Abstract
Oxidative modification to the side chain of histidine can noticeably change the collision-induced dissociation (CID) pathways of peptides containing this oxidized residue. In cases where an oxidized peptide consists of two or more isomers differing only in the site of modification, oxidation to histidine usually causes the other oxidized sites to be mis-assigned in CID spectra. These spectral mis-assignments can sometimes be avoided by using multiple stages of MS/MS (MSn) or via specially-optimized liquid chromatographic separation conditions. In this manuscript, we demonstrate that these mis-assignments can be more readily and easily avoided by using electron-transfer dissociation (ETD) to dissociate the oxidized peptides. Furthermore, we find that the relative insensitivity of ETD to side chain chemistry allows the extent of oxidative modification to be determined readily for peptide isomers having more than one site of oxidation. The current results along with previous studies of oxidized peptides suggest that ETD is probably a better technique than CID for obtaining correct sequence and modification information for oxidized peptides.
Keywords: electron transfer dissociation, oxidized peptides, MS/MS, peptide sequencing, protein oxidation
Introduction
Mass spectrometry (MS) is the technique of choice to determine amino acid modifications in peptides and proteins. Numerous studies have used MS to identify and locate protein post-translational modifications,1-3 and increasingly MS is being used in combination with covalent labeling reactions to study protein structures. An emerging set of methods to study protein structures are those that rely on oxidative modifications as indicators of structure. These methods use radicals (e.g. •OH) to modify solvent-exposed4-8 or metal-bound amino acids.9-20 After oxidation, modifications to amino acid side chains are usually identified by digesting the protein of interest with a proteolytic enzyme and subjecting the modified peptides to tandem MS (MS/MS) analyses. The pattern of amino acid modifications is then used to obtain information about the protein structure or changes in its structure upon binding a ligand or another protein. During the MS/MS analyses, determination of the modified amino acids relies on finding product ions whose m/z ratios are shifted from expected values.
Determining the individual amino acid modifications is usually straightforward because oxidative modifications often do not change peptide ion dissociation patterns, but there are several examples in which they do. Oxidative modifications to cysteine and methionine residues have very noticeable effects on peptide ion dissociation patterns. For instance, oxidation of cysteine to cysteic acid can lead to very selective peptide dissociation in some cases and to more efficient overall peptide dissociation in other cases.21-24 The presence of cysteic acid enhances peptide bond dissociation at its C-terminal side and can allow mobilization of an additional proton that initiates cleavages more efficiently at other peptide bonds as well. Oxidation of cysteine to cysteine sulfinic acid also gives rise to selective dissociation on the C-terminal side of this residue when the number of arginine residues equals or exceeds the peptide charge state.25, 26 The prevalence of selective dissociations can ultimately limit the structural information obtained from CID studies. Oxidation of methionine to methionine sulfoxide can also have a notable effect on peptide ion dissociation patterns.27-32 When the number of protons on the peptide does not exceed the number of basic residues, product ion spectra of peptides containing methionine sulfoxide are dominated by a neutral loss of methane sulfenic acid (CH3SOH).31 Indeed, in many cases no other sequence information is present, highlighting the effect that this oxidative modification can have.
While cysteine and methionine residues are readily oxidized, other amino acids such as those with aromatic side chains are also susceptible to oxidation.33 Histidine, in particular, can be easily oxidized, and our group recently reported that oxidation of histidine to 2-oxo-histidine can change peptide dissociation patterns very noticeably.34 In some cases, we have found that histidine oxidation can result in tandem mass spectra with misleading information, causing oxidized amino acids to be incorrectly identified. Clearly, such misleading information is very problematic for methods that rely on oxidative modifications as indicators of protein structure. These incorrect assignments during CID analyses could have broader significance as well. Oxidized histidine residues are commonly found in proteins from cells that have undergone oxidative stress.35-37 2-oxo-histidine is even considered to be a marker of cellular oxidative stress.38
Given the spectral interpretation difficulties that oxidized histidines can cause when peptides containing this oxidized residue are subjected to CID, we set out to explore the merits of using electron transfer dissociation (ETD) as an alternative. The success of ETD, and the analogous technique electron-capture dissociation, in obtaining sequence information from post-translationally modified peptides is well established.39-44 We have also previously shown that ETD provides more useful sequence information for peptides containing oxidized cysteine or methionine residues.45 Thus, we predicted that for peptides with oxidized histidines ETD would be able to provide product ion spectra that are less prone to misinterpretation. In addition, for peptides in which more than one residue is modified, we were interested in investigating whether ETD could provide accurate, semi-quantitative data about the degree to which individual sites are modified. Such data are very difficult to obtain with CID but could be useful for improving the accuracy of protein structural analysis methods that rely on oxidative labeling.
Experimental
Materials
Hydrogen peroxide (30%), formic acid, tris(hydroxymethyl)-aminomethane (Tris), and tris(hydroxymethyl)aminomethane hydrochloride (Tris-HCl) were obtained from EM Science (Gladstone, NJ). Ascorbic acid and copper(II) sulfate were purchased from the Sigma-Aldrich Corporation (St. Louis, MO). Acetic acid and HPLC-grade methanol were obtained from Fisher Scientific (Fair Lawn, NJ). All reagents were used as provided. Distilled, deionized water was generated with a Millipore (Burlington, MA) Simplicity 185 water purification system.
The peptides angiotensin I (DRVYIHPFHL), OVA peptide 323-339 (ISQAVHAAHAEINEAGR), β-Amyloid 10-20 (YEVHHQKLVFF), Myelin Proteolipid Protein 139-151 (HSLGKWLGHPDKF), and Brain Derived Acidic Fibroblast Growth Factor 102-111 (HAEKHWFVGL) were obtained from American peptide company (Sunnyvale, CA).
Peptide oxidation
All peptides were oxidized using metal-catalyzed oxidation (MCO) reactions as described previously.12-17 Briefly, MCO reactions were performed at room temperature in aqueous solutions containing 250 μM peptide, 250 μM CuSO4, 10 mM ascorbate, 2 mM H2O2, and 50 mM Tris-HCl/Tris, buffered to a pH of 7.4. In all cases the reactions were initiated by the addition of ascorbate, H2O2, or both. The reactions were stopped after 30 minutes by the addition of 1% (by volume) glacial acetic acid.
Instrumentation
Mass spectral analyses were performed on a Bruker (Billerica, MA) Esquire LC quadrupole ion trap mass spectrometer and a Bruker HCTultra PTM Discovery System quadrupole ion trap mass spectrometer. On both mass spectrometers the source conditions were chosen to maximize the ion signal of the desired ions. For direct injection experiments on the Esquire LC, the sample was delivered at 1 μL/min using a syringe pump. HPLC-MS analyses on this mass spectrometer were conducted using an HP1100 (Agilent, Wilmington DE) system with a Discovery C18 column (2.1 × 150 mm; Supelco). The LC effluent was split in a 1:4 ratio with the smaller outlet being fed into the electrospray ionization (ESI) source of the mass spectrometer. For separation of the oxidized peptide isomers, isocratic separations were performed. The two mobile phases that were used were (A) water with 0.1% formic acid and (B) methanol with 0.1% formic acid. The mobile phase composition during the separation of the oxidized isomers consisted of between 30 and 40% of mobile phase B and 60 to 70% of mobile phase A, depending on the peptide to be separated.
Tandem mass spectrometry (MS/MS) experiments using electron transfer dissociation (ETD) were carried out on a Bruker HCTultra PTM Discovery quadrupole ion trap mass spectrometer. Samples were infused at a flow rate of 1 to 2 μL/min using a Cole Palmer syringe pump (Vernon Hills, IL). ETD was performed using total reaction times between 50 and 200 ms and low mass cutoffs ranging from m/z 30 to 200; these values were chosen to maximize dissociation efficiency. Accumulation times for the fluoranthene radical anion, which was used as the ETD reagent, were optimized for each experiment but were typically between 3 ms and 5 ms. In some cases, collisional activation was used to aid the ETD of doubly-charged ions. When this procedure was performed, the collision activation process used a low-amplitude resonance excitation voltage to resonantly excite the singly-charged radical cations that did not dissociate upon ETD of their doubly-charged precursors in a manner similar to previously described.46
Results and Discussion
Histidine oxidation: misassignments of oxidized residues using CID but correct assignments using ETD
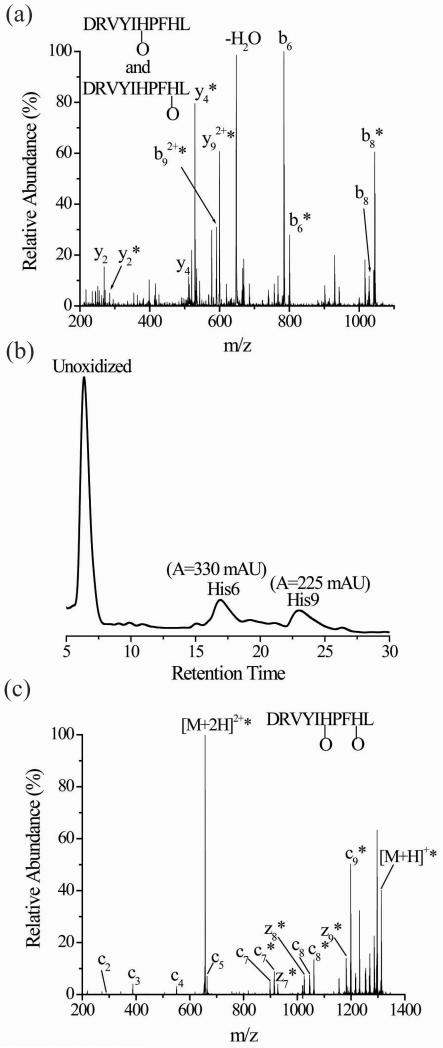
Oxidation of histidine can lead to very noticeable changes in the dissociation patterns of peptide ions containing this residue as we demonstrated recently.33 The preferential cleavage at the C-terminal side of histidine, which is often observed in product ion spectra of peptides with basic residues that equal or exceed the number of protons on the peptide,47-52 is reduced or eliminated upon oxidation of histidine. Furthermore, tandem mass spectra of peptides with oxidized histidine residues can easily be misinterpreted, especially when the oxidized peptide is a mixture of more than one isomer differing in the site of oxidation.34 For example, by comparing the percentages of oxidized forms for the product ions from CID (Figure 1a) of oxidized angiotensin I (DRVYIHPFHL), one concludes that His6, Pro7, Phe8, and His9 are all likely oxidation sites (Table 1). A series of MSn experiments on the y4+O, (b9+O)2+, and the b8+O product ions determines that oxidation is limited to only His6 and His9 (Figures S1, S2, and S3 in Supplemental Information). Separation of the oxidized isomers by LC and subsequent MS/MS analysis also confirms the presence of only two isomers with oxidation at His6 and His9, respectively (Figure S4, Supplemental Information). It should be noted that isocratic separation conditions are necessary to separate these two isomers (Figure 1b). Such conditions are atypical for most LC-MS studies of peptides from, for example, protein digests. The misleading interpretation of the initial CID spectrum (i.e. data in Table 1) arises because oxidative modifications at His6 and His9 suppress the normally prominent dissociation pathways adjacent to these residues. Shut down of these preferential dissociation pathways occurs at different places in the two different oxidized isomers, which is what causes the spectral misinterpretation.34
Figure 1.
(a) CID of singly oxidized angiotensin I (M+O+2H)2+ with prior LC separation. (b) UV trace of the two oxidized isomers after LC separation. The His6 and His9 labels in the inset correspond to peptides that are oxidized at these residues. (c) ETD of singly oxidized angiotensin I (M+O+2H)2+ after simultaneously subjecting the isomeric His6- and His9-oxidized forms to ETD without prior separation. The asterisks indicate the product ions that are oxidized.
Table 1.
Percentage of oxidized product ions observed in the CID spectrum of the doubly charged ion of oxidized angiotensin peptide (M+O+2H)2+
| Product Ion | m/zunoxidizeda | m/zoxidizeda | Percentage oxidizedb,c |
|---|---|---|---|
| y2 | 269 | 285 | 25 ± 2 |
| y3 | 416 | 432 | 15 ± 1 |
| y4 | 513 | 529 | 90 ± 5 |
| y5 | 649 | 665 | 100 |
| y6 | 763 | 779 | 100 |
| y7 | 926 | 942 | 100 |
| y8 | 1025 | 1041 | 100 |
| y92+ | 591 | 599 | 100 |
| b2 | 272 | 288 | 0 |
| b3 | 371 | 387 | 0 |
| b4 | 534 | 550 | 0 |
| b5 | 647 | 663 | 0 |
| b6 | 784 | 800 | 20 ± 6 |
| b7 | 881 | 897 | 45 ± 3 |
| b8 | 1028 | 1044 | 80 ± 5 |
| b92+ | 583 | 591 | 100 |
These m/z ratios correspond to the nominal m/z ratio of the observed product ions.
The percentage oxidized is obtained by dividing the ion abundance of the oxidized product ion by the sum of the oxidized and unoxidized product ions.
The error associated with oxidized percentage value corresponds to the standard deviation from at least three separate measurements
Because the dissociation pathways in ETD are less sensitive to side-chain chemistry,45 we predicted that ETD of oxidized angiotensin I might avoid misassignments of the oxidized residues. So, ETD was performed on the doubly protonated version of oxidized angiotensin I without prior LC separation of the oxidized isomers. This ETD spectrum shows unoxidized c2-c5 ions, unoxidized and oxidized forms of the c7 and c8 product ions, and only an oxidized form of the c9 product ion in addition to only oxidized forms of the z7-z9 product ions (Figure 1c). By comparing the percentages of oxidized forms for each product ion, it is clear that the same misassignments that arise during interpretation of the CID spectrum (Figure 1a and Table 1) do not arise upon interpretation of the ETD spectrum (Table 2). The oxidation percentages indicate that His6 and His9 are oxidized. The ring structure of proline at residue 7 prevents the formation of a c6 product ion, so the actual ETD data itself cannot alone confirm whether His6 or Pro7 is oxidized, but in this case we know that His6 is oxidized from the previous LC-CID-MS/MS data. Moreover, the percent oxidation of the c7 and c8 ions indicate that oxidation at His6 accounts for about 65% of the oxidized peptide, which is consistent with the LC data shown in Figure 1b. The integrated peak areas for the two oxidized isomers from the UV chromatogram indicate that His6 accounts for about 60% of the oxidation. As expected, oxidation of His6 and His9 does not significantly influence the extent to which the c7-c9 product ions are formed from the two different isomers.
Table 2.
Percentage of oxidized product ions observed in the ETD spectrum of the doubly charged ion of the oxidized angiotensin I peptide (M+O+2H)2+
| Product Ion | m/zunoxidizeda | m/zoxidizeda | Percentage oxidizedb,c |
|---|---|---|---|
| c2 | 289 | 305 | 0 |
| c3 | 388 | 404 | 0 |
| c4 | 551 | 567 | 0 |
| c5 | 664 | 680 | 0 |
| c7 | 898 | 914 | 66 ± 2 |
| c8 | 1045 | 1061 | 64 ± 1 |
| c9 | 1182 | 1198 | 100 |
| z7 | 910 | 926 | 100 |
| z8 | 1009 | 1025 | 100 |
| z9 | 1165 | 1181 | 100 |
These m/z ratios correspond to the nominal m/z ratio of the observed product ions.
The percentage oxidized is obtained by dividing the ion abundance of the oxidized product ion by the sum of the oxidized and unoxidized product ions.
The error associated with oxidized percentage value corresponds to the standard deviation from at least three separate measurements
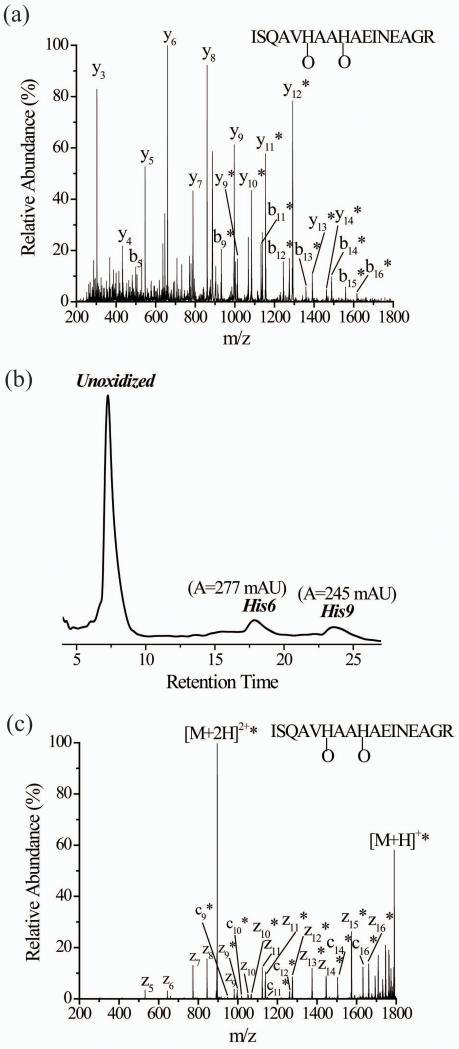
Another example of the effect of oxidized histidine on peptide CID patterns involves the OVA peptide (ISQAVHAAHAEINEAGR). The CID spectrum of the (M+O+2H)2+ species of the OVA peptide is difficult to interpret correctly (Figure 2a). Upon examining the percentages of oxidized forms for each product ion in the y- and b-series, there appears to be a mixture of perhaps four isomeric peptide ions differing by the site of oxidation. This conclusion arises from the series of y ions from y9 to y12 and b ions from b6 to b9 that have increasing oxidation percentages (Table S1, Supplemental Information). Complete oxidation (100%) of the product ions from y12 to y14 and b9 to b16, along with increasing oxidation percentages in the b6, b7, and b8 ions and the y9, y10, and y11 ions suggest that His6, Ala7, Ala8, and His9 are all oxidized. LC-MS data of the oxidized OVA peptide, again using atypical isocratic conditions, provides evidence for only two oxidized isomers (Figure 2b). MS/MS of the two individual oxidized isomers after LC separation clarifies that oxidation is limited to only His6 and His9 (Figures S5a and b, Supplemental Information). Presumably the difficulty in correctly interpreting the CID data arises from a similar effect observed with angiotensin I.
Figure 2.
(a) CID of singly oxidized OVA peptide (M+O+2H)2+ after simultaneously subjecting the isomeric His6- and His9- oxidized forms to CID without prior separation. (b) The UV trace of the two oxidized isomers of singly oxidized OVA peptide after LC separation. The His6 and His9 labels correspond to peptides that are oxidized at these residues. (c) ETD of singly oxidized OVA peptide (M+O+2H)2+ after simultaneously subjecting the isomeric His6- and His9-oxidized forms to ETD without prior separation. The asterisks indicate the product ions that are oxidized.
The ETD spectrum of the unseparated (M+O+2H)2+ species of the OVA peptide shows unoxidized z5-z8 product ions, unoxidized and oxidized forms of z9-z11 product ions, and oxidized z12-z16 and c9-16 product ions (Figure 2c). Comparing the percentages of the oxidized product ions, it becomes clear that only His6 and His9 are oxidized and Ala7 and Ala8 are not (Table 3). Moreover, the percent oxidation of the z9, z10, and z11 product ions indicate that oxidation at His9 accounts for about 46% of the oxidized peptide, which is consistent with the LC data shown in Figure 2b, which indicates His9 accounts for about 47% of the oxidation. The ETD spectrum of the triply-charged version of the oxidized peptide also shows very similar oxidation percentages, indicating that the charge state has little effect on the information that is gathered (Figure S6, Supplemental Information). Just as in the case of angiotensin I, ETD appears to provide more accurate and straightforward information than CID for peptides containing oxidized histidine residues.
Table 3.
Percentage of oxidized product ions observed in the ETD spectrum of the doubly charged ion of the oxidized Ova peptide (M+O+2H)2+
| Product Ion | m/zunoxidizeda | m/zoxidizeda | Percentage oxidizedb,c |
|---|---|---|---|
| z5 | 530 | 546 | 0 |
| z6 | 643 | 659 | 0 |
| z7 | 772 | 788 | 0 |
| z8 | 843 | 859 | 0 |
| z9 | 980 | 996 | 46.1 ± 0.3 |
| z10 | 1051 | 1067 | 45 ± 3 |
| z11 | 1122 | 1138 | 46 ± 1 |
| z12 | 1259 | 1275 | 100 |
| z13 | 1358 | 1374 | 100 |
| z14 | 1429 | 1445 | 100 |
| z15 | 1557 | 1573 | 100 |
| z16 | 1644 | 1660 | 100 |
| c9 | 932 | 948 | 100 |
| c10 | 1003 | 1019 | 100 |
| c11 | 1132 | 1148 | 100 |
| c12 | 1245 | 1261 | 100 |
| c13 | 1359 | 1375 | 100 |
| c14 | 1488 | 1504 | 100 |
| c15 | 1559 | 1575 | 100 |
| c16 | 1616 | 1632 | 100 |
These m/z ratios correspond to the nominal m/z ratio of the observed product ions.
The percentage oxidized is obtained by dividing the ion abundance of oxidized and unoxidized product ions.
The error associated with oxidized percentage value corresponds to the standard deviation from at least three separate measurements.
Misassignments of other oxidized residues can be corrected using ETD
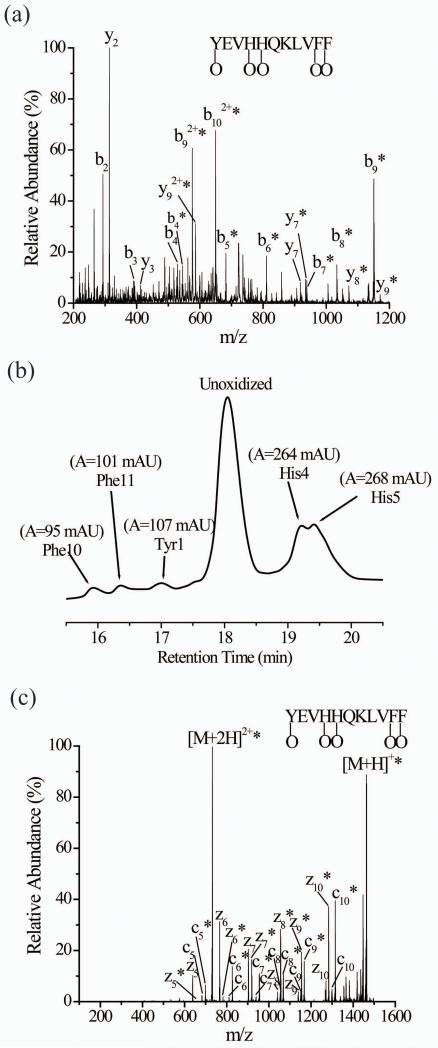
Even when residues other than histidine are oxidized, CID data is often ambiguous with regards to the sites of oxidation. An example is the CID data of singly-oxidized β-amyloid peptide (YEVHHQKLVFF) (Figure 3a and Table S2, Supplemental Information). The oxidation percentages for the y2, b9, b92+, and b102+ product ions indicate that Phe10 and Phe11 are oxidized and even suggest that oxidation at these two residues accounts for about 10-15% of the peptide’s oxidation. The failure to observe oxidized versions of the y3, y5, and y6 ions, however, bring this conclusion into question. Similarly confusing are the changes in the oxidation percentages of the b-series of ions from the b9 ion to the b5 ion. The oxidation percentage drops by about 30% from the b9 to b8 but the percentage is successively higher (73%, 78%, and 83%) for the b7, b6, and b5 ions, respectively; note that the oxidation percentage for the b9 and b5 ions are essentially the same. These reproducibly inconsistent oxidation percentages, along with the unoxidized y3, y5, and y6, make it impossible to confidently conclude which residues are oxidized in the β-amyloid peptide. LC-MS data of the oxidized peptide, however, provide evidence for five different oxidized isomers (Figure 3b). Just like in the examples above, atypical LC conditions were necessary to fully separate these isomers. MS/MS of each individual oxidized isomer after LC separation indicates that oxidation occurs at Tyr1, His4, His5, Phe10 and Phe11 (Figure S7, Supplemental Information).
Figure 3.
(a) CID of singly oxidized β-amyloid peptide (M+O+2H)2+ after simultaneously subjecting the isomeric oxidized forms to CID without prior separation. (b) The UV trace of the five oxidized isomers of singly oxidized β-amyloid peptide after LC separation. The Phe10, Phe11, Tyr1, His4, and His5 labels correspond to peptides that are oxidized at these residues. (c) ETD of singly oxidized β-amyloid peptide (M+O+2H)2+ after simultaneously subjecting the isomeric oxidized forms to ETD without prior separation. The asterisks indicate the product ions that are oxidized.
The ETD spectrum of oxidized β-amyloid shows unoxidized and oxidized forms of z5-z10 ions and c5-c10 product ions (Figure 3c). The presence of unoxidized and oxidized z5-z10 and c5-c10 product ions and increasing percentages of oxidized forms indicate that Tyr1, His4, His5, Phe10 and Phe11 are oxidized (Table 4). For example, 84% of the z10 product ions are oxidized, indicating that the extent of oxidation at Tyr1 is 16%. Similarly, 91% of the c10 product ions are oxidized, indicating that the extent of oxidation at Phe11 is 9%. If a similar analysis is done for the other residues, the oxidation percentages for Phe10 (9%), His4 (30%), and His5 (36%) can be determined. The oxidation percentages obtained from the ETD data are consistent with the UV absorbances from LC (Figure 3b). The ETD spectrum of the triply-charged version of oxidized β-amyloid (Figure S8, Supplemental Information) also gives very similar extents of oxidation for each of the oxidized residues.
Table 4.
Percentage of oxidized product ions observed in the ETD spectrum of the doubly charged ion of the oxidized β-amyloid peptide (M+O+2H)2+
| Product Ion | m/zunoxidizeda | m/zoxidizeda | Percentage oxidizedb,c |
|---|---|---|---|
| z5 | 637 | 653 | 18 ± 1 |
| z6 | 765 | 781 | 19 ± 2 |
| z7 | 902 | 918 | 48 ± 2 |
| z8 | 1039 | 1055 | 84 ± 2 |
| z9 | 1138 | 1154 | 83 ± 4 |
| z10 | 1267 | 1283 | 84 ± 4 |
| c5 | 683 | 699 | 82 ± 3 |
| c6 | 811 | 827 | 81 ± 2 |
| c7 | 939 | 955 | 83 ± 1 |
| c8 | 1052 | 1068 | 82 ± 1 |
| c9 | 1151 | 1167 | 81.0 ± 0.4 |
| c10 | 1298 | 1314 | 91 ± 1 |
These m/z ratios correspond to the nominal m/z ratio of the observed product ions.
The percentage oxidized is obtained by dividing the ion abundance of the oxidized product ion by the sum of the oxidized and unoxidized product ions.
The error associated with oxidized percentage value corresponds to the standard deviation from at least three separate measurements
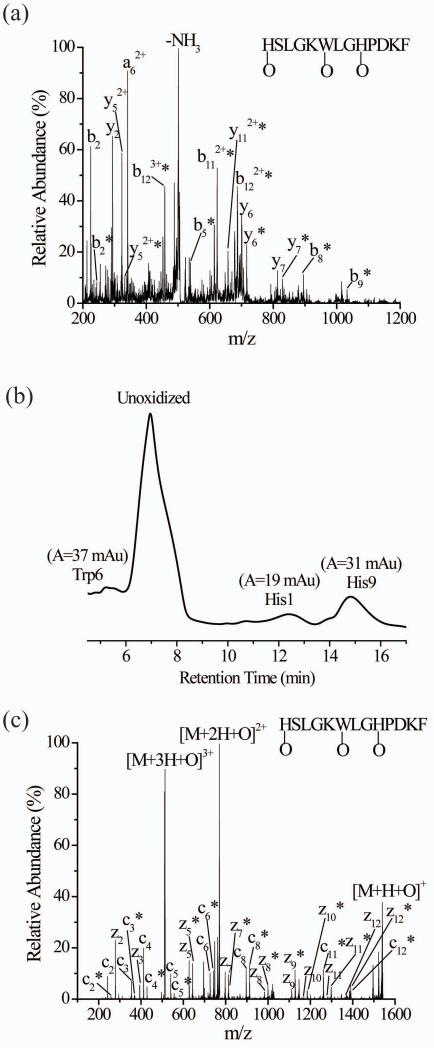
Another example in which CID experiments are unable to provide confident assignment of the oxidation sites involves the Myelin Proteolipid Protein (MPP) 139-151 (HSLGKWLGHPDKF). The CID spectrum of the triply-charged oxidized peptide (Figure 4a and Table S3), without prior LC separation, suggests a number of oxidation sites, with definitive assignments difficult to make. LC-MS data of the oxidized peptide indicate that three oxidized isomers exist (Figure 4b), and MS/MS of each isomer points to His1, Trp6, and His9 as the oxidized residues. Unlike the CID spectrum, the ETD spectrum of triply-charged oxidized peptide from an unseparated sample correctly identifies the three oxidation sites (Figure 4c) and provides the correct oxidation percentages for each residue. The presence of unoxidized and oxidized z5-z12 and c2-c8 product ions and increasing percentages of these oxidized forms provide clear evidence that His1, Trp6, and His9 are modified (Table 5). Moreover, the degree of oxidation at each residue (i.e. His1 = 20%; Trp6 = 30%; His9 = 50%) is consistent with the LC data for this peptide (Figure 4b). Yet again, ETD more accurately identifies oxidation sites than CID. We have obtained similar results for other peptides that contain both oxidized histidine residues and other oxidized residues. These peptides include HAEKHWFVGL and WGQGGGTHNQ.
Figure 4.
(a) CID of singly oxidized MPP peptide (M+O+3H)3+ after simultaneously subjecting the isomeric oxidized forms to CID without prior separation. (b) The UV trace of the three oxidized isomers of the singly oxidized MPP peptide after LC separation. The Trp6, His1, and His9 labels correspond to peptides that are oxidized at these residues. (c) ETD of singly oxidized MPP peptide (M+O+3H)3+ after simultaneously subjecting the isomeric oxidized forms to ETD without prior separation. The asterisks indicate the product ions that are oxidized.
Table 5.
Percentage of oxidized product ions observed in the ETD spectrum of the triply charged ion of the oxidized MPP peptide (M+O+3H)3+
| Product Ion | m/zunoxidizeda | m/zoxidizeda | Percentage oxidizedb,c |
|---|---|---|---|
| z2 | 278 | 294 | 0 |
| z3 | 393 | 409 | 0 |
| z5 | 627 | 643 | 48.3 ± 0.3 |
| z7 | 797 | 813 | 47 ± 1 |
| z8 | 983 | 999 | 79 ± 2 |
| z9 | 1111 | 1127 | 78 ± 1 |
| z10 | 1168 | 1184 | 79 ± 2 |
| z11 | 1281 | 1297 | 80 ± 6 |
| z12 | 1368 | 1384 | 79 ± 2 |
| c2 | 242 | 258 | 21 ± 1 |
| c3 | 355 | 371 | 19 ± 2 |
| c4 | 412 | 428 | 20 ± 1 |
| c5 | 540 | 556 | 21 ± 1 |
| c6 | 726 | 742 | 51.8 ± 0.4 |
| c7 | 839 | 855 | 52 ± 5 |
| c8 | 896 | 912 | 53 ± 1 |
| c10 | 1130 | 1146 | 100 |
| c11 | 1245 | 1261 | 100 |
| c12 | 1373 | 1289 | 100 |
These m/z ratios correspond to the nominal m/z ratio of the observed product ions.
The percentage oxidized is obtained by dividing the ion abundance of the oxidized product ion by the sum of the oxidized and unoxidized product ions.
The error associated with oxidized percentage value corresponds to the standard deviation from at least three separate measurements
Conclusions
Histidine oxidation influences peptide dissociation patterns in such a way that CID data from these peptides often leads to incorrect assignments of oxidized residues. ETD, however, overcomes this problem because dissociation patterns from this technique are relatively insensitive to side chain chemistry. This relative insensitivity to side-chain chemistry also allows the amount of oxidation occurring at given residues to be accurately determined from ETD spectra of peptides that have more than one oxidation site. In contrast, CID spectra of such peptides rarely reflect the accurate oxidation extent. LC-MS can help overcome this problem, but atypical LC conditions are needed to fully separate the oxidized isomers. ETD provides a way to get the correct information by dissociating the oxidized isomers simultaneously. With this dissociation technique, typical LC conditions can be used without having to be concerned about the inability to separate oxidized isomers. Finally, with an increasing number of researchers using oxidative surface mapping to study protein structure, ETD would appear to be the dissociation technique of choice to avoid misidentification and mis-quantitation of oxidation sites that are quite common with CID data.
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Institutes of Health (RO1 GM075092).
References
- 1.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nature Methods. 2007;4:798. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 2.Dalpathado DS, Desaire H. Glycopeptide analysis by mass spectrometry. Analyst. 2008;133:731. doi: 10.1039/b713816d. [DOI] [PubMed] [Google Scholar]
- 3.Temporini C, Callerli E, Massolini G, Caccialanza G. Integrated analytical strategies for the study of phosphorylation and glycosylation in proteins. Mass Spectrometry Reviews. 2008;27:207. doi: 10.1002/mas.20164. [DOI] [PubMed] [Google Scholar]
- 4.Maleknia SD, Downard K. Radical approaches to probe protein structure, folding, and interactions by mass spectrometry. Mass Spectrometry Reviews. 2001;20:388. doi: 10.1002/mas.10013. [DOI] [PubMed] [Google Scholar]
- 5.Guan JQ, Chance MR. Structural proteomics of macromolecular assemblies using oxidative footprinting and mass spectrometry. Trends in Biochemical Sciences. 2005;30:583. doi: 10.1016/j.tibs.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Downard KM. Ions of the interactome: The role of MS in the study of protein interactions in proteomics and structural biology. Proteomics. 2006;6:5374. doi: 10.1002/pmic.200600247. [DOI] [PubMed] [Google Scholar]
- 7.Takamoto K, Chance MR. Radiolytic protein footprinting with mass Spectrometry to probe the structure of macromolecular complexes. Annual Review of Biophysics and Biomolecular Structure. 2006;35:251. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 8.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical Reviews. 2007;107:3514. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 9.Kurahashi T, Miyazaki A, Suwan S, Isobe M. Extensive investigations on oxidized amino acid residues in H2O2-treated Cu,Zn-SOD protein with LC-ESI-Q-TOF-MS, MS/MS for the determination of the copper-binding site. Journal of American Chemical Society. 2001;123:9268. doi: 10.1021/ja015953r. [DOI] [PubMed] [Google Scholar]
- 10.Hovorka SW, Williams TD, Schöneich C. Characterization of the metal-binding site of bovine growth hormone through site-specific metal catalyzed oxidation and high-performance liquid chromatography-tandem mass spectrometry. Analytical Biochemistry. 2002;300:206. doi: 10.1006/abio.2001.5447. [DOI] [PubMed] [Google Scholar]
- 11.Schöneich C, Williams TD. Cu(II)-catalyzed oxidation of β-amyloid peptide targets His13 and His14 over His6: detection of 2-oxo-histidine by HPLC-MS/MS. Chemical Research in Toxicology. 2002;15:717. doi: 10.1021/tx025504k. [DOI] [PubMed] [Google Scholar]
- 12.Lim J, Vachet RW. Development of a methodology based on metal-catalyzed oxidation reactions and mass spectrometry to determine the metal binding sites in copper metalloproteins. Analytical Chemistry. 2003;75:1164. doi: 10.1021/ac026206v. [DOI] [PubMed] [Google Scholar]
- 13.Lim J, Vachet RW. Using mass spectrometry to study copper-protein binding under native and non-native conditions. Analytical Chemistry. 2004;76:3498. doi: 10.1021/ac049716t. [DOI] [PubMed] [Google Scholar]
- 14.Bridgewater JD, Vachet RW. Metal-catalyzed oxidation reactions and mass spectrometry: the roles of ascorbate and different oxidizing agents in determining Cu-protein-binding sites. Analytical Biochemistry. 2005;41:122. doi: 10.1016/j.ab.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 15.Bridgewater JD, Vachet RW. Using microwave-assisted metal-catalyzed oxidation reactions and mass spectrometry to increase the rate at which the copper-binding sites of a protein are determined. Analytical Chemistry. 2005;77:4649. doi: 10.1021/ac0502551. [DOI] [PubMed] [Google Scholar]
- 16.Bridgewater JD, Lim J, Vachet RW. Transition metal-peptide binding studied by metal-catalyzed oxidation reactions and mass spectrometry. Analytical Chemistry. 2006;78:2432. doi: 10.1021/ac051983r. [DOI] [PubMed] [Google Scholar]
- 17.Bridgewater JD, Lim J, Vachet RW. Using metal-catalyzed oxidation reactions and mass spectrometry to identify amino acid residues within 10 Ǻ of the metal in Cu-binding proteins. Journal of the American Society for Mass Spectrometry. 2006;17:1552. doi: 10.1016/j.jasms.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Sadineni V, Galeva NA, Schöneich C. Characterization of the metal-binding site of human prolactin by site-specific metal-catalyzed oxidation. Analytical Biochemistry. 2006;358:208. doi: 10.1016/j.ab.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Kowalik-Jankowska T, Rajewska A, Jankowska E, Wisniewska K, Grzonka Z. Products of Cu(II)-catalyzed oxidation of the N-terminal fragments of α-synuclein in the presence of hydrogen peroxide. Journal of Inorganic Biochemistry. 2006;100:1623. doi: 10.1016/j.jinorgbio.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Srikanth R, Wilson J, Burns CS, Vachet RW. Identification of Cu(II) Coordinating Residues in the Prion Protein by Metal-Catalyzed Oxidation Mass Spectrometry: Evidence for Multiple Isomers at Low Cu(II) Loadings. Biochemistry. 2008;47:9258. doi: 10.1021/bi800970m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burlet O, Yang C-Y, Gaskell SJ. Influence of cysteine to cysteic acid oxidation on the collision-activated decomposition of protonated peptides: evidence for intraionic interactions. Journal of the American Society for Mass Spectrometry. 1992;3:337. doi: 10.1016/1044-0305(92)87061-3. [DOI] [PubMed] [Google Scholar]
- 22.Cox KA, Gaskell SJ, Morris M, Whiting A. Role of the site of protonation in the low-energy decompositions of gas-phase peptide ions. Journal of the American Society for Mass Spectrometry. 1996;7:522. doi: 10.1016/1044-0305(96)00019-0. [DOI] [PubMed] [Google Scholar]
- 23.Summerfield SG, Cox KA, Gaskell SJ. The Promotion of d-type ions during the low energy collision-induced dissociation of some cysteic acid-containing peptides. Journal of the American Society for Mass Spectrometry. 1997;8:25. [Google Scholar]
- 24.Tsaprailis G, Nair H, Somogyi A, Wysocki VH, Zhong W, Futrell JH, Summerfield SG, Gaskell SJ. Influence of secondary structure on the fragmentation of protonated peptides. Journal of American Chemical Society. 1999;121:5142. [Google Scholar]
- 25.Wang Y, Vivekananda S, Men L, Zhang Q. Fragmentation of protonated ions of peptides containing cysteine, cysteine sulfinic acid, and cysteine sulfonic acid. Journal of the American Society for Mass Spectrometry. 2004;15:697. doi: 10.1016/j.jasms.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Men L, Wang Y. Further studies on the fragmentation of protonated ions of peptides containing aspartic acid, glutamic acid, cysteine sulfinic acid, and cysteine sulfonic acid. Rapid Communications in Mass Spectrometry. 2005;19:23. doi: 10.1002/rcm.1748. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, Smith JB, Abraham EC. Identification of a MS-MS fragment diagnostic for methionine sulfoxide. Journal of Mass Spectrometry. 1996;31:1309. [Google Scholar]
- 28.Lagerwerf FM, van de Weert M, Heerma W, Haverkamp J. Identification of oxidized methionine in peptides. Rapid Communications in Mass Spectrometry. 1996;10:1905. doi: 10.1002/(SICI)1097-0231(199612)10:15<1905::AID-RCM755>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 29.Qin J, Chait BT. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Analytical Chemistry. 1997;69:4002. doi: 10.1021/ac970489n. [DOI] [PubMed] [Google Scholar]
- 30.O’Hair RAJ, Reid GE. Neighboring group versus cis-elimination mechanisms for side chain loss from protonated methionine, methionine sulfoxide and their peptides. European Journal of Mass Spectrometry. 1999;5:325. [Google Scholar]
- 31.Reid GE, Roberts KD, Kapp EA, Simpson RJ. Statistical and mechanistic approaches to understanding the gas-phase fragmentation behavior of methionine sulfoxide containing peptides. Journal of Proteome Research. 2004;3:751. doi: 10.1021/pr0499646. [DOI] [PubMed] [Google Scholar]
- 32.Lioe H, Laskin J, Reid GE, O’Hair RAJ. Energetics and dynamics of the fragmentation reactions of protonated peptides containing methionine sulfoxide or aspartic acid via energy- and time-resolved surface induced dissociation. Journal of Physical Chemistry A. 2007;111:10580. doi: 10.1021/jp073040z. [DOI] [PubMed] [Google Scholar]
- 33.Buxton GV, Greenstock CL, Helman WP, Ross AB. Critical review of rate constants for reactions of hydrated electrons, hydrogen atoms, and hydroxyl radicals in aqueous solution. Journal of Physical and Chemical Reference Data. 1988;17:513. [Google Scholar]
- 34.Bridgewater JD, Srikanth R, Lim J, Vachet RW. The effect of histidine oxidation on the dissociation patterns of peptide ions. Journal of the American Society for Mass Spectrometry. 2007;18:553. doi: 10.1016/j.jasms.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chemical Research in Toxicology. 1997;10:485. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 36.Schöneich C. Reactive oxygen species and biological aging: a mechanistic approach. Experimental Gerontology. 1999;34:19. doi: 10.1016/s0531-5565(98)00066-7. [DOI] [PubMed] [Google Scholar]
- 37.Davies MJ. The oxidative environment and protein damage. Biochimica et Biophysica Acta. 2005;1703:93. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Uchida K, Kawakishi S. 2-Oxo-histidine as a novel biological marker for oxidatively modified proteins. FEBS Letters. 1993;332:208. doi: 10.1016/0014-5793(93)80632-5. [DOI] [PubMed] [Google Scholar]
- 39.Zubarev RA, Kelleher N, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. Journal of American Chemical Society. 1998;120:3265. [Google Scholar]
- 40.Stensballe A, Jensen ON, Olsen JV, Haselmann KF, Zubarev RA. Electron capture dissociation of singly and multiply phosphorylated Peptides. Rapid Communications in Mass Spectrometry. 2000;14:1793. doi: 10.1002/1097-0231(20001015)14:19<1793::AID-RCM95>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 41.Bakhtiar R, Guan Z. Electron capture dissociation mass spectrometry in characterization of post-translational modifications. Biochemical and Biophysical Research Communications. 2005;334:1. doi: 10.1016/j.bbrc.2005.05.138. [DOI] [PubMed] [Google Scholar]
- 42.Mirgorodskaya E, Roepstorff P, Zubarev RA. Localization of O-glycosylation sites in peptides by electron capture dissociation in a fourier transform mass spectrometer. Analytical Chemistry. 1999;71:4431. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 43.Bakhtiar R, Guan Z. Electron capture dissociation mass spectrometry in characterization of peptides and proteins. Biotechnology Letters. 2006;28:1047. doi: 10.1007/s10529-006-9065-z. [DOI] [PubMed] [Google Scholar]
- 44.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochimica et Biophysica Acta. 2006;1764:1811. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srikanth R, Wilson J, Bridgewater JD, Numbers JR, Lim J, Olbris MR, Kettani A, Vachet RW. Improved sequencing of oxidized cysteine and methionine containing peptides using electron transfer dissociation. Journal of the American Society for Mass Spectrometry. 2007;18:1499. doi: 10.1016/j.jasms.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Analytical Chemistry. 2007;79:477. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Wysocki VH, Tabb DL, Yates JR. The influence of histidine on cleavage C-terminal to acidic residues in doubly-protonated tryptic peptides. International Journal of Mass Spectrometry. 2002;219:233. [Google Scholar]
- 48.Tabb DL, Smith LL, Breci LA, Wysocki VH, Lin D, Yates JR. Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Analytical Chemistry. 2003;75:1155. doi: 10.1021/ac026122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: A framework for understanding peptide dissociation. Journal of Mass Spectrometry. 2000;35:1399. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 50.Farrugia JM, Taverner T, O’Hair RAJ. Side-chain involvement in the fragmentation reactions of the protonated methyl esters of histidine and its peptides. International Journal of Mass Spectrometry. 2001;209:99. [Google Scholar]
- 51.Farrugia JM, O’Hair RAJ, Reid GE. Do all b2 ions have oxazolone structures? Multistage mass spectrometry and ab initio studies on protonated N-acyl amino acid methyl ester model systems. International Journal of Mass Spectrometry. 2001;210/211:71. [Google Scholar]
- 52.Tsaprailis G, Nair H, Zhong W, Kuppannan K, Futrell JH, Wysocki VH. A mechanistic investigation of the enhanced cleavage at histidine in the gas-phase dissociation of protonated peptides. Analytical Chemistry. 2004;76:2083. doi: 10.1021/ac034971j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.