Abstract
Microarray studies have shown that individual synthetic small interfering RNAs (siRNAs) can have substantial off-target effects. Pools of siRNAs, produced by incubation of dsRNAs with recombinant Dicer or RNase III, can also be used to silence genes. Here we show that diced siRNA pools are highly complex, containing hundreds of different individual siRNAs. This high complexity could either compound the problem of off-target effects, since the number of potentially problematic siRNAs is high, or it could diminish the problem, since the concentration of any individual problematic siRNA is low. We therefore compared the off-target effects of diced siRNAs to chemically synthesized siRNAs. In agreement with previous reports, we found that two chemically synthesized siRNAs targeted against p38α MAPK (MAPK14) induced off-target changes in the abundance of hundreds of mRNAs. In contrast, three diced siRNA pools against p38α MAPK had almost no off-target effects. The off-target effects of a synthetic siRNA were reduced when the siRNA was diluted 3-fold in a diced pool and completely alleviated when it was diluted 30- or 300-fold, suggesting that when problematic siRNAs are present within a diced pool, their absolute concentration is too low to result in significant off-target effects. These data rationalize the observed high specificity of RNA interference in C. elegans and D. melanogaster, where gene suppression is mediated by endogenously-generated diced siRNA pools, and provide a strategy for improving the specificity of RNA interference experiments and screens in mammalian cells.
Keywords: RNAi, siRNA, d-siRNA, e-siRNA, gene silencing, microarray, mitogen activated protein kinase (MAPK)
INTRODUCTION
RNA interference (RNAi) is a powerful method for carrying out loss-of-function studies in diverse organisms and cell types (Fire et al, 1998; Meister and Tuschl, 2004; Silva et al, 2004; Tomari and Zamore, 2005). Its usefulness and reliability is exemplified by the fact that RNAi is frequently used for loss-of-function screening in organisms, like C. elegans (Gonczy et al, 2000; Ashrafi et al, 2003; Kamath and Ahringer, 2003; Kamath et al, 2003; Lee et al, 2003; Pothof et al, 2003; Simmer et al, 2003; Vastenhouw et al, 2003) and D. melanogaster (Kiger et al, 2003; Boutros et al, 2004; Dasgupta and Perrimon, 2004; Eggert et al, 2004; Cherry et al, 2005), that are also amenable to classical forward and reverse genetic approaches. Moreover, RNAi has allowed loss-of-function experiments and screens to be performed in vertebrate cells where alternative approaches are much more limited (Elbashir et al, 2001a; Brummelkamp et al, 2003; Kittler et al, 2004; Paddison et al, 2004).
In vertebrate systems, RNAi is generally accomplished by the transfection or expression of small interfering RNAs (siRNAs), which avoids the non-specific suppression of protein synthesis induced by larger dsRNAs in many vertebrate cell types (Elbashir et al, 2001a). The antisense strand of the siRNA guides RNA-induced silencing complexes (RISC) to complementary mRNAs via nucleic acid interactions (Hammond et al, 2000; Elbashir et al, 2001b) and subsequently the mRNAs are cleaved by an endonuclease (Zamore et al, 2000; Liu et al, 2004; Song et al, 2004) and digested to single nucleotides by an exonuclease (Caudy et al, 2003).
Because base pairing is ordinarily very precise, the target specificity of siRNAs could be very high, and initial indications were that this was the case. In C. elegans RNAi phenotypes correlate well with true genetic null phenotypes for many genes (Fire et al, 1998). In mammalian cells, studies of luciferase reporter genes indicated high specificity of siRNAs for their intended targets as well; single base pair changes could abolish an siRNA's ability to silence its target (Elbashir et al, 2001a).
Recently, microarray analysis has been used to look more comprehensively for siRNA off-target effects. One group showed that an exogenous green fluorescent protein gene could be silenced with only small variations in the levels of endogenous mRNAs (mostly >2-fold) (Chi et al, 2003). Because these changes were inconsistent between replicates, they were attributed to experimental noise (Chi et al, 2003). A second group silenced several endogenous targets and analyzed global mRNA changes by microarray analysis (Semizarov et al, 2003). As long as the siRNA concentration was less than 20 nM, different siRNAs to the same target were found to produce similar mRNA changes (Semizarov et al, 2003). This argued that most of the changes in mRNA levels were secondary to the reduction of the intended target, rather than unwanted off-target effects (Semizarov et al, 2003).
However, a third group found that siRNAs could trigger off-target effects. Each of eight siRNAs designed to suppress p38α MAPK (also termed MAPK14) was found to also cause changes in the abundances of many other mRNAs (Jackson et al, 2003). Each siRNA affected a different subset of mRNAs, implying that the changes were sequence-specific off-target effects rather than non-sequence-specific toxic effects or secondary effects of p38α MAPK suppression (Jackson et al, 2003). Off-target effects could not be avoided by simply decreasing the dose of siRNA or confining the experiment to a particular time window (Jackson et al, 2003). Similar results were found for a second target, IGF1R (insulin-like growth factor receptor) (Jackson et al, 2003). These findings indicated that off-target effects might be fairly common in siRNA-treated cells. This conclusion was particularly worrisome in light of the exploding popularity of siRNA approaches for loss-of-function experiments and screens.
The prevailing hypothesis is that many of the off-target effects of siRNAs are mediated by cross-hybridization due to stretches of limited identity between the siRNAs and the off-target genes. This suggests a number of potential strategies for reducing cross-hybridization and off-target effects. Off-target effects resulting from the passenger siRNA strand can potentially be eliminated by modifications that prevent its incorporation into the RISC. For example, chemically modified siRNAs and locked nucleic acids have been used to prevent passenger siRNA strand mediated RNA cleavage (Elmen et al, 2005; Judge et al, 2006).
Recent studies on the sequence requirements for RNAi-mediated gene silencing have suggested ways of decreasing off-target effects that are the result of cross-hybridization between a transcript and either the guide or passenger strand of the siRNA. Both the number and position of mismatches between the siRNA strands and a cross-hybridizing gene affects the likelihood of off-target silencing (Du et al, 2005; Holen et al, 2005). In general, mismatches within the core of the sequence are more detrimental to silencing than mismatches at either the 5′- or 3′-end, although there are some disparities regarding the importance of the 5′-end (Du et al, 2005; Holen et al, 2005).
Additional studies found that 6 or 7 consecutive matches between the siRNA guide strand and the off-target RNA as well as the context of the matching sequence within the off-target RNA are important parameters that determine whether or not an off-target mRNA is cleaved (Lin et al, 2005; Birmingham et al, 2006). In concert with in silico analyses of siRNA specificity (Huesken et al, 2005; Qiu et al, 2005) these empirical measures of specificity have resulted in siRNA design algorithms that avoid particular siRNA sequences that may decrease off-target transcript abundance.
In addition, there may be off-target effects that do not depend upon cross-hybridization between the siRNA and the off-target genes (Bridge et al, 2003; Sioud and Sorensen, 2003; Sledz et al, 2003; Kariko et al, 2004a; Kariko et al, 2004b; Persengiev et al, 2004; Judge et al, 2005). These may include aptamer-like effects or other poorly understood sequence-specific, cross-hybridization-independent mechanisms.
We and others have shown that complex pools of siRNAs can be produced by incubating in vitro transcribed long double stranded RNAs (∼500 bp) with recombinant bacterial RNase III (Yang et al, 2002; Kittler et al, 2005) or recombinant human Dicer (Provost et al, 2002; Zhang et al, 2002; Kawasaki et al, 2003; Myers et al, 2003). These pools compare favorably with chemically synthesized siRNAs in terms of their effectiveness (Myers et al, 2003; Myers and Ferrell, 2005a; Myers and Ferrell, 2005b), and they have been successfully applied to both large scale screens (Liou et al, 2005) and functional studies of individual proteins (Fink et al, 2003; Myers et al, 2003; Jones et al, 2004; Chi et al, 2006).
Here we address the issue of off-target changes in mRNA abundance for Dicer-generated siRNA pools (d-siRNAs) and RNase III-generated siRNA pools (e-siRNAs). A priori, it seemed possible that the pools might be more problematic than single siRNAs, since a typical diced pool contains many different siRNAs, each of which might contribute off-target effects. On the other hand, the low absolute concentration of any individual problematic siRNAs in a pool might render their off-target effects insignificant, particularly if each problematic siRNA affected a different subset of genes. To distinguish between these two possibilities, we have examined the complexity of diced siRNA pools, and compared the off-target effects of d-siRNAs and e-siRNAs to those of three chemically synthesized siRNAs previously shown to elicit a range of off-target effects (Jackson et al, 2003a). We found that d-siRNAs and e-siRNAs produce minimal off-target effects as compared to chemically synthesized siRNAs.
MATERIALS AND METHODS
Preparation of siRNAs, d-siRNAs, and e-siRNAs
The 14-1, 14-5, and 14-8 siRNAs (Table 1) were purchased from Ambion (Austin, TX) as deprotected, desalted duplexes. The d-siRNAs were prepared as previously described (Myers and Ferrell, 2005b). Briefly, PCR was used to generate in vitro transcription templates. The T7 promoter (underlined in Table 1) was added to the 5′-end of both the sense and antisense strand via PCR. Using MegaScript reagents, T7 polymerase (Cat. No. 1334, Ambion) was used to transcribe the sense and antisense RNA strands, which anneal during transcription. Recombinant human Dicer (Myers and Ferrell, 2005b) was used to cleave the large dsRNA into d-siRNA by incubation for 16 hr at 37°C. Recombinant bacterial RNase III (Cat. No. 2290, Ambion) was used to cleave large dsRNA into e-siRNA by incubation for 2 hr at 37°C as described by the manufacturer (Kittler et al, 2005). The Micro-to-midi RNA purification system (Cat. No. 12183-018, Invitrogen; Carlsbad, CA) was used to purify the d-siRNAs and e-siRNAs as described previously (Myers and Ferrell, 2005b).
Table 1.
siRNAs, d-siRNAs, and e-siRNAs
| siRNA2 | Sense strand | Antisense strand | Position relative to start codon |
|---|---|---|---|
| 14-1 | 5′-CCUACAGAGAACUGCGGUUdTdT-3′ | 5′-AACCGCAGUUCUCUGUAGGdTdT-3′ | 203-221 |
| 14-5 | 5′-CCAGUGGCCGAUCCUUAUGdTdT-3′ | 5′-CAUAAGGAUCGGCCACUGGdTdT-3′ | 952-970 |
| 14-8 | 5′-GGCCUUUUCACGGGAACUCdTdT-3′ | 5′-GAGUUCCCGUGAAAAGGCCdTdT-3′ | 1129-1147 |
| GL3 | 5′-CUUACGCUGAGUACUUCGAdTdT-3′ | 5′-UCGAAGUACUCAGCGUAAGdTdT-3′ | 155-173 |
| Diced pools | Sense primer | Antisense strand | Position relative to start codon |
|---|---|---|---|
| d-14-1 | 5′-GCGTAATACGACTCACTATAGG- | 5′-GCGTAATACGACTCACTATAGG- | 1-551 |
| ATGTCTCAGGAGAGGCCCAC-3′ | GCCGCGTAGCCTGTCATTTC-3′ | ||
| d-14-5 | 5′-GCGTAATACGACTCACTATAGG- | 5′-GCGTAATACGACTCACTATAGG- | 552-1082 |
| CACTAGGTGGTACAGGGCTC-3′ | CAGGACTCCATCTCTTCTTGG-3′ | ||
| d-14-8/ | 5′-GCGTAATACGACTCACTATAGG- | 5′-GCGTAATACGACTCACTATAGG- | 1087-1638 |
| e-14-8 | CCTGGTTTCTGTTCTGTTGATCCC-3′ | TAAGCAAGTTAATCACAGGCTAGTGC-3′ | |
| GL3 | 5′-GCGTAATACGACTCACTATAGG- | 5′-GCGTAATACGACTCACTATAGG- | 114-615 |
| AACAATTGCTTTTACAGATGC-3′ | AGGCAGACCAGTAGATCC-3′ |
Cloning individual d-siRNAs
d-siRNAs for human cyclin B1 were generated from bases 820-1479 of the open reading frame as described (Myers and Ferrell, 2005b). d-siRNAs were cloned using a strategy adapted from Elbashir et al (Elbashir et al, 2001b). 2 μg of d-siRNAs were gel purified, extracted from a 12% (w/v) urea gel and precipitated with absolute ethanol. Purified products were ligated to 5 μM of 3′ adapter oligonucleotides (Dharmacon, Lafayette CO) with T4 RNA ligase (NEB), purified, gel extracted from a 12% (w/v) urea gel and subjected to phosphorylation with T4 polynucleotide kinase (NEB). Phosphorylated products were ligated to 5 μM of 5′ adapter oligonucleotides (Dharmacon) and purified by phenol/chloroform extraction followed by precipitation with 10 M ammonium acetate. Ligated products were subjected to reverse transcription (Superscript III First-Strand Synthesis Kit, Invitrogen) using 3′ oligo (5′GACTAGCTGGAATTCAAGGATGCGGTTAAA), and then amplified by PCR using 5′ oligo (5′CAGCCAACGGAATTCATACGACTCACTAAA) and 3′ oligo (5′GACTAGCTGGAATTCAAGGATGCGGTTAAA). Amplified products were then cloned into pCR 2.1-TOPO (Invitrogen), and positive clones were screened by PCR and sequenced using M13 forward and reverse primers.
Cell culture and transfections
HeLa cells (Cat. No. CCL-2, ATCC; Manassas, VA) were cultured at 37°C and 5% (v/v) CO2 in Dulbecco's modified Eagle's media (DMEM, Cat. No. 11995-65, Invitrogen) containing 10% (v/v) fetal bovine serum (FBS, Cat. No. 26140-079, Invitrogen), 100 U/ml penicillin, 100 μg/ml streptomycin, and additional 4 mM glutamine (Cat. No. 10378-016, Invitrogen). HeLa cells were plated at ∼30% confluence whenever a density of ∼80% confluence was attained in order to maintain logarithmic growth. Transfections were carried out with GeneSilencer (Cat. No. T500750, Genlantis; San Diego, CA) as previously described (Myers and Ferrell, 2005b). Briefly, ∼8 × 104 HeLa cells were plated per well in 6-well dishes 18-24 hr prior to transfection. Shortly before transfection, serum-containing medium was removed, cells were rinsed twice with 2 ml of Opti-mem I (31985-070, Invitrogen) followed by addition of 1 ml of Opti-mem I. For most experiments, siRNAs, d-siRNAs, and e-siRNAs were mixed with GeneSilencer transfection reagents and added to cells such that the final concentration was 10 nM. Other final concentrations of siRNAs/d-siRNAs/e-siRNAs were used for the experiments shown in Figure 1. Three hours and 30 min after addition of transfection mix, DMEM containing 20% (v/v) FBS, 200 U/ml penicillin, 200 μg/ml streptomycin, and additional 8 mM glutamine was added to each well to restore each component to normal concentrations. Cells were also treated with transfection reagent alone, subjected to the media change to Opti-mem I, or left untreated.
Figure 1.
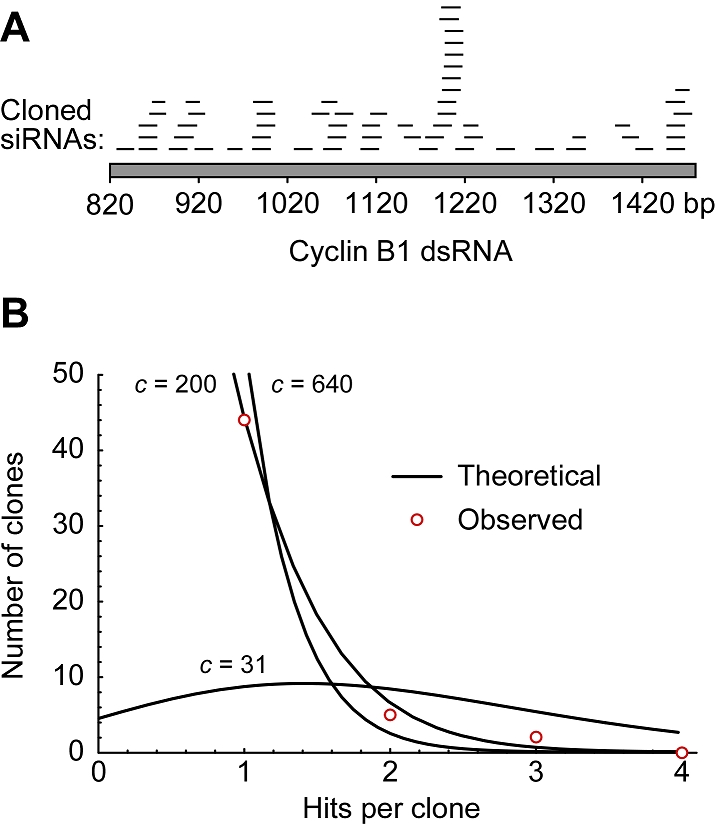
Complexity of d-siRNA pool. (A) A 660 bp fragment of the human cyclin B1 dsRNA was digested in vitro with human recombinant Dicer. Sixty of the resulting siRNAs were cloned and sequenced. (B) The predicted frequencies of cloning individual siRNAs once, twice, three times, or four times, out of a sample of 60 clones, for three different assumed pool complexities (c=31, 200, 640). The observed frequencies (open circles) fit best to a pool complexity of 200.
Preparation of cellular extracts and purification of total RNA
The PARIS kit (Cat. No. 1921, Ambion) was used to produce total cellular extract and purify total RNA as described by the manufacturer. About 48 hr after transfection, cells were lysed with cell disruption buffer (PARIS) containing protease inhibitors (Myers and Ferrell, 2005b). One-third of the lysate was saved for analysis of protein. Total RNA was isolated from the remaining two-thirds.
SDS-PAGE and western blotting
The total amount of protein in each cell lysate was measured with the BCA protein assay kit (Cat. No. 23225, Pierce Biotechnology; Rockford, IL). The volume of each lysate was adjusted with cell disruption buffer (PARIS) containing protease inhibitors such that the total protein concentration of each lysate was equal. After addition of Laemmli sample buffer about 15 μg of total protein per lane was electrophoresed in a 10% (w/v), 29:1 (acrylamide:bisacrylamide) SDS polyacrylamide gel (Myers and Ferrell, 2005b). Proteins were transferred to Immobilon-P polyvinylidine fluoride membrane (PVDF, Millipore, Billerica, MA) in 25 mM Tris, 192 mM glycine, and 10% (v/v) methanol. Antibodies were diluted in Dulbecco's phosphate-buffered saline (D-PBS, Cat. No. 14040-182, Invitrogen) containing 1% (w/v) non-fat milk powder and 0.01% (v/v) Tween-20 (Cat. No. P1379 Sigma-Aldrich; St. Louis, MO). Affinity purified p38α MAPK IgG (Cat. No. SC-535, Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:1000 dilution (0.2 μg/ml), MEK1 antiserum 662 (Hsiao et al, 1994) was used at 1:2000, and affinity purified IgG raised against eIF2α phosphorylated at serine 51 (cat. No. 44-728G, Biosource International; Camarillo, CA) was used at a 1:500 dilution (0.5 μg/ml). Horseradish peroxidase (HRP) conjugated goat IgG raised against rabbit IgG (Cat. No. 81-6120, Zymed, San Francisco, CA) was used at a 1:5000 dilution (0.3 μg/ml) for detecting MEK1 and p38α MAPK or a 1:2500 dilution (0.6 μg/ml) for detection of phosphorylated eIF2α. Immun-Star HRP chemiluminescence kit (Cat. No. 170-5040, BioRad, Hercules, CA), a GelDoc (BioRad), and the QuantityOne software package (BioRad) were used for visualizing and quantifying MEK1, p38α MAPK, and phosphorylated eIF2α abundance.
Microarray hybridization and data analysis
Fluorescently-labeled cDNA was synthesized from RNA isolated from cells transfected with one of the siRNAs, d-siRNAs, or e-siRNAs. A reference set of fluorescently-labeled cDNAs was prepared from RNA that was isolated from HeLa cells treated with the GeneSilencer transfection reagent only. Comparative hybridization to a cDNA microarray with 41,421 spots from 24,472 unique putative genes (based on cluster ID) was then used to identify mRNAs whose levels were different in siRNA-treated cells compared to the reference control.
To obtain sufficient quantities of mRNA for microarray hybridization, polyadenylated transcripts from all samples, including the reference, were amplified using the MessageAmp aRNA kit (Cat. No. 1750, Ambion) after any residual DNA was removed from isolated total RNA with RNase-free DNase provided in the DNA-free kit (Cat. No. 1906, Ambion). Using standard protocols the amplified RNA (aRNA) made from cells transfected with one of the small dsRNAs was subjected to reverse-transcription in the presence of random hexamers (Cat. No. 27-2166-01, Amersham, Piscataway, NJ) and Cy5-labeled d-UTP (Cat. No. 55022, Amersham) to produce fluorescently labeled cDNA probes. Similarly the reference aRNA was reverse-transcribed in the presence of random hexamers and Cy3-labeled d-UTP (Cat. No. 53022, Amersham) to produce a second set of fluorescently labeled cDNA probes. The fluorescent probes were then comparatively hybridized (without fragmentation) to cDNA micorarrays as previously described (DeRisi et al, 1997; Wang et al, 2000).
For each of the 54 microarrays, mRNA abundance measurements were independently normalized within each array such that the mean Cy5:Cy3 ratio was 1. Measurements of mRNA abundance were considered good if the regression correlation between the Cy5 and Cy3 fluorescence intensity across the pixels comprising the scanned image of the corresponding array elements was greater than 0.6, if the mean intensity of Cy3 was 2-fold greater than the median background Cy3 intensity, and if the Cy5 normalized mean intensity was 2-fold greater than the median background Cy5 intensity.
Significance analysis of microarrays (SAM) (Tusher et al, 2001) was used to identify mRNAs for which consistent differences were observed between treatments. For Figure 4A, the p38αMAPK siRNAs/d-siRNAs/e-siRNAs were taken as one class and were compared to a second class comprising the GL3 controls. For Figure 4B, ten separate SAM analyses were performed, with each individual treatment considered to be one class and compared to a second class comprising all of the other treatments. Cluster analysis (Eisen et al, 1998) was used to organize the heat maps.
Figure 4.
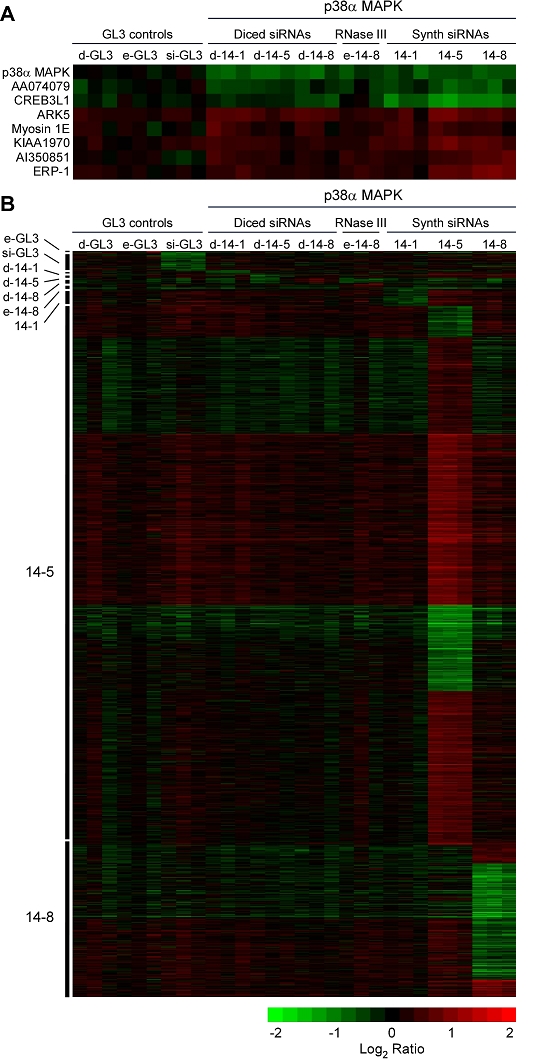
Secondary and off-target effects in cells treated with siRNAs, d-siRNAs, and e-siRNAs. Cells were transfected with various siRNAs and pools at a final concentration of 10 nM and were subjected to microarray analysis. (A) Putative secondary effects. The samples whose p38α MAPK mRNA was suppressed were compared to the GL3 control samples by significance analysis of microarrays (SAM) (Tusher et al, 2001) in order to identify mRNAs whose expression is regulated by p38α MAPK. (B) Off-target effects. Ten SAM analyses were carried out, comparing each of the treatments (3 experiments) to the other nine (27 experiments). No significant mRNA changes were detected for one of the treatments (d-GL3). For the other treatments, the mRNAs that changed significantly are represented as heat maps. The treatment to which each heat map corresponds is indicated on the left; for example, the comparison of e-GL3 to the other treatments is shown at the top, and the largest heat map compares the 14-5-treatment to the others. Within each heat map, the mRNAs are organized by clustering.
Complete microarray data can be found in the Stanford Microarray Database (http://genome-www5.stanford.edu/).
RESULTS
d-siRNA pools are highly complex
We began by estimating the complexity of a diced pool of siRNAs. Filpowicz and co-workers have shown that recombinant human Dicer prefers to cleave dsRNA substrates at their free termini (Zhang et al, 2002). Thus, after short periods of digestion the complexity of a diced pool might be relatively low. However, for gene silencing experiments, in vitro dicing reactions are generally carried out for long periods of time to maximize the yield of 21 bp products (Myers and Ferrell, 2005b). Under these conditions, the complexity of the pool could be high.
We diced to completion (16 hr) a 660 bp dsRNA fragment derived from the human cyclin B1 open reading frame. The d-siRNAs were cloned using a strategy adapted from Elbashir et al, and 60 clones were isolated and sequenced. The higher the complexity of the 660 bp pool, the lower the odds are of obtaining any individual siRNA sequence more than once. If the 60 clones sequenced represent a random sample of the pool, the Poisson distribution describes the odds of obtaining any individual siRNA sequence, once, twice, or more than twice, for any assumed complexity. If the complexity is assumed to be 31 (the minimum number of 21-mers required to span 660 bp), then 9/60 sequences would be expected to be obtained once, 8/60 twice, 5/60 three times, and so on (Figure 1B). On the other hand, if the complexity is assumed to be 640 (the maximum number of 21-mers that can be obtained from a 660 bp dsRNA), then 55/60 would be expected to be obtained once, and 2 or 3 twice (Figure 1B). By fitting Poisson distribution curves to the experimentally observed distribution of sequences and minimizing the variance between the observed and predicted distributions, an estimate can be obtained for the complexity of the pool. If some siRNAs in the pool are easier to clone than others, then the estimate will be a lower bound for the true complexity.
As shown in Figure 1A and Supplementary Table 1, the 60 sequences were distributed throughout the 660 bp dsRNA. There was a “hot spot” about 380 nt from the 5′ end of the dsRNA, from which 13 of the 60 sequences were cloned. This could represent a region of preferred Dicer cleavage or a region whose sequences were most easily cloned. Most of the sequences were 21 nt in length (60%), and most of the sequences were obtained only once: 44 were isolated once, 5 were isolated twice, 2 were isolated three times, and none were isolated more than three times. This distribution yields an estimated complexity of 200 different siRNAs in the pool. Note that if the cloning process was biased towards particular sequences (like the 13 “hot spot” sequences), the actual complexity would be higher than the estimated complexity. We carried out a similar analysis on a second 660 bp dsRNA, and obtained similar results. Thus, diced siRNA pools are highly complex.
Potencies of the synthetic, diced, and RNase III-generated siRNAs
We purchased three p38α MAPK siRNAs that had been studied in previously published work (Jackson et al, 2003): 14-5, which was reported to alter the levels of a large number of off-target mRNAs; 14-8, which was intermediate in terms of the number of mRNAs affected and whose off-target profile was qualitatively distinct from that of 14-5; and 14-1, which produced the least severe off-target effects (Table 1, Figure 2A). We also transcribed three ∼550 bp p38α MAPK dsRNAs, (Table 1, Figure 2A) and subjected them to digestion with recombinant Dicer. In addition, one of the three ∼550 bp dsRNAs was subjected to digestion with RNase III. These ∼550 bp regions encompassed each of the three synthetic siRNAs (Figure 2A) and contained stretches of identity to various p38α MAPK-related genes and to some unrelated genes as well. For example, the 551 bp region containing siRNA 14-1 included 26 consecutive nucleotides of identity to p38β MAPK, 20 nucleotide stretches of identity to p38γ MAPK and p38δ MAPK, 17 nucleotides to the alkaline phosphatase ALPI and to MAP3K7; the 531 bp region containing siRNA 14-5 included 20 consecutive nucleotides of identity to p38β MAPK, 17 nucleotides to the trithorax-related gene MLL, and 16 nucleotides to ERK1 and p38δ MAPK; and the 552 bp region containing siRNA 14-8 included 19 consecutive nucleotides of identity to MLL, 17 nucleotides to CD44 and the putative amidase FLJ31204, 12 nucleotides to p38β MAPK, and 11 nucleotides to the angiomotin-like gene AMOTL1. Thus each long dsRNA region has the potential to produce the same off-target effects as one of the three synthetic siRNAs, plus additional off-target effects due to cross-hybridization to a subset of these ten other genes.
Figure 2.
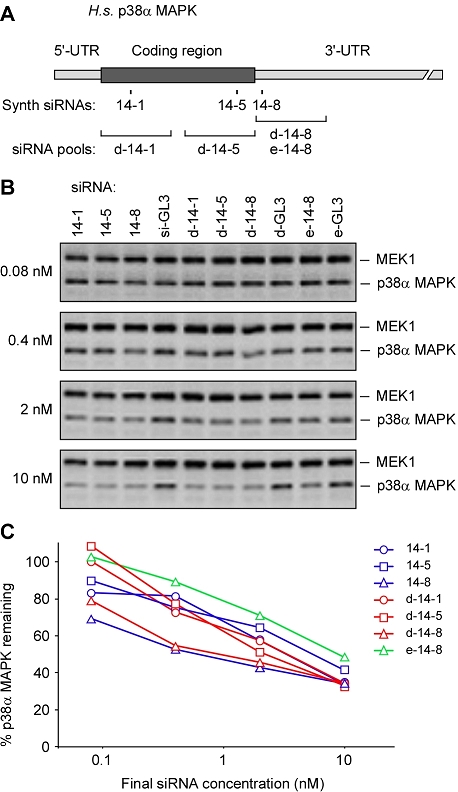
Silencing p38α MAPK expression with synthetic siRNAs, d-siRNAs, and e-siRNAs. (A) Schematic of the p38α MAPK cDNA and the sequences targeted by the siRNAs, d-siRNAs, and e-siRNA employed here. The lengths of the cDNA, siRNAs, and pools are drawn to scale. (B) Dose-dependent decreases in p38α MAPK protein levels in HeLa cells transfected with siRNAs, d-siRNAs, and e-siRNAs. Equivalent amounts of whole cell lysates were subjected to SDS-PAGE and immunoblotted with MEK1 antiserum (a loading control) or a p38α MAPK antibody. (C) Band intensity was quantified, normalized to the respective control (a GL3 luciferase-derived siRNA as the synthetic control, and Dicer- or RNase III-generated luciferase d-siRNAs/e-siRNAs as the other controls), and plotted as dose/inhibition curves.
Dicer produced a mix of diced siRNAs (d-siRNAs) of approximately 21-22 bp, whereas RNase III produced a mix of siRNAs (e-siRNAs) with a more heterogenous size distribution (15-30 bp) (data not shown). Various concentrations of the synthetic siRNAs and these d-siRNA/e-siRNA pools were transfected into HeLa cells. As controls, we transfected a synthetic GL3 luciferase siRNA (si-GL3), an in vitro diced GL3 control (d-GL3), or an RNase III-generated GL3 control (e-GL3). Cells were lysed 48 hr after transfection and lysates were immunoblotted with antibodies against p38α MAPK and MEK1, a constant-abundance protein of similar molecular mass used as a loading control.
As shown in Figures 2B and C, the potencies of the various siRNAs and siRNA pools were similar, with synthetic siRNA 14-8 and diced pool d-14-8 being the most potent and RNase III-treated pool e-14-8 being the least potent. At a final concentration of 10 nM the various siRNAs reduced p38α MAPK protein levels by ∼50-70%. Since this amount of silencing was comparable to those attained in previous studies of siRNA off-target effects (Jackson et al, 2003) and higher siRNA doses may be problematic (Semizarov et al, 2003), we decided to assess the off-target profiles of all of the siRNAs and pools at this concentration.
Global analysis of mRNA changes
HeLa cells were transfected in triplicate with p38α MAPK siRNAs (14-1, 14-5 or 14-8), d-siRNAs (d-14-1, d-14-5 or d-14-8), e-siRNAs (e-14-8), or control luciferase-derived siRNAs. Lysates were aliquoted for immunoblot analysis (for p38α MAPK, MEK1, and eIF2α, a marker of non-specific repression of translation) and for global analysis of mRNA levels. Each of the p38α MAPK siRNAs and pools produced a ∼50-70% decrease in p38α MAPK protein levels and a ∼30-40% decrease in p38α MAPK mRNA levels (Figure 3A). No significant changes in p38α MAPK protein or mRNA levels were seen in the control-treated cells (Figure 3A). None of the treatments elevated the levels of eIF2α phosphorylation (Figure 3A), arguing that the siRNA-induced decreases in p38α MAPK levels were not due to phospho-eIF2α-mediated global inhibition of translation.
Figure 3.
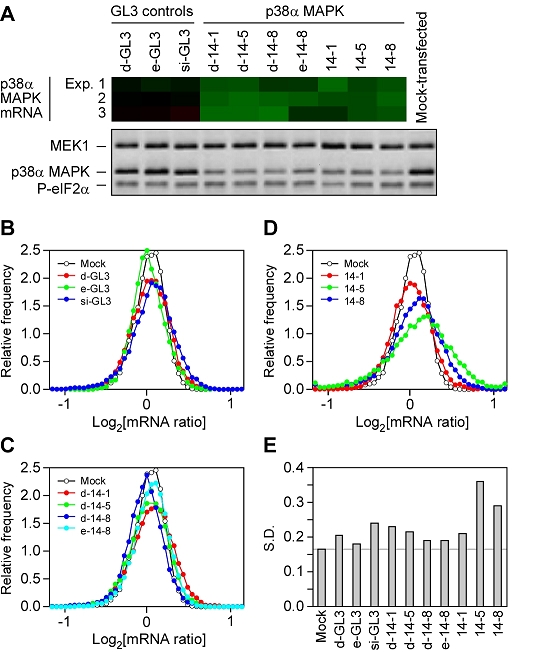
Global changes in gene expression in siRNA-treated cells. (A) Decreases in p38α MAPK mRNA and protein levels. mRNA levels were measured by microarray analysis and data from three experiments are shown. Protein levels were also assessed in triplicate; one representative blot, showing p38α MAPK levels, MEK1 levels, and eIF2α phosphorylation, is shown here. (B-D) Distributions of mRNA levels. (E) Standard deviations of the mRNA distributions.
In the microarray analysis, a total of 34,944 spots were considered well-measured (green channel mean intensity >2-fold over green channel median background intensity; red channel mean intensity >2-fold over red channel median background intensity; regression correlation between red and green pixels, for each spot, >0.6). Of these, 9538 spots, corresponding to 7587 different genes, were measured in all 30 arrays (10 siRNA treatments assessed in triplicate). We also carried out a triplicate analysis of mock-transfected cells in a separate experiment, with 7257 out of the 9538 mRNA spots considered well-measured in all three replicates. As shown in Table 2 and Figure 3B-E, the mock-transfected cells showed little variation in mRNA levels. None of the 7257 mRNAs changed by more than 2-fold (Table 2), and the log-ratio distribution showed a narrow, symmetrical peak centered close to zero (Figure 3B-D). The standard deviation of the log-ratio distribution was 0.17, which means that 68% of the mRNAs were measured at between 89% and 112% of their baseline levels. This can be regarded as the intrinsic variability of the mRNA isolation and microarray quantification procedures.
Table 2.
Global changes in mRNA levels in cells treated with diced siRNAs, RNase-III siRNAs and synthetic siRNAs
| GL3 controls | Diced siRNAs | RNase III | Synthetic siRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mock | d-GL3 | e-GL3 | si-GL3 | d-14-1 | d-14-5 | d-14-8 | e-14-8 | 14-1 | 14-5 | 14-8 | |
| Number of mRNAs that changed more than 2-fold | 0 | 0 | 0 | 13 | 5 | 4 | 1 | 0 | 1 | 123 | 57 |
| Number of mRNAs that decreased more than p38α MAPK | 1820 | 3653 | 3330 | 3329 | 32 | 23 | 18 | 93 | 47 | 428 | 171 |
| Number of mRNAs that changed significantly by SAM | - | 0±0 | 4±1 | 33±1 | 7±1 | 9±2 | 12±1 | 11±4 | 35±1 | 1105±1 | 322±1 |
A total of 9538 mRNA spots (corresponding to 7587 different genes) were considered well-measured in all 30 siRNA-treated samples. Of those, 7257 spots were also well-measured in the three mock-transfected samples. The ranges shown for the number of mRNAs that changed significantly by SAM are calculated from the estimated false discovery rate.
All of the cells treated with siRNAs showed somewhat greater variation in mRNA levels, as indicated by a slight broadening of the log-ratio distribution (Figure 3B-D) and an increase in the distributions' standard deviations (Figure 3E). These changes were most noticeable in the cells treated with two of the synthetic siRNAs, 14-5 and 14-8 (Figure 3D-E and Table 2). Whereas none of the mRNAs changed by more than 2-fold in the mock-treated cells and the cells treated with control d-GL3 and e-GL3 siRNA pools, a total of 123 mRNA spots out of 9538 changed by more than 2-fold in the 14-5-treated cells (Table 2). Moreover, 428 mRNAs out of 9538 were suppressed to a greater extent than the intended target, p38? MAPK (Table 2). Synthetic siRNA 14-8 induced fewer, but still substantial, changes in mRNA levels; 57 mRNAs changed by more than 2-fold and 171 mRNAs decreased more than p38α MAPK.
The remaining synthetic siRNA (14-1) and all of the diced siRNAs exhibited minimal global changes in gene expression. The number of mRNAs suppressed more than p38α MAPK ranged from 18 (for diced siRNA d-14-8) to 47 (for synthetic siRNA 14-1) out of 9538, and the number of mRNAs varying more than 2-fold ranged from one to five (Table 2). Thus, in comparison to siRNAs 14-5 and 14-8, the changes in mRNA levels produced by the diced pools were minimal. The RNase III-generated siRNA pool (e-14-8) had mild off-target effects (no mRNAs changed by more than 2-fold), but because it was the least effective at silencing the target p38α MAPK gene, a relatively high number of mRNAs were suppressed more than the target was (93 out of 9538; Table 2).
Secondary effects and off-target effects
Some of the changes in mRNA levels could be secondary to the suppression of p38α MAPK. Such secondary effects would be expected to occur in all of the samples treated with p38α MAPK siRNAs/d-siRNAs/e-siRNAs, and not in the samples treated with luciferase siRNAs/d-siRNAs/e-siRNAs. To identify potential secondary changes, we divided the data into two classes, with the d-GL3-, e-GL3-, and si-GL3-treated cells (9 experiments) constituting one class and the d-14-1-, d-14-5-, d-14-8-, e-14-8-, 14-1-, 14-5-, and 14-8-treated cells (21 experiments) constituting the other. We then applied the significance analysis of microarrays (SAM) method (Tusher et al, 2001), a modified t-test that compares how much an mRNA changes between the two classes to its variability within each class, thereby identifying mRNAs whose between-class changes are statistically significant (Tusher et al, 2001). The mRNAs identified by SAM should include the intended target mRNA (p38α MAPK) and any secondary mRNAs.
Figure 4A shows the mRNAs that decreased (top) and increased (bottom) significantly, where we took a maximum SAM q-value of 25% as our cutoff for significant changes. The mRNAs identified included the intended target, p38α MAPK, two other suppressed mRNAs and five induced mRNAs (Figure 4A). At least two of the mRNAs (ARK5 and CREB3L1) are connected to the p38α MAPK pathway in ways that suggest possible feedback roles (positive and negative, respectively). All in all, the number of candidate secondary mRNAs was small, amounting to 7 out of the 9538 mRNAs deemed well-measured.
To identify siRNA-specific off-target effects, we performed ten SAM analyses, with each treatment (3 experiments) compared to all other treatments (27 experiments) and the p38α MAPK mRNA and the putative secondary mRNAs excluded from consideration. The results are shown in Table 2 and Figure 4B. One of the treatments, the d-GL3 control, produced no significant changes in mRNA levels. Moreover, none of the d-siRNAs or e-siRNAs caused more than twelve significant changes (Table 2). None of the ten mRNAs mentioned above as potentially cross-hybridizing to one or more of the siRNA pools (p38β MAPK, p38γ MAPK, p38δ MAPK, MLL, ALPI, MP3K7, ERK1, AMOTL1, CD44, and FLJ31204) was found to change significantly.
In contrast, the synthetic siRNAs affected larger numbers of mRNA levels. The si-GL3 control produced 33 significant changes and the p38α MAPK siRNAs produced 35 changes (14-1), 1105 changes (14-5), and 322 changes (14-8). There was a small degree of overlap between the mRNAs affected by 14-5 and 14-8, but for the most part the profiles of mRNAs were distinct, arguing that the effects were specific to particular siRNA sequences rather than shared by synthetic siRNAs in general. For those features we found significant by SAM, there was a reasonable correlation between the changes in mRNA abundance seen here and those reported previously (Jackson et al, 2003), with Pearson correlation coefficients of 0.64, 0.54, and 0.71 for 14-1, 14-5, and 14-8 respectively. Note that different luciferase-derived siRNAs were used in the two studies and therefore results were not compared.
To test the idea that off-target silencing of the siRNAs was due to short stretches of sequence identity with the off-target mRNAs, we characterized the degree of identity between the four synthetic siRNAs and the 173 most highly-suppressed off-target mRNAs. Almost all of the suppressed mRNAs possessed a stretch of at least 6 nucleotides of identity (97%) with the relevant siRNA, and almost none of the suppressed mRNAs possessed a stretch of 11 or more (1%), with an average of 7.4 + 1.1 (S.D.) consecutive nucleotides. This is consistent with the possibility that short stretches of identity mediate the off-target effects. However, a control group of 172 unaffected mRNAs showed the same extent of identity to the siRNAs: 98% had a stretch of at least 6 nucleotides, 1% had a stretch of 11 or more, and the average was 7.3 ± 1.1 (S.D.). Thus, as a group, the suppressed mRNAs were no more likely to possess a long stretch of sequence identity with the relevant siRNAs than the unaffected mRNAs were.
Others have also recently shown that exact or near-exact matches between either strand of an siRNA and off-target mRNAs could not explain the unintentional reduction in nontargeted mRNAs (Birmingham et al, 2006). Instead one or more exact matches between the hexamer or hepatamer seed region of the siRNA (positions 2-7 or 2-8 from the 5′-end) and the 3′-UTR of the off-target mRNAs occurred at a greater frequency than matches between the seed region and the 3′-UTR of untargeted mRNAs. Using the Birmingham et al. siRNA seed locator (Birmingham et al, 2006), we found that this model can explain at least some of the off-target changes we observed. Treatment with 14-5 and 14-8 reproducibly decreased the abundance of 239 and 216 different transcripts, respectively (Table 3). 42 of the 239 transcripts unintentionally reduced by 14-5 contained one seed match in the 3′-UTR whereas only 19 of 240 randomly chosen transcripts contained a seed match (P < 0.01 using χ2 test). Of these 42 mRNAs, 10 were present in a previous study of the effects of 14-5 (Jackson et al, 2003), and 7 of the 10 decreased in that study as well as ours (Supplementary Figure 1). Additionally, 47 of the 216 mRNAs reduced by 14-8 contained one seed match in the 3′-UTR in contrast to 20 of the 240 untargeted transcripts (P < 0.001 using χ2 test). Of these 47 mRNAs, 31 were present in a previous study of the effects of 14-8 (Jackson et al, 2003), and 20 of the 31 decreased in that study as well as ours (Supplementary Figure 1). Treatment with the GL3 and 14-1 siRNAs reproducibly decreased the abundance of smaller numbers of transcripts, 33 and 35 respectively (Table 3). In each case three of these untargeted transcripts contained seed matches, which is not significantly different from the number of matches in 33 or 35 random sequences. Finally multiple seed matches in the 3′-UTRs of off-target transcripts were enriched relative to untargeted transcripts only in the cells treated with 14-8 siRNA (Table 3).
Table 3.
siRNA seed matches to 3′-UTR can explain some but not all siRNA induced off-target effects.
| siRNA | ||||
|---|---|---|---|---|
| si-GL3 | 14-1 | 14-5 | 14-8 | |
| Number of single seed matches to 3′UTR human genome | 1187 | 1463 | 4460 | 4967 |
| Number of multiple seed matches to 3′UTR human genome | 55 | 124 | 927 | 1012 |
| Number of mRNAs that were significantly reduced (SAM) | 34 | 35 | 239 | 216 |
| Number of SAM mRNAs with single seed match | 3 | 3 | 42 | 47 |
| Number of random mRNAs with single seed match | 1 | 1 | 19 | 20 |
| P-value (χ2 test) | 1 | 1 | 0.01 | 0.001 |
| Number of SAM mRNAs with multiple seed match | 0 | 0 | 10 | 16 |
| Number of random mRNAs with multiple seed match | 0 | 0 | 7 | 4 |
| P-value (χ2 test) | 1 | 1 | 1 | 0.01 |
In summary, a range of off-target effects were seen with the synthetic siRNAs; two had relatively minor effects (the si-GL3 control and 14-1) and two had marked effects (14-5 and 14-8). These findings agree qualitatively with previously published findings (Jackson et al, 2003). In contrast, all of the d-siRNA and e-siRNA pools examined here were nearly free of off-target effects.
Diluting siRNAs into a d-siRNA pool minimizes off-target effects
We hypothesized that the d-siRNA and e-siRNA pools had such minimal off-target effects because each unique siRNA within a pool is present at a low absolute concentration. If, for example, sequence-specific off-target effects are mediated by the binding of an siRNA to an eight-nucleotide stretch of mRNA sequence, only 15 out of the 529 possible 22-mers that could be derived from digestion of a 550 bp dsRNA (< 3% of the total) would be expected to also match that eight-nucleotide sequence. Thus at a total concentration of 10 nM d-siRNA, the concentration of siRNAs complementary to a given eight-nucleotide sequence would be less than 0.3 nM. This might be too low to result in detectable off-target effects. If more than eight consecutive nucleotides of identity are required for off-target effects, the absolute concentration of potentially cross-hybridizing 22-mers would be even lower.
To test these ideas, we added the synthetic siRNA with the most prominent off-target effects (14-5) to the corresponding diced pool (d-14-5) at dilutions of 1:3, 1:30, or 1:300, and then transfected these mixtures in triplicate into HeLa cells at a final total RNA concentration of 10 nM. We also added the 14-5 synthetic siRNA to a diced luciferase siRNA pool at the same dilutions. We then analyzed p38α MAPK levels, MEK1 levels, and eIF2α phosphorylation levels in all of the samples, and subjected cells transfected with 14-5, 14-5/d-14-5 mixtures, and various control siRNAs to microarray analysis.
Consistent with previous experiments (Figures 3 and 4), 14-5, 14-8 and d-14-5 were found to reduce p38α MAPK protein levels by 60-70% relative to treatment with d-GL3 or transfection reagent alone (Figure 5A). Similar silencing of p38α MAPK was seen in the samples treated with mixtures of 14-5 and d-14-5 (Figure 5A). In contrast, silencing was reduced when 14-5 was diluted with an irrelevant diced luciferase control (Figure 5A). The level of eIF2α phosphorylation was similar to mock transfected cells in all cases (Figure 5A).
Figure 5.
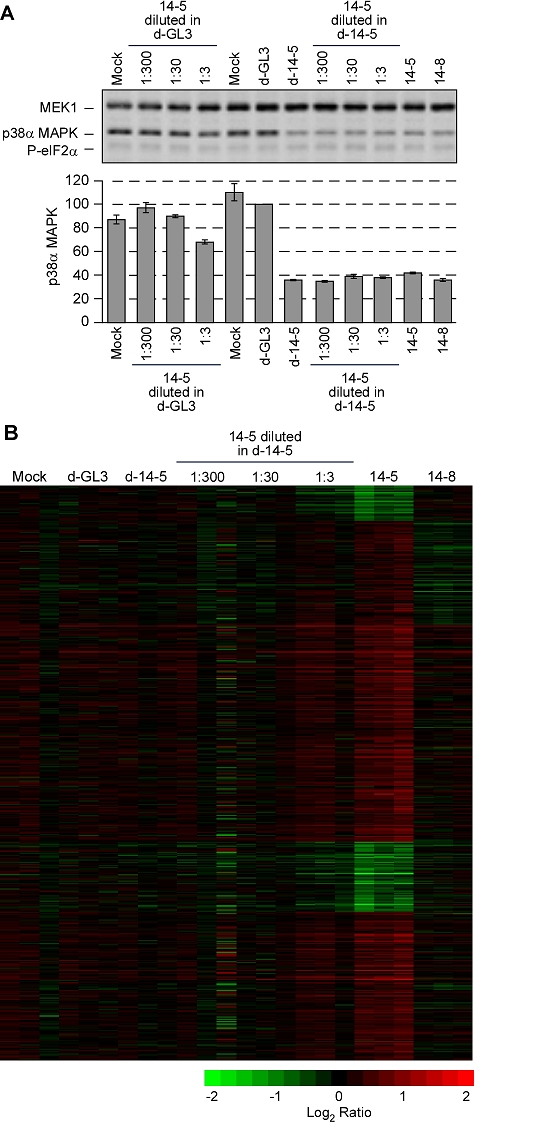
Off-target effects are titratable. (A) Changes in p38α MAPK protein levels. HeLa cells were transfected in triplicate with the siRNAs and pools shown, all at a final concentration of 10 nM. One representative immunoblot, showing MEK1 levels (loading control), p38α MAPK levels, and eIF2α phosphorylation is shown along with quantitative p38α MAPK data from all three replicates. Data are plotted as means ± S.D. (B) Fluorescence intensity data from the microarray experiments. The 1105 mRNAs identified as changing significantly in the SAM analysis of 14-5 vs. the other treatments (shown in Figure 4B) were used for the heat map shown here. Genes that were not well-measured in one or more samples are omitted from the heat map.
We then carried out microarray analysis and focused on the mRNAs whose levels had been previously shown to change significantly in the 14-5-treated samples (Figure 4B). The pattern of mRNA changes induced by 14-5 (Figure 5B) was qualitatively similar to that seen in Figure 4B. Dilution of the 14-5 siRNA by a factor of 3 with d-14-5 decreased, but did not eliminate, this pattern of off-target changes (Figure 5B) without decreasing the amount of on-target suppression the p38α MAPK mRNA (Figure 5A). Dilution of the 14-5 siRNA by a factor of 30 or 300 with d-14-5 rendered the off-target signature undetectable (Figure 5B), again without decreasing the amount of suppression of p38α MAPK. We noted that in one of the three 1:300 dilution experiments there were some changes in mRNA levels (Figure 5B), but the changes were irreproducible and distinct from the 14-5 off-target signature and therefore were attributed to experiment-to-experiment variation. Thus, the minimal off-target effects seen with diced siRNAs can be rationalized on the basis of dilution of individual problematic siRNAs to insignificant absolute concentrations.
DISCUSSION
Here we have confirmed previous studies (Jackson et al, 2003) showing that individual siRNAs can produce substantial off-target changes in mRNA abundance (Figures 4-5). Some of the observed decreases in mRNA abundance may be due to cross-hybridization to seed sequences in the off target mRNA 3′-UTRs (Table 3), as previously hypothesized (Birmingham et al, 2006). In support of this idea, the seed sequences from the more problematic individual siRNAs (14-5 and 14-8) had more matches to 3′-UTR sequences than did the least problematic individual siRNA (14-1). Other mechanisms might explain the observed increases in some mRNAs and the changes in the abundances of mRNAs that had no obvious cross-hybridization potential. Because these other mechanisms are incompletely understood, it is not possible to develop siRNA design algorithms that would ensure siRNA specificity.
Irrespective of the exact mechanism(s) of off-target effects of individual siRNAs, complex d-siRNA pools are essentially free of off-target effects (Figure 4), even when the diced pool is known to contain a problematic siRNA (Figure 5). The simplest explanation for the low off-target effects of d-siRNAs is that although an siRNA pool contains hundreds of different siRNAs (Figure 1), all of which would be expected to hybridize with the intended target, very few of them would be expected to cross-hybridize to any particular off-target gene or to exert some other sequence-specific off-target effect such as an aptamer effect. A 1:3 dilution of a problematic individual siRNA caused a decrease in the magnitude of its off-target effects, but higher dilutions were needed to eliminate them (Figure 5) suggesting that the complexity of pools is essential to attaining specific gene silencing.
The low incidence of off-target effects in HeLa cells treated with diced siRNAs agrees well with the general impression of high specificity in C. elegans and Drosophila gene silencing, where large dsRNAs, which act after being converted to complex pools of siRNAs by the endogenous Dicer protein, are routinely used. The minimal off-target effects of d-siRNAs make them a particularly attractive method for the transient suppression of gene expression in applications like large scale screening experiments where it might not be feasible to assess the off-target effects of every siRNA or siRNA pool used.
Finally, it should be emphasized that even when an siRNA or siRNA pool produces no off-target changes in mRNA abundance, it is still possible that it will affect the translation of off-target mRNAs. Thus, although freedom from off-target mRNA effects is certainly desirable for gene silencing experiments, it is not sufficient to guarantee specificity. The use of multiple siRNAs or siRNA pools, and the rescue of an RNAi-induced phenotype with an RNAi-resistant construct (e.g., a 3′-UTR-directed d-siRNA pool (like d-14-8) combined with a rescue construct lacking the 3′-UTR) remain important approaches for establishing the specificity of an RNAi-induced phenotype.
CONCLUSIONS
Individual siRNAs can produce substantial off-target changes in mRNA abundance as a result of cross-hybridization between the seed region of the siRNA and the 3′UTR of the off-targeted transcript.
Other cross-hybridization independent mechanisms might explain the increases in off-target mRNA abundance but are incompletely understood and therefore cannot necessarily be avoided by adding a feature to siRNA design algorithms.
Complex d-siRNA pools are essentially free of off-target effects even when the diced pool is known to contain a problematic siRNA.
The minimal off-target effects of d-siRNAs make them a particularly attractive method for large scale, loss-of-function screening experiments.
Supplementary Table 1. Sequences of cloned siRNAs
| Nucleotide Position | Sequence | Times isolated |
|---|---|---|
| 828 | GTTGTTGCAGGAGACCATGT | 1 |
| 853 | ACTGTCTCCATTATTGATCGG | 1 |
| 865 | ATTGATCGGTTCATGCAGA | 1 |
| 868 | GATCGGTTCATGCAG | 1 |
| 887 | ATTGTGTGCCCAAGAAGATGC | 1 |
| 893 | TGCCCAAGAAGATGCTGCAGCT | 1 |
| 897 | CAAGAAGATGCTGCAGCTGGTT | 1 |
| 906 | GCTGCAGCTGGTTGGT | 3 |
| 909 | GCAGCTGGTTGGTGTCACT | 1 |
| 916 | GTTGGTGTCACTGCCATGTTT | 1 |
| 953 | AAGAAATGTACCCTCCA | 1 |
| 981 | CTTTGCTTTTGTGACTGACAA | 1 |
| 982 | TTTGCTCTTGTGACTGACAACA | 1 |
| 986 | CTTTTGTGACT | 1 |
| 1028 | AGATGGAAATGAAGGTTCTAA | 1 |
| 1048 | AGAGCTCTAAACTTTGGTCT | 1 |
| 1058 | ACTTTGGTCTGGGTCGGCC | 1 |
| 1061 | TTGGTCTGGGTCGGCCTCTAC | 2 |
| 1066 | CTGGGTCGGCCTCTACCTTTG | 2 |
| 1067 | TGGGTCGGCCTCTACCTTTGC | 1 |
| 1074 | GCCTCTACCTTTGCACTTCCT | 1 |
| 1103 | CATCTAAGATTGGAGAGGTTG | 1 |
| 1105 | TCTAAGATTGGAGAGGTTGA | 1 |
| 1105 | TCTAAGATTGGAGAGGTTGATG | 1 |
| 1114 | GGAGAGGTTGATGTCGAGC | 1 |
| 1145 | CCAAATACCTGATGGAA | 1 |
| 1152 | CCTGATGGAACTAACTATG | 1 |
| 1169 | TGTTGGACTATGACATGGTGC | 1 |
| 1176 | CTATGACATGGTGCACTTTCC | 1 |
| 1185 | GGTGCACTTTCCTCCTTCTCA | 1 |
| 1190 | ACTTTCCTCCTTCTCAAATTG | 2 |
| 1192 | TTTCCTCCTTCTCAAATTGCAG | 1 |
| 1194 | TCCTCCTTCTCAAATTGCAGCA | 1 |
| 1195 | CCTCCTTCTCAAATTGCAGCA | 2 |
| 1197 | TCCTTCTCAAATTGCAGCAGG | 1 |
| 1198 | CCTTCTCAAATTGCAGCAGG | 1 |
| 1198 | CCTTCTCAAATTGCAGCAGGA | 1 |
| 1210 | GCAGCAGGAGCTTTTTGCTTA | 1 |
| 1219 | GCTTTTTGCTTAGCACT | 1 |
| 1220 | CTTTTTGCTTAGCACTGAAAA | 1 |
| 1256 | AATGGACACCAACTCTACAAC | 1 |
| 1300 | TCTCTTCTTCCAGTTATGCAG | 1 |
| 1335 | TGTAGTCATGGTAAATCAAGGA | 1 |
| 1342 | ATGGTAAATCAAGGA | 1 |
| 1390 | GCCACATNGAAGCATGC | 1 |
| 1398 | GAAGCATGCTAAGATCAGC | 2 |
| 1413 | CAGCACTCTACCACAGCTGA | 1 |
| 1447 | CAAGATTTAGCCAAGGCTGTGG | 1 |
| 1448 | AAGATTTAGCCAAGGCTGTGG | 3 |
| 1456 | GCCAAGGCTGTGGCAAAGGTG | 1 |
| 1458 | CAAGGGTGTGGCAAAG | 1 |
Nucloetide position is relative to start codon as defined by human cyclin B1 sequence (NM 031966).
Supplementary Figure 1. Comparison of off-target effects to previous data. mRNAs whose abundance decreased and had a match in their 3′-UTR to the seed region of a particular siRNA were selected from our data and previous data and are represented in the form of a heat map (Jackson et al 2003).
Acknowledgments
This work was supported by NIH grants GM46383 (to J.E.F.) and CA77097 (to P.O.B.). P.O.B. is an investigator of the Howard Hughes Medical Institute. We thank Gil Chu for helpful discussions, and Delquin Gong, Josh Jones, Jason Casolari, Daniel Riordan, Zach Serber, Jeff Ubersax, and Tom Wehrman for comments on the manuscript. We thank the Stanford Microarray Database and Janos Demeter for enabling us to better analyze microarray data.
LIST OF ABBREVIATIONS
- d-siRNA
Diced siRNAs
- e-siRNA
RNase III-generated siRNAs
- MAPK
Mitogen-activated protein kinase
STATEMENT OF COMPETING INTERESTS
J.W.M. and J.E.F., Jr. declare financial competing interest. Recombinant Dicer is sold by several private companies in concordance with an agreement between the companies and J.W.M., J.E.F., Jr. and Stanford University.
REFERENCES
- Ashrafi K, Chang FY, Watts JL, et al. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, et al. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- Cherry S, Doukas T, Armknecht S, et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JT, Chang HY, Wang NN, Chang DS, Dunphy N, Brown PO. Genomewide view of gene silencing by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100:6343–6346. doi: 10.1073/pnas.1037853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi JT, Wang Z, Nuyten DS, et al. Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta R, Perrimon N. Using RNAi to catch Drosophila genes in a web of interactions: insights into cancer research. Oncogene. 2004;23:8359–8365. doi: 10.1038/sj.onc.1208028. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert US, Kiger AA, Richter C, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Thonberg H, Ljungberg K, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink CC, Bayer K-U, Myers JW, Ferrell JE, Jr., Schulman H, Meyer T. Selective regulation of neurite extension and synapse formation by the b but not the a isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Holen T, Moe SE, Sorbo JG, Meza TJ, Ottersen OP, Klungland A. Tolerated wobble mutations in siRNAs decrease specificity, but can enhance activity in vivo. Nucleic Acids Res. 2005;33:4704–4710. doi: 10.1093/nar/gki785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KM, Chou SY, Shih SJ, Ferrell JE., Jr. Evidence that inactive p42 mitogen-activated protein kinase and inactive Rsk exist as a heterodimer in vivo. Proc Natl Acad Sci U S A. 1994;91:5480–5484. doi: 10.1073/pnas.91.12.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jones JT, Myers JW, Ferrell JE, Meyer T. Probing the precision of the mitotic clock with a live-cell fluorescent biosensor. Nat Biotechnol. 2004;22:306–312. doi: 10.1038/nbt941. [DOI] [PubMed] [Google Scholar]
- Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kariko K, Bhuyan P, Capodici J, et al. Exogenous siRNA mediates sequence-independent gene suppression by signaling through toll-like receptor 3. Cells Tissues Organs. 2004a;177:132–138. doi: 10.1159/000079987. [DOI] [PubMed] [Google Scholar]
- Kariko K, Bhuyan P, Capodici J, Weissman D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J Immunol. 2004b;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- Kawasaki H, Suyama E, Iyo M, Taira K. siRNAs generated by recombinant human Dicer induce specific and significant but target site-independent gene silencing in human cells. Nucleic Acids Res. 2003;31:981–987. doi: 10.1093/nar/gkg184. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kiger A, Baum B, Jones S, et al. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Heninger AK, Franke K, Habermann B, Buchholz F. Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat Methods. 2005;2:779–784. doi: 10.1038/nmeth1005-779. [DOI] [PubMed] [Google Scholar]
- Kittler R, Putz G, Pelletier L, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, et al. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou J, Kim ML, Heo WD, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Myers JW, Ferrell JE., Jr. In: Appasani K (Eds) RNA interference technology: from basic biology to drug development. New York, NY: Cambridge University Press; 2005a.. Dicer in RNAi: its roles in vivo and utility in vitro. [Google Scholar]
- Myers JW, Ferrell JE., Jr. In: Carmichael G (Eds) RNA Silencing: Methods and Protocols. Totowa, NJ: Humana Press; 2005b. Silencing gene expression with Dicer-generated siRNA pools. [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE., Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nature Biotechnology. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Silva JM, Conklin DS, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) Rna. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothof J, van Haaften G, Thijssen K, et al. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. Embo J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S, Adema CM, Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semizarov D, Frost L, Sarthy A, Kroeger P, Halbert DN, Fesik SW. Specificity of short interfering RNA determined through gene expression signatures. Proc Natl Acad Sci U S A. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Mizuno H, Brady A, Lucito R, Hannon GJ. RNA interference microarrays: high-throughput loss-of-function genetics in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:6548–6552. doi: 10.1073/pnas.0400165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, Van Der Linden AM, et al. Genome-Wide RNAi of C. elegans Using the Hypersensitive rrf-3 Strain Reveals Novel Gene Functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M, Sorensen DR. Cationic liposome-mediated delivery of siRNAs in adult mice. Biochem Biophys Res Commun. 2003;312:1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Fischer SE, Robert VJ, et al. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr Biol. 2003;13:1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]


