Abstract
Wolbachia endosymbionts are ubiquitously found in diverse insects including many medical and hygienic pests, causing a variety of reproductive phenotypes, such as cytoplasmic incompatibility, and thereby efficiently spreading in host insect populations. Recently, Wolbachia-mediated approaches to pest control and management have been proposed, but the application of these approaches has been hindered by the lack of genetic transformation techniques for symbiotic bacteria. Here, we report the genome and structure of active bacteriophages from a Wolbachia endosymbiont. From the Wolbachia strain wCauB infecting the moth Ephestia kuehniella two closely related WO prophages, WOcauB2 of 43,016 bp with 47 open reading frames (ORFs) and WOcauB3 of 45,078 bp with 46 ORFs, were characterized. In each of the prophage genomes, an integrase gene and an attachment site core sequence were identified, which are putatively involved in integration and excision of the mobile genetic elements. The 3′ region of the prophages encoded genes with sequence motifs related to bacterial virulence and protein-protein interactions, which might represent effector molecules that affect cellular processes and functions of their host bacterium and/or insect. Database searches and phylogenetic analyses revealed that the prophage genes have experienced dynamic evolutionary trajectories. Genes similar to the prophage genes were found across divergent bacterial phyla, highlighting the active and mobile nature of the genetic elements. We suggest that the active WO prophage genomes and their constituent sequence elements would provide a clue to development of a genetic transformation vector for Wolbachia endosymbionts.
Members of the genus Wolbachia are endosymbiotic bacteria belonging to the Alphaproteobacteria and infecting a wide range of arthropods, including over 60% of insect species, and some filarial nematodes. They are vertically transmitted through the maternal germ line of their host and are known to distort host reproduction by causing cytoplasmic incompatibility (CI), parthenogenesis, male killing, or feminization. The ability of Wolbachia to cause these reproductive phenotypes is thought to be responsible for their efficient and rapid spread into host populations (5, 21, 35, 51).
Recently, Wolbachia-mediated pest control approaches have been proposed. A number of insect pests that have important medical and hygienic consequences, such as tsetse flies and mosquitoes that vector devastating human pathogens including African sleeping disease trypanosomes, malaria plasmodia, dengue viruses, Japanese encephalitis viruses, and others, often also carry Wolbachia infections (8, 24, 25, 34). In theory, if maternally transmitted genetic elements coinherited with a CI-inducing Wolbachia, such as mitochondria, the Wolbachia itself, or other coinfecting endosymbionts, are transformed with a gene of interest (like a gene that confers resistance of the vector insect against the pathogen infection), the genetic trait is expected to be spread and fixed in the host insect population, driven by the symbiont-induced reproductive phenotype (1, 2, 10, 11, 13, 32, 43, 44). The paratransgenesis and Wolbachia-driven population replacement approaches are, although potentially promising in controlling such insect-borne diseases, still at a conceptual stage mainly because no technique has been available for Wolbachia transformation.
For genetic transformation of bacteria, mobile genetic elements such as plasmids, bacteriophages, and transposons have been used successfully. For example, pUC plasmids, λ phages, and transposons have been widely utilized for transforming Escherichia coli and other model bacterial species (38). While few plasmids and transposons have been reported from Wolbachia, a family of bacteriophages, called WO phages, has been detected from a diverse array of Wolbachia strains (3, 6, 7, 12, 17, 18, 31, 39, 49). For example, in the genomes of the Wolbachia strains wMel from the fruit fly Drosophila melanogaster and wPip from the mosquito Culex quinquefasciatus, three and five WO prophages are present, respectively (26, 52). Many of the prophages are pseudogenized and inactive while some are active and capable of producing phage particles (4, 7, 15, 17, 30, 40). Such active WO phage elements may provide tools for genetic transformation of Wolbachia endosymbionts.
λ phage and many other temperate bacteriophages alternate between lytic phase and lysogenic phase in their life cycles. In the lytic phase, phage particles are produced and released via host cell lysis for infection to new host cells. In the lysogenic phase, the phage genome is integrated into the host genome via a site-specific recombination process, and the integrated phage genome, called prophage, is maintained in the host genome and multiplies together with the host DNA replication (38). Upon infection and lysogenic integration of λ phage, both ends of the linear phage genomic DNA are connected by DNA ligase, and the resultant circular phage genome is inserted into the E. coli genome by site-specific recombination at a region containing a core sequence of an attachment (att) site (28). att sites on the phage genome and the bacterial genome are called attP (phage att site) and attB (bacterial att site), respectively. After integration, attP and attB are located on both ends of the prophage, called attL (left prophage att site) and attR (right prophage att site), respectively. The integration and excision processes are mediated by a site-specific recombinase, called λ integrase, encoded in the phage genome (see Fig. S1 in the supplemental material) (27, 50). Hence, the att site and the integrase are the pivotal functional elements that mediate site-specific integration and excision of λ phage. Considering the structural similarity between λ phage and WO phage (31), identification of the att site and integrase from WO phage is of interest in that these elements could be utilized for delivering foreign genes into the Wolbachia genome.
In order to identify a functional att site and integrase of WO phage, the complete genome sequences of active prophage elements producing phage particles should be determined. Here, the Wolbachia strain wCauB derived from the almond moth Cadra cautella was investigated because wCauB was reported to actively produce phage particles, and a partial genome sequence of its WO phage has been determined (15). In the original host insect, C. cautella, wCauB coexists with another Wolbachia strain wCauA, and both cause CI phenotypes and produce phage particles (15, 41). Not to be confounded by the coinfecting Wolbachia strains, we used a transfected line of the Mediterranean flour moth Ephestia kuehniella infected with wCauB only, which was generated by interspecific ooplasm transfer (42). It should be noted that a mass preparation procedure for WO phage particles by centrifugation has been established for the wCauB-infected E. kuehniella (15).
In this study, we determined the complete genome sequences of two active WO prophages, named WOcauB2 and WOcauB3, that are capable of producing phage particles and that are located on the genome of the Wolbachia strain wCauB. Furthermore, we identified core sequences of att sites and integrase genes of these WO phages that are putatively involved in integration of the genetic elements into the Wolbachia genome.
MATERIALS AND METHODS
Insect materials.
A transfected line of E. kuehniella harboring the Wolbachia strain wCauB and a Wolbachia-free line of E. kuehniella of the same genetic background as a control were maintained on an artificial diet at 25°C under a long-day regimen (16 h of light-8 h of darkness) as previously described (15).
Preparation of Wolbachia genomic DNA.
A concentrated fraction of wCauB cells was prepared by the method of Sun et al. (47) with modification. Adult insects of wCauB-infected E. kuehniella were homogenized by using a mortar and pestle in an extraction buffer (90 mM KCl, 55 mM CaCl2, 15 mM MgSO4, 30 mM NaCl, 250 mM sucrose), and the homogenate was filtered through a 95-μm-pore-size nylon mesh. The filtrate was centrifuged at a maximum g (gmax) of 200 for 10 min, the supernatant was centrifuged at 3,000 × gmax for 10 min, and the supernatant was further centrifuged at 5,000 × gmax for 10 min. The resultant pellet containing concentrated Wolbachia cells was subjected to DNA extraction by using DNAzol (Molecular Research Center, Inc.).
Preparation of phage genomic DNA.
A concentrated fraction of WO phage particles was prepared according to the method of Fujii et al. (15) with modification. At the final ultracentrifugation step in the method, 1 volume of the supernatant containing suspended phage particles was layered on 2 volumes of a solution containing 40% glycerol, 50 mM Tris-HCl (pH 7.4), and 10 mM MgSO4. The resultant pellet containing concentrated WO phage particles was subjected to DNA extraction as previously described (15).
Genomic library construction and screening.
A genomic library of wCauB was constructed by using a CopyControl Fosmid Library Production Kit (Epicentre). The fosmid clones were transfected into competent cells of E. coli, and the transformed cells were grown on nylon membranes (9 cm by 11 cm) placed on chloramphenicol-containing LB agar plates at a density ranging from 103 to 105 colonies per membrane. The transformant colonies on replica membranes were lysed with 0.5 N sodium hydroxide, neutralized with 1 M Tris-HCl (pH 7.6), and baked at 80°C overnight, whereby their DNA was fixed on the membranes. The fosmid library was screened for clones containing WO prophage genome regions by using colony hybridization and PCR. The phage particle DNA was amplified by using an Illustra Genomiphi DNA Amplification Kit (GE Healthcare) and was radiolabeled with [α-32P]dCTP by a random priming method. The membranes were hybridized with the labeled probe, whereby putative positive clones were identified by autoradiography. The putative positive colonies were picked from the original membranes and subjected to PCR targeting a minor capsid protein gene (orf7) of the prophage with the primers orf7Fw (5′-GAA ATG CTT GTT CAG CTA ATA GC-3′) and orf7Rv (5′-ATA AAT TCT CCT ATT TTT TCT GGC A-3′). The PCR-positive clones were regarded as true positives and subjected to shotgun sequencing.
Shotgun sequencing, assembly, and annotation.
Fosmid DNA was prepared from each of the PCR-positive clones and subjected to shotgun sequencing. The obtained sequences were assembled by the programs phred, phrap, and consed (14, 19). Open reading frames (ORFs) consisting of at least 50 codons and starting with either ATG, GTG, or TTG were considered putative genes. Homology searches were performed by using NCBI's BLASTn and BLASTp systems (http://www.ncbi.nlm.nih.gov/BLAST/).
Cloning and sequencing of the attP region.
The genomic region of the WOcauB2 phage particle containing attP was amplified by PCR with the primers WOcauB2 attP Fw (5′-TAC ACC TTT ACA CCT AGC AGC AGA GAA TG-3′) and WOcauB2 attP Rv (5′-GAT GTA ACA GAG CTA CTT GAG GGT GAA AT-3′) and LA-Taq DNA polymerase (TaKaRa) under a temperature profile of 94°C for 3 min, 35 cycles of 98°C for 10 s and 62°C for 30 s, and a final extension of 68°C for 10 min. The PCR product, around 12 kb in size, was subjected to shotgun sequencing as described above. The genomic region of the WOcauB3 phage particle containing attP was amplified by PCR with the primers WOcauB3 attP Fw (5′-CAT ATT TTG ATT ACG ACG AGA GAC CAA CGA-3′) and WOcauB3 attP Rv (5′-CTA TGC CAA GGG GTT TAT AAG CCA AGA AGA-3′) as described above. The PCR products, 7.3 kb, 3.7 kb, and 1.9 kb in size, were subjected to TA cloning and sequencing as previously described (23).
Alignments and phylogenetic analyses.
Prior to phylogenetic analyses, homology searches by BLASTp analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were performed on the nonredundant protein database at the website of the National Center for Biotechnology Information using deduced amino acid sequences as queries. The output E-values of the searches were used as criteria for data parsing. All phylogenetic analyses were conducted on the basis of deduced amino acid sequences. Multiple alignments were generated using the program packages MAFFT, version 5.8 (22), and/or Clustal X (48), followed by manual refinement using the program Seeview (16). In the case of analyses of ORF7 and another 15 WO phage gene products, aligned sites that included alignment gap(s) were omitted from the phylogenetic analysis. In the case of serine recombinase, catalytic domain sequences predicted by SMART programs (http://smart.embl-heidelberg.de/) were subjected to phylogenetic analysis, and all alignment gap(s) were removed as previously described (45). Phylogenetic trees were constructed by the neighbor-joining method (37) using Clustal X. Bootstrap tests were performed with 1,000 resamplings.
Southern blotting of WO prophages.
The Wolbachia genomic DNA was digested by restriction endonucleases, separated in 1% agarose gels, and transferred and fixed to Immobilon-Ny+ membranes (Millipore). The following probes were prepared by PCR from the phage particle DNA: a 203-bp ankyrin motif gene fragment (B2gp44) amplified with the primers ANKB2gp44Fw (5′-CGA GCA AGA CCT TTG CAT TC-3′) and ANKB2gp44Rv (5′-AAC GTT TGC TCC CGC TTC A A-3′), a 386-bp SpvB motif gene fragment (B3gp44) amplified with the primers SpvBFw (5′-AGA CGC ATA GCA GAA TGG TTA CTT GAG GAA-3′) and SpvBRv (5′-GGT AAT CGT TGG TCT CTC GTC GTA ATC A-3′), a 1,267-bp integrase gene fragment (B3gp1) amplified with the primers B3RecFw (5′-TCT TCT TGG CTT ATA AAC CCC TTG GCA TAG-3′) and B3RecRv (5′-TCA TCA AGA CAA CAG GCA CAG GA-3′), a 562-bp patatin-like gene fragment (B2gp43) amplified with the primers B2PatatinFw (5′-AGC AGA GAC AAA AAT AAA AAA GTG GCA AAG TA −3′) and B2PatatinRv (5′-GCA AAC ACT CCG CCA TCT ATC AAC ACT −3′), and a 295-bp minor capsid protein gene (orf7) fragment amplified with the primers B2orf7Fw (5′-GAA ATG CTG GTT GAG CTA ATA GC −3′) and B2orf7Rv (5′-ATA AAT TCT CCT ATC TTT TCT GGC A −3′). PCR was performed under the temperature profile of 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 2 min. The PCR products were electrophoresed in agarose gels, excised and purified by using a QIAquick Gel Extraction Kit (Qiagen), and radiolabeled with [α-32P]dCTP by a random priming method. The labeled probes were hybridized to the membranes as described previously (38).
PCR-RFLP.
A segment of the discrepant region between WOcauB1 and WOcauB2 (see Fig. S2 and S3 in the supplemental material) was amplified by PCR with the primers B2orf7Fw (5′-GAA ATG CTG GTT GAG CTA ATA GC-3′) and RFLPprimerRv (5′-GTG CAT CAA AAA ACT CAG GGC TTA CC-3′) under a temperature profile of 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 2 min. The PCR product was purified by a PCR purification kit (Qiagen) and cloned with TA cloning vector pT7Blue (Takara) as described previously (23). For restriction fragment length polymorphism (RFLP) genotyping, 59 clones were subjected to digestion with the restriction endonuclease DraI or SmaI and analyzed in 1% agarose gels.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study were deposited in the DDBJ (DNA Data Bank of Japan) under accession numbers AB478515 and AB478516.
RESULTS AND DISCUSSION
Identification of two Wolbachia genomic regions containing WO prophages.
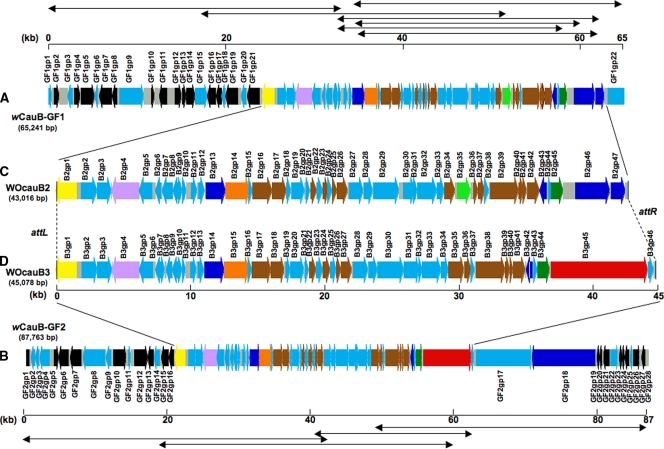
By colony hybridization and PCR screening of the genomic library of the Wolbachia strain wCauB, 11 positive fosmid clones were obtained. Shotgun sequencing and assembly of the fosmid clones resulted in two large contigs, wCauB-GF1 consisting of seven fosmid clones and wCauB-GF2 consisting of four fosmid clones, without ambiguity. wCauB-GF1 was 65,241 bp in size and encoded 69 ORFs (Fig. 1A; see also Tables S1 and S2 in the supplemental material) while wCauB-GF2 was 87,763 bp in size and contained 74 ORFs (Fig. 1B; see also Tables S1 and S3 in the supplemental material). These DNA sequences contained phage-related genes in the central region, whereas Wolbachia genomic genes were preferentially detected in the flanking regions (Fig. 1). Hence, we expected that wCauB-GF1 and wCauB-GF2 contain the full-length sequence of a WO prophage.
FIG. 1.
Structure of WO prophages identified from a genomic library screening of the Wolbachia strain wCauB. (A) The contig wCauB genome fragment 1 (wCauB-GF1) consisting of seven fosmid clones. (B) The contig wCauB genome fragment 2 (wCauB-GF2) consisting of four fosmid clones. (C) The complete WOcauB2 prophage region encoded in wCauB-GF1. (D) The complete WOcauB3 prophage region encoded in wCauB-GF2. Bidirectional arrows at top and bottom show the fosmid clones assembled. ORFs are shown by thick arrows while noncoding regions are gray boxes. Colors of ORFs indicate their functional categories as follows: black, genes with significant similarity to known bacterial genes including Wolbachia genes; brown, phage-related genes; yellow, DNA recombinase genes; light green, transposase genes; orange, terminase large subunit genes; lavender, regulatory protein gene repA; dark blue, ankyrin motif genes; dark green, patatin-like genes; red, SpvB motif genes; light blue, hypothetical genes.
Homology-based estimation of prophage regions.
Each of the putative ORFs of wCauB-GF1 and wCauB-GF2 was subjected to homology searches in the DNA databases. In the 5′ region of wCauB-GF1, a gene similar to WD0721 of the Wolbachia strain wMel, encoding a putative Mg2+ chelate-related protein (GF1gp21), was adjacent to a gene encoding a Ser-type recombinase (B2gp1) putatively involved in phage integration (Fig. 1A). In the 5′ region of wCauB-GF2, a gene similar to WD0820 of wMel, encoding a putative Sna5/YciO/YrdC/YwlC family protein (GF2gp16), was also adjacent to a Ser-type recombinase gene (B3gp1) (Fig. 1B). These arrangements suggested that the Ser-type recombinase gene is located at an end of the prophage regions on the wCauB genome. Actually, a number of phage-related genes encoding a terminase, capsid proteins, tail proteins, etc., and also many hypothetical genes of unknown function were located in the downstream regions of the Ser-type recombinase gene, which are likely to represent the WO prophage regions. Meanwhile, the 3′ ends of the prophage regions were difficult to infer on the basis of such sequence comparisons.
Cloning and sequencing of the attP site of the phage particle genome.
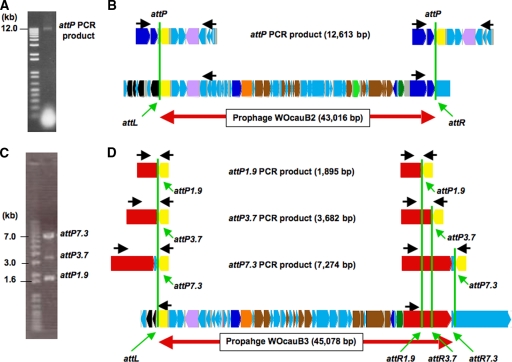
If a prophage region is active and producing phage particles, PCR with outward primers on both ends of the prophage is expected to yield a product containing the attP site derived from the self-ligated circular phage particle genome (see Fig. S1 in the supplemental material). By comparing the attP site sequence and the prophage sequence, both ends of the prophage regions can be determined. Hence, we prepared a fraction of concentrated WO phage particles from wCauB-infected E. kuehniella by centrifugation; phage particle DNA was extracted from the fraction, the DNA was subjected to PCR for amplifying the attP-containing region of the WO phage particle genomes, and the PCR products were cloned and sequenced.
Determination of WOcauB2 prophage region.
For the phage particle genome originating from the WO prophage encoded in the wCauB-GF1 region, a 12-kb DNA fragment was amplified with a pair of outward primers (Fig. 2A), indicating that the prophage is certainly active. On the DNA fragment, a Ser-type recombinase gene in the 5′ region (B2gp1) and an ankyrin motif gene in the 3′ region (B2gp47) were joined to each other, indicating that these genes comprise the 5′ and 3′ ends of the prophage region (Fig. 2B). The prophage, named WOcauB2, was 43,016 bp in size and encoded 47 ORFs (Fig. 1C and 2B).
FIG. 2.
Determination of WOcauB2 and WOcauB3 prophage regions. (A) A 12-kb PCR product containing the WOcauB2 attP region. (B) Alignment of the attP PCR product sequence and the wCauB-GF1 sequence. (C) Three PCR products, 7.3 kb, 3.7 kb, and 1.9 kb in size, containing the WOcauB3 attP region. (D) Alignment of the attP PCR product sequences and the wCauB-GF2 sequence. Black arrows show the locations of PCR primers. Green vertical lines indicate the putative recombination sites. Colors of ORFs are as described in the legend of Fig. 1.
Determination of the WOcauB3 prophage region.
For the phage particle genome derived from the WO prophage encoded in the wCauB-GF2 region, unexpectedly, three DNA fragments of different sizes (7.3 kb, 3.7 kb, and 1.9 kb) were amplified with a pair of outward primers (Fig. 2C). On all the DNA fragments, a Ser-type recombinase gene in the 5′ region (B3gp1) and genes of unknown function in the 3′ region (B3gp45/B3gp46) were joined to each other. The size differences between the three fragments were attributed to truncations in a gene containing a Salmonella virulence plasmid protein B motif (SpvB motif gene; B3gp45) (Fig. 2D). When the 7.3-kb fragment was regarded as the full-length fragment, the prophage, named WOcauB3, was 45,078 bp in size and encoded 46 ORFs (Fig. 1D and 2D).
Structural similarity between WOcauB2 and WOcauB3.
The prophages WOcauB2 and WOcauB3 were structurally similar to each other (see Fig. S2 in the supplemental material). The overall nucleotide sequence similarity was 80.0% (37,361/46,694, including alignment gaps) between them. When three variable regions were excluded, the nucleotide sequence similarity rose to 92.7% (33,617/36,281, including alignment gaps). Not only the nucleotide sequences but also arrangements of the ORFs were conserved between them. These patterns strongly suggested that WOcauB2 and WOcauB3 comprise a family of closely related bacteriophages.
Structural discrepancy between WOcauB2 and WOcauB3 prophages.
Meanwhile, the homology plot between the prophages WOcauB2 and WOcauB3 revealed three regions that exhibited little sequence similarity to each other (see Fig. S2 in the supplemental material). Region 1 was small (682 bp for WOcauB2 and 1,082 bp for WOcauB3), consisting of several hypothetical protein genes of unknown function. Region 2 was also small (1,392 bp), due to a transposase gene (B2gp35) insertion in WOcauB2. Region 3 was relatively large (4,879 bp for WOcauB2 and 7,076 bp for WOcauB3). In WOcauB2, the region consisted of two ORFs with ankyrin motifs. In WOcauB3, the region contained an ORF with an SpvB motif. These results indicated that WOcauB2 and WOcauB3 are certainly similar but distinct phage elements.
Relationship of WOcauB2 and WOcauB3 to the previously described WOcauB1.
In a previous study by Fujii et al. (15), a partial sequence of WO prophage (20,484 bp) associated with wCauB was determined, which was named WOcauB1. WOcauB2 showed very high sequence similarity (98.8%; 20,251/20,501 bp including alignment gaps) to WOcauB1 except for a small discrepant region (see Fig. S3 in the supplemental material), whereas WOcauB3 was less similar (82.7%; 17,330/20,952 bp including alignment gaps) to WOcauB1. When the discrepant region was excluded, the sequence similarity between WOcauB1 and WOcauB2 rose to 99.3% (20,105/20,250 bp including alignment gaps). In the discrepant region, the similarity value was 58.2% (146/251 bp including alignment gaps) (see Fig. S4 in the supplemental material). These results indicated that WOcauB2 is very similar but slightly different from WOcauB1. Since our wCauB-infected E. kuehniella line was derived from the line studied by Fujii et al. (15), the sequence discrepancies were unexpected. The following possibilities are conceivable to account for the sequence discrepancies: (i) the insect was initially polymorphic in having both WOcauB1 and WOcauB2 elements, while the insect had lost WOcauB1 during maintenance; (ii) the insect originally possessed WOcauB1, and mutations occurred during maintenance to form WOcauB2; or (iii) the sequence discrepancies are due to sequencing or assembling errors in the previous study. PCR-RFLP assays confirmed that our insect line contains only WOcauB2-type sequence but no WOcauB1-type sequence (see Table S4 in the supplemental material). Hence, we concluded that, at least currently, the wCauB-infected E. kuehniella line harbors WOcauB2 and WOcauB3 but not WOcauB1.
Relationship of WOcauB2 and WOcauB3 to WO phages of other Wolbachia strains from diverse insects.
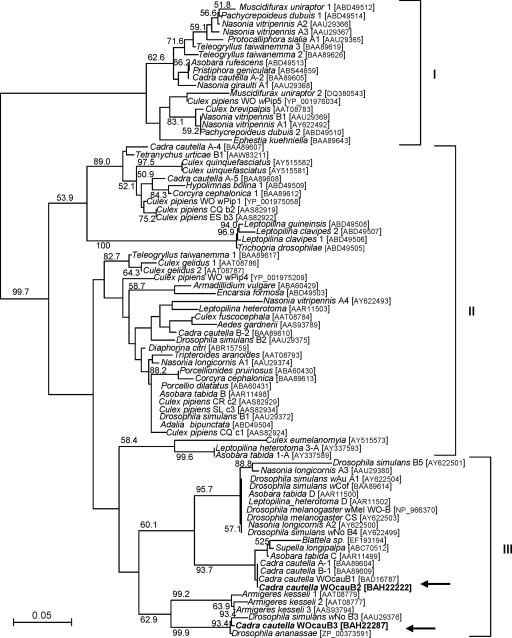
Molecular phylogenetic analysis of the amino acid sequences of the orf7 gene product, ORF7, a putative capsid protein, encoded by B2gp17 in WOcauB2 and by B3gp18 in WOcauB3 was performed together with 80 ORF7 sequences retrieved from the databases. Bordenstein and Wernegreen (3) showed that ORF7 sequences of diverse WO phages form three major clusters, which were also identified in our analysis. The sequences of WOcauB2 and WOcauB3 were placed in group III of the phylogeny and belonged to distinct clades (Fig. 3). These results indicated that WOcauB2 and WOcauB3 are certainly related to each other but are distinct bacteriophages.
FIG. 3.
Molecular phylogenetic analysis of WOcauB2 and WOcauB3 on the basis of ORF7 sequences. In addition to our sequences, 80 ORF7 sequences from 39 insect species were retrieved from the databases and subjected to analysis. A neighbor-joining tree inferred from 85 aligned amino acid sites is shown. Bootstrap values higher than 50% are shown at the nodes. The scientific name of the host insect, acronym for the sequence/Wolbachia/WO phage strain, and sequence accession number are indicated. I, II, and III represent the ORF7 clusters identified by Bordenstein and Wernegreen (3). Arrows indicate the placements of the WOcauB2 and WOcauB3 sequences.
Phylogenetic diversity of genes in WOcauB2 and WOcauB3.
Molecular phylogenetic analyses were also performed for the other 15 genes of WOcauB2 and WOcauB3 that were commonly found in WO phage sequences from the Wolbachia strains wMel, wPip, and wKue (26, 31, 52) available in the DNA databases (see Fig. S5 in the supplemental material). In 12 of the 15 trees, WOcauB2 and WOcauB3 formed a cluster. In 10 trees, the clades were supported by bootstrap values of over 90%. In the remaining three trees, WOcauB2 and WOcauB3 did not form a cluster. These results suggested that, although phylogeny and synteny of the genes are largely conserved between WOcauB2 and WOcauB3 (see Fig. S2 in the supplemental material), some of the genes have been considerably diversified between them, and this is probably mediated by the dynamic gene flux between WO phages through coinfection, lateral transfer, and/or recombination (3).
Comparison of WOcauB2 and WOcauB3 with WO phages in the Wolbachia strains wMel and wPip.
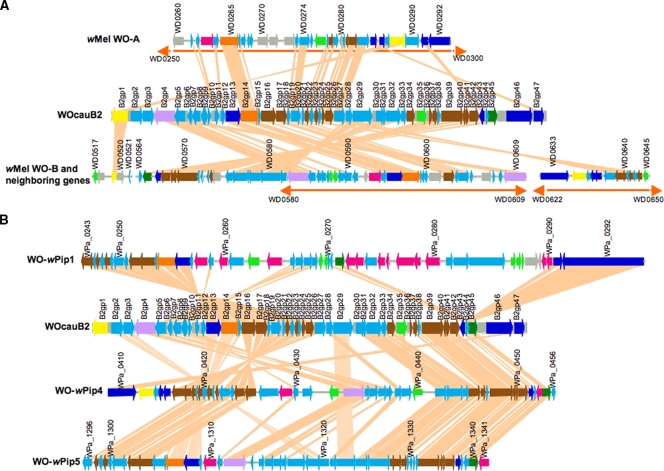
The gene content and order of WOcauB2 were compared with those of WO prophages encoded in the Wolbachia strain wMel from the fruit fly D. melanogaster. Proportions of common ORFs were 34.0% (16/47) from WOcauB2 to WO-A and 74.4% (35/47) from WOcauB2 to WO-B. The gene orders were conserved only partially between WOcauB2 and WO-A/WO-B (Fig. 4A). When the gene content and order of WOcauB2 were compared with those of WO prophages encoded in the Wolbachia strain wPip from the mosquito C. quinquefasciatus, proportions of common ORFs were 48.9% (23/47) from WOcauB2 to WO-wPip1, 74.4% (35/47) from WOcauB2 to WO-wPip4, and 78.7% (37/47) from WOcauB2 to WO-wPip5. The gene orders were also only partially conserved between WOcauB2 and WO-wPip1, -wPip4, and -wPip5 (Fig. 4B). These results clearly indicated that a number of inversion/translocation/recombination events must have taken place in the evolutionary course of these WO phages.
FIG. 4.
Gene order comparisons between WO prophages. (A) Comparisons between WOcauB2 and wMel prophages WO-A and WO-B. (B) Comparisons between WOcauB2 and wPip prophages WO-wPip1, WO-wPip4, and WO-wPip5. ORFs encoded in WOcauB2 were subjected to BLASTp searches against the wMel and wPip genomes, and matching ORFs with E-values less than 1e-9 are connected by tangerine lines. Orange bidirectional arrows represent the regions of wMel WO-A and WO-B assigned by Wu et al. (52). Colors of ORFs are as described in the legend of Fig. 1.
Evolutionary dynamics of WO phage genes across bacterial phyla.
Where do WOcauB2 and WOcauB3 come from? What evolutionary histories have the WO prophages experienced before getting into the Wolbachia strain wCauB? In an attempt to address these questions, database searches were performed using each of the WOcauB2 ORFs and several WOcauB3-specific ORFs as queries. Figure 5 summarizes the results, wherein microbial species and their affiliations representing up to the 10 top hits with E-values less than 1e-20 are compiled. For detailed information on the top hits, also refer to Table S1 in the supplemental material. These analyses unveiled several unexpected and intriguing patterns, as follows.
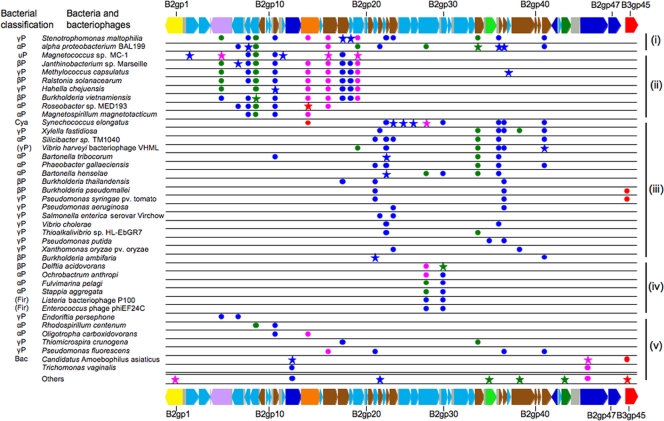
FIG. 5.
Distribution of genes allied to ORFs encoded in WOcauB2 and WOcauB3 among diverse bacterial taxa. For each of the ORFs, up to 10 top hits with E-value less than 1e-20 were collected from the databases by BLASTp searches. Stars highlight the first top hit for each of the ORFs, and circles show the other top hits. Colors of stars/circles indicate the similarity levels: blue, E-values from 1e-20 to 1e-60; green, E-values from 1e-61 to 1e-100; pink, E-values from 1e-101 to 1e-160; and red, E-values of 0. On the top and bottom are all ORFs of WOcauB2 and B3gp45 of WOcauB3 that were subjected to the database searches. For the bacterial classification at left, the following abbreviations were used: αP, the Alphaproteobacteria; βP, the Betaproteobacteria; γP, the Gammaproteobacteria; uP, the Proteobacteria, unclassified; Cya, the Cyanobacteria: Fir, the Firmicutes; and Bac, the Bacteroidetes. Parentheses show bacteriophages derived from the bacterial taxa. Note that T. vaginalis near the bottom is not a bacterium but a protozoan. On the right side are the gene detection patterns i to v as described in the text. Colors of ORFs are as described in the legend of Fig. 1.
First, genes homologous to those of WOcauB2/WOcauB3 were detected from phylogenetically diverse bacteria, representing the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, the Firmicutes, the Cyanobacteria, and the Bacteroidetes. Even when focused on the first top hits (Fig. 5), Alphaproteobacteria (Bartonella and Roseobacter, e.g.), Betaproteobacteria (Burkholderia and Delfia, e.g.), Gammaproteobacteria (Stenotrophomonas and Hahella, e.g.), a cyanobacterium (Synechococcus), and a Bacteroidetes (“Candidatus Amoebophilus”) were identified to harbor WO phage-related genes. These results strongly suggested that genes constituting the WO phages have been moving around across the diverse bacterial phyla.
Second, the detection patterns of the WO phage-related genes in these bacterial species suggest that they occur not at random but, interestingly, in blocks. Roughly speaking, we recognized the following five patterns in WO phage genomes: (i) the presence of many ORFs ranging from B2gp4 to B2gp42, (ii) the presence of ORFs ranging from B2gp4 to B2gp19, (iii) the presence of ORFs ranging from B2gp21 to B2gp42, (iv) the presence of the two specific ORFs B2gp29 and B2gp30, and (v) the presence of a few ORFs with no obvious pattern (Fig. 5). These results suggested that there are several evolutionarily coherent gene sets in the WO phage genomes. Although their biological functions are currently unknown, the conserved gene sets might have some important roles in the life cycle of the mobile genetic elements.
Third, the trichomonad protozoan Trichomonas vaginalis contained two WO phage-related genes, suggesting lateral gene transfer from a bacteriophage to the eukaryote. Putative lateral gene transfers from bacteria to T. vaginalis have already been reported (9, 29). Active phagocytotic incorporation of bacteria by the unicellular protozoan (11) and occurrence of an endocellular bacterial symbiont like Mycoplasma hominis (36) might have predisposed T. vaginalis to such lateral gene transfers.
Taken together, all these evolutionary analyses consistently indicated that WOcauB2 and WOcauB3 are certainly mobile genetic elements that have experienced quite dynamic and complicated evolutionary trajectories.
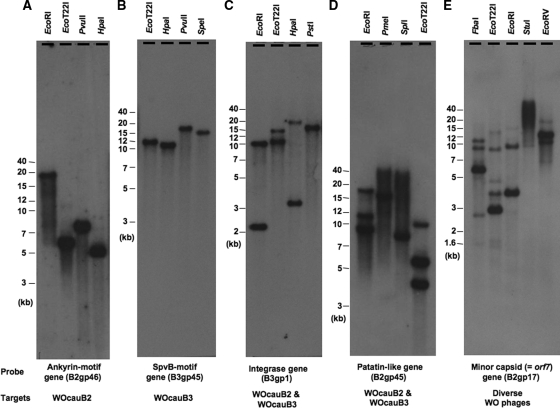
Southern blot detection of WO prophages in the wCauB genome.
Fosmid genomic library screening (Fig. 1) and PCR cloning of the attP region (Fig. 2) revealed that the Wolbachia strain wCauB possesses at least two copies of active WO prophages, WOcauB2 and WOcauB3, that are capable of producing phage particles. In order to confirm whether they exist as single copies and whether other WO prophage elements are present in the wCauB genome, a series of Southern blot analyses was performed. Using an ankyrin motif gene probe that targets WOcauB2 specifically, a single band was consistently detected with different restriction endonucleases (Fig. 6A), confirming that there is a single copy of WOcauB2. Using an SpvB motif gene probe that targets WOcauB3 specifically, a single band was consistently observed (Fig. 6B), indicating that WOcauB3 is also a single-copy prophage. Using an integrase gene probe that targets both WOcauB2 and WOcauB3, two bands were consistently observed (Fig. 6C). Using a patatin-like gene probe that recognizes both WOcauB2 and WOcauB3, more than two bands were observed with several restriction endonucleases (Fig. 6D), suggesting the presence of WO prophage(s) other than WOcauB2 and WOcauB3. Using an orf7 gene probe that is conserved among diverse WO phages, up to six bands were detected with different restriction endonucleases (Fig. 6E), revealing several other WO prophage copies in the wCauB genome that are probably degenerative and incapable of producing phage particles.
FIG. 6.
Southern blot detection of WO prophages in the genome of Wolbachia strain wCauB. (A) Specific detection of WOcauB2 targeting an ankyrin motif gene (B2gp46). (B) Specific detection of WOcauB3 targeting an SpvB motif gene (B3gp45). (C) Collective detection of WOcauB2 and WOcauB3 using an integrase gene (B2gp1) as a probe. (D) Collective detection of WOcauB2 and WOcauB3 using a patatin-like gene (B2gp45) as a probe. (E) Collective detection of diverse WO phages using the orf7 gene (B2gp17) as a probe. Restriction endonucleases used are given at the top, and probes and target WO phages are shown at the bottom of each of the autoradiograms.
Identification of sequence elements required for integration of WO phages: integrases.
Integrase is one of the essential components that allows temperate phages to be integrated into and excised from host bacterial genomes (see Fig. S1 in the supplemental material). Diverse integrases are categorized into two families, the tyrosine recombinases and the serine recombinases, on the basis of their evolutionary and mechanistic features (20, 33, 46). Both B2gp1 of WOcauB2 and B3gp1 of WOcauB3 exhibited significant similarities to the serine recombinases. A domain annotation program, SMART, identified the catalytic domain typical of the serine recombinases, which contains a serine residue as an active site and is located at the N terminus, in both B2gp1 and B3gp1 (data not shown). Smith and Thorpe (45) conducted a molecular phylogenetic analysis of 72 serine recombinases on the basis of their catalytic domain of around 150 amino acid residues, whereby three broad groups, namely resolvase/invertase, IS607-like, and φC31-like, were identified. Our analysis placed B2gp1 and B3gp1 in the φC31-like group that mainly consists of phage integrases (see Fig. S6 in the supplemental material). In many phages like λ, φC31, and TP901-1, the integrase genes are found adjacent to att sites (33). In WOcauB2 and WOcauB3, the serine recombinases were certainly located adjacent to attL (Fig. 1 and 2). From all these results taken together, it was concluded that WOcauB2 and WOcauB3 prophages possess a structurally intact φC31-like integrase gene at the 5′ terminus next to the attL site. The integrase gene product seems likely to be involved in integration/excision of the mobile genetic elements although its biochemical properties and expression patterns are to be examined in future studies.
Identification of sequence elements required for integration of WO phages: core sequences.
The core sequence of the att site is the region where two DNA strands are recombined during site-specific recombination reactions upon phage integration and excision (see Fig. S1 in the supplemental material) (45). By comparing attP, attL, and attR sequences, we attempted to identify the core sequences of WOcauB2 and WOcauB3. For WOcauB2, only a single nucleotide T was identical among the att sites (see Fig. S7A in the supplemental material). For WOcauB3, the trinucleotides TTG were consistently found among the att sites (see Fig. S7B to D in the supplemental material). Hence, these sequences were inferred to be the candidate core sequences for WOcauB2 and WOcauB3, respectively. Thus far, a number of core sequences have been identified from diverse bacteriophages, and these sequences exhibit several common features: (i) they are recognized as a common nucleotide sequence among attP, attL, attR, and attB sites, (ii) they consist of two or more consecutive nucleotides, and (iii) they are flanked by a pair of inverted repeat sequences, potentially forming a palindromic stem-loop structure (45). Under these criteria, however, the candidate core sequences of WOcauB2 and WOcauB3 are anomalous in two respects: they consist of a single nucleotide in WOcauB2 and are not associated with flanking inverted repeats in either WOcauB2 or WOcauB3 (see Fig. S7 in the supplemental material). Future studies should experimentally confirm whether the serine recombinases of WOcauB2 and WOcauB3 can utilize the candidate core sequences for recombination reactions without flanking inverted repeats.
Biological relevance of WO phages?
Because of the ubiquity of WO phages in the genomes of diverse Wolbachia strains (3) and the prevalence of Wolbachia infection in diverse insects (21), it is conceivable that WO phages may affect various biological aspects of the endosymbiotic bacteria and, consequently, may affect the fitness and phenotype of the host insects. However, previous studies have mostly focused on the phylogenetics and the diversity of WO phages on the basis of a limited number of gene sequences. Physiological, functional, and molecular biological aspects of WO phages have been very poorly explored. To our knowledge, the only work on biological relevance of WO phage was reported by Bordenstein et al. (4), wherein infection densities of a Wolbachia strain in the wasp Nasonia vitripennis were negatively correlated with infection densities of a WO phage, which may lead to attenuated costs of Wolbachia infection to the host insect, including lower intensity of CI. In this study, we determined the complete sequences of two closely related but distinct WO phages, which provided insights into candidate molecular mechanisms of WO phages to affect their host bacteria and insects. Although WOcauB2 and WOcauB3 are structurally very similar, their 3′ end regions are quite divergent (see Fig. S2, region 3, in the supplemental material), and we identified several intriguing genes containing so-called “effector” motifs, such as ankyrin motif genes (Fig. 1C, B2gp46 and B2gp47), an SpvB motif gene (Fig. 1D, B3gp45), and patatin-like genes (Fig. 1C and D, B2gp45 and B3gp44, respectively). Here, we suggest the possibility that, although speculative, WO phages might be regarded as “effector carriers” that encode functional molecular cassettes on their 3′ ends and vector them across different Wolbachia strains via coinfection, lateral transfer, and/or recombination. Detailed arguments on the issue are presented in the supplemental material (see Fig. S8 to S10 and the text in the supplemental material). Expression patterns, biochemical properties, and biological activities of these candidate effector genes are to be examined in future studies.
Potential of WO phages for transforming the Wolbachia genome.
Finally, we point out that the complete sequencing and structural analyses of the WOcauB2 and WOcauB3 prophage genomes would provide a clue to development of a genetic transformation system for Wolbachia endosymbionts. The active WO phages can be candidate vectors to deliver foreign genes into Wolbachia genomes. The att sites and integrase genes would enable controlled and site-directed Wolbachia transformation. The 3′ end of WO prophages may provide a location for introducing multiple-cloning sites, expression cassettes, and/or selection markers to accommodate foreign genes of interest. Of course, there are still many hurdles to be overcome, including verification of the functioning of the att site-integrase pairs in E. coli or other model bacteria, production of genetically modified WO phage particles, transformation of cultured/concentrated Wolbachia cells with the engineered WO phage particles, etc., which deserve future studies.
Supplementary Material
Acknowledgments
This work was supported by the Grand Challenges in Global Health program Modifying Mosquito Population Age Structure To Eliminate Dengue Transmission from the U.S. National Institutes of Health.
Footnotes
Published ahead of print on 10 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Beard, C. B., S. L. O'Neill, R. B. Tesh, F. F. Richards, and S. Aksoy. 1993. Modification of arthropod vector competence via symbiotic bacteria. Parasitol. Today 9:179-183. [DOI] [PubMed] [Google Scholar]
- 2.Beard, C. B., R. V. Durvasula, and F. F. Richards. 1998. Bacterial symbiosis in arthropods and the control of disease transmission. Emerg. Infect. Dis. 4:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordenstein, S. R., and J. J. Wernegreen. 2004. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21:1981-1991. [DOI] [PubMed] [Google Scholar]
- 4.Bordenstein, S. R., M. L. Marshall, A. J. Fry, U. Kim, and J. J. Wernegreen. 2006. The tripartite associations between bacteriophage, Wolbachia, and arthropods. PLoS Pathog. 2:e43. doi: 10.1371/journal.ppat.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourtzis, K., and T. A. Miller. 2003. Insect symbiosis. CRC Press, New York, NY.
- 6.Braquart-Varnier, C., P. Grève, C. Félix, and G. Martin. 2005. Bacteriophage WO in Wolbachia infecting terrestrial isopods. Biochem. Biophys. Res. Commun. 337:580-585. [DOI] [PubMed] [Google Scholar]
- 7.Chauvatcharin, N., A. Ahantarig, V. Baimai, and P. Kittayapong. 2006. Bacteriophage WO-B and Wolbachia in natural mosquito hosts: infection incidence, transmission mode and relative density. Mol. Ecol. 15:2451-2461. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Q., T. D. Ruel, W. Zhou, S. K. Moloo, P. Majiwa, S. L. O'Neill, and S. Aksoy. 2000. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 4:44-50. [DOI] [PubMed] [Google Scholar]
- 9.de Koning, A. P., F. S. L. Brinkman, S. J. M. Jones, and P. J. Keeling. 2000. Lateral gene transfer and metabolic adaptation in the human parasite Trichomonas vaginalis. Mol. Biol. Evol. 17:1769-1773. [DOI] [PubMed] [Google Scholar]
- 10.Dobson, S. L. 2003. Reversing Wolbachia-based population replacement. Trends Parasitol. 19:128-133. [DOI] [PubMed] [Google Scholar]
- 11.Doolittle, W. F. 1998. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14:307-311. [DOI] [PubMed] [Google Scholar]
- 12.Duron, O., P. Fort, and M. Weill. 2006. Hypervariable prophage WO sequences describe an unexpected high number of Wolbachia variants in the mosquito Culex pipiens. Proc. Biol. Sci. 273:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durvasula, R. V., A. Gumbs, A. Panackal, O. Kruglov, S. Aksoy, R. B. Merrifield, F. F. Richards, and C. B. Beard. 1997. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc. Natl. Acad. Sci. USA 94:3274-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 15.Fujii, Y., T. Kubo, H. Ishikawa, and T. Sasaki. 2004. Isolation and characterization of the bacteriophage WO from Wolbachia, an arthropod endosymbiont. Biochem. Biophys. Res. Commun. 317:1183-1188. [DOI] [PubMed] [Google Scholar]
- 16.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 17.Gavotte, L., F. Vavre, H. Henri, M. Ravallec, R. Stouthamer, and M. Boulétreau. 2004. Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol. Biol. 13:147-153. [DOI] [PubMed] [Google Scholar]
- 18.Gavotte, L., H. Henri, R. Stouthamer, D. Charif, S. Charlat, M. Boulétreau, and F. Vavre. 2007. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol. Biol. Evol. 24:427-435. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 20.Grindley, N. D., K. L. Whiteson, and P. A. Rice. 2006. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 75:567-605. [DOI] [PubMed] [Google Scholar]
- 21.Hilgenboecker, K., P. Hammerstein, P. Schlattmann, A. Telschow, and J. H. Werren. 2008. How many species are infected with Wolbachia?—a statistical analysis of current data. FEMS Microbiol. Lett. 281:215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katoh, K., K. Kuma, H. Toh, and T. Miyata. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi, Y., and T. Fukatsu. 2003. Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl. Environ. Microbiol. 69:6082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kittayapong, P., K. J. Baisley, V. Baimai, and S. L. O'Neill. 2000. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J. Med. Entomol. 37:340-345. [DOI] [PubMed] [Google Scholar]
- 25.Kittayapong, P., V. Baimai, and S. L. O'Neill. 2002. Field prevalence of Wolbachia in the mosquito vector Aedes albopictus. Am. J. Trop. Med. Hyg. 66:108-111. [DOI] [PubMed] [Google Scholar]
- 26.Klasson, L., T. Walker, M. Sebaihia, M. J. Sanders, M. A. Quail, A. Lord, S. Sanders, J. Earl, S. L. O'Neill, N. Thomson, S. P. Sinkins, and J. Parkhill. 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25:1877-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landy, A. 1989. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu. Rev. Biochem. 58:913-949. [DOI] [PubMed] [Google Scholar]
- 28.Landy, A., and W. Ross. 1977. Viral integration and excision: structure of the lambda att sites. Science 197:1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Markos, A., A. Miretsky, and M. Müller. 1993. A glyceraldehyde-3-phosphate dehydrogenase with eubacterial features in the amitochondriate eukaryote, Trichomonas vaginalis. J. Mol. Evol. 37:631-643. [DOI] [PubMed] [Google Scholar]
- 30.Masui, S., H. Kuroiwa, T. Sasaki, M. Inui, T. Kuroiwa, and H. Ishikawa. 2001. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem. Biophys. Res. Commun. 283:1099-1104. [DOI] [PubMed] [Google Scholar]
- 31.Masui, S., S. Kamoda, T. Sasaki, and H. Ishikawa. 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51:491-497. [DOI] [PubMed] [Google Scholar]
- 32.McMeniman, C. J., R. V. Lane, B. N. Cass, A. W. Fong, M. Sidhu, Y. F. Wang, and S. L. O'Neill. 2009. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323:141-144. [DOI] [PubMed] [Google Scholar]
- 33.Nash, H. A. 1996. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments, p. 2363-2376. In F. C. Niedhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 34.O'Neill, S. L., R. H. Gooding, and S. Aksoy. 1993. Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med. Vet. Entomol. 7:377-383. [DOI] [PubMed] [Google Scholar]
- 35.O'Neill, S. L., A. A. Hoffmann, and J. H. Werren. 1997. Influential passengers: Inherited microorganisms and arthropod reproduction. Oxford University Press, New York, NY.
- 36.Rappelli, P., M. F. Addis, F. Carta, and P. L. Fiori. 1998. Mycoplasma hominis parasitism of Trichomonas vaginalis. Lancet 352:1286. [DOI] [PubMed] [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Sanogo, Y. O., and S. L. Dobson. 2004. Molecular discrimination of Wolbachia in the Culex pipiens complex: evidence for variable bacteriophage hyperparasitism. Insect Mol. Biol. 13:365-369. [DOI] [PubMed] [Google Scholar]
- 40.Sanogo, Y. O., and S. L. Dobson. 2006. WO bacteriophage transcription in Wolbachia-infected Culex pipiens. Insect. Biochem. Mol. Biol. 36:80-85. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki, T., and H. Ishikawa. 2000. Transinfection of Wolbachia in the Mediterranean flour moth, Ephestia kuehniella, by embryonic microinjection. Heredity 85:130-135. [DOI] [PubMed] [Google Scholar]
- 42.Sasaki, T., T. Kubo, and H. Ishikawa. 2002. Interspecific transfer of Wolbachia between two lepidopteran insects expressing cytoplasmic incompatibility: a Wolbachia variant naturally infecting Cadra cautella causes male killing in Ephestia kuehniella. Genetics 162:1313-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinkins, S. P., C. F. Curtis, and S. L. O'Neill. 1997. The potential application of inherited symbiont systems to pest control, p. 155-175. In S. L. O'Neill, A. A. Hoffmann, and J. H. Werren (ed.), Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, New York, NY.
- 44.Sinkins, S. P., and F. Gould. 2006. Gene drive systems for insect disease vectors. Nat. Rev. Genet. 7:427-435. [DOI] [PubMed] [Google Scholar]
- 45.Smith, M. C., and H. M. Thorpe. 2002. Diversity in serine recombinases. Mol. Microbiol. 44:299-307. [DOI] [PubMed] [Google Scholar]
- 46.Stark, W. M., M. R. Boocock, and D. J. Sherratt. 1992. Catalysis by site-specific recombinases. Trends Genet. 8:432-439. [PubMed] [Google Scholar]
- 47.Sun, L. V., J. M. Foster, G. Tzertzinis, M. Ono, C. Bandi, B. E. Slatko, and S. L. O'Neill. 2001. Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J. Bacteriol. 183:2219-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaishampayan, P. A., D. P. Dhotre, R. P. Gupta, P. Lalwani, H. Ghate, M. S. Patole, and Y. S. Shouche. 2007. Molecular evidence and phylogenetic affiliations of Wolbachia in cockroaches. Mol. Phylogenet. Evol. 44:1346-1351. [DOI] [PubMed] [Google Scholar]
- 50.Weisberg, R. A., and A. Landy. 1983. Site-specific recombination in phage lambda, p. 211-250. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Werren, J. H. 1997. Biology of Wolbachia. Annu. Rev. Microbiol. 42:587-609. [DOI] [PubMed] [Google Scholar]
- 52.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.