Abstract
Both ovarian and pituitary hormones are required for the pubertal development of the mouse mammary gland. Estradiol directs ductal elongation and branching, while progesterone leads to tertiary branching and alveolar development. The purpose of this investigation was to identify estrogen-responsive genes associated with pubertal ductal growth in the mouse mammary gland in the absence of other ovarian hormones and at different stages of development. We hypothesized that the estrogen-induced genes and their associated functions at early stages of ductal elongation would be distinct from those induced after significant ductal elongation had occurred. Therefore, ovariectomized prepubertal mice were exposed to 17β-estradiol from two to twenty-eight days, and mammary gland global gene expression analyzed by microarray analysis at various times during this period. We found that: a) gene expression changes in our estrogen-only model mimic those changes that occur in normal pubertal development in intact mice, and b) both distinct and overlapping gene profiles were observed at varying extents of ductal elongation, and c) cell proliferation, the immune response, and metabolism/catabolism were the most common functional categories associated with mammary ductal growth. Particularly striking was the novel observation that genes active during carbohydrate metabolism were rapidly and robustly decreased in response to estradiol. Lastly, we identified mammary estradiol-responsive genes that are also co-expressed with Estrogen Receptor α in human breast cancer. In conclusion, our genomic data support the physiological observation that estradiol is one of the primary hormonal signals driving ductal elongation during pubertal mammary development.
Keywords: microarray, genomic, mouse, estrogen receptor
Introduction
Estrogens regulate physiological processes in various tissues, including but not limited to the uterus, mammary gland, bone, prostate, colon, lung, heart and brain. The mammary gland has long been recognized as an estrogen-responsive tissue, and estrogen is implicated in the development of breast cancer, based on data from both clinical and animal studies. While estrogens are not required for the prenatal development of the rodent mammary gland, they are required to initiate its development during puberty (Fisher et al. 1998), and in the adult for ductal maintenance and the minor changes in side branching that occur during estrus (Hennighausen and Robinson 2005). Prior to birth, a rudimentary ductal structure consisting of epithelial cells forms, and this structure remains quiescent until puberty, at which time terminal end buds appear at the tips of growing ducts, and begin ductal elongation through the stroma, which primarily consists of adipose and connective tissue. 17β-Estradiol (Fisher et al. 1998) or Estrogen Receptor alpha (ERα) (Bocchinfuso et al. 2000) is critical for this outgrowth of mammary ducts, and ovariectomy of adult female mice causes regression of terminal end buds (Daniel et al. 1987), which can be restored by estradiol treatment.
Estradiol stimulates a number of physiological responses in the mammary gland. These include DNA synthesis in mammary epithelial cells and proliferation of stromal cells (Shyamala and Ferenczy 1984). In mammals, estrogens also increase mammary gland glucose metabolism (Shyamala and Ferenczy 1982), progesterone receptor levels (Toft and O’Malley 1972; Xu et al. 2005; Zucchi et al. 2004), lipid synthesis, histamine release (Shelesnyak 1959; Zeppa and Womack 1962), and immune cell recruitment (Gouon-Evans et al. 2002; Kayisli et al. 2004). Given the well-documented role that estrogen plays in the progression of human breast cancer, the identification of estradiol-regulated genes in developing mouse mammary gland may allow for the identification of novel effectors of estrogen action and/or biomarkers for the progression of estrogen-dependent cancers and other estrogen-driven diseases.
Estradiol treatment can rescue mammary development in ovariectomized, but not hypophysectomized rodents, indicating a role for both estradiol and pituitary hormones in mammary morphogenesis. It is known that insulin-like growth factor-1 (IGF-1) can have a minor independent effect on mammary development, but estradiol is required for full ductal elongation (Ruan et al. 1995). In the female mouse, both estrogen and progesterone contribute to mammary gland development, although estrogen is generally considered to be responsible for ductal outgrowth and minor branching, while progesterone promotes tertiary branching and alveolar development that completes the ductal tree. The set of genes expressed during pubertal development of the mammary gland in the intact mouse has been published (Master et al. 2002; McBryan et al. 2007), although the differential effects of estrogen and progesterone were not distinguished. Therefore, to identify estradiol-regulated genes during pubertal mammary development, we have mimicked the normal pubertal developmental process by using a model in which ovariectomized prepubertal mice are treated with estradiol.
Therefore, we ovariectomized prepubertal mice and exposed them to estradiol for up to four weeks to allow ductal elongation through the mammary fat pad. We investigated the gene expression profiles by microarray analysis as early as two days after treatment and as late as 28 days after, at a time when up to 70% of the fat pad was filled with ducts. Our analysis indicates that the genes regulated by estradiol in early stages of ductal growth include those that are unique to these early stages as well as others that are present during the entire four weeks of treatment. Functional analysis of estrogen-regulated genes indicated that metabolism, cell proliferation, and immune function were consistently represented at all developmental time-points, while some functional groups were unique to early (2-5 days estradiol) or later (14-28 days estradiol) stages of ductal growth. Finally, we identified genes regulated by estrogen in the mouse mammary gland that are genes know to co-express with ERα in breast tumor specimens, suggesting the possible involvement of these genes in estrogen-dependent breast cancer.
Results
Temporal genomic profiling of estradiol-induced mammary gland development
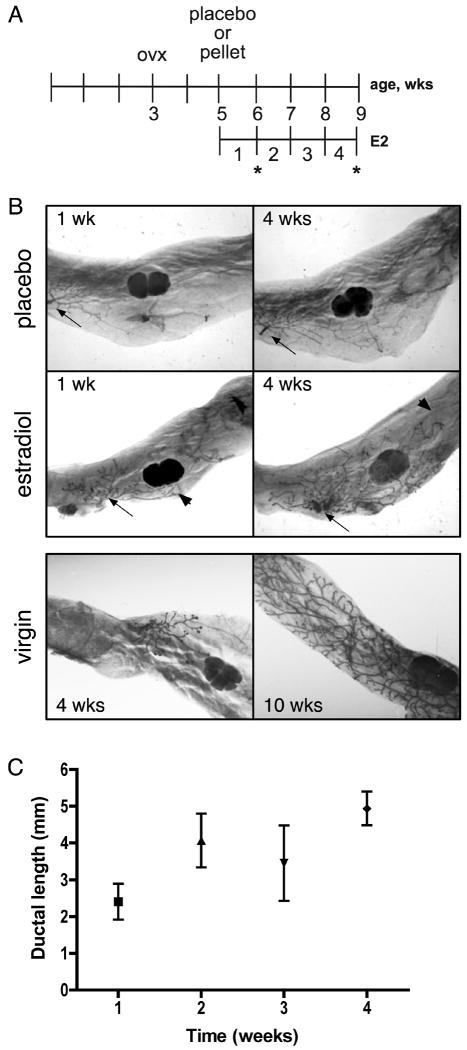
To identify estradiol-regulated genes involved in various stages of pubertal mouse mammary development, we used an ablation/replacement model (Flux 1954) in which prepubertal ovariectomized mice were exposed to estrogens to induce mammary gland growth. Mice were ovariectomized at 21 days old, allowed to rest for two weeks, and then placebo or 17β-estradiol pellets were implanted subdermally (Figure 1A). Our goal was to induce ductal elongation at approximately the same rate as would be observed in intact virgin mice. Normal ductal elongation requires approximately six weeks (between four and ten weeks of age) (Hennighausen and Robinson 1998). Our pilot studies (data not shown) indicated that approximately 75% of maximal ductal elongation occurred by four weeks of estradiol exposure—a rate that mimicked natural ductal growth. Based on these data, and because we were particularly interested in molecular events during early elongation, we selected 7, 14, and 28 days for our microarray analysis. At these time-points, mammary tissue was obtained for RNA isolation and the contralateral gland inspected by whole-mount analysis for ductal growth. Estradiol induced visual growth of the prepubertal epithelial ductal rudiment as early as 7 days of treatment, and this growth continued until approximately 50-70% of the fat pad was filled by 28 days of treatment (Figure 1B), a length of time which approximates the rate of ductal development in a normal virgin mouse (Figure 1B, lower panel), in which ductal growth begins around 28 days of age and ends by 10 weeks (Hennighausen and Robinson 1998). Ductal length was measured relative to the edge of the lymph node, and ductal length increased as the duration of estradiol treatment increased (Figure 1C). Ductal length in the placebo control throughout the time course was minimal (data not shown).
Figure 1.
Estradiol treatment of ovariectomized mice induces ductal growth and elongation in mouse mammary gland. A. Schematic of treatment strategy. Twenty-one-day old mice were ovariectomized, allowed to rest for two weeks, then exposed to either placebo or estradiol. Mammary glands were collected after two, five, seven, fourteen or twenty-eight days of treatment. Asterisks indicate the weeks represented in (B). B. (Upper panel) Whole-mounts of mammary glands from placebo or estradiol-treated mice after one or four weeks of treatment. (Lower panel) Whole-mounts of mammary glands from virgin mice at 4 and 8 weeks of age. Arrows indicate approximate origin of ductal growth. Arrowheads indicate limit of ductal growth. C. Ductal length in the mammary gland over the time course of estradiol treatment. Points represent the average ductal length measured from 3-5 mammary glands, ± SEM.
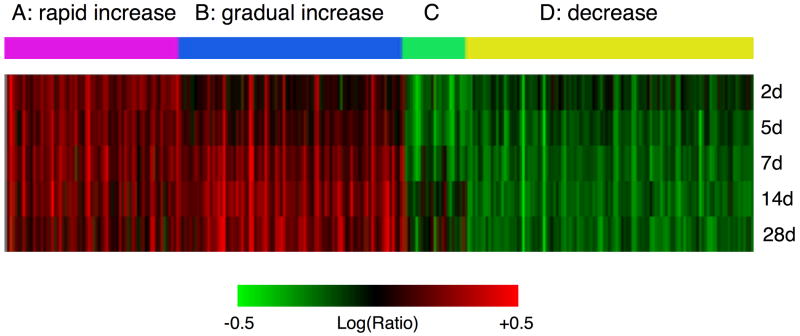
Mammary tissue was isolated from placebo and estradiol-treated mice, five mice per group, after 7, 14, and 28 days of treatment. RNA from individual mammary glands was isolated, pooled by group, prepared for microarray analysis, then labeled and hybridized pair-wise on multiple chip replicates including dye swaps (see Materials & Methods for details). Differentially expressed genes were clustered, with red and green representing up-regulated and down-regulated genes, respectively (Figure 2). Because many genes were regulated after 7 days of estradiol treatment, the mammary gene expression profile after 2 and 5 days of estradiol was obtained to determine at which point gene regulation could first be detected (Figure 2). As little as two days of estradiol treatment resulted in significant gene regulation. Interestingly, while overlap was observed between various time-points, genes unique to each time-point were also observed, indicating that the gene expression profile varied with the extent of estradiol exposure and ductal length.
Figure 2.
Clustering by self-organizing maps (SOM analysis) of mouse mammary gland gene expression after estradiol treatment. A. Red and green represent up- and down-regulated genes, respectively. Combined cluster representing gene expression changes from two, five, seven, fourteen or twenty-eight days of treatment with 17β-estradiol (p ≤ 0.001 in both biological replicates for d7, d14, and d28, or in the one biological replicate for d2 and d5). The 4-colour bar labeled A, B, C, and D indicates gene expression patterns identified by SOM analysis.
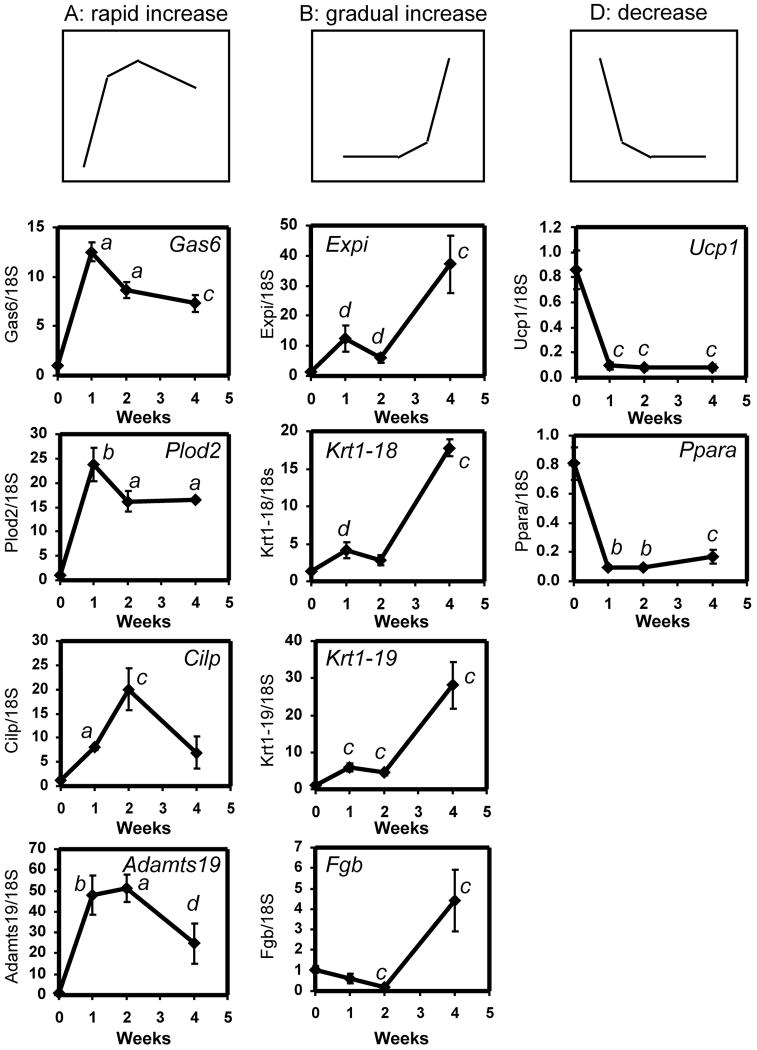
These variations in gene expression profile were represented by four gene expression patterns (three major, one minor) that were observed over the course of treatment as determined by clustering the data as Self-Organizing Maps (Figure 2 and 3, Patterns A, B, C, and D, and Supplementary Table 1). The three major patterns represented genes whose expression: A) increased rapidly after 7 days, then dropped slightly after 28 days of treatment (“rapid increase” genes), B) increased gradually at 7 days and 14 days, but then increased dramatically by 28 days (“gradual increase” genes), and D) decreased rapidly at 7 days, which continued up and to including 28 days (“decreased” genes). (Pattern C did not resolve into an identifiable pattern but represented a variety of patterns that did not fit into A, B, or D). Examples of genes in each of the three major patterns were chosen for confirmation (7, 14, and 28d time-points). Table 2 lists the fold gene expression changes of these genes as determined by microarray analysis, and Figure 3 shows the confirmation of these genes by quantitative PCR analysis. These genes were chosen based on their previously-established association with mammary development, involution, tumourigenesis, cellular proliferation, and/or regulation by estradiol. “Rapid Increase” genes (A): Growth arrest-specific 6 (Gas6) is mitogenic in a mammary cell line (Goruppi et al. 2001; Varnum et al. 1995) and its expression is regulated by estradiol in mouse mammary epithelial cells (Mo et al. 2007). The expression of procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (Plod2), which catalyzes the hydroxylation of lysyl residues in collagen-like peptides, increases during pubertal mammary development (McBryan et al. 2007) and is elevated in mammary tumors in WAP-SVT/t transgenic mice (Klein et al. 2005). The extracellular matrix protein, cartilage intermediate layer protein (Cilp) is upregulated by estradiol in ERα-expressing osteosarcoma cells (Klein et al. 2005). A disintegrin and metalloproteinase domain, with thrombospondin type-I modules 19 (Adamts19) is a member of the ADAMTS family of proteins whose expression is frequently dysregulated in human breast carcinoma (Porter et al. 2004). “Gradual increase genes” (B): Extracellular proteinase inhibitor (Expi) is upregulated during mouse mammary gland involution (Baik et al. 1998) and in mammary epithelium in a model of mammary epithelial preneoplastic progression (Aldaz et al. 2002). Keratin complex 1, acidic, gene 18 (Krt1-18) is a marker for ductal epithelial cells (Kouros-Mehr and Werb 2006; Rudland and Hughes 1989) and is present in the inner cells in benign breast lesions but comparatively fewer such cells in breast carcinoma (Rudland et al. 1993). Keratin complex 1, acidic, gene 19 (Krt1-19) is found in mammary TEBs and ducts (Kouros-Mehr and Werb 2006) and is a known estradiol-regulated gene (Frasor et al. 2004). Fibrinogens are extracellular matrix proteins that are found at the tumor-host interface in various tumor types, and their deposition in breast tumor stroma is a hallmark of breast carcinoma ((Rybarczyk and Simpson-Haidaris 2000) and references therein). Fibrinogen B beta polypeptide (Fgb) is induced in estradiol-treated MCF-7 breast cancer cells (Pentecost et al. 2005). Finally, genes whose expression decreased (D) included uncoupling protein 1 (Ucp1) and peroxisome proliferative activated receptor alpha (PPARa), both of which are downregulated during mammary gland pubertal development (Master et al. 2002). In each case, the trend observed by quantitative PCR confirmed the trend observed by microarray analysis (Figure 3 and Table 2).
Figure 3.
Confirmation by quantitative PCR of estrogen regulation of selected genes observed in microarray analysis (Table 2). RNA was prepared from one #4 mammary gland from each of 4-5 mice and cDNA was prepared and analyzed from individual mammary glands. Each graph was generated from one of two independent time-course experiments. Genes representative of each of the three major patterns identified by SOM clustering (A, B, and D) were confirmed. Results are expressed as the ratio of the variable gene to the 18S control, ± SEM. a, p < 0.0001 vs. placebo; b, p < 0.001 vs. placebo; c, p < 0.01 vs. placebo; d, p < 0.05 vs. placebo (unpaired, two-tailed student’s t-test).
Table 2.
Microarray data indicating fold gene expression changes of selected genes in mouse mammary gland after ovariectomized prepubertal mice were treated with estradiol for 1, 2 and 4 weeks (values of two independent experiments). These data were confirmed using quantitative PCR (Figure 3).
| Gene name | 1 wk (7d) | 2 wks (14d) | 4 wks (28d) | |||
|---|---|---|---|---|---|---|
| Adamts19 | 17 | 19 | 29 | 22 | 7.8 | 13 |
| Cilp | 13 | 13 | 21 | 18 | 6.3 | 7.3 |
| Expi | 5.0 | 7.2 | 4.5 | 4.9 | 5.7 | 7.9 |
| Fgb | -2.7 | -2.5 | -2.8 | -1.2 | 4.8 | 2.8 |
| Gas6 | 13 | 11 | 11 | 9.5 | 4.8 | 6.6 |
| Krt18 | 4.1 | 5.3 | 4.1 | 4.6 | 8.3 | 9.1 |
| Krt19 | 5.9 | 11 | 6.7 | 9.3 | 12 | 18 |
| Plod2 | 7.8 | 8.7 | 9.5 | 7.1 | 2.9 | 4.5 |
| Ppara | -3.8 | -3.9 | -4.5 | -4.6 | -3.8 | -4.4 |
| Ucp1 | -18 | -2.8 | -19 | -17 | -16 | -20 |
Validation of an estrogen-specific pubertal mammary gland outgrowth model
Genes shown previously to be regulated by estradiol or involved in mammary gland development were observed in our microarray analysis. These observations confirmed that estrogen responsiveness was represented in our model, and that genes previously associated with pubertal mammary development were also represented.
Confirmation of estrogen-regulated genes
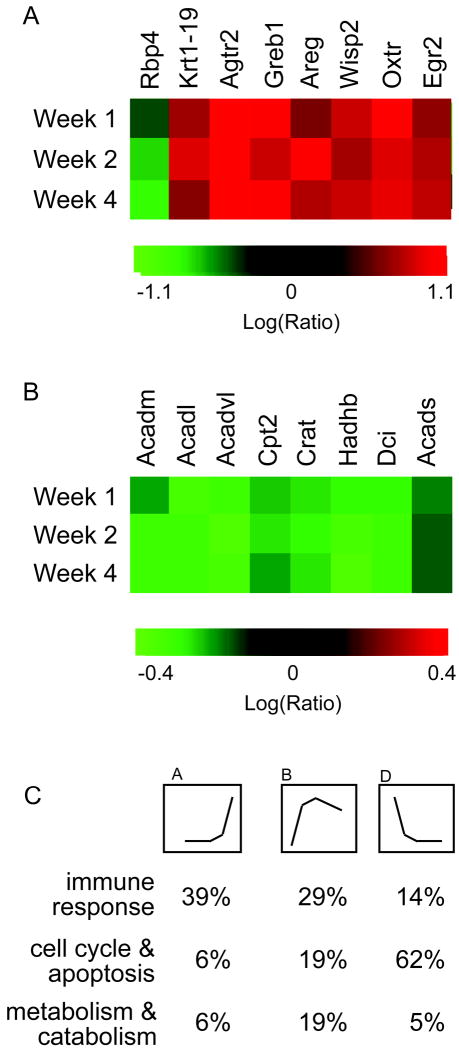
Known estrogen-regulated genes (Figure 4A) included: (but were not limited to): a. retinol binding protein 4, plasma (Rbp4) which was downregulated in our study, consistent with its downregulation by estradiol in the rat uterus (Bucco et al. 1996). In addition, seven genes previously shown to be upregulated by estradiol were also upregulated in our study: keratin complex 1, acidic, gene 19 (Krt1-19) (Choi et al. 2000) (Kian Tee et al. 2004), angiotensin II receptor, type 2 (Agtr2) (Ivanga et al. 2007), gene regulated by estrogen in breast cancer protein (Greb1) (Ghosh et al. 2000; Rae et al. 2005), amphiregulin (Areg) (Martinez-Lacaci et al. 1995), WNT1 inducible signaling pathway protein 2 (Wisp2) (Kian Tee et al. 2004), oxytocin receptor (Oxtr) (Bale and Dorsa 1997), and early growth response 2 (Egr2) (Pedram et al. 2002). We also compared our dataset to that of Frasor et al., who identified estradiol-regulated genes in a human MCF-7 breast cancer cell line (Frasor et al. 2004). Of the ~121 genes identified by Frasor et al. as estradiol-regulated in MCF-7 cells, we found that 39 of these (32%) (Table 3) were also regulated in mammary glands of estradiol-treated mice (3). Of these 39 genes also regulated by estradiol in our study, 15 (38.5%) were regulated in the same direction (either up- or down-regulated) as in MCF-7 cells. These differences in the direction of gene regulation between the two datasets, that is, whether gene expression increased or decreased, are likely due to differences in the model systems. These differences include species (mouse vs. human), disease state (normal mammary gland vs. breast adenocarcinoma), sample origin (whole organ (multiple cell types) vs. single (epithelial) cell type), and method of estradiol exposure (whole animal vs. in vitro culture).
Figure 4.
Microarray data includes expected genomic responses to estradiol in mouse mammary gland. A. Heat map showing microarray data of estrogen-regulated genes identified in other studies: keratin complex 1, acidic, gene 19 (Krt1-19), angiotensin II receptor, type 2 (Agtr2), gene regulated by estrogen in breast cancer protein (Greb1), retinol binding protein 4, plasma (Rbp4), amphiregulin (Areg), WNT1 inducible signaling pathway protein 2, oxytocin receptor (Oxtr), and early growth response 2 (Egr2). B. Expression of several genes involved in fatty acid metabolism is decreased. Medium chain acyl-CoA dehydrogenase (Acadm), Very-long-chain acyl-CoA dehydrogenase (Acadvl), Carnitine palmitoyltransferase II (Cpt2), Long chain acyl-CoA dehydrogenase (Acadl), 3-hydroxyacyl CoA dehydrogenase (Hadh), (Carnitine acetyltransferase (Crat), Dodecenoyl-CoA Δ-isomerase (Dci), and Short chain acyl-CoA dehydrogenase (Acads). C. Percent of genes associated with the immune response, cell cycle/apoptosis, and metabolism/catabolism grouped by gene expression pattern.
Table 3.
Genes regulated by estradiol in developing mouse mammary gland and in the estradiol-treated human breast cancer cell line, MCF-7 (Frasor et al. 2004).
| MCF-7 | Mammary gland | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Gene Description | 8h | 48h | 2d | 5d | 7d | 14d | 28d |
| ABCG1 | ATP binding cassette, subfamily G (WHITE), member 1 | 0.35 | 0.47 | - | - | 0.77 | 1.3 | - |
| ADCY9 | Adenylate cyclase 9 | 1.83 | 2.53 | - | - | - | 1.3 | - |
| ALDH4A1 | Aldehyde dehydrogenase 4, member A1 | 0.64 | 0.38 | - | 0.7 | - | - | 0.3 |
| ANXA3 | Annexin A3 | 0.71 | 0.28 | - | 1.3 | 2.3 | 2.4 | 1.7 |
| AREG | Amphiregulin | 6.81 | 7.49 | 4.7 | 5.0 | 11.3 | 6.8 | 10.9 |
| BIK | BCL2-interacting killer | 0.33 | 0.21 | - | - | 1.3 | - | 1.2 |
| CALCR | Calcitonin receptor | 7.45 | 7.91 | 1.8 | - | - | - | - |
| CBX6 | Chromobox homologue 6 | 0.69 | 0.31 | - | - | 1.1 | 1.2 | - |
| CD24 | CD24 antigen | 0.74 | 0.27 | - | - | 1.4 | - | 1.3 |
| CTSH | Cathepsin H | 0.78 | 0.34 | - | 1.5 | - | 1.8 | 1.4 |
| CYP1B1 | Cytochrome P450, Cyp1B1 | 5.23 | 3.17 | 0.63 | 0.73 | 1.5 | - | - |
| DBN1 | Drebrin 1 | 0.69 | 0.36 | - | - | 1.5 | 1.6 | 1.4 |
| DUSP1 | Dual specificity phosphatase 1 | 0.37 | 0.31 | - | - | 0.77 | 0.77 | 0.77 |
| DUSP4 | Dual specificity phosphatase 4 | 0.38 | 0.32 | 1.9 | 2.2 | 1.4 | 1.5 | 2.3 |
| ECH1 | Enoyl Coenzyme A hydratase 1, peroxisomal | 0.67 | 0.29 | - | - | 0.71 | 0.67 | - |
| ENC1 | Ectodermal-neural cortex (with BTB-like domain) | 0.49 | 0.28 | - | - | - | - | 1.2 |
| GNS | Glucosamine (N-acetyl)-6-sulfatase | 0.84 | 0.34 | 1.3 | 1.3 | 1.3 | 1.4 | 1.3 |
| GREB1 | GREB1 protein | 6.14 | 7.61 | 6.1 | 7.6 | 25.3 | 26 | 28 |
| IER3 | Immediate early response 3 | 0.48 | 0.29 | - | 1.4 | 2.0 | 2.4 | 2.4 |
| IFI30 | IFN {gamma}-inducible protein 30 | 0.76 | 0.35 | 1.7 | 1.8 | - | 1.4 | 1.6 |
| IL1R1 | Interleukin 1 receptor, type I | 0.22 | 0.04 | 1.5 | 1.5 | - | 1.4 | 1.6 |
| INHBB | Inhibin, βB | 0.73 | 0.18 | 0.60 | 0.63 | 1.4 | 1.7 | 1.7 |
| IRF6 | IFN regulatory factor 6 | 0.37 | 0.33 | - | - | 2.1 | - | - |
| KRT7 | Keratin 7 | 0.72 | 0.21 | 2.2 | 2.2 | 6.3 | 3.6 | 4.5 |
| LAMB1 | Laminin, β1 | 0.6 | 0.17 | - | 1.4 | 1.6 | 1.6 | 1.4 |
| MATN2 | Matrilin 2 | 0.54 | 0.25 | - | - | 1.2 | - | - |
| MMD | Monocyte to macrophage differentiation-associated | 0.26 | 0.29 | - | - | 0.7 | 0.8 | - |
| PRPS1 | Phosphoribosyl pyrophosphate synthetase 1 | 0.6 | 0.32 | 1.8 | 1.8 | 1.3 | 1.2 | 1.6 |
| PTGES | Prostaglandin E receptor 3 (EP3) | 1.42 | 2.62 | - | 0.60 | 0.63 | 0.59 | 0.67 |
| PVALB | Parvalbumin | 0.66 | 0.13 | 0.69 | 0.46 | 0.37 | 0.37 | 0.37 |
| RAB31 | RAB31 | 7.22 | 4.95 | 1.8 | 1.8 | 1.6 | 2.1 | 2.0 |
| RASGRP1 | RAS guanyl releasing protein 1 | 3.81 | 2.35 | - | - | - | - | 0.63 |
| SDC2 | Syndecan 2 | 2.5 | 2.41 | - | 0.76 | - | - | 0.59 |
| SLIT2 | Slit homologue 2 (Drosophila) | 0.52 | 0.27 | - | - | 3.9 | 1.9 | - |
| SMPD1 | Sphingomyelin phosphodiesterase 1, acid lysosomal | 0.44 | 0.32 | - | - | 0.83 | 0.83 | - |
| SNCG | Synuclein, {gamma} (breast cancer-specific protein 1) | 0.69 | 0.26 | - | - | - | - | 1.6 |
| TGFB2 | Transforming growth factor, β2 | 0.64 | 0.34 | - | - | 1.9 | 2.0 | - |
| TGFB3 | Transforming growth factor, β3 | 0.53 | 0.4 | 1.4 | 1.5 | 2.3 | 1.8 | 1.9 |
| WISP2 | WNT1 inducible signaling pathway protein 2 | 2.39 | 4.9 | 4.5 | 4.1 | 4.2 | 10.9 | 6.2 |
Confirmation of genes regulated during normal mammary gland development
Previous microarray analysis of mammary gland development in intact mice (in which other ovarian hormones including estrogens would impact gene expression) (Master et al. 2002) revealed that genes involved in fatty-acid metabolism ((Medium chain acyl-CoA dehydrogenase (Acadm), Long chain acyl-CoA dehydrogenase (Acadl), Very-long-chain acyl-CoA dehydrogenase (Acadvl), Carnitine palmitoyltransferase II (Cpt2), (Carnitine acetyltransferase (Crat), 3-hydroxyacyl CoA dehydrogenase (Hadh), Dodecenoyl-CoA Δ-isomerase (Dci), Short chain acyl-CoA dehydrogenase (Acads) and PPARa) were highly expressed in neonatal mice (when circulating levels of estradiol are low), but dropped during pubertal mammary gland development (when estradiol levels increase). We observed similar reductions in expression of these genes (Figure 4B and 3). Finally, we compared our dataset to a second microarray study which identified genes associated with pubertal mouse mammary development in intact mice (McBryan et al. 2007). We used the “master list of pubertal genes” generated by McBryan et al. from gene expression analysis of mammary tissue of 3-7 week old intact mice (McBryan et al. 2007). Of the 930 unique genes listed, 879 were represented on the Agilent microarray chip used in our study, and of these genes, 383 (or 44% of those on the array) were identified in our estrogen-induced mammary growth model (Supplementary Table 2). These data confirm that our model identifies estradiol-regulated genes and mimics gene expression of the normal developing mouse mammary gland.
Several genes known to be involved in mammary cell proliferation were also represented in our dataset, and were temporally regulated by estradiol (Table 4). These genes included Tgfb1 (transforming growth factor β (TGFβ)), c-myc (myelocytomatosis oncogene (c-myc)), Cdkn1a or p21 (cyclin-dependent kinase inhibitor 1a), and Egfr (epidermal growth factor receptor (EGFR)). Interestingly, Tgfb1 and c-myc were regulated early (d2-d7) in ductal development, while Egfr gene expression increased only between d7-d28 of estradiol (Table 4). Cdkn1a levels were increased throughout the time-course.
Table 4.
Temporal expression of genes known to be involved in the regulation of mammary gland development or breast cancer cell proliferation.
| Gene | Days of E2 exposure | Fold increase | p-value |
|---|---|---|---|
| Tgfβ | 2 | 1.4 | 2.0E-04 |
| c-myc | 2 | 1.3 | 1.8E-03 |
| 5 | 1.3 | 7.9E-03 | |
| 7 | 1.3 | 1.0E-05 | |
| Egfr | 7 | 1.3 | 1.0E-04 |
| 14 | 1.3 | 4.0E-04 | |
| 28 | 1.3 | 1.0E-10 | |
| Cdkn1a | 2 | 3.4 | ~0 |
| 5 | 2.5 | 8.5E-19 | |
| 7 | 2.3 | 2.0E-09 | |
| 14 | 2.1 | 2.8E-28 | |
| 28 | 1.5 | 5.0E-04 |
Because TGFβ is critical for maintaining the ductal structure in the developing mammary gland (Ewan et al. 2002; Nelson et al. 2006) and regulates gene expression during pubertal mammary gland development (McBryan et al. 2007), we determined which estradiol-regulated genes identified in our dataset were also regulated by TGFβ (Supplementary Tables 3-8). Comparison of our dataset to a database containing 3269 TGFβ-responsive genes (http://actin.ucd.ie/tgfbeta/tgfbeta_2.php) indicated that, averaged over all time-points, 586 (18%) of genes with changing expression profiles in our mammary dataset are known TGFβ-responsive genes (Supplementary Table 3), suggesting that the estradiol-mediated increase in TGFβ expression from d2 to d7 may have resulted in the downstream regulation of TGFβ-responsive genes. Of the genes held in common between the two datasets, 25 to 41% of genes downregulated by TGFβ were also downregulated by estradiol in our dataset. Similarly, 58 to 76% of genes upregulated by TGFβ were also upregulated by estradiol in our dataset. Further comparison of the 586 known TGFβ-responsive genes identified in our dataset with the TGFβ-responsive genes identified by McBryan et al. during pubertal development indicated that 25 to 40% of TGFβ-responsive genes identified during pubertal development are also identified in our dataset (Supplementary Table 3). Taken together, these results further support the notion that our model mimics gene expression of the normal developing mouse mammary gland.
Functional annotation of genes regulated during estradiol-induced mammary gland development
To understand the functions of the genes regulated during estradiol-induced mammary gland development, functional annotation was determined using two approaches. First, we analyzed the function of genes at each individual time-point, to understand how the function of all genes changed over time (Supplementary Table 9). Second, we analyzed the functions of genes grouped according to each of the three major gene expression patterns identified by SOM analysis (Figure 2 and Supplementary Tables 10-12) to determine if specific functions were characteristic of these patterns. The biological functions for both analyses were generated using the Database for Annotation, Visualization and Integrated Discovery 2.1 (DAVID 2.1) Functional Annotation tool (Dennis et al. 2003) (see Materials & Methods).
When individual time-points were analyzed (Supplementary Table 9), we found that many functional categories were significantly over-represented (p ≤ 0.01) throughout the time course (eg. “adaptive immune response” and “cell differentiation”). However, many groups were significant only after 7 days of treatment (“macromolecule biosynthetic process” and “cell migration”), while some functional groups were not significant until day 28 of treatment (eg. “tube development” and “morphogenesis of a branching structure”).
Gene functions were then analyzed based on their gene expression pattern (A: “rapid increase”, B: “gradual increase” and D: “decrease” in Figure 2). “Gradual increase” genes represented 70 different Gene Ontology Biological Process 4 (BP4) categories, which included a wide variety of cellular functions (Supplementary Table 10), including “metabolism”, “inflammatory response”, “signal transduction”, “cytoskeletal organization and biogenesis”, “epithelial cell differentiation”, “proliferation”, “endocytosis”, and “programmed cell death”. However, the largest category of genes (39%) were involved in the immune response (Figure 4C). “Rapid increase” genes were represented by 62 different GO BP4 categories (Supplementary Table 11) and these categories were comparable to those found in the “gradual increase” genes. Again, the immune response represented a large number of genes (29%) (Figure 4C). In contrast, 62% of “decrease” genes (Figure 4C and Supplementary Table 12) were metabolic/catabolic genes and in particular those involved in carbohydrate metabolism (glycolysis, citrate cycle, and oxidative phosphorylation). The three most common gene functions across all three gene groups were “immune response”, “metabolism/catabolism”, and “cell cycle/apoptosis” (Figure 4C).
Pathway analysis of genes regulated during estradiol-induced mammary gland development
Pathway analysis was undertaken using the same two approaches as for functional analysis. We again used the DAVID 2.1 web application, and used it to identify molecular interaction and reaction networks found in the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (Kanehisa et al. 2004). Only pathways significantly (p ≤ 0.01) represented were included in each analysis. First, we identified pathways that were representative of each individual time-point (Supplementary Table 13). As predicted from the functional analysis, metabolic pathways were represented (eg. “glutathione metabolism”, “citrate cycle”, “PPARα (peroxisome proliferator activated receptor α) signaling”, “fatty acid metabolism”, and “glycolysis”), as were immune-related pathways (“complement and coagulation cascades” and “hematopoietic cell lineage”) and those related to extracellular matrix (ECM)-cell and cell-cell interactions (“ECM-receptor interaction”, “focal adhesion”, and “cell adhesion molecules”. Some pathways were present at all time-points (eg. ECM-receptor interaction) while others were present only at later time-points (eg. “glycolysis/gluconeogensis” and “focal adhesion”). When pathways were identified based on expression pattern (“rapid increase”, “gradual increase”, and “decrease”), we found that each group represented unique pathways (Supplementary Table 14). For example, pathways identified from genes in the “decrease” category were exclusively metabolic, while genes in the “rapid increase” category included ECM-receptor, immune, and metabolic pathways.
Estradiol-mediated mammary development and tumourigenesis
To evaluate a potential role for estrogen-mediated regulation of breast tumorigenesis and/or progression, we compared mouse estrogen-regulated mammary genes identified in our analysis with genes shown to be co-expressed with ERα in human breast tumors found in the ONCOMINE™ Cancer Profiling Database (www.oncomine.org) (Rhodes and Chinnaiyan 2004). ONCOMINE™ is a proprietary database of published cancer-related microarray data that can be searched using various online tools.
Microarray data obtained from ER-positive breast tumor tissue was searched specifically for genes from our 14d dataset (the time-point with the greatest number of differentially regulated genes). Genes whose ONCOMINE™ correlation values with ESR1 were greater than or equal to 0.4 (see Materials & Methods for details) were chosen for further analysis, and our mammary microarray array data searched (GeneSpring software) for significant estrogen-dependent regulation of these genes. A list was compiled of ONCOMINE™ genes that were also identified in our database and were increased at least 1.5-fold or decreased by at least 50% by estradiol treatment compared to the placebo control and p ≤ 0.01 (Table 5). Thirty-three genes met these criteria, several of which have been previously shown to co-express with ESR1 in breast cancer, such as GATA3 and FOXA1 (also known as HNF3A) (Lacroix and Leclercq 2004).
Table 5.
Genes co-expressed with Estrogen Receptor alpha in human breast tumor samples, p ≤ 0.01, averaged 1.5-fold or greater change in two independent experiments in at least one time-point. Ns=not significant.
| Fold change after estradiol treatment compared to placebo |
||||||
|---|---|---|---|---|---|---|
| Gene | Gene Description | Accession # | 7d | 14d | 28d | Study |
| Adamts15 | A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 15 | BC043308 | 4.1 | 4.4 | 3.2 | a |
| Bst2 | RIKEN cDNA 2310015I10 gene | NM_198095 | 2.2 | 1.9 | 1.8 | e |
| Cish | cytokine inducible SH2-containing protein | NM_009895 | 1.3 | -1.6 | -1.6 | a |
| Crip1 | Cysteine-rich protein 1 (intestinal) | AK003075 | 1.6 | 1.8 | 1.6 | bd |
| Dusp4 | Dual specificity phosphatase 4 | AK012530 | 2.3 | 1.5 | 1.4 | d |
| Fdft1 | farnesyl diphosphate farnesyl transferase 1 | NM_010191 | -2.0 | -1.8 | -1.4 | f |
| Fer1l3 | Fer-1-like 3, myoferlin (C. elegans) | BC025649 | 2.3 | 3.0 | 2.2 | a |
| Foxa1 | forkhead box A1 | NM_008259 | ns | ns | 2.5 | cde |
| G1p2 | interferon-stimulated protein (15 kDa) (Isg15) | NM_015783 | 2.7 | 3.1 | 2.8 | e |
| Gadd45g | growth arrest and DNA-damage-inducible 45 gamma | NM_011817 | 2.2 | 1.5 | 1.2 | a |
| Galnt6 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 | NM_172451 | -1.6 | -1.4 | -1.7 | ac |
| Gata3 | GATA-binding protein 3 | NM_008091 | 1.4 | 1.2 | 1.9 | abcdeghij |
| Gli3 | GLI-Kruppel family member GLI3 | AK037232 | ns | 2.4 | ns | a |
| Greb1 | gene regulated by estrogen in breast cancer protein | NM_015764 | 27.9 | 26.0 | 25.3 | abi |
| Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 | NM_145942 | ns | -1.7 | -1.5 | f |
| Hpn | hepsin | NM_008281 | ns | 1.4 | ns | h |
| Ifit1 | interferon-induced protein with tetratricopeptide repeats 1 | NM_008331 | 1.9 | 2.2 | 2.5 | e |
| Ifit2 | interferon-induced protein with tetratricopeptide repeats 2 | NM_008332 | ns | 1.6 | 1.7 | e |
| Krt1-18 | keratin complex 1, acidic, gene 18 | NM_010664 | 4.4 | 4.5 | 9.0 | d |
| Krt1-19 | keratin complex 1, acidic, gene 19 | NM_008471 | 8.6 | 7.8 | 14.9 | d |
| Letmd1 | RIKEN cDNA 1110019O13 gene | NM_134093 | -1.6 | -1.8 | -1.8 | a |
| Lgals3bp | lectin, galactoside-binding, soluble, 3 binding protein | NM_011150 | 1.7 | 1.6 | 1.8 | e |
| Maged2 | Melanoma antigen, family D, 2 | NM_030700 | 1.4 | 1.8 | 1.4 | abci |
| Plekha6 | Pleckstrin homology domain containing, family A member 6 | BC025841 | ns | 2.8 | 3.4 | c |
| Stc2 | stanniocalcin 2 | NM_011491 | 2.8 | 2.0 | 3.5 | aj |
| Tjp3 | tight junction protein 3 | NM_013769 | 1.3 | ns | 1.8 | i |
| Tmem30b | RIKEN cDNA 9130011B11 gene | NM_178715 | 3.0 | 2.2 | 5.1 | b |
| Tspan1 | RIKEN cDNA 9030418M05 gene | NM_133681 | ns | ns | 2.9 | b |
Discussion
In this study, we show that pubertal development of mouse mammary ducts can be induced in vivo by exposing prepubertal ovariectomized female mice to estradiol. The main difference between our estrogen-only model and other published mammary development models conducted with intact mice is the presence of other ovarian hormones. As expected, our selective estrogen profile displays overlap with the genes and their functions identified in two other microarray analyses of pubertal ductal development in intact mice (Master et al. 2005; McBryan et al. 2007). Master et al. (Master et al. 2002) analyzed gene expression changes in the developing mammary gland of intact mice from two weeks of age (before puberty) through to pregnancy, lactation, and post-lactational involution. The degree of ductal development observed after 7 days of estradiol exposure in our model approximates that observed in their intact five-week-old virgin mice. Despite the presence of the ovarian hormones progesterone and prolactin in their model, we observe overlap in the regulated genes and their associated functions in our estrogen-only study. Similarly, McBryan et al. determined the gene expression changes in mouse mammary tissue in intact mice 3 to 7 weeks of age (McBryan et al. 2007) from which they created a “master list” of pubertal genes. Approximately 40% of the genes on this list were also regulated in our model. In summary, our new results complement these previous reports by indicating the changes identified from previous studies that are likely to be estrogen-specific actions.
While the use of the whole mammary gland limited our ability to identify the cellular location of the estrogen-responsive genes in this study, many of the genes that we observed in our profile are known to be expressed exclusively in the epithelial cells of the mammary ductal tree. This structure includes a basal layer of myoepithelial cells, which surround the luminal cells. Examples of epithelial-specific genes whose expression changed in response to estradiol in our study included Krt1-5, which is expressed in the myoepithelial cells, Krt8 and Krt1-18, which are expressed primarily in the luminal epithelia, and Krt1-14, which is found primarily in the myoepithelial but also in luminal cells (Mikaelian et al. 2006). We were also able to identify genes whose expression is enriched in terminal end buds (TEBs) and ducts. Using microdissection, Kouros-Mehr et al. identified genes enriched in either the TEB or mature ducts (Kouros-Mehr et al. 2006; Kouros-Mehr and Werb 2006). In their study, some of the most highly enriched genes in both the TEBs and ducts included Krt1-18, Krt1-19, and Expi, all of which we have identified as upregulated by estradiol in our study. We also identified genes that were overexpressed exclusively in TEBs in their study: amphiregulin (Areg) and FXYD domain-containing ion transport regulator 3 (Fxyd3). In summary, use of our model allowed us to detect gene expression changes specific to mature ducts or TEBs, and distinct epithelial subtypes of the ductal tree.
One of the main goals of our study was to compare gene expression profiles at early stages of ductal elongation with profiles after significant ductal elongation had occurred. We originally hypothesized that the genes active early in ductal development, ie. when ductal growth was minimal, would be distinct from those in later development when the ductal tree was extensive. Our data indicates that while many genes were regulated at both early and later stages, 10 to 30% of the genes were regulated only at one time-point, indicating that a subset of genes were indeed active early in ductal development, while others were active later in development. These temporal differences in gene expression were also reflected in the gene functions and pathways identified at early and later development.
Physiological role of estrogen in mammary gland outgrowth during puberty
In the mammary gland, estradiol increases: DNA synthesis in mammary epithelial cells, proliferation of stromal cells (62), glucose metabolism (63), progesterone receptor levels (72, 83, 86), lipid synthesis, histamine release (61, 84), and recruitment of migrant immune response cells (25, 36). The gene functions and pathways identified in our model correlate with these physiological responses, demonstrating that estradiol in the absence of other ovarian or pituitary hormones is capable of triggering many signal transduction pathways characteristic of normal mammary development. Three of the major functions of the estradiol-regulated genes identified in our study included metabolism, the immune response, and cellular proliferation.
Metabolism
One of the most striking observations resulting from our study was the estrogen-stimulated rapid and sustained decrease in genes associated with oxidative phosphorylation, the tricarboxylic acid (TCA) cycle, glycolysis, and fatty acid metabolism. Master et al. (Master et al. 2002) report decreased expression of genes associated with “fatty acid metabolism”, “the mitochondrion”, and “oxidoreductase” during pubertal development; however only a few genes associated with fatty acid metabolism were reported. We have now identified a large number of genes involved in carbohydrate and fatty acid metabolism that decrease in response to estradiol and are likely to decrease during the mammary gland transition from neonate to adult. To our knowledge, a thorough biochemical study of the metabolic changes within the mammary gland during ductal outgrowth has not been conducted; however our results indicate that a reduction in ATP production may occur. In support of this, inspection of available genomic data (McBryan et al. 2007) indicates that many of the oxidative phosphorylation-related genes downregulated in our estrogen-outgrowth model are also downregulated during puberty in intact mice. Although the reason for this decrease is unclear, Master et al. (Master et al. 2002) suggest that the elevated use of fatty acid metabolism in the neonatal gland is due to increased thermogenesis characteristic of brown fat required to maintain body temperature during the neonatal period. Based on this hypothesis, our data suggests that this increased neonatal thermogenesis may also require greater use of carbohydrates during the neonatal period, with the result that this elevated carbohydrate metabolism decreases in pubertal or adult mice. Our hypothesis is supported by studies showing that expression of select genes in the glycolytic pathway, TCA cycle and electron transport chain increase when young rodents are exposed to the cold, suggesting that glucose utilization is also stimulated during thermogenesis in brown fat (Daikoku et al. 2000; Scarpace et al. 1999).
Immune response
One of the most highly-enriched and rapidly-activated functional groups observed after estradiol treatment involved immune function. The increased expression of immune-related genes in the mouse mammary gland during puberty has been observed previously (Gouon-Evans et al. 2002; Gouon-Evans et al. 2000; Master et al. 2005), although the genes involved in this response have not been well-characterized. During ductal growth, migratory leukocytes are present in the mammary stroma, and both macrophages and eosinophils accumulate around TEBs (Gouon-Evans et al. 2002). Depletion of leukocytes impairs ductal development and formation of TEBs, suggesting that macrophage and eosinophils are critical for ductal morphogenesis (Gouon-Evans et al. 2000). Macrophage-colony-stimulating-factor 1 (Csf1) is a growth factor and cytokine that controls the production, differentiation, and function of macrophages and other cells of the mononuclear phagocytic lineage through its interaction with the Csf1 receptor (Csf1r). Csf1-null mice, which lack many types of macrophages and monocytes, demonstrate impaired ductal outgrowth during puberty (Gouon-Evans et al. 2000). This phenotype is similar to mice lacking Csf1r (Dai et al. 2002), which is expressed exclusively in macrophages. We observe an increase in Csf1 and Csf1r expression after estradiol treatment. While it is possible that estradiol increases the transcription of these genes directly, it is more likely that the estrogen-stimulated immune cell influx would impact transcript levels within the gland by altering the cellular composition of the tissue rather than reflecting estradiol-mediated regulation of transcription. In addition to macrophages, eosinophils also migrate to TEBs during ductal growth. Transcript levels of eotaxin, an eosinophilic chemoattractant, are increased in the prepubertal mammary gland and coincide with eosinophil recruitment (Gouon-Evans et al. 2000). We observe an increase in eotaxin mRNA after 28 days of estradiol treatment. Overall, our data suggests that estrogen plays a role in mammary ductal development by recruiting and/or stimulating migratory leukocytes.
Cellular proliferation
Estrogen stimulates the proliferation of mammary epithelial and stromal cells, and this proliferation is critical for mammary outgrowth (62). Of the many estradiol-regulated genes we identified, at least four were well-known regulators of cellular proliferation, including Tgfb1, c-myc, Cdkn1a (p21), and Egfr. First, TGFβ is critical for maintaining the ductal structure in the developing mammary gland, and inhibits both lateral branching and ductal growth (Ewan et al. 2002; Nelson et al. 2006). TGFβ is thought to counteract estrogen-induced ductal morphogenesis (Lanigan et al. 2007), and Ewan et al. (Ewan et al. 2002) propose a model in which estrogen increases TGFβ activity, which limits proliferation in response to estradiol. Second, we observed increased expression of c-Myc, an oncogene and transcription factor that mediates cell cycle progression, apoptosis, and DNA synthesis and plays a significant role in estradiol-induced cellular mitogenesis (Musgrove et al. 2008). This observation correlates with previous studies showing that c-myc is increased by estradiol in human breast cancer cells (Lazennec et al. 2001), the rodent uterus (Hewitt et al. 2003; Murphy et al. 1987), and primate mammary epithelium (Dimitrakakis et al. 2006). Third, cyclin-dependent kinase inhibitor, Cdkn1a (p21) expression was increased throughout the time-course, and may be reflective of the rapid cellular proliferation that occurs during ductal growth. Estradiol has been reported to both increase (Lazennec et al. 2001; Levenson et al. 2002; Thomas et al. 1998) and decrease (Foster et al. 2001; Prall et al. 2001) Cdkn1a expression in breast cancer cell lines, although its effect on Cdkn1a in mammary gland gene expression is less clear. Interestingly, estradiol treatment also increases cdkn1a levels in the mammary glands of ovariectomized heifers, suggesting estradiol-mediated regulated of Cdkn1a may occur in other species (Li and Capuco 2008). Finally, we observed an estradiol-mediated increase in expression of the epidermal growth factor receptor (Egfr), one of the four members of the EGFR (ERBB gene) family. No changes in expression of Erbb2, Erbb3, or Erbb4 were observed, despite the recent evidence for their role in mammary gland development (Muraoka-Cook et al. 2008; Stern 2003; Stern 2008). EGFR is essential for ductal growth and branching during pubertal mammary gland development (Wiesen et al. 1999; Xie et al. 1997) and is thought to be a critical mediator of estradiol action during puberty. Estradiol activates Egfr in ovariectomized mice (Sebastian et al. 1998), and the expression of amphiregulin, the major EGFR ligand expressed in mammary epithelial cells and TEBs during puberty, is robustly increased by estradiol in our model. The regulation of Egfr and its ligand in our estrogen-only model further supports the claim that estradiol enhances the actions of these hormones on mammary development.
Mechanisms of estrogen-mediated gene regulation in the mammary gland
One of the limitations of our model is that it does not allow us to distinguish between estradiol-mediated and developmental effects on gene expression. For example, gene expression changes at late time-points may reflect an increase in epithelial structures in the estradiol-exposed mammary gland, rather than the direct action of estradiol on these “late-effect” genes. However, a direct role for estrogen action in mouse mammary gland development is supported by studies in which estradiol pellets were implanted directly into mammary tissue of immature mice (Daniel et al. 1987; Haslam 1988; Silberstein et al. 1994). Ductal growth occurred only in the vicinity of the implants in ovariectomized females, and in intact animals, normal growth was blocked locally by implants containing antiestrogens, suggesting a direct role for estrogen via the estrogen receptor alpha (ERα) on mammary growth (Daniel et al. 1987; Silberstein et al. 1994). Studies in mice lacking ERα (αERKO) also support a requirement for both ovarian and pituitary hormones in mammary ductal morphogenesis. The absence of pubertal ductal branching in αERKO mice (Bocchinfuso and Korach 1997) has been shown to arise from the lack of estrogen actions both directly on the mammary gland and indirectly on the circulating levels of progesterone and prolactin (Bocchinfuso et al. 2000). Estrogen has been shown to directly regulate many genes identified in our analysis, including Krt1-19, Greb1, Areg, and Wisp2 (Frasor et al. 2004; Kian Tee et al. 2004). In summary, it is likely that ERα directly mediates a subset of genes identified in our microarray analysis, and that indirect actions of Erα or developmental effects are also responsible for the gene expression changes we observe.
ER-regulated genes in normal human breast and in breast cancer
Our study also suggests that a subset of the estrogen-responsive genes identified in our model may also be estrogen-dependent in human breast tissue. A recent microarray analysis identified estradiol-regulated genes in human breast epithelium in a model in which normal human breast tissue was transplanted into mice, which were then treated with estradiol (Wilson et al. 2006). Interestingly, we observe that 46% of the 56 differentially-regulated genes observed in response to estrogen in the human tissue are also observed as estrogen-regulated in our mouse mammary gland model (Table 6), with 67% of theses common genes regulated in the same direction (either up or down) in both models. These results suggest that these genes may be estrogen-dependent and involved in the development of both human and mouse mammary gland. Many of these genes have not previously been associated with mammary development. For example, chemokine (C-X-C motif) ligand 9 (CXCL9) is a chemokine involved in immune cell trafficking, and its levels decrease in both human and mouse mammary gland after estradiol treatment. How this decreased expression contributes to mammary development remains to be determined.
Table 6.
Genes regulated during estradiol-induced mouse mammary gland development and by estradiol in human breast tissue transplanted into female nude mice (Wilson et al. 2006).
| Human | Mouse mammary gland | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Gene Description | 7d | 2d | 5d | 7d | 14d | 28d |
| AREG | Amphiregulin | 4.2 | 4.7 | 5.0 | 11.3 | 6.8 | 10.9 |
| BGN | Biglycan | 0.3 | - | - | - | 1.3 | - |
| CCL2 | Chemokine (C-C motif) ligand 2 | 0.6 | - | 1.3 | 2.2 | 1.8 | - |
| COL6A1 | Collagen, type VI, a1 | 0.6 | 1.6 | 1.7 | 1.8 | 1.62 | - |
| CSPG2 | Chondroitin sulphate proteoglycan 2 (versican) | 0.5 | - | 1.3 | 1.6 | 2.0 | 1.4 |
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 0.5 | -1.9 | -1.8 | -1.9 | -1.5 | -1.7 |
| DPT | Dermatopontin | 0.5 | -1.9 | -2.1 | - | -1.1 | - |
| EMILIN1 | Elastin microbril interfacer 1 | 0.6 | - | - | - | -1.6 | -1.9 |
| ENPEP | Glutamyl aminopeptidase | 0.5 | -1.5 | - | - | - | -1.5 |
| FN1 | Fibronectin 1 | 0.5 | 1.4 | 1.9 | 2.6 | 4.2 | 4.1 |
| FXYD3 | FXYD domain-containing ion transport regulator 3 | 1.4 | 1.2 | - | 2.2 | 1.5 | 2.2 |
| GATA3 | GATA binding protein 3 | 0.6 | - | - | 1.4 | - | 1.9 |
| GREB1 | gene regulated by estrogen in breast cancer protein 1 | 2.8 | 15 | 17 | 28 | 26 | 25 |
| IGJ | Immunoglobulin J polypeptide | 0.5 | 1.7 | -1.3 | -1.3 | 1.6 | 2.2 |
| KRT19 | Keratin 19 | 1.5 | - | - | 8.6 | 7.8 | 14.9 |
| PRG4 | Proteoglycan 4 | 1.3 | 6.1 | 4.8 | 7.4 | 7.2 | - |
| RGS5 | Regulator of G-protein signalling 5 | 0.5 | -1.6 | -2.0 | -1.3 | -1.3 | -2.0 |
| S100A8 | S100 calcium-binding protein A8 | 0.6 | 1.8 | 1.4 | - | -1.7 | - |
| SERPINH1 | Serine (or cysteine) proteinase inhibitor, clade H | 0.5 | - | - | 1.2 | 1.3 | 1.2 |
| TACSTD1 | Tumour-associated calcium signal transducer | 1.5 | 2.1 | 1.9 | 2.5 | 2.5 | 5.2 |
| XBP1 | X-box binding protein 1 | 1.4 | 1.6 | - | 1.3 | - | - |
The growth of many breast tumors is estrogen-dependent, so it is interesting that several microarray studies of human breast tumor specimens have identified genes that are coexpressed with ESR1. Why this coexpression occurs and how it relates to tumor development is not well understood, and there are several possible explanations. First, the product of these coexpressed genes may directly or indirectly affect ESR1 transcription or message stability. Secondly, the ER may directly or indirectly regulate the transcription of these genes. In order to further understand these possible mechanisms, we compared the estrogen-regulated genes from our mouse mammary study with those co-expressing with ESR1 in breast tumor microarray profiles obtained from the ONCOMINE™ database. Both GATA3 and FOXA1 (HNF3A), which are known to co-express with ERα in human breast tumors, were identified by this approach. Gata3 is critical for luminal epithelial differentiation in the mouse mammary gland, and Gata3 regulates transactivation of the Foxa1 promoter. Despite the correlation between ER and GATA3 expression in human breast tumors, there is no evidence that estradiol directly regulates the levels of GATA3 and FOXA1, or that they in turn regulate the levels of ESR1, although FOXA1 binding sites are frequently located near ERE sites in the human genome (Carroll et al. 2005). However, our results suggest that, at least in mouse mammary gland, the expression of Gata3 and Foxa1 is increased in response to estradiol treatment, and together with previous studies suggest an indirect effect of estradiol action on these genes. We have also identified several genes known to be involved in metastasis and invasion (Tactsd1) (Basak et al. 1998; Wurfel et al. 1999) and breast carcinogenesis (Fgb) ((Rybarczyk and Simpson-Haidaris 2000) and references therein), neither of which was previously shown to be regulated by estrogens, which may now reveal their involvement in estrogen-dependent breast cancer cell proliferation and tumor progression.
In summary, we have shown that estradiol treatment alone mimics at the genomic level normal mammary gland development in intact pubertal mice. Interestingly, we have identified carbohydrate metabolism as a major pathway whose activity appears to be dramatically decreased during pubertal mammary development. Finally, we have identified genes regulated by estrogen treatment in both mouse and human mammary glands, and those that co-express with Estrogen Receptor α in human breast cancer. In conclusion, our genomic data support the physiological observation that estradiol is one of the primary hormonal signals driving ductal elongation during pubertal mammary development involving differential patterns of gene expression.
Materials and Methods
Animals, treatments, and tissue collection
All animals were handled according to NIH guidelines and in compliance with an NIEHS approved animal protocol. Female C57 Bl/6 mice ovariectomized prior to puberty (21 days of age) were purchased from Charles River (Raleigh, NC), and housed for two weeks to allow for clearance of endogenous steroids. After two weeks, estradiol pellets (17β-estradiol, 90-day pellets, 0.1 mg/pellet) or placebo pellets (placebo, 90-day pellets, 0.1 mg/pellet) were implanted subdermally with a trocar as described by the supplier (Innovative Research of America, Catalog numbers NE-121 (17β-estradiol) and NE-111 (placebo)). Previous studies from our laboratory (Bocchinfuso et al. 2000) also used 0.1 mg 17β-estradiol pellets to induce ductal elongation, and resulted in serum estradiol levels of approximately 330 pg/mL/pellet.
Mammary gland isolation and analysis
At various time-points after the pellets were implanted, ie. 2, 5, 7, 14, and 28 days, four to five animals per time-point were sacrificed using CO2 asphyxiation and one #4 mammary gland (Hedrich 2004) was collected. Our pilot studies (data not shown) indicated that approximately 75% of maximal ductal elongation occurred by four weeks of estradiol exposure—a rate that mimicked natural ductal growth. Based on these data, and because we were particularly interested in molecular events during early elongation, we selected 2, 5, 7, 14, and 28 days for our microarray analysis. Ductal growth was confirmed by whole-mount analysis of the #4 gland, and the contralateral #4 gland was excised for RNA isolation and snap-frozen in liquid nitrogen. The entire four week time-course was repeated to provide a completely independent set of mammary tissue to serve as a biological replicate; however, two and five-day treatments were not duplicated. Ductal lengths were measured by image analysis of whole mounts using Image Pro software (SciMeasure, Decatur, GA) and a Leica dissecting microscope (Bannockburn, IL). The distance from the outermost edge of the lymph node (side nearest the nipple) to the furthest terminal end bud beyond the node was measured.
Microarray analysis
For each time-course experiment, one frozen #4 mammary gland was individually pulverized from four to five animals per treatment group, then homogenized in 3mL Trizol (Invitrogen, Carlsbad, CA) and RNA was prepared according to the manufacturer’s protocol. The RNA from individual animals was then pooled for each treatment group (four to five animals per group) and further purified using the QIAGEN (Valencia, CA) RNeasy Mini kit (Cat. No. 74104) clean-up protocol.
Gene expression analysis was conducted using Agilent Mouse Oligo arrays (pattern number 011978) (Agilent Technologies, Palo Alto, CA). Total RNA was amplified using the Agilent Low RNA Input Fluorescent Linear Amplification Kit protocol. Starting with 500ng of total RNA, Cy3 or Cy5 labeled cRNA was produced according to manufacturer’s protocol. For each two color comparison, 750 ng of each Cy3 and Cy5 labeled cRNAs were mixed and fragmented using the Agilent In Situ Hybridization Kit protocol. In each case, samples from estradiol-treated animals were co-hybridized with a placebo sample. Hybridizations were performed for 17 hours in a rotating hybridization oven using the Agilent 60-mer oligo microarray processing protocol. Slides were washed as indicated in this protocol and then scanned with an Agilent Scanner. Data was obtained using the Agilent Feature Extraction software (v7.5), using defaults for all parameters.
The Agilent Feature Extraction Software performed error modeling, adjusting for additive and multiplicative noise. The resulting data were processed using the Rosetta Resolver® system (version 7.1) (Rosetta Biosoftware, Kirkland, WA) and the associated Rosetta error model (Weng et al. 2006). The Resolver system combines ratio profiles to create ratio experiments using an error-weighted average as described in Stoughton and Dai (Stoughton and Dai 2002; Weng et al. 2006). P-values are generated and propagated throughout the system and represent the probability that a given gene is significantly differentially expressed. Rosetta Resolver identified potentially significant changes in the mean expression level of approximately 2000 (2, 5, 7, and 28d) or 3000 (14d) candidate genes. The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/, and all MIAME-compliant related information regarding these studies are accessible through GEO Series accession number GSE4647 (Edgar et al. 2002).
Functional analysis
The Database for Annotation, Visualization and Integrated Discovery 2.1 (DAVID 2.1) Functional Annotation tool (Dennis et al. 2003) was used to determine Gene Ontology (GO) functions. All categories are derived from the GO Biological Process category, Level 4, and all analyses were conducted with Maximum EASE Score/p-value set to 0.01.
ONCOMINE™ analysis (www.oncomine.org) (Rhodes and Chinnaiyan 2004)
The “Gene Search” tool was used to identify studies in which ESR1 was represented. The Co/Ex tool was then used to identify genes that were co-expressed with ESR1 in breast tissue-related studies (van ’t Veer et al. 2002), (Wang et al. 2005), (Zhao et al. 2004), (West et al. 2001), (Perou et al. 1999), (Perou et al. 2000), (Sorlie et al. 2001), (Sorlie et al. 2003), (van de Vijver et al. 2002), (Gruvberger et al. 2001). Genes whose ONCOMINE™ correlation values with ESR1 were greater than or equal to 0.4 were chosen for further analysis. This value was chosen by inspection of the correlation values in order to maximize the number of significantly correlated genes while minimizing the output of genes whose correlation was known a priori likely not to be biologically relevant. We then searched the mammary 14d estradiol p ≤ 0.001 dataset (the dataset with the largest number of significantly-regulated genes) for the genes that met these ONCOMINE™ criteria. The list generated from ONCOMINE™ was imported into the Rosetta Resolver® system (version 5.1) (Rosetta Biosoftware, Kirkland, WA) and the fold changes associated with the genes were then exported as a spreadsheet.
Reverse transcription and Quantitative PCR analysis
RNA was isolated from mammary glands of individual mice (five mice per treatment group), treated with DNaseI, then reverse-transcribed using Superscript II (Invitrogen Corporation) according to the manufacturer’s protocol. cDNA levels were detected using quantitative PCR with the ABI PRISM 7900 Sequence Detection System (Applied Biosystems, Foster City, CA) and SYBR Green I dye, and primers were created using Applied Biosystems Primer Express Software version 2.0 (Table 1) as previously described (Hewitt et al. 2005). Fold expression or repression was determined by quantitation of cDNA from target (treated) samples relative to a calibrator sample (placebo). For all samples, the gene for 18S was used as the endogenous control for normalization of initial RNA levels. Expression ratios were calculated according to the mathematical model described by Pfaffl (45), where ratio=(Etarget)ΔCt(target)/(E18s)ΔCt(18s) and E=efficiency of the primer set, calculated from the slope of a standard curve of log (ng of cDNA) vs. Ct value for a sample that contains the target according to the formula E=10-(1/slope) and ΔCt=Ct(vehicle)-Ct(treated sample). These expression ratios were calculated for each treatment group as described above, and average and SEM calculated. The data were analyzed for statistical significance using an unpaired, two-tailed student’s t-test comparing estradiol-treated samples to placebo-treated controls.
Table 1.
Sequences for quantitative PCR primers.
| Gene symbol | Accession # | Primer range | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|---|---|
| Adamts19 | NM_175506 | 851-927 | GGTCACCCGCACCGTTTAT | TGGCGGTGAACAGCTGAGT |
| Areg | NM_009704 | 743-817 | CACAGCGAGGATGACAAGGA | GAGGATGATGGCAGAGACAAAGA |
| Cilp | AK081544 | 635-702 | CCGAGTCACTGCTGCTGACA | CGCGGTTATTCCCCATGTAC |
| Expi | NM_007969 | 133-209 | TTTGTTCTGGTAGCTTTGATTTTCA | GCGCCAGGTTTTTCTTTGG |
| Gas6 | NM_019521 | 199-260 | TGGGCACTGCGCTTCTG | CCGCAACAGCACAGTGTGA |
| Greb1 | NM_015764 | 203-284 | CGGGCTTTGTGAGGAGTCA | CCGGAGCACAAAGAAGACAGA |
| Krt1-18 | NM_010664 | 508-595 | CTTGCCGCCGATGACTTTA | CCTTGCGGAGTCCATGGA |
| Krt1-19 | M28698 | 467-542 | GAGGACTTGCGCGACAAGA | GGCGAGCATTGTCAATCTGTAG |
| Plod2 | NM_011961 | 448-525 | ATAAGCGGCTGGCAGACAAG | CATAGCCAATAAAGCCTCCAGAA |
| Ppara | X89577 | 231-306 | CCATACAGGAGAGCAGGGATTT | TTACCTACGCTCAGCCCTCTTC |
| Fgb | NM_181849 | 679-755 | GCACTGTCAGCTGCAACATTC | GGATGTCTCACCTCCCTTCCT |
| Ucp1 | BC012701 | 398-471 | AAAGGTGTCCTAGGGACCATCA | GCAGGCAGACCGCTGTACA |
| 18S | X56974 | 1271-1351 | GAAACTGCGAATGGCTCATTAA | GAATTACCACAGTTATCCAAGTAGGA |
Supplementary Material
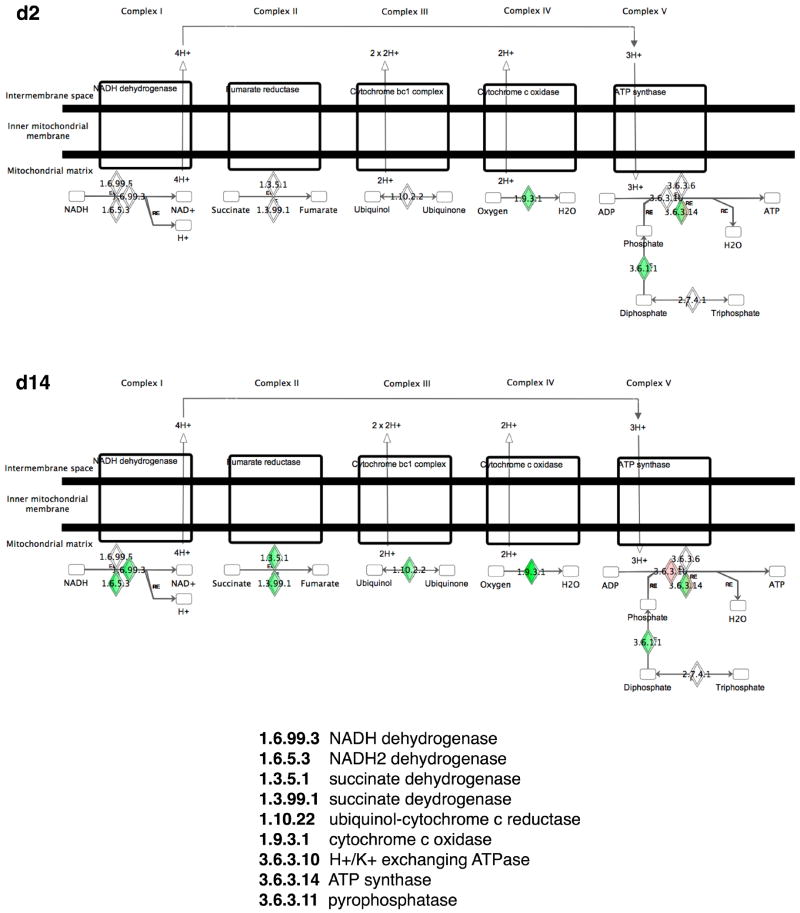
Figure 5.
Graphic summary of genes in the oxidative phosphorylation pathway in mouse mammary gland after treatment for two days or two weeks with estradiol. Red and green represent increased and decreased gene expression; the intensity of the colour reflects the degree of expression.
Acknowledgments
We are grateful to Dr. Julie Hall for providing whole mount samples from intact mice, and to Dr. Andrew Fernandes (The University of Western Ontario) for his bioinformatic analysis to identify TGFbeta-regulated genes in our estrogen-regulated mammary dataset. We would also like to thank James Clark, Page Myers, and Clay Rouse for assistance with the implantation of estradiol pellets. We are also grateful to Jie Liu (NIEHS) for providing the primers for Ucp1, PPARa, and Fgb. Thanks also to Harriet Kinyamu and Wendy Jefferson for critical reading of the manuscript, and to the members of the Korach lab for their input throughout the course of this study.
Funding: This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences.
Footnotes
Declarations of Interest: The authors having nothing to declare.
References
- Aldaz CM, Hu Y, Daniel R, Gaddis S, Kittrell F, Medina D. Serial analysis of gene expression in normal p53 null mammary epithelium. Oncogene. 2002;21(41):6366–6376. doi: 10.1038/sj.onc.1205816. [DOI] [PubMed] [Google Scholar]
- Baik MG, Lee MJ, Choi YJ. Gene expression during involution of mammary gland (review) Int J Mol Med. 1998;2(1):39–44. [PubMed] [Google Scholar]
- Bale TL, Dorsa DM. Cloning, novel promoter sequence, and estrogen regulation of a rat oxytocin receptor gene. Endocrinology. 1997;138(3):1151–1158. doi: 10.1210/endo.138.3.4998. [DOI] [PubMed] [Google Scholar]
- Basak S, Speicher D, Eck S, Wunner W, Maul G, Simmons MS, Herlyn D. Colorectal carcinoma invasion inhibition by CO17-1A/GA733 antigen and its murine homologue. J Natl Cancer Inst. 1998;90(9):691–697. doi: 10.1093/jnci/90.9.691. [DOI] [PubMed] [Google Scholar]
- Bocchinfuso WP, Korach KS. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia. 1997;2(4):323–334. doi: 10.1023/a:1026339111278. [DOI] [PubMed] [Google Scholar]
- Bocchinfuso WP, Lindzey JK, Hewitt SC, Clark JA, Myers PH, Cooper R, Korach KS. Induction of Mammary Gland Development in Estrogen Receptor-{alpha} Knockout Mice. Endocrinology. 2000;141(8):2982–2994. doi: 10.1210/endo.141.8.7609. [DOI] [PubMed] [Google Scholar]
- Bucco R, Zheng W, Wardlaw S, Davis J, Sierra-Rivera E, Osteen K, Melner M, Kakkad B, Ong D. Regulation and localization of cellular retinol-binding protein, retinol-binding protein, cellular retinoic acid-binding protein (CRABP), and CRABP II in the uterus of the pseudopregnant rat. Endocrinology. 1996;137(7):3111–3122. doi: 10.1210/endo.137.7.8770937. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Choi I, Gudas LJ, Katzenellenbogen BS. Regulation of keratin 19 gene expression by estrogen in human breast cancer cells and identification of the estrogen responsive gene region. Mol Cell Endocrinol. 2000;164(1-2):225–237. doi: 10.1016/s0303-7207(00)00197-0. [DOI] [PubMed] [Google Scholar]
- Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99(1):111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- Daikoku T, Shinohara Y, Shima A, Yamazaki N, Terada H. Specific elevation of transcript levels of particular protein subtypes induced in brown adipose tissue by cold exposure. Biochim Biophys Acta. 2000;1457(3):263–272. doi: 10.1016/s0005-2728(00)00107-9. [DOI] [PubMed] [Google Scholar]
- Daniel CW, Silberstein GB, Strickland P. Direct action of 17 beta-estradiol on mouse mammary ducts analyzed by sustained release implants and steroid autoradiography. Cancer Res. 1987;47(22):6052–6057. [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- Dimitrakakis C, Zhou J, Wang J, Matyakhina L, Mezey E, Wood JX, Wang D, Bondy C. Co-expression of estrogen receptor-alpha and targets of estrogen receptor action in proliferating monkey mammary epithelial cells. Breast Cancer Res. 2006;8(1):R10. doi: 10.1186/bcr1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewan KB, Shyamala G, Ravani SA, Tang Y, Akhurst R, Wakefield L, Barcellos-Hoff MH. Latent transforming growth factor-beta activation in mammary gland: regulation by ovarian hormones affects ductal and alveolar proliferation. Am J Pathol. 2002;160(6):2081–2093. doi: 10.1016/s0002-9440(10)61158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CR, Graves KH, Parlow AF, Simpson ER. Characterization of mice deficient in aromatase (ArKO) because of targeted disruption of the cyp19 gene. Proc Natl Acad Sci U S A. 1998;95(12):6965–6970. doi: 10.1073/pnas.95.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flux DS. Growth of the mammary duct system in intact and ovariectomized mice of the CHI strain. J Endocrinol. 1954;11(3):223–237. doi: 10.1677/joe.0.0110223. [DOI] [PubMed] [Google Scholar]
- Foster JS, Henley DC, Bukovsky A, Seth P, Wimalasena J. Multifaceted regulation of cell cycle progression by estrogen: regulation of Cdk inhibitors and Cdc25A independent of cyclin D1-Cdk4 function. Mol Cell Biol. 2001;21(3):794–810. doi: 10.1128/MCB.21.3.794-810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64(4):1522–1533. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60(22):6367–6375. [PubMed] [Google Scholar]
- Goruppi S, Chiaruttini C, Ruaro ME, Varnum B, Schneider C. Gas6 induces growth, beta-catenin stabilization, and T-cell factor transcriptional activation in contact-inhibited C57 mammary cells. Mol Cell Biol. 2001;21(3):902–915. doi: 10.1128/MCB.21.3.902-915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Lin EY, Pollard JW. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002;4(4):155–164. doi: 10.1186/bcr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127(11):2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- Gruvberger S, Ringner M, Chen Y, Panavally S, Saal LH, Borg A, Ferno M, Peterson C, Meltzer PS. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61(16):5979–5984. [PubMed] [Google Scholar]
- Haslam SZ. Local versus systemically mediated effects of estrogen on normal mammary epithelial cell deoxyribonucleic acid synthesis. Endocrinology. 1988;122(3):860–867. doi: 10.1210/endo-122-3-860. [DOI] [PubMed] [Google Scholar]
- Hedrich HJ, editor. The laboratory mouse. New York: Elsevier Academic Press; 2004. [Google Scholar]
- Hennighausen L, Robinson GW. Think globally, act locally: the making of a mouse mammary gland. Genes Dev. 1998;12(4):449–455. doi: 10.1101/gad.12.4.449. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6(9):715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Collins J, Grissom S, Deroo B, Korach KS. Global uterine genomics in vivo: microarray evaluation of the estrogen receptor alpha-growth factor cross-talk mechanism. Mol Endocrinol. 2005;19(3):657–668. doi: 10.1210/me.2004-0142. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Deroo BJ, Hansen K, Collins J, Grissom S, Afshari CA, Korach KS. Estrogen receptor-dependent genomic responses in the uterus mirror the biphasic physiological response to estrogen. Mol Endocrinol. 2003;17(10):2070–2083. doi: 10.1210/me.2003-0146. [DOI] [PubMed] [Google Scholar]
- Ivanga M, Labrie Y, Calvo E, Belleau P, Martel C, Luu-The V, Morissette J, Labrie F, Durocher F. Temporal analysis of E2 transcriptional induction of PTP and MKP and downregulation of IGF-I pathway key components in the mouse uterus. Physiol Genomics. 2007;29(1):13–23. doi: 10.1152/physiolgenomics.00291.2005. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayisli UA, Guzeloglu-Kayisli O, Arici A. Endocrine-immune interactions in human endometrium. Ann N Y Acad Sci. 2004;1034:50–63. doi: 10.1196/annals.1335.005. [DOI] [PubMed] [Google Scholar]
- Kian Tee M, Rogatsky I, Tzagarakis-Foster C, Cvoro A, An J, Christy RJ, Yamamoto KR, Leitman DC. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors alpha and beta. Mol Biol Cell. 2004;15(3):1262–1272. doi: 10.1091/mbc.E03-06-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Guhl E, Zollinger R, Tzeng YJ, Wessel R, Hummel M, Graessmann M, Graessmann A. Gene expression profiling: cell cycle deregulation and aneuploidy do not cause breast cancer formation in WAP-SVT/t transgenic animals. J Mol Med. 2005;83(5):362–376. doi: 10.1007/s00109-004-0625-1. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127(5):1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev Dyn. 2006;235(12):3404–3412. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol. 2004;219(1-2):1–7. doi: 10.1016/j.mce.2004.02.021. [DOI] [PubMed] [Google Scholar]
- Lanigan F, O’Connor D, Martin F, Gallagher WM. Molecular links between mammary gland development and breast cancer. Cell Mol Life Sci. 2007;64(24):3159–3184. doi: 10.1007/s00018-007-7386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. ER beta inhibits proliferation and invasion of breast cancer cells. Endocrinology. 2001;142(9):4120–4130. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson AS, Svoboda KM, Pease KM, Kaiser SA, Chen B, Simons LA, Jovanovic BD, Dyck PA, Jordan VC. Gene expression profiles with activation of the estrogen receptor alpha-selective estrogen receptor modulator complex in breast cancer cells expressing wild-type estrogen receptor. Cancer Res. 2002;62(15):4419–4426. [PubMed] [Google Scholar]
- Li RW, Capuco AV. Canonical pathways and networks regulated by estrogen in the bovine mammary gland. Funct Integr Genomics. 2008;8(1):55–68. doi: 10.1007/s10142-007-0055-6. [DOI] [PubMed] [Google Scholar]
- Martinez-Lacaci I, Saceda M, Plowman GD, Johnson GR, Normanno N, Salomon DS, Dickson RB. Estrogen and phorbol esters regulate amphiregulin expression by two separate mechanisms in human breast cancer cell lines. Endocrinology. 1995;136(9):3983–3992. doi: 10.1210/endo.136.9.7649107. [DOI] [PubMed] [Google Scholar]
- Master SR, Hartman JL, D’Cruz CM, Moody SE, Keiper EA, Ha SI, Cox JD, Belka GK, Chodosh LA. Functional microarray analysis of mammary organogenesis reveals a developmental role in adaptive thermogenesis. Mol Endocrinol. 2002;16(6):1185–1203. doi: 10.1210/mend.16.6.0865. [DOI] [PubMed] [Google Scholar]
- Master SR, Stoddard AJ, Bailey LC, Pan TC, Dugan KD, Chodosh LA. Genomic analysis of early murine mammary gland development using novel probe-level algorithms. Genome Biol. 2005;6(2):R20. doi: 10.1186/gb-2005-6-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryan J, Howlin J, Kenny PA, Shioda T, Martin F. ERalpha-CITED1 co-regulated genes expressed during pubertal mammary gland development: implications for breast cancer prognosis. Oncogene. 2007;26(44):6406–6419. doi: 10.1038/sj.onc.1210468. [DOI] [PubMed] [Google Scholar]
- Mikaelian I, Hovick M, Silva KA, Burzenski LM, Shultz LD, Ackert-Bicknell CL, Cox GA, Sundberg JP. Expression of terminal differentiation proteins defines stages of mouse mammary gland development. Vet Pathol. 2006;43(1):36–49. doi: 10.1354/vp.43-1-36. [DOI] [PubMed] [Google Scholar]
- Mo R, Tony Zhu Y, Zhang Z, Rao SM, Zhu YJ. GAS6 is an estrogen-inducible gene in mammary epithelial cells. Biochem Biophys Res Commun. 2007;353(1):189–194. doi: 10.1016/j.bbrc.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook RS, Feng SM, Strunk KE, Earp HS., 3rd ErbB4/HER4: role in mammary gland development, differentiation and growth inhibition. J Mammary Gland Biol Neoplasia. 2008;13(2):235–246. doi: 10.1007/s10911-008-9080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LJ, Murphy LC, Friesen HG. Estrogen induction of N-myc and c-myc proto-oncogene expression in the rat uterus. Endocrinology. 1987;120(5):1882–1888. doi: 10.1210/endo-120-5-1882. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Sergio CM, Anderson LR, Inman CK, McNeil CM, Alles MC, Gardiner-Garden M, Ormandy CJ, Butt AJ, Sutherland RL. Identification of downstream targets of estrogen and c-myc in breast cancer cells. Adv Exp Med Biol. 2008;617:445–451. doi: 10.1007/978-0-387-69080-3_43. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Vanduijn MM, Inman JL, Fletcher DA, Bissell MJ. Tissue geometry determines sites of mammary branching morphogenesis in organotypic cultures. Science. 2006;314(5797):298–300. doi: 10.1126/science.1131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Aitkenhead M, Hughes CC, Levin ER. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J Biol Chem. 2002;277(52):50768–50775. doi: 10.1074/jbc.M210106200. [DOI] [PubMed] [Google Scholar]
- Pentecost BT, Bradley LM, Gierthy JF, Ding Y, Fasco MJ. Gene regulation in an MCF-7 cell line that naturally expresses an estrogen receptor unable to directly bind DNA. Mol Cell Endocrinol. 2005;238(1-2):9–25. doi: 10.1016/j.mce.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96(16):9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, Girling AC, Ball RY, Edwards DR. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10(7):2429–2440. doi: 10.1158/1078-0432.ccr-0398-3. [DOI] [PubMed] [Google Scholar]
- Prall OW, Carroll JS, Sutherland RL. A low abundance pool of nascent p21WAF1/Cip1 is targeted by estrogen to activate cyclin E*Cdk2. J Biol Chem. 2001;276(48):45433–45442. doi: 10.1074/jbc.M104752200. [DOI] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92(2):141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Chinnaiyan A. Bioinformatics Strategies for Translating Genome-Wide Expression Analyses into Clinically Useful Cancer Markers. Ann NY Acad Sci. 2004;1020(1):32–40. doi: 10.1196/annals.1310.005. [DOI] [PubMed] [Google Scholar]
- Ruan W, Catanese V, Wieczorek R, Feldman M, Kleinberg DL. Estradiol enhances the stimulatory effect of insulin-like growth factor-I (IGF-I) on mammary development and growth hormone-induced IGF-I messenger ribonucleic acid. Endocrinology. 1995;136(3):1296–1302. doi: 10.1210/endo.136.3.7867584. [DOI] [PubMed] [Google Scholar]
- Rudland PS, Hughes CM. Immunocytochemical identification of cell types in human mammary gland: variations in cellular markers are dependent on glandular topography and differentiation. J Histochem Cytochem. 1989;37(7):1087–1100. doi: 10.1177/37.7.2471725. [DOI] [PubMed] [Google Scholar]
- Rudland PS, Leinster SJ, Winstanley J, Green B, Atkinson M, Zakhour HD. Immunocytochemical identification of cell types in benign and malignant breast diseases: variations in cell markers accompany the malignant state. J Histochem Cytochem. 1993;41(4):543–553. doi: 10.1177/41.4.8450194. [DOI] [PubMed] [Google Scholar]
- Rybarczyk BJ, Simpson-Haidaris PJ. Fibrinogen assembly, secretion, and deposition into extracellular matrix by MCF-7 human breast carcinoma cells. Cancer Res. 2000;60(7):2033–2039. [PubMed] [Google Scholar]
- Scarpace PJ, Matheny M, Tumer N. Differential down-regulation of beta3-adrenergic receptor mRNA and signal transduction by cold exposure in brown adipose tissue of young and senescent rats. Pflugers Arch. 1999;437(3):479–483. doi: 10.1007/s004240050804. [DOI] [PubMed] [Google Scholar]
- Sebastian J, Richards RG, Walker MP, Wiesen JF, Werb Z, Derynck R, Hom YK, Cunha GR, DiAugustine RP. Activation and function of the epidermal growth factor receptor and erbB-2 during mammary gland morphogenesis. Cell Growth Differ. 1998;9(9):777–785. [PubMed] [Google Scholar]
- Shelesnyak MC. Histamine releasing activity of natural estrogens. Proc Soc Exp Biol Med. 1959;100(4):739–741. doi: 10.3181/00379727-100-24762. [DOI] [PubMed] [Google Scholar]
- Shyamala G, Ferenczy A. The nonresponsiveness of lactating mammary gland to estradiol. Endocrinology. 1982;110(4):1249–1256. doi: 10.1210/endo-110-4-1249. [DOI] [PubMed] [Google Scholar]
- Shyamala G, Ferenczy A. Mammary fat pad may be a potential site for initiation of estrogen action in normal mouse mammary glands. Endocrinology. 1984;115(3):1078–1081. doi: 10.1210/endo-115-3-1078. [DOI] [PubMed] [Google Scholar]
- Silberstein GB, Van Horn K, Shyamala G, Daniel CW. Essential role of endogenous estrogen in directly stimulating mammary growth demonstrated by implants containing pure antiestrogens. Endocrinology. 1994;134(1):84–90. doi: 10.1210/endo.134.1.8275973. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DF. ErbBs in mammary development. Exp Cell Res. 2003;284(1):89–98. doi: 10.1016/s0014-4827(02)00103-9. [DOI] [PubMed] [Google Scholar]
- Stern DF. ERBB3/HER3 and ERBB2/HER2 duet in mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2008;13(2):215–223. doi: 10.1007/s10911-008-9083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoughton RS, Dai H. Statistical combining of cell expression profiles. 6351712 USA patent. 2002
- Thomas TJ, Faaland CA, Adhikarakunnathu S, Watkins LF, Thomas T. Induction of p21 (CIP1/WAF1/SID1) by estradiol in a breast epithelial cell line transfected with the recombinant estrogen receptor gene: a possible mechanism for a negative regulatory role of estradiol. Breast Cancer Res Treat. 1998;47(2):181–193. doi: 10.1023/a:1005925931215. [DOI] [PubMed] [Google Scholar]
- Toft D, O’Malley BW. Target tissue receptors for progesterone: the influence of estrogen treatment. Endocrinology. 1972;90(4):1041–1045. doi: 10.1210/endo-90-4-1041. [DOI] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Varnum BC, Young C, Elliott G, Garcia A, Bartley TD, Fridell YW, Hunt RW, Trail G, Clogston C, Toso RJ, et al. Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth-arrest-specific gene 6. Nature. 1995;373(6515):623–626. doi: 10.1038/373623a0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22(9):1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, Zuzan H, Olson JA, Jr, Marks JR, Nevins JR. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98(20):11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesen JF, Young P, Werb Z, Cunha GR. Signaling through the stromal epidermal growth factor receptor is necessary for mammary ductal development. Development. 1999;126(2):335–344. doi: 10.1242/dev.126.2.335. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Sims AH, Howell A, Miller CJ, Clarke RB. Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocr Relat Cancer. 2006;13(2):617–628. doi: 10.1677/erc.1.01165. [DOI] [PubMed] [Google Scholar]
- Wurfel J, Rosel M, Seiter S, Claas C, Herlevsen M, Weth R, Zoller M. Metastasis-association of the rat ortholog of the human epithelial glycoprotein antigen EGP314. Oncogene. 1999;18(14):2323–2334. doi: 10.1038/sj.onc.1202542. [DOI] [PubMed] [Google Scholar]
- Xie W, Paterson AJ, Chin E, Nabell LM, Kudlow JE. Targeted expression of a dominant negative epidermal growth factor receptor in the mammary gland of transgenic mice inhibits pubertal mammary duct development. Mol Endocrinol. 1997;11(12):1766–1781. doi: 10.1210/mend.11.12.0019. [DOI] [PubMed] [Google Scholar]
- Xu J, Fan S, Rosen EM. Regulation of the Estrogen-Inducible Gene Expression Profile by the Breast Cancer Susceptibility Gene BRCA1. Endocrinology. 2005;146(4):2031–2047. doi: 10.1210/en.2004-0409. [DOI] [PubMed] [Google Scholar]
- Zeppa R, Womack NA. The role of histamine release in chronic cystic mastitis. Surgery. 1962;52:195–202. [PubMed] [Google Scholar]
- Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, Jeffrey SS. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15(6):2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi I, Mento E, Kuznetsov VA, Scotti M, Valsecchi V, Simionati B, Vicinanza E, Valle G, Pilotti S, Reinbold R, Vezzoni P, Albertini A, Dulbecco R. Gene expression profiles of epithelial cells microscopically isolated from a breast-invasive ductal carcinoma and a nodal metastasis. PNAS. 2004;101(52):18147–18152. doi: 10.1073/pnas.0408260101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.