Abstract
Context
The incidence of recurrent miscarriage (RM; ≥3 consecutive pregnancy losses) is estimated as 1%-2% in fertile couples. Familial clustering of RM has suggested the contribution of a genetic component.
Objective
Low level of HCG in maternal serum during the first trimester of the pregnancy is a clinically accepted risk factor for miscarriage. We sought to study whether variation in Chorionic Gonadotropin beta subunit genes (CGB) expressed in placenta may contribute to the risk of RM.
Design
Resequencing of CGB5 and CGB8, the two most actively transcribed loci of the four HCG beta duplicate genes.
Setting
A case-control study involving two sample sets, from Estonia (n=194) and from Finland (n=185).
Patients
RM patients (n=184) and fertile controls (n=195).
Results
From 71 identified variants in CGB5 and CGB8, 48 SNPs were novel. Significant protective effect was associated with two SNPs located at identical positions in intron 2 in both CGB5 (p=0.007, OR=0.53) and CGB8 (p=0.042, OR=0.15); and with four CGB5 promoter variants (p<0.03; OR=0.54-0.58). The carriers of minor alleles had reduced risk of RM. The haplotype structure of the CGB8 promoter was consistent with balancing selection; a rare mutation in CGB8 initiator element was detected only among patients (n=3). In addition, three rare non-synonymous substitutions were identified among RM cases as possible variants increasing the risk of recurrent pregnancy loss.
Conclusions
The findings encourage studying the functional effect of the identified variants on CGB expression and HCG hormone activity to further elucidate the role of CGB variation in RM.
Keywords: recurrent miscarriage, HCG beta genes, resequencing, association study
Introduction
Recurrent miscarriage (RM) or habitual abortion is defined as three or more consecutive pregnancy losses before 22 gestational weeks or the spontaneous abortion of an embryo/fetus weighing less than 500g. The occurrence of RM is estimated as 1%-2% of fertile couples (1, 2). Although the patients with RM undergo multiple diagnostic tests to detect parental chromosomal anomalies, maternal thrombophilic, endocrine, or immunological disorders, over 50% of the RM cases are classified as idiopathic (3). An increased prevalence of miscarriage among first-degree relatives of the women suffering from RM (4) suggests genetic contribution in recurrent pregnancy loss. Possible candidates include genes regulating the development of maternal immunotolerance and inflammatory response, coagulation, angiogenesis, vascular tone, and apoptosis. Prime candidates of the molecular causes of RM have been various trombophilic gene mutations (5-7). Convincing data has also been reported on the association between the miscarriage rates and the polymorphisms in HLA-G gene expressed on the surface of the invading cytotrophoblasts (8).
So far, major interest has focused on the physiological response of the mother to the pregnancy. Less attention has been paid to the placental proteins coded by the fetal genome with contribution from both maternal and paternal genes and their variants. One of the first proteins produced by the conceptus is human chorionic gonadotropin (HCG), also known as “the pregnancy hormone” due to its essential role in human reproduction. The main function of HCG is to delay the apoptosis of the corpus luteum during the first trimester of pregnancy. HCG has several paracrine effects in the process of implantation (9), angiogenesis and placentation (10, 11), and development of maternal immunotolerance (12). Low level and non-exponential increase of HCG in maternal serum during the first trimester of the pregnancy is a clinically accepted risk factor for miscarriage (13-15).
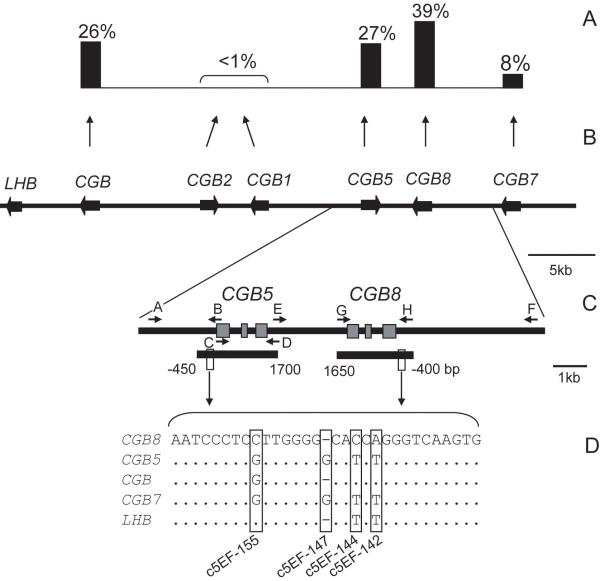
The hormone-specific HCG beta-subunit is expressed by syncytiotrophoblasts of placenta and is encoded by four Chorionic Gonadotropin Beta genes (CGB, CGB5, CGB7 and CGB8) located within the LHB/CGB gene cluster at 19q13.3 (Fig. 1B). Among the four HCG beta duplicate genes CGB8 and CGB5 are the most actively transcribed and contribute together 62-82% to the total pool of beta-subunit mRNA transcripts (Fig.1A, 16-18). Our previous data on the HCG beta genes showed that (i) their diversity level is one of the highest reported for human genes; (ii) there is high interindividual and intergenic difference in expression and (iii) mRNA transcription level is significantly lower in cases of RM compared to normal first trimester pregnancies (18-20). Now we have addressed the question whether particular variants in these genes may contribute to pregnancy failure. High genetic variation in the LHB/CGB region and the aim to capture both rare and common variation prompted us to choose resequencing instead of traditional genotyping. We analyzed CGB5 and CGB8 in Estonian and Finnish RM cases (n=184) and fertile women (n=195) by comparing variation and haplotype patterns between the two groups. Consistent with hypothesis of the study, we identified genetic variants in HCG beta genes either significantly increasing or reducing a subject’s risk to experience recurrent pregnancy loss.
Figure 1.
Genomic and expressional context for the design of the association study targeting HCG beta genes. (A) The contribution of each individual gene into the total mRNA transcript pool of all six CGB genes (18). (B) Schematic presentation of the LHB/CGB gene cluster with genes marked as black wide arrows in the direction of transcription on sense strand. (C) The position of long-range PCR primers (black arrows) and extent of resequenced CGB5 and CGB8 regions (short black bars). Gene exons are depicted with grey boxes. Capital letters correspond to the primer sequences listed in Supplementary Table S1. (D) The aligned consensus sequences of the 5′upstream element of LHB/CGB genes. The nucleotide positions distinctive for each HCG beta and LHB gene co-localizing with CGB5 SNPs c5EF-155, c5EF-147, c5EF-144 and c5EF-142 (Table 1) are highlighted.
All positions are given relative to mRNA transcription start site.
Subjects and Methods
Study subjects
The study was approved by the Ethics Committees of the University of Tartu, Estonia (protocols no 117/9, 16.06.03, 126/14, 26.04.2004) and the Department of Obstetrics and Gynecology, Helsinki University Central Hospital Outpatient Clinic for women with recurrent miscarriage (protocol no 298/E2/2000). Subjects were recruited and blood samples for the DNA extraction were collected at the Women’s Clinic of Tartu University Hospital and Nova Vita Clinic, Centre for Infertility Treatment and Medical Genetics, Tallinn, Estonia in 2003-2007; and in the Department of Gynaecology and Obstetrics of the Helsinki University Hospital in Finland during 2001-2004. Written informed consent was obtained from every study participant. In both participating centers patients with at least ≥3 abortions during the first trimester of pregnancy were recruited (n=184; age 18-40 yrs). As maternally and paternally derived gene variants contribute equally to the function of a fetal genome, the patient group included both, the women and their partners, who had experienced recurrent pregnancy losses. In Estonian sample collection the patient group consisted of 32 couples and 29 females with RM, and additional 3 couples with ≥3 unsuccessful in vitro fertilization procedures. In Finnish sample collection the RM group consisted of 40 couples and 5 females with RM (detailed description in 21, 22). The control group (n=195) consisted of age-matched fertile women with no history of miscarriage and either at least one normal pregnancy (the Finnish subjects, n=100) or more stringently, ≥3 successful deliveries (the Estonian subjects, n= 95). The control group was designed under the assumption that fertile women with no history of spontaneous abortions are carrying gene variants supporting successful pregnancies. Their male partners were not recruited into the control group as detailed reliable information on their past reproductive history was unavailable.
All patients had a normal karyotype tested from peripheral blood lymphocyte cultures. Female patients having uterine anomalies were excluded by ultrasonography or hysterosonogram.
Amplification and resequencing of CGB5 and CGB8
DNA was extracted from peripheral blood using a protocol based on the salting-out method for DNA extraction. The CGB5 (~ 1.7 kb fragment) and CGB8 (long-range PCR ~ 8.3 kb; nested PCR ~2.5 kb fragment) genomic regions (Fig. 1C) were amplified and resequenced using previously described primers and conditions (19). The resequenced region involving CGB8 covered 2050 bp including the entire CGB8 (1474 bp), 400 bp of 5′ upstream region. The resequenced region for CGB5 (1468 bp) covered the full genic region and part of 3′downstream region (Fig. 1C). Additional primers were designed for the analysis of the 5′upstream region of the CGB5 gene (450 bp) using the Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Specificity of the PCR products was verified in three steps: (1) design of unique primer pairs for specific amplification of only one of the seven duplicated genes; (2) verification of monomorphic status of gene-specific positions used as markers for each individual gene (Supplementary Fig. S1); (3) test for Hardy-Weinberg Equilibrium (HWE) for each identified SNP. Primer sequences for PCR and resequencing are listed in Supplementary Table S1. The sequences were resolved using either ABI 3730 X1 or ABI 3730 XL DNA Analyzer (Applied Biosystems) and assembled into a contig as described (19). Polymorphisms were identified using the PolyPhred program (Version 6.02) (http://www.phrap.org/phredphrapconsed.html) (23) and confirmed by manual checking. A genetic variant was verified only if it was observed in both forward and reverse orientations. In case of indel heterozygosity the genotype of the subject was confirmed using two independent forward and two reverse primers. The nomenclature of the identified polymorphisms was based on the GenBank reference sequences: NM_033043.1 GI:15451747 for CGB5, NM_033183.2 GI:146229337 for CGB8.
Data analysis
Allele frequencies were estimated and conformance to HWE was calculated (α = 0.05). In total 8 rare SNPs in 5′upstream region of CGB5 were found to be deviating from HWE, as one individual was homozygous for minor allele of all these SNPs.
Haplotypes were inferred from unphased genotype data using the Bayesian statistical method in the program PHASE 2.1.1 (http://www.stat.washington.edu/stephens/; 24), applying the model allowing recombination. The running parameters were: number of iterations = 1000, thinning interval = 1, burn-in = 100; the - X10 parameter was used for increasing the number of iterations of the final run of the algorithm.
Sequence diversity parameters and neutrality tests were calculated using DnaSP (ver. 4.0; http://www.ub.es/dnasp/; 25) with the most probable phased haplotypes as an input sequence. The direct estimate of per-site heterozygosity (π) was derived from the average pairwise sequence differences, while Watterson’s θ represents an estimate of the expected per-site heterozygosity based on the number of segregating sites (S). The basis of the Tajima’s D statistic (26) is the difference between the π and θ estimates: under neutral conditions π = θ and DT = 0. The Ewens-Watterson homozygosity test implemented in Arlequin 2.000 software (http://cmpg.unibe.ch/software/arlequin3/; 27) was used to test the hypothesis that haplotypes are selectively neutral. An excess of rare variants (= homozygosity excess) indicates directional selection, while an excess of intermediate frequency variants (= homozygosity deficiency) indicates balancing selection. The relationship between inferred haplotypes was analyzed with NETWORK 4.201 software (http://www.fluxus-technology.com) using the Median-Joining network algorithm (28). Haplotype networks of CGB5 and CGB8 were calculated using (i) SNPs located in genic region from the transcription initiation site until the end of the mRNA and (ii) promoter SNPs located 5′upstream of the genic region. Singleton polymorphisms were excluded from network calculations (cannot be reliably phased) performed with default parameters. The descriptive statistics of linkage disequilibrium (LD), r2 was calculated for pairs of markers and summarized by Haploview software (29).
The significance of the association between the identified SNPs in CGB5 and CGB8 genes and occurrence of RM was tested using Cochran-Armitage test for trend implemented in statistical analysis package JMP® 6.0.3 with Genomics module 2.0.6 (http://www.jmp.com/software/genomics/). The same test was applied to address the interpopulation (Estonians, Finns) differentiation. Odds ratio (OR) with 95% confidence intervals (CI) were calculated to show the strength and direction of the association. In all tests, p<0.05 was considered statistically significant.
Results
Resequencing of CGB5 and CGB8
We sequenced the entire genic and 5′upstream regions of CGB5 and CGB8 genes in a sample collection consisting of Finnish and Estonian patients with recurrent miscarriage (RM) (n=184; n=85 Finns, n=99 Estonians) and fertile controls (n=195; n=100 Finns, n=95 Estonians). For every subject the entire sequenced region covered 4.3 kbp (Fig. 1B-C). In total 71 variants were identified: 29 and 19 SNPs in the genic part of CGB5 and CGB8, respectively; 18 and 3 SNPs in the 5′ upstream regions of CGB5 and CGB8, respectively; and 2 SNPs 3′downstream of CGB5 (Table 1). Among the 71 detected SNPs 48 (68%) were novel variants, previously not described in dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and literature. Neither CGB5 nor CGB8 has been covered by the most recent version of HAPMAP (http://www.hapmap.org/; release March 2008). The diversity parameter π that describes the mean nucleotide diversity per bp differed in the genic and 5′ upstream regions (Table 2). Among fertile women the diversity of CGB5 (π=2.71×10-3) and CGB8 (~π=2.01×10-3) 5′ upstream regions was approximately twofold higher compared to the genic regions of CGB5 (~π=1.69×10-3) and CGB8 (~π=9×10-4) (Table 2).
TABLE 1.
Characteristic of SNPs identified in CGB5 and CGB8 in Estonian and Finnish sample sets
| SNP codea | Position relative to ATG | Location | Alleleb major/minor Aminoacid changec | Minor allele frequency in a subsample (%) | population difference p-valued |
rs numbere | |
|---|---|---|---|---|---|---|---|
| Estonian (n=194) | Finnish (n = 185) | ||||||
| Variants in CGB5 genomic region | |||||||
| c5F-447 | -812 | 5′ up-stream | T/G | 0 | S (Co) | 0.304 | |
| c5F-399 | -764 | T/C | 0 | 0.82 | 0.167 | ||
| c5EF-322 | -687 | T/C | 0.52 | 0.82 | 0.667 | ||
| c5EF-315 | -680 | T/G | S (Pa) | 1.9 | 0.071 | ||
| c5EF-314 | -679 | C/A | S (Pa) | 1.09 | 0.234 | ||
| c5EF-309 | -674 | C/T | S (Pa) | 1.09 | 0.234 | ||
| c5EF-306 | -671 | T/G/C | S (Pa) | 2.45 | 0.037 | ||
| c5EF-291 | -656 | C/T | 21.65 | 17.66 | 0.162 | rs4801789 | |
| c5F-204 | -569 | A/G | 0 | 1.36 | 0.051 | ||
| c5F-191 | -557 | T/C | 0 | 1.36 | 0.051 | ||
| c5EF-155 | -520 | G/C | 10.57 | 9.24 | 0.543 | ||
| c5EF-147 | -512 | G/del | 10.57 | 8.7 | 0.373 | ||
| c5EF-144 | -509 | T/C | 10.57 | 10.6 | 0.989 | ||
| c5EF-142 | -507 | T/A | 10.57 | 10.6 | 0.989 | ||
| c5EF-82 | -447 | G/A | 1.8 | 1.63 | 0.853 | ||
| c5EF-30 | -395 | G/C | 0.77 | 1.36 | 0.429 | ||
| c5E-28 | -393 | C/T | 1.55 | 0 | 0.016 | ||
| c5EF-1 | -366 | A/G | S (Co) | S (Co) | 0.973 | ||
| c5E101 | -265 | 5′UTR | C/T | 0.52 | 0 | 0.166 | |
| c5EF138 | -228 | A/G | 12.11 | 6.76 | 0.011 | rs710899 | |
| c5E157 | -209 | C/T | 0.52(Pa) | 0 | 0.166 | ||
| c5F206 | -160 | C/T | 0 | S (Co) | 0.305 | ||
| c5F324 | -42 | G/A | 0 | 0.54 (Co) | 0.146 | ||
| c5EF345 | -21 | G/C | 22.16 | 16.76 | 0.055 | rs12610392 | |
| c5F354 | -12 | G/del | 0 | 1.08 | 0.040 | ||
| c5E519 | 154 | intron1 | G/T | 0.52 | 0 | 0.166 | |
| c5E525 | 160 | A/G | 0.52 | 0 | 0.166 | rs35621293 | |
| c5E527 | 162 | G/A | 0.52 | 0 | 0.166 | ||
| c5E529 | 164 | G/A | S (Pa) | 0 | 0.328 | ||
| c5EF544 | 179 | T/G | 33.25 | 25.68 | 0.024 | rs3956245 | |
| c5E551 | 186 | C/T | 0.77 | 0 | 0.089 | rs4002422 | |
| c5F553 | 188 | T/C | 0 | 0.54 (Co) | 0.146 | ||
| c5EF580 | 215 | G/A | 21.65 | 17.03 | 0.104 | rs34524624 | |
| c5F660 | 295 | A/C | 0 | S (Pa) | 0.305 | ||
| c5EF666 | 301 | C/T | 32.73 | 25.41 | 0.027 | rs35871536 | |
| c5EF789 | 424 | exon2 | G/A p.Pro24Pro |
21.13 | 17.03 | 0.148 | rs35133942 |
| c5E912 | 547 | intron2 | G/C | 0.77 | 0 | 0.089 | |
| c5E918 | 553 | C/G | 0.52 | 0 | 0.166 | rs33933429 | |
| c5EF1038 | 673 | C/T | 11.86 | 10.81 | 0.651 | rs34335161 | |
| c5EF1069 | 704 | A/G | 1.55 | 2.43 | 0.376 | rs33976607 | |
| c5F1111 | 746 | A/T | 0 | 0.54 (Co) | 0.146 | ||
| c5EF1115 | 750 | C/T | 3.35 | 0.54 | 0.005 | rs34935416 | |
| c5F1178 | 813 | exon3 | G/C p.Val76Leu |
0 | S (Pa) | 0.305 | |
| c5EF1258 | 893 | C/T p.Tyr102Tyr |
4.64 | 0.81 (Co) | 0.001 | rs35756580 | |
| c5E1390 | 1025 | C/A p.Pro146Pro |
S (Co) | 0 | 0.328 | ||
| c5EF1402 | 1037 | T/C p.Ser150Ser |
S (Co) | S (Pa) | 0.973 | ||
| c5EF1426 | 1061 | G/A p.Ser158Ser |
S (Co) | S (Pa) | 0.973 | ||
| c5E1501 | 1136 | 3′ down-stream | T/C | S (Pa) | 0 | 0.328 | |
| c5EF1660 | 1295 | A/T | 1.29 | 1.08 | 0.502 | ||
| Variants in CGB8 genomic region | |||||||
| c8EF-287 | -659 | 5′ up-stream | T/C | 26.49 | 23.85 | 0.417 | rs4801790 |
| c8EF-186 | -558 | G/T | 40.21 | 39.08 | 0.754 | rs8102901 | |
| c8EF-4 | -376 | T/A | 0.52 (Pa) | S (Pa) | 0.627 | ||
| c8EF105 | -268 | 5′UTR | G/C | 4.12 | 0.57 (Co) | 0.003 | rs34212754 |
| c8EF108 | -265 | C/T | 39.95 | 39.08 | 0.808 | rs13345685 | |
| c8EF301 | -72 | T/A | 5.41 | 6.32 | 0.597 | rs35930240 | |
| c8EF432 | 60 | intron1 | A/C | 0.52 | 0.57 (Co) | 0.913 | |
| c8F461 | 89 | T/C | 0 | S (Pa) | 0.290 | ||
| c8EF523 | 151 | G/T | S (Co) | 0.86 (Pa) | 0.264 | ||
| c8F526 | 154 | T/G | 0 | S (Co) | 0.290 | rs2387591 | |
| c8EF541 | 169 | G/C | 39.69 | 39.37 | 0.928 | rs13345575 | |
| c8F551 | 179 | G/T | 0 | S (Co) | 0.290 | ||
| c8F558 | 186 | T/C | 0 | S (Co) | 0.290 | ||
| c8EF673 | 301 | T/C | S (Co) | S (Co) | 0.938 | ||
| c8E806 | 434 | exon2 | C/T p.Arg28Trp |
S (Pa) | 0 | 0.343 | |
| c8EF869 | 497 | G/A p.Val49Ile |
2.32 | S (Pa) | 0.017 | ||
| c8EF925 | 553 | intron2 | G/C | 2.06 | 3.16 | 0.341 | rs2303050 |
| c8EF1045 | 673 | C/T | 0.52 | 1.72 (Co) | 0.112 | rs33943298 | |
| c8EF1076 | 704 | G/A | 1.8 | 2.01 | 0.836 | ||
| c8EF1122 | 750 | T/C | 1.8 | 2.01 | 0.836 | ||
| c8E1237 | 865 | exon3 | C/G p.Pro93Arg |
S (Pa) | 0 | 0.343 | |
| c8E1418 | 1046 | A/T pArg153Arg |
S (Pa) | 0 | 0.343 | ||
a SNP code includes gene and sample name (e.g.c5=CGB5; E=Estonians, F=Finns), location relative to mRNA start site; GenBank references: NM_033043.1 GI:15451747 for CGB5, NM_033183.2 GI:146229337 for CGB8; non-synonymous changes detected only in patients are underlined
alleles at the coding strand
coding from ATG including signal protein
Cochran-Armitage test for trend
Variants originally described by Hallast et al., 2005 (19) are highlighted in bold. All variants identified in the current study have been submitted to dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/), accession numbers ss105106983 - ss105107053.
S - singleton SNP; Co - only among fertile women with no miscarriages; Pa - only among RM patients
TABLE 2.
Diversity parameters and neutrality tests of the CGB5 and CGB8 genes in fertile women and patients of recurrent miscarriages (RM)
| CGB5 | CGB8 | |||||
|---|---|---|---|---|---|---|
| Full regiona | 5′upstream regionb | Genic regionc | Full regiona | 5′upstream regionb | Genic regionc | |
| No of SNPs | 46 | 17 | 29 | 23 | 3 | 20 |
| Fertile women | ||||||
| Diversity (π)d | 0.00193 | 0.00271 | 0.00169 | 0.00119 | 0.00201 | 0.00095 |
| Tajima De | -1.12980 | -1.16944 | -0.88132 | -0.35277 | 2.29389 * | -0.96141 |
| P-value of Ewens-Watterson F statisticf | ns | ns | ns | ns | 0.007 | ns |
| RM Patients | ||||||
| Diversity (π)d | 0.00172 | 0.00195 | 0.0016 | 0.00115 | 0.00199 | 0.0009 |
| Tajima De | -1.23744 | -1.23550 | -0.91052 | -0.56306 | 1.26879 | -1.06361 |
| P-value of Ewens-Watterson F statisticf | ns | ns | ns | ns | ns | ns |
SNPs in 5′upstream and genic regions
SNPs located in the region of −435 bp up to −1 bp from the start site of mRNA sequence
SNPs located in mRNA seqence: +1 bp up to +1082 bp from the start site of mRNA sequence
Estimate of nucleotide diversity (π) per basepair from average pairwise differences among individuals
The basis of the Tajima’s D statistics (DT) is the difference between observed (π) and expected (θ) diversity estimates: under neutral conditions π = θ and DT = 0
This statistic tests the observed allele frequency spectrum with the expected allele frequency spectrum under the neutral model (Hardy-Weinberg Equilibrium).
p=0.0169; ns - non-significant (p>0.05)
Two thirds of the identified variants (n=41; 58%) were shared by Estonian and Finnish sample collections. In both sample collections there were 15 population-specific SNPs represented as single or low frequency variants (<2%). Majority of the shared SNPs showed no differences (p>0.05) among the two study populations. Significant difference in allele frequencies was detected for 8 out of 71 SNPs, most being rare variants (Table 1). Linkage disequilibrium (LD) between the identified SNPs in the resequenced region was nearly absent in both population samples (Fig. 2).
Figure 2.
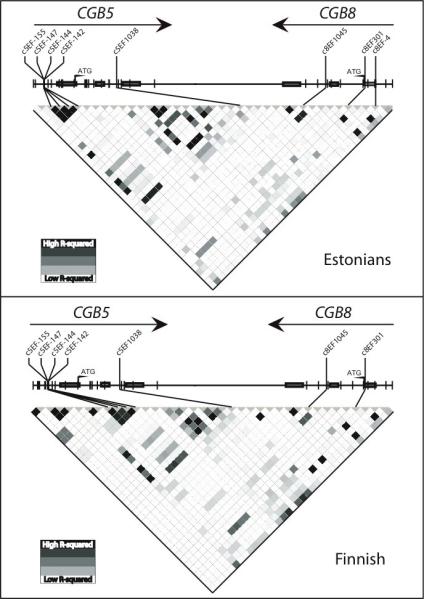
LD structure of the resequenced CGB5-CGB8 genomic region in Estonian and Finnish sample sets. The plot has been drawn based on r2 statistic and polymorphisms are ordered as located on the sense strand of genomic DNA sequence. Singleton variants have been excluded for reliable estimation of LD pattern. The direction of transcription of CGB5 and CGB8 genes is depicted with arrows. Polymorphisms with a significant association to recurrent miscarriages (Table 3) are given above the LD plot.
Four SNPs represented non-synonymous amino acid changes: CGB5 p.Val76Leu in a single Finnish RM patient, CGB8 p.Arg28Trp and CGB8 p.Pro93Arg in single Estonian patients, and CGB8 p.Val49Ile in one Finnish and two Estonian patients, and also seven Estonian fertile women (Table 1). Further experimental studies have to be conducted before drawing any conclusions about their effect on the hormone function.
CGB5 and CGB8 variants lowering the risk for recurrent pregnancy loss
A case-control study targeting the association of identified CGB5 and CGB8 genetic variants with RM was carried out separately for the Estonian (RM cases n=99; fertile women defined as controls n=95) and the Finnish subjects (cases n=85; controls n=100) as well as for the joint dataset. The comparison of single marker and haplotype distribution in the two sample sets revealed low population stratification (Table 1, Supplementary Fig. S2) facilitating the joint analysis in order to increase the statistical power of the study.
In the full case-control sample set a significant association with RM was detected for the CGB5 5′upstream polymorphisms (c5EF-155, c5EF-147, c5EF-144, c5EF-142) (p<0.03; OR=0.58 [95% CI 0.35-0.93]; Cochran-Armitage trend test) (Table 3). Analysis of the Estonian (p=0.083; OR=0.54 [95% CI 0.27-1.1]) and the Finnish (p≤0.131; OR=0.58 [95% CI 0.29-1.19]) subsamples supported trend for association in both study populations independently, but the p-values did not reach statistical significance (p>0.05) due to reduced samples sizes. The significant association with all four CGB5 promoter polymorphisms results from higher minor allele frequency (MAF) in fertile women (12.05%-13.08%) compared to RM group (7.10%-7.92%). This difference between the control group and RM cases was consistent in both study populations: 13.16% compared to 8.08 % and 11-13% compared to 5.95-7.74%, in Estonians and Finns, respectively (Table 3). On the haplotype network of CGB5 upstream region the promoter variants carrying the minor alleles of the four polymorphisms form a remote clade (H1-H2, H10-H11; Fig. 3A).
TABLE 3.
Variants in CGB5 and CGB8 genes significantly associated with recurrent miscarriage (RM). Association P-values and odds ratio (OR) with 95% confidence interval (CI) was calculated by Cochran-Armitage test for trend
| Estonians (n=194) | Finns (n=185) | All individuals (n=379) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF (%) | p-value | OR (95%CI) |
MAF (%) | p-value | OR (95%CI) |
MAF (%) | p-value | OR (95%CI) |
||||
| SNP | Fertile women n=95 |
RM patients n=99 |
Fertile women n=100 |
RM patients n=85 |
Fertile women n=195 |
RM patients n=184 |
||||||
| c5EF-155 | 13.16 | 8.08 | 0.083 | 0.54 (0.27-1.1) |
11.50 | 6.55 | 0.129 | 0.58 (0.29-1.19) |
12.31 | 7.38 | 0.024 | 0.57 (0.35-0.93) |
| c5EF-147 | 13.16 | 8.08 | 0.083 | 0.54 (0.27-1.1) |
11.00 | 5.95 | 0.094 | 0.52 (0.24-1.13) |
12.05 | 7.10 | 0.018 | 0.54 (0.32-0.91) |
| c5EF-144 | 13.16 | 8.08 | 0.083 | 0.54 (0.27-1.1) |
13.00 | 7.74 | 0.131 | 0.61 0.31-1.17) |
13.08 | 7.92 | 0.023 | 0.58 (0.36-0.93) |
| c5EF-142 | 13.16 | 8.08 | 0.083 | 0.54 (0.27-1.1) |
13.00 | 7.74 | 0.131 | 0.61 (0.31-1.17) |
13.08 | 7.92 | 0.023 | 0.58 (0.36-0.93) |
| c5EF1038 | 14.47 | 9.09 | 0.079 | 0.57 (0.30-1.08) |
14.00 | 7.06 | 0.036 | 0.48 (0.24-0.97) |
14.36 | 8.15 | 0.007 | 0.53 (0.32-0.85) |
| c8EF301* | 5.79 | 5.05 | 0.740 | 0.86 (0.35-2.12) |
8.85 | 3.21 | 0.034 | 0.35 (0.12-0.96) |
7.33 | 4.24 | 0.072 | 0.55 (0.29-1.06) |
| c8EF1045* | 0.53 | 0.51 | 0.977 | 0.97 (0.06-15.6) |
3.13 | 0 | 0.025 | na | 1.83 | 0.28 | 0.042 | 0.15 (0.02-1.03) |
significant association (p<0.05) only in one population
MAF - minor allele frequency; na - not applicable as monomorphic among fertile women
Figure 3.
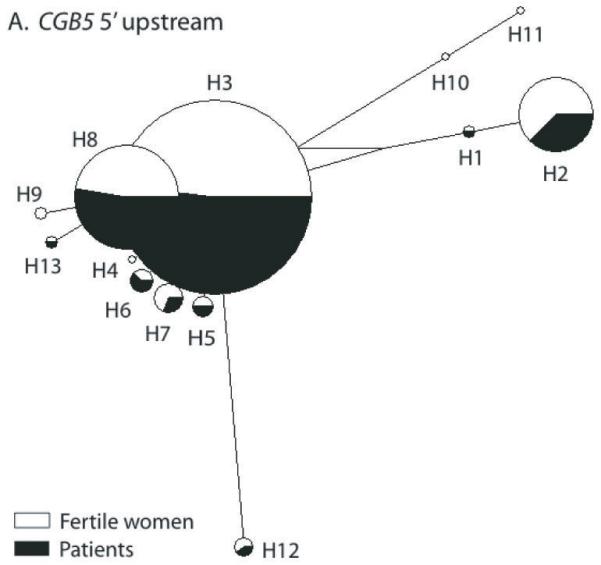
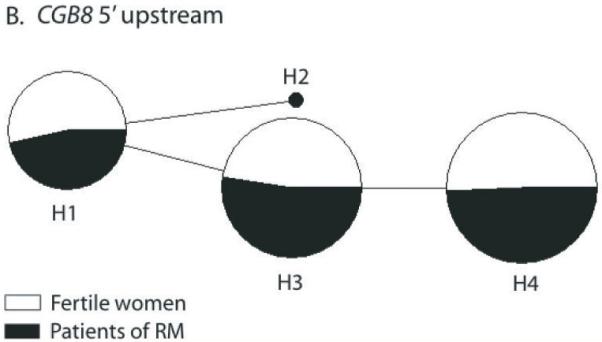
Median-Joining (MJ) networks of predicted haplotypes in the 5′ upstream region of CGB5 (A) and CGB8 (B). Singleton polymorphisms were excluded from the analysis, because of unreliable phasing. The size of each node is proportional to the haplotype frequency in the total dataset. The relative distribution of each haplotype among the recurrent miscarriage (RM) cases (black) and fertile controls (white) is indicated. Haplotype nomenclature is shown in Supplementary Table S2. (A) The carrier status of haplotypes H1-H2 and H10-H11 lowered 1.7-fold the risk of RM. (B) Haplotype H2 defined by the minor allele of a proximal promoter mutation c8EF-4 was exclusively identified in RM patients in both study populations, Estonians and Finns.
MJ networks of predicted haplotypes in the genic regions of CGB5 and CGB8 are shown in Supplementary Fig. S3.
Among the CGB5 genic SNPs a strong protective effect was detected for the minor allele of intron 2 c5EF1038 (p<0.007; OR=0.53 [95% CI 0.32-0.85]), represented with the frequency 14.36% in fertile women compared to 8.15% in the RM group (Table 3). This effect reached statistical significance in the separate analysis of the Finnish subjects (p=0.036; OR=0.48 [95% CI 0.24-0.97]) and showed a trend for association in the Estonian (p=0.079; OR=0.57 [95% CI 0.30-1.08]) subsample. No increase in protection towards RM was detected for the combination of the minor alleles of the CGB5 5′upstream and the intronic SNP (data not shown).
Notably, the association of four CGB5 SNPs with the protective effect towards pregnancy loss was sufficiently robust to remain significant even when only the female RM patients (n=109) were considered as cases (Table 4). A separate analysis of male RM patients revealed similar trends for association and protective effect sizes as compared to female RM cases, although the p-values were non-significant possibly due to smaller sample size (n=75) that reduced the statistical power. However, the inclusion of both sexes gave a stronger effect than gender-specific analysis in all but one SNP (c5EF1038; Table 3, 4) further supporting the contribution of both, maternal and paternal genes in the reproductive success.
TABLE 4.
Case-control analysis after subdividing recurrent miscarriage (RM) patients by gender. Cochran-Armitage test for trend was performed for the variants in CGB5 and CGB8 genes showing significant association with RM in the full sample set (Table 3)
| Fertile women n=195 | Female RM patients n=109 | Male RM patients n=75 | |||||
|---|---|---|---|---|---|---|---|
| SNP | MAF % | MAF % | p-value | OR (95%CI) | MAF % | p-value | OR (95%CI) |
| c5EF-155 | 12.31 | 7.34 | 0.058 | 0.59 (0.32-1.03) | 7.43 | 0.105 | 0.57 (0.26-1.13) |
| c5EF-147 | 12.05 | 6.88 | 0.039 | 0.53 (0.28-0.98) | 7.43 | 0.115 | 0.57 (0.28-1.15) |
| c5EF-144 | 13.08 | 7.34 | 0.031 | 0.53 (0.29-0.95) | 8.78 | 0.171 | 0.64 (0.34-1.22) |
| c5EF-142 | 13.08 | 7.34 | 0.031 | 0.53 (0.29-0.95) | 8.78 | 0.171 | 0.64 (0.34-1.22) |
| c5EF1038 | 14.36 | 6.88 | 0.006 | 0.45 (0.25-0.81) | 10 | 0.182 | 0.66 (0.36-1.22) |
MAF - minor allele frequency; OR - odds ratio; CI - confidence interval
Population-specific associations were detected in the Finnish sample collection with two rare SNPs (MAF <10%) in CGB8: c8EF301 (p=0.034) and c8EF1045 (p=0.025) (Table 3). Interestingly, the protective variant in the intron 2 of CGB8 (c8EF1045) is located at the same position within the gene as the CGB5 intronic variant (c5EF1038).
Rare CGB8 promoter variants increase the susceptibility to recurrent miscarriages
The resequenced CGB8 5′upstream region stands out with only 3 SNPs (two common and one rare) compared to the respective region for CGB5 with 18 SNPs (Table 1). The rare allele A of SNP c8EF-4 was solely represented in patients, one from Finland and two from Estonia (Cochran-Armitage trend test, p=0.071). This polymorphism is located within the AP1-like sequence overlapping the HCG beta initiator element critical for basal transcription and downstream of the Ets-2 binding site acting as a major enhancer of HCG beta gene expression (30).
We applied two neutrality tests to explore observed versus expected distribution of SNPs and haplotypes in the 5′upstream region of CGB8. Both, the Tajima’s D statistic (DT=2.29, p<0.05; Table 2) as well as Ewens-Watterson homozygosity test (p=0.007) indicated a possible scenario of balancing selection driving the three apparently most efficient CGB8 promoter variants (H1, H3, H4) to high frequency in both populations (Fig. 3B; Supplementary Table S2: Supplementary Fig. S2). The rare variant H2 carried the minor allele of c8EF-4, identified solely in patients. Notably, the haplotype combining the minor alleles of c8EF-287 (C; MAF=25.2%) and c8EF-186 (T; MAF=39.7%) is expected to be present with the frequency of 10%, but was not observed in the current study (Fig. 3B; Supplementary Table S2).
Discussion
Here we report the first case-control study targeting the variation in HCG beta genes in association with recurrent miscarriages (RM). Most association studies on RM have so far focused on susceptibility variants of maternal genes involved in physiological adaptation to pregnancy, such as development of immunotolerance at feto-maternal interface or alterations in fibrinolytic and coagulation pathways. As these genes also contribute to complex diseases, the role of their variants in susceptibility to RM may not be specific (5, 8, 22, 31). HCG beta genes are expressed in blastocysts shortly after fertilization (20, 32) and are essential for successful implantation. Thus, a genetic variant of these genes is more likely to have an effect on pregnancy outcome. Our study focused on CGB8 and CGB5 that provide the major fraction of HCG beta mRNA transcripts and the resequencing method was chosen instead of genotyping.
The human CGB8 and CGB5 genes are located among the seven duplicate genes within the LHB/CGB gene cluster. Major complications in targeting duplicated genes in association studies are high sequence similarity (>92%), high diversity, large number of population-specific variants and low LD due to high gene conversion activity (19, 33). These characteristics make it technically challenging to select reliable tag-SNPs and establish genotyping methods capable of targeting unique SNPs in duplicated genes. In the 379 subjects we identified only 14 out of 30 (47%) SNPs in CGB5 present in a public SNP database NCBI dbSNP and 9 out of 44 (20%) in CGB8. Several of the variants not observed in our study have been predicted in silico or by using high-throughput methods and may actually be multisite or paralogous gene variants (34). Alternatively, some of these SNPs could indeed represent variants specific to other than Estonian or Finnish populations. For example, an amino acid substitution Val79Met (nomenclature based on mature protein; from ATG p.Val99Met) in CGB5 exon 3 has been reported at carrier frequency 4.2% in a random population from the Midwest of the United States (35) but it was absent in a 580 DNA samples originating from five European populations (36). In the current study, in relatively large samples sets drawn from two neighboring populations, one third of the identified variants (MAF<2%) were found in only one population, although the sample size was sufficient to identify all common variants (MAF>5%) originally described in a large mutation screening of LHB/CGB genes (Table 1; 19). Full resequencing data collected in this study enabled to identify several rare non-synonymous and promoter variants and to conduct haplotype analysis.
Consistent with the hypothesis of the study, we identified genetic variants in HCG beta genes significantly increasing or reducing the risk of RM. A protective effect was detected for the minor alleles of two SNPs (c5EF1038 and c8EF1045) located at the identical positions in intron 2 in both CGB5 and CGB8 and for four CGB5 promoter variants (c5EF-155; c5EF-147; c5EF-144; c5EF-142). The carrier status of the minor alleles of these six SNPs reduced the risk of RM 1.7-fold in comparison to the wildtype carriers. Interestingly, the “protective” alleles of the CGB5 promoter SNPs form a motif (C-del-C-A; H2 on Fig. 3A) identical to the promoter sequence of CGB8 (Fig. 1D), which has been shown to be most actively transcribed HCG beta gene (18). The actual contribution of these sequence variants to mRNA transcription and splicing efficiency is still to be explored.
The current data suggest the CGB8 and especially its promoter region to be under stronger functional constraint compared to CGB5 in spite of high DNA sequence similarity (98-99%) between the two genes (Supplementary Fig. S1). Firstly, we detected > 2 times less polymorphisms in CGB8 genomic region (n=22) compared to CGB5 (n=49). Secondly, three rare CGB8 variants that may exhibit an effect on hormone action were present exclusively in RM patients (p.Arg28Trp, p.Pro93Arg and a c8EF-4 within proximal promoter) compared to only one such SNP in CGB5 (p.Val76Leu). Thirdly, the applied neutrality tests indicated a balancing selection in the promoter region of CGB8, but not of CGB5. Additionally, we identified only three of the four predicted major CGB8 promoter haplotypes (Fig. 3B). The haplotype combining the minor alleles of c8EF-287 (MAF 25.2 %) and c8EF-186 (MAF 39.7%) was absent in the current dataset in spite of the relatively high minor allele frequency. The discrepancy between observed (0%) and expected (10%) frequency may be explained by the localization of these SNPs within Sp1/AP-2 binding sites (37) residing in the critical region for the trophoblast-specific expression as well as cAMP-responsiveness of the HCG beta gene transcription (38, 39). Functional studies should reveal whether these sequence variants indeed possess a combinatory effect influencing the binding of the AP-2 and Sp1 transcription factors to the promoter of HCG beta and alter the transcription of genes.
One of the key factors in obtaining reliable results in a case-control study is a clearly defined study group and replication of the results in an independent dataset. We applied parallel analysis of case-control sample sets collected from two neighboring countries in order to confirm the robustness of the association across populations. As the stratification was low between these populations we also conducted a joint analysis of the two sample sets in order to raise the statistical power. Although there were minor differences in subject recruitment, the obtained results were concordant in two populations and the strength of detected associations increased in the analysis of the pooled dataset. In addition we identified two gene variants lowering the risk of RM in the Finnish dataset only possibly owing to the specific demographic history of the Finnish population (40).
In conclusion, these data from two populations provide the first evidence for the role of the variation in HCG beta genes in contributing to the susceptibility of RM. The findings encourage further studies addressing the functional effect of the identified promoter, intronic and rare protein-altering variants on HCG beta gene expression and HCG hormone activity. The diagnostic application of our findings may facilitate the improvement of early and preventive treatment of RM.
Supplementary Material
Acknowledgements
We thank the participating patients, prof. Helle Karro and Dr. Andres Salumets for providing facilities for patient material collection, and dr. Pille Hallast for advice in resequencing of the LHB/CGB genes.
Funding: M.L. is the Howard Hughes Medical Institute International Scholar (grant #55005617) and Wellcome Trust International Senior Research Fellow in Biomedical Science in Central Europe (grant 070191/Z/03/Z). In addition, funding for the work was partially provided by Estonian Science Foundation grants no. 5796 and 7471 (K.R., L.N., P.K., M.L.) and Estonian Ministry of Education and Science core grants no. 0182721s06 (M.L., K.R., L.N., T.M., P.K.) and 0182641s04 (K.R.); as well as by Sigrid Juselius Foundation & Finnish State Fund (V.M.U., M.K., K.A) and Sanofi-Aventis (V.M.U.). K.R. and L.N. have been awarded by fellowships from Graduate School in Biomedicine and Biotechnology (1.0101-0167).
Footnotes
Author disclosure statement: The authors have nothing to disclose.
References
- 1.Berry CW, Brambati B, Eskes TK, Exalto N, Fox H, Geraedts JP, Gerhard I, Gonzales Gomes F, Grudzinskas JG, Hustin J, Jouppila P, Lindblom BKA, Mantoni N, Montenegro N, Nogales Fernandes F, O’Rahilly R, Pedersen JF, Peters PWJ, Regan L, Rushton DI, van Straaten HWM, Tarlatzis BC, Wells M. The Euro-Team Early Pregnancy (ETEP) protocol for recurrent miscarriage. Hum Reprod. 1995;10:1516–1520. doi: 10.1093/humrep/10.6.1516. [DOI] [PubMed] [Google Scholar]
- 2.Jauniaux E, Farquharson RG, Christiansen OB, Exalto N. Evidence-based guidelines for the investigation and medical treatment of recurrent miscarriage. Hum Reprod. 2006;21:2216–2222. doi: 10.1093/humrep/del150. [DOI] [PubMed] [Google Scholar]
- 3.Li TC, Makris M, Tomsu M, Tuckerman E, Laird S. Recurrent miscarriage: aetiology, management and prognosis. Hum Reprod Update. 2002;8:463–481. doi: 10.1093/humupd/8.5.463. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen OB. A fresh look at the causes and treatments of recurrent miscarriage, especially its immunological aspects. Hum Reprod Update. 1996;2:271–293. doi: 10.1093/humupd/2.4.271. [DOI] [PubMed] [Google Scholar]
- 5.Goodman CS, Coulam CB, Jeyendran RS, Acosta VA, Roussev R. Which thrombophilic gene mutations are risk factors for recurrent pregnancy loss? Am J Reprod Immunol. 2006;56:230–236. doi: 10.1111/j.1600-0897.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 6.Rey E, Kahn SR, David M, Shrier I. Thrombophilic disorders and fetal loss: a meta-analysis. Lancet. 2003;361:901–908. doi: 10.1016/S0140-6736(03)12771-7. [DOI] [PubMed] [Google Scholar]
- 7.Robertson L, Wu O, Langhorne P, Twaddle S, Clark P, Lowe GDO, Walker ID, Greaves M, Brenkel I, Regan L, Greer IA. Thrombophilia in pregnancy: a systematic review. Br J Haematol. 2006;132:171–196. doi: 10.1111/j.1365-2141.2005.05847.x. [DOI] [PubMed] [Google Scholar]
- 8.Hviid TV. HLA-G in human reproduction: aspects of genetics, function and pregnancy complications. Hum Reprod Update. 2006;12:209–232. doi: 10.1093/humupd/dmi048. [DOI] [PubMed] [Google Scholar]
- 9.Licht P, Fluhr H, Neuwinger J, Wallwiener D, Wildt L. Is human chorionic gonadotropin directly involved in the regulation of human implantation? Mol Cell Endocrinol. 2007;269:85–92. doi: 10.1016/j.mce.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Toth P, Lukacs H, Gimes G, Sebestyen A, Pasztor N, Paulin F, Rao CV. Clinical importance of vascular LH/hCG receptors - a review. Reprod Biol. 2001;1:5–11. [PubMed] [Google Scholar]
- 11.Herr F, Baal N, Reisinger K, Lorenz A, McKinnon T, Preissner KT, Zygmunt M. HCG in the regulation of placental angiogenesis. Results of an in vitro study. Placenta. 2007:85–93. doi: 10.1016/j.placenta.2007.02.002. S. [DOI] [PubMed] [Google Scholar]
- 12.Kayisli UA, Selam B, Guzeloglu-Kayislim O, Demir R, Arici A. Human chorionic gonadotropin contributes to maternal immunotolerance and endometrial apoptosis by regulating Fas-Fas ligand system. J Immunol. 2003;171:2305–2313. doi: 10.4049/jimmunol.171.5.2305. [DOI] [PubMed] [Google Scholar]
- 13.Buyalos RP, Glassman LM, Rifka SM, Falk RJ, Macarthy PO, Tyson VJ, DiMattina M. Serum beta-human chorionic gonadotropin, estradiol and progesterone as early predictors of pathologic pregnancy. J Reprod Med. 1992;37:261–266. [PubMed] [Google Scholar]
- 14.Dumps P, Meisser A, Pons D, Morales MA, Anguenot JL, Campana A, Bischof P. Accuracy of single measurements of pregnancy-associated plasma protein-A, human chorionic gonadotropin and progesterone in the diagnosis of early pregnancy failure. Eur J Obstet Gynecol Reprod Biol. 2002;100:174–180. doi: 10.1016/s0301-2115(01)00470-5. [DOI] [PubMed] [Google Scholar]
- 15.Tong S, Wallace EM, Rombauts L. Association between low day 16 hCG and miscarriage after proven cardiac activity. Obstet Gynecol. 2006;107:300–304. doi: 10.1097/01.AOG.0000196505.09016.f3. [DOI] [PubMed] [Google Scholar]
- 16.Bo M, Boime I. Identification of the transcriptionally active genes of the chorionic gonadotropin beta gene cluster in vivo. J Biol Chem. 1992;267:3179–3184. [PubMed] [Google Scholar]
- 17.Miller-Lindholm AK, LaBenz CJ, Ramey J, Bedows E, Ruddon RW. Human chorionic gonadotropin-beta gene expression in first trimester placenta. Endocrinology. 1997;138:5459–5465. doi: 10.1210/endo.138.12.5618. [DOI] [PubMed] [Google Scholar]
- 18.Rull K, Laan M. Expression of beta-subunit of human chorionic gonadotropin genes during the normal and failed pregnancy. Hum Reprod. 2005;20:3360–3368. doi: 10.1093/humrep/dei261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallast P, Nagirnaja L, Margus T, Laan M. Segmental Duplications and Gene Conversion: Human Luteinizing Hormone/ Chorionic Gonadotropin Beta Gene Cluster. Genome Res. 2005;15:1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rull K, Hallast P, Uusküla L, Jackson J, Punab M, Salumets A, Campbell RK, Laan M. Fine-scale quantification of HCG beta gene transcription in human trophoblastic and non-malignant non-trophoblastic tissues. Mol Hum Reprod. 2008;14:23–31. doi: 10.1093/molehr/gam082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulander VM. Ph.D.thesis. University of Helsinki, Faculty of Medicine, Institute of Clinical Medicine; 2007. Venous thromboembolism during pregnancy and the impact of thrombophilia in pregnancy complications. http://urn.fi/URN:ISBN:978-952-10-3671-2. [Google Scholar]
- 22.Kaare M, Ulander VM, Painter JN, Ahvenainen T, Kaaja R, Aittomäki K. Variations in the thrombomodulin and endothelial protein C receptor genes in couples with recurrent miscarriage. Hum Reprod. 2007;22:864–868. doi: 10.1093/humrep/del436. [DOI] [PubMed] [Google Scholar]
- 23.Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat Genet. 2006;38:1457–1462. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- 24.Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Gen. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 26.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider S, Roessli D, Excoffier L. Arlequin ver. 2.000: A software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva; Switzerland: 2000. [Google Scholar]
- 28.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh D, Ezashi T, Ostrowski MC, Roberts RM. A central role for Ets-2 in the transcriptional regulation and cyclic adenosine 5′-monophosphate responsiveness of the human chorionic gonadotropin-beta subunit gene. Mol Endocrinol. 2003;17:11–26. doi: 10.1210/me.2002-0223. [DOI] [PubMed] [Google Scholar]
- 31.Dosiou C, Giudice LC. Natural Killer Cells in Pregnancy and Recurrent Pregnancy Loss: Endocrine and Immunologic Perspectives. Endocr Rev. 2005;26:44–62. doi: 10.1210/er.2003-0021. [DOI] [PubMed] [Google Scholar]
- 32.Jurisicova A, Antenos M, Kapasi K, Meriano J, Casper RF. Variability in the expression of trophectodermal markers beta-human chorionic gonadotrophin, human leukocyte antigen-G and pregnancy specific beta-1 glycoprotein by the human blastocyst. Hum Reprod. 1999;14:1852–1858. doi: 10.1093/humrep/14.7.1852. [DOI] [PubMed] [Google Scholar]
- 33.Sedman L, Padhukasahasram B, Kelgo P, Laan M. Complex signatures of locus-specific selective pressures and gene conversion on human growth hormone/chorionic somatomammotropin genes. Hum Mutat. 2008:1–13. doi: 10.1002/humu.20767. 0. DOI 10.1002/humu.20767 [Epub ahead of print, May 12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredman D, White SJ, Potter S, Eichler EE, Den Dunnen JT, Brookes AJ. Complex SNP-related sequence variation in segmental genome duplications. Nat Genet. 2004;36:861–866. doi: 10.1038/ng1401. [DOI] [PubMed] [Google Scholar]
- 35.Miller-Lindholm AK, Bedows E, Bartels CF, Ramey J, Maclin V, Ruddon RW. A naturally occurring genetic variant in the human chorionic gonadotropin-beta gene 5 is assembly inefficient. Endocrinology. 1999;140:3496–3506. doi: 10.1210/endo.140.8.6915. [DOI] [PubMed] [Google Scholar]
- 36.Jiang M, Savontaus ML, Simonsen H, Williamson C, Müllenbach R, Gromoll J, Terwort N, Alevizaki M, Huhtaniemi I. Absence of the genetic variant Val79Met in human chorionic gonadotropin-beta gene 5 in five European populations. Mol Hum Reprod. 2004;10:763–766. doi: 10.1093/molehr/gah098. [DOI] [PubMed] [Google Scholar]
- 37.Johnson W, Jameson JL. AP-2 (activating protein 2) and Sp1 (selective promoter factor 1) regulatory elements play distinct roles in the control of basal activity and cyclic adenosine 3′,5′-monophosphate responsiveness of the human chorionic gonadotropin-beta promoter. Mol Endocrinol. 1999;13:1963–1975. doi: 10.1210/mend.13.11.0386. [DOI] [PubMed] [Google Scholar]
- 38.Albanese C, Kay TW, Troccoli NM, Jameson JL. Novel cyclic adenosine 3′,5′-monophosphate response element in the human chorionic gonadotropin beta-subunit gene. Mol Endocrinol. 1991;5:693–702. doi: 10.1210/mend-5-5-693. [DOI] [PubMed] [Google Scholar]
- 39.Steger DJ, Hecht JH, Mellon PL. GATA-binding proteins regulate the human gonadotropin alpha-subunit gene in the placenta and pituitary gland. Mol Cell Biol. 1994;14:5592–5602. doi: 10.1128/mcb.14.8.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norio R, Nevanlinna HR, Perheentupa J. Hereditary diseases in Finland; rare flora in rare soul. Ann Clin Res. 1973:109–141. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.