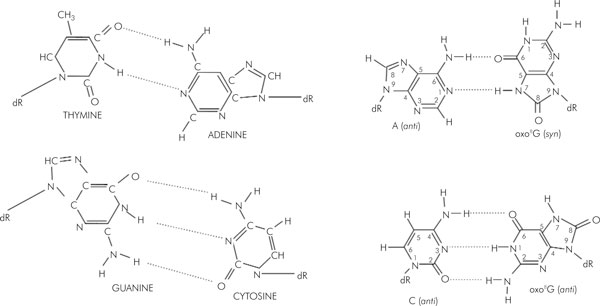
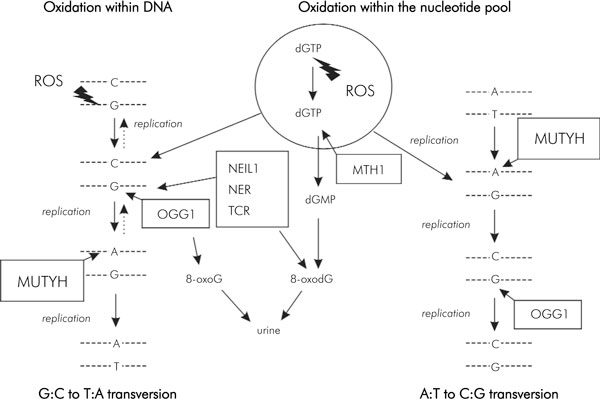
The primary function of MUTYH as a BER DNA glycosylase is to excise adenines or 2-hydroxy-adenines (2-OH-A) misincorporated opposite 7,8-dihydro-8-oxo-guanine (8-oxoG or GO) [70]. GO is one of the most stable products of DNA damage resulting from reactive oxygen species (ROS) [71] because this oxidized form of the guanine base can pair with adenine as well as cytosine with equal frequency during DNA replication and thus has the potential to cause a high rate of G:C to T:A transversions [72,73]. In E. coli MutY, together with MutM and MutT (formamidopyrimidine-DNA glycosylase: FPG), play important roles in reducing the mutagenic effects of GO lesions [74,75]. MutM removes GO paired with cytosine and introduces a single strand gap as a result of the accompanying AP-lyase activity [75]; whereas MutT hydrolyzes 8-oxo-dGTP and depletes it from the nucleotide pool [73]. Similar DNA error avoiding mechanisms have also been shown in human cells, whereby MTH1 (MutT homologue), OGG1 (MutM homologue), and MUTYH (MutY homologue) have been proposed to function in the reduction of GO in human genome [76].
Yang and co-workers demonstrated that the cloned cDNA of MUTYH [37,77] complement the mutator phenotype of a MutY E. coli strain and prevented G:C to T:A transversions [78]. Suppressive activities of MUTYH against G:C to T:A transversions have also been shown in human cells in vivo [79].
Unlike MTH1 or OGG1, MUTYH possesses no detectable AP-lyase activity [78]. The authors also demonstrated that APE1 catalytic activity is required for the formation of cleaved AP DNA and stimulation of MUTYH glycosylase activity by increasing the formation of the MUTYH-DNA complex. Similar findings were also reported by Parker and co-workers, who showed that MUTYH interacts with APE1, PCNA, and RPA, suggesting a role in long-patch BER [69]. Despite the structural homology between MUTYH and its bacterial counterpart, the human MUTYH protein efficiently removes 2-OH-A from 2-OH-A:G mismatches [38], while the bacterial protein removes 2-OH-A from the substrate containing 2-OH-A:G pairs very poorly [80] (Fig. 2, 3).
Figure 2.
Base pairing properties of oxo8G residues in DNA.
Figure 3.
Different pathways for repairing damaged G residues caused by reactive oxygen species (adapted from [123]).
Recent findings from experiments employing full-length structure of MutY cross-linked to DNA containing 8-oxoG have shed light on the mechanism of recognition and removal of 8-oxoG:A mismatched by MutY. Although the precise mechanism is still unclear, the results of these experiments suggest that MutY relies heavily on the recognition of GO to locate A bases for excision. For example, Fromme and co-workers [68] demonstrated that in the X-ray crystal structure of an inactive variant (Asp144Asn) of B. stearothermophilus MutY there are extensive contacts with 8-oxoG but minimal contacts with adenine. In another instance Bernards and co-workers [82] conducted time-resolved fluorescence experiments of the MutY A-excision reaction using 8-oxo·GA substrates and found a multiphase reaction profile, with a fast process being associated with changes at 8-oxoG and a slower process associated with altering the environment of the adenine. MutY exposes its target by deeply penetrating the DNA helix, interrupting helical stacking on both strands, encircling the DNA with its catalytic core and MutT-like domains, and rotating the phosphodiester bonds surrounding the nucleotide, causing the target base adenine to be flipped out of the DNA helix [68,83]. The iron-sulphur cluster [4Fe-4S] contained in the catalytic core of MutY plays a significant role in facilitating DNA binding with the damaged region and catalyzing the removal of adenine [84]. This iron-sulphur cluster plays a DNA-dependent electron transfer role which enables MutY to quickly and efficiently seek out damaged sites in the genome using DNA-mediated charge transport [85].
Cross-talk between MUTYH and other DNA repair enzymes
Evidence is accumulating in the literature implicating the interactions of DNA glycosylases and non-BER pathways, although in most cases the details of the mechanism are not well understood [86]. In addition to their participation in BER, DNA glycosylases have been reported to interact with nucleotide excision repair (NER) and mismatch repair (MMR).
MUTYH-initiated BER and MMR pathways share common features in terms of function and timing of action. Functionally, both pathways participate in repairing DNA lesions resulting from DNA oxidation. Both pathways take place immediately after DNA replication to increase the fidelity of DNA replication and bear a task to distinguish newly synthesized DNA strands from their parental counterparts [37,87-93].
The molecular evidence of the interaction of MUTYH and MMR proteins was first obtained from an elegant study by Gu and co-workers [94]. In their experiment they found that hMUTYH directly interacts with hMSH6 in the hMSH2/hMSH6 (hMutSα) heterodimer, which functions to bind to the mismatches and initiate the repair on the daughter DNA strands [95]. They also observed that this physical protein interaction can occur in the absence of DNA. Based on their findings, the authors further hypothesized that proteins involved in DNA replication, mismatch repair and base excision repair may exist as a multiple-protein complex and that hMUTYH may be orientated in the replication fork to recognize 8-oxoG on the parental strands and to excise misincorporated A on the daughter strand [94]. This hypothesis is supported by some evidence. Both hMUTYH and hMSH6 interact with replication proteins PCNA and RPA [69,96,97]. Moreover, both proteins show overexpression during S phase and colocalize with PCNA at replication foci [69,87,96].
Although to date there is no direct evidence for the physical interactions between MUTYH and proteins involved in NER, there is evidence indicating that DNA glycosylases are coupled to the NER pathway, more specifically to the subpathway transcription-coupled repair (TCR).
One example is the repair of thymine glycols which are normally repaired by thymine glycol DNA glycosylase. Leadon and Cooper [98] reported that thymine glycols generated in NER- and BER-proficient human cells following ionizing radiation exposure are removed in a biased fashion from the transcribed strand of an expressed gene. In this case, TCR of thymine glycols exists in the same locus in cells where BER is active, suggesting that the thymine glycol-DNA glycosylase is coupled to TCR. It is logical therefore that the question arises whether such coupled repair exists for other lesions that are normally repaired by BER glycosylases.
It has been proposed that the crucial event in transcription-coupled NER (TC-NER) is the stalling of an elongating RNA polymerase II upon a lesion, which recruits the repair proteins to the damage site [99-102]. Therefore, the ability of a lesion on the transcribed strand to block the RNA polymerase transcription complex has been assumed to be crucial for TCR. Kathe and co-workers [103] found that DNA base damage does not block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts, but single-strand breaks do. It is known that single-strand breaks are common BER processing intermediates. It is therefore tempting to speculate that a strand break produced by a DNA glycosylase at an oxidative lesion in a transcription bubble would serve as a block to the RNA polymerase apparatus which in turn will signal the TC-NER to take place.
MUTYH variants and their role in carcinogenesis
Cheadle and Sampson [104] have presented a comprehensive review of MUTYH variants and their diagnostic implications. They reported that as of the end of 2006, 30 mutations that are predicted to produce truncated proteins have been reported in MUTYH, consisting of 11 nonsense, 9 small insertion/deletions and 10 splice site variants. Moreover, 52 missense variants and three small inframe insertion/deletions have been reported that are distributed throughout the gene [104]. To date, as presented in Table 2, the list of reported MUTYH variants has grown. Of the reported variants, Y165C and G382D together account for approximately 73% of reported MUTYH variants [104] and have been commonly identified in Caucasian populations, including American, British, Danish, Finnish, Dutch, Italian, and Portuguese (reviewed in refs. [61,105]). Beside these two variants, the occurrence of the rest of the variants is rare, although recurrent variants have been observed in some populations and will be discussed later in this section.
Table 2.
List of reported MUTYH variants
| Type of mutation | Name of mutation/variant | References |
| Nonsense | R83X | [106] |
| Y90X | [107] | |
| E182X | [108] | |
| Q196X | ||
| R233X | ||
| Q300X | [108] | |
| E466X | [109] | |
| Insertions/deletions | 199delGT | |
| 252delG | ||
| 263insAG | ||
| 379delC | ||
| 817delG | ||
| 1059insC | ||
| 1103–1105delC | ||
| 1186–1187insGG | ||
| 1419delC | ||
| Splice | IVS1(36)+1G>A | [110] |
| IVS1(36)+5G.C | [111] | |
| IVS4(347)-1G>C | ||
| IVS4(347)-1G>A | ||
| IVS5(421)-1G>C | ||
| IVS8(649)-1G>A | ||
| IVS10(891)+3A>C | ||
| IVS10(892)-2A>G | [112] | |
| IVS12(1145)-2A>G | ||
| IVS13(1281)+25del30 | [108] | |
| IVS15(1476)+2T>C | ||
| Missense and inframe ins/dels | M15V | |
| P18L | [113] | |
| V22M | [114] | |
| G25D | [113] | |
| V61E | [115] | |
| D91N | [116] | |
| Y114H | ||
| W117R | ||
| 137insIW | ||
| P143L | ||
| D147H | ||
| Y165C | ||
| Y166S | ||
| R168H | ||
| R168L | ||
| R68C | ||
| R171Q | ||
| G175E | ||
| I209V | ||
| V220M | ||
| R227W | ||
| R231C | ||
| R231H | ||
| V232F | ||
| R260Q | ||
| R260W | ||
| M269V | ||
| C276W | ||
| P281L | ||
| R295C | ||
| Q324H | [114] | |
| Q324R | ||
| F344Y | ||
| P345T | ||
| P366T | [116] | |
| L374P | ||
| G382D | ||
| P391S | ||
| P391L | ||
| Q400R | ||
| A405T | ||
| L406M | ||
| R412C | ||
| R423P | [117] | |
| R423Q | [117] | |
| R423R | [117] | |
| A459D | ||
| A459T | ||
| 1391delAGG | ||
| 1395delGGA (466delE) | ||
| V479F | ||
| Q484H | ||
| S501F | [114] | |
| P502L | ||
As seen in Table 2, most of the variants reported, as is usually the case with most genes, are categorized as single nucleotide polymorphisms (SNPs). SNPs are characterized by single base changes in genes and other DNA sequences and are discovered by DNA re-sequencing, single strand conformation polymorphism (SSCP), or nucleotide probing technologies, such as pyrosequencing, real time PCR and multiplex ligation-dependent probe amplification (MLPA). These single base changes give rise to missense and nonsense variants and may or may not result in an obvious phenotypic trait.
Over the past decade the determination of SNPs has become an important method of characterizing the individual differences in genetic makeup, as compared to the "normal" genome sequence. Of potentially greater importance are the SNPs that may increase an individual's susceptibility toward malignancy because of subtle differences in the polymorphic protein products due to nonsense mutations, especially when such variations are present in key structural areas.
Evidence supporting a role for defective MUTYH in human carcinogenesis is accumulating. The first study to establish such a role was that of Al-Tassan and co-workers [114], who described a British family in which three siblings with colorectal cancer and adenomatous polyposis were compound heterozygous for two germline mutations in the MUTYH gene that result in MUTYH proteins containing amino acid substitutions Tyr165Cys (Y165C) and Gly382Asp (G382D). Unaffected family members were either homozygous normal or heterozygous for one of the mutations, thus suggesting an autosomal recessive pattern of inheritance for the phenotype of multiple adenomas. The authors further showed that proteins containing these substitutions in E. coli lead to severe impairment of base excision repair. The Y165C mutation is located in the HhH motif, which is highly conserved in all mammalian MutY as well as in E. coli MutY proteins [62,118,119]. Functionally, it completely abolishes adenine glycosylase activity toward the A:8-oxoG mispair. The Gly382Asp mutation resides in the MutT-like domain and also gives rise to decreased adenine removal [114].
Since this discovery, which is the first to demonstrate a direct link between a defective human DNA repair gene and predisposition to colorectal cancer, considerable work has established the relationship between mutations in MUTYH and colorectal adenomas and carcinomas, and this disorder is now referred to as MUTYH-associated polyposis (MAP) [105,107,120]. To date, testing for MUTYH variants is recommended for patients who have clinical features of familial adenomatous polyposis (FAP) but either do not have inherited mutations in APC or have a family history consistent with recessive inheritance, as is the case for MAP [121].
As seen in Table 2 there are many variants of MUTYH reported in the literature, in addition to Y165C and G382D. These variants range from simple one nucleotide substitutions to insertions/deletions (indels) affecting more than one nucleotide. Some of these variants may give rise to non-functional MUTYH. Many of these variants, however, are silent, causing missense changes, or are small in-frame indels. Although without functional analyses and/or segregation analyses it is problematic to determine the effect of these variants, on the basis of the type of mutation, the location in the sequence of MUTYH, and the corresponding position in the structure it of MUTYH, is possible however, to make a prediction about the consequences of some of these types of mutation in some cases.
Among examples of the variants in which the consequence of MUTYH function may be predicted by their corresponding position in the structure of MUTYH are the P18L [113], V22M [114], and G25D [113] variants. These three SNPs are present in the RPA binding site in the N-terminus of MUTYH and could interfere with the localization of MUTYH to the site of DNA replication. In other instances, the variants E466X [109] and Y90X [107], which were found in individuals of Indian and Pakistani descent, respectively, represent truncated MUTYH protein. In these variants, X indicates a stop codon which results in premature termination of protein synthesis and in turn gives rise to non-functional MUTYH protein.
Nonetheless, the consequences of most of the missense variants are not obvious. Moreover, some missense variants are not conserved in the bacterial MutY enzymes, so it is difficult to make predictions (reviewed in ref. [122]). This, coupled with their collective frequency and lack of functional data, poses major difficulties for molecular diagnostics since many will be benign polymorphisms.
Mutations in the MUTYH gene and defective MUTYH activities are just beginning to be identified in human cancers. Consequently, it is obvious that more information about the clinical and molecular properties of MAP is needed to aid in the diagnosis and treatment of affected patients and family members. Such knowledge might also provide insight into how MUTYH mutations contribute more globally to malignancies other than colorectal cancer.
References
- Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- Loeb L, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- Cuniasse P, Fazakerly GV, Guschlbauer W, Kaplan B, Sowers LC. The abasic site as a challenge to DNA polymerase. A nuclear magnetic resonance study of G, C and T opposite a model abasic site. J Mol Biol. 1990;213:303–314. doi: 10.1016/S0022-2836(05)80192-5. [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:9315–9348. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Wood RD. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu Rev Biophys Biomol Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- Nilsen H, Krokan HE. Base excision repair in a network of defense and tolerance. Carcinogenesis. 2001;22:987–998. doi: 10.1093/carcin/22.7.987. [DOI] [PubMed] [Google Scholar]
- Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means. Mutat Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. ASM PRESS Washington DC; 1995. pp. 208–70. [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/S0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, Golemis EA, Genuardi M, Neri G. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci USA. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TR, Laval F. Isolation and structure of a cDNA expressing a mammalian 3-methyladenine-DNA glycosylase. EMBO J. 1990;9:3337–3342. doi: 10.1002/j.1460-2075.1990.tb07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Ibeanu GC, Tano K, Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J Biol Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- Engelward BP, Weeda G, Wyatt MD, Broekhof JL, de Wit J, Donker I, Allan JM, Gold B, Hoeijmakers JH, Samson LD. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc Natl Acad Sci USA. 1997;94:13087–13092. doi: 10.1073/pnas.94.24.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor EE, Wilson JJ, Wyatt MD. Effects of substrate specificity on initiating the base excision repair of N-methylpurines by variant human 3-methyladenine DNA glycosylases. Chem Res Toxicol. 2005;18:87–94. doi: 10.1021/tx049822q. [DOI] [PubMed] [Google Scholar]
- Haushalter KA, Todd Stukenberg MW, Kirschner MW, Verdine GL. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr Biol. 1999;9:174–185. doi: 10.1016/S0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Tanaka T, Terato H, Ohmae E, Izumi S, Katayanagi K, Ide H. Mutational analysis of the damage-recognition and catalytic mechanism of human SMUG1 DNA glycosylase. Nucleic Acids Res. 2004;32:5291–5302. doi: 10.1093/nar/gkh859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinari P, Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- Neddermann P, Gallinari P, Lettieri T, Schmid D, Truong O, Hsuan JJ, Wiebauer K, Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- Muller SJ, Caradonna S. Cell cycle regulation of a human cyclin-like gene encoding uracil-DNA glycosylase. J Biol Chem. 1993;268:1310–1319. [PubMed] [Google Scholar]
- Caradonna S, Ladner R, Hansbury M, Kosciuk M, Lynch F, Muller S. Affinity purification and comparative analysis of two distinct human uracil-DNA glycosylases. ExpCell Res. 1996;222:345–359. doi: 10.1006/excr.1996.0044. [DOI] [PubMed] [Google Scholar]
- Olsen LC, Aasland R, Wittwer CU, Krokan HE, Helland DE. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989;8:3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Markussen FH, Olsen LC, Aasland R, Aarsaether N, Bakke O, Krokan HE, Helland DE. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178:3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo T, Nishioka K, Imaiso Y, Iwai S, Shimokawa H, Oda H, Fujiwara T, Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/S1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker AH, Seki S, Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M, Fukasawa K, Yoshinari T, Inoue H, Ohtsuka E, Nishimura S. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- Bennett RA, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc Natl Acad Sci USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH. Properties and applications of human DNA repair genes. Mutat Res. 1991;247:213–219. doi: 10.1016/0027-5107(91)90017-i. [DOI] [PubMed] [Google Scholar]
- Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. 8-oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat Res. 2003;531:127–139. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J Biol Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- Gary R, Kim K, Cornelius HL, Park MS, Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J Biol Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- Pascucci B, Stucki M, Jónsson ZO, Dogliotti E, Hübscher U. Long patch base excision repair with purified human proteins. DNA ligase I as patch size mediator for DNA polymerases delta and epsilon. J Biol Chem. 1999;274:33696–33702. doi: 10.1074/jbc.274.47.33696. [DOI] [PubMed] [Google Scholar]
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/S0301-472X(03)00083-3. [DOI] [PubMed] [Google Scholar]
- Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol. 2000;10:919–922. doi: 10.1016/S0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- Takao M, Zhang QM, Yonei S, Yasui A. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine: 8-oxoguanine DNA glycosylase. Nucl Acids Res. 1999;27:3638–3644. doi: 10.1093/nar/27.18.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A, Gu Y, Lu AL. Purification and characterization of a mammalian homolog of Escherichia coli MutY mismatch repair protein from calf liver mitochondria. Nucleic Acids Res. 2000;28:3206–3215. doi: 10.1093/nar/28.17.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinoe A, Behmanesh M, Tominaga Y, Ushijima Y, Hirano S, Sakai Y, Tsuchimoto D, Sakumi K, Wake N, Nakabeppu Y. Identification and characterization of two forms of mouse MUTYH proteins encoded by alternatively spliced transcripts. Nucleic Acids Res. 2004;32:477–487. doi: 10.1093/nar/gkh214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Wang C, Hu Z, Greeley GH, Jr, Makalowski W, Hellmich HL, Englander EW. Hypoxia induces mitochondrial DNA damage and stimulates expression of a DNA repair enzyme, the Escherichia coli MutY DNA glycosylase homolog (MYH), in vivo, in the rat brain. J Neurochem. 2002;80:928–937. doi: 10.1046/j.0022-3042.2002.00774.x. [DOI] [PubMed] [Google Scholar]
- Parker AR, Eshleman JR. Human MutY: gene structure, protein functions and interactions, and role in carcinogenesis. Cell Mol Life Sci. 2003;60:2064–2083. doi: 10.1007/s00018-003-3053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel RC, Lloyd RS. Cloning, overexpression, and biochemical characterization of the catalytic domain of MutY. Biochemistry. 1997;36:11140–11152. doi: 10.1021/bi9709708. [DOI] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd RS, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1996. p. 5. [DOI] [PubMed]
- Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CF, McRee DE, Fisher CL, O'Handley SF, Cunningham RP, Tainer JA. Atomic structure of the DNA repair (4-iron-4-sulfur) enzyme endonuclease III. Science. 1992;258:434–440. doi: 10.1126/science.1411536. [DOI] [PubMed] [Google Scholar]
- Noll DM, Gogos A, Granek JA, Clarke ND. The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine; adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- Chmiel NH, Golinelli MP, Francis AW, David SS. Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Res. 2001;29:553–564. doi: 10.1093/nar/29.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- Au KG, Cabrera M, Miller JH, Modrich P. Escherichia coli mutY gene product is required for specific A-G–C.G mismatch correction. Proc Natil Acad Sci USA. 1988;85:9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek A, Berardini M, Fishel R. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J Biol Chem. 2002;277:8260–8266. doi: 10.1074/jbc.M111269200. [DOI] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- Tchou J, Michaels ML, Miller JH, Grollman AP. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993;268:26738–26744. [PubMed] [Google Scholar]
- Boiteux S, Radicella JP. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie. 1999;81:59–67. doi: 10.1016/S0300-9084(99)80039-X. [DOI] [PubMed] [Google Scholar]
- McGoldrick JP, Yeh YC, Solomon M, Essigmann JM, Lu AL. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol Cell Biol. 1995;15:989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29:743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane A, Shinmura K, Sunaga N, Saitoh T, Yamaguchi S, Shinmura Y, Yoshimura K, Murakami H, Nojima Y, Kohno T, Yokota J. Suppressive activities of OGG1 and MYH proteins against G:C to T:A mutations caused by 8-hydroxyguanine but not by benzo[a]pyrene diol epoxide in human cells in vivo. Carcinogenesis. 2003;24:1031–1037. doi: 10.1093/carcin/bgg056. [DOI] [PubMed] [Google Scholar]
- Hashiguchi K, Zhang QM, Sugiyama H, Ikeda S, Yonei S. Characterization of 2-hydroxyadenine DNA glycosylase activity of Escherichia coli MutY protein. Int J Radiat Biol. 2002;78:585–592. doi: 10.1080/09553000210130560. [DOI] [PubMed] [Google Scholar]
- Loft S, Møller P. Oxidative DNA damage and human cancer: need for cohort studies. Antioxid Redox Signal. 2006;8:1021–1031. doi: 10.1089/ars.2006.8.1021. [DOI] [PubMed] [Google Scholar]
- Bernards AS, Miller JK, Bao KK, Wong I. Flipping Duplex DNA Inside Out. Flipping duplex DNA inside out: a double base-flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J Biol Chem. 2002;277:20960–20964. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Curr Opin Chem Biol. 2005;9:145–151. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Boal AK, Yavin E, Barton JK. DNA repair glycosylases with a [4Fe-4S] cluster: a redox cofactor for DNA-mediated charge transport? J Inorg Biochem. 2007;101:1913–1921. doi: 10.1016/j.jinorgbio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair (Amst) 2007;6:517–529. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Boldogh I, Milligan D, Lee MS, Bassett H, Lloyd RS, McCullough AK. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine: 8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181:6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Yasui A. DNA repair initiated by glycosylases in the nucleus and mitochondria of mammalian cells; how our cells respond to a flood of oxidative DNA damage. J Dermatol Sci Suppl. 2005;1:S9–19. doi: 10.1016/j.descs.2005.06.001. [DOI] [Google Scholar]
- Gu Y, Parker A, Wilson TM, Bai H, Chang DY, Lu AL. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J Biol Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- Gradia S, Acharya S, Fishel R. The role of mismatched nucleotides in activating the hMSH2–hMSH6 molecular switch. J Biol Chem. 2000;275:3922–3930. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon SA, Cooper PK. Preferential repair of ionizing radiation-Induced damage in the transcribed strand of an active human gene is defective in cockayne syndrome. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. Nucleotide excision repair. II: From yeast to mammals. Trends Genet. 1993;9:211–217. doi: 10.1016/0168-9525(93)90121-W. [DOI] [PubMed] [Google Scholar]
- Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- Garfinkel DJ, Bailis AM. Nucleotide excision repair, genome stability, and human disease: new insight from model systems. J Biomed Biotech. 2002;2:55–60. doi: 10.1155/S1110724302201023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–8956. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- Kathe SD, Shen GP, Wallace SS. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J Biol Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- Cheadle JP, Sampson JR. MUTYH-associated polyposis – from defect in base excision repair to clinical genetic testing. DNA Repair (Amst) 2007;6:274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Jones S, Dolwani S, Cheadle JP. MutYH (MYH) and colorectal cancer. Biochem Soc Trans. 2005;33:679–683. doi: 10.1042/BST0330679. [DOI] [PubMed] [Google Scholar]
- Aretz S, Uhlhaas S, Goergens H, Siberg K, Vogel M, Pagenstecher C, Mangold E, Caspari R, Propping P, Friedl W. MUTYH-associated polyposis: 70 of 71 patients with biallelic mutations present with an attenuated or atypical phenotype. Int J Cancer. 2006;119:807–814. doi: 10.1002/ijc.21905. [DOI] [PubMed] [Google Scholar]
- Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, Maynard J, Pigatto F, Shaw J, Cheadle JP. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 2003;362:39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- Eliason K, Hendrickson BC, Judkins T, Norton M, Leclair B, Lyon E, Ward B, Noll W, Scholl T. The potential for increased clinical sensitivity in genetic testing for polyposis colorectal cancer through the analysis of MYH mutations in North American patients. J Med Genet. 2005;42:65–66. doi: 10.1136/jmg.2004.025973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C --> T:A mutations. Hum Mol Genet. 2002;11:2961–2967. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- Niessen RC, Sijmons RH, Ou J, Olthof SG, Osinga J, Ligtenberg MJ, Hogervorst FB, Weiss MM, Tops CM, Hes FJ, de Bock GH, Buys CH, Kleibeuker JH, Hofstra RM. M208UTYH and the mismatch repair system: partners in crime? Hum Genet. 2006;119:206–211. doi: 10.1007/s00439-005-0118-5. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Shinmura K, Saitoh T, Takenoshita S, Kuwano H, Yokota J. A single nucleotide polymorphism at the splice donor site of the human MYH base excision repair gene results in reduced translation efficiency of its transcripts. Genes Cells. 2002;7:461–474. doi: 10.1046/j.1365-2443.2002.00532.x. [DOI] [PubMed] [Google Scholar]
- Tao H, Shinmura K, Hanaoka T, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y, Ozawa T, Tsujinaka T, Li Z, Yamaguchi S, Yokota J, Sugimura H, Tsugane S. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis. 2004;25:1859–1866. doi: 10.1093/carcin/bgh206. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Yamaguchi S, Saitoh T, Kohno T, Yokota J. Somatic mutations and single nucleotide polymorphisms of base excision repair genes involved in the repair of 8-hydroxyguanine in damaged DNA. Cancer Letters. 2001;166:65–69. doi: 10.1016/S0304-3835(01)00435-9. [DOI] [PubMed] [Google Scholar]
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nature Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- Halford SE, Rowan AJ, Lipton L, Sieber OM, Pack K, Thomas HJ, Hodgson SV, Bodmer WF, Tomlinson IP. Germline mutations but not somatic changes at the MYH locus contribute to the pathogenesis of unselected colorectal cancers. Am J Pathol. 2003;162:1545–1548. doi: 10.1016/S0002-9440(10)64288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Franken PF, Reinards TH, Weiss MM, Wagner A, Klift H van der, Kloosterman S, Houwing-Duistermaat JJ, Aalfs CM, Ausems MG, Brocker-Vriends AH, Gomez Garcia EB, Hoogerbrugge N, Menko FH, Sijmons RH, Verhoef S, Kuipers EJ, Morreau H, Breuning MH, Tops CM, Wijnen JT, Vasen HF, Fodde R, Hes FJ. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP) J Med Genet. 2005;42:e54. doi: 10.1136/jmg.2005.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XL, Djureinovic T, Werelius B, Lindmark G, Sun XF, Lindblom A, Swedish LR. Germline mutations in the MYH gene in Swedish familial and sporadic colorectal cancer. Genet Test. 2005;9:147–151. doi: 10.1089/gte.2005.9.147. [DOI] [PubMed] [Google Scholar]
- Manuel RC, Hitomi K, Arvai AS, House PG, Kurtz AJ, Dodson ML, McCullough AK, Tainer JA, Lloyd RS. Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase. J Biol Chem. 2004;279:46930–46939. doi: 10.1074/jbc.M403944200. [DOI] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd RS, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- Dolwani S, Sampson J, Jones S, Eccles D, Ellis A, Evans G, Frayling I, Mak T, Pigatto F, Jordan S, Maher E, Maynard J, Shaw J, Cheadle J. MYH polyposis: a new autosomal recessive form of familial adenomatous polyposis due to defective base excision repair-reappraisal of genetic risk and family management. Gastroenterology. 2003;124(Suppl 1):A46–198. doi: 10.1016/S0016-5085(03)80226-3. [DOI] [Google Scholar]
- Chow E, Thirlwell C, Macrae F, Lipton L. Colorectal cancer and inherited mutations in base-excision repair. Lancet Oncol. 2004;5:600–606. doi: 10.1016/S1470-2045(04)01595-5. [DOI] [PubMed] [Google Scholar]
- David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loft S, Møller P. Oxidative DNA damage and human cancer: need for cohort studies. Antioxid Redox Signal. 2006;8:1021–31. doi: 10.1089/ars.2006.8.1021. [DOI] [PubMed] [Google Scholar]