Abstract
RNases are involved in critical aspects of RNA metabolism in all organisms. Two classes of RNases that digest RNA from an end (exo-RNases) are known: RNases that use water as a nucleophile to catalyze RNA degradation (hydrolytic RNases) and RNases that use inorganic phosphate (phosphorolytic RNases). It has been shown previously that the absence of the two known Escherichia coli phosphorolytic RNases, polynucleotide phosphorylase and RNase PH, leads to marked growth and ribosome assembly defects. To investigate the basis for these defects, a screen for growth suppressors was performed. The majority of suppressor mutations were found to lie within nsrR, which encodes a nitric oxide (NO)-sensitive transcriptional repressor. Further analysis showed that the suppressors function not by inactivating nsrR but by causing overexpression of a downstream gene that encodes a hydrolytic RNase, RNase R. Additional studies revealed that overexpression of another hydrolytic RNase, RNase II, similarly suppressed the growth defects. These results suggest that the requirement for phosphorolytic RNases for robust cellular growth and efficient ribosome assembly can be bypassed by increased expression of hydrolytic RNases.
Many aspects of RNA metabolism are performed by RNases, a class of enzymes defined by their ability to cleave phosphodiester bonds in RNA (13, 30). Most organisms contain multiple RNases, and the crucial cellular functions of these enzymes include a role in RNA processing and maturation, turnover of unstable RNAs, and degradation of defective RNAs (14, 15). Genetic analysis has shown that several of these enzymes are essential for cell viability, whereas in other cases the absence of certain combinations of RNases can cause synthetic defects (15, 29).
For many years, Escherichia coli has been an important model organism for studying RNase function. E. coli is known to harbor about 20 RNases, a number that is likely to increase since the RNases responsible for several processes are still unknown (15). The RNases can be classified into two groups: endo-RNases and exo-RNases. Endo-RNases cleave RNA internally and generate RNA fragments. Exo-RNases cleave one terminal phosphodiester bond at a time, either from the 5′ or the 3′ end, releasing mononucleotides as a product.
Currently, E. coli has eight known exo-RNases, each of which possesses 3′-to-5′ activity (38). Six of these enzymes, RNase II, RNase D, RNase T, RNase R, RNase BN, and oligoribonuclease, are hydrolytic RNases that use water as a nucleophile, generating nucleotide monophosphates as a product. Two exo-RNases, polynucleotide phosphorylase (PNPase) and RNase PH, use inorganic phosphate as a nucleophile and release nucleotide diphosphates during RNA digestion. These enzymes are referred to as phosphorolytic RNases. PNPase, which was identified over 50 years ago, is implicated in the digestion of mRNA fragments generated during mRNA turnover and of structured RNAs with the assistance of helicases to open RNA duplexes (18, 32). In addition, PNPase mutants exhibit increased antibiotic sensitivity, cold-sensitive growth, and altered turnover of regulatory RNAs (1, 28). In several pathogenic bacteria, PNPase is also important for infectivity (33, 36). RNase PH, in contrast, participates in the maturation of transfer and other small RNAs through removal of precursor sequences at the 3′ end (23, 25). Although both PNPase and RNase PH have important cellular roles, neither is essential under standard laboratory growth conditions because of the presence of backup enzymes that perform similar functions.
Because of the different nucleophilic requirements of phosphorolytic and hydrolytic enzymes, it has been unclear whether this has any influence on the cellular function of the RNases. In this context, it was shown several years ago that about 10% of the mononucleotides released during RNA turnover in E. coli are diphosphates generated by phosphorolytic RNases and that the remaining mononucleotides are monophosphates generated by hydrolytic RNases (9, 19). Thus, the phosphorolytic RNases appear to have a minor role in overall RNA turnover compared to the hydrolytic RNases. Despite their smaller contribution, however, mutations in both PNPase and RNase PH have been found to cause marked cell growth and ribosome assembly defects (23, 37). In contrast, cells lacking RNase II, the major hydrolytic RNase, display very few growth defects (16, 31). These observations suggest that phosphorolytic RNases might catalyze a set of key processes important for cell growth. In order to address this issue, mutations that could suppress the growth defect of a strain lacking both PNPase and RNase PH were identified. Identification and analysis of the suppressors are described here.
MATERIALS AND METHODS
Strains and plasmids.
Strain MG1655 has been described previously (5). An rph+ derivative of MG1655 (MG1655*) was constructed by Donald Court (National Cancer Institute, Bethesda, MD) and was obtained from Kenneth Rudd (University of Miami). Derivatives of these strains containing nonpolar Δrph or ΔnsrR alleles or a Δpnp::kan allele were constructed by transduction of mutants derived from the Keio strain collection (3). MG1655 Δrph Δpnp was constructed by excision of the kan gene in MG1655 Δrph::kan by recombination of frt sites that flank the kan gene, followed by transduction of the Δpnp::kan allele into the derivative strain. Introduction of a multicopy pnp plasmid into this strain restored normal growth (data not shown), indicating that the growth defect observed in this strain was not due to a polar effect.
Plasmid pDK24 is a derivative of pBR322 that overexpresses RNase II (17). A multicopy nsrR plasmid (pJP07) and a control plasmid (p2975) were provided by Stephen Spiro (University of Texas, Dallas). An RNase R-expressing plasmid, pMPM-A5/RNR, was constructed by subcloning a 2.7-kb XmnI-DraIII chromosomal DNA fragment containing the entire rnr coding sequence between the EcoRI and DraIII sites in pMPM-A5, a cloning vector that contains a PBAD promoter (27).
Measurement of cell growth.
For each strain, three or four individual colonies were grown to saturation at 37°C in LB medium supplemented with 0.2% glucose and 1 mM MgSO4, and this was followed by subculturing of 100 μl of the saturated culture in 5 ml of fresh medium. Cell density was measured spectrophotometrically at 600 nm four or five times during exponential growth. The doubling time and standard deviation for each strain were determined by linear regression. The correlation coefficient for each doubling time measurement was at least 0.99.
Transposon mutagenesis and mapping of insertion sites.
Transposon mutagenesis of MG1655 Δrph Δpnp was performed by infection with λ1324, a replication-defective phage that harbors a mini-Tn10 cam transposon (24). Following transposition and outgrowth for 1 h, cells were plated on selective plates containing chloramphenicol (Cam). The plates were incubated at 37°C overnight and screened for growth suppressors, which were visualized as large colonies against a background of small colonies. Suppressors were purified by restreaking on LB medium plates containing Cam. To confirm that a transposon insertion is responsible for the suppressor phenotype, the Camr marker was transduced to MG1655 Δrph Δpnp, and the resulting transductants were retested for the suppressor phenotype.
To determine the site of transposon insertion, chromosomal DNA was directly sequenced using a 32P-labeled outwardly directed primer (5′ GCTGCCTCCCAGAGCCTGAT 3′) specific for the mini-Tn10 cam transposon. The site of insertion was determined by aligning the chromosomal sequences that followed transposon-specific sequences with the MG1655 sequence. In some cases, DNA corresponding to the mini-transposon and associated flanking DNA was first subcloned into a multicopy plasmid. Chromosomal DNA derived from the suppressors was digested with BstUI and BstBI and ligated to pT7/T3α-18 (BRL) predigested with restriction enzymes SmaI and AccI, and the ligation product was introduced into electrocompetent cells. The desired clones were identified by selection for ampicillin- and Cam-resistant plasmid derivatives. The plasmids were purified and sequenced as described above.
Western blot analysis.
Cell extracts were prepared by pelleting exponentially growing cells, resuspending the cells in sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate, 10% glycerol, 0.001% bromophenol blue), and heating them at 100°C for 8 to 10 min. Twenty micrograms of total protein per sample was electrophoresed on a NuPage Novex bis-Tris 4 to 12% polyacrylamide gradient gel (Invitrogen), transferred to a nitrocellulose membrane, and probed with anti-RNase R primary antibody and horseradish peroxidase-conjugated anti-rabbit antibody (GE Healthcare). The immunoreactive bands were visualized using Supersignal West Femto substrate (Pierce). Preliminary experiments with unpurified RNase R antibody revealed the presence of nonspecific cross-reacting bands. Therefore, the antibody was purified by binding to purified RNase R adsorbed on nitrocellulose, removal of nonspecific antibodies by washing, and elution of RNase R-specific antibody with glycine-HCl (pH 2.5).
Ribosome and RNA analysis.
Ribosomal profiles were generated by ultracentrifugation of cell extracts on a 14 to 32% sucrose gradient, as described previously (22). RNA was isolated using the hot phenol method and was analyzed by primer extension using a 23S rRNA-specific primer, as described previously (35).
Real-time reverse transcription (RT)-PCR.
For each sample, 1 μg of DNase I-treated total RNA was reversed transcribed with 50 fmol of an rnr-specific oligonucleotide (5′ GCCGCCTTTTTTACCTGCAT 3′) using primer extension conditions, as described previously (35). One-fifth of the resulting cDNA was used to prepare 20-μl reaction mixtures containing 1× DyNAmo HS SYBR green master mixture (Finnzymes Oy) and 200 nM rnr-specific oligonucleotides (5′ GCCGCCTTTTTTACCTGCAT 3′ and 5′ GGGCAACGCCTGATGGGGG 3′). The reactions were performed using a Bio-Rad CFX96 real-time thermocycler, and cycle threshold values were determined. Relative rnr expression levels were calculated based on differences in the cycle threshold values.
RESULTS
Strains lacking phosphorolytic RNases.
To verify that the loss of both phosphorolytic RNases leads to growth defects, Δrph or Δpnp deletion mutations were transduced into MG1655, the first sequenced E. coli strain (5), or into a derivative strain, MG1655*, that contains a wild-type rph allele. This yielded a set of isogenic strains that contained both phosphorolytic RNases (MG1655*), lacked one phosphorolytic RNase (MG1655* Δpnp and MG1655 Δrph), or lacked both phosphorolytic RNases (MG1655 Δrph Δpnp). Growth measurements indicated that the growth of MG1655 Δrph was normal and that the growth rate of MG1655* Δpnp was 15% lower. However, the double mutant grew significantly slower, and its growth rate was twofold lower (Fig. 1A and 1C). These results confirm that the loss of both enzymes has a substantially greater negative effect on cell growth than the loss of either enzyme alone.
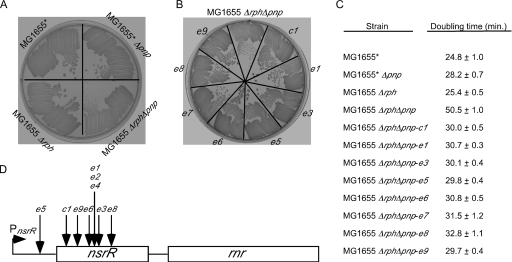
FIG. 1.
Growth of strains containing Δpnp and/or Δrph alleles and suppressors of the Δrph Δpnp growth defect. (A) MG1655* and derivatives containing Δpnp and/or Δrph alleles were streaked on an LB agar plate and incubated overnight at 37°C. (B) MG1655 Δrph Δpnp and eight distinct growth suppressors (see below) were streaked on an LB agar plate and grown at 37°C. Each suppressor is indicated by a different designation. Two suppressors (e2 and e4) were not streaked because they were found to harbor an insertion at the same location as e1. (C) Doubling times for MG1655*, derivatives containing Δpnp and/or Δrph alleles, and the MG1655 Δrph Δpnp suppressor strains determined in rich medium at 37°C. (D) Schematic diagram of the nsrR operon and transposon insertion sites for the MG1655 Δrph Δpnp suppressors. The nsrR gene and the downstream rnr coding regions are indicated by rectangles. The nsrR transcription start is indicated by a right-facing arrow, and the 5′ and 3′ untranslated regions flanking the nsrR coding region are indicated by a line. The location of each of mapped insertion site is indicated. The insertion sites in the 423-bp nsrR coding region are positions 31 (c1), 82 (e9), 147 (e6), 156 (e1, e2, and e4), 161 (e3), and 242 (e8). The e5 mutation maps to 32 bp upstream of the coding region. The insertion site for the e7 mutation could not be determined and hence is not shown.
Screen for insertional suppressors of the MG1655 Δrph Δpnp slow-growth phenotype.
To gain further insight into the impaired growth of MG1655 Δrph Δpnp, a transposon mutagenesis screen was performed to identify suppressors of the growth defect. Accordingly, a culture of MG1655 Δrph Δpnp was infected with λ1324, a defective λ vehicle that harbors a mini-Tn10 cam transposon (24), and transposon insertions were selected on plates containing LB medium with Cam. Screening of approximately 10,000 Camr colonies led to identification of approximately two dozen mutants that showed enhanced growth. To determine whether the transposon insertions were directly responsible for the suppressor effect, the Camr marker was transduced into a fresh MG1655 Δrph Δpnp background. In this manner, 10 strains that displayed a suppressor phenotype resulting from a mini-Tn10 cam transposon insertion were identified (Fig. 1B). To characterize the suppressor effect quantitatively, the cell doubling times for MG1655 Δrph Δpnp and each different suppressor strain were determined. Each suppressor strain exhibited more rapid growth, and in most cases the doubling time was restored to nearly that of the wild-type strain (Fig. 1C).
The MG1655 Δrph Δpnp suppressor mutations map to the nsrR gene.
The locations of the transposition insertion sites for the different suppressor mutants were determined by sequencing chromosomal DNA abutting the transposon inserts (see Materials and Methods). For 9 of the 10 suppressors, the transposon insertion site was found to map to the nsrR coding region or between the nsrR promoter and the coding region (Fig. 1D). The nsrR gene encodes an NO-sensitive transcriptional repressor that controls a regulon involved in the cellular response to reactive nitrogen molecules (6, 20). Seven distinct nsrR mutations were isolated; six of these mutations were isolated once, and the seventh was identified three times.
Insertions into nsrR rescue ribosomal defects.
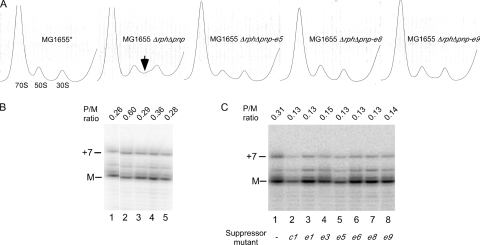
A notable defect associated with the absence of PNPase and RNase PH is the accumulation of defective or incompletely formed ribosomes, especially when the growth temperature is less than 37°C (37). To investigate whether nsrR suppressor mutants can correct such ribosomal defects, the ribosome profiles of MG1655 Δrph Δpnp and three representative mutants (MG1655 Δrph Δpnp-e5, MG1655 Δrph Δpnp-e8, and MG1655 Δrph Δpnp-e9) were assessed. As a control, the ribosome profile of MG1655* was also generated. The initial efforts to grow the strains at 30°C were hampered by the extremely slow growth of MG1655 Δrph Δpnp; therefore, the strains were first grown at 37°C and then shifted to 30°C for 2 h. Cell extracts were prepared from each strain, and ribosomes were fractionated by sucrose density gradient ultracentrifugation under subunit association conditions (100 mM NH4Cl and 10 mM Mg2+). As expected, the ribosomal profile for MG1655* consisted of a major 70S ribosomal peak and minor peaks comprising the individual 50S and 30S ribosomal subunits (Fig. 2A). For MG1655 Δrph Δpnp, in addition to the 70S, 50S, and 30S peaks, increased absorbance at ∼40S was also observed. The accumulation of such particles is usually correlated with ribosome maturation defects and has been previously noted in an rph pnp strain background (4, 10, 11, 21, 37). However, when the ribosomal profiles of the suppressor strains were generated, the abundance of particles sedimenting at ∼40S was significantly reduced. These results indicate that each of the three suppressor mutations can substantially correct the ribosomal defects caused by a lack of phosphorolytic RNases.
FIG. 2.
Suppression of ribosomal defects by transposon insertions into nsrR. (A) Strains MG1655*, MG1655 Δrph Δpnp, MG1655 Δrph Δpnp-e5, MG1655 Δrph Δpnp-e8, and MG1655 Δrph Δpnp-e9 were grown at 37°C to early log phase and then shifted to 30°C for 2 h. When necessary, prewarmed medium was added to cultures to maintain log-phase growth. Ribosomes were analyzed by sucrose density gradient ultracentrifugation of cell extracts. The positions of the peaks corresponding to 70S ribosomes, as well as the individual 50S and 30S ribosomal subunits, are indicated. The increase in absorbance at ∼40S observed for MG165 Δrph Δpnp is indicated by an arrow. (B and C) Analysis of 23S rRNA processing. (B) Strains were grown as described above for panel A. Total RNA was isolated from MG1655* (lane 1), MG1655 Δrph Δpnp (lane 2), MG1655 Δrph Δpnp-e5 (lane 3), MG1655 Δrph Δpnp-e8 (lane 4), or MG1655 Δrph Δpnp-e9 (lane 5) and analyzed by primer extension using a 23S rRNA-specific labeled primer (5′ CCTTCATCGCCTCTGACTGCC 3′). The positions of the mature 5′ end (M) and a precursor containing a 7-nt 5′ extension (+7) are indicated. The ratio of the precursor containing a 7-nt 5′ extension to the mature RNA (P/M ratio) is also indicated. (C) RNA was isolated from MG1655 Δrph Δpnp (lane 1) or from the seven distinct nsrR suppressors grown at 37°C and was analyzed as described above for panel B.
To characterize ribosome biogenesis at the RNA level, the processing of 23S rRNA was assessed. Defects in ribosome assembly are frequently correlated with the accumulation of immature 23S rRNA within 40S particles that contain 3 to 7 nucleotides (nt) of unprocessed precursor sequences at the 5′ end and 7 to 9 nt at the 3′ end (34). To gauge the extent of 23S rRNA processing, total RNA was isolated from MG1655*, MG1655 Δrph Δpnp, and the three suppressor strains grown as described above, which was followed by primer extension using a 23S rRNA-specific primer. The analysis of the products showed that there were significantly larger amounts of unprocessed 23S rRNA in MG1655 Δrph Δpnp than in MG1655*, but the e5, e8, and e9 mutations reduced the level of unprocessed 23S rRNA to nearly that of MG1655* (Fig. 2B). Therefore, along with restoration of a normal ribosomal profile, each of the three suppressor mutations also reduced the amount of unprocessed 23S rRNA. Additional experiments performed with MG1655 Δrph Δpnp and the seven suppressor mutants grown at 37°C indicated that each mutation reduced the amount of unprocessed 23S rRNA at this temperature as well (Fig. 2C).
Transposon insertions in nsrR increase expression of RNase R.
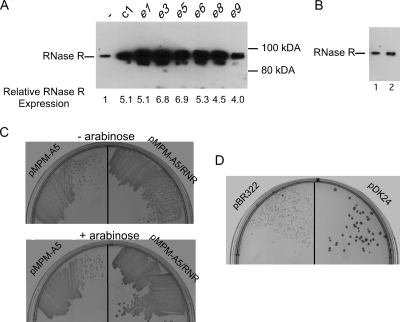
Microarray experiments have indicated that the NsrR regulon comprises ∼20 E. coli genes distributed over nine operons (20). However, none of these genes is believed to play any role in RNA metabolism. Therefore, it was unclear why transposon insertion into nsrR suppressed a defect caused by the absence of phosphorolytic RNases. Interestingly, nsrR is the first gene in an operon that includes rnr and rlmB, in which rnr encodes RNase R, one of six known hydrolytic exo-RNases, and rlmB encodes an RNA methyltransferase that methylates a unique residue in 23S rRNA (8, 26). Based on the hypothesis that transposon insertions into nsrR could affect RNase R expression, the RNase R levels of MG1655 Δrph Δpnp and the suppressor strains were compared using Western blot analysis. Significantly, the RNase R level was considerably increased in each suppressor strain (Fig. 3A). Quantification of rnr expression was performed using real-time RT-PCR. These experiments indicated that each suppressor strain exhibited significantly increased (4.0- to 6.9-fold) rnr expression compared to MG1655 Δrph Δpnp.
FIG. 3.
RNase R overexpression and RNase II overexpression suppress the MG1655 Δrph Δpnp slow-growth phenotype. (A) Transposon insertions in nsrR increase RNase R expression. Cell extracts from MG1655 Δrph Δpnp (indicated by a dash), as well as derivatives containing seven different transposon insertions in nsrR, were analyzed by Western blotting using RNase R antibodies. The positions of the RNase R immunoreactive band and molecular weight markers are indicated. A minor band apparent in some lanes could be a proteolytic product derived from full-length RNase R. (B) A nonpolar ΔnsrR mutation does not affect RNase R expression. Cell extracts from MG1655 Δrph Δpnp (lane 1) and MG1655 Δrph Δpnp ΔnsrR (lane 2) were analyzed by Western blotting, as described above for panel A. (C) Plasmid-directed overexpression of RNase R suppresses the growth defect of Δrph Δpnp strains. MG1655 Δrph Δpnp was transformed with pMPM-A5 or pMPM-A5/RNR, and the transformed cells were restreaked on an LB agar plate containing ampicillin (upper plate) or an LB agar plate containing ampicillin and 0.2% arabinose (lower plate) and incubated overnight at 37°C. (D) MG1655 Δrph Δpnp was transformed with either a control plasmid (pBR322) or a multicopy RNase II-expressing plasmid (pDK24). The transformed strains were plated on an LB agar plate supplemented with ampicillin and incubated overnight at 37°C.
One explanation for the increased RNase R levels could be that nsrR autoregulates the expression of the nsrR-rnr-rlmB operon. Thus, inactivation of nsrR would be expected to increase expression of the downstream genes. Although the levels of the nsrR-rnr-rlmB operon RNA are regulated by other RNases (7), there is no evidence suggesting that the nsrR-rnr-rlmB operon is autoregulated. Nor is a recognizable NsrR operator sequence located at the beginning of this operon. To directly test whether NsrR regulates RNase R, a Western blot analysis was performed using cell extracts derived from MG1655 Δrph Δpnp and a derivative that contains a nonpolar ΔnsrR deletion, and no significant differences in the RNase R levels were found (Fig. 3B). Corroborating this result, using real-time RT-PCR, no significant differences in rnr levels were found between a strain having a ΔnsrR deletion and a strain lacking a ΔnsrR deletion or between a strain containing a multicopy nsrR plasmid and a strain containing a control plasmid (data not shown). Thus, NsrR does not appear to regulate RNase R expression. A second, more likely possibility is that the transposon insertions, through the presence of outwardly directed promoters, provide read-through transcription that elevates rnr expression. Further studies are required to ascertain whether this hypothesis is correct.
Ectopic overexpression of RNase R or RNase II suppresses the MG1655 Δrph Δpnp growth defect.
To directly test whether enhanced RNase R expression is sufficient to explain the suppressor phenotype of the nsrR transposon insertions, a plasmid containing the rnr coding region downstream of an arabinose-inducible PBAD promoter, pMPM-A5/RNR, was constructed. This plasmid, along with a control plasmid (pMPM-A5), was introduced into MG1655 Δrph Δpnp, and the transformed cells were restreaked on plates either lacking or containing arabinose. Under conditions in which PBAD expression was not induced, each strain grew poorly (Fig. 3C). However, when arabinose-supplemented plates were used, cells containing pMPM-A5/RNR grew significantly better than cells of the control strain. These experiments suggest that the suppressor effects of the mutants identified in this study can be directly attributed to increased expression of RNase R.
Because defects in MG1655 Δrph Δpnp could be suppressed by RNase R overexpression, it was of interest to investigate whether overexpression of any other hydrolytic RNase could have the same effect. To examine this, MG1655 Δrph Δpnp was transformed with a multicopy plasmid encoding RNase II (pDK24) or with a control plasmid (pBR322), and growth of the transformed strains was evaluated at 37°C. Interestingly, the strain harboring the RNase II plasmid grew significantly better than the control strain (Fig. 3D). Therefore, as was the case with RNase R, RNase II overproduction suppresses the growth defects of strains lacking phosphorolytic enzymes, suggesting that there is a common basis for suppression.
DISCUSSION
E. coli harbors several exo-RNases, six of which use water as a nucleophile to catalyze phosphodiester cleavage and two of which (PNPase and RNase PH) use inorganic phosphate. Whether the nucleophile used influences the cellular functions of these exo-RNases has been unclear in the past. To gain further insight into the phosphorolytic enzymes, a genetic screen was performed using transposon mutagenesis to identify suppressors of the growth defect of Δrph Δpnp strains. A number of distinct suppressors were found, and the majority of them mapped to nsrR (Fig. 1).
NsrR is a transcriptional repressor that contains typical DNA binding motifs and recognizes a ∼20-bp operator site. Genome-wide studies using DNA microarrays have been used to identify the genes regulated by NsrR (20). In many cases these genes correspond to genes or operons that contain a recognizable NsrR operator and/or are involved in the cellular response to reactive nitrogen species. In contrast, there is no evidence that nsrR regulates any gene involved in RNA metabolism. Therefore, it appeared to be unlikely that transposon inactivation of nsrR could be directly responsible for the improved growth of strains lacking phosphorolytic RNases. A more likely possibility was that the insertions could indirectly affect expression of rnr, which lies downstream of nsrR. This prediction was borne out by Western blot and real-time RT-PCR analyses, which showed that each of the seven distinct nsrR suppressors substantially increased the RNase R level in the cell (Fig. 3A), possibly due to enhanced rnr transcription by transposon-specific promoters. In contrast, a nonpolar ΔnsrR mutation had no effect on RNase R expression and was unable to suppress the growth defect of MG1655 Δrph Δpnp (Fig. 3B and data not shown). Additional experiments using an RNase R plasmid indicated that plasmid-directed overexpression of RNase R is sufficient to mediate the suppressor effect (Fig. 3C).
To further investigate whether the suppressor effect is specific to RNase R, Δrph Δpnp cells were transformed with a multicopy RNase II plasmid. It was observed that increased RNase II expression can also rescue the growth defects (Fig. 3D). Thus, overexpression of at least two hydrolytic RNases can suppress defects caused by the lack of phosphorolytic RNases. Interestingly, overexpression of RNase II was also recently found to suppress the growth defect of cells lacking PNPase at low temperatures (2).
The growth defects resulting from a lack of PNPase and RNase PH are reminiscent of the defects that occur in strains lacking either PNPase and RNase II or PNPase and RNase R. PNPase and RNase II together are believed to be collectively important for degrading RNA fragments generated by endo-RNases during RNA turnover, whereas PNPase and RNase R are required to digest structured RNA fragments, such as those that are formed during the degradation of defective rRNA and tRNA (12, 18). In either situation, loss of either pair of enzymatic functions causes cell lethality, indicating that the degradation of each class of RNA products is crucial for normal cellular function. Although the absence of PNPase and RNase PH has a less severe effect, a similar line of reasoning may apply to these enzymes. Thus, phosphorolytic RNases collectively could play an important role in the metabolism of yet another class of RNAs. Such RNAs could be functional RNAs whose turnover or processing is normally carried out by phosphorolytic RNases or a class of RNA fragments that are preferentially degraded by these enzymes. The studies described here suggest that although such RNAs would be preferred substrates for the phosphorolytic RNases, they can be significantly processed or degraded by RNase R or RNase II when these hydrolytic RNases are overexpressed.
In summary, the findings presented here help clarify certain aspects of RNase function. Based on the evidence, the phosphorolytic RNases collectively appear to be engaged in the metabolism of a class of RNAs that remains to be identified. However, the growth defects due to a lack of these enzymes can be suppressed by overexpression of at least two hydrolytic enzymes, which suggests that the catalytic mechanism of the phosphorolytic enzymes is not responsible for any unique cellular function. The growth defects of Δrph Δpnp strains and their suppression are correlated with increased and decreased abundance of defective ribosomes, respectively, which suggests that the primary cause of the growth defects is defective ribosome biogenesis. However, the mechanism through which the phosphorolytic RNases regulate ribosome biogenesis and cell growth remains to be elucidated.
Acknowledgments
I thank Nancy S. Gutgsell and Kevin Jagessar for assistance with ribosome analysis, Murray Deutscher and Chenglu Chen (University of Miami) for providing RNase R antibodies and purified RNase R, Kenneth Rudd (University of Miami) for providing strains, and Stephen Spiro (University of Texas, Dallas) for providing a multicopy nsrR plasmid.
This work was supported by start-up funds from the Lucille P. Markey Foundation and by a grant from the National Institutes of Health (GM-081735).
Footnotes
Published ahead of print on 17 July 2009.
REFERENCES
- 1.Andrade, J. M., and C. M. Arraiano. 2008. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA 14543-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awano, N., C. Xu, H. Ke, K. Inoue, M. Inouye, and S. Phadtare. 2007. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 1895808-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 22006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharat, A., M. Jiang, S. M. Sullivan, J. R. Maddock, and E. D. Brown. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 1887992-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairrao, F., and C. M. Arraiano. 2006. The role of endoribonucleases in the regulation of RNase R. Biochem. Biophys. Res. Commun. 343731-737. [DOI] [PubMed] [Google Scholar]
- 8.Cairrao, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 501349-1360. [DOI] [PubMed] [Google Scholar]
- 9.Chaney, S. G., J. J. Duffy, and P. D. Boyer. 1972. Patterns of oxygen interchange between water, substrates, and phosphate compounds of Escherichia coli and Bacillus subtilis. J. Biol. Chem. 2472145-2150. [PubMed] [Google Scholar]
- 10.Charollais, J., M. Dreyfus, and I. Iost. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 322751-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charollais, J., D. Pflieger, J. Vinh, M. Dreyfus, and I. Iost. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 481253-1265. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, Z. F., and M. P. Deutscher. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17313-318. [DOI] [PubMed] [Google Scholar]
- 13.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 6255-108. [DOI] [PubMed] [Google Scholar]
- 14.Condon, C. 2007. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 10271-278. [DOI] [PubMed] [Google Scholar]
- 15.Deutscher, M. P. 2006. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 34659-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutscher, M. P., and N. B. Reuven. 1991. Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 883277-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donovan, W. P., and S. R. Kushner. 1983. Amplification of ribonuclease II (rnb) activity in Escherichia coli K-12. Nucleic Acids Res. 11265-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donovan, W. P., and S. R. Kushner. 1986. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffy, J. J., S. G. Chaney, and P. D. Boyer. 1972. Incorporation of water oxygens into intracellular nucleotides and RNA. I. Predominantly non-hydrolytic RNA turnover in Bacillus subtilis. J. Mol. Biol. 64565-579. [DOI] [PubMed] [Google Scholar]
- 20.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 1894410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutgsell, N. S., M. P. Deutscher, and J. Ofengand. 2005. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 111141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jain, C. 2008. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA 14381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly, K. O., N. B. Reuven, Z. Li, and M. P. Deutscher. 1992. RNase PH is essential for tRNA processing and viability in RNase-deficient Escherichia coli cells. J. Biol. Chem. 26716015-16018. [PubMed] [Google Scholar]
- 24.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204139-180. [DOI] [PubMed] [Google Scholar]
- 25.Li, Z., S. Pandit, and M. P. Deutscher. 1998. 3′ Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. USA 952856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovgren, J. M., and P. M. Wikstrom. 2001. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 1836957-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer, M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 16341-46. [DOI] [PubMed] [Google Scholar]
- 28.McMurry, L. M., and S. B. Levy. 1987. Tn5 insertion in the polynucleotide phosphorylase (pnp) gene in Escherichia coli increases susceptibility to antibiotics. J. Bacteriol. 1691321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91457-466. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson, A. W. 1999. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 23371-390. [DOI] [PubMed] [Google Scholar]
- 31.Piedade, J., R. Zilhao, and C. M. Arraiano. 1995. Construction and characterization of an absolute deletion mutant of Escherichia coli ribonuclease II. FEMS Microbiol. Lett. 127187-193. [DOI] [PubMed] [Google Scholar]
- 32.Py, B., C. F. Higgins, H. M. Krisch, and A. J. Carpousis. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381169-172. [DOI] [PubMed] [Google Scholar]
- 33.Rosenzweig, J. A., G. Weltman, G. V. Plano, and K. Schesser. 2005. Modulation of yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280156-163. [DOI] [PubMed] [Google Scholar]
- 34.Sirdeshmukh, R., and D. Schlessinger. 1985. Ordered processing of Escherichia coli 23S rRNA in vitro. Nucleic Acids Res. 135041-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slagter-Jager, J. G., L. Puzis, N. S. Gutgsell, M. Belfort, and C. Jain. 2007. Functional defects in transfer RNAs lead to the accumulation of ribosomal RNA precursors. RNA 13597-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ygberg, S. E., M. O. Clements, A. Rytkonen, A. Thompson, D. W. Holden, J. C. Hinton, and M. Rhen. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 741243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, Z., and M. P. Deutscher. 1997. An essential function for the phosphate-dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J. Bacteriol. 1794391-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 291017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]