Abstract
The advent of genomic and proteomic technologies in this post-genomic era has urged the researchers to develop novel research strategies against cancer by targeting the human genes that would greatly facilitate to identify more promising treatment and to develop accurate early diagnosis for cancer. To harness the power of cancer genetic information towards better treatment we have developed a cancer gene database called CanGeneBase (CGB). It is a comprehensive data collection of cancer-related genes with the intention of helping the researchers to stay on a single platform to gain exclusive information on the genes of their interest. According to the Cancer Gene Data Curation Project, about 4,700 genes have been identified as being related to cancer. The present CanGeneBase covers about 12 different types of cancer which includes 190 unique gene entries. Each entry encompasses about 33 useful parameters to provide detailed information about specific gene. CanGeneBase is made in such a way that it can be easily accessed by either gene symbol or by the type of cancer.
Availability
The database is freely available at http://122.165.25.137/bioinfo/cancerdb/
Keywords: Cancer, database, drug list, oncogenes, tumour suppressor genes, cancer types, target, molecular descriptors
Background
Due to the development of advanced technologies and highly validated research methodologies, many new genes have been identified whose roles in cancer have also been elucidated. The mysteries of cancer mechanism are being uncovered and the secrets disclosed have to be maintained as a comprehensive collection in order to gain access to unrevealed facts. The genetic abnormalities associated with cancer seem to be multifaceted in nature. The correlation of facts about different types of cancer and the genes involved in each type gives deeper insights into the disease mechanism which leads to new ventures aimed at more promising treatments. According to American Cancer Society, 7.6 million people died of cancer in the world during the year 2007 [1]. There are about 4,700 cancer causing genes have been identified [2]. The feasibility of better cancer treatments for all cancer types based on a single reference database has not been developed yet, though there are databases available to provide information on the cancer genes. Although there are databases specifically meant for specific cancer types, no comprehensive database is available as an all inclusive source of information on genes specific for different types of cancer. To address these challenges, we have created a cancer gene database (CanGeneBase) which caters researchers with the comprehensive valuable information aimed at fostering discoveries in the field of oncology and to relate all the gene information pertaining to various types of cancers. There are about 200 types of cancers and its information are available across the published literature [3–5], the current release of CanGeneBase focuses on more specific cancer types that are considered to be the most fatal form. Additionally, we would be updating the gene information regularly to include new findings so that the researchers could remain well equipped with the information using CanGeneBase.
Methodology
Database construction
The data in CanGeneBase were collected from various reputed journals and databases such as Entrez [6], Swiss-Prot [7] and DrugBank [8]. Molinspiration, a cheminformatics software suite requires either drug structure or SMILES notation to calculate molecular descriptors for a given drug molecule. The SMILES notations were obtained from DrugBank [8] and PubChem [9] and submitted to the Molinspiration property calculation web interface. For drug entries without the SMILES notation, the two dimensional structures were drawn in ACD ChemSketch [10] and the SMILES notations were generated by using ChemSketch toolbox. The database was constructed using MySQL, a powerful open source SQL based database package. The database interface was made using PHP integrated with HTML through Apache web server to produce a user-friendly interface.
Features of CanGeneBase
The CanGeneBase database provides user-friendly accessibility with the help of search options using the gene symbol as an entity or using specific cancer type. This search facilitates to retrieve information about the genes related to queried cancer type. The gene search provides a list of about 33 unique parameters which includes the Gene Symbol, Gene Name, Aliases, Gene type, Description, DNA Size, Chromosomal location, Transcripts, Isoforms, Organism, Taxonomy, Journal, Title, Author, Post Translational Modifications, Disorders, Drugs, HIV-1 Protein Interactions, Protein Motifs / Domains / Families, Functions, Signalling Pathways and Tissue Specificity. Besides that, each entry provides links to Entrez Gene, HomoloGene, PubMed, SNP, Probe, OMIM [6], Ensembl [11], PharmGKB [12], Genecards [13], GenBank [14] and Swiss-Prot [7] to gain more knowledge about the cancer gene. It also provides details of the drugs, targeting the genes with its three dimensional structure including molecular descriptors with its external link to PubChem [9] and KEGG [15]. These capabilities aid in retrieving information about the particular gene including list of drugs available and other properties of the gene on a single platform.
User-friendly interface
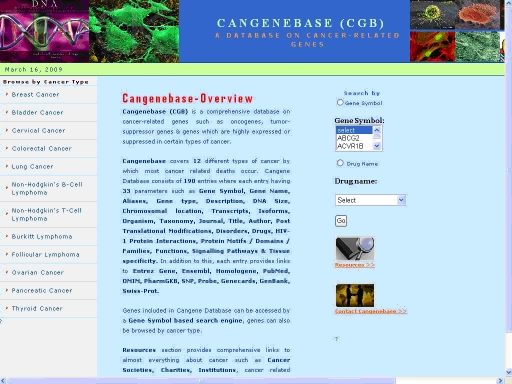
The homepage of the CanGeneBase (Figure 1) displays options for selection of gene by gene symbol, selection of drug by drug names and selection of any particular type of cancer from the list of various types. The user can access the required information by following any of the three selections. The selection of a gene directed to gene information page, where 33 important parameters are listed with links to various repositories. The selection of cancer type lists all of genes associated with the particular cancer and each being linked to the gene information page. The drug selection connects to a page where the molecular descriptors of the drug chosen can be accessed with a link to download the three dimensional structure of the drug.
Figure 1.
Screenshot of CanGeneBase homepage with options for selection of gene/drug/cancer type.
Caveats and future development
Not all genes published in literature are included in the current release. The database would be validated further and updated periodically so as to provide complete and more authentic information.
Conclusion
The current release of CanGeneBase seeks to provide valuable gene and drug information on the major types of cancer. The service intended to be rendered by the current release would be very useful resource for the scientific community in developing more targeted treatments for this multifaceted cancer disease. The unique advantage of CanGeneBase is that it provides flexible search criteria that enable the user to query the database with either the gene or the drug identities. Besides the molecular descriptors of drugs for 12 different types of cancer, more than 6270 unique features of gene have been made available in this current version.
Footnotes
Citation:Kumar et al, Bioinformation 3(10): 422-424 (2009)
References
- 1. http://www.medicinenet.com/script/main/art.asp?articlekey=85892.
- 2. http://ncicb.nci.nih.gov/projects/cgdcp.
- 3. http://www.tumor-gene.org/
- 4. http://cgap.nci.nih.gov/
- 5. http://www.cancer.gov/cancertopics/alphalist/
- 6. http://www.ncbi.nlm.nih.gov/
- 7.Bairoch, Apweiler R. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David S, et al. Nucleic acid Res. 2006;34:668–672. [Google Scholar]
- 9. http://www.ncbi.nlm.nih.gov/sites/entrez?db=pccompound.
- 10.Hunter, Allen D. J Chem Educ. 1997;74:905. [Google Scholar]
- 11. http://www.ensembl.org.
- 12.Klein TE, et al. Pharmacogenomics. 2001;1(3):167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- 13.Rebhan M, et al. Trends in Genetics. 1997;13:163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 14.Benson DA, et al. Nucleic Acids Res. 2008:25–30. [Google Scholar]
- 15. http://www.genome.jp/kegg/