Abstract
The statistical properties of the cross-correlation between two time series has been studied. An analytical expression for the cross-correlation function’s variance has been derived. Based on these results, a statistically robust method has been proposed to detect the existence and determine the direction of cross-correlation between two time series. The proposed method has been characterized by computer simulations. Applications to single-molecule fluorescence spectroscopy are discussed. The results may also find immediate applications in fluorescence correlation spectroscopy (FCS) and its variants.
1 Introduction
While the range of topics being addressed by optical single-molecule spectroscopy has been expanding with an astonishing speed,1 several fundamental issues pertaining to the theoretical basis of data interpretation remain unresolved. Here, we discuss issues related to the objective assessment of the quality of single-molecule time traces.
In the field of single-molecule spectroscopy, it is common to reject invalid time traces and not include them in further data analysis. For example, when Förster resonance energy transfer (FRET) is used to study the time-dependent conformational changes in a macromolecule (e.g., proteins, DNA, or RNA), a pair of fluorescent donor and acceptor probes is attached to the molecule of interest to provide distance information. In the case of proteins, however, the probes are usually linked to the macromolecule non-selectively such that one may have molecules labeled with two donors, two acceptors, or only a single (donor or acceptor) probe. Data collected on molecules with any of these configurations will have to be rejected prior to data analysis lest they adversely impact subsequent interpretation. In diffusion-type experiments, the alternating-laser excitation scheme has been proposed to help remove these constructs from the ensemble of molecules.2 In experiments investigating immobilized molecules, one typically selects single-molecule traces that exhibit anti-correlated donor and acceptor emission pattern based on visual inspection. For the latter example, selection (and rejection) of single-molecule traces based on the subjective visual inspection alone can be ambiguous.
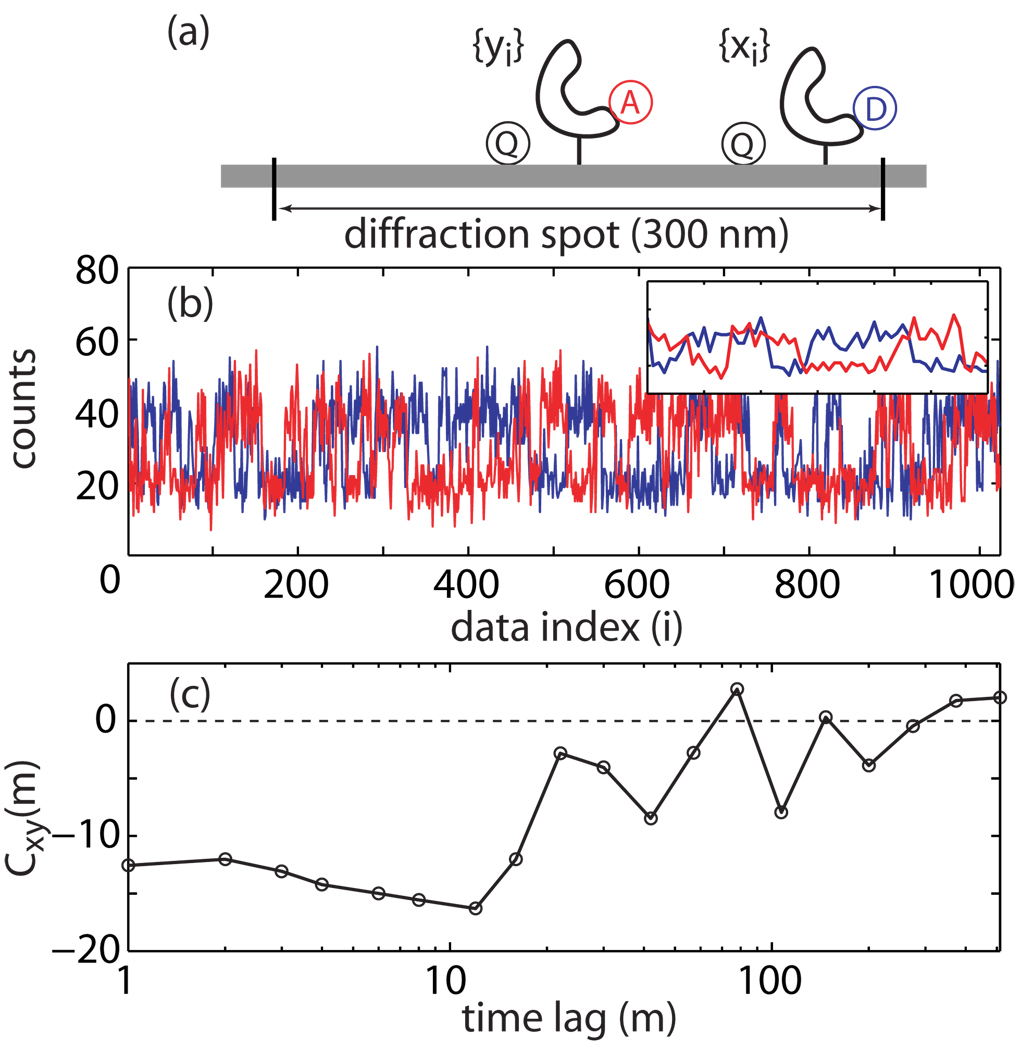
This problem can be illustrated by the following example. Consider a FRET experiment in which both the donor and the acceptor can be quenched non-specifically via a quencher in the vicinity of the macromolecule under investigation. The quenching is time dependent because of the slow conformational fluctuations of the macromolecule. As illustrated in Fig. la, it is possible that two singly labeled macromolecules co-localize within the same diffraction-limited detection spot (diameter ~300 nm), where the acceptor-macromolecule is at the center (better direct excitation and photon collection efficiency) and the donor-macromolecule is at the edge (reduced excitation and photon collection efficiency). To an experimental observer, the FRET intensity trace from this configuration cannot be distinguished from a true donor-acceptor doubly labeled molecule without further analysis. This difficulty is illustrated in Fig. 1b where the simulated donor (blue line) and acceptor (red line) traces appear as if they indeed come from FRET; the inset further shows how they can appear to be anti-correlated. One might think that cross-correlation analysis will help to resolve this problem; yet, without a quantitative assessment, visual examination combined with inappropriate data presentation can further exacerbate the problem. Fig. 1c displays a cross-correlation curve for the traces shown in Fig. 1b with log averaging. It appears anti-correlated even though, by construction, the donor and acceptor signals should be uncorrelated. Therefore, Fig. 1b–c clearly demonstrate the difficulties of evaluating single-molecule time traces based on visual assessment alone.
Figure 1.
Identification of anti-correlated FRET traces based on visual inspection alone can be misleading, (a) A scenario that can lead to seemingly anti-correlated FRET traces which, in fact, should be uncorrelated. (b) Simulated intensity time traces for the donor (blue) and the acceptor (red) appear as if they are anti-correlated. The inset displays a zoom-in for the traces between the 230–290 index range, (c) A cross-correlation analysis of the donor and acceptor traces, and presented after log10 averaging—a practice not encouraged. Without a quantitative assessment, the cross-correlation curve may lead one to believe that the two traces in (b) are anti-correlated.
This work is intended to provide a practical solution to problems of this nature. More specifically, one focuses on making an objective and statistically robust statement about the existence of cross-correlation between two time series and the direction of correlation. An important criterion for the solution is that it be general, independent of an explicit knowledge of the distributions of the two time series or their time-dependent variations. This problem is recast as finding an appropriate test statistic. This work is a continuation of previously published analysis of the variance of auto-correlation functions, in which correlated fluctuations within a single time-series are considered.3 The present manuscript is concerned with correlated fluctuations between two time series, such as those observed in single-molecule FRET experiments, and aims to provide a much needed evaluation of the variance present in such cross-correlation functions. While the motivations and applications discussed here involve FRET-type single-molecule experiments, the proposed solution, Eq. 4, is general and is expected to be applicable to other areas of research including fluorescence correlation spectroscopy (FCS), computer dynamics simulations and evolution genomics, to name a few. The critical region in Eq. 5 is useful for evaluating time series with vanishing auto-correlation whereas the critical region in Eq. 6 is useful for time series with non-vanishing auto-correlation. The performance of these tests was characterized using computer simulations and was found to be satisfactory for practical applications.
2 Basic Considerations
Consider a series of N pairs of experimental observables, {(x1, y1),…, (xN, yN)}, discretely sampled at a fixed time interval, δt ≡ ti+1 − ti, with xi ≠ yi. One is interested in knowing if x and y are correlated and, if so, whether they are positively or negatively correlated (anti-correlated). Conventionally, Pearson’s correlation coefficient, , is used to assess the correlation between the X and Y variables.4 The major limitation of using the well-established statistical identifiers such as the Pearson’s coefficient is that they have been developed based on the assumption that there is no measurement noise. When there is significant measurement noise—as is usually the case with low-signal experiments such as single-molecule spectroscopy and imaging—the correlation statistic becomes ill defined, making it difficult to evaluate the correlation quantitatively. In fact, the distribution of measurement noise is generally not known and may be difficult to characterize. An alternative method for the identification of correlated pair observables will be needed. Here, one considers characterizing the correlation of X and Y by cross-correlation between them. This approach only requires that the measurement noise is not correlated in time, so that the noise does not contribute to correlation time lags greater than 0.
Let X denote the stationary stochastic process that generates the observable x. The elements in {xi} do not have to be independent of each other; therefore, the ensemble-averaged auto-correlation of the {xi} series is not necessarily zero.
That is,
where 〈…〉 denotes ensemble averaging and δx ≡ x − 〈x〉. For an event series of finite size (N « ∞), the ensemble averaging is replaced by sample-averaged expectation value, denoted by E{…}. For example, the correlation function is approximated by,
where m = |ti − tj|/δt. The above approximation assumes the periodic condition, xi = xi+N. In practical applications, this assumption allows one to compute correlation functions using the discrete Fourier transformation. Similarly, the elements in {yi} are considered to be from a stationary stochastic process, Y. Since the elements in {yi} do not have to be uncorrelated, the auto-correlation of y is Cyy (|tk − tl|) = 〈δykδyl〉 ≥ 0. The two stochastic processes, X and Y, do not have to have the same statistical properties. For example, X could be a Gaussian process whereas Y could be a Poisson processes.
Following a recently developed statistical test for auto-correlation,3 the first step for testing the existence of cross-correlation in a time series is to derive an expression for the uncertainties (in the form of variance) in cross-correlation under the condition in which X and Y are uncorrelated. The X–Y cross-correlation is expressed as,
| (1) |
where δyi ≡ yi – 〈y〉 and the periodic condition for both X and Y has been assumed. It is important that the formulation be able to deal with finite-length time series, and that the expression be general, independent of the underlying distributions in {xi} and {yi}.
3 Uncertainties in Cross-Correlation
To evaluate the statistical significance of a cross-correlation, one starts with a simple case in which X and Y are assumed to be independent. More general cases dealing with correlated X and/or Y will be discussed in the next section. One further assumes that the elements in {xi} are mutually independent; as are the elements in {yi}. These assumptions serve the purpose of quantifying how the stochastic noise contributes to the resulting cross-correlation. The statistical uncertainties are evaluated by the variance for the cross-correlation, var{Cxy}.
Following a similar procedure for deriving the variance in auto-correlation,3 the variance for cross-correlation is,
| (2) |
Eq. 2 is the first major result of this work (see Appendix A for derivation). Note that, because of the periodic condition imposed on the calculation, the variance is independent of the index lag, m. Large-number principles (the Central-Limit Theorem) predict that Cxy should behave as a Gaussian random variable, regardless of the distributions underlying {xi} and {yi}. As expected, its variance, var{Cxy}, scales approximately as N−1/2 for large N. Eq. 2 allows one to calculate the statistical uncertainties in a cross-correlation from the sample without explicitly knowing the underlying distributions in X and Y. By comparing the previously developed expression for variance of an auto-correlation function3 with Eq. 2, it is apparent that the the functional form of the variance is substantially different for auto- and cross-correlations.
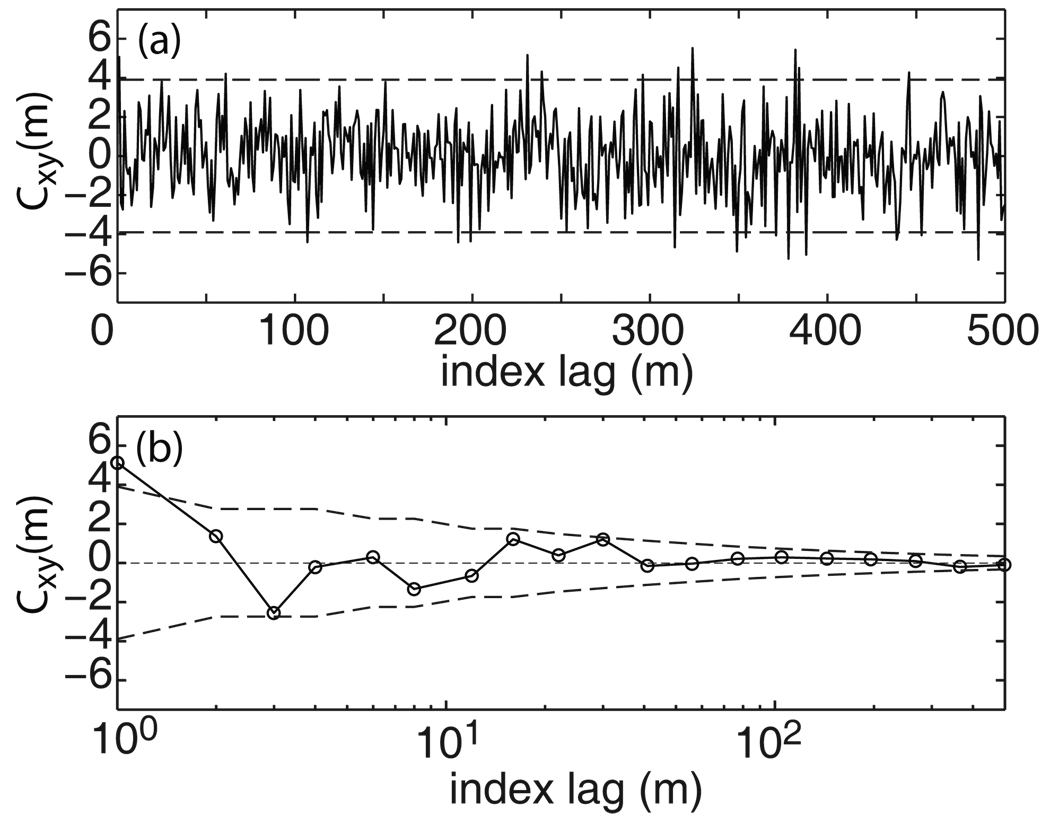
To illustrate the results, Fig. 2 displays the cross-correlation trace between random variables X and Y. X was sampled from a Gaussian distribution with a probability density function, , whereas Y was sampled from a Poisson distribution with a probability function, fp(y) = yλ exp[−λ]/y!. Using these two probability density functions, a total of 1,000 {(xi, yi)} pairs were generated (Matlab R2006b with Statistics Toolbox, The Mathworks, Natick, MA) with the following parameters: μx = 10, σx = 20, and λ = 10. The cross-correlation was calculated using discrete Fourier transformation. Of the 500 index lags included in the figure, 22 of them (~4.4%) exceed the 95% confidence intervals.
Figure 2.
Comparison of the analytical expression (Eq. 2) for expected variance of Cxy(m) with computer simulations for independent {xi} and {yi}. The dashed lines denote expected 95% confidence intervals (~3.9 for this data set). (a) The results plotted on the linear index lag (m) scale, (b) The trace in (a) averaged over log-m scale.
The same trace was averaged on the log10 scale and plotted in Fig. 2b. The log-averaged trace appears visually pleasing and exhibits an apparent positive correlation with a reasonable decay. Such an appearance is in fact an artifact arising from the way the plot is prepared. Since there should be no cross-correlation by construction, Fig. 2b clearly demonstrates another example showing that visual assessment alone, in particular when combined with log-averaging, can be greatly misleading when interpreting results of correlation analysis. The next section describes a rigorous way of evaluating the existence and direction of cross-correlation.
4 Existence and Direction of Cross-Correlation
The problem of testing the existence of cross-correlation and the determination of the correlation direction is recast as a two-sided statistical test problem. The null hypothesis, H0, is the case in which there is no cross-correlation. There are two alternative hypotheses, H1 and H−1, in which the former denotes positive cross-correlation and the latter negative. Since each Cxy(m) is an average over N pairs of random variable products, δxiδyi+m, Cxy(m) is also a random variable itself. For large N (typically N > 25, valid under almost all experimental conditions), the probability distribution for Cxy(m) under the null hypothesis (H0) can be very well approximated by a Gaussian with zero mean and variance vax{Cxy} (the Central-Limit Theorem). The probability density function is,
| (3) |
where is calculated using Eq. 2. In other words, when there is no cross-correlation between X and Y, each lag in Cxy(m) can be viewed as a stochastic step of a Brownian random walker with a mean step size of σxy.
With this understanding, one may formulate a statistical test for the existence of cross-correlation: The null hypothesis that there is no cross-correlation between two time series of arbitrary distribution is rejected with a false-positive error rate of α when the test statistic, ZN, exceeds the critical value cα of confidence level (1 – α):
| (4) |
where nt is number of time lags included in the test. Eq. 4 is the second major result of this work. The critical region can be calculated using
| (5) |
where Erfc−1(α) is the inverse complementary error function and can be computed numerically. For example, the confidence intervals for α = 0.31, 0.1, and 0.05 are σxy, 1.64σxy and 1.96σxy, respectively. If the null hypothesis is rejected, the sign of ZN gives the direction of cross-correlation. As an example, the trace shown in Fig. 2 was found to have a ZN = 0.27 and was categorized as exhibiting ”no correlation” within 95% false-positive confidence interval.
4.1 Characterization of the Test for Observables with Vanishing Auto-Correlation
The proposed test for the existence of cross-correlation was characterized using computer simulations. The results are summarized in Table 1. In these simulations, 25 time lags (nt = 25) were used for the test. The results show that the proposed test performs very well, even for short time series. Increasing the test sample size (greater nt) will decrease the statistical noise in the test; however, it will also reduce the power of the test. In fact, the general characteristics of the false-negative rate and the power of the test will depend on the specific type of cross-correlation in the data. Before turning the discussion to the power of the test, one further characterizes Eq. (2) and Eq. (4) for cases where there are non-vanishing auto-correlations, Cxx > 0 and/or Cyy > 0.
Table 1.
False-positive rate (probability) for the statistical test of correlation in a time series characterized by computer simulations. The statistics were obtained from 10,000 simulations of independently identically distributed Gaussian random variables for X and Poisson random variables for Y. The parameters used to generate the simulated data are the same as those used to generate Fig. 2.
| N = 200 | N = 400 | N = 800 | N = 1600 | N = 3200 | |
|---|---|---|---|---|---|
| 1 × σxy | 0.32 | 0.32 | 0.32 | 0.32 | 0.31 |
| 1.64 × σxy | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 |
| 1.96 × σxy | 0.05 | 0.05 | 0.05 | 0.05 | 0.05 |
4.2 Characterization of the Test for Observables with Non-Vanishing Auto-Correlation
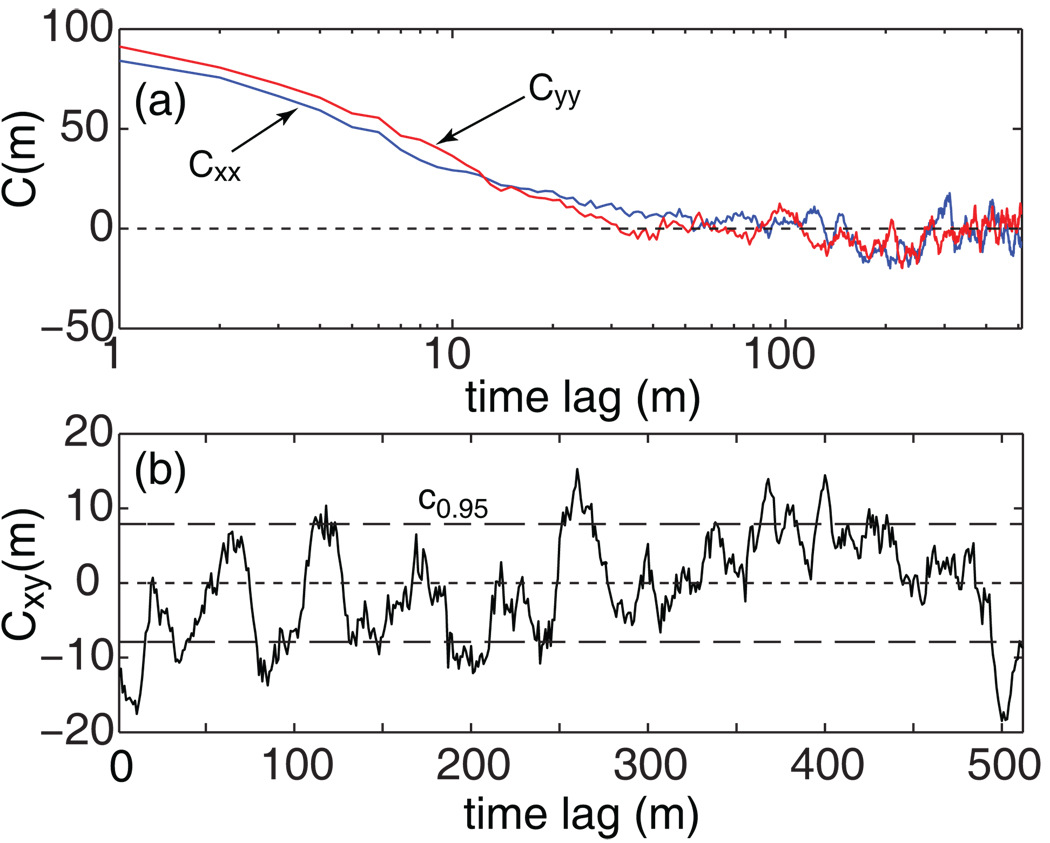
When there is correlation among {xi} (or among {yi}), different time lags in a cross-correlation function, say Cxy(m) and Cxy(m'), are no longer independent even when X and Y are uncorrelated (cf. Fig. 3a). These correlations, in turn, will result in increased uncertainties in the cross-correlation function. In other words, applying the unsealed statistical test in Eq. 4 to such data streams will result in a greater false-positive rate α than the confidence region c1–α would have allowed. This point is illustrated in Fig. 3b, which displays the cross-correlation function of the time series displayed in Fig. 1b on a linear scale without log10 averaging. It also shows how the fluctuations can be correlated (cf. Fig. 2a for an uncorrelated case), resulting in greater uncertainties in the cross-correlation.
Figure 3.
(a) The auto-correlation functions of the donor (Cxx) and the acceptor (Cyy) traces for the traces shown in Fig. 1b, showing that {xi} and {yi} are not mutually independent within their respective set. (b) The cross-correlation, Cxy. It is clear that the unscaled confidence region, c1-α at α = 0.05, does not encompass the cross-correlation function because the different time lags (m) in Cxy(m) are no longer independent.
Currently, an analytical expression to quantify such an increase does not seem to be readily obtainable for general cases when the form of the autocorrelation functions, Cxx and Cyy, are unknown. Nevertheless, it is possible to devise an empirical way of correcting for the correlation-related uncertainties. Following Zwanzig and Ailawadi5 and Schenter et al.,6 the idea is to rescale the confidence region by taking into account the correlations. When there is no correlation, the c1–α in Eq. 5 is calculated using Eq. 2. When there is correlation, there will be fewer number of effectively independent time lags. Assuming that the correlation in Cxx and Cyy decays with a constant and , respectively, then the number of independent lags can be approximated by Neff ≃ N/mτ, where . In an application, mτ can be obtained empirically from fitting the Cxx and Cyy functions to an exponential model. This idea leads to the scaled confidence interval for the test in Eq. 4,
| (6) |
where is calculated using Eq. 2 but replacing N with N eff.
The performance of this scaled test was studied using computer simulations, in which both X and Y exhibit non-vanishing auto-correlation (with a constant of ) but with no cross-correlation between them. The results are summarized in Table 2. It is clear that the unsealed critical region, Eq. 5, leads to a much greater false-positive error rate than the confidence interval α would have indicated. On the other hand, the scaled critical region, Eq. 6, is able to reproduce the expected error rate quite well, especially for larger-size samples, suggesting the practical usefulness of the proposed test.
Table 2.
Computer-simulation characterization of false-positive rates for the statistical test (Eq. 6) of cross-correlation between two time series, {xi} and {yi}, both of which have non-vanishing auto-correlations. The statistics were obtained from 10,000 simulations of a two-state model randomly switching between two photon-counting levels; that is, both X and Y follow Poisson statistics. Numbers in the parentheses are results using the test for uncorrelated observables (Eq. 4). Simulation details are in Appendix B.
| N = 200 | N = 400 | N = 800 | N = 1600 | N = 3200 | |
|---|---|---|---|---|---|
| 1 × σxy | 0.28 (0.72) | 0.28 (0.74) | 0.31 (0.75) | 0.31 (0.75) | 0.32 (0.75) |
| 1.64 × σxy | 0.08 (0.56) | 0.09 (0.58) | 0.09 (0.59) | 0.10 (0.60) | 0.10 (0.60) |
| 1.96 × σxy | 0.04 (0.49) | 0.05 (0.51) | 0.04 (0.52) | 0.05 (0.53) | 0.05 (0.54) |
5 Power of the Cross-Correlation Test: The Single-Exponential Model
The power of a statistical test (detection power) is the probability of rejecting the null hypothesis (H0) when the alternative hypothesis (H1 or H−1) is true. For the present problem, the power will depend on the form of the cross-correlation such as the amplitude and the relaxation rate, as well as on the conditions used in the test such as the false-positive rate (α) and the number of time lags used. In order for the test to be practical, it is important to characterize how the detection power depends on theses parameters. To this end, computer simulations of FRET traces from a simple two-state jump model are used to examine the performance. The simulation details are included in Appendix B.
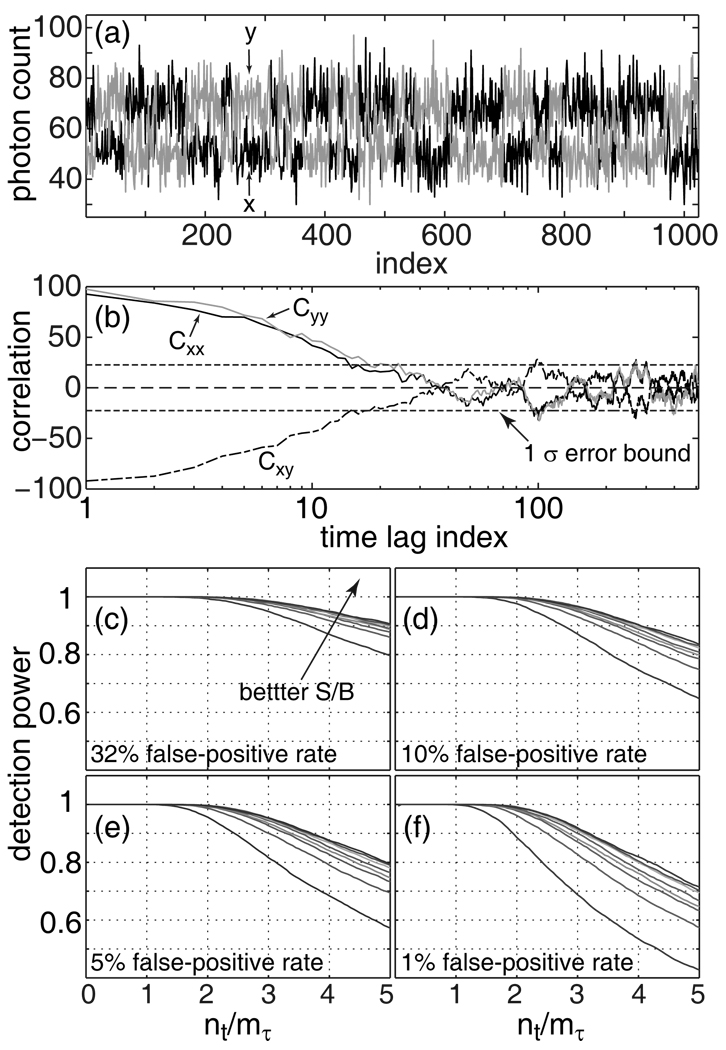
Fig. 4a displays a pair of typical FRET intensity traces from the simulation. The auto-correlations, Cxx and Cyy, as well as their cross-correlation, Cxy, all exhibit non-vanishing correlation, as shown in Fig. 4b. The relaxation rate in the auto-correlation gives an mτ ~ 10, which is in turn used to calculate the for the test statistic, Eq. 6. The power of the test, denned as the ratio between the number of simulations with successfully detected cross-correlation and the total number of simulations (10,000), was calculated as a function of the signal-to-background ratio (S/B) and the number of time lags in the test (nt/mτ) at various false-positive rates (α). As shown in Fig. 4c–f, several general observations can be made. It is apparent that the probability of detecting cross-correlation improves with better signal-to-background ratio. The detection power appears to be close to unity for test lengths, nt, shorter than the relaxation length, mτ. Longer test lengths tend to result in degraded detection power; this is because the correlation will vanish at longer time lags (large m), which in turn will reduce the numerical value of the test statistic, ZN, through averaging (cf. Eq. 4). Finally, while a tighter confidence interval (smaller α) guarantees less frequent false-positive identification, it also decreases the detection power. Overall, these computer simulations indicate that the proposed quantitative statistical test is very powerful (as a statistical test), and that a reasonable test length, nt, in the case of non-vanishing auto-correlation can be set to nt = mτ.
Figure 4.
Power of the statistical test for an exponential model, (a) Typical FRET donor (blue, x) and acceptor (red, y) traces from computer simulations. (b) Auto-correlations, Cyy (red) and Cxx (blue), and the cross-correlation, Cxy (dashed-dot), of the data in (a). Panels (c) to (f) displays the detection power of the statistical test in Eq. 4 and Eq. 6 plotted as a function of signal-to-background ratio (S/B = 1.2, 1.4, 1.6, 1.8, 2.0, 3.0, 4.0, 5.0) and the test length (nt/mτ).
6 Case Studies: Applications to Single-Molecule FRET of Polyproline and Enzyme
Single-molecule fluorescence spectroscopy has recently been used to shed new light on many biological systems (for recent reviews see1,7,8). While it has the unique ability to monitor the time-dependent behavior of molecules without ensemble averaging, single-molecule spectroscopy requires extremely high sensitivity which leads to challenges in data collection and processing. Namely, these experiments are frequently short, due to photo-degradation of the fluorescent probes, and they commonly have a low signal-to-background ratio, characterized by a large amount of background noise in the raw intensity data. These issues make a thorough characterization of the contribution of noise to the analysis of single-molecule data a requirement for an accurate interpretation of results. This section presents two distinct practical applications of the newly developed statistical test for the existence and direction of cross-correlation: the elimination of non-ideal FRET trajectories from single-molecule data sets and the identification of trajectories with significant anti-correlation between emission from donor and acceptor probes (which signifies the presence of intra-molecular conformational changes).
6.1 Un-correlated fluctuations in a model compound: Polyproline
A quantitative assessment of the uncertainty in cross-correlation functions is a critical issue for FRET-based experiments since one is typically comparing the intensity fluctuations between two short, low signal-to-background ratio intensity-vs.-time traces: one for the acceptor probe and the other for the donor. Since the intensity of acceptor emission is dependent on the inverse sixth power of the distance between the donor and acceptor probes, an ideal FRET experiment should have uncorrelated or anti-correlated intensity fluctuations between the two observed signals. Deviations from ideal behavior, however, are commonly seen in single-molecule fluorescence trajectories. For instance, “blinking” of the probes is a major concern; physically this arises from a transition to a dark state of the probe, commonly a triplet state or non-emissive isomer.9,10 Blinking of the donor probe in a single-molecule FRET experiment will also cause a concomitant blinking of the acceptor since energy transfer pathways between the probes are non-existent if the donor is in a dark state. The experimentalist needs to remove those trajectories that display blinking from the data set before data analysis, since such trajectories could lead to erroneous conclusions. If the timescale of blinking is on the order of seconds this can be accomplished with reasonable accuracy by visual inspection of intensity traces; however, since the lifetime of the dark state should be exponentially distributed, visual inspection will miss many more blinking events than it identifies. A single-molecule FRET trajectory with donor blinking will have positively cross-correlated intensity fluctuations, distinguishing it from the ideal case in which intensity fluctuations should be uncorrelated or anti-correlated. Such non-ideal behavior in single-molecule FRET trajectories can be identified quantitatively using the new statistical test for the existence of cross-correlation between two time series.
Here, the newly proposed statistical test for the existence of cross-correlation is utilized to analyze single-molecule FRET data collected on fluorescently labeled polyproline peptides in order to test for non-ideal positive cross-correlation of intensity fluctuations. Polyproline peptides have previously been used as a “spectroscopic ruler” to calibrate FRET experiments11 since they are believed to prefer a relatively rigid left-handed type-two helix.12 Recently, single-molecule FRET experiments have cast doubt on the accuracy of polyproline spectroscopic ruler, these deviations from ideal behavior have been attributed to miss-estimation of persistence length of the proline helix13 and cis-trans isomerization of prolines residues.14–16 Nevertheless, polyproline peptides remain an important model for the fundamental understanding of the unfolded state in proteins.17 They are expected to be relatively rigid on the typical time scales probed by single-molecule FRET experiments, milliseconds – minutes.15 By contrast, bending of short polyprolines due to thermal fluctuations should be small in magnitude and relatively fast while cis-trans isomerization is expected to occur on the time scale of minutes. Thus, one would expect to observe no significant cross-correlation in intensity fluctuations of single-molecule FRET trajectories when short polyproline peptides are studied under ideal energy transfer conditions.
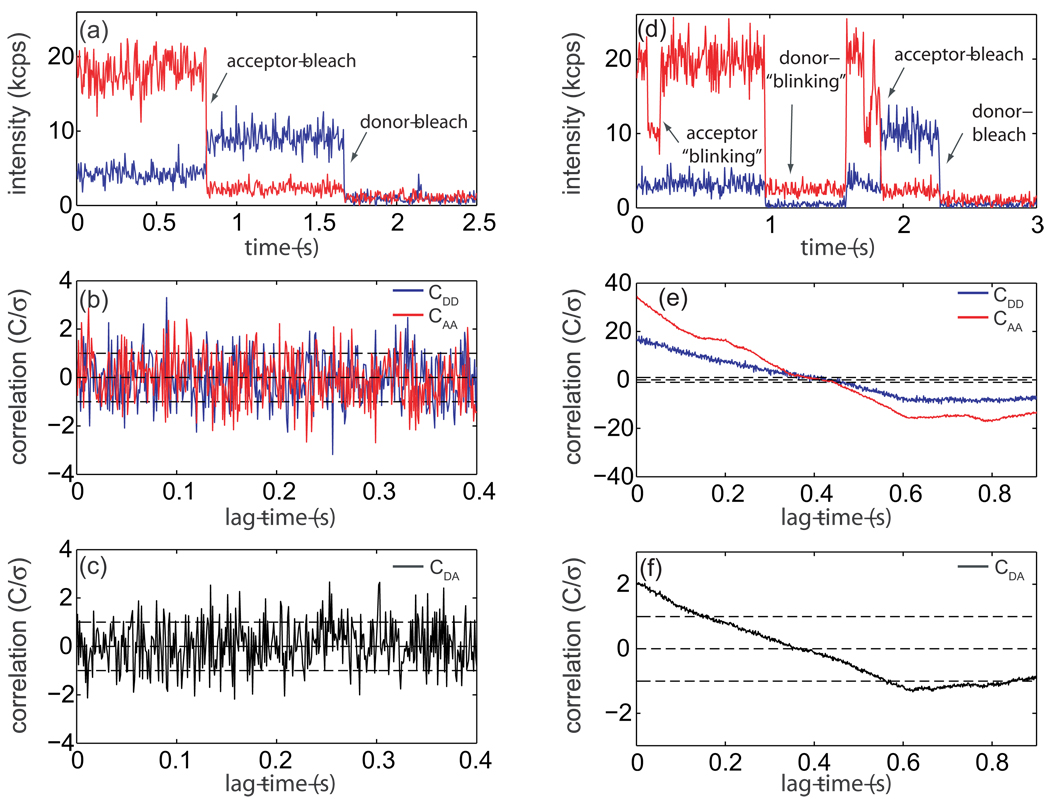
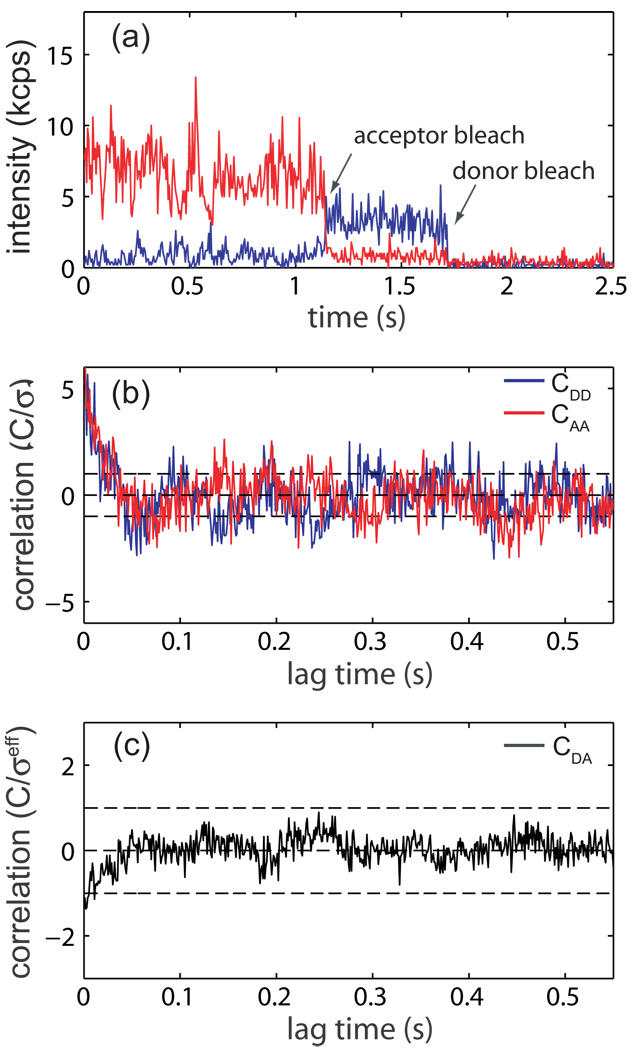
Single-molecule FRET experiments were carried out on a polyproline peptide with the sequence P15CG3K(biotin), as described previously.14 The donor probe, AlexaFluor 555 C5-succinimidyl ester (Invitrogen), was attached to the N-terminus of the peptide while the acceptor probe, AlexaFluor 647 C2-maleimide (Invtrogen), was attached to the cysteine residue. Labeled peptides were immobilized on a biotin-PEG derivitized quartz cover slip through biotin-streptavidin chemistry and experiments were performed on a single-molecule confocal microscope.14 A sample intensity-vs.-time trajectory collected on a labeled polyproline molecule is displayed in Fig. 5a. All subsequent analysis is concerned with the region of the trajectory before the acceptor probe photo-bleach in which energy transfer is occurring. The first step in determining whether the acceptor and donor channel have significant correlation is to test each individually for significant auto-correlation. This can be achieved by applying the previously developed statistical test for auto-correlation in a time-series.3 Intensity auto-correlation functions for donor (blue line) and acceptor (red line) were calculated by discrete Fourier transform with a bin-size of 1 ms and are plotted in Fig 5b. The previously developed test statistic for auto-correlation indicates that both correlation functions in Fig 5b are uncorrelated to 95% confidence3 (Donor: test statistic = 1.8 × 10−3, critical region = 6.6 × 10−2; Acceptor: test statistic = 2.4 × 10−1, critical region = 3.3 × 10−1). Only the first nt = 25 time lags from each auto-correlation function were used in the test. Since both time-series display vanishing auto-correlation, the new test for cross-correlation can be applied directly. A discrete Fourier transform intensity cross-correlation function with a bin size of 1 ms has been calculated for the trajectory in Fig. 5a and is displayed in Fig. 5c. Applying Eq. 5 to the first nt = 25 time lags in this cross-correlation function gives a test statistic ZN = 4.3 × 10−2 (Eq. 4) and a critical region with a false-positive rate of 5% of c0.05 = 1.5 · 10−1 (Eq. 5). Accordingly, this demonstrates to 95% confidence that the trajectory in Fig. 5a has no significant cross-correlation in the region before the acceptor bleaches for timescales longer than 1 ms, as expected for a polyproline molecule under ideal energy transfer conditions.
Figure 5.
Single-molecule polyproline FRET trajectory and cross-correlation analysis, (a) Intensity-vs.-time trajectory for a single polyproline molecule binned at 5 ms. Acceptor emission is in red and donor emission in blue. Photo-degradation of the probes is indicated by arrows, all subsequent analysis of this trajectory is performed on the region before the acceptor probe bleaches, (b) Discreet Fourier transform auto-correlation function for donor (CDD, blue) and acceptor (CAA, red) from the region of the trajectory from a with a bin size of 1 ms. A previously developed statistical test for the existence of auto-correlation in a time series3 allows us to determine that neither probe has significant auto-correlation with 95% confidence. Error bars are plotted to one standatrd deviation.3 Correlation functions have been normalized by their standard devieations. (c) Discreet Fourier transform cross-correlation (CDA) for the trajectory in a with a bin size of 1 ms. According to the newly proposed test for the existance of cross-correlation, the test statistic |ZN| = 4.3 × 10−2 (Eq. 4) while the critical region with a false-positive rate of 5% is c0.05 = 1.5 × 10−1 (Eq. 5), demonstrating that this trajectory has no significant cross-correlation to 95% confidence, (d) Intensity-vs.-time trajectory for a single-molecule displaying non-ideal photo-physics with a 5 ms bin size. Arrows indicate the ”blinking” of each probe, (e) Auto-correlation function for each trajectory in a with a 1 ms bin size showing significant correlation, (f) Cross-correlation function for the trajectory in a with a 1 ms bin size. Application of the statistical test for cross-correlation (Eq. 4 and Eq. 6) demonstrates that this trajectory has significant positive correlation to 95% confidence.
To illustrate the usefulness of cross-correlation analysis in data selection, a non-ideal trajectory has been displayed and analyzed in Fig. 5d–f. The intensity-vs.-time trajectory in Fig. 5d (5 ms bin size) displays both donor and acceptor “blinking” events (indicated by arrows). While this trajectory could easily be eliminated by visual inspection, it was chosen to demonstrate the types of non-ideal photo-physical behavior commonly seen in single-molecule fluorescence experiments. Auto-correlation analysis of each intensity trace with a 1 ms bin size reveals that both have significant correlation (Fig. 5e).3 Each auto-correlation was fit to a single exponential in order to determine the number of independent observations for use in the scaled critical region (Eq. 6; donor relaxation time lags, acceptor relaxation time lags). A discrete Fourier transform cross-correlation function with a 1 ms bin size is plotted in Fig. 5f. Error bars have been calculated according to Eq. 2 with N = N eff = N/mt, where . According to Eq. 4 and Eq. 6, the test statistic ZN = 7.6 while the scaled critical region with a false positive rate of 5% . Since |ZN| > cα and ZN > 0, this indicates that the trajectory in 5d is positively correlated to 95% conficdence. Thus, this single-molecule trajectory could rigorusly be eliminited from further data analysis and interpretation due to non-ideal photophysical effects of the probes during the FRET measurements since the interpretation of these blinking events as distance changes which would result in erronious results.
6.2 Anti-correlated fluctuations due to conformational dynamics: Adenylate Kinase
Correlation function analysis can also play an important role in analysis of single-molecule FRET trajectories since the technique is frequently employed in systems that are believed to have time dependent behavior.18–20 The experimentalist is interested in two questions regarding the time scale of the process under investigation: (1) What is the average rate? (2) What is the molecule-to-molecule variation of the rates? Since distance fluctuations lead to anti-correlated intensity fluctuations in single-molecule experiments, one is now interested in testing for significant anti-correlation in the intensity fluctuations before the data can be fit to a model describing the underlying motions, an application for which the statistical test for cross-correlation is well suited.
As an example, cross-correlation analysis is applied to an Adenylate Kinase (AK) enzyme undergoing conformational fluctuations. AK serves as a model system for the functional role of conformational dynamics in enzymes.21 This enzyme’s active site is covered by a lid domain which undergoes a large amplitude conformational transition from open to closed that has been proposed to be an elementary step in AK’s reaction mechanism.22 A His6-tagged, dual cysteine mutant AK was prepared, labeled with AlexaFluor 555/647 C2-maleimide (Invitrogen), and immobilized on a quartz cover slip as described previously.22 A single-molecule fluorescence intensity-vs.-time trajectory collected on substrate-free E. Coli AK is presented in Fig 6a. Fig. 6b shows intensity auto-correlation functions calculated for both donor (blue) and acceptor (red) time series individually with a bin size of 1 ms. Here, both trajectories show significant auto-correlation to 95% confidence3 (donor: test statistic = 8.6 × 10−2, critical region = 1.3 × 10−2; acceptor: test statistic = 8.5 × 10−1, critical region = 1.1 × 10−1). In order to calculate the number of effective independent observations for use in the scaled critical region (Eq. 6), both auto-correlation functions in Fig. 6b were fit to a single exponential, yielding a donor relaxation time of time lags and an acceptor relaxation time of time lags. A discrete fourier transform intensity cross-correlation function for the trajectory in Fig. 6a is displayed in Fig. 6c. Error bars are calculated according to Eq. 2 with N = Neff = N/mt, where . Accordingly, the test statistic for the existence of cross-correlation gives ZN = −3.2 × 10−1 (Eq. 4) while the scaled 95% confidence region is (Eq. 6). Since and ZN < 0, the trajectory in Fig. 6a displays significant anti-correlation with 95% confidence, as expected for a protein undergoing conformational fluctuations. A fit the cross-correlation function in Fig. 6c to a single exponential yields a relaxation time for conformational fluctuations of 18 ms for the trajectory in Fig. 6a. Even though the single-exponential model is a gross simplification of complicated protein dynamics, it should capture the basic features of the protein movements. Indeed, the relaxation time simmilar to the the average interconversion time of 2.9 ± 0.7 ms predicted by the simplified two-state motional-narrowing model previously used to measure the mean opening and closing rates of AK’s lid domain.22
Figure 6.
Single-molecule FRET trajectory on substrate-free Adenylate Kinase (AK) and cross-correlation analysis, (a) Intensity-vs.-time trajectory for a single substrate-free AK molecule binned at 5 ms. Acceptor emission is in red and donor emission in blue. Photo-degradation of the probes is indicated by arrows, all subsequent analysis of this trajectory is performed on the region before the acceptor probe bleaches, (b) Discreet Fourier transform auto-correlation function for donor (CDD, blue) and acceptor (CAA, red) from the region of the trajectory from panel a before the acceptor probe bleaches with a bin size of 1 ms. Both probes show significant auto-correlation to 95% conficence.3 Each correlation function was fit to a single-exponential decay giving a donor relaxation of time lags and an acceptor relaxation of time lags. Error bars are plotted to one standatrd deviation.3 Correlation functions have been normalized by their standard devieation. (c) Discreet Fourier transform cross-correlation (CDA) for the trajectory in panel a with a bin size of 1 ms. According to the newly proposed test for the existance of cross-correlation, the test statistic ZN = −3.2 × 10−1 (Eq. 4) while the critical region with a false-positive rate of 5% is (Eq. 6). Since |ZN| > cα and ZN < 0, we conclude that this trajectory has significant anti-correlation with 95% confidence.
7 Concluding Remarks
Analytical expressions have been derived for the variance in a cross-correlation function, based on which a statistical test has been proposed for the existence and direction of the correlation. An empirical test has also been proposed for time series with non-vanishing auto-correlation and verified by computer simulation. This test is general—independent of explicit knowledge of the processes under investigation—and capable of dealing with a short, low signal-to-background ratio time series. The new test is particularly useful to the field of single-molecule spectroscopy, where all of these experimental challenges are frequently encountered. Two applications to single-molecule FRET have been demonstrated. Cross-correlation function analysis can be used to identify single-molecule trajectories with non-ideal energy transfer due to changes in emissive properties in the probes, blinking for instance. Such non-ideal trajectories are seen in every single-molecule data set and, whether or not it is reported in the literature, their removal is a critical element of subsequent data analysis. Identification is frequently accomplished by visual inspection, which tends to be biased to events that occur on a time scale of seconds and will vary from person to person; thus, a standard, non-biased criteria for removal of non-ideal trajectories will allow greater reliability and consistency in single-molecule data interpretation. Cross-correlation function analysis is also frequently applied to single-molecule FRET trajectories in order to characterize the time scale at which the process under investigation is occurring. The newly developed statistical test for the existence of cross-correlation allows one to rigorously determine whether a significant correlation exists before subsequent analysis is performed. A frequent finding in single-molecule experiments on biological systems is that the behavior of individual molecules is often quite heterogeneous.23 While some molecules may be highly dynamic, others may be relatively static under the period of investigation. This phenomenon is no doubt due to the complexity in the energy landscape of bio-molecules and its characterization is one of the motivations for performing single-molecule experiments. For structure-function dynamics studies, the statistical test for the existence of cross-correlation could be used to group molecules into classes based on whether or not they display significant anti-correlated intensity fluctuations. Though motivations for and applications of the methods described in this article are focused on single-molecule FRET, they are expected to be widely applicable whenever a cross-correlation must be analyzed.
Acknowledgments
This work was supported by the National Science Foundation and the National Institutes of Health. HY is an Alfred P. Sloan Fellow.
A Variance of Cross-Correlation for Observables with Vanishing Auto-Correlation
In the following derivation, the elements in {xi} are assumed to be mutually independent; that is, for i ≠ j. Similarly, {yi} are assumed to be mutually independent, to give for i ≠ j. Finally, X and Y are assumed to be independent, giving . When there is correlation among {xi} or {yi} or both, the above reduction becomes invalid. The difficulty mainly arises from such terms as the xkxiyk+myi+m term in Eq. 8 and the , and terms in Eq. 10.
The cross-correlation function is defined by,
where m = 0,1,…., N – 1. The variance of the cross-correlation function is evaluated by,
| (7) |
The first term in Eq. 7 is,
| (8) |
The second term in Eq. 7 is,
| (9) |
The last term in Eq. 7 is,
| (10) |
B Simulation Details for a Two-State Stochastic Switching Model
Consider a single molecule stochastically switching from two states, X and Y, following the reaction scheme,
in which the molecule gives a low FRET signal (greater donor intensity) when it is at the X state and a high FRET signal (greater acceptor intensity) when it is at the Y state. The rejection method was used to simulate the switching dynamics. Basically, a random number r between 0 and 1 was generated and compared with kf or kb. For the system staring from the X state, a jump is said to occur when r < kf (here the time unit is assumed to be 1); otherwise, the system will stay at the X state. The same procedure was used for the Y → X transition.
The traces in Fig. 1 were generated using kf = kb = 1/20 and a signal-to-background ratio of 2 for both channels. The background is assumed to have, on average, 20 photon counts per time unit. The actual number of counts was generated using the Poisson random number generator that comes with the Statistical Toolbox in Matlab. The donor and acceptor traces were generated separately so that they are independent of each other. The traces in Fig. 4 were generated in the same way, except that the acceptor trace was generated so that it is exactly out of phase with respect to the donor trace, mimicking a FRET signal. All simulations were done using Matlab.
References
- 1.Moerner WE. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright R, Weiss S. Biophys. J. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson JA, Yang H. J. Chem. Phys. 2008;128:214101–214106. doi: 10.1063/1.2931943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson K. Philos. Trans. R. Soc. Lond. A. 1903;200:1–66. [Google Scholar]
- 5.Zwanzig R, Ailawadi NK. Phys. Rev. 1969;182:280–283. [Google Scholar]
- 6.Schenter GK, Lu HP, Xie XS. J. Phys. Chem. A. 1999;103:10477–10488. [Google Scholar]
- 7.Tinnefeld P, Sauer M. Angew. Chem. Int. Ed. 2005;44:2642–2671. doi: 10.1002/anie.200300647. [DOI] [PubMed] [Google Scholar]
- 8.Michalet X, Weiss S, Jager M. Chem. Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Widengren J, Schwille P. J. Phys. Chem. A. 2000;104:6416–6428. [Google Scholar]
- 10.Fureder-Kitzmuller E, Hesse J, Ebner A, Gruber H, Schutz G. Chem. Phys. Lett. 2005;404:13–18. [Google Scholar]
- 11.Stryer L, Haugland R. Proc. Natl. Acad. Sci. U.S.A. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowan P, McGavin S. Nature. 1955;176:501–503. doi: 10.1038/1761062a0. [DOI] [PubMed] [Google Scholar]
- 13.Schuler B, Lipman E, Steinbach P, Kumke M, Eaton W. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins LP, Chang H, Yang H. J. Phys. Chem. 2006;110:5191–5203. doi: 10.1021/jp055886d. [DOI] [PubMed] [Google Scholar]
- 15.Doose S, Neuweiler H, Barsch H, Sauer M. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17400–17405. doi: 10.1073/pnas.0705605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Best RB, Merchant KA, Gopich IV, Schuler B, Bax A, Eaton WA. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18964–18969. doi: 10.1073/pnas.0709567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi ZS, Chen K, Liu ZG, Kallenbach NR. Chem. Rev. 2006;106:1877–1897. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 18.Ha T, Ting AY, Caldwell WB, Deniz AA, Chemla DS, Schultz PG, Weiss S. Proc. Nat. Acad. Sci. USA. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talaga DS, Lau WL, Roder H, Tang J, Jia Y, DeGrado WF, Hochstrasser RM. Proc. Nat. Acad. Sci. USA. 2000;97:13021–13026. doi: 10.1073/pnas.97.24.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Hu D, Vorpagel ER, Lu HP. J. Phys. Chem. B. 2003;107:7947–7956. [Google Scholar]
- 21.Yan HG, Tsai MD. Advances in Enzymology. Vol. 73. John Wiley and Sons Inc.; New York NY 10016 USA: 1999. Nucleoside monophosphate kinases: Structure, mechanism, and substrate specificity. [DOI] [PubMed] [Google Scholar]
- 22.Hanson JA, Duderstadt K, Watkins LP, S B, Brokaw J, Chu JW, Yang H. Proc. Natl. Acad. Sci., U.S.A. 2007;104:18055–18060. doi: 10.1073/pnas.0708600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cosa G, Zeng YN, Liu HW, Landes CF, Makarov DE, Musier-Forsyth K, Barbara PF. J. Phys. Chem. B. 2006;110:2419–2426. doi: 10.1021/jp054189i. [DOI] [PubMed] [Google Scholar]