Abstract
Collagens are members of one of the most important families of structural proteins in higher organisms. There are 28 types of collagens encoded by 43 genes in humans that fall into several different functional protein classes. Mutations in the major fibrillar collagen genes lead to osteogenesis imperfecta (COL1A1 and COL1A2 encoding the chains of Type I collagen), chondrodysplasias (COL2A1 encoding the chains of Type II collagen), and vascular Ehlers-Danlos syndrome (COL3A1 encoding the chains of Type III collagen). Over the last two decades, mutations in these collagen genes have been catalogued, in the hopes to understand the molecular etiology of diseases caused by these mutations, characterize the genotype-phenotype relationships, and develop robust models predicting the molecular and clinical outcomes. To better achieve these goals, it is necessary to understand the natural patterns of variation in collagen genes in human populations. We screened exons, flanking intronic regions, and conserved non-coding regions for variations in COL1A1, COL1A2, COL2A1 and COL3A1 in 48 individuals from each of four ethnically diverse populations. We identified 459 single nucleotide polymorphisms (SNPs), more than half of which were novel and not found in public databases. Of the 52 SNPs found in coding regions, 15 caused amino acid substitutions while 37 did not. Although the four collagens have similar gene and protein structures, they have different molecular evolutionary characteristics. For example, COL1A1 appears to have been under substantially stronger negative selection than the rest. Phylogenetic analysis also suggests that the four genes have very different evolutionary histories among the different ethnic groups. Our observations suggest that the study of collagen mutations and their relationships with disease phenotypes should be performed in the context of the genetic background of the subjects.
Introduction
Collagen is the most abundant protein in mammals, constituting a quarter of the total protein weight [1; 2]. Collagens are grouped into families based on their structural and functional features. Types I, II, and III collagens, the major components of fibrillar collagens, account for 70% of the total body collagens. Type I collagen is the major protein in bone, skin, tendon, ligament, sclera and cornea tissues, blood vessels, and hollow organs. Type II collagen is found in articular cartilage. Type III collagen is often associated with type I collagen and is a major protein in skin, vessels, intestine, and the uterus.
Fibrillar collagens are the end product of synthesis, assembly, secretion and processing of proα chains that assemble into trimeric proteins (procollagens) that are transported through the cells, and in the extracellular matrix are converted by proteolysis to collagens which assemble into fibrillar arrays. Type I procollagen consists of two proα1(I) chains (encoded by COL1A1) and one proα2(I) chain (encoded by COL1A2); type II procollagen consists of three identical proα1(II) chains (encoded by COL2A1); and type III procollagen consists of three identical proα1(III) chains (encoded by COL3A1). Mutations in these collagen genes lead to osteogenesis imperfecta (OI) (COL1A1 and COL1A2) [2; 3], chondrodysplasis (COL2A1), and vascular Ehlers-Danlos Syndrome (EDS) (COL3A1) [4].
Although mutations in these genes and the resulting clinical phenotypes are widely known, the relationships between genotypes and clinical phenotypes are not well defined. For example, the severity of the OI phenotype that results from mutations that substitute glycine residues in the triple helical domains of the proα chains of type I procollagen depends on the gene in which the mutation occurs, the location of the mutation, and the nature of the substituting amino acid [3]. However, beyond that general statement, it is difficult to identify precise rules for prediction of phenotype for a new, previously unseen, mutation. Understanding the physiochemical properties of native and altered collagens should provide the basis to define the molecular etiology of the clinical phenotypes. However, phenotype variation observed in patients carrying the same mutation suggests an influence of the genetic background on which the specific mutation occurs (the altered allele, the other allele, and the background interacting genes).
As a first step to assembling these data, we have resequenced all the coding regions and significant portions of the intronic, and 5′ and 3′ untranslated regions of COL1A1, COL1A2, COL2A1, and COL3A1 in 48 individuals from each of 4 ethnic groups (African American, European, Mexican, and Chinese) in the SOPHIE (Studies of Pharmacogenetics in Ethnically Diverse Populations) cohort. We identified 459 SNPs in the four collagen genes, 273 of which were not found in dbSNP. Examination of the molecular evolutionary characteristics of these naturally occurring variations revealed significant differences among populations. Correlating collagen mutations with background variations in the future should provide additional insights into understanding the molecular etiology of collagen disorders and towards predicting the molecular and clinical outcomes of mutations.
Materials and Methods
Subject population
DNA samples were collected from a cohort of 192 unrelated individuals (48 African Americans, 48 European Americans, 48 Mexican Americans, and 48 Chinese Americans; with equal representation from both genders) from the San Francisco Bay Area enrolled in the SOPHIE project (Studies of Pharmacogenetics in Ethnically Diverse Populations). These individuals had no known genetic or acquired disorders at the time of sampling and had all four grandparents of the same population origin.
Variant identification and verification
The structures of the four collagen genes share an intriguing feature: most of the exons are 54 bp long and the rest are either 2 times 54, 3 times 54, or a combination of 45 and 54 bp [1]. Due to their small sizes and the close proximity of many exons to each other, we designed amplicons with an average size of 1.2 kbp. Introns located between such nearby exons were completely sequenced. We also used the VISTA tool [5] to align each human collagen gene with its mouse counterpart and looked for non-coding regions with sequence conservation greater than 70%. Intron 1 (between the first and the second exons) in COL1A1, COL1A2, and COL2A1 is several kbp in size and contains several clusters of highly conserved sequences. These regions were also included in our resequencing effort.
We sequenced all coding regions, intron sequences for at least 100 bp on either side of each exon and the evolutionarily conserved domains in non-coding regions, covering a total of 80 kbp of the total 140 kbp genomic length of all four genes (COL1A1, COL1A2, COL2A1, and COL3A1). Typically, fragments of 1 to 1.5 kbp were amplified with primers designed to include the largest amount of coding sequence. Except for fragments of sizes smaller than 700 bp and were sequenced in one direction, the rest were sequenced in both directions. Primer sequences, PCR conditions and sequence coordinates of each fragment are available in the supplementary materials (Suppl. Table 1). Primers were designed using the program Primer3 [6], or were previously designed and tested. All primers were synthesized by Integrated DNA Technologies (Coralville, IA). Each PCR reaction included 4ng of genomic DNA from a pool of 2 individuals (of the same ethnicity and gender) and 1μM each of the forward and reverse primers. PCR protocol was based on the PlatinumTaq kit from Invitrogen (Carlsbad, CA) with slight modifications. The PCR cycling conditions were as follows: 2-minute incubation at 95°C to denature the genomic DNA and activate the polymerase, followed by 35 cycles of amplification: denaturation for 10 s at 92°C, primer annealing for 20 s at appropriate temperature, and extension for 1 min at 72°C. The samples were held for 10 min at 72° C for final extension. PCR products were purified by the clean-up reagent ExoSAP-IT (USB Corp., Cleveland, Ohio) to remove unincorporated dNTPs and PCR primers. The purified products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA). The sequencing products were filter-purified using genCLEAN dye terminator clean-up plates (Genetix USA Inc., Boston, MA), and read in the ABI 3730×l DNA analyzer (Applied Biosystems, Foster City, CA). Sequence traces were aligned with the reference sequence retrieved from the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway) [7], and scored in Sequencher v4.7 (Gene Codes Corp., Ann Arbor, MI).
All singleton SNPs were resequenced. Amplicons containing one or more SNP with a minor allele frequency (MAF) greater than 5% were resequenced in individuals to generate data for analysis. All variants identified in this study have been submitted to the public databases PharmGKB (http://www.pharmgkb.org) and dbSNP (www.ncbi.nlm.nih.gov/projects/SNP/).
Population genetics parameters
Under the assumption of an infinite-sites neutral-allele model, the population genetic parameter (θ̂) and the average heterozygosity (π̂) estimators of nucleotide diversity were calculated [8] for each gene and separately for coding and non-coding regions in each population. We also used Tajima's D statistics to measure departure from the expected patterns of neutral variation [9]. To test for the presence of positive selection, we employed the E-test, which is especially sensitive to high-frequency variants [10].
Phylogenetic analysis
Genetic distances are used to measure the global genetic difference between two populations. Reynolds distance assumes that the differences between any two populations have arisen primarily due to genetic drift [11]. For each of the four collagen COL1A1, COL1A2, COL2A1 and COL3A1 regions we used Reynolds distance to compute the genetic distance from a set of gene frequencies in the different ethnic groups.
Trees are the most common method of phylogenetic analysis. The tree topology and branch lengths represent historical events such as splitting and separations. We used the Fitch and Margoliash method, which assumes that the amount of evolution between two groups as depicted by a branch length is directly proportional to the genetic distance observed between them [12]. We used the gendist program from the PHYLIP package version 3.6 (http://evolution.genetics.washington.edu/phylip.html) to generate the distance matrix for the four ethnic groups using the genotype data, and followed by the fitch program to generate the trees.
Results
SNP discovery
We identified 458 bi-allelic SNPs and 1 tri-allelic site (in COL2A1) in almost 80 kbp of genomic sequence (Table 1). Compared with build 126 of dbSNP, 273 of the 459 SNPs are novel. Sixty of the novel SNPs (22%) are common, with > 5% MAF in at least one population. There are 183 SNPs reported in dbSNP but not found in our sample set. Of those, 160 are single submissions that have not been validated or have no associated allele frequency information, suggesting that they are either very rare or due to errors [13]. The remaining 23 SNPs were missed due to technical reasons: SNPs in close proximity to an insertion/deletion (indel) were masked by the overlapping peak patterns, and SNPs too close to the beginning or the end of a sequencing trace were embedded in poor sequence.
Table 1. Summary of SNPs identified in the four collagen genes.
| Total number of nucleotides sequenced (bp) | 79,858 | ||||
| Coding | Non-coding | ||||
| COL1A1 | 4,395 | 10,095 | |||
| COL1A2 | 4,101 | 19.243 | |||
| COL2A1 | 4,067 | 16,824 | |||
| COL3A1 | 4,185 | 16,948 | |||
| No. of polymorphic sites detected | |||||
| Total | Novel a | Novel with MAF ≥ 5% a, b | Listed in dbSNP but not seen in data set a, c | Only one submitter and/or no freq. information a, d | |
| COL1A1 | 106 | 75 | 17 | 38 | 36 |
| COL1A2 | 98 | 49 | 11 | 44 | 40 |
| COL2A1 | 141 | 83 | 21 | 59 | 54 |
| COL3A1 | 114 | 66 | 11 | 42 | 30 |
Based on dbSNP build 126.
In at least one population.
Mutations (amino acid changes that involve glycine or proline substitutions) listed in dbSNP are excluded for comparison as we do not expect to find any in our healthy cohort.
SNPs listed in dbSNP as a single submission without any confirmation.
Ethnic differences in the molecular evolution of the collagen genes
Of the 459 SNPs identified, 208 were population-specific (Suppl. Table 2), with 110 (53%) found only in the African American population, 40 (19%) found only in Chinese Americans, 35 (17%) only in Mexican American subjects, and 23 (11%) only in European Americans. Most of the population-specific SNPs we identified were novel (187 out of 208) and their minor allele frequencies were low. Overall, 16 / 110 (15%) African American-specific variants had MAF ≥ 0.05; 5 / 40 (12.5%) Chinese American-specific variants had MAF ≥ 0.05; and only one European American-specific variant (out of 23) had MAF ≥ 0.05. None of the Mexican American-specific variants had MAF ≥ 0.05.
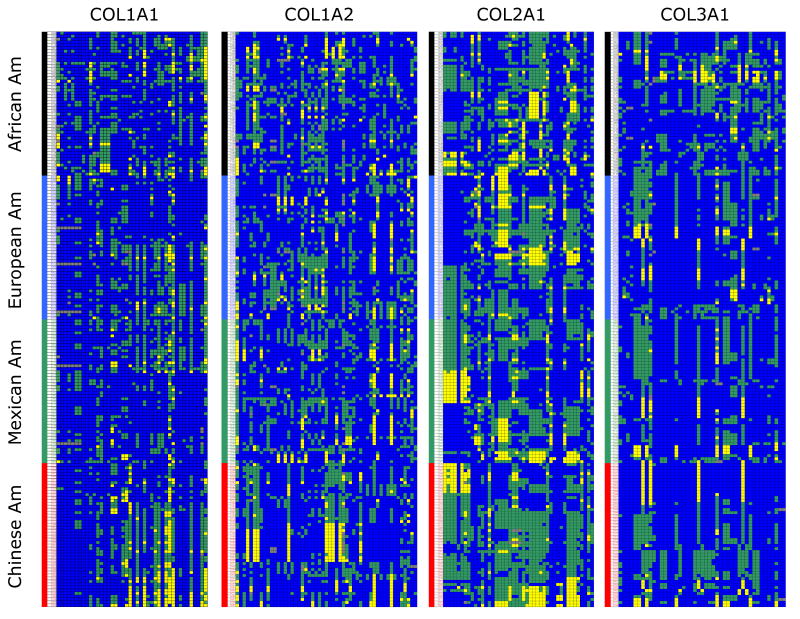
Figure 1 is a visual overview of all the common SNPs (MAF ≥ 0.05) found in the four genes. Samples within each ethnic group were clustered based on the genotype data using the VG2 program (http://pga.gs.washington.edu/VG2.html). (Clustered maps that include all variants found in each of the four collagen genes are included as Suppl. Figure 1a-d.) We noticed that (i) the European American and the Mexican American datasets shared very similar allelic structures in COL1A1, COL1A2, and COL3A1; (ii) the African American and the Chinese American datasets showed very different structures from the other two groups; and (iii) COL2A1 was markedly different from the other three collagen genes, as it uniquely exhibited high heterozygosity across all four ethnicities.
Figure 1. A visual genotype map of all common alleles (MAF ≥ 0.05) in the four collagen genes.
SNPs in each gene were arranged from the 5′ (left) to 3′ (right). Each row represents an individual in one of the four ethnic groups: African American (black), European American (blue), Mexican American (green), and Chinese American (red). Homozygous major alleles are represented in blue, heterozygous loci in green, and homozygous minor alleles in yellow. Within each ethnic group, individuals were further clustered by samples using the VG2 program available from the Nickerson group at SeattleSNP (http://pga.gs.washington.edu/VG2.html).
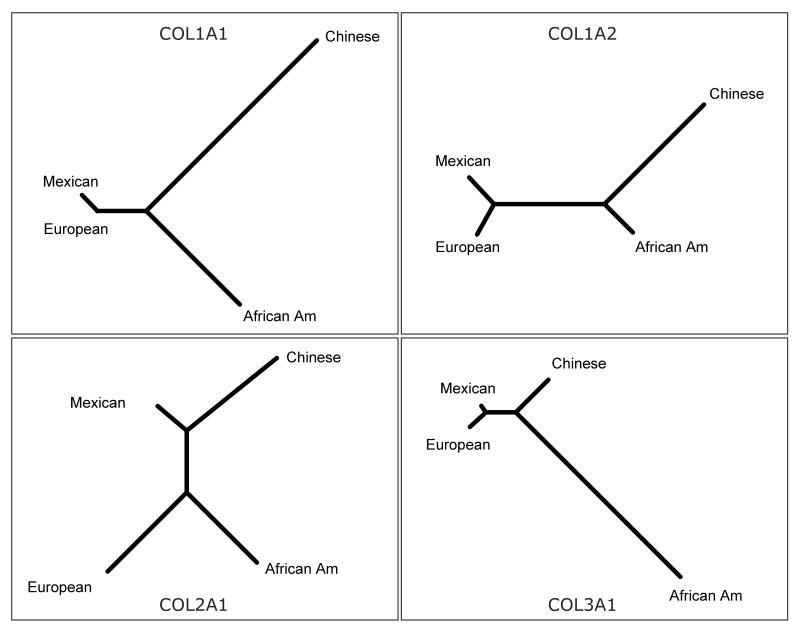
To verify our initial observations, we used the allele frequencies at all polymorphic sites with MAF ≥ 0.05 to calculate the Reynolds distances between the four ethnic groups for each gene, and used PHYLIP to generate the phylogenetic trees in Figure 2. Results shown in Figure 2 confirmed our initial observations. In addition, for the type I collagen genes COL1A1 and COL1A2 the Chinese Americans were most distinct from all other groups. This was not the case for COL3A1, in which the African Americans were evolutionarily separated from the rest. The phylogenetic tree for COL2A1 had a unique topology compared to the other three collagen genes, suggesting that COL2A1 had an evolutionary history which differentiated the four ethnic groups most clearly.
Figure 2. Phylogenetic trees for the four collagen genes.
Genotype data from Figure 1 were used to determine the four-way Reynolds distances and phylogenetic trees were generated using the PHYLIP software package.
Selection characteristics of the collagen genes
To further understand the patterns of natural variation in the four collagen genes, we computed two descriptors for nucleotide diversity, the average heterozygosity (π̂), and the population genetic parameter (θ̂) [8]. These two distinct numerical descriptors, one measuring the allele frequency spectrum and the other the rate of polymorphism, were used to calculate Tajima's D, a statistic which detects deviations from the neutral mutation model [9]. The beta distribution of Tajima's D (between -1.782 ∼ 2.072 for n = 96 and at 95% confidence interval) assumes that polymorphism ascertainment is independent of allele frequency spectrum, i.e. no selection is in force. A highly positive Tajima's D value indicates an excess of common variations in a region, and suggests a possible heterozygote advantage, whereas a negative Tajima's D is indicative of an excess of rare variants, consistent with negative selection. These parameters were calculated for the entire sequenced region as well as for individual sequence classes (coding versus non-coding and synonymous versus non-synonymous changes) and the results were grouped either by ethnicity (Suppl. Table 2) or by gene (Suppl. Table 3). In summary, only the coding region in COL1A1 had significantly negative Tajima's D values, which ranged from -2.61 to -2.38 (highlighted bold in Suppl. Table 2), suggesting that the coding region of COL1A1 is under negative selective pressure.
The Tajima's D test was designed to test whether there are too few or too many rare variants than common ones, but it does not address the relative abundance of those that are of high- and low-frequency classes. Zeng et al. recently proposed a new statistical test, the E-test, to probe for positive selection by contrasting high- and low-frequency variants [10]. In the Chinese American dataset as well as in the gene COL2A1 we observed the presence of many high-frequency variants, suggesting that a positive selection might be at work. We employed the E-test to detect positive selection for all four collagen genes in each of the four ethnic groups (Suppl. Table 4). Basically, at p value = 0.001, none of the values from the E-test indicated a significant positive selection force is in place. However, the COL2A1 gene in Chinese Americans showed the highest E-test value. It was particularly obvious in some of the regions of the gene that we examined (data not shown).
The ratio of non-synonymous to synonymous changes (πNS/πS) provides a way to assess the degree of negative selection in the coding region of a gene, on the assumption that deleterious alleles are in low abundance and that synonymous changes reflect selection [14]. For COL1A1 and COL1A2 the ratios were significantly less than 1, suggesting that they are under negative selection (Suppl. Table 3). The ratios for COL2A1 and COL3A1 were close to 1, suggesting that they are under little or no negative selective pressure.
All the coding SNPs identified in this study were listed in Table 2. As a recurring theme in our observations, COL1A1 was devoid of common variants in the coding region, whereas relatively common non-synonymous SNPs (MAF ≥ 0.05) could be found in the other three collagen genes: A549P (COL1A2), S9T, G1336S (both in COL2A1), and T698A (COL3A1). Genome-wide codon usage in human has been previously assessed by determining the relative synonymous codon usage (RSCU [15]) values from more than 80,000 human coding sequences (http://www.kazusa.or.jp/codon). In our dataset, a majority of the synonymous changes with significant alterations in RSCU values – greater than 50% changes and in bold types in Table 2 – involved the glycine residue. Based on these global RSCU values, almost all of them except for Gly543 in COL2A1 and Gly748 in COL3A1 changed a more commonly used codon to the least used codon, GGU. Some of these variants had average MAF ≥ 0.05, such as Val626 and Gly1054 in COL1A2, and Gly696 in COL2A1.
Table 2. Coding SNPs in the four collagen genes.
| Coding SNPs a |
Syn. codon changes b |
RSCU changes c | AA d | MEX d | CHI d | EUR d | rs number (dbSNP) e |
|
|---|---|---|---|---|---|---|---|---|
| COL1A1 | R59R |
CG →
CG →
CG
|
11.5 → 4.6 | 0.05 | 1057297 | |||
| G154G |
GG →
GG →
GG
|
22.3 → 10.8 | 0.01 | – | ||||
| V607V |
GU →
GU →
GU
|
14.5 → 11.0 | 0.01 | – | ||||
| N705N |
AA →
AA →
AA
|
19.1 → 16.9 | 0.01 | – | ||||
| G725G |
GG →
GG →
GG
|
22.3 → 10.8 | 0.01 | – | ||||
| A1075T | – | 0.06 | 0.01 | 0.01 | 1800215 | |||
| V1081V |
GU →
GU →
GU
|
11.0 → 14.5 | 0.01 | 1800217 | ||||
| R1141Q | – | 0.01 | – | |||||
| D1153D |
GA →
GA →
GA
|
21.8 → 25.2 | 0.10 | 0.01 | 1800218 | |||
| V1177I | – | 0.01 | – | |||||
| K1371K |
AA →
AA →
AA
|
31.9 → 24.3 | 0.02 | – | ||||
| S1393S |
UC →
UC →
UC
|
17.7 → 15.1 | 0.01 | 0.01 | 1800219 | |||
| T1419T |
AC →
AC →
AC
|
18.9 → 13.1 | 0.01 | – | ||||
| COL1A2 | T29T |
AC →
AC →
AC
|
18.9 → 13.1 | 0.34 | 0.05 | 0.45 | 0.02 | 1801182 |
| D82D |
GA →
GA →
GA
|
21.8 → 25.2 | 0.60 | 0.13 | 0.49 | 0.19 | 1800222 | |
| P482P |
CC →
CC →
CC
|
16.9 → 19.8 | 0.41 | 0.40 | 0.27 | 0.37 | 412777 | |
| N528S | – | 0.01 | – | |||||
| A549P | – | 0.18 | 0.26 | 0.09 | 0.30 | 42524 | ||
| A564T | – | 0.01 | – | |||||
| V626V |
GU →
GU →
GU
|
28.2 → 11.0 | 0.01 | 0.07 | 0.46 | 0.02 | 1800238 | |
| F850F |
UU →
UU →
UU
|
20.4 → 17.5 | 0.01 | – | ||||
| G1054G |
GG →
GG →
GG
|
22.3 → 10.8 | 0.07 | 0.05 | 0.06 | 0.09 | 1800248 | |
| COL2A1 | S9T | – | 0.31 | 0.42 | 0.50 | 0.19 | 3803183 | |
| E73D | – | 0.02 | 0.03 | 0.03 | – | |||
| G99G |
GG →
GG →
GG
|
22.3 → 16.5 | 0.16 | 0.21 | 0.32 | 0.17 | 3737548 | |
| R446R |
CG →
CG →
CG
|
10.5 → 6.2 | 0.01 | – | ||||
| G543G |
GG →
GG →
GG
|
10.8 → 22.3 | 0.01 | – | ||||
| T569I | – | 0.04 | – | |||||
| D613D |
GA →
GA →
GA
|
25.2 → 21.8 | 0.01 | – | ||||
| G696G |
GG →
GG →
GG
|
22.3 → 10.8 | 0.03 | 0.01 | 0.13 | 0.04 | 2276454 | |
| N731N |
AA →
AA →
AA
|
16.9 → 19.1 | 0.44 | 0.27 | 0.50 | 0.30 | 1635553 | |
| G759G |
GG →
GG →
GG
|
16.4 → 22.3 | 0.01 | 0.02 | 1793940 | |||
| G822G |
GG →
GG →
GG
|
22.3 → 16.4 | 0.07 | 0.03 | – | |||
| P931P |
CC →
CC →
CC
|
6.9 → 16.9 | 0.09 | 0.01 | 1793947 | |||
| A982T | – | 0.01 | – | |||||
| V1262I | – | 0.01 | 0.02 | 0.02 | 0.07 | 12721427 | ||
| G1287G |
GG →
GG →
GG
|
22.3 → 10.8 | 0.16 | 0.02 | 0.01 | 17122498 | ||
| P1299P |
CC →
CC →
CC
|
19.8 → 17.5 | 0.01 | 0.01 | 12721379 | |||
| G1336S | – | 0.04 | 0.38 | 0.45 | 0.11 | 2070739 | ||
| G1356G |
GG →
GG →
GG
|
22.3 → 10.8 | 0.01 | – | ||||
| P1414P |
CC →
CC →
CC
|
6.9 → 16.9 | 0.02 | – | ||||
| COL3A1 | G177G |
GG →
GG →
GG
|
22.3 → 16.5 | 0.01 | – | |||
| A419A |
GC →
GC →
GC
|
27.9 → 18.5 | 0.07 | 0.01 | – | |||
| P553P |
CC →
CC →
CC
|
17.5 → 16.9 | 0.02 | – | ||||
| L643L |
 UG →
UG →  UG
UG
|
12.9 → 39.8 | 0.01 | – | ||||
| A679T | – | 0.01 | 0.01 | – | ||||
| T698A | – | 0.09 | 0.31 | 0.23 | 0.26 | 1800255 | ||
| G748G |
GG →
GG →
GG
|
10.8 → 22.3 | 0.30 | 0.17 | 0.05 | 0.24 | 1801184 | |
| N986N |
AA →
AA →
AA
|
19.1 → 16.9 | 0.01 | – | ||||
| V1205I | – | 0.01 | 0.05 | 2271683 | ||||
| P1218P |
CC →
CC →
CC
|
6.9 → 17.5 | 0.03 | – | ||||
| Q1353H | – | 0.02 | 1516446 | |||||
Nonsynonymous SNPs are in bold, novel SNPs are in italics.
Only codons for synonymous amino acid changes were listed.
RSCU (Relative Synonymous Codon Usage) values for the human genome were obtained from the Codon Usage
Database (http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=Homo+sapiens+[gbpri]). Changes in RSCU of more than 50% (either increase or decrease) are highlighted in bold.
Allele frequency for each of the four populations: AA (African American); MEX (Mexican); CHI (Chinese); and EUR (European).
Reference SNP cluster identifier (http://www.ncbi.nlm.nih.gov/projects/SNP/).
Insertion-deletion polymorphisms
We observed 26 non-coding indels (or about 5% of the total polymorphisms identified) in the non-coding sequences only.
Discussion
The collagens encoded by the four genes we studied are some of the most important structural proteins in humans and other higher organisms. Our data showed that the four collagen genes we resequenced have different molecular evolutionary characteristics. In addition, phylogenetic analysis suggests that the four genes have very different evolutionary histories in the four ethnic groups we examined. Careful consideration of these characteristics may lead to new insights into the genotype-phenotype relationship in patients with collagen gene mutations and the evolutionary history of these structural genes in different populations.
Rare variants, because they arose recently [16], are more likely to be population-specific and can serve as sensitive indicators of recent migration and gene flow among various populations [17]. In our study, the Mexican American population shared a substantial number of rare variants in the four collagen genes with the African-Americans and the European Americans (Suppl. Table 5). By comparison, very few rare variants were shared between the Chinese Americans and the other three populations. This pattern of relatedness is consistent with the history of the four ethnic groups in the United States, where the Mexican Americans and the African Americans are both admixed with European descent [18; 19]. We also observed (Figure 2) a relative closeness between the Mexican Americans and the European Americans in COL1A1, COL1A2, and COL3A1. The phylogenetic tree of COL3A1 reflects the current understanding of human migration patterns: our common ancestors evolved over time and migrated out of Africa, and eventually settled in geographically separate areas of the European, Asian and finally American continents [20]. This pattern was not seen in the other three collagen genes. Intriguingly, for COL1A1 and COL1A2, the Chinese Americans appeared as a much more distant group when compared to the African Americans. Thus, even though the four collagen genes are highly similar in terms of gene and protein structures, they appear to have evolved in separate ways. These findings point to a hypothetical scenario that the ethnic background could play an important role in affecting the variable expressivity of the same mutation among different patients, such as a preexisting difference in gene functions (level of gene expression for example) that are embedded in the ethnic background of the patients.
For members of a class of highly conserved proteins, COL2A1 and COL3A1 had unexpectedly high πNS/πS ratios (0.83 and 0.97, respectively). For example, the average πNS/πS for the ABC transporters and the SLC transporters are 0.15 and 0.28, respectively [18]. The much higher πNS/πS ratios in COL2A1 and COL3A1 can be accounted for by the few non-synonymous SNPs that exist at high frequencies in our sample set, e.g., G1336S in COL2A1 (average MAF = 0.24) and T698A in COL3A1 (average MAF = 0.22) (Table 2). Although it is known that common non-synonymous changes are unlikely to have any deleterious effect [14], the presence of common amino acid substitutions in a major structural protein in the body suggests that the protein is under less negative selective pressure than expected.
Although both COL1A1 and COL1A2 had low πNS/πS ratios (0.38 and 0.21, respectively), only the COL1A1 gene showed significantly negative Tajima's D values in the coding region (Suppl. Table 2). COL1A2, on the other hand, had positive Tajima's D values across all four ethnic groups. The apparent discrepancy can be accounted for by the many synonymous SNPs that exist at high frequencies in COL1A2 coding region (Table 2), thereby inevitably driving down the πNS/πS ratio. In addition, the Tajima's D is designed to detect whether there is an excess of rare variants in the region-of-interest, and since all but two coding SNPs in COL1A2 were common (MAF ≥ 0.05), this test is not particularly sensitive. We also noticed both in COL1A1 and COL1A2 each with an amino acid substitution presented at relatively high frequencies (A1075T in COL1A1; A549P in COL1A2). Both amino acid changes are located in regions that were previously mapped to be important for ligand bindings [21]. A549P in particular, is in the region at which many lethal OI mutations on the proα2(I) chain were found. The fact that an amino acid change that could introduce a significant alteration to the protein structure (alanine to proline) is present at high frequency within a healthy population should provide important insights to the study of the relationships between collagen mutations and their clinical manifestations.
Synonymous changes in coding regions, i.e. changes in codon usage, are usually overlooked, as the resultant amino acid chain remains the same, and the variant should not manifest any discernable effect. However, at least two recent independent studies reported that synonymous changes can affect either the mRNA stability in the human dopamine receptor D2 (DRD2) gene, or substrate specificity of the ABC transporter ABCB1 (encoded by the gene MDR1) [22; 23]. Codon usage in the collagen genes is particularly intriguing when we consider the genome-wide codon usage in humans. The G nucleotide is rarely used as a third base for the four codons that encode glycine (GGN) and the four that encode proline (CCN) – G occurs in only 11 of the 390 (2.8%) glycine codons in COL1A1 protein and is not found in any of the 278 proline codons. The CCG proline codon is the least used codon genome-wide (RSCU = 6.9; compared to RSCUCCU = 17.5). In contrast, unlike for proline, the codon GGG that codes for glycine is in fact more commonly used than GGU in the human genome (RSCUGGG = 16.4; RSCUGGU = 10.8). 45.1% of the glycine codons in COL1A1 and 50.9% in COL1A2 are GGU (40.9% in COL2A1; 43.6% in COL3A1); this marked preference for U as the third base in codons can also be seen in proline residues [24]. This phenomenon is most evident in codons for the proline occupying the Y-position in the Gly-X-Y triplets characteristic of the triple helical domain, in which the Y residue precedes glycine. The excess of U at the third position likely represents the accumulation of changes that occurred at historical CCC proline codon followed by a GGN glycine codon. If the CpG dinucleotide was altered by cytosine methylation and deamination, a T would be generated in place of the C. If the alteration of the C occurred on the non-coding strand equivalent, the effect would be to alter that glycine codon, a highly deleterious mutation. With their unique coding sequence, the collagen genes may therefore have a unique evolutionary history as seen in our study.
Large-scale screens have devoted very little attention to indels as natural variation until a recent study by Mills et al. [25], which reported a whole-genome map of indels identified by DNA resequencing in 36 individuals. That study reported 10 non-coding indels in the four collagen genes. In contrast, our study observed 26 non-coding indels (or about 5% of the total polymorphisms identified). Although we observed more indels, the number was still low when compared to previous studies on other genes. For example, Wang et al. reported 10 indels out of 71 SNPs (∼14%) from screening the multidrug resistance-associated protein MRP1 in 142 samples from four different populations, even though they observed an evolutionary constraint on the gene [26]. In another study, Leabman et al. reported 8 indels in the coding regions from a set of 24 membrane transporter genes [18]. Unlike membrane transporters and other non-structural protein genes, the four collagen genes encode for collagen propeptides that form fibrils, and therefore require proper structures and functions of all units. Variants that disrupt the integrity of the coding sequence, even at a heterozygous state, can act as dominant negative mutations. The complete absence of coding indels in our data set is consistent with the detrimental consequences of any frame-shift or truncation to the collagen protein structures.
Recently, several groups reported that a deficiency in prolyl 3-hydroxylase 1, responsible for a critical post-translational modification in the fibrillar collagen chains, results in a recessive form of OI [27; 28; 29]. These recent findings demonstrated that, despite the use of the collagen protein as the classic textbook example of a structural protein for more than two decades, much of the biology that determines its synthesis, chain assembly, and functions remain understood only in outlines. Our study begins with a systematic investigation into allelic variations in the collagen genes. The evolutionary characteristics in these genes may help to explain and understand phenotypic effects that are difficult to comprehend solely on the basis of mutations.
Supplementary Material
Acknowledgments
The work was funded by the National Institutes of Health Grant AR051582 (TEK, principal investigator). The pooled genotype data for COL1A1, COL1A2, COL2A1, and COL3A1 are available at the PharmGKB (www.pharmgkb.org; PS206641). The authors wish to thank Kathleen Giacomini (University of California – San Francisco) (NIH GM61374) for providing us with access to the SOPHIE collection samples, Dale Bodian (Stanford University), and Ludmila Pawlikowska (University of California – San Francisco) for helpful comments with this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chu ML, Prockop DJ. Collagen: Gene Structure. In: Royce PM, Steinmann B, editors. Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects. Wiley-Liss; New York, NY: 2002. pp. 223–248. [Google Scholar]
- 2.Kielty CM, Grant ME. The Collagen Family: Structure, Assembly, and Organization in the Extracellular Matrix. In: Royce PM, Steinmann B, editors. Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects. Wiley-Liss; New York, NY: 2002. pp. 159–221. [Google Scholar]
- 3.Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, Milgrom S, Hyland JC, Korkko J, Prockop DJ, De Paepe A, Coucke P, Symoens S, Glorieux FH, Roughley PJ, Lund AM, Kuurila-Svahn K, Hartikka H, Cohn DH, Krakow D, Mottes M, Schwarze U, Chen D, Yang K, Kuslich C, Troendle J, Dalgleish R, Byers PH. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat. 2007;28:209–21. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pepin M, Schwarze U, Superti-Furga A, Byers PH. Clinical and genetic features of Ehlers-Danlos syndrome type IV, the vascular type. N Engl J Med. 2000;342:673–80. doi: 10.1056/NEJM200003093421001. [DOI] [PubMed] [Google Scholar]
- 5.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–9. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 7.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press, Inc.; New York: 2000. [Google Scholar]
- 9.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–9. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–84. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 13.Platzer M, Hiller M, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Huse K. Sequencing errors or SNPs at splice-acceptor guanines in dbSNP? Nat Biotechnol. 2006;24:1068–70. doi: 10.1038/nbt0906-1068b. [DOI] [PubMed] [Google Scholar]
- 14.Fay JC, Wyckoff GJ, Wu CI. Positive and negative selection on the human genome. Genetics. 2001;158:1227–34. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–43. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watterson GA, Guess HA. Is the most frequent allele the oldest? Theor Popul Biol. 1977;11:141–160. doi: 10.1016/0040-5809(77)90023-5. [DOI] [PubMed] [Google Scholar]
- 17.Slatkin M. Rare alleles as indicators of gene flow. Evolution. 1985;39:53–65. doi: 10.1111/j.1558-5646.1985.tb04079.x. [DOI] [PubMed] [Google Scholar]
- 18.Leabman MK, Huang CC, DeYoung J, Carlson EJ, Taylor TR, de la Cruz M, Johns SJ, Stryke D, Kawamoto M, Urban TJ, Kroetz DL, Ferrin TE, Clark AG, Risch N, Herskowitz I, Giacomini KM. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci U S A. 2003;100:5896–901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, Duan J, Carr JL, Lee MS, Koshy B, Kumar AM, Zhang G, Newell WR, Windemuth A, Xu C, Kalbfleisch TS, Shaner SL, Arnold K, Schulz V, Drysdale CM, Nandabalan K, Judson RS, Ruano G, Vovis GF. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–93. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 20.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- 21.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. Mapping the Ligand-binding Sites and Disease-associated Mutations on the Most Abundant Protein in the Human, Type I Collagen. J Biol Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 22.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–16. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 23.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 24.Tromp G, Kuivaniemi H, Stacey A, Shikata H, Baldwin CT, Jaenisch R, Prockop DJ. Structure of a full-length cDNA clone for the prepro alpha 1(I) chain of human type I procollagen. Biochem J. 1988;253:919–22. doi: 10.1042/bj2530919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–90. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Sew PH, Ambrose H, Ryan S, Chong SS, Lee EJ, Lee CG. Nucleotide sequence analyses of the MRP1 gene in four populations suggest negative selection on its coding region. BMC Genomics. 2006;7:111. doi: 10.1186/1471-2164-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnes AM, Chang W, Morello R, Cabral WA, Weis M, Eyre DR, Leikin S, Makareeva E, Kuznetsova N, Uveges TE, Ashok A, Flor AW, Mulvihill JJ, Wilson PL, Sundaram UT, Lee B, Marini JC. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N Engl J Med. 2006;355:2757–64. doi: 10.1056/NEJMoa063804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morello R, Bertin TK, Chen Y, Hicks J, Tonachini L, Monticone M, Castagnola P, Rauch F, Glorieux FH, Vranka J, Bachinger HP, Pace JM, Schwarze U, Byers PH, Weis M, Fernandes RJ, Eyre DR, Yao Z, Boyce BF, Lee B. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127:291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 29.Cabral WA, Chang W, Barnes AM, Weis M, Scott MA, Leikin S, Makareeva E, Kuznetsova NV, Rosenbaum KN, Tifft CJ, Bulas DI, Kozma C, Smith PA, Eyre DR, Marini JC. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat Genet. 2007;39:359–65. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.