Abstract
Autism spectrum disorders (ASD) are neurodevelopmental disorders characterized by delayed/abnormal language development, deficits in social interaction, repetitive behaviors and restricted interests. The heterogeneity in clinical presentation of ASD, likely due to different etiologies, complicates genetic/biological analyses of these disorders. DNA microarray analyses were conducted on 116 lymphoblastoid cell lines (LCL) from individuals with idiopathic autism who are divided into three phenotypic subgroups according to severity scores from the commonly used Autism Diagnostic Interview-Revised questionnaire and age-matched, nonautistic controls. Statistical analyses of gene expression data from control LCL against that of LCL from ASD probands identify genes for which expression levels are either quantitatively or qualitatively associated with phenotypic severity. Comparison of the significant differentially expressed genes from each subgroup relative to the control group reveals differentially expressed genes unique to each subgroup as well as genes in common across subgroups. Among the findings unique to the most severely affected ASD group are 15 genes that regulate circadian rhythm, which has been shown to have multiple effects on neurological as well as metabolic functions commonly dysregulated in autism. Among the genes common to all three subgroups of ASD are 20 novel genes mostly in putative noncoding regions, which appear to associate with androgen sensitivity and which may underlie the strong 4:1 bias toward affected males.
Keywords: autism phenotypes, gene expression profiling, circadian rhythm, novel genes
Introduction
Autism spectrum disorders (ASD) collectively refer to a group of related neurodevelopmental disorders characterized by delayed or abnormal language development, deficits in social interaction, and restricted, repetitive behaviors and interests. The heterogeneity in clinical presentation of ASD, thought to be due to different etiologies, complicates genetic and molecular analyses of these disorders. Recently, several types of cluster analyses of data from ADI, ADI-R, and ADOS test instruments have revealed distinct, albeit somewhat different, components within the ASD spectrum [Lord, Leventhal, & Cook, 2001; Silverman et al., 2001; Stevens et al., 2000; Tadevosyan-Leyfer et al., 2003; Tanguay, Robertson, & Derrick, 1998]. One study, which analyzed 98 items on the ADI/ADI-R from 292 individuals, identified six clusters of variables that include spoken language, social intent, compulsions, developmental milestones, savant skills, and sensory aversions [Tadevosyan-Leyfer et al., 2003]. By selecting for the “savant skills” phenotype, Nurmi et al. demonstrated greatly increased linkage to chromosome 15q11–q13 (HLOD∼2.6) relative to that obtained with an unsegregated ASD population with an HLOD of ∼0.8 [Nurmi et al., 2003]. Similarly, incorporating language phenotypes into linkage analyses identified markers on chromosome regions 7q and 13q [Bradford et al., 2001]. In a similar vein, stratifying the ASD population according to the severity of a specific core symptom (e.g., repetitive behavior) allowed quantitative correlations with altered neurotransmitter levels (5-HT and oxytocin) [Hollander et al., 2000]. Interestingly, deficient oxytocin levels were also linked to deficits in social recognition (a common symptom in ASD) in a study focused on the molecular basis of “social memory” in an animal model [Winslow & Insel, 2002]. In addition, quantitative trait loci (QTL) can be clearly associated with severely language-impaired and nonverbal communication-impaired endophenotypes of ASD [Alarcon, Yonan, Gilliam, Cantor, & Geschwind, 2005; Chen, Kono, Geschwind, & Cantor, 2006]. Most recently, several QTL were identified based upon severity scores on the Social Responsiveness Scale [Duvall et al., 2007], which was designed to quantitate deficits in social functioning typical of ASD, but which are present within the broader population as a social endophenotype [Constantino et al., 2003]. Thus, a strong argument has been made for subtyping or stratifying ASD subjects according to phenotype or severity within a specific domain as well as for including related individuals expressing the “broad autism phenotype” in genetic and molecular analyses. By reducing the variability in this heterogeneous population, a “phenotypic approach” is expected to facilitate the identification of genes and pathways contributing to, or associated with, a specific behavioral/symptomatic phenotype.
As described in the accompanying article [Hu & Steinberg, 2009], we have applied several clustering algorithms to scored data from the Autism Diagnostic Interview-Revised (ADIR) questionnaires from nearly 2000 individuals with idiopathic autism in an attempt to divide the autistic probands into phenotypic subgroups based upon severity across 123 ADIR items. Our approach differs significantly from that employed by other investigators in that our subgroups are defined by multiple items within different behavioral or functional categories, including spoken language, nonverbal communication, social skills, play skills, physical attributes and sensitivities, aggression, and savant skills, while many other studies utilize at most several item scores within a single category to define subgroups of individuals. Another aspect of our approach that differs from previous analyses is that we employ multiple clustering algorithms to the data, which results in a clearer and more intuitive phenotypic description of the subgroups. Using these combined methods to identify both severe and mild subgroups of ASD individuals as well as those with notable savant skills, we now demonstrate discrimination of autistic from nonautistic individuals based upon gene expression profiles. In addition, both qualitative and quantitative differences in gene expression are observed between the subgroups. Furthermore, we show that the several phenotypes of idiopathic autism can also be distinguished by pathway analyses, corroborating the distinct biological phenotypes of ASD.
Materials and Methods
Sample Selection and Exclusionary Criteria
Lymphoblastoid cell lines (LCL) for gene expression analyses were selected based upon phenotype analyses and exclusionary criteria described in detail in the accompanying manuscript [Hu & Steinberg, 2009]. In brief, autistic probands were clustered into four groups based upon 123 scores from the ADIR questionnaire. Representative samples from three of the groups were selected after excluding all females, individuals with cognitive impairment (Raven's scores <70), those with known genetic or chromosomal abnormalities (e.g., Fragile X, Retts, tuberous sclerosis, chromosome 15q11–q13 duplication), those born prematurely (<35 weeks gestation), and those with diagnosed comorbid psychiatric disorders (e.g., bipolar disorder, obsessive compulsive disorder, severe anxiety). Impairment in spoken language was additionally confirmed based upon low standard scores (<80) on the Peabody Picture Vocabulary Test.
Cell Culture
The LCL were cultured as previously described [Hu, Frank, Heine, Lee, & Quackenbush, 2006] according to the protocol specified by the Rutgers University Cell and DNA Repository, which maintains the Autism Genetic Research Exchange (AGRE) collection of biological materials from autistic individuals and relatives. Briefly, cells are cultured in RPMI 1640 supplemented with 15% fetal bovine serum, and 1% penicillin/streptomycin. Cultures are split 1:2 every 3−4 days and cells are typically harvested for RNA isolation 3 days after a split while the cultures are in logarithmic growth phase.
Gene Expression Analyses on Spotted DNA Microarrays
Gene expression profiling is accomplished using TIGR 40K human arrays as previously described [Hu et al., 2006]. The annotation file for this microarray platform is GPL3427 in the Gene Expression Omnibus (GEO) archive. Total RNA was isolated from LCL using the TRIzol (Invitrogen, USA) isolation method according to the manufacturer's protocols, and cDNA was synthesized, labeled, and hybridized to the microarrays as described in our earlier study, with the exception that cDNA from each sample was labeled with Cy-3 dye and hybridized against Cy-5 labeled reference cDNA prepared from Universal human RNA (Stratagene, USA). This “reference” design allows the flexibility to perform different comparisons among the samples since all expression values are against a common reference. After hybridization, washing of the arrays, and laser scanning to elicit dye intensities for each element on the array, the raw intensity data was normalized and filtered using the Lowess normalization and standard deviation (variance) regularization programs [Quackenbush, 2002] contained within MIDAS [Saeed et al., 2003]. Prior to and following normalization and filtering of microarray data, LOWESS plots were visually inspected for skewness and artifacts. Additional microarray quality control assessment included principal components analysis (PCA) of the entire microarray data set to identify potential outliers. As no outlier experiments were detected, the entire data set was included for statistical analysis. The normalized intensities were then uploaded into MEV [Saeed et al., 2003], and a 70% data filter was applied, which removes genes for which log2 intensity values are missing in >30% of the samples (>35 out of 116 samples). This filtering step, when applied to data from 116 samples, reduces the number of transcripts from 41,472 to 25,221, which are then used for the statistical analyses in MEV. Significant differentially expressed genes were identified using the Significance Analysis of Microarrays (SAM) [Chu, Weir, & Wolfinger, 2002; Tusher, Tibshirani, & Chu, 2001] module within MEV for both 2-class and 4-class analyses, with a maximum false discovery rate (FDR) of 5%. Among our control subjects, there were 6 who are siblings of individuals in the savant group and 1 who is a sibling of an individual in the mild group. Because genotype has been shown to influence gene expression profiles [Cheung & Spielman, 2002; Cheung et al., 2003; Morley et al., 2004], we removed all controls who were siblings of the respective autistic probands prior to each SAM analyses.
Quantitative PCR Analysis
Select genes were confirmed by real-time RT-PCR on an ABI Prism 7300 Sequence Detection System using Invitrogen's Platinum SYBR Green qPCR SuperMix-UDG with ROX. Total RNA (same preparations used in microarray analyses) was reverse transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Briefly, 1 μg of total RNA was added to a 20 μl reaction mix containing reaction buffer, magnesium chloride, dNTPs, an optimized blend of random primers and oligo (dT), an RNase inhibitor and a MMLV RNase H+reverse transcriptase. The reaction was incubated at 25°C for 5 min followed by 42°C for 30 min and ending with 85°C for 5 min. The cDNA reactions were then diluted to a volume of 50 μl with water and used as a template for quantitative PCR.
Quantitative RT-PCR (qRT-PCR) primers for genes identified by microarray analysis as differentially expressed were selected for specificity by the National Center for Biotechnology Information Basic Local Alignment Search Tool of the human genome, and amplicon specificity was verified by first-derivative melting curve analysis with the use of software provided by PerkinElmer (Emeryville, CA) and Applied Biosystems (Foster City, CA). Sequences of primers used for the real-time RT-PCR are given in Supplemental Table I.
qRT-PCR analyses were performed on all samples, with quantification and normalization of relative gene expression using the comparative threshold cycle (Delta-delta-Ct) method [Livak & Schmittgen, 2001] as described previously [Hu et al., 2006]. The expression of the “housekeeping” genes MDH1, ARF1, and ACSL5 were used for normalization as these genes did not exhibit differential expression in our microarray assays. The qRT-PCR reactions were done in triplicate.
Treatment of LCL With Dihydroxytestosterone
Lymphoblastoid cells from a nonautistic individual (HI0813) were cultured in RPMI media with 15% FBS and 1% antibiotics at 37°C and 5% CO2. Prior to treatment, the cells were transferred to phenol red free RPMI media with 15% charcoal dextran-treated serum and 1% antibiotics for 24 hr to deplete the medium of endogenous hormones [Arnold, Le, McFann, & Blackman, 2005]. Dihydrotestosterone (DHT) was dissolved in ethanol (final ethanol concentration not exceeding 0.001%) and added at a final concentration of 10 nM. The cells were harvested 2 days after treatment with DHT and RNA was isolated using TRIzol reagent (Invitrogen). Quantitative PCR was performed as described above using primers for seven novel transcripts (Supplemental Table I). The qRT-PCR reaction for each gene was done in triplicate and average expression values were calculated relative to the vehicle-treated samples. In this study, ARF1, ACSL5, and MDH1 were used for normalization of the queried genes. All log2(fold change) values for the seven genes are statistically significant with P-value <0.05.
Gene Ontology and Pathway Analyses
The data sets of differentially expressed genes between autistic probands and unrelated controls were analyzed using Ingenuity Pathway Analysis (IPA) and Pathway Studio 5 to identify relational gene networks, high-level functions, and small molecules associated with the gene regulatory networks. For pathway analyses using IPA, the Fisher Exact Test was used to identify significant pathways and functions. To determine P-values, the number of genes with a particular function within the experimental data set is compared to the number of genes with that function in Ingenuity's Knowledge Base, which is a comprehensive database containing manually curated and published information about the functions and interactions of millions of genes and gene products. Unless otherwise noted, we applied an expression cutoff of log2 (ratio)≥±0.3 to the data before pathway analyses as we are generally able to confirm by qRT-PCR analyses average expression values of this magnitude.
Other Statistical Analyses
The P-values for the enrichment of differentially expressed genes in QTL were calculated based on the hypergeometric test [Johnson, Kotz, & Kemp, 1992], which was chosen because of its computational efficiency to calculate the statistical significance of an observed number of overlapped genes given the number of selected genes and the reference pattern. This test has been widely used in large-scale biomedical studies to evaluate whether an observation is significantly enriched when compared to a reference pattern [Chen et al., 2007; Falcon & Gentleman, 2007]. Only genes with complete chromosome start/end information were included in this analysis. An overlap between a gene and a QTL was determined based on the following criteria: (1) they are on the same chromosome; (2) the gene's start position is between the QTL's start and end positions, or the QTL's start position is between the gene's start and end positions. Of the 40,032 transcripts on the microarray, 29,723 have complete chromosome start/end information and 9,734 of these genes overlap with the QTLs according to our stated criteria.
Data Access
All microarray data is reported according to the MIAME standards and has been submitted to the GEO repository for public access. The GEO accession number is GSE15402.
Results
In the accompanying manuscript [Hu & Steinberg, 2009], we report on a novel clustering method for stratifying autistic individuals according to phenotypes, which encompass 123 scores on 63 distinct items on the ADIR questionnaire, most of which are represented by two separate scores related to “current” (existing) or “ever” (previously exhibited) behaviors. In this manuscript, the gene expression profiles of LCL of individuals from three of the four phenotypic subgroups that resulted from the cluster analyses of ADIR scores were analyzed and were shown to demonstrate different functions overrepresented within the different subgroups that are suggestive of distinct “biological phenotypes.” To test the concept that the phenotypic subgroups can be differentiated from each other by gene expression profiles, we analyzed the subgroup with severe language impairment and high severity scores across most of the ADIR items used for clustering (except for savant skills), the mild subgroup comprised of individuals many of whom were clinically diagnosed with PDD-NOS or Asperger's Syndrome who exhibited distinctly lower severity ADIR item scores, and the subgroup of individuals with noticeably high scores in the savant skills categories to identify genes that may be associated with this unusual and interesting trait. Included within the savant group were some individuals who, despite exhibiting pronounced savant skills, were also language impaired. The demographic profile of the individuals whose LCL were included in this study is provided in Supplemental Table II, with additional information (e.g., scores on the Peabody Picture Vocabulary Test and Raven's Test) provided in the accompanying manuscript. The intermediate subgroup was not included in this study because we wanted to be able to first demonstrate differences between phenotypic groups at the extreme ends of the spectrum.
DNA Microarray Analyses of ASD Phenotypic Subgroups Show Quantitative and Qualitative Differences in Gene Expression
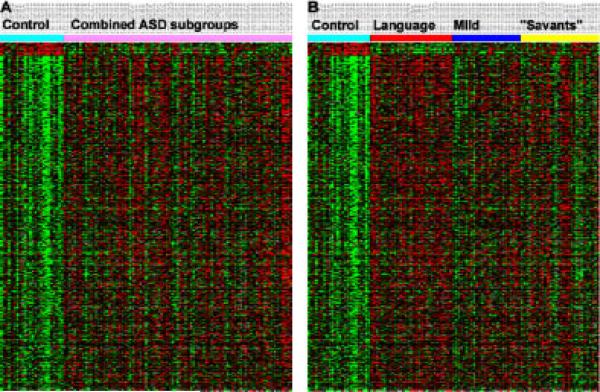
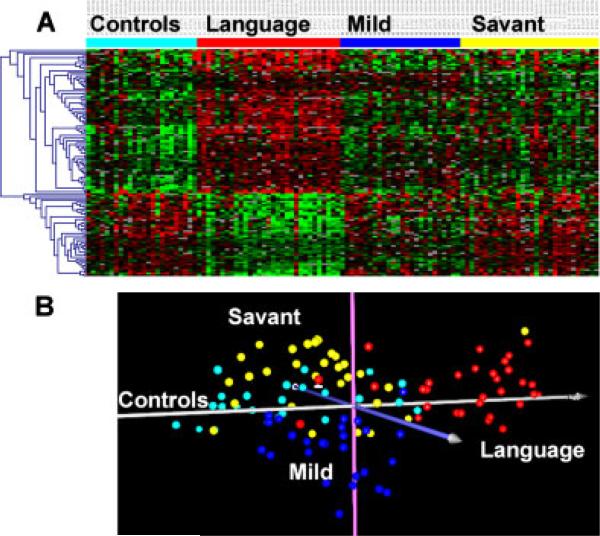
Gene expression profiles of LCL from each of the autistic individuals studied and age-matched controls were obtained by cDNA microarray analyses. A 2-class analysis of the data reveals a set of significant differentially expressed genes (FDR≤5%) that distinguish controls from all autistic samples (Fig. 1A and Supplemental Table III). Interestingly, when the samples from the autistic individuals are grouped according to phenotype, the gene expression matrix from this analysis reveals a gradient in differential gene expression for some genes in which the level of gene expression reflects the overall severity of the ASD phenotype relative to controls (Fig. 1B). Separation of the three ASD phenotypes from each other as well as from controls was also revealed by a 4-class SAM analysis of the microarray data (FDR≤0.1%) from all individuals. An expression matrix of the top 123 most significant genes from this 4-class analysis illustrates quantitative as well as qualitative differences in gene expression profiles that relate to “phenotype” (Fig. 2A, Supplemental Table IV). PCA was applied to this set of significant genes to reduce the dimensionality of the microarray data with respect to individual samples (Fig. 2B), and confirms that phenotypic subgroups can be differentiated from each other as well as from nonautistic controls, although there is still some mixing of the phenotypes, particularly between the “savant” group (yellow) and controls (turquoise), which are most similar in terms of gene expression profiles (Fig. 2A). The differences in gene expression profiles among several ASD phenotypes thus emphasize the need to identify and utilize more homogeneous samples for biological analyses.
Figure 1.
Gene expression analyses of LCL from controls and autistic individuals in which the samples from the autistic individuals are shown in no specific order (A) and grouped according to ASD phenotype derived from cluster analyses of ADIR scores (B). Top 530 significant genes identified by a 2-class SAM analysis with a false discovery rate (FDR) <5%, which compared gene expression between controls and all autistic individuals. The expression level of each gene (row) in this “heat map” is shown relative to that of RNA from a Universal human RNA standard (Stratagene). cDNA from the reference standard was labeled with Cy-5, while the cDNA of the samples was labeled with Cy-3. The heat map represents the log2(ratio) of Cy5/Cy3 fluorescence, with green showing decreased gene expression in Cy5 relative to Cy3 and red indicating the reverse. Thus, a change in color from green to red in this figure represents a decrease in relative expression.
Figure 2.
(A) Gene expression matrix of the top 123 significant genes from 4-class SAM analysis of gene expression data from the three ASD groups and nonautistic control group, with an expression cutoff of log2(ratio)≥±0.3. A more stringent FDR (<0.1%) was employed in this analysis to maximize gene discrimination among the groups although the separation of groups is still obvious with 673 genes (Supplemental Fig. 1). Color coding: Turquoise = nonautistic individuals; red = ASD with severe language impairment; blue = mild ASD; yellow = ASD with savant skills. Again, the heat map represents the log2(ratio) of Cy5/Cy3 fluorescence, with green showing decreased gene expression in Cy5 relative to Cy3 and red indicating the reverse. (B) Principal components analysis of samples on the basis of the 123 significant differentially expressed genes identified by the 4-class SAM analyses described above. The one savant individual (yellow) that clearly associates with the language subgroup (red) also has a severe language impairment.
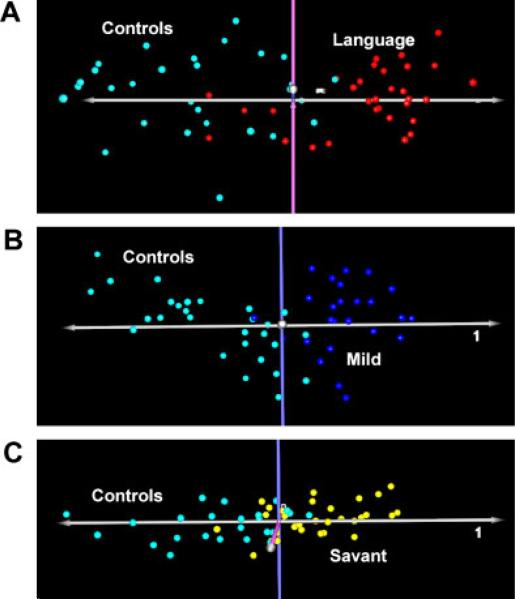
Toward this goal, we treated each ASD subgroup as a separate class and performed 2-class statistical analyses on the gene expression data obtained from each of the groups in comparison to nonautistic controls to identify the differentially expressed genes that were specific to each subgroup. Figure 3A–C show the PCA plots demonstrating separation of individuals from each of the ASD subgroups and controls on the basis of gene expression profile. Lists of the genes included in the respective PCA analyses are provided in Supplemental Tables V–VII.
Figure 3.
Separation of phenotypic variants of ASD from controls based on principal components analysis (PCA) of the differentially expressed genes identified by 2-class SAM analyses for each of the ASD subgroups and controls. (A) Separation of individuals with severe language impairment (L) and controls (C) based on 602 differentially expressed genes (FDR<0.1%). Turquoise-colored points represent control samples in all figures, with the second color representing the ASD samples. (B) Separation of individuals with mild ASD (M) and controls based on 502 differentially expressed genes (FDR<0.5%). (C) Separation of ASD “savants” (S) and controls based on 127 differentially expressed genes (FDR<0.5%). Differentially expressed genes for the respective ASD groups are listed in Supplemental Tables V–VII.
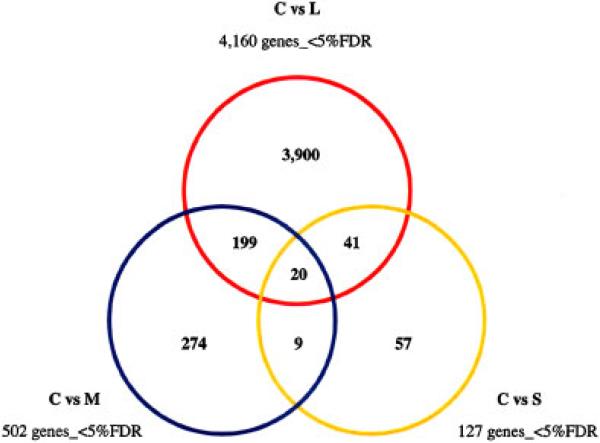
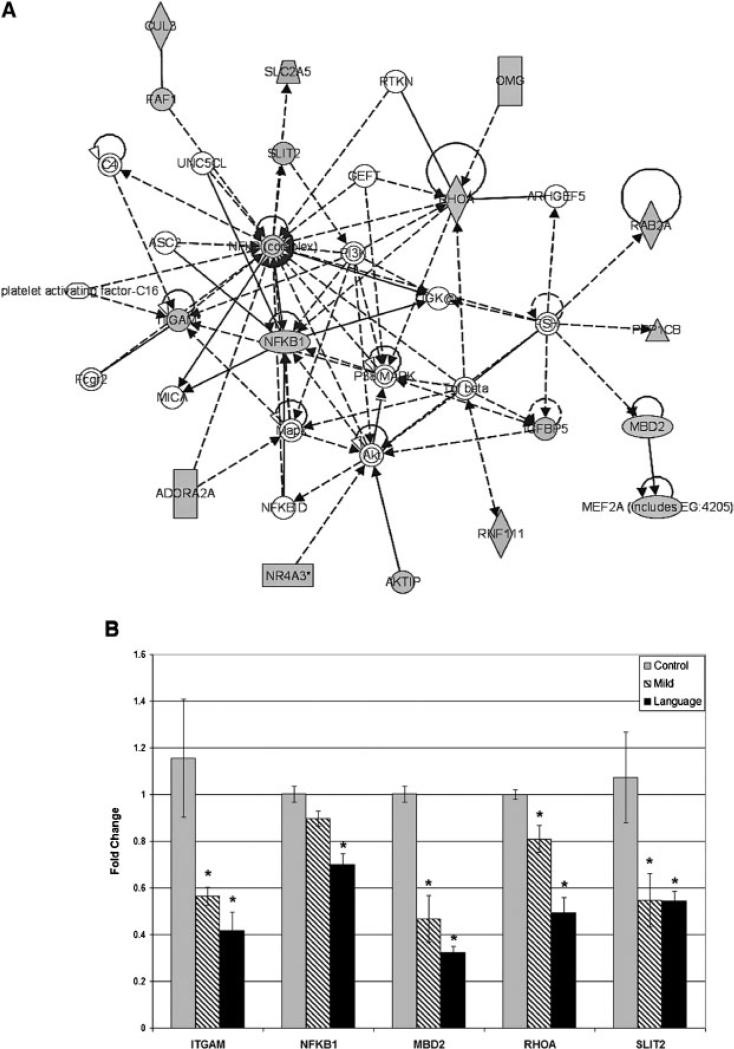
Overlapping as Well as Unique Genes are Associated With Each ASD Subgroup
The Venn diagram in Figure 4 displays the overlap in differentially expressed genes among the three ASD subgroups. Pathway analysis of the overlapping genes between the L and M subgroups with Pathway Studio 5 software reveals a network of genes that affect common functional targets, such as synaptic transmission, neurogenesis, neurulation, long-term potentiation (learning), protein ubiquitination, and brain function that have been identified as dysfunctional in ASD (Supplemental Fig. 2). Of additional interest are the disorders associated with this set of genes, including inflammation, epilepsy, muscle function (hypotonia), disorders of intestinal absorption, and diabetes mellitus, which have been reported in subsets of individuals with ASD [Herbert et al., 2006; Hollander & Nowinski, 2003; Levy et al., 2007; Palmen, van Engeland, Hof, & Schmitz, 2004; Pardo, Vargas, & Zimmerman, 2005; Polleux & Lauder, 2004; Schultz et al., 2008]. Ingenuity Pathways Analysis software was used to identify the top multigene network (Fig. 5), with the most significant functions, pathways, and associated disorders listed in Table I. Interestingly, many of the genes in the network shown in Figure 5A are involved in neurological functions and disorders (Table II), with the predominant functions being neurite outgrowth, axon pathfinding, neurogenesis, and brain morphology. Thus, qRT-PCR was used to confirm several of the genes in this network (Fig. 5B). Table III lists the overlapping significantly differentially expressed transcripts across all three ASD subgroups (see Fig. 4). What is intriguing about this set of genes is that all 20 transcripts are novel and uncharacterized transcribed sequences, 19 of which map to intronic or intergenic regions. The majority of these transcripts also appear to be associated with cellular response to androgens as revealed by gene expression studies on Androgen Insensitivity Syndrome and androgen-sensitive and -insensitive prostate tumors, which are reported in GEO [Holterhus, Hiort, Demeter, Brown, & Brooks, 2003; Zhao et al., 2005]. To examine the possibility that some of these genes are androgen responsive, LCL were treated with dihydroxytestosterone, a potent metabolite of testosterone, and the levels of these transcripts were quantitated relative to vehicle-treated cells. As shown in Figure 6, all seven of the tested transcripts exhibited a response to DHT treatment. Further studies are needed to identify and fully characterize the nature of these possibly noncoding transcripts.
Figure 4.
Venn diagram showing overlap of differentially expressed genes relative to controls among three subgroups of ASD. Significant genes were determined for each group by 2-class SAM analysis of the data from each ASD group and that of the nonautistic controls, with FDR≤5%. Because of the quantitative differences in gene expression that appeared to correlate with severity of the phenotype, no expression cutoff was applied in order to observe the maximum overlap of differentially expressed genes among the ASD groups. The complete list of differentially expressed genes between the L group and controls is given in Supplemental Table VIII. [Color figure can be viewed online at www.interscience.wiley.com]
Figure 5.
(A) Top multigene network associated with the data set of differentially expressed genes (FDR≥5%) in both the severely language-impaired (L) and mild (M) ASD groups, as determined by Ingenuity Pathway Analysis. An expression cutoff of log2(ratio)≥±0.3 was applied prior to the analysis. (B) qRT-PCR analyses on five representative samples each from the L and M subgroups as well as the control group, focusing on hub genes in Figure 5A as well as genes of relevance to inflammation (NFKB1) and epigenetic regulation (MBD2), which are both implicated in autism. Data represent fold-changes±SE calculated using the delta-delta-Ct method and with respect to average Ct values obtained from the control group (n = 5). *P<0.05
Table I.
High-Level Functions, Pathways, and Disorders Associated With Differentially Expressed Genes Shared Between the Severely Language-Impaired (L) and Mild (M) ASD Subgroups
| Top biological functions | P-values* | # genes/category |
|---|---|---|
| Top molecular and cellular functions | ||
| Cell morphology | 1.29E–03 t0 4.81E–02 | 10 |
| Cellular assembly and organization | 1.29E–03 to 4.81E–02 | 10 |
| Cell cycle | 1.54E–02 to 4.64E–02 | 9 |
| Cell death | 2.78E–03 to 4.94E–02 | 13 |
| Cell-to-cell signaling and interaction | 3.95E–03 to 3.88E–02 | 3 |
| Top canonical pathways | ||
| IL-8 signaling | 4.48E–03 | 4/185 |
| Regulation of actin-based motility by Rho | 4.87E–03 | 3/94 |
| Integrin signaling | 6.63E–03 | 4/203 |
| Semaphorin signaling in neurons | 1.72E–02 | 2/52 |
| Chemokine signaling | 3.71E–02 | 2/77 |
| Physiological system development and function | ||
| Embyonic development | 1.9E–04 to 4.17E–02 | 11 |
| Nervous system development and function | 1.29E–03 to 4.26E–02 | 13 |
| Connective tissue development and function | 2.78E–03 to 4.64E–02 | 6 |
| Cardiovascular system development and function | 3.95E–03 to 2.35E–02 | 2 |
| Cell-mediated immune response | 3.95E–03 to 3.12E–02 | 6 |
| Diseases and disorders | ||
| Inflammatory disease | 6.74E–04 to 4.26E–02 | 10 |
| Inflammatory response | 6.74E–04 to 4.64E–02 | 6 |
| Infection mechanism | 1.55E–03 to 4.26E–02 | 4 |
| Genetic disorder | 1.55E–03 to 4.26E–02 | 24 |
| Neurological disease | 2.09E–03 to 3.12E–02 | 11 |
P-values indicate the probability that the indicated function, pathway, or disorder is associated with the data set of differentially expressed genes by random chance, as determined by the Fisher Exact Test within Ingenuity Pathway Analysis.
Table II.
Neurological Functions and Disorders Associated With Differentially Expressed Genes Shared Between the Severely Language-Impaired (L) and Mild (M) ASD Subgroups
| Top neurological functions and disorders | P-value* | Molecules |
|---|---|---|
| Nervous system development and function | ||
| Outgrowth of neurites | 1.29E–03 | ADORA2A, EXT1, NFKB1, OMG, RHOA, SLIT2 |
| Coalignment of neurofilaments | 3.95E–03 | DST |
| Morphology of dendritic trees | 3.95E–03 | RHOA |
| Growth of axons | 4.72E–03 | EXT1, OMG, SLIT2 |
| Growth of neurons | 7.03E–03 | AKTIP, SLIT2 |
| Development to neurites | 8.19E–03 | ADORA2A, DST, RHOA, SLIT2 |
| Neurogenesis | 1.04E–02 | CRIM1, EXT1, FNBP1, OMG, RHOA, SLIT2 |
| Differentiation of microglia | 1.18E–02 | ITGAM |
| Long-term memory of rats | 1.57E–02 | NFKB1 |
| Branching of neurites | 2.71E–02 | FNBP1, SLIT2 |
| Chemorepulsion of axons | 2.73E–02 | SLIT2 |
| Development of central nervous system | 2.94E–02 | ADORA2A, AKTIP, CHD7, SERPINI1 |
| Morphology of brain | 3.12E–02 | AKTIP |
| Neurological disease | ||
| Huntington's disease | 2.09E–03 | ADORA2A, ANKRD11, CRIM1, NFKB1, PPP1CB, RERE, SERPINI1, SLIT2 |
| Dysmyelination of peripheral nervous system | 3.95E–03 | DST |
| Spinocerebellar ataxia, type 1 | 7.88E–03 | IGFBP5 |
| Neurodegeneration of striatum | 1.18E–02 | ADORA2A |
| Collapse of growth cone | 1.48E–02 | RHOA, SLIT2 |
| Leakage of blood-brain barrier | 1.57E–02 | SERPINI1 |
| Damage of pyramidal neurons | 1.96E–02 | NFKB1 |
| Catalepsy of mice | 3.12E–02 | ADORA2A |
P-value indicates the probability that the indicated function or disorder is not associated with the data set of significantly differentially expressed genes as determined by the Fisher Exact Test as implemented by Ingenuity Pathway Analysis software.
Table III.
Differentially Expressed Transcripts Across All three ASD Subgroups Analyzed
| GenBank # | Associated gene | Map region | log2(L/C) | log2(M/C) | log2(S/C) | log2(A/C) | Adj P-value* |
|---|---|---|---|---|---|---|---|
| T65857 | Unknown | Intergenic | −0.878 | −0.510 | −0.745 | −0.825 | 2.21E–04 |
| N47010 | KIAA1432 | Intronic | −0.802 | −0.486 | −0.747 | −0.763 | 4.52E–03 |
| AI076295 | MEMO1 | Intronic | −0.547 | −0.555 | −0.634 | −0.627 | 1.54E–03 |
| H97875 | DENND5B | Intronic | −0.361 | −0.488 | −0.465 | −0.477 | 9.43E–04 |
| AA704941 | LARP5 | Intergenic | −0.507 | −0.304 | −0.411 | −0.470 | 2.60E–04 |
| AA907052 | SMA4, GUSBP1 | Intronic | −0.307 | −0.462 | −0.496 | −0.438 | 3.04E–04 |
| H56961 | JMJD2C | Intronic | −0.456 | −0.289 | −0.430 | −0.436 | 2.30E–04 |
| AA995108 | CUL3 | Intronic | −0.373 | −0.386 | −0.351 | −0.422 | 3.67E–04 |
| H73587 | XTP2, BAT2D1 | Intronic | −0.383 | −0.336 | −0.329 | −0.390 | 2.16E–03 |
| H63175 | USP47 | Intronic | −0.232 | −0.302 | −0.370 | −0.357 | 6.94E–04 |
| H25019 | ZZZ3 | Intronic | −0.239 | −0.285 | −0.444 | −0.349 | 4.04E–03 |
| AA406078 | ZEB1 | Intronic | −0.268 | −0.279 | −0.363 | −0.342 | 9.01E–03 |
| AA906454 | MUDENG | Intronic | −0.346 | −0.230 | −0.304 | −0.340 | 2.42E–04 |
| AA026388 | SENP6 | Intronic | −0.224 | −0.230 | −0.460 | −0.320 | 1.46E–04 |
| N73227 | PARG | Intronic | −0.256 | −0.249 | −0.332 | −0.317 | 5.24E–03 |
| AI276056 | ATP13A3 | Intronic | −0.206 | −0.261 | −0.280 | −0.284 | 2.08E–03 |
| R11217 | FBXW7 | Intronic | −0.218 | −0.250 | −0.272 | −0.262 | 4.89E–04 |
| N26823 | RBBP6 | −0.259 | −0.185 | −0.206 | −0.249 | 2.27E–05 | |
| AA700707 | ATP11B | Intergenic | −0.206 | −0.216 | −0.272 | −0.245 | 3.37E–03 |
| H85885 | KIAA0999 | Intronic | −0.171 | −0.225 | −0.238 | −0.232 | 5.47E–03 |
L: severely language impaired; M: mildly affected; S: with notable savant skills; A: all autistic groups combined; C: nonautistic control group.
Statistical significance of unpaired t-test comparing controls vs. all autistic probands (A). The adjusted P-value was obtained using a standard Bonferroni correction for multiple testing.
Figure 6.
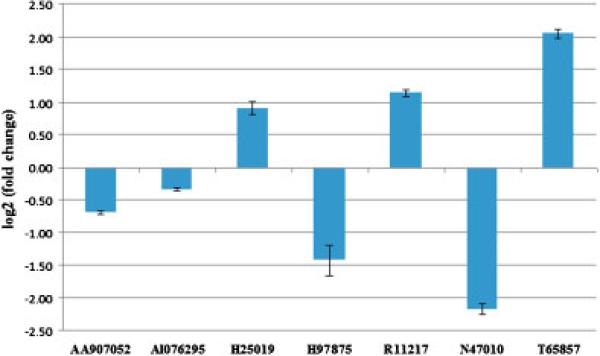
Response of seven novel transcripts to 10 nM dihydroxytestosterone (DHT). The data shows the relative expression of the transcripts in DHT-treated cells to that in vehicle (ethanol-treated) cells after a 2-day exposure to the hormone. P-values for triplicate analyses were all <0.05.
Functional Analyses of the Different Subgroups of ASD on the Basis of Gene Expression Profiles: Evidence for Distinct Biological Phenotypes
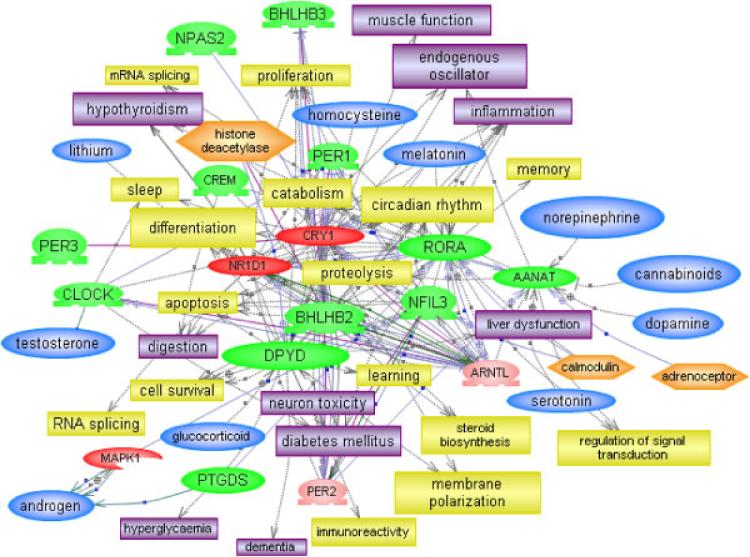
To understand the differences in the pathways and functions that are affected in each of the phenotypic subgroups, separate pathway and functional analyses were conducted on the gene data sets derived from comparison of the respective phenotypes vs. controls. Table IV summarizes the results obtained from IPA for each group in terms of categories of molecular and cellular functions, canonical pathways, and toxicity that are significantly enriched within the differentially expressed gene data set. It is clear from this summary that biological functions and pathways are most altered in the severely language-impaired subgroup and least altered in the “savant” subgroup. Among the genes relating to molecular and cellular functions, cell death genes are overwhelmingly overrepresented in the subgroup with severe language deficits, while genes involved in small molecule biochemistry, free radical scavenging, and cellular function and maintenance are overrepresented in the differentially expressed gene data set of the mild phenotype. It is interesting that RNA posttranscriptional modification is a highly significant molecular function enriched in the savant phenotype, as variation in RNA splicing has been noted to be associated with autism [Talebizadeh et al., 2006]. Among the genes involved in specific canonical pathways are those related to liver toxicity (hepatic fibrosis and stellate cell activation), which are enriched in both the severely language-impaired and the mild subgroups, but with more genes and hepatic cholestasis also affected in the severe subgroup. It is proposed that the dysregulation of at least some of these genes may be responsible for gastrointestinal disorders that are often associated with autism. Further comparison of the severe and mild subgroups on the basis of genes that are significantly enriched for neurological functions and disorders revealed not only differences in the number of genes associated with cell death in the severely language-impaired subgroup, but also a greater number of differentially expressed genes involved with various neurological disorders commonly associated with autism, such as allodynia, catalepsy, hypernocieption, and epilepsy (Table V). Particularly noteworthy are the 15 genes that are involved in the regulation of circadian rhythm, which also affect many of the neurological functions and disorders commonly associated with ASD, such as synaptic plasticity, learning, memory, inflammation, cytokine production, and digestion (Fig. 7) [Herbert et al., 2006; Palmen et al., 2004; Pardo et al., 2005; Persico & Bourgeron, 2006; Polleux & Lauder, 2004]. Differential expression of these genes was observed only in the samples from severely language-impaired individuals (L subgroup), with each individual showing altered expression of multiple (but not all 15) genes. We have confirmed differential expression of 6 genes (out of 6 tested) by qRT-PCR (Table VI).
Table IV.
Pathway and Functional Analyses of Differentially Expressed Genes From Three ASD Subgroups
| Severely language-impaired | Mildly autistic | “Savant” group |
|---|---|---|
| Molecular and cellular functions (P-value) [#genes] | ||
| Cell death (1.54E–10 to 5.99E–03) [83] | Small molecule biochemistry (2.53E–04 to 3.58E–02) [16] | RNA post-transcriptional modification (1.63E–04 to 1.76E–02) [4] |
| Cellular development (1.58E–08 to 5.61E–03) [60] | Free radical scavenging (4.20E–04 to 9.79E–03) [5] | Cellular growth and proliferation (2.75E–04 to 4.99E–02) [5] |
| Cellular movement (7.40E–08 to 5.78E–03) [48] | Cellular function and maintenance (1.07E–03 to 3.13E–02) [8] | Cell cycle (4.61E–04 to 1.14E–02) [2] |
| Cell growth and proliferation (2.00E–06 to 5.95E–03) [78] | DNA replication, recombination, repair (1.23E–03 to 2.70E–02) [5] | Cellular assembly and organization (1.04E–03 to 4.79E–02) [3] |
| Cell signaling (1.01E–05 to 1.79E–03) [32] | Cell signaling (1.25E–03 to 3.58E–02) [6] | Cellular development (1.04E–03 to 3.29E–02) [3] |
| Top canonical pathways (P-value) [#genes/genes in category] | ||
| cAMP-mediated signaling (3.19E–03) [8/159] | IL-8 signaling (7.39E–03) [4/185] | Glutamate receptor signaling (6.28E–02)* [1/67] |
| Hepatic fibrosis/stellate cell activation (4.06E–03) [7/131] | Hepatic fibrosis/stellate cell activation (1.96E–02) [3/135] | |
| Hepatic cholestasis (6.01E–03) [7/162] | Cell cycle: G1/S checkpoint regulation (2.67E–02) [2/59] | |
| B cell receptor signaling (7.97E–03) [7/148] | Wnt/β-catenin signaling (3.62E–02) [3/166] | |
| NF-kB signaling (8.6E–03) [7/143] | ||
| Top toxicity lists (P-value) [#genes/genes in category] | ||
| Hepatic stellate cell activation (2.74E–04) [5/35] | Positive acute phase response proteins (3.29E–02) [1/32] | |
| Hepatic fibrosis (3.01E–03) [6/85] | ||
| Hepatic cholestasis (7.66E–03) [7/135] | ||
| NF-kB signaling pathway (1.14E–02) [6/112] | ||
| Gene regulation by PPARa (2.18E–02) [5/95] | ||
Ingenuity Pathway Analysis software was used to analyze the data sets of significantly differentially expressed genes (FDR<5%) for functions and pathways that were statistically enriched, that is, not likely to be identified by chance. The Fisher Exact Test is used to reveal functions/pathways with P-values<0.05, which indicates that there is less than a 5% probability that the association between the identified genes and the respective pathway or function is due to random chance.
The exception to this P-value cutoff of 0.05 is the P-value for glutamate receptor signaling which is 0.063.
Table V.
Neurological Functions and Disorders Associated With Differentially Expressed Genes From the Language, Mild and Savant Subgroups
| Subgroup-associated neurological disorders/functions | Genes |
|---|---|
| Language subgroup | |
| Neurological disorders | |
| Apoptosis/cell death of neuroglia, astrocytes, neurons | BTG1, QK1, TNFSF10, GAS6, CD44, PTGDS, TLR4, MYC, EDN1, MAPK1, HDAC9, ITGA2, CREM, FOX03, MAP1B, TGFB2, CTF1, ADORA2A, DAPK1, GLRX, SH3RF1, TP53BP2, MAP3K5, BID, FN1, INSR, MAOA, NOVA1, NR1D1, SH3RF1, IL1RN, PDCD6IP, APBB1 |
| Catalepsy | ADORA2A, CNR1, PRKAR2B |
| Allodynia | GAL, IL1RN, PRKCG, PTGDS, TLR4 |
| Seizures (mice) | GAL, IL1RN |
| Hypernociception | EDN1, IL15, IL1RN |
| Nervous system functions | |
| Circadian rhythm | AANAT, BHLHB2, BHLHB3, CLOCK, CREM, CRY1, DPYD, MAPK1, NFIL3, NPAS2, NR1D1, PER1, PER3, PTGDS, RORA |
| Generation of neuronal progenitors | ASCL1, CNR1 |
| Neurological process | CTF1, GAL, NR3C1, PLAU, SERPINE2, ADORA2A, ASCL1, CNR1, CREM, CYBB, GM2A, GNAI1, NOVA1, OPHN1, PRKAR2B, PRKCG |
| Olfactory memory | GAL, PLAU |
| Mild subgroup | |
| Neurological disorders | |
| Damage of striatum | ADORA2A, SOD2 |
| Familial encephalopathy with neuroserpin inclusion bodies | SERPINI1 |
| Invasion of neuroepithelial cells | MYH10 |
| Spinocerebellar ataxia, type 1 | IGFBP5 |
| Neurodegeneration | ADORA2A, SOD2, TGFBR2 |
| Hydrocephalus of brain | MYH10 |
| Leakage of blood-brain barrier | SERPINI1 |
| Schizophrenia | GNAS, MARCKS (includes EG:4082), SERPINI1, SOD2, UFD1L |
| Gilosarcoma | MGMT |
| Amyotrophic lateral sclerosis of mice | SOD2 |
| Nervous system functions | |
| Outgrowth of neurites | ADORA2A, GNAS, MARCKS (includes EG:4082), MYH10, OMG, SLIT2 |
| Cytostasis of neurites | MYH10 |
| Elongation of sensory axons | SLIT2 |
| Olfactory response of organism | GNAS, SLIT2 |
| Branching of neurites | FNBP1, SLIT2 |
| Neurogenesis of embryo | NEUROD2 |
| Development of central nervous system | ADORA2A, MARCKS (includes EG:4082), MYH10, SERPINI1, TGFBR2 |
| Differentiation of microglia | ITGAM |
| Volume of brain | SOD2 |
| Contact repulsion of axons | SLIT2 |
| Savant subgroup | |
| Neurological disorders | |
| Amyotrophic lateral sclerosis | SOD2, TARDBP |
| Damage of striatum | SOD2 |
| Nervous system functions | |
| Volume of brain | SOD2 |
Each data set containing significantly differentially expressed genes from a 2-class SAM (FDR<5%) for each of the subgroups was analyzed using Ingenuity Pathway Analysis network prediction software, using an expression cutoff of log2(ratio) of ±0.3 for functional/pathway analyses. Fisher Exact P-values for enrichment of genes associated with the specified disorders and functions < 0.05, which means that there is less than a 5% likelihood that the association between the indicated neurological disorder or function is due to random chance.
Figure 7.
Differentially expressed circadian rhythm genes in the severe ASD phenotype affect multiple processes commonly associated with ASD pathology. The color coding of the entities within this relational gene/molecular network are as follows: Red—genes that show increased expression in autistic individuals on average relative to controls; Green—genes that show decreased expression in autistic individuals on average relative to controls; Pink—additional circadian regulators inserted by Pathway Studio 5. Blue—small molecules including melatonin, serotonin, testosterone, and nitric oxide (NO); Yellow—cell processes; Lavender—disorders associated with these genes.
Table VI.
Quantitative RT-PCR Confirmation of Six of the Circadian Rhythm Genes
| Gene | qPCR log2 | SE | Microarray log2 | SE |
|---|---|---|---|---|
| AANAT | −2.115 | 0.31 | −0.468 | 0.03 |
| BHLHB2 | 0.913 | 0.28 | 0.851 | 0.12 |
| CRY1 | 1.202 | 0.36 | 0.865 | 0.10 |
| DPYD | −2.135 | 0.79 | −1.080 | 0.52 |
| NPAS2 | −0.350 | 0.70 | −0.657 | 0.08 |
| PER3 | −1.279 | 0.52 | −1.102 | 0.27 |
Five representative samples each were selected from both the control and severely language impaired groups for qRT-PCR analyses (in triplicate) of AANAT, BHLHB2, CRY1, DPYD, NPAS2, and PER3. The average expression values from microarray and qRT-PCR analyses are shown for comparison along with the standard error of the mean (SE) for each set of analyses on the representative samples.
Many Differentially Expressed Genes are Associated With Autism QTL Identified by Genetic Analyses
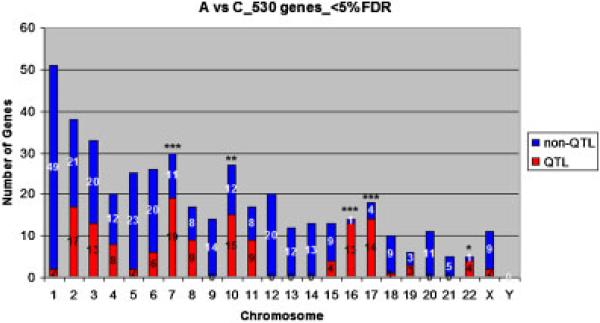
Gene expression analyses indicate that there are hundreds to thousands of genes that are differentially expressed between LCL of nonautistic individuals and those of each of the three ASD subgroups studied. To investigate whether these genes bear any relationship to genetically identified autism susceptibility loci, we mapped the differentially expressed genes to QTL reported by seven laboratories [Alarcon et al., 2005; Bailey et al., 1998; Chen et al., 2006; Duvall et al., 2007; Philippe et al., 1999; Szatmari, Paterson, Zwaigenbaum, Roberts, Brian, Liu et al., 2007b; Weiss et al., 2008]. On average, about 27−33% of the differentially expressed genes are associated with autism QTL across all subgroups and the autistic samples combined (Fig. 8). The hypergeometric distribution test was applied to determine whether there is statistically significant enrichment of differentially expressed genes in QTLs within the combined autistic sample (A) as well as within the individual ASD subgroups. The analyses showed that there is indeed enrichment of differentially expressed genes in the selected QTLs for all groups, except for the language (L) subgroup, where the P-values were 0.001, 0.266, 0.001, and 0.003, for the A, L, M, and S gene data sets reported in Supplemental Tables III–VI, respectively. With respect to distribution of differentially expressed genes in QTL on specific chromosomes, there is significant enrichment (P<0.025) of differentially expressed genes in QTLs on chromosomes 7, 10, 16, 17, and 22 for the combined autistic group (530 genes at <5% FDR), as shown in Figure 8. In addition, there is enrichment in QTLs on chromosomes 2, 4, 7, 8, 10, 16, 17, 19, and 22 for the L subgroup (P<0.05), chromosomes 7, 16, and 17 for the M group (P = 0.0001), and on chromosomes 7 and 16 for the S subgroup (P<0.0015) according to Chi square tests. It is notable that all of these chromosomes have undergone intensive genetic analyses as “hot spots” with respect to autism [Auranen et al., 2000; Balciuniene et al., 2007; Buxbaum et al., 2001; Kumar et al., 2008; McCauley et al., 2005; Santangelo et al., 2000; Schellenberg et al., 2006; Stone, Merriman, Cantor, Geschwind, & Nelson, 2007; Wassink et al., 2008]. Thus, the layering of gene expression data onto genetic data may be a useful means of prioritizing candidate genes within the candidate susceptibility loci for further functional and genetic analyses.
Figure 8.
Association of differentially expressed genes in the combined autistic sample (A) with quantitative trait loci for ASD identified by genetic analyses. For in silico mapping of our differentially expressed genes in autism-related QTLs described in the literature, a QTL interval was defined as two LOD units around the linkage peak, provided that mapping information on multiple genetic markers in the vicinity of the peak was given. Otherwise, the interval was defined by 10−15 megabases on either side of a lone genetic marker defining the QTL. Red: differentially expressed genes within identified QTL; Blue: differentially expressed genes not associated with QTL. Chi square analyses were used to identify significant enrichment of differentially expressed genes on a particular chromosome. ***P<0.0005; **P<0.015; *P<0.025. P-values for enrichment of differentially expressed genes in QTLs on specific chromosomes for the ASD subgroups are provided in the text.
Discussion
Genetic and other biological analyses of idiopathic autism, which makes up at least 70−80% of ASD cases have been hampered by the inherent heterogeneity of presentation of ASD in different individuals which, in turn, increases the noise in the experimental data. We therefore sought to reduce the phenotypic heterogeneity of clinical samples obtained through the AGRE/NIMH tissue repository by subgrouping/stratifying individuals based upon cluster analyses of 123 of their respective ADIR scores on 63 items, some of which are queried twice, with respect to current behaviors and previously exhibited behaviors. While other studies have utilized several ADIR item scores within a specific domain (e.g., spoken language, nonverbal communication, social skills, or repetitive behaviors) to stratify ASD individuals for genetic analyses [Alarcon et al., 2005; Chen et al., 2006; Duvall et al., 2007; Hollander et al., 2000], this is the first study to subgroup individuals on the basis of ADIR item scores that reflect the full range of deficits commonly associated with ASD. We demonstrate that the gene expression profiles associated with each of the 3 ASD phenotypes that were selected for DNA microarray analyses show both qualitative and quantitative differences, which are dependent on ASD phenotype. We also show overlap of some of the differentially expressed genes among subgroups, which indicates common underlying biological deficits in ASD as well as differences that suggest dysregulation of specific pathways or functions in a particular subgroup of ASD.
ASD Phenotypic Subgroups can be Distinguished on the Basis of Gene Expression Profiling
We demonstrate here that the gene expression profiles associated with each of the three ASD phenotypes that were selected for DNA microarray analyses show both quantitative and qualitative differences, which are dependent on ASD phenotype. The quantitative differences that were revealed in a 2-class analysis of the gene expression profiles of all autistic probands vs. controls were particularly surprising and likely identify genes that influence the severity of ASD. Such genes would thus serve as good candidates for expression QTL (eQTL) analyses, which, in turn, will help to prioritize genes for in-depth genetic association and linkage studies. It should be pointed out that the gradient of gene expression in Figure 1B is only apparent when the samples are clustered according to ASD subtype, thus validating the value of our clustering methods, which were applied to 123 selected ADIR item scores. Genes whose expression levels are qualitatively, but not quantitatively, dependent on subgroups (as represented by subgroup-specific genes in the Venn diagram of Fig. 4) also present a strong case for subtyping ASD individuals according to our methods since averaging gene expression values across all samples would dampen the overall expression differences from controls and obscure the biological differences between the subgroups. It is therefore suggested that such clustering of individuals to reduce the phenotypic heterogeneity of the study groups will also be of value to genetic and other biological analyses of ASD.
Overlapping Differentially Expressed Genes May Underlie Basic Deficits in ASD
The Venn diagram in Figure 4 summarizes the number of overlapping differentially expressed genes among the three ASD subgroups, with the largest overlap occurring between the severe (L) and mild (M) subgroups. Among the major functions associated with this set of overlapping genes are apoptosis and inflammation, as well as many neurological and metabolic processes commonly associated with ASD, such as central nervous system development, synaptic transmission, brain function, neuronal death, protein ubiquitination, RNA splicing, and oxidative stress (Supplemental Fig. 2). Genes, which were confirmed by qRT-PCR analyses (ITGAM, NFKB1, RHOA, SLIT2, and MBD2) are all strong candidates for further evaluation of their role in ASD (Fig. 5). ITGAM is involved in synapse formation and neuron toxicity, and is associated with chronic neural inflammation and microglial activation, which has been indicated in previous reports [Pardo et al., 2005; Vargas, Nascimbene, Krishnan, Zimmerman, & Pardo, 2005]. Similarly, the transcription factor NFKB1 is also a key regulator of inflammatory responses, which have been associated with ASD [DeFelice et al., 2003; Jyonouchi, Sun, & Le, 2001; Zimmerman et al., 2005]. RHOA and SLIT2 are components of the synpatogenesis/axon guidance pathway, which is strongly implicated in ASD [Jamain et al., 2003; Matzke et al., 2007; Persico & Bourgeron, 2006; Szatmari, Paterson, Zwaigenbaum, Roberts, Brian, Liu et al., 2007a]. These biological processes (inflammation, axon guidance) as well as others shown in Supplemental Figure 2 (e.g., apoptosis, neurogenesis, cell survival, and sex determination) replicate those identified in our previous gene expression studies of monozygotic twins discordant in diagnosis or severity of autism [Hu et al., 2006] and autistic–nonautistic sib pairs [Hu, Nguyen, Kim, Steinberg, Sarachana et al., 2009]. Altered expression of MBD2, a methyl-CpG binding protein, suggests the role of epigenetic factors in ASD. Indeed, several mutations have been identified in this family of transcriptional regulator proteins in autistic patients [Li, Yamagata, Mori, Yasuhara, & Momoi, 2005] and MeCP2, in particular, is responsible for Rett's Syndrome, a genetically defined ASD [Amir et al., 1999; Van Den Veyver & Zoghbi, 2001]. Our previous observation of gene expression differences between monozygotic twins discordant in diagnosis or severity of autism further supports the role of epigenetic regulation in ASD [Hu et al., 2006].
The most intriguing of the overlapping genes are the 20 novel genes that are shared by all three ASD groups because of their potential importance to core symptoms or pathology of ASD (Table III). As mentioned earlier, most if not all of these highly significant differentially expressed genes have been observed to be differentially regulated within the context of androgen insensitivity [Holterhus et al., 2003; Zhao et al., 2005]. This, in itself, is very interesting because of the hypothesis that higher levels of fetal testosterone may be a risk factor for ASD [Baron-Cohen, Knickmeyer, & Belmonte, 2005; Knickmeyer, Baron-Cohen, Raggatt, & Taylor, 2005; Knickmeyer & Baron-Cohen, 2006]. In fact, there is experimental support for this hypothesis, both from analysis of serum levels of androgens in individuals with ASD [Geier & Geier, 2006; Ingudomnukul, Baron-Cohen, Wheelwright, & Knickmeyer, 2007], as well as from our own studies [Hu et al., 2009], which show dysregulation of genes within the steroid hormone biosynthetic pathway in LCL from ASD probands as well as higher testosterone levels in their LCL extracts relative to their respective nearly age-matched siblings. From the location of these genes in the intronic regions of known genes and intergenic regions of the DNA, it is highly possible that at least some of these transcripts are noncoding RNA, some of which are known to play critical roles in neurodevelopment and function [Kapsimali et al., 2007; Kosik, 2006; Rogaev, 2005]. Although differentially expressed microRNA has been observed in both brain [Abu-Elneel et al., 2008] and LCL [Talebizadeh, Butler, & Theodoro, 2008; Sarachana et al., unpublished data] from autistic probands vs. nonautistic individuals, the novel transcripts do not include these microRNA and have not been found in several microRNA databases. The nature of the genes with which these transcripts are associated may reveal some of the functions impacted by autism, if the intronic transcripts are regulators of the respective genes. One example of a gene containing such a transcript is the Jumonji-domain-containing histone demethylase JMJD2C (GASC1) [Cloos et al., 2006; Pollard et al., 2008], which has been identified as an androgen receptor coregulator whose transcript is modestly affected by DHT [Urbannuci, Waltering, Suikki, Helenius, & Visakorpi, 2008]. Interestingly, deletions of chromosome 9p24 in the region of this gene (as well as DMRT1 and DMRT3) have been associated with autism or autistic-like behaviors [Vinci et al., 2007]. Because all of the novel transcripts are decreased in LCL from autistic individuals relative to controls, another possible explanation to consider is defective splicing [Talebizadeh et al., 2006] or excision of introns, which may be related to an overall decrease in expression of multiple splicing factors observed in our studies (Supplemental Tables VI–VIII). Clearly, more research is needed to identify and characterize these novel transcripts as well as to demonstrate their function within the context of ASD.
Subgroup-Specific Genes Suggest Dysregulation of Specific Pathways Associated With the Respective ASD Phenotypes
Subtyping of ASD individuals prior to gene expression analyses also revealed differentially expressed genes that were unique to each subgroup. Fifteen circadian rhythm regulatory or responsive genes were among the genes identified as differentially expressed in the most severe (L) subgroup but not in the mild or savant subgroups, suggesting a connection between dysregulation of circadian rhythm and the severity of this phenotype. In 2002, Wimpory et al. proposed a relationship between social timing, “clock” (circadian rhythm) genes, and autism [Wimpory, Nicholas, & Nash, 2002], and more recently demonstrated genetic association of PER1 and NPAS2, but not other circadian rhythm genes, with autistic disorder [Nicholas et al., 2007]. This very interesting hypothesis is based in part upon the prevalence of sleep disorders in ASD, which suggest deficits in the regulation of circadian rhythm [Johnson & Malow, 2008; Malow, 2004]. A recent report that Fragile X-related proteins regulate transcriptional activity of the clock genes provides additional experimental support for the involvement of circadian rhythm in ASD [Zhang et al., 2008]. Bourgeron has further proposed a connection between clock and synaptic genes (NLGN3, NLGN4, NRXN1, and SHANK3) in ASD [Bourgeron, 2007]. He also pointed out the importance of gene dosage in the balance of excitatory and inhibitory signaling at the synapse and suggested the possible importance of the circadian rhythm in controlling such signaling and hence the severity of ASD. The significance of gene dosage effects (which can be manifested by altered gene expression) as contributors to ASD are emphasized by recent studies, which show that copy number variants can be associated with both familial and spontaneous forms of ASD [Sebat et al., 2007; Szatmari, Paterson, Zwaigenbaum, Roberts, Brian, Liu et al., 2007b]. Our network analysis of the 15 circadian rhythm genes that are differentially expressed only in the severe ASD group shows the relationships between these genes and many neurological functions as well as disorders typically observed in ASD (Fig. 7). It should be mentioned that multiple genes (though not all 15) are differentially expressed in each individual, suggesting a multi-hit mechanism of dysregulation of the circadian rhythm in the most severe phenotype of ASD.
Among the genes confirmed by qRT-PCR are AANAT, BHLBH2 (DEC1), CRY1, NPAS2, PER3, and DPYD. A significant decrease is observed for AANAT, an enzyme which catalyzes the rate-limiting first step of the biochemical conversion of serotonin to melatonin, a key regulator hormone of the circadian cycle. A reduction in this enzyme would be consistent with the abnormally low levels of melatonin, which have been reported in a number of studies of autistic patients [Nir et al., 1995], which may be associated with the high incidence of sleep disorders reported in ASD [Johnson & Malow, 2008]. Related to the decrease in melatonin in ASD, Melke et al. have recently reported two polymorphisms in the promoter of the gene for acetylserotonin methyltransferase (ASMT), the last enzyme in the synthesis of melatonin, which are associated with decrease in ASMT transcripts and reduced ASMT activity [Melke et al., 2008]. Overexpression of BHLHB2/DEC1, which regulates the expression of the master circadian regulator genes CLOCK and BMAL1, has also been shown to delay the phase of several clock genes (e.g., DEC1, DEC2, and PER1), which contain E boxes in their regulatory regions [Nakashima et al., 2008; Sato et al., 2004]. CRY1 and PER3 are also transcriptional modulators of CLOCK/BMAL1, while NPAS2 is a CLOCK analog expressed primarily in brain tissues [Bertolucci et al., 2008; Van Der Schalie, Conte, Marz, & Green, 2007]. While not directly involved in the control of circadian rhythm, DPYD is a major target of the clock genes and a particularly important gene with respect to neurological functions. In fact, DPYD deficiency leads most frequently to epilepsy, mental and motor retardation (all symptoms frequently associated with autism), and other developmental disorders, with 18% of DPYD-deficient individuals receiving a diagnosis of autism [Van Kuilenburg et al., 1999]. Metabolically, DPYD catalyzes the breakdown of uracil to β-alanine, which activates both GABAA and glycine receptors with the same efficacy as their respective natural ligands [Horikoshi, Asanuma, Yanagisawa, Anzai, & Goto, 1988; Wu, Gibbs, & Farb, 1993]. Thus, a deficiency in DPYD or the resultant subnormal levels of β-alanine can be predicted to lead to decreased inhibitory signaling activity at the synapse. Interestingly, anti-convulsant medications, which are often prescribed as a therapeutic regimen for epilepsy associated with DPYD deficiency are also efficacious in improving behaviors in a subgroup of ASD individuals, even without apparent seizures. It is therefore suggested that evaluation of DPYD status, β-alanine levels, or circadian rhythm function in ASD individuals might be helpful in identifying those patients that would most benefit from this type of medication. Overall, the net effect of the observed changes in gene expression is the dysregulation of circadian rhythm in this most severely affected subgroup of ASD individuals. Since the circadian rhythm affects not only neurological but also endocrine, gastrointestinal, and cardiovascular functions, dysregulation of these genes can also have a systemic impact on affected individuals, causing many of the symptoms that are often associated with ASD. Thus, it may be proposed that interventions aimed at normalizing the circadian “clock” may ameliorate some of the symptoms associated with ASD for this subgroup.
Relevance of Gene Expression Profiles in Peripheral Tissues and Cell Lines to ASD
While the first large-scale gene expression analysis of autism was performed on postmortem brain tissues from autistic and nonautistic individuals [Purcell, Jeon, Zimmerman, Blue, & Pevsner, 2001], several subsequent gene expression studies, including our earlier study on monozygotic twins discordant for autism, have utilized LCL or primary blood lymphocytes from autistic probands and controls with the goal of identifying expressed biomarkers for diagnostic purposes [Baron, Liu, Hicks, & Gregg, 2006; Gregg et al., 2008; Hu et al., 2006; Nishimura et al, 2007a]. It is further postulated that these surrogate experimental models of autism may reveal clues to the pathobiology of this neurobiological disorder [Walker, Segal, & Aschner, 2006]. Indeed, gene expression profiles of different brain regions have been shown to exhibit the highest similarity to whole blood [Sullivan, Fan, & Perou, 2006], while a meta-analysis of studies performed in blood and postmortem brain demonstrated convergent gene expression changes [Middleton et al., 2005]. Although there is great variability in the specific differentially expressed genes identified by the different studies on peripheral cells from autistic individuals vs. controls, in part due to different microarray platforms and experimental design, common biological themes (including inflammation, immune system dysregulation, neurite outgrowth, and disruption of axon guidance) emerge from these studies and reflect neuropathological findings [Amaral, Schumann, & Nordahl, 2008; Palmen et al., 2004; Pardo et al., 2005; Zimmerman et al., 2005]. In addition to providing further support for the involvement of these processes in the pathophysiology of ASD, this study also identifies circadian rhythm dysregulation as a prominent aberration in the subtype of ASD characterized by severe language impairment, which is consistent with recent studies implicating the involvement of “clock” genes and circadian metabolism in autism [Bourgeron, 2007; Melke et al., 2008; Nicholas et al., 2007] and related disorders, such as Fragile X [Zhang et al., 2008]. Of particular interest for future studies are the novel genes of unknown function, the majority of which are in noncoding regions, that are significantly differentially expressed across all three subgroups of ASD identified here. Because of their apparent sensitivity to androgens based upon our pilot studies on DHT-treated LCL (Fig. 6) as well as gene expression data deposited into the GEO repository for data from large-scale microarray analyses [Holterhus et al., 2003; Zhao et al., 2005], these genes may underlie the prominent 4:1 male-to-female sex bias in susceptibility to ASD.
Summary
This study demonstrates the value of subdividing individuals with ASD on the basis of cluster analyses of ADIR scores that incorporate all three core domains of ASD as described in the accompanying manuscript. Stratifying the sample by cluster analyses revealed quantitative differences in gene expression that appear to correlate with severity of ASD phenotype as well as gene expression profiles for each subtype that associate a “biological phenotype” (i.e., gene expression profile) to the respective functional/behavioral phenotype. The biological phenotypes reveal differences in some of the biological functions affecting individuals with ASD, such as circadian rhythm dysregulation in the severe (L) phenotype, suggesting possible therapeutic interventions specific to this subgroup. On the other hand, overlapping genes among the phenotypes indicate dysregulation of genes controlling both neurological and metabolic functions that may lie at the core of ASD.
In summary, our studies show that:
The level of expression of some genes relates directly to the severity of the phenotype, and may serve as useful candidates for eQTL analyses.
Network analysis of genes that are shared between the severely language-impaired and mild ASD subgroups reveals a set of genes that are probably critical with respect to the neurological and metabolic abnormalities of ASD.
A set of differentially expressed genes, most of which lie in noncoding regions, are shared among all three ASD subgroups examined.
Differences between affected functions and pathways among the different phenotypic groups may be responsible for the differences in symptom severity observed in autism.
Finally, our results suggest that some of the neurological manifestations of ASD are at least in part the result of dysregulated signaling and metabolic pathways that are reflective of a systemic disorder which, once identified, may be treatable. The implications of these findings as well as those of others who have identified gene signatures of psychiatric disorders in lymphoblasts [Baron et al., 2006; Gregg et al., 2008; Iwamoto, Kakiuchi, Bundo, Ikeda, & Kato, 2004; Kakiuchi et al., 2003, 2004, 2008; Kato et al., 2005; Maes et al., 2007; Nishimura et al., 2007b; Tang, Lu, Aronow, Wagner, & Sharp, 2002; Tang, Nee, Lu, Ran, & Sharp, 2003] support the use of non-neuronal tissues, including patient-derived LCL and primary peripheral cells, to investigate the pathobiology of ASD.
Supplementary Material
Acknowledgments
We are grateful for the contributions of laboratory interns, Mandana Manzari for identifying genes associated with reported QTLs, Xinyi Zhou and Jonathan Chang for literature searches and preliminary qRT-PCR analyses. This work was supported by NIMH Grant # R21 MH073393 (V. W. H) and Autism Speaks, Grant #2381 (V. W. H). We also gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium* and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). We especially thank Dr.Vlad Kustanovich of AGRE for providing us with additional information about the samples, which were not easily retrievable in the database. *The AGRE Consortium: Dan Geschwind, M.D., Ph.D., UCLA, Los Angeles, CA; Maja Bucan, Ph.D., University of Pennsylvania, Philadelphia, PA; W.Ted Brown, M.D., Ph.D., F.A.C.M.G., N.Y.S. Institute for Basic Research in Developmental Disabilities, Long Island, NY; Rita M. Cantor, Ph.D., UCLA School of Medicine, Los Angeles, CA; John N. Constantino, M.D., Washington University School of Medicine, St. Louis, MO; T.Conrad Gilliam, Ph.D., University of Chicago, Chicago, IL; Martha Herbert, M.D., Ph.D., Harvard Medical School, Boston, MA; Clara Lajonchere, Ph.D, Cure Autism Now, Los Angeles, CA; David H. Ledbetter, Ph.D., Emory University, Atlanta, GA; Christa Lese-Martin, Ph.D., Emory University, Atlanta, GA; Janet Miller, J.D., Ph.D., Cure Autism Now, Los Angeles, CA; Stanley F. Nelson, M.D., UCLA School of Medicine, Los Angeles, CA; Gerard D. Schellenberg, Ph.D., University of Washington, Seattle, WA; Carol A. Samango-Sprouse, Ed.D., George Washington University, Washington, D.C.; Sarah Spence, M.D., Ph.D., UCLA, Los Angeles, CA; Matthew State, M.D., Ph.D., Yale University , New Haven, CT. Rudolph E. Tanzi, Ph.D., Massachusetts General Hospital, Boston, MA. Contributions to this manuscript: VWH conceived and designed the experiments, performed data analyses, and wrote the manuscript. TS and KSK performed the microarray experiments and TS also assisted in the microarray data analysis. AN and MES performed qRT-PCR analyses, SK conducted the analyses of novel transcripts in DHT-treated cells, TL was responsible for printing and quality control of the DNA microarray slides, and YL performed the hypergeometric test on the genes in QTL, and NHL performed the χ2 analyses to determine the significance of gene distribution in QTLs on specific chromosomes as well as participated in the discussion and editing of the manuscript.
Additional Supporting Information may be found in the online version of this article.
Grant sponsor: National Institute of Mental Health, NIH; Grant number: R21 MH073393; Grant sponsor: Autism Speaks; Grant number: 2381.
Footnotes
Present address of Kyung Soon Kim is Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN 55905
Present address of Mara E. Steinberg is Department of Hearing and Speech Sciences, University of Maryland; College Park, MD 20742
References
- Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics. 2008;9:153–161. doi: 10.1007/s10048-008-0133-5. [DOI] [PubMed] [Google Scholar]
- Alarcon M, Yonan AL, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and ordered-subsets analysis of autism endophenotypes support language QTLs. Molecular Psychiatry. 2005;10:747–757. doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends in Neurosciences. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genetics. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arnold JT, Le H, McFann KK, Blackman MR. Comparative effects of DHEA vs. testosterone, dihydrotestosterone, and estradiol on proliferation and gene expression in human LNCaP prostate cancer cells. American Journal of Physiology—Endocrinology and Metabolism. 2005;288:E573–E584. doi: 10.1152/ajpendo.00454.2004. [DOI] [PubMed] [Google Scholar]
- Auranen M, Nieminen T, Majuri S, Vanhala R, Peltonen L, Järvelä I. Analysis of autism susceptibility gene loci on chromosomes 1p, 4p, 6q, 7q, 13q, 15q, 16p, 17q, 19q and 22q in finnish multiplex families. Molecular Psychiatry. 2000;5:320–322. doi: 10.1038/sj.mp.4000708. [DOI] [PubMed] [Google Scholar]
- Bailey A, Hervas A, Matthews N, Palferman S, Wallace S, et al. A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Human Molecular Genetics. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- Balciuniene J, Feng N, Iyadurai K, Hirsch B, Charnas L, et al. Recurrent 10q22–q23 deletions: A genomic disorder on 10q associated with cognitive and behavioral abnormalities. American Journal of Human Genetics. 2007;80:938–947. doi: 10.1086/513607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron CA, Liu SY, Hicks C, Gregg JP. Utilization of lymphoblastoid cell lines as a system for the molecular modeling of autism. Journal of Autism and Developmental Disorders. 2006;36:973–982. doi: 10.1007/s10803-006-0134-x. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen S, Knickmeyer RC, Belmonte MK. Sex differences in the brain: Implications for explaining autism. Science. 2005;310:819–823. doi: 10.1126/science.1115455. [DOI] [PubMed] [Google Scholar]
- Bertolucci C, Cavallari N, Colognesi I, Aguzzi J, Chen Z, et al. Evidence for an overlapping role of CLOCK and NPAS2 transcription factors in liver circadian oscillators. Molecular and Cellular Biology. 2008;28:3070–3075. doi: 10.1128/MCB.01931-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron T. The possible interplay of synaptic and clock genes in autism spectrum disorders. Cold Spring Harbor Symposia on Quantitative Biology. 2007;72:645–654. doi: 10.1101/sqb.2007.72.020. [DOI] [PubMed] [Google Scholar]
- Bradford Y, Haines J, Hutcheson H, Gardiner M, Braun T, et al. Incorporating language phenotypes strengthens evidence of linkage to autism. American Journal of Medical Genetics—Neuropsychiatric Genetics. 2001;105:539–547. [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Smith CJ, Kilifarski M, Reichert J, et al. Evidence for a susceptibility gene for autism on chromosome 2 and for genetic heterogeneity. American Journal of Human Genetics. 2001;68:1514–1520. doi: 10.1086/320588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GK, Kono N, Geschwind DH, Cantor RM. Quantitative trait locus analysis of nonverbal communication in autism spectrum disorder. Mol Psychiatry. 2006;11:214–220. doi: 10.1038/sj.mp.4001753. [DOI] [PubMed] [Google Scholar]
- Chen Y, Blackwell TW, Chen J, Gao J, Lee AW, States DJ. Integration of genome and chromatin structure with gene expression profiles to predict c-MYC recognition site binding and function. PLoS Computational Biology. 2007;3(4):e63. doi: 10.1371/journal.pcbi.0030063. doi:10.1371/journal.pcbi.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Spielman RS. The genetics of variation in gene expression. Nature Genetics. 2002;32:522–525. doi: 10.1038/ng1036. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen K-Y, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nature Genetics. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Chu T-M, Weir B, Wolfinger R. A systematic statistical linear modeling approach to oligonucleotide array experiments. Mathematical Biosciences. 2002;176:35–51. doi: 10.1016/s0025-5564(01)00107-9. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, et al. The putative oncogene GASC1 demethylates triand dimethylated lysine 9 on histone H3. Nature. 2006;442:307. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, et al. Validation of a brief quantitative measure of autistic traits: Comparison of the social responsiveness scale with the autism diagnostic interview-revised. Journal of Autism and Developmental Disorders. 2003;33:427–433. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- DeFelice ML, Ruchelli ED, Markowitz JE, Strogatz M, Reddy KP, et al. Intestinal cytokines in children with pervasive developmental disorders. American Journal of Gastroenterolology. 2003;98:1777–1782. doi: 10.1111/j.1572-0241.2003.07593.x. [DOI] [PubMed] [Google Scholar]
- Duvall JA, Lu A, Cantor RM, Todd RD, Constantino JN, Geschwind DH. A quantitative trait locus analysis of social responsiveness in multiplex autism families. American Journal of Psychiatry. 2007;164:656–662. doi: 10.1176/ajp.2007.164.4.656. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Geier DA, Geier MR. A clinical and laboratory evaluation of methionine cycle-transsulfuration and androgen pathway markers in children with autistic disorders. Hormone Research. 2006;66:182–188. doi: 10.1159/000094467. [DOI] [PubMed] [Google Scholar]
- Gregg JP, Lit L, Baron CA, Hertz-Picciotto I, Walker W, et al. Gene expression changes in children with autism. Genomics. 2008;91:22–29. doi: 10.1016/j.ygeno.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Herbert MR, Russo JP, Yang S, Roohi J, Blaxill M, et al. Autism and environmental genomics. Neurotoxicology. 2006;27:671–684. doi: 10.1016/j.neuro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Hollander E, Nowinski CV. Core symptoms, related disorders, and course of autism. Marcel Dekker, Inc.; New York, NY: 2003. p. 15. [Google Scholar]
- Hollander E, Novotny S, Allen A, Aronowitz B, Cartwright C, DeCaria C. The relationship between repetitive behaviors and growth hormone response to sumatriptan challenge in adult autistic disorder. Neuropsychopharmacology. 2000;22:163–167. doi: 10.1016/S0893-133X(99)00121-9. [DOI] [PubMed] [Google Scholar]
- Holterhus PM, Hiort O, Demeter J, Brown PO, Brooks JD. Differential gene-expression patterns in genital fibroblasts of normal males and 46,XY females with androgen insensitivity syndrome: Evidence for early programming involving the androgen receptor. Genome Biology. 2003;4(6):R37. doi: 10.1186/gb-2003-4-6-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi T, Asanuma A, Yanagisawa K, Anzai K, Goto S. Taurine and b-alanine act on both GABA and glycine receptors in xenopus oocyte injected with mouse brain messenger RNA. Molecular Brain Research. 1988;4:97–105. doi: 10.1016/0169-328x(88)90002-2. [DOI] [PubMed] [Google Scholar]
- Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Nguyen A, Kim KS, Steinberg ME, Sarachana T, Scully MA, Soldin SJ, Luu T, Lee NH. Gene expression profiling of lymphoblasts from autistic and nonaffected sib pairs: Altered pathways in neuronal development and steroid biosynthesis. PLoS ONE. 2009 doi: 10.1371/journal.pone.0005775. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu VW, Steinberg ME. Novel clustering of items from the Autism Diagnostic Interview-Revised to define phenotypes within autism spectrum disorders. Autism Research. 2009;2:67–77. doi: 10.1002/aur.72. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingudomnukul E, Baron-Cohen S, Wheelwright S, Knickmeyer R. Elevated rates of testosterone-related disorders in women with autism spectrum conditions. Hormones and Behavior. 2007;51:597–604. doi: 10.1016/j.yhbeh.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Molecular Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nature Genetics. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KP, Malow BA. Sleep in children with autism spectrum disorders. Current Neurology and Neuroscience Reports. 2008;8:155–161. doi: 10.1007/s11910-008-0025-y. [DOI] [PubMed] [Google Scholar]
- Johnson NL, Kotz S, Kemp AW. Univariate discrete distributions. Wiley; New York, NY: 1992. [Google Scholar]
- Jyonouchi H, Sun S, Le H. Proinflammatory and regulatory cytokine production associated with innate and adaptive immune responses in children with autism spectrum disorders and developmental regression. Journal of Neuroimmunology. 2001;120:170–179. doi: 10.1016/s0165-5728(01)00421-0. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, et al. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nature Genetics. 2003;35:171–175. doi: 10.1038/ng1235. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C, Ishiwata M, Umekage T, Tochigi M, Kohda K, et al. Association of the XBP1 −116C/G polymorphism with schizophrenia in the japanese population. Psychiatry and Clinical Neurosciences. 2004;58:438–440. doi: 10.1111/j.1440-1819.2004.01280.x. [DOI] [PubMed] [Google Scholar]
- Kakiuchi C, Ishiwata M, Nanko S, Ozaki N, Iwata N, et al. Up-regulation of ADM and SEPX1 in the lymphoblastoid cells of patients in monozygotic twins discordant for schizophrenia. American Journal of Medical Genetics, Part B: Neuropsychiatric Genetics. 2008;147:557–564. doi: 10.1002/ajmg.b.30643. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RH, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Iwayama Y, Kakiuchi C, Iwamoto K, Yamada K, et al. Gene expression and association analyses of LIM (PDLIM5) in bipolar disorder and schizophrenia. Molecular Psychiatry. 2005;10:1045–1055. doi: 10.1038/sj.mp.4001719. [DOI] [PubMed] [Google Scholar]
- Knickmeyer R, Baron-Cohen S, Raggatt P, Taylor K. Foetal testosterone, social relationships, and restricted interests in children. Journal of Child Psychology and Psychiatry. 2005;46:198–210. doi: 10.1111/j.1469-7610.2004.00349.x. [DOI] [PubMed] [Google Scholar]
- Knickmeyer RC, Baron-Cohen S. Fetal testosterone and sex differences in typical social development and in autism. J Child Neurol. 2006;21:825–845. doi: 10.1177/08830738060210101601. [DOI] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nature Reviews. Neuroscience. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kumar RA, Karamohamed S, Sudi J, Conrad DF, Brune C, et al. Recurrent 16p11.2 microdeletions in autism. Human Molecular Genetics. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- Levy SE, Souders MC, Ittenbach RF, Giarelli E, Mulberg AE, Pinto-Martin JA. Relationship of dietary intake to gastrointestinal symptoms in children with autistic spectrum disorders. Biological Psychiatry. 2007;61:492–497. doi: 10.1016/j.biopsych.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Li H, Yamagata T, Mori M, Yasuhara A, Momoi MY. Mutation analysis of methyl-CpG binding protein family genes in autistic patients. Brain and Development. 2005;27:321–325. doi: 10.1016/j.braindev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lord C, Leventhal BL, Cook EH., Jr. Quantifying the phenotype in autism spectrum disorders. American Journal of Medical Genetics. 2001;105:36–38. [PubMed] [Google Scholar]
- Maes OC, Xu S, Yu B, Chertkow HM, Wang E, Schipper HM. Transcriptional profiling of alzheimer blood mononuclear cells by microarray. Neurobiology of Aging. 2007;28:1795–1809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Malow BA. Sleep disorders, epilepsy, and autism. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10:122–125. doi: 10.1002/mrdd.20023. [DOI] [PubMed] [Google Scholar]
- Matzke A, Sargsyan V, Holtmann B, Aramuni G, Asan E, et al. Haploinsufficiency of c-met in CD44−/− mice identifies a collaboration of CD44 and c-met in vivo. Molecular and Cellular Biology. 2007;27:8797–8806. doi: 10.1128/MCB.01355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley JL, Li C, Jiang L, Olson LM, Crockett G, et al. Genome-wide and ordered-subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Medical Genetics. 2005;6:1. doi: 10.1186/1471-2350-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melke J, Goubran Botros H, Chaste P, Betancur C, Nygren G, et al. Abnormal melatonin synthesis in autism spectrum disorders. Molecular Psychiatry. 2008;13:90–98. doi: 10.1038/sj.mp.4002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Pato CN, Gentile KL, McGann L, Brown AM, et al. Gene expression analysis of peripheral blood leukocytes from discordant sib-pairs with schizophrenia and bipolar disorder reveals points of convergence between genetic and functional genomic approaches. American Journal of Medical Genetics—Neuropsychiatric Genetics. 2005;136B:12–25. doi: 10.1002/ajmg.b.30171. [DOI] [PubMed] [Google Scholar]
- Morley M, Birnbaum HG, Morley MA, Sisitsky T, Greenberg PE, Wolfe F. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;12:430. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, et al. DEC1 modulates the circadian phase of clock gene expression. Molecular and Cellular Biology. 2008;28:4080–4092. doi: 10.1128/MCB.02168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas B, Rudrasingham V, Nash S, Kirov G, Owen MJ, Wimpory DC. Association of Per1 and Npas2 with autistic disorder: Support for the clock genes/social timing hypothesis. Molecular Psychiatry. 2007;12:581–592. doi: 10.1038/sj.mp.4001953. [DOI] [PubMed] [Google Scholar]
- Nir I, Meir D, Zilber N, Knobler H, Hadjez J, Lerner Y. Brief report: Circadian melatonin, thyroid-stimulating hormone, prolactin, and cortisol levels in serum of young adults with autism. Journal of Autism and Developmental Disorders. 1995;25:641–654. doi: 10.1007/BF02178193. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Human Molecular Genetics. 2007a;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, et al. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Human Molecular Genetics. 2007b;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Nurmi EL, Dowd M, Tadevosyan-Leyfer O, Haines JL, Folstein SE, Sutcliffe JS. Exploratory subsetting of autism families based on savant skills improves evidence of genetic linkage to 15q11–q13. Journal of the American Academy of the Child and Adolescent Psychiatry. 2003;42:856–863. doi: 10.1097/01.CHI.0000046868.56865.0F. [DOI] [PubMed] [Google Scholar]
- Palmen SJ, van Engeland H, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004;127:2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- Pardo CA, Vargas DL, Zimmerman AW. Immunity, neuroglia and neuroinflammation in autism. International Review of Psychiatry. 2005;17:485–495. doi: 10.1080/02646830500381930. [DOI] [PubMed] [Google Scholar]
- Persico AM, Bourgeron T. Searching for ways out of the autism maze: Genetic, epigenetic and environmental clues. Trends in Neurosciences. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille C, Rastam M, Sponheim E, et al. Genome-wide scan for autism susceptibility genes. Paris autism research international sibpair study. Human Molecular Genetics. 1999;8:505–512. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- Pollard PJ, Loenarz C, Mole DR, McDonough MA, Gleadle JM, et al. Regulation of jumonji-domain-containing histone demthylases by hypoxia-inducible factor (HIF)-1alpha. The Biochemical Journal. 2008;416:387. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Polleux F, Lauder JM. Toward a developmental neurobiology of autism. Mental Retardation and Developmental Disabilities Research Reviews. 2004;10:303–317. doi: 10.1002/mrdd.20044. [DOI] [PubMed] [Google Scholar]
- Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/wnl.57.9.1618. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nature Genetics. 2002;32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Rogaev EI. Small RNAs in human brain development and disorders. Biochemistry (Mosc) 2005;70:1404–1407. doi: 10.1007/s10541-005-0276-z. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Santangelo S, Ashley-Koch A, Pericak-Vance M, Silverman J, Smith CJ, Buxbaum J. Combined analysis of data on chromosome 7q from three autism genome scans. American Journal of Medical Genetics—Neuropsychiatric Genetics. 2000;96:480. [Google Scholar]
- Sato F, Kawamoto T, Fujimoto K, Noshiro M, Honda KK, et al. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation: Interaction with BMAL1. European Journal of Biochemistry. 2004;271:4409–4419. doi: 10.1111/j.1432-1033.2004.04379.x. [DOI] [PubMed] [Google Scholar]
- Schellenberg GD, Dawson G, Sung YJ, Estes A, Munson J, et al. Evidence for multiple loci from a genome scan of autism kindreds. Molecular Psychiatry. 2006;11:1049–1060. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- Schultz SF, MacFabe DF, Ossenkopp KP, Scratch S, Whelan J, et al. Intracerebroventricular injection of propionic acid, an enteric bacterial metabolic end-product, impairs social behavior in the rat: Implications for an animal model of autism. Neuropharmacology. 2008;54:901–911. doi: 10.1016/j.neuropharm.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JM, Smith CJ, Schmeidler JM, Buxbaum JD, Lawlor BA, Fitzgerald M. Symptom domains in autism and related conditions: Evidence for familiality. American Journal of Medical Genetics—Neuropsychiatric Genetics. 2001;105:593. doi: 10.1002/ajmg.10048. [DOI] [PubMed] [Google Scholar]
- Stevens MC, Fein DA, Dunn M, Allen D, Waterhouse LH, et al. Subgroups of children with autism by cluster analysis: A longitudinal examination. Journal of the American Academy of Child and Adolescent Psychiatry. 2000;39:346–352. doi: 10.1097/00004583-200003000-00017. [DOI] [PubMed] [Google Scholar]
- Stone JL, Merriman B, Cantor RM, Geschwind DH, Nelson SF. High density SNP association study of a major autism linkage region on chromosome 17. Human Molecular Genetics. 2007;16:704–715. doi: 10.1093/hmg/ddm015. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. American Journal of Medical Genetics—Neuropsychiatric Genetics. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genetics. 2007a;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genetics. 2007b;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadevosyan-Leyfer O, Dowd M, Mankoski R, Winklosky B, Putnam S, et al. A principal components analysis of the autism diagnostic interview-revised. Journal of the American Academy of Child and Adolescent Psychiatry. 2003;42:864–872. doi: 10.1097/01.CHI.0000046870.56865.90. [DOI] [PubMed] [Google Scholar]
- Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. Journal of Medical Genetics. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebizadeh Z, Butler MG, Theodoro MF. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Research. 2008;1:307. doi: 10.1002/aur.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Lu A, Aronow BJ, Wagner KR, Sharp FR. Genomic responses of the brain to ischemic stroke, intra-cerebral haemorrhage, kainate seizures, hypoglycemia, and hypoxia. European Journal of Neuroscience. 2002;15:1937–1952. doi: 10.1046/j.1460-9568.2002.02030.x. [DOI] [PubMed] [Google Scholar]
- Tang Y, Nee AC, Lu A, Ran R, Sharp FR. Blood genomic expression profile for neuronal injury. Journal of Cerebral Blood Flow and Metabolism. 2003;23:310–319. doi: 10.1097/01.WCB.0000048518.34839.DE. [DOI] [PubMed] [Google Scholar]
- Tanguay PE, Robertson J, Derrick A. A dimensional classification of autism spectrum disorder by social communication domains. Journal of the American Academy of Child and Adolescent Psychiatry. 1998;37:271–277. doi: 10.1097/00004583-199803000-00011. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbannuci A, Waltering KK, Suikki HE, Helenius MA, Visakorpi T. Androgen regulation of the androgen receptor coregulators. BMC Cancer. 2008;8:219. doi: 10.1186/1471-2407-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Veyver IB, Zoghbi HY. Mutations in the gene encoding methyl-CpG-binding protein 2 cause rett syndrome. Brain and Development. 2001;23:S147–S151. doi: 10.1016/s0387-7604(01)00376-x. [DOI] [PubMed] [Google Scholar]
- Van Der Schalie EA, Conte FE, Marz KE, Green CB. Structure/function analysis of xenopus cryptochromes 1 and 2 reveals differential nuclear localization mechanisms and functional domains important for interaction with and repression of CLOCK-BMAL1. Molecular and Cellular Biology. 2007;27:2120–2129. doi: 10.1128/MCB.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kuilenburg ABP, Vreken P, Abeling NGGM, Bakker HD, Meinsma R, et al. Genotype and phenotype in patients with dihydropyrimidine dehydrogenase deficiency. Human Genetics. 1999;104:1–9. doi: 10.1007/pl00008711. [DOI] [PubMed] [Google Scholar]
- Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Annals of Neurology. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- Vinci G, Chantot-Bastaraud S, El Houate B, Lortat-Jacob S, Brauner R, McElreavey K. Association of deletion 9p,46,XY gonadal dysgenesis and autistic spectrum disorder. Molecular Human Reproduction. 2007;13:685. doi: 10.1093/molehr/gam045. [DOI] [PubMed] [Google Scholar]
- Walker SJ, Segal J, Aschner M. Cultured lymphocytes from autistic children and non-autistic siblings up-regulate heat shock protein RNA in response to thimerosal challenge. Neurotoxicology. 2006;27:685–692. doi: 10.1016/j.neuro.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Wassink TH, Vieland VJ, Sheffield VC, Bartlett CW, Goedken R, et al. Posterior probability of linkage analysis of autism dataset identifies linkage to chromosome 16. Psychiatric Genetics. 2008;18:85–91. doi: 10.1097/YPG.0b013e3282f9b48e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, et al. Association between microdeletion and microduplication at 16p11.2 and autism. New England Journal of Medicine. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- Wimpory D, Nicholas B, Nash S. Social timing, clock genes and autism: A new hypothesis. Journal of Intellectual Disability Research. 2002;46:352–358. doi: 10.1046/j.1365-2788.2002.00423.x. [DOI] [PubMed] [Google Scholar]
- Winslow JT, Insel TR. The social deficits of the oxytocin knockout mouse. Neuropeptides. 2002;36:221–229. doi: 10.1054/npep.2002.0909. [DOI] [PubMed] [Google Scholar]
- Wu F-S, Gibbs TT, Farb DH. Dual activation of GABA(A) and glycine receptors by β-alanine: Inverse modulation by progesterone and 5α-pregnan-3α-ol-20-one. European Journal of Pharmacology—Molecular Pharmacology Section. 1993;246:239–246. doi: 10.1016/0922-4106(93)90037-a. [DOI] [PubMed] [Google Scholar]
- Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. American Journal of Human Genetics. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kim Y, Wang P, Lapointe J, Tibshirani R, et al. Genome-wide characterization of gene expression variations and DNA copy number changes in prostate cancer cell lines. Prostate. 2005;63:187–197. doi: 10.1002/pros.20158. [DOI] [PubMed] [Google Scholar]
- Zimmerman AW, Jyonouchi H, Comi AM, Connors SL, Milstien S, et al. Cerebrospinal fluid and serum markers of inflammation in autism. Pediatric Neurology. 2005;33:195–201. doi: 10.1016/j.pediatrneurol.2005.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.