Abstract
Here, the class I polyhydroxyalkanoate synthase (PhaC) from Ralstonia eutropha was investigated regarding the functionality of its conserved C-terminal region and its ability to tolerate translational fusions to its C terminus. MalE, the maltose binding protein, and green fluorescent protein (GFP) were considered reporter proteins to be translationally fused to the C terminus. Interestingly, PhaC remained active only when a linker was inserted between PhaC and MalE, whereas MalE was not functional. However, the extension of the PhaC N terminus by 458 amino acid residues was required to achieve a functionality of MalE. These data suggested a positive interaction of the extended N terminus with the C terminus. To assess whether a linker and/or N-terminal extension is generally required for a functional C-terminal fusion, GFP was fused to the C terminus of PhaC. Both fusion partners were active without the requirement of a linker and/or N-terminal extension. A further reporter protein, the immunoglobulin G binding ZZ domain of protein A, was translationally fused to the N terminus of the fusion protein PhaC-GFP and resulted in a tripartite fusion protein mediating the production of polyester granules displaying two functional protein domains.
Polyhydroxyalkanoates (PHAs) are biopolyesters synthesized by many bacteria and some archaea in times of unbalanced nutrient availability (7, 14-16, 22). These polyesters are stored as water-insoluble inclusions inside the cells and serve as energy and carbon storage (11, 29, 30). PHA synthases catalyze the stereoselective conversion of (R)-3-hydroxyacyl-coenzyme A (CoA) to PHAs while CoA is released and intracellular PHA granules are formed (32). The PHA synthase remains covalently attached to the PHA granule surface and has been targeted by protein engineering, i.e., translational fusion to the dispensable and variable N terminus, to enable the display of various protein functions without affecting the synthase activity (8, 26). PHA granules displaying certain functionalities have been considered as biobeads for biotechnological and medical applications (11).
PHA synthases can be divided into four classes. Class I and class II enzymes consist of only one subunit (PhaC) (28) and produce short-chain-length PHAs (class I) or medium-chain-length PHAs (class II), respectively (30, 33). Polyester synthases belonging to class III consist of two subunits, PhaC and PhaE, and produce short-chain-length PHAs (20, 21). Class IV PHA synthases are similar to enzymes belonging to class III. The synthases of this class comprise the two subunits PhaC and PhaR (23, 24).
It was previously shown that the N terminus of PhaC is a highly variable region and not essential for PHA synthase activity (30, 35). In contrast, the C terminus is a rather conserved region among class I and class II PHA synthases and is essential for enzyme activity (31). Alignments of the amino acid sequences of different PHA synthases revealed that the C terminus of these enzymes is hydrophobic and was therefore suggested to interact with the hydrophobic core of PHA granules (30). The PhaC subunits of class III and class IV PHA synthases do not show a high hydrophobicity for their C- terminal regions. Previous studies showed that the PhaC subunit of the class IV PHA synthase from Bacillus megaterium tolerates fusions to its C terminus without a loss in activity as long as the hydrophobic second subunit, PhaR, is present as well (23).
The aim of this study was to assess the effect of the conserved hydrophobic C terminus of PhaC on enzyme activity with regard to the possibility of translationally fusing protein functions for display at the PHA granule surface. This will be of interest for the display of proteins that require their free C terminus for activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. Escherichia coli XL1 Blue cultures were grown at 37°C, and E. coli BL21(DE3) cultures were grown at 25°C. The following antibiotics were added: ampicillin at 75 μg/ml, tetracycline at 12.5 μg/ml, and chloramphenicol 50 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| E. coli strain, plasmid, or oligonucleotide | Genotype, description, or sequence | Reference or source |
|---|---|---|
| Strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 lac[F′ proAB lacIqlacZΔM15 Tn10 (Tcr)] | 9 |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pMCS69 | pBBR1MCS derivative containing the genes phaA and phaB of Ralstonia eutropha colinear to the lac promoter | 17 |
| pET-14b | Apr; T7 promoter | Novagen |
| pETC | pET-14b derivative coding for the phaC wild type under T7 promoter control | 27 |
| pCWE | pBluescript SK(−) derivative containing the NdeI/BamHI-inserted phaC gene from R. eutropha | 26 |
| pMAL-c2G | Plasmid for expression and purification of maltose binding protein fusions | New England Biolabs |
| pCWESpeGFP | pCWESpe containing the SpeI-inserted gfp gene | 25 |
| pET-14b ZZ(−)phaC | pET-14b containing the XbaI/BamHI fragment comprising the ZZ-phaC gene | 8 |
| pET-14b PhaC-linker-MalE | pET-14b derivative containing malE fused to the 3′ end of phaC via a linker sequence | This study |
| pET-14b PhaC-MalE | pET-14b derivative containing malE directly fused to the 3′ end of phaC | This study |
| pET-14b M-PhaC-linker-MalE | pET-14b PhaC-linker-MalE derivative containing the mpl sequence fused to the 5′ end of phaC | This study |
| pET-14b M-PhaC-MalE | pET-14b PhaC-MalE derivative containing the mpl sequence fused to the 5′ end of phaC | This study |
| pMAL-Mpl-EC | pMAL-c2x derivative containing the mpl sequence | 36 |
| pCWESpeMpl-EC | pCWE derivative containing the mpl sequence fused to the 5′ end of phaC | This study |
| pET-14b M-PhaC | pETC derivative containing the mpl sequence fused to the 5′ end of phaC | This study |
| pET-14b PhaC-linker-SG-linker-MalE | pET-14b PhaC-linker-MalE derivative containing the SG linker sequence upstream of malE | This study |
| pET-14b PhaC-linker-GFP | pET-14b derivative containing gfp fused to the 3′ end of phaC via a linker sequence | This study |
| pET-14b PhaC-linker-SG-linker-GFP | pET-14b PhaC-linker-GFP derivative containing the SG linker sequence upstream of gfp | This study |
| pET-14b PhaC-GFP | pET-14b derivative containing gfp directly fused to the 3′ end of phaC | This study |
| pET-14b ZZ-PhaC-GFP | pET-14b PhaC-GFP derivative containing the ZZ sequence 5′ of phaC | This study |
| Oligonucleotides | ||
| linker I | 5′-P-TAT GGT GCT GGC GGT GGC GAT TGA TAA ACG CGG AGG CGG TGG AGG CC-3′ | This study |
| linker II | 5′-P-TCG AGG CCT CCA CCG CCT CCG CGT TTA TCA ATC GCC ACC GCC AGC ACC A-3′ | This study |
| MalE XhoI | 5′-CTC GAG ATG AAA ATC GAA GAA GGT AAA CTG GTA ATC-3′ | This study |
| MalE BamHI | 5′-GGA TCC TTA CTT GGT GAT ACG AGT CTG CGC GTC TTT CAG GGC TTC ATC GAC-3′ | This study |
| PhaC XbaI | 5′-TCT AGA AAT AAG GAG ATA CTA GTA TGG CGA CCG GCA AAG GCG CGG CAG CTT CCA CGC AG-3′ | This study |
| PhaC no stop | 5′-CAT ATG TGC CTT GGC TTT GAC GTA TCG CCC AGG-3′ | This study |
| GFP no start XhoI | 5′-ATG ACC TCG AGA GTA AAG GAG AAG AAC TTT TCA CTG GAG TTG TC-3′ | This study |
| GFP stop BamHI | 5′-GGA TCC TCA TTT GTA TAG TTC ATC CAT GCC ATG TGT AAT-3′ | This study |
| SG linker I | 5′-P-TCG AGA GCG GCG GTG GCG GTA GCG GTG GCG GTG GCA GCG GCG GTG GCG GTA GCC CCG GGC-3′ | 12 (modified) |
| SG linker II | 5′-TCG AGC CCG GGG CTA CCG CCA CCG CCG CTG CCA CCG CCA CCG CTA CCG CCA CCG CCG CTC-3′ | 12 (modified) |
| AscI PhaC | 5′-GGC GCG CCG TGC GCG CTG CT-3′ | This study |
| XhoI PhaC no stop | 5′-CTC GAG TGC CTT GGC TTT GAC GTA T-3′ | This study |
Construction of plasmids encoding C-terminal synthase fusions.
General cloning procedures and isolation of DNA were performed as described elsewhere previously (34). Deoxynucleoside triphosphate, T4 DNA ligase, and Pfx polymerase were purchased from Invitrogen; primers were purchased from Sigma-Aldrich (St. Louis, MO). DNA sequences of new plasmid constructs were confirmed by DNA sequencing according to the chain termination method using an automatic sequencer (3730 DNA analyzer; Applied Biosystems).
The nucleic acid sequence of phaC was PCR amplified using oligonucleotides PhaC XbaI and PhaC no stop and plasmid pCWE as the template. The sequence of MalE was amplified from vector pMAL-c2G by using the oligonucleotides MalE XhoI and MalE BamHI. The green fluorescent protein (GFP) sequence was amplified from plasmid pCWESpeGFP using the oligonucleotides GFP no start XhoI and GFP stop BamHI. To obtain the sequences of the complete fusion proteins, all respective fragments were ligated step by step into the respective restriction enzyme sites of vector pET-14b. The two linker sequences were encoded in the respective forward and reverse primers and used for direct primer-dimer ligation (VLAVIDKRGGGGG [linker] and SGGGSGGGSGGGGS [SG-linker]). Hence, plasmids pET-14b PhaC-linker-MalE, pET-14b PhaC-linker-SG-linker-MalE, pET-14b PhaC-linker-GFP, and pET-14b PhaC-linker-SG-linker-GFP were obtained. To construct a direct linker-free fusion, the 3′ part of phaC was amplified by PCR using oligonucleotides AscI PhaC and XhoI PhaC no stop. The respective fragment was exchanged to create plasmids pET-14b PhaC-MalE and pET-14b PhaC-GFP. In order to further expose the MalE protein, a nonfunctional N-terminal extension to PhaC was obtained. The sequence of this M extension was amplified from plasmid pMAL-Mpl-EC (gift from Peilin Xu), and plasmid pCWESpeMpl-EC was obtained. Further subcloning led to plasmids pET-14b M-PhaC-MalE, pET-14b M-PhaC-linker-MalE, and pET-14b M-PhaC. Plasmid pET-14b ZZ-PhaC-GFP codes for the double-functionalized synthase. The ZZ fragment was subcloned from plasmid pET-14b ZZ(−)phaC and inserted into the respectively prepared plasmid pET-14b PhaC-GFP. All plasmids and oligonucleotides are listed in Table 1. The respective plasmids were used to transform competent E. coli BL21(DE3) cells harboring plasmid pMCS69. Plasmid pMCS69 carries the β-ketothiolase (phaA) and the acetoacetyl-CoA reductase (phaB) genes, which are required for the formation of the precursor R-3-hydroxybutyryl-CoA, the substrate of the polyester synthase (1).
PHB granule isolation, polyester, and protein analysis.
Polyhydroxybutyrate (PHB) granule isolation was carried out as described previously (18), and polyester production was confirmed by gas chromatography (GC)/mass spectrometry (MS) (7). PHB granule protein profiles were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described elsewhere previously (19). The gels were stained with Coomassie brilliant blue G250.
MALDI-TOF MS.
The bead proteins were separated by SDS-PAGE, and protein bands of interest were cut off the gel. Proteins were subjected to matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS. Identification of tryptic peptides was performed by collision-induced dissociation tandem MS and enabled the identification of fusion proteins.
Microscopy.
Solutions of cells containing PHB granules and isolated granules were examined for GFP fluorescence as described previously (25).
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were normalized according to the protein concentration, which was determined by the Bradford method (6). The wells of microtiter plates were coated with 100 μl of a granule suspension and incubated overnight at 4°C. After blocking with 3% (wt/vol) bovine serum albumin, each well was washed with phosphate-buffered saline containing 0.05% Tween 20. The wells were then incubated with mouse monoclonal antibody to maltose binding protein (horseradish peroxidase) conjugate (Abcam, Cambridge, United Kingdom) for 1 h. After extensive washing, 100 μl of an o-phenylenediamine substrate solution was added to each well, and after 15 min, the reaction was stopped by adding 100 μl of 1 N H2SO4 to the mixture. Substrate conversion was measured at 490 nm using a microtiter plate reader (Biotek ELX808; Biostrategy, Auckland, New Zealand). The immunoglobulin G (IgG) binding ability was determined as described previously (18).
Maltose binding assay.
PHB granule suspensions were normalized to the protein concentration. Protein (2 mg/ml) was incubated with 0.6 mg/ml maltose for 4 h at 25°C in small shaking Erlenmeyer flasks. The assay solution was centrifuged, and the maltose concentration of the supernatant was determined using a colorimetric assay from Sigma (A3403).
RESULTS
The specifically designed linker is required to allow translational fusions to the C terminus of the PHA synthase.
E. coli BL21(DE3) cells harboring plasmids pMCS69 and pET-14b PhaC-MalE were not able to produce detectable amounts of PHB. However, cells containing plasmids pMCS69 and pET-14b PhaC-linker-MalE produced PHB, as confirmed by GC/MS analysis. The linker sequence was designed to maintain the hydrophobic environment close to the carboxy terminus of the synthase. Hydrophobic amino acid residues are followed by charged amino acid residues (VLAVIDKRGGGGG) to facilitate the proper surface display of the fusion partner. Isolated PHB granules displayed the fusion protein, as shown by SDS-PAGE analysis and ELISA. Despite the detection of MalE by ELISA, no maltose binding could be shown using the maltose binding assay. Hence, the linker region between PhaC and MalE was further extended, and plasmid pET-14b PhaC-linker-SG-linker-MalE, encoding the fusion protein, was constructed. The second linker sequence was inserted behind the original linker and consisted of a repeated serine-glycine motive (SGGGSGGGSGGGGS), which was already previously successfully employed as a linker between PhaC and a fusion partner (12). This plasmid also mediated PHB granule formation in recombinant E. coli (pMCS69), and the PHB granules displayed the fusion protein (see Fig. S1 in the supplemental material). However, the activity of the fusion partner MalE could not be confirmed.
An N-terminal extension of the PHA synthase restores its activity when directly fused to MalE.
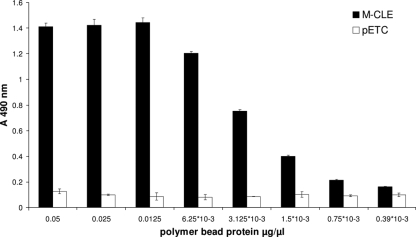
A 458-amino-acid-long nonfunctional extension, designated M, of the N terminus of the synthase protein restored PHB granule-forming activity even when the C terminus of the synthase was directly fused to MalE. Cells harboring plasmid pET-14b M-PhaC-MalE produced detectable amounts of PHB in recombinant E. coli (pMCS69). The respective fusion protein was attached to the PHB granules (see Fig. S1 in the supplemental material) (ELISA data not shown) and identified by MALDI-TOF MS analysis (data not shown). Although PhaC was active, the activity of the fusion partner MalE could not be demonstrated with the maltose binding assay. A slightly modified hybrid gene encoding a fusion protein with the N-terminal extension and the linker sequence between PhaC and MalE mediated the functionality of both fusion partners. Cells harboring plasmids pMCS69 and pET-14b M-PhaC-linker-MalE produced PHB granules, as verified by GC/MS analysis. The fusion protein was identified by MALDI-TOF MS (see Table ST1 in the supplemental material), and the isolated PHB granules displayed the fusion protein, as demonstrated by anti-MalE ELISA (Fig. 1; see Fig. S1 in the supplemental material). The maltose binding activity of the MalE protein was demonstrated using the maltose binding assay. These beads reduced the maltose concentration by up to 20%; control beads included in the experiment did not show any binding.
FIG. 1.
Display of the maltose binding protein assessed by ELISA. PHB granules were isolated from BL21(DE3)(pMCS69) cells harboring pET-14b M-PhaC-linker-MalE (M-CLE) (black columns) or pETC (white columns). All measurements were conducted in triplicates; standard deviations are indicated.
The activity of the entire fusion protein is dependent on the C-terminal fusion partner.
In order to assess whether the fusion protein data described above can be applied to various fusion partners, GFP was chosen as an alternative C-terminal fusion partner. Recombinant E. coli (pMCS69) harboring plasmid pET-14b PhaC-linker-GFP did not produce PHB, but cells showed nonlocalized green fluorescence, as observed by fluorescence microscopy (data not shown). Extending the linker region between both proteins resulted in detectable synthase and GFP activities. Recombinant E. coli (pMCS69) cells harboring plasmid pET-14b PhaC-linker-SG-linker-GFP were able to synthesize PHB granules, which displayed the fusion protein (see Fig. S2 in the supplemental material). Green fluorescence of the isolated PHB beads as well as of cells containing granules displaying the fusion protein could be observed (data not shown). The fusion protein was identified by MALDI-TOF MS (see Table ST1 in the supplemental material).
To assess whether a linker would be required for the functionality of both fusion partners, a direct fusion of the reporter protein GFP to the C terminus of the PHA synthase was constructed. This enabled the production of the fusion protein, which was identified by MALDI-TOF MS (see Table ST1 in the supplemental material). Recombinant E. coli (pMCS69) harboring plasmid pET-14b PhaC-GFP produced PHB granules, as confirmed by GC/MS analysis. SDS-PAGE analysis (see Fig. S2 in the supplemental material) and fluorescence microscopy confirmed the display and functionality of the respective fusion protein at the granule surface (data not shown).
Translational fusions to the N and C termini of the PHA synthase mediate the simultaneous display of two protein functions at the bead surface.
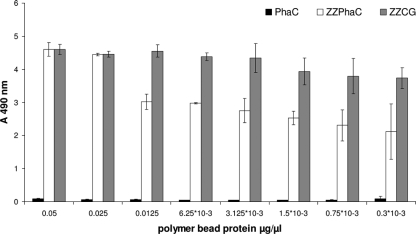
After the successful engineering of the synthase C terminus for the display of one protein function, the possibility of the simultaneous display of two protein functions by generating one tripartite fusion protein was assessed. Therefore, plasmid pET-14b ZZ-PhaC-GFP was constructed, and recombinant E. coli BL21(pMCS69) cells harboring this plasmid were able to produce PHB beads, as verified by GC/MS. The fusion protein comprising the IgG binding domain ZZ, the PHA synthase PhaC, and the GFP protein was displayed at the polyester bead surface (see Fig. S3 and Table ST1 in the supplemental material). These multifunctional PHB beads showed green fluorescence (data not shown) and were also able to bind IgG, as shown by ELISA (Fig. 2).
FIG. 2.
IgG binding ability of the tripartite synthase fusion protein assessed by ELISA. PHB granules were isolated from BL21(DE3)(pMCS69) cells harboring either pETC (black columns), pET-14b ZZ(−)phaC (white columns), or pET-14b ZZ-PhaC-GFP (gray columns). All measurements were conducted in triplicates; standard deviations are indicated.
DISCUSSION
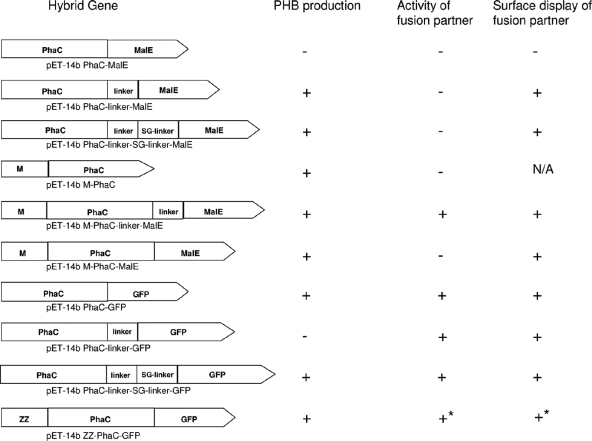
This study provided insights into a new mode of functional display at the PHA granule surface, employing the C terminus of the class I PHA synthase as a fusion target. The nature of the protein translationally fused to the C terminus of PhaC affects the activities of both fusion partners. Maintaining the hydrophobic environment around the C terminus of the synthase was found to be required to retain activity (30, 31). In contrast to N-terminal fusions, some C-terminal fusions were inactivating the PHA synthase (Fig. 3). MalE directly fused to the synthase C terminus led to inactivation, while a direct fusion of GFP to the C terminus of PhaC led to PHB granule formation and the display of the functional fusion partner. A hydrophobicity analysis of the N-terminal regions of MalE and GFP showed that GFP comprises an extended hydrophobic region similar to the designed linker region (5, 10), whereas the N-terminal region of MalE was found to be hydrophilic. This suggested that the hydrophobic N terminus of GFP, when directly fused to PhaC, does not interfere with the proposed anchor function of the hydrophobic C terminus of the PHA synthase. However, MalE was shown to be compatible with various proteins when translationally fused to the N and C termini of the respective proteins (13). Intriguingly, the insertion of the designed linker between PhaC and GFP inactivated PhaC, whereas a further extension of the linker by an additional SG linker did not interfere with PHA synthase activity. These data implied that the N terminus of GFP inherently provides the hydrophobic environment needed by the synthase protein to remain active and that the linker length inserted between PhaC and GFP is critical for PHA synthase activity.
FIG. 3.
Schematic overview of hybrid genes used in this study. PHB production was assessed by GC/MS analysis. The activity of the fusion partner was assessed by either fluorescence microscopy for GFP or maltose binding for MalE. The surface display of the fusion partner MalE was assessed by ELISA. An asterisk indicates that the IgG binding activity as well as the surface display of ZZ were assessed by ELISA. N/A, not applicable due to the inactivity of the fusion partner.
Previously, the immobilization of proteins which needed a free carboxy terminus for activity on the surface of PHA granules was possible only when fused to the granule-associated structural protein PhaP (2-4). In contrast to the phasin proteins, the PHA synthase, which is the only essential protein for granule formation, is covalently attached to the polyester granule surface. Therefore, utilizing the PhaC protein to fuse proteins of interest either N or C terminally would allow a variety of proteins to be displayed on the PHA granule surface. In addition, simultaneous fusions to the N and C termini, as shown with the tripartite fusion protein ZZ-PhaC-GFP, enable the display of two functionalities without the need for any additional protein anchor.
Supplementary Material
Acknowledgments
This study was supported by the New Zealand Foundation for Research Science and Technology and Massey University. Anika Jahns received a Technology for Industry doctoral fellowship.
We gratefully acknowledge the construction of the plasmid pCWESpeMpl-EC by V. Peters (Massey University).
Footnotes
Published ahead of print on 6 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amara, A. A., and B. H. A. Rehm. 2003. Replacement of the catalytic nucleophile cysteine-296 by serine in class II polyhydroxyalkanoate synthase from Pseudomonas aeruginosa-mediated synthesis of a new polyester: identification of catalytic residues. Biochem. J. 374:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwood, J. A., and B. H. A. Rehm. 2009. Protein engineering towards biotechnological production of bifunctional polyester beads. Biotechnol. Lett. 31:131-137. [DOI] [PubMed] [Google Scholar]
- 3.Bäckström, B. T., J. A. Brockelbank, and B. H. A. Rehm. 2007. Recombinant Escherichia coli produces tailor-made biopolyester granules for applications in fluorescence activated cell sorting: functional display of the mouse interleukin-2 and myelin oligodendrocyte glycoprotein. BMC Biotechnol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banki, M. R., T. U. Gerngross, and D. W. Wood. 2005. Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association. Protein Sci. 14:1387-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, K., M. Grey, and L. Bulow. 2008. Probing protein surface accessibility of amino acid substitutions using hydrophobic interaction chromatography. J. Chromatogr. A 1215:152-155. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brandl, H., R. A. Gross, R. W. Lenz, and R. C. Fuller. 1988. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl. Environ. Microbiol. 54:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockelbank, J. A., V. Peters, and B. H. A. Rehm. 2006. Recombinant Escherichia coli strain produces a ZZ domain displaying biopolyester granules suitable for immunoglobulin G purification. Appl. Environ. Microbiol. 72:7394-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-BLUE—a high-efficiency plasmid transforming RecA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376. [Google Scholar]
- 10.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 11.Grage, K., A. C. Jahns, N. Parlane, R. Palanisamy, I. A. Rasiah, J. A. Atwood, and B. H. A. Rehm. 2009. Bacterial polyhydroxyalkanoate granules: biogenesis, structure, and potential use as nano-/micro-beads in biotechnological and biomedical applications. Biomacromolecules 10:660-669. [DOI] [PubMed] [Google Scholar]
- 12.Grage, K., and B. H. A. Rehm. 2008. In vivo production of scFv-displaying biopolymer beads using a self-assembly-promoting fusion partner. Bioconjug. Chem. 19:254-262. [DOI] [PubMed] [Google Scholar]
- 13.Ha, J.-S., J. J. Song, Y.-M. Lee, S.-J. Kim, J.-H. Sohn, C.-S. Shin, and S.-G. Lee. 2007. Design and application of highly responsive fluorescence resonance energy transfer biosensors for detection of sugar in living Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 73:7408-7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J., Q. Lu, L. Zhou, J. Zhou, and H. Xiang. 2007. Molecular characterization of the phaECHm genes, required for biosynthesis of poly(3-hydroxybutyrate) in the extremely halophilic archaeon Haloarcula marismortui. Appl. Environ. Microbiol. 73:6058-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hezayen, F. F., B. H. A. Rehm, R. Eberhardt, and A. Steinbüchel. 2000. Polymer production by two newly isolated extremely halophilic archaea: application of a novel corrosion-resistant bioreactor. Appl. Microbiol. Biotechnol. 54:319-325. [DOI] [PubMed] [Google Scholar]
- 16.Hezayen, F. F., A. Steinbüchel, and B. H. A. Rehm. 2002. Biochemical and enzymological properties of the polyhydroxybutyrate synthase from the extremely halophilic archaeon strain 56. Arch. Biochem. Biophys. 403:284-291. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann, N., A. A. Amara, B. B. Beermann, Q. Qi, H. J. Hinz, and B. H. A. Rehm. 2002. Biochemical characterization of the Pseudomonas putida 3-hydroxyacyl ACP:CoA transacylase, which diverts intermediates of fatty acid de novo biosynthesis. J. Biol. Chem. 277:42926-42936. [DOI] [PubMed] [Google Scholar]
- 18.Jahns, A. C., R. G. Haverkamp, and B. H. A. Rehm. 2008. Multifunctional inorganic-binding beads self-assembled inside engineered bacteria. Bioconjug. Chem. 19:2072-2080. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Liebergesell, M., B. Schmidt, and A. Steinbüchel. 1992. Isolation and identification of granule-associated proteins relevant for poly(3-hydroxyalkanoic acid) biosynthesis in Chromatium vinosum D. FEMS Microbiol. Lett. 78:227-232. [DOI] [PubMed] [Google Scholar]
- 21.Liebergesell, M., and A. Steinbüchel. 1992. Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur. J. Biochem. 209:135-150. [DOI] [PubMed] [Google Scholar]
- 22.Lu, Q., J. Han, L. Zhou, J. Zhou, and H. Xiang. 2008. Genetic and biochemical characterization of the poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthase in Haloferax mediterranei. J. Bacteriol. 190:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCool, G. J., and M. C. Cannon. 2001. PhaC and PhaR are required for polyhydroxyalkanoic acid synthase activity in Bacillus megaterium. J. Bacteriol. 183:4235-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCool, G. J., and M. C. Cannon. 1999. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J. Bacteriol. 181:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, V., D. Becher, and B. H. A. Rehm. 2007. The inherent property of polyhydroxyalkanoate synthase to form spherical PHA granules at the cell poles: the core region is required for polar localization. J. Biotechnol. 132:238-245. [DOI] [PubMed] [Google Scholar]
- 26.Peters, V., and B. H. A. Rehm. 2005. In vivo monitoring of PHA granule formation using GFP-labeled PHA synthases. FEMS Microbiol. Lett. 248:93-100. [DOI] [PubMed] [Google Scholar]
- 27.Peters, V., and B. H. A. Rehm. 2008. Protein engineering of streptavidin for in vivo assembly of streptavidin beads. J. Biotechnol. 134:266-274. [DOI] [PubMed] [Google Scholar]
- 28.Qi, Q., and B. H. A. Rehm. 2001. Polyhydroxybutyrate biosynthesis in Caulobacter crescentus: molecular characterization of the polyhydroxybutyrate synthase. Microbiology 147:3353-3358. [DOI] [PubMed] [Google Scholar]
- 29.Rehm, B. H. A. 2006. Genetics and biochemistry of polyhydroxyalkanoate granule self-assembly: the key role of polyester synthases. Biotechnol. Lett. 28:207-213. [DOI] [PubMed] [Google Scholar]
- 30.Rehm, B. H. A. 2003. Polyester synthases: natural catalysts for plastics. Biochem. J. 376:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehm, B. H. A., R. V. Antonio, P. Spiekermann, A. A. Amara, and A. Steinbüchel. 2002. Molecular characterization of the poly(3-hydroxybutyrate) (PHB) synthase from Ralstonia eutropha: in vitro evolution, site-specific mutagenesis and development of a PHB synthase protein model. Biochim. Biophys. Acta 1594:178-190. [DOI] [PubMed] [Google Scholar]
- 32.Rehm, B. H. A., and A. Steinbüchel. 1999. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int. J. Biol. Macromol. 25:3-19. [DOI] [PubMed] [Google Scholar]
- 33.Ren, Q., G. De Roo, B. Kessler, and B. Witholt. 2000. Recovery of active medium-chain-length-poly-3-hydroxyalkanoate polymerase from inactive inclusion bodies using ion-exchange resin. Biochem. J. 349:599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 35.Schubert, P., N. Krüger, and A. Steinbüchel. 1991. Molecular analysis of the Alcaligenes eutrophus poly(3-hydroxybutyrate) biosynthetic operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and identification of the promoter. J. Bacteriol. 173:168-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Q., R. M. Pan, Y. C. Ge, and P. Xu. 2004. Expression of the soluble extracellular domain of human thrombopoietin receptor using a maltose-binding protein-affinity fusion system. Biol. Pharm. Bull. 27:219-221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.