Abstract
Replication protein A (RPA) is a heterotrimeric protein consisting of RPA1, RPA2 and RPA3 subunits that binds to ssDNA with high affinity. The response to replication stress requires the recruitment of RPA and the MRE11/RAD50/NBS1 (MRN) complex. RPA bound to ssDNA stabilizes stalled replication forks by recruiting checkpoint proteins involved in fork stabilization. MRN can bind DNA structures encountered at stalled or collapsed replication forks, such as ssDNA-dsDNA junctions or breaks and promote the restart of DNA replication. Here, we demonstrate that RPA2 phosphorylation regulates the assembly of DNA damage-induced RPA and MRN foci. Using purified proteins, we observe a direct interaction between RPA with both NBS1 and MRE11. By utilizing RPA bound to ssDNA, we demonstrate that substituting RPA with phosphorylated RPA or a phosphomimetic decreases the interaction with the MRN complex. Also, the N-terminus of RPA1 is a critical component of the RPA-MRN protein-protein interaction. Deletion of the N-terminal oligonucleotide-oligosaccharide binding fold (OB-fold) of RPA1 abrogates RPA interactions with MRN and individual proteins of the MRN complex. Further identification of residues critical for MRN binding in the N-terminus of RPA1 show that substitution of Arg31 and Arg41 with alanines disrupts the RPA-MRN interaction and alters cell cycle progression in response to DNA damage. Thus, the N-terminus of RPA1 and phosphorylation of RPA2 regulate RPA-MRN interactions and are important in the response to DNA damage.
Introduction
Replication Protein A (RPA) was originally purified as a replication factor that stimulated helicase strand displacement and polymerase synthesis (1). RPA is now known to be essential for many aspects of DNA metabolism, including initiating DNA damage checkpoint signaling and repair of DNA damage (2-6). RPA facilitates activities in replication and repair through its interactions with other proteins. Encountering a lesion that stalls the replication fork, leads to the uncoupling of helicase and polymerase activities which generates long stretches of ssDNA that are stabilized by RPA binding (7, 8). RPA recruits and interacts with ATRIP and ATR to facilitate the activation of ATR signaling (9). Following ATR recruitment, ATR phosphorylates RPA and a number of other substrates including, CHK1, MCM2, MCM7, RAD9 and RAD17, at the replication fork, presumably to stabilize the fork and promote restart of replication (10).
Another protein complex involved in ATR and ATM signaling pathways is the MRN complex. This complex consists of MRE11, NBS1 and RAD50 and functions in DNA replication, DNA damage recognition, cell cycle checkpoint activation, and DNA repair (11). The MRN complex prevents DSBs during replication in Xenopus extracts (12, 13) and recruits ATM to double strand breaks (DSBs) through interaction with NBS1 (14, 15). In addition, the nuclease activities of MRE11 facilitate UV-induced ATR signaling (16). In fact, RPA and the MRN complex co-localize to discrete foci and interact in response to DNA replication fork blockage induced by UV (17). It was recently shown that this protein-protein interaction involves an acidic alpha helix peptide in MRE11 that binds to the DNA Binding Domain (DBD) F located at the N-terminus of RPA1 (18). This RPA-MRN interaction was shown to be important for suppressing DNA replication initiation following DNA damage (19).
The functional role of RPA in DNA metabolism relies on DBDs that bind to ssDNA and disrupt duplex DNA structure (reviewed in (20). These DBDs are OB folds that are present in each of the subunits of RPA, which includes RPA1 (p70), RPA2 (p32) and RPA3 (p14) subunits. RPA1 contains DBD A and B, which are responsible for RPA’s high affinity binding to ssDNA. DBD C is located in the C-terminus of RPA1 and DBD F is located in the N-terminus of RPA1. DBD D is in the central region of RPA2 and DBD E is within RPA3. The DBD F domain has a basic cleft region that has recently been suggested to control checkpoint protein-protein interactions (21, 22). Multiple checkpoint proteins interact with the DBD F domain, including P53, ATRIP, RAD17, RAD9 and MRE11 (18, 21, 22). The key to these protein interactions involves the basic cleft region of the DBD F domain binding to an acidic alpha helix domain located within the checkpoint proteins. Following DNA damage, RPA is hyperphosphorylated on the N-terminus of RPA2 (23). This phosphorylation has the potential to regulate RPA protein interactions through the binding of the negatively charged N-terminus of RPA2 to the basic cleft region of DBD F. Indeed, when small peptides of the N-terminus of RPA2 were mutated by introducing negatively charged amino acids designed to mimic phosphorylated residues, the acidic peptide interacted with the basic cleft of the DBD F domain (21, 24).
Previous published results have shown that the RPA binding surface on MRE11 mapped to amino acids 521-569 and that an MRE11 peptide consisting of amino acids 539-553 bound to the N-terminus of RPA1 (18, 19). In contrast to the previous published results, we directly compare wt-RPA1 and mutant RPA1 defective for MRN binding in cells with the complete MRN complex. In this manner, we more specifically implicate the N-terminus of RPA1 in the binding to the MRN complex. Additionally, we have determined that phosphorylation of RPA2 regulates RPA-MRN interactions and the assembly of RPA and NBS1 foci. Unlike the previous publications, we identify an interaction between RPA and NBS1 using purified proteins. Our data and previous results suggest that the N-terminus of RPA1 interacts with the MRN complex in unperturbed cells and phosphorylation of RPA2 disrupts RPA-MRN interactions in response to DNA damage.
Experimental Procedures (Materials and Methods)
Materials
DNA oligonucleotides were purchased from IDT DNA Technologies (IDT), Inc. The RPA2 antibodies were purchased from Thermo Fisher Scientific and Bethyl Laboratories, and the V5 agarose immobilized and RPA1 antibodies were from Bethyl Laboratories. MRE11 and NBS1 antibodies were from Genetex Inc. and Bethyl Laboratories, and the RAD50 antibody was from Genetex Inc. The HA antibody was purchased from Covance and the γ-H2AX antibody was purchased from Thermo Fisher Scientific. The Ultra TMB ELISA solution was from Pierce. The 96-well streptavidin plates were from Thermo Fisher Scientific. All other reagents and chemicals were from standard suppliers.
Cell Culture
All cells were maintained at 37 °C and 5% CO2 in Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone), 100 U/ml penicillin and 100 μg/ml streptomycin. UMSCC38 cells are oral squamous cell carcinoma cells established at the University of Michigan and generously provided by Dr. Thomas Carey, University of Michigan. For etoposide treatment, cells were incubated in growth medium containing etoposide for 2 h, and then replenished with fresh growth medium and incubated for 6 and 24 h post removal of etoposide.
Retroviral Gene Expression
Wild-type and alanine-substituted RPA32 cDNAs were inserted into the retroviral vector pQCXIH (Clontech). RPA2 DNA sequence representing amino acids 18-271 were amplified from pET-11d vectors containing the wild-type and alanine-subsituted (RPA2: 4,8,11,12,13,21,23,29,33,39) RPA heterodimer (generously supplied by Marc Wold, University of Iowa, IA) and were digested with BamHI and NaeI. Amino acids 1-17 were created by the ligation of two phosphorylated, double-stranded oligonucleotides with AgeI and NaeI overhangs. The N-terminal double-stranded oligonucleotides and the amplified C-terminus containing a HA-tag sequence were ligated into AgeI/BamHI-digested pQCXIH. Wild-type and mutated RPA70 cDNAs were inserted into the retroviral vector pQCXIP (Clontech). RPA70 was initially PCR amplified from the RPA-containing pET-11d vector using a NotI site-containing sense oligonucleotide and nested oligonucleotides coding for a C-terminal V5 tag containing an EcoRI site and ligated to the cloning vector pGEM-t-easy (Promega). Truncated RPA70 (amino acids 1-167) was similarly created using an internal sense site. Aspartic acid to Alanine substitutions at amino acids 31 and 41 were created by site-directed mutagenesis (New England Biolabs). Wild-type and mutated RPA70 were excised from pGEM-t-easy using restriction enzymes NotI and EcoRI and ligated into the retroviral expression vector pQCXIP. For retroviral infection, Phoenix A cells were plated overnight in 60-mm culture dishes at 5×106 cells/ml and transfected with 24 μg of plasmid using Lipofectamine 2000 (Invitrogen). After 48 h at 37°C, cells were incubated for an additional 24 h at 32°C. Supernatants were collected, centrifuged at 2000 g for 10 min and added to 25% confluent UMSCC38 cells in the presence of 10 μg/ml polybrene (Sigma). After 48 h, UMSCC38 cells were selected with 40 μg/ml hygromycin or 500 ng/ml puromycin. Surviving cells were grown to near confluence and tested for expression of recombinant RPA by western blot analysis.
Immunofluorescence
Cells were grown on 22-mm coverslips overnight prior to drug treatment. After an initial wash with PBS, cells were extracted with PBS containing 0.5% Triton X-100 for 5 min on ice, and fixed with 4% paraformaldehyde for 15 min. Next, the coverslips were blocked with 15% FBS and then incubated with HA, NBS1 or γ-H2AX antibodies diluted in PBS containing 2% BSA overnight. The coverslips were then washed with PBS and incubated with Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 568-conjugated anti-rabbit antibodies for 1 h. Nuclei were stained with a 1 μg/ml 4′,6-diamidino-2-phenylindole solution. Immunofluorescent images were captured digitally with a Zeiss axiovert 200M microscope.
Protein Purification
RPA and RPA mutants were purified as previously described using the expression vectors kindly provided by Marc Wold (25). Hyperphosphorylated RPA was purified as previously described (26). Briefly, hyperphosphorylated RPA was purified by taking E. coli extracts containing RPA and fractionating on a ssDNA cellulose column. Bound RPA protein was eluted and the resulting RPA fraction was approximately 80% pure. The RPA was dialyzed and mixed with HeLa extracts. The mixture was supplemented with an ATP regenerating system, ssDNA and phosphatase inhibitors to inhibit phosphatase activity within the cell extracts. This reaction allows for endogenous kinases within the HeLa extract to phosphorylate RPA. Following incubation for 3 h at 30 °C, the hyperphosphorylated RPA was purified identically to recombinant RPA (25). MRN was purified using a procedure similar to that previously described (27). Briefly, 300 ml of Sf9 cells at 1.25 × 106 cells/ml were infected with MRE11, RAD50 and NBS1 baculovirus (kindly provided by James Carney). The MRE11 and NBS1 proteins contain a 6X-His-tag. Cells were incubated 48-72 h and harvested by centrifugation. Cells were dounce homogenized and sonicated followed by centrifugation to remove cellular debris. The supernatant was loaded onto a 1 ml nickel column equilibrated in 10 mM imidazole. The gel matrix was washed with 10 mM imidazole followed by 20 and 30 mM imidazole washes. Bound MRN was eluted using 250 mM imidazole. In some instances, a mono Q column was also utilized to achieve greater purity (27). MRN and RPA purifications are shown in the Supporting Information section as Supplemental Fig.S1 and Fig. S2, respectively.
Immunoprecipitations
For immunoprecipitation (IP) reactions with purified proteins, 300 or 500 ng of purified RPA and 2-4 μg of individual or partially purified MRN complex was allowed to interact at 4° C for 30 min in PBS, 0.01% NP-40 and 1 mg BSA. The proteins were then pulled-down for 1h with either 1μg of specific antibody with sepharose beads or Ni-NTA agarose beads. Reactions were washed with PBS with 0.01% NP-40 and analyzed by western blot. For the IPs using ssDNA, RPAs were pre-incubated with 42-mer ssDNA and then MRN protein was added and assessed as described above. For IPs with cell lysates, cells were washed with phosphate-buffered saline and resuspended in cell lysis buffer (50 mM Tris-HCl, [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO4, and protease inhibitor cocktail) for 30 min on ice. Cell lysates were centrifuged for 20 min at 20,000 × g. The supernatants were incubated with anti-V5 affinity gel (Sigma) at 4°C overnight. The beads were washed three times with phosphate buffered saline containing 0.1% Nonidet P-40. The beads were resuspended in SDS sample buffer, boiled and separated by SDS-PAGE.
Immunoblotting
Cell lysates and immunoprecipitates were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were immunoblotted using the following primary antibodies: RPA1, RPA2, MRE11 NBS1 and RAD50. Secondary antibodies were Alexa Fluor 680-conjugated anti-rabbit (Invitrogen), DyLight 800-conjugated anti-mouse (ThermoFisher Scientific). Blots were visualized using infrared fluorescence (LI-COR Inc.) or chemiluminescence.
ELISAs
For 2 h at 37°C, 100ng of protein was bound to the bottom of a 96 well plate, blocked for 1h at 37° in PBS with 5% Nonfat Milk and 0.1% BSA, and then 2.5 pmols of purified RPA was added for 1h. Next, the primary polyclonal RPA2 antibody (1:4000 dilution) was added followed by washing. The secondary goat anti-rabbit antibody (1:3000 dilution) was then added and wells washed. Finally, 100uL of 1-Step Ultra TMB-ELISA solution was added and the change in absorbance at 610 nm was monitored at 1 min intervals for 20 min and analyzed using SoftMax Pro software (Molecular Devices Corp.). Relative binding activity is based on HRP activity that is determined as the change in absorbance at 610 nm/min. The reactions displayed linear kinetics for the duration of the assay (data not shown). For reactions with ssDNA, 2.5 pmols of RPA was incubated with 2.5 pmols of 42-mer ssDNA on ice before being added to the wells coated with 5 pmols of MRN, MRE11 or NBS1. For the streptavidin-biotin-ssDNA ELISA, 5 pmols of biotinylated-ssDNA was added to the streptavidin coated wells. RPAs were subsequently added and allowed to bind the biotinylated-ssDNA. The wells were then washed to remove any inactive and unbound RPA and blocked with BSA and biotin before MRN was added to the wells. MRE11 primary and HRP-conjugated secondary antibodies were used for detection similar to that described above.
Chromatin isolation
Chromatin fractionation was performed as previously described with slight modifications (Mendez and Stillman, 2000). Briefly, to isolate chromatin, the cells were washed once with PBS and incubated on ice in buffer A (10 mM HEPES, [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO4, and phosphatase inhibitor cocktail) with 0.1 % Triton X-100 for 5 min. Nuclei were collected by centrifugation (10 min, 1300 × g, 4°C) and washed once with buffer A and lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO4, and phosphatase inhibitor cocktail (Calbiochem) for 30 min on ice. Insoluble chromatin was collected by centrifugation (5 min, 1700 × g, 4°C). The final chromatin pellet was resuspended in SDS sample buffer, boiled and separated by SDS-PAGE.
Results
Phosphorylation of RPA2 regulates co-localization of RPA and MRN foci
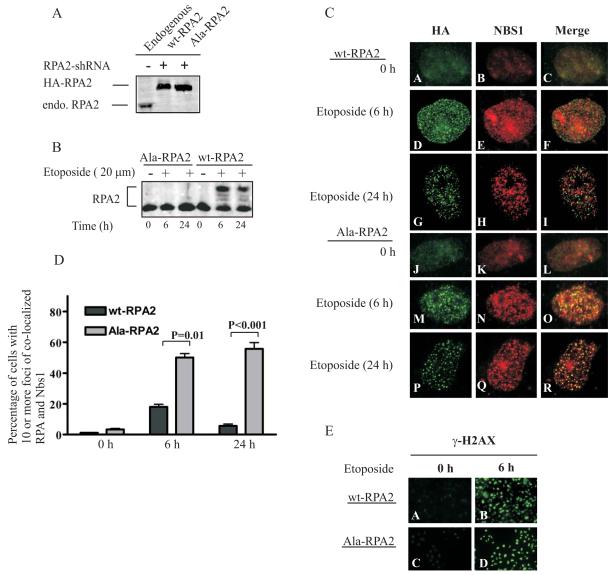
At drug concentrations used in this study, most etoposide-stabilized cleavage complexes contain high levels of ssDNA breaks rather than dsDNA breaks (28-30). These ssDNA breaks can stall replication forks and if not stabilized lead to replication fork collapse and subsequent dsDNA breaks. The increased foci formation of RPA is consistent with the presence of etoposide-associated ssDNA breaks. NBS1 has been shown to associate with ssDNA lesions (31). It was previously reported that NBS1 and RPA do not co-localize immediately following 3 h of etoposide treatment (32). To determine whether the phosphorylation of RPA2 alters the assembly of RPA and MRN foci, we utilized UMSCC38 cells that express wt-RPA2 or Ala-RPA2 (Fig. 1). Ala-RPA2 contains alanine substitutions at S4, S8, S11, S12, S13, T21, S23, S29, S33, and S39 within the N-terminus of RPA2 (Fig. 1B). Endogenous RPA2 in both cell lines was down-regulated by stably expressing shRNA targeting the 3′-UTR of RPA2 (Fig. 2A). Treatment with etoposide induced phosphorylation of RPA2 in the cells expressing wt-RPA2, however; no phosphorylation was detected in the cells expressing Ala-RPA2 (Fig. 2B). DNA damage caused the assembly of NBS1 and RPA to intranuclear foci and phosphorylation of H2AX (Fig. 2C and E). In the cells expressing wt-RPA2, NBS1 and RPA foci co-localized to a minor extent at 6 h which decreased by 24 h after etoposide treatment (Fig. 2C, panels D-I and Fig. 2D). Cells expressing Ala-RPA2 contained RPA and MRN foci that co-localized at 6 h and continued to co-localize at 24 h (Fig. 2C, panels M-R). Thus, while the phosphorylation of RPA2 is not required to recruit and assemble components of the MRN complex to sites of DNA damage, it does contribute to co-localization of MRN with RPA and the regulation of this process.
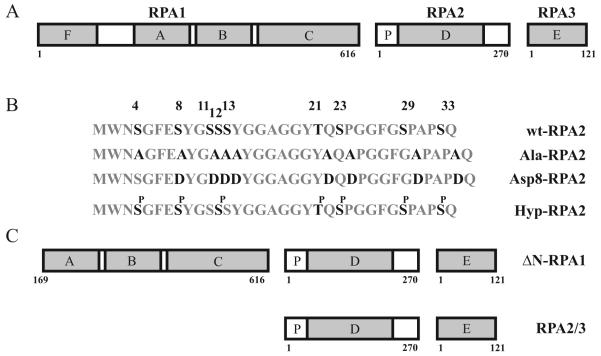
Fig. 1.
Diagram of RPA and RPA mutants used in this study. A, Organization of the RPA heterotrimer. B. Amino acid sequences of the RPA2 N-terminal domain used in this study, including wildtype RPA2 (wt-RPA2), alanine and aspartic acid substituted RPA2 (Ala-RPA2, Asp8-RPA2), as well as the predicted residues affected by hyperphosphorylation (Hyp-RPA2). C, RPA constructs containing either truncated (ΔN-RPA1) or omitted (RPA2/3) RPA1.
Fig. 2.
Phosphorylation of RPA2 restricts interactions between NBS1 and RPA. A, Protein expression levels in UMSCC38 cells stably expressing HA-tagged wt-RPA2 and Ala-RPA2, and shRNA-RPA2 constructs were analyzed by separating cell lysates by SDS-PAGE and immunoblotting with a general anti-RPA2 antibody. B, C and E, UMSCC38 cells stably expressing wt-RPA2 or Ala-RPA2 with downregulation of endogenous RPA2 were not treated (0 h) or treated with etoposide (20 μM for 2 h) followed by 4 h and 22 h recovery. B, Cell lysates were separated by SDS-PAGE and immunoblotted with RPA2 antibodies. C, Fixed cells were stained with anti-HA and anti-NBS1 antibodies. D, Cells were scored for RPA-MRN co-localization to DNA damage foci. Two hundred cells were scored per experiment. Error bars indicate standard errors (n = 3). P values were calculated using an unpaired, two-tailed t test. E, Fixed cells were stained with anti-γ-H2AX antibodies.
DBD-F of RPA interacts with the MRN complex and phosphorylation of RPA2 decreases the interaction
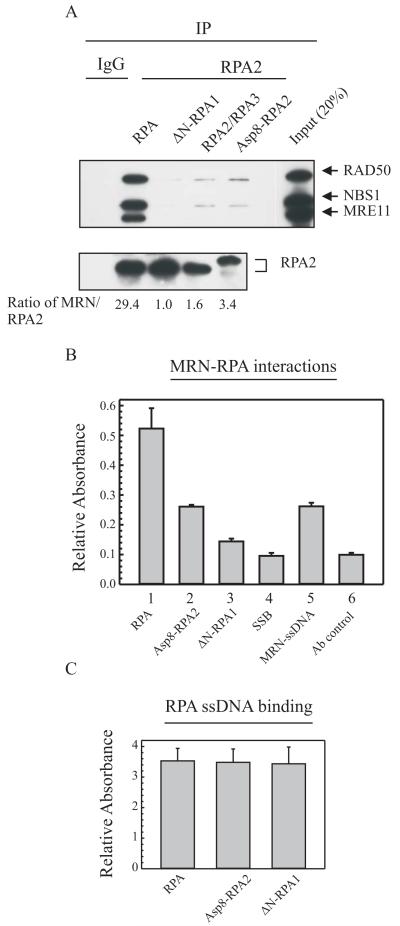
The persistent co-localization of the phospho-mutant RPA with NBS1 suggested that the phosphorylation of wild-type RPA2 modulates RPA-MRN interaction. Therefore, we initially utilized co-immunoprecipitations (co-IPs) and purified proteins to test whether phosphorylation of RPA2 alters the protein interaction with the MRN complex. We performed co-IPs with RPA bound to ssDNA, which more closely resembles RPA at DNA lesion sites (Fig. 3A). In this experiment, we also utilized various mutant RPAs to identify the domains required for the MRN interaction (Fig. 1B and C). The Asp8-RPA2 mutant is a phosphomimetic that mimics hyperphosphorylated RPA containing aspartic acid substitutions at seven serines and one threonine within amino acids 1-40 of the N-terminus of RPA2 and ΔN-RPA1 is a 1-168 N-terminal deletion of RPA1 (24, 33). The RPA2/3 consists of RPA2 and RPA3, respectively, and does not contain RPA1. We observed a 95% decrease in the interaction between the partially purified MRN complex and the RPA2/3 protein complex indicating the primary interaction occurs with RPA1 (Fig. 3A). When compared with the RPA-MRN interaction, both the Asp8-RPA2 and ΔN-RPA1 displayed an 88% and 97%, decrease, respectively, in binding to the MRN complex when bound to ssDNA.
Fig. 3.
RPA interaction with the MRN complex requires the N-terminus of RPA1 and is regulated by RPA2 phosphorylation. A, Various purified RPAs were used to assess the protein-protein interactions. RPAs (2.5 pmol) were incubated with ssDNA (5 pmol) prior to the addition of the MRN (5 pmol) complex expressed and purified from baculovirus preparations. RPA2 antibodies were used to immunoprecipitate wt-RPA and the RPA mutants. The MRN and RPA2 levels were quantitated with NIH image software. The numbers are the ratios of MRN to RPA2 normalized to the ratio for the ΔNRPA1 sample. RPA, wt RPA; ΔN-RPA1, N-terminal deletion of RPA1 (1-168); RPA2/RPA3, RPA without RPA1 and Asp8-RPA2, a phosphomimetic of RPA2. B and C, Biotin-labeled ssDNA bound to streptavidin coated 96 well plates was incubated with wild-type and mutant RPAs. B, Wt, Asp8-RPA2 and ΔN-RPA1 were added to wells coated with biotin-labeled ssDNA bound to streptavidin. The wells were washed to remove unbound RPA and blocked with BSA and biotin. Purified MRN (5 pmol) was subsequently added to the wells and protein interactions were detected using MRE11 and HRP-conjugated antibodies. The SSB control represents the E. coli single-strand binding protein coating the biotin-labeled DNA prior to MRN addition and the MRN-ssDNA sample represents MRN binding without prior RPA addition. C, The amount of purified RPA (2.5-3.0 pmol) bound to ssDNA was detected with RPA2 and HRP-conjugated antibodies and normalized based on absorbance at 610 nm.
During the protein purification of RPA, a small amount of the protein becomes inactive, likely from the stringency of the chromatography conditions, and loses the ability to bind ssDNA. To eliminate the inactive pool of protein and account for possible differences in RPA and mutant RPA ssDNA binding activity in the co-IP experiments, we quantified the effects of RPA2 phosphorylation and the N-terminus of RPA1 on RPA-MRN interactions using ELISAs with streptavidin (SA) bound plates and biotinylated-ssDNA. In this assay, once active RPA and mutant RPA binds to ssDNA, the inactive protein forms are washed out of the wells. There is a decrease in binding of Asp8-RPA2 to the MRN complex compared with RPA binding to MRN (Fig. 3B, lane 2 compared with lane 1). Also, ΔN-RPA1 binds weakly to MRN when bound to ssDNA and the relative absorbance is only just above the SSB and antibody controls (Fig. 3B, lanes 3, 4 and 6, respectively). In the absence of RPA, we detect MRN binding to ssDNA (Fig. 3B, lane 5). As a control, we used E. coli SSB to bind and block the MRN complex from binding ssDNA (Fig. 3B, lane 4). Due to possible differences in RPA and mutant RPA ssDNA binding activities, the amount of active RPA proteins was determined for each protein and normalized to ensure equal amounts of RPA, Asp8-RPA2 and ΔN-RPA1 were bound to ssDNA (Fig. 3C). The control reactions were performed using kinetic as well as endpoint analysis to ensure equal protein binding. These data support a role for the phosphorylation of RPA2 in modulating the RPA-MRN interaction when RPA is bound to ssDNA and also indicates that the N-terminus of RPA1 plays a critical role in the interaction with the MRN complex.
MRE11 and NBS1 are sufficient for RPA-MRN interactions
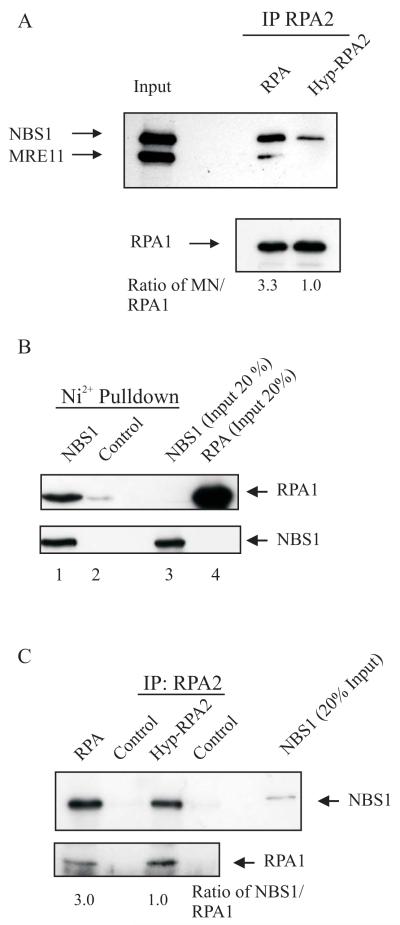
We have previously demonstrated the capability to produce and purify biologically relevant hyperphosphorylated RPA by incubating partially purified recombinant RPA with a HeLa extract in the presence of phosphatase inhibitors. We identified the sites phosphorylated using mass spectrometry and phospho-specific antibodies, and verified that the same RPA2 sites phosphorylated by HU- and UV-induced damage were phosphorylated with our HeLa extract method of phosphorylation (26). Having shown that RPA interacts with MRN, we wanted to further assess the interaction between RPA and components of the MRN complex. Here, we immunoprecipitated purified recombinant unphosphorylated and hyperphosphorylated RPA using an antibody directed against RPA2 to pull down the purified MRE11/NBS1 (MN) complex (Fig. 4A). Both unphosphorylated RPA and hyperphosphorylated RPA interacted with MN; however, hyperphosphorylation of RPA2 decreased binding to MN by 70% (Fig. 4A, RPA compared to HypRPA).
Fig. 4.
RPA binds to MRE11/NBS1 and to NBS1. A, Purified RPA and hyperphosphorylated RPA (HypRPA) (2.5 pmol) were incubated with MRE11/NBS1 (5 pmol) followed by addition of RPA2 antibodies and protein G-agarose beads. Proteins bound to the beads were eluted and separated by SDS-PAGE, and immunoblotted with RPA1, NBS1 and MRE11 antibodies. The numbers are the ratios of MN to RPA1 and normalized to the ratio for the HypRPA sample. B, His-NBS1 (5 pmol) was incubated with RPA (2.5 pmol), and then pulled down with Ni2+ matrix. Proteins bound to the beads were eluted and separated by SDS-PAGE, and immunoblotted with RPA1 and NBS1 antibodies. C, Purified RPA (2.5 pmol) and HypRPA (2.5 pmol) were incubated with purified NBS1 (5 pmol). Immunoprecipitated proteins were separated by SDS-PAGE followed by immunoblotting with NBS1 and RPA1 antibodies. The numbers are the ratios of NBS1 to RPA1 and normalized to the ratio for the HypRPA sample.
RPA has previously been shown to interact with MRE11 (17). Our co-IP results with the MN complex suggest RPA has a higher binding affinity for NBS1 than MRE11. To further investigate the RPA-NBS1 interaction, we tested whether RPA binds directly to NBS1. We utilized nickel pulldowns (NBS1 is His-tagged) to assess the NBS1 interaction with RPA. NBS1 was allowed to interact with RPA and then proteins were bound to nickel matrix. Using the nickel pull-down, RPA interacted with NBS1 (Fig. 4B, lane 1). In contrast, RPA alone interacted minimally with the nickel matrix (Fig. 4B, lane 2). To assess the effect of RPA phosphorylation on RPA-NBS1 interactions, RPA and hyperphosphorylated RPA interactions with NBS1 were examined. Purified RPA did interact with NBS1 and phosphorylation of RPA2 decreased this interaction 67% (Fig. 4C). Overall, these data support an interaction between RPA and NBS1 and are consistent with our previous results that show phosphorylation of RPA2 decreases interaction between RPA and the MRN complex.
RPA bound to ssDNA interacts with NBS1 and MRE11
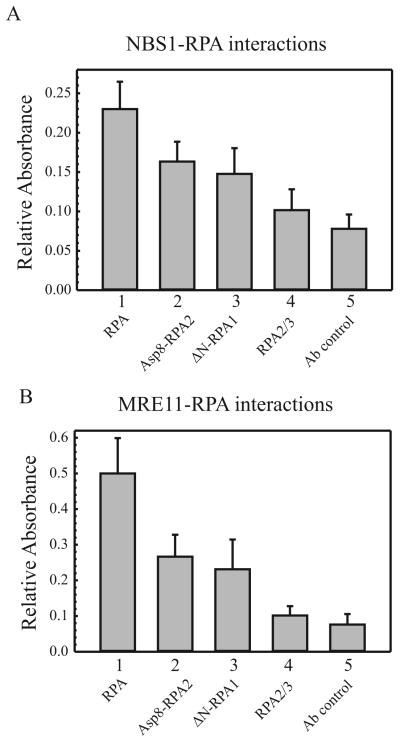
RPA binds to both MRE11 and NBS1, in the absence of ssDNA possibly at multiple OB binding surfaces on RPA. To determine whether ssDNA competes for these binding surfaces, we performed ELISAs using purified MRE11 and NBS1 with RPA and RPA mutants (Fig. 5). We were unable to detect an interaction between RPA2/3 and NBS1 or MRE11 as indicated by the lack of signal above the Ab control (Fig. 5A and B, compare lanes 4 and lane 5). This indicates that the RPA1 subunit is critical for the interaction with both NBS1 and MRE11. Due to comparable binding properties of Hyp-RPA2 and Asp8-RPA2 to the MRN complex and ssDNA, we used Asp8-RPA2 for the protein interactions with MRE11 and NBS1. For both NBS1 and MRE11, we observed a decreased interaction with Asp8-RPA2 and ΔN-RPA1 compared with RPA in the presence of ssDNA (Fig. 5A and B, lanes 1-3) consistent with the RPA-MRN binding results. This data supports a role for RPA-NBS1 and RPA-MRE11 interactions when RPA is bound to ssDNA.
Fig. 5.
RPA interacts with both MRE11 and NBS1. A and B, a modified ELISA was performed as described under Material and Methods. Purified RPA, Asp8-RPA2, ΔN-RPA1 and RPA2/3 (2.5 pmol) were pre-incubated with ssDNA (2.5 pmol) before addition to the 96 well plates coated with purified (A) NBS1 or (B) MRE11 (5 pmol/well). Primary antibodies directed against RPA2 were added followed by the addition of HRP-conjugated secondary antibodies. The absorbance at 610 nm was measured over a period of 20 min. The relative absorbance values represent the affinity of the RPA forms for NBS1 and MRE11.
MRN interaction with RPA requires the basic cleft of the N-terminus of RPA1. The N-terminus of RPA70 is important in the cellular response to DNA damage
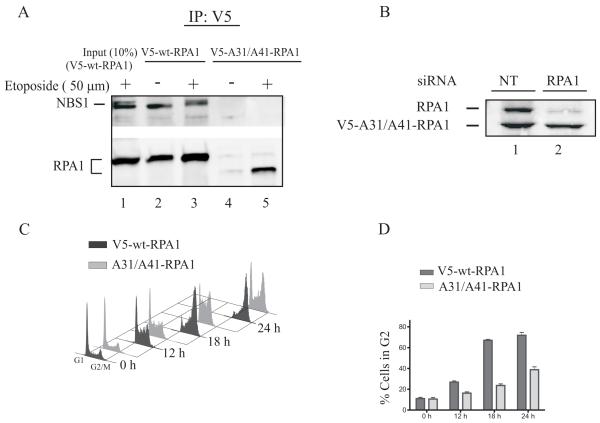
Our data indicate the N-terminus of RPA1 is necessary for binding to the MRN complex when RPA is bound to ssDNA. Therefore, we hypothesized that mutating basic residues in the cleft of the N-terminal domain would disrupt the interaction between RPA and the MRN complex. To test this hypothesis, an RPA1 mutant was generated with alanine substitutions for residues Arg31 and Arg41 which are located in the basic cleft of the N-terminal domain. V5-wt-RPA1 and V5-Ala31/Ala41-RPA1 were expressed in UMSCC38 cells. Immunoprecipitation of V5-wt-RPA1 protein complexes from chromatin fractions indicated they contained endogenous NBS1 (Fig. 6A, lanes 2 and 3). After etoposide treatment, an increase in the wild-type form of RPA1 was immunoprecipitated from chromatin while the mutant form only associated with chromatin after etoposide treatment (Fig. 6A, lanes 3 and 5). The increase in V5-wt-RPA1 immunoprecipitated from chromatin was not accompanied by an increased amount of endogenous NBS1 associated with V5-wt-RPA1 (Fig. 6A, lane 3). It is worthwhile to note that these IP reactions were performed immediately after the 2 h etoposide incubation without any recovery time. In contrast to V5-wt-RPA1, immunoprecipitated V5-Ala31/Ala41-RPA1 from etoposide treated cells did not contain detectable endogenous NBS1 (Fig. 6A, lane 5). These results confirm that the conserved basic cleft in the N-terminus of RPA1 is required for stable RPA-MRN interactions.
Fig. 6.
The basic cleft of the N-terminal domain of RPA1 is required for RPA-MRN interaction. Cells stably expressing V5-RPA1 or V5-A31A41-RPA1 were not treated or treated with etoposide (50 μM for 2 h). Chromatin lysates were prepared and incubated with anti-V5 agarose beads. In A, the immunoprecipitated proteins were separated by SDS-PAGE, followed by immunoblotting with antibodies to NBS1 and RPA1. B, Cells are treated with RPA1-siRNA or non-targeting siRNA (NT) for 48 h. Cell lysates were separated by SDS-PAGE and immunoblotted with RPA1 antibodies. C, UMSCC38 cells and cells expressing V5-wt - and V5-Ala31/Ala41-RPA1 were treated with RPA1-siRNA or non-targeting siRNA (NT) for 48 h. Cells were then exposed to 50 μM etoposide for 2 h or not treated (0 h) followed by 10, 16, and 22 h recovery. DNA content was measured by flow cytometry. The results are representative of two independent experiments. In D, the quantification of the percent of cells in G2 is presented.
To test the importance of the N-terminus of RPA1 in the DDR, cells were treated with 50 μM etoposide for 2 h and the drug was washed out. RPA1 siRNA effectively downregulated endogenous RPA1 without affecting the resistant exogenous form (Fig. 6B). Progression through the cell cycle was detected by propidium iodide staining of DNA content and analyzed by flow cytometry. The cells expressing V5-wt-RPA1 arrested at G2 by 24 h consistent with the parent UMSCC38 cells. In contrast, the V5-Ala 31/Ala 41-RPA1 mutants accumulated more in S-phase and less in G2 (Fig. 6C and D). These data provide evidence that the N-terminal domain of RPA1 functions by interacting with multiple proteins, including the MRN complex, in the cellular response to replication stress.
Discussion
When cells are treated with such diverse agents as etoposide, cisplatin or UV, it has been assumed the unique DNA adducts that form are responsible for the activation of the DDR pathway. However, recent work has suggested that these DNA adducts are converted into a common structure that involves uncoupling of the replication helicase and polymerases, generating long stretches of ssDNA (8, 34-36). Consistent with this idea, a primed ssDNA substrate with 5′- and 3′-junctions and ssDNA gaps mimicked a stalled replication fork and activated ATR checkpoint signaling in Xenopus extracts (37). A single free 5′-junction on a primed template was shown to induce ATR-dependent phosphorylation of Chk1 and Rad1 in Xenopus extracts (37). This is of interest because RPA binds ssDNA with a defined polarity (38, 39). The orientation of an RPA trimer is in a 5′-3′ direction with the N-terminus of RPA1 at the 5′-end and the C-terminus of RPA1 as well as RPA2 at the 3′ -end. This would place the N-terminus of RPA1 at the 5′-ds/ss junction. Many important proteins bind to the N-terminus of RPA1, including Rad9, part of DNA damage-activated Rad9-Hus1-Rad1 clamp, and the ATR interacting partner, ATRIP (18, 22). A recent analysis showed that cells expressing an RPA1 R41E/R43E mutant caused a defect in CHK1 phosphorylation following UV radiation (18). Consistent with this analysis, our data show cells expressing the RPA1 basic cleft mutant, A31A41-RPA1, do not accumulate exclusively in G2/M following etoposide treatment.
The N-terminus of RPA1 is attached to the rest of RPA1 through a flexible linker (40). This flexibility may be important to allow RPA to recruit different DNA damage response proteins and the DNA binding polarity of RPA would ensure these proteins are directed to the lesion site for further activation of the DDR. Moreover, our data suggest that proteins that bind to the N-terminus of RPA1 are potentially assembled or disassembled from protein complexes by DNA damage-induced phosphorylation of RPA2; this implicates both the N-terminus of RPA1 and RPA2 in directing and controlling protein interactions at the site of the lesion. If the N-terminus of RPA1 binds ssDNA and also functions as a protein interaction domain, what role would RPA2 phosphorylation have in modulating these functions? One intriguing possibility is that the phosphorylation of RPA2 competes with proteins bound to RPA1 thus displacing them from RPA1. This is consistent with the interaction that was observed between a phosphomimetic RPA2 and the N-terminus of RPA1 (21, 24). For example, our data demonstrates that the N-terminus of RPA1 is required for NBS1 and MRE11 binding and phosphorylation of RPA2 decreases these protein interactions. We propose that before RPA2 is phosphorylated, RPA interacts with NBS1 and MRE11 during unperturbed DNA replication and then, following DNA damage-induced phosphorylation of RPA2, RPA2 displaces NBS1 and MRE11 from RPA1. This would allow the MRN complex to act with additional factors to assist in stabilization of the lesion or relocate to other sites in the genome. In Xenopus laevis egg extracts, the MRN complex relocates to restarting replication forks to promote recovery of replication following DNA damage (13). In summary, our data suggest that the molecular lever like action of RPA2 interacting with the N-terminus of RPA1 allows RPA to orchestrate multiple interactions at sites of DNA damage in a dynamic process that stabilizes the site of the lesion and assists in its repair.
Supplementary Material
Acknowledgment
We would like to thank Anbarasi Kothandapani, Sanjeevani Arora and Venkata Srinivas Mohan Nimai Dangeti for critical reading of the manuscript.
This study has been supported by research grants from the American Cancer Society (ACS, RSG-06-163-01-GMC) and the Ohio Cancer Research Associates, Inc. to S.M.P, National Institutes of Health Grant P20-RR018759 and the Department of Human and Health Services of Nebraska to G.G.O.
Abbreviations and Textual Footnotes
- Ala-RPA2
mutant RPA2 with alanine substitutions at serine and threonine phosphorylation sites located within amino acids 1-33 of RPA2
- Asp8-RPA2
mutant RPA2 with aspartic acid substitutions at serine and threonine phosphorylation sites located within amino acids 1-33 of RPA2
- ATR
ataxia telangiectasia-related
- ATRIP
ATR interacting protein
- bp
base pairs
- co-IPs
co-immunoprecipitations
- DBD
DNA Binding Domains
- DDR
DNA damage response
- DSBs
double strand breaks
- ELISAs
enzyme linked immunosorbent assays
- HU
hydroxyurea
- MRN
MRE11, RAD50 and NBS1
- ΔN-RPA1
mutant RPA1 containing a deletion of amino acids 1-168
- NER
nucleotide excision repair
- OB
oligosaccharide/oligonucleotide binding
- RPA
Replication protein A
- SA
streptavidin
- SSB
single-strand DNA binding protein
- ssDNA
single-stranded DNA
- UV
ultraviolet
References
- 1.Wobbe CR, Weissbach L, Borowiec JA, Dean FB, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA in vitro with purified proteins. Proc.Natl.Acad.Sci.U.S.A. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramilo C, Gu L, Guo S, Zhang X, Patrick SM, Turchi JJ, Li GM. Partial reconstitution of human DNA mismatch repair in vitro: characterization of the role of human replication protein A. Mol Cell Biol. 2002;22:2037–2046. doi: 10.1128/MCB.22.7.2037-2046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park MS, Ludwig DL, Stigger E, Lee SH. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J.Biol Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 4.Golub EI, Gupta RC, Haaf T, Wold MS, Radding CM. Interaction of human rad51 recombination protein with single- stranded DNA binding protein, RPA. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrault R, Cheong N, Wang H, Wang H, Iliakis G. RPA facilitates rejoining of DNA double-strand breaks in an in vitro assay utilizing genomic DNA as substrate. Int J Radiat Biol. 2001;77:593–607. doi: 10.1080/09553000110036773. [DOI] [PubMed] [Google Scholar]
- 6.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 7.Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science. 2000;289:2133–2137. doi: 10.1126/science.289.5487.2133. [DOI] [PubMed] [Google Scholar]
- 8.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 10.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 12.Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol.Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 13.Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. Embo J. 2006;25:1764–1774. doi: 10.1038/sj.emboj.7601045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 15.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM Activation and Its Recruitment to Damaged DNA Require Binding to the C Terminus of Nbs1. Mol.Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 Nuclease Activity Has Essential Roles in DNA Repair and Genomic Stability Distinct from ATM Activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J.Biol.Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- 18.Xu X, Vaithiyalingam S, Glick GG, Mordes DA, Chazin WJ, Cortez D. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol Cell Biol. 2008;28:7345–7353. doi: 10.1128/MCB.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol. 2007;27:6053–6067. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu.Rev.Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 21.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, Bochkarev A. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 24.Binz SK, Lao Y, Lowry DF, Wold MS. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J.Biol.Chem. 2003;278:35584–35591. doi: 10.1074/jbc.M305388200. [DOI] [PubMed] [Google Scholar]
- 25.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J.Biol Chem. 1994;269:11121–11132. published erratum appears in J Biol Chem 1994 Jun 10;269(23):16519. [PubMed] [Google Scholar]
- 26.Patrick SM, Oakley GG, Dixon K, Turchi JJ. DNA damage induced hyperphosphorylation of replication protein A. 2 Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry. 2005;44:8438–8448. doi: 10.1021/bi048057b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JH, Paull TT. Purification and biochemical characterization of ataxia-telangiectasia mutated and Mre11/Rad50/Nbs1. Methods Enzymol. 2006;408:529–539. doi: 10.1016/S0076-6879(06)08033-5. [DOI] [PubMed] [Google Scholar]
- 28.Wozniak AJ, Ross WE. DNA damage as a basis for 4′-demethylepipodophyllotoxin-9-(4,6-O-ethylidene-beta-D-glucopyranoside) (etoposide) cytotoxicity. Cancer Res. 1983;43:120–124. [PubMed] [Google Scholar]
- 29.Bromberg KD, Burgin AB, Osheroff N. A two-drug model for etoposide action against human topoisomerase IIalpha. J Biol Chem. 2003;278:7406–7412. doi: 10.1074/jbc.M212056200. [DOI] [PubMed] [Google Scholar]
- 30.Long BH, Musial ST, Brattain MG. Single- and double-strand DNA breakage and repair in human lung adenocarcinoma cells exposed to etoposide and teniposide. Cancer Res. 1985;45:3106–3112. [PubMed] [Google Scholar]
- 31.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robison JG, Lu L, Dixon K, Bissler JJ. DNA lesion-specific co-localization of the Mre11/Rad50/Nbs1 (MRN) complex and replication protein A (RPA) to repair foci. J Biol Chem. 2005;280:12927–12934. doi: 10.1074/jbc.M414391200. [DOI] [PubMed] [Google Scholar]
- 33.Gomes XV, Wold MS. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 34.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Stokes MP, Van Hatten R, Lindsay HD, Michael WM. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J Cell Biol. 2002;158:863–872. doi: 10.1083/jcb.200204127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 37.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 39.de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG, Hoeijmakers JH. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev. 1998;12:2598–2609. doi: 10.1101/gad.12.16.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs DM, Lipton AS, Isern NG, Daughdrill GW, Lowry DF, Gomes X, Wold MS. Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J Biomol NMR. 1999;14:321–331. doi: 10.1023/a:1008373009786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.