Abstract
Despite the fact that closely related bacteria can cause different levels of disease, the genetic changes that cause some isolates to be more pathogenic than others are generally not well understood. We use a combination of approaches to determine which factors contribute to the increased virulence of a Bordetella bronchiseptica lineage. A strain isolated from a host with B. bronchiseptica-induced disease, strain 1289, was 60-fold more virulent in mice than one isolated from an asymptomatically infected host, strain RB50. Transcriptome analysis and quantitative reverse transcription-PCR showed that the type III secretion system (TTSS) genes were more highly expressed by strain 1289 than strain RB50. Compared to strain RB50, strain 1289 exhibited greater TTSS-mediated cytotoxicity of a mammalian cell line. Additionally, we show that the increase in virulence of strain 1289 compared to that of RB50 was partially attributable to the TTSS. Using multilocus sequence typing, we identified another strain from the same lineage as strain 1289. Similar to strain 1289, we implicate the TTSS in the increased virulence of this strain. Together, our data suggest that the TTSS is involved in the increased virulence of a B. bronchiseptica lineage which appears to be disproportionately associated with disease. These data are consistent with the view that B. bronchiseptica lineages can have different levels of virulence, which may contribute to this species' ability to cause different severities of respiratory disease.
Although the disease caused by different strains of pathogenic bacteria is known to vary, the molecular basis for these differences has been difficult to disentangle from the many other genetic changes that occur as strains diverge. Recently, a growing number of studies have identified factors that contribute to increased virulence of bacterial lineages by using a combination of genome-wide analyses, phylogenetics, mutational analysis, and host infection models (15, 46, 49-51, 56, 58). Horizontal gene transfer of novel virulence factors, phage integration, phenotypic variation, gene loss, and mutation have been shown to alter the phenotype or severity of disease (15, 51, 56).
Bordetella bronchiseptica is a gram-negative respiratory pathogen that infects a wide range of mammals and is closely related to Bordetella pertussis and Bordetella parapertussis, the causative agents of whooping cough in humans (18, 33, 36). Colonization of hosts by B. bronchiseptica can lead to a range of diseases, from lethal pneumonia to asymptomatic infection (18), which is thought to be caused by differences in host immune status, polymicrobial infection, and/or bacterial strain variation (18, 33). However, in inbred and specific-pathogen-free mice, the 50% lethal dose (LD50) can still differ by up to 100,000-fold between bacterial strains, suggesting that substantial differences in virulence may be due to strain variation alone (5, 19, 20).
While the population structure of these bacteria appears to be clonal, B. pertussis and B. parapertussis are more monomorphic than B. bronchiseptica strains, and isolates of this species can be related more distantly to each other than to either of the human-associated pathogens (11, 36, 37, 60). Previously, it was shown that differences in gene regulation between B. bronchiseptica strains can correlate with phylogenetic lineage (17) and strains can differ in virulence factor expression (3, 19, 31, 36, 47). Recently, we showed that phylogenetic lineages can differ in virulence factor expression and virulence, as a lineage of B. bronchiseptica was found to lack expression of adenylate cyclase toxin and exhibit decreased virulence (5).
B. bronchiseptica strains express many virulence factors, including adhesins, secretion systems, autotransporters, and toxins, that are globally regulated by the BvgAS two-component signal transduction system (7, 53). In the nonvirulent, or Bvg− phase, which occurs at 25°C or in the presence of chemical modulators such as MgSO4 or nicotinic acid, BvgAS is unable to activate virulence-associated genes (10, 35, 38). During the virulent, or Bvg+ phase, which occurs when the bacteria are grown at 37°C and in the absence of chemical modulators, BvgAS activates the expression of a large set of virulence factors (7, 10, 38). The Bvg+ phase is both necessary and sufficient for colonization of the respiratory tract (1, 8). Among the Bvg-regulated genes are those encoding a type III secretion system (TTSS) which is similar to others shown to directly translocate effector proteins through a needle-like injection apparatus directly into eukaryotic cells, disrupt host cell signaling, and induce necrotic-like cell death (27, 62). Under Bvg+ conditions, the btr regulatory locus (including btrS, btrU, btrW, and btrV) is transcribed (34). BtrS is an ECF sigma factor that is necessary and sufficient for activating the more than 20 TTSS-related genes (bsc, bop, bsp, and bte) (34). BscN is the putative ATPase that provides energy for the secretion of effector proteins and is required for the function of the TTSS apparatus (62). TTSS gene products BopB, BopC, and BteA have been shown to be secreted and required for cytotoxicity in mammalian cells (28, 29, 41). In a murine model of infection, the TTSS increases host interleukin-10 production and enhances persistence of B. bronchiseptica in the lower respiratory tract (44, 61, 62).
While correlations between the level of virulence or disease caused by B. bronchiseptica strains and specific bacterial factors have been made (5, 39, 47), limited studies have directly tested whether these factors cause some strains to be more virulent than others (4) and whether these characteristics are associated with a particular phylogenetic lineage. Here, we use genome-wide analyses, phylogenetics, allelic exchange, and a murine model of infection to determine the bacterial factors that contribute to the increased virulence of a B. bronchiseptica lineage. We compared the relative virulence, as measured by LD50, of B. bronchiseptica strains from a diseased (strain 1289) or asymptomatically infected (strain RB50) host (8). Strain 1289 was approximately 60-fold more virulent than strain RB50. Transcriptome analysis showed that TTSS-related genes were more highly expressed by strain 1289 than RB50. The TTSS-mediated cytotoxicity and virulence of 1289 was greater than that of strain RB50. Using multilocus sequence typing (MLST) analysis, we identified another strain from the same sequence type (ST) as strain 1289 and showed that, similar to strain 1289, the increased virulence of this strain was partially attributable to the TTSS. Together, our data indicate that the TTSS is involved in the increased virulence of a B. bronchiseptica lineage. This is consistent with the idea that different phylogenetic lineages can differentially regulate their virulence factors to modulate their overall level of virulence, which may contribute to the ability of B. bronchiseptica strains to cause different severities of respiratory disease.
MATERIALS AND METHODS
Bacterial strains and growth.
B. bronchiseptica isolates, sources, locations, dates, anatomical sites of isolation, references, and all available health and necropsy reports (strains 1289, S308, and S314) are included herein or in Table S1 in the supplemental material or have been previously described (strain RB50) (8). In cases where there was an available health report, strains were grouped as those from diseased hosts (i.e., having lower respiratory tract infection requiring medical treatment or causing death) or strains isolated from a nondiseased host (i.e., having an asymptomatic infection) (see Table S1 in the supplemental material). For the rest of the isolates studied here, a more detailed clinical report was not available. The effect of in vitro passaging on B. bronchiseptica strains has been kept to a minimum, as all the isolates used in our murine model of infection in this study have been minimally passaged (estimated to be less than five times). All strains were maintained on Bordet-Gengou (BG) agar (Difco, Sparks, MD) containing 10% sheep's blood (Hema Resources, Aurora, OR) with 20 μg/ml streptomycin (Sigma, St. Louis, MO). For inoculation, bacteria were grown overnight at 37°C in Stainer-Scholte (SS) broth (51a) to logarithmic phase, bacterial density was measured by optical density read at 600 nm (OD600), and bacteria were diluted in sterile phosphate-buffered saline (Omnipur, Gibbstown, NJ) to the appropriate concentration. Inocula were confirmed by plating dilutions on BG agar and counting the resulting colonies after 2 days of incubation at 37°C, as previously described (22, 26). Because the Bvg+ phase is required for colonization of the respiratory tract, only strains that appeared Bvg+ (domed and β-hemolytic) were used in the murine model of infection. Strains that exhibited rough colony morphology and lacked β-hemolysis, an indication of avirulent Bvg− mutants which commonly occur upon in vitro passaging, were excluded from this type of analysis (52). For this reason, strains 1973 and 1987 were not analyzed in vivo.
Animal experiments.
Four- to 6-week-old C57BL/6 mice were obtained from Jackson Laboratories at The Pennsylvania State University. All experiments were completed in accordance with institutional guidelines. For inoculation, mice were lightly sedated with 5% isofluorane (IsoFlo; Abbott Laboratories) in oxygen and the indicated number of CFU of the appropriate B. bronchiseptica strain in 50 μl of phosphate-buffered saline was gently pipetted onto the external nares as previously described (22, 26). For survival curves, groups of three or four mice were inoculated with the indicated dose and percent survival was monitored over a 28-day period. Mice with lethal bordetellosis, indicated by ruffled fur, labored breathing, and diminished responsiveness, were euthanized to alleviate unnecessary suffering (22, 32, 43). Statistical significance was calculated using a Fisher's exact test where groups of mice were compared in terms of survival or death at a similar dose of two different strains, as previously described (5). To quantify the number of bacteria in the lungs, trachea, and nasal cavity, groups of three or four mice were inoculated with a sublethal dose (1 × 104 CFU) and sacrificed at the time points indicated below. Bacterial numbers in all respiratory organs were quantified as previously described (22, 25). The mean ± standard error of the results was determined for each treatment group. Statistical significance in bacterial load between strains was calculated by using analysis of covariance in Minitab (version 13.30; Minitab., Inc.). The explanatory variable used was the bacterial strain, and a covariate for day was fitted to control for the change in load over time (5). We report F values, the test statistic for analyses of covariance, as well as P values, for full statistical disclosure (40). A P value of ≤0.05 was taken as statistically significant. All animal experiments were repeated at least twice with similar results.
Expression arrays and statistical analysis.
The expression array was carried out as previously described (5, 38). Briefly, bacteria were grown in SS broth, subcultured at a starting OD600 of 0.02 into 50 ml of SS broth, grown at 37°C for 24 h with shaking, and harvested in log phase (OD600 of 1.0). Total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA), treated with RNase-free DNase I (Invitrogen, Carlsbad, CA), and purified using RNeasy columns (Qiagen, Valencia, CA) according to the manufacturer's instructions. RNA was isolated from two independent biological replicates of strains RB50 and 1289. A two-color hybridization format was used, and dye swap experiments were performed. For each reaction mixture, 5 μg of cDNA was fluorescently labeled. The two differentially labeled reaction mixtures to be compared were combined and hybridized to a B. bronchiseptica strain RB50-specific long-oligonucleotide microarray (5, 38). The slides were then scanned using a GenePix 4000B microarray scanner and analyzed with GenePix Pro software (Axon Instruments, Union City, CA). The spots were assessed visually to identify those of low quality, and the arrays were normalized so that the median of the ratio across each array was equal to 1.0. Spots of low quality were identified and were filtered out prior to analysis. Ratio data from the two biological replicates were compiled and normalized based on the total Cy3 percent intensity and Cy5 percent intensity to eliminate slide-to-slide variation. Gene expression data were then normalized to the expression of 16S rRNA. The statistical significance of the gene expression changes observed was assessed by using the significance analysis of microarrays (SAM) program (59). A one-class unpaired SAM analysis using a false discovery rate of 0.30% (<0.1%) was performed. All microarray expression data are available in Table S2 in the supplemental material.
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) was completed as previously described (5, 38), and RNA was extracted as described for the microarray experiment. One microgram of RNA from each biological replicate was reverse transcribed using 300 ng of random oligonucleotide hexamers and SuperScript III RTase (Invitrogen, Carlsbad, CA). The resulting cDNA was diluted 1:1,000, and 1-μl amounts were used in qRT-PCR mixtures containing 300 nM primers that were designed with Primer Express software (Applied Biosystems, Foster City, CA) and 2× SYBR green PCR master mix (Applied Biosystems, Foster City, CA). To confirm the lack of DNA contamination, reactions of mixtures without reverse transcriptase were completed. Dissociation curve analysis was performed to verify product homogeneity. Threshold fluorescence was established within the geometric phase of exponential amplification, and the cycle threshold (CT) determined for each reaction mixture. The CT from all biological replicates for each strain was compiled, and the 16S RNA amplicon was used as an internal control for data normalization. The change in transcript level was determined by using the relative quantitative CT method (ΔΔCT) (48). All primer sequences and changes in gene expression analyzed by qRT-PCR are available (see Table S2 in the supplemental material).
CGH and statistical analysis.
Comparative genomic hybridization (CGH) analysis was completed as previously described (5). Briefly, strains RB50 and 1289 were grown in SS broth at 37°C with shaking overnight and genomic DNA was isolated from bacterial cultures by using a DNA extraction kit (Qiagen, Valencia, CA) and digested with DpnII. For each labeling reaction, 2 μg of digested genomic DNA was randomly primed using Cy5 and Cy3 dye-labeled nucleotides with BioPrime DNA labeling kits (Invitrogen, Carlsbad, CA), and the two differentially labeled reaction mixtures to be compared were combined and hybridized to a B. bronchiseptica RB50-specific long-oligonucleotide microarray (5, 38). Dye swap experiments were performed. Statistical analysis of CGH data was performed with SAS software version 9.1.3 (SAS Institute, Inc., Cary, NC). A MODECLUS procedure (PROC MODECLUS) based on nonparametric density estimation was used to cluster genes into divergent or nondivergent groups. All CGH data are available in Table S3 in the supplemental material.
Deletion of bscN in B. bronchiseptica.
The gene encoding the ATPase that provides energy for secretion of proteins via the TTSS, bscN, was deleted from strain RB50 as previously described (62). The deletion of bscN from strains 1289 and S308 was completed as follows. The 419 base pairs upstream and the first three codons of the bscN gene were PCR amplified using primers flanked with EcoRI on the 5′ end and BamHI on the 3′ end (F-ATCGAATTCCGGATCAGGCGGAGAAGA and R-TAAGGATCCCTGACGCATGCCCCTATC, respectively). The 420 base pairs downstream and the last three codons of the bscN gene were PCR amplified using primers flanked with BamHI on the 5′ end and EcoRI on the 3′ end (F-CGCGGATCCGAATCCTAATGGACCTGG and R-TAGGAATTCTCCAGGCTCTCGCGCAAG, respectively). The PCR conditions were 95°C for 5 min; 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 1 min; and 72°C for 5 min. These fragments were PCR purified (Qiagen, Valencia, CA), BamHI digested (New England Biolabs), gel purified (Qiagen, Valencia, CA), and ligated overnight at 4°C (New England Biolabs). The ligation product was then amplified with the 5′ F and 3′ R primers as described above. The 846-bp product was ligated into the TOPO-TA vector and transformed into Mach1 DH5α cells (Invitrogen, Carlsbad, CA). The presence of the insert in TOPO-TA was screened for by the loss of β-galactosidase activity and EcoRI digestion of the plasmid from resulting transformants. The 838-bp insert was digested from TOPO-TA, gel purified, and ligated overnight into the EcoRI-digested pSS4245, a new Bordetella allelic exchange vector (S. Stibitz, unpublished data). The ligation product was transformed as described above. The presence of the insert in pSS4245 was screened for by extracting the plasmid from the resulting transformants and digesting it with EcoRI. The resulting positive clone was named pSS4245ΔbscN. The positive clones were sequenced after insertion into TOPO-TA and pSS4245 to ensure that PCR-induced mutations did not occur. DH5α harboring pSS4545ΔbscN or a plasmid competent for mating, pSS1827 (54), and the appropriate B. bronchiseptica strain grown under Bvg− conditions by growth on BG plus 50 mM MgSO4 was mated for 4 h on a BG-10 mM MgCl2-50 mM MgSO4 plate at 37°C. Then, B. bronchiseptica containing pSS4245ΔbscN was positively selected for by using BG-streptomycin-kanamycin-50 mM MgSO4 plates and incubated for 5 days at 37°C. The resulting B. bronchiseptica colonies were streaked onto BG-streptomycin-kanamycin-50 mM MgSO4 and grown at 37°C for 4 days. The resulting colonies were streaked onto BG plates and incubated for 2 days at 37°C, which resulted in colonies lacking pSS4245 and containing either the wild-type or knockout gene. Colonies were screened for the presence of either the wild-type or knockout gene by using screening primers (F-ATCGACTACTTCGCGGGTATCGAGAA and R-GAGCAGCTGGATTTCATGCTCGTG) which detected either the wild-type bscN gene (2,003 bp) or the bscN deletion (678 bp) with PCR conditions of 95°C for 5 min; 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 1 min; and 72°C for 5 min. To further confirm the presence and absence of the bscN gene, screening primers which amplify the middle of the bscN gene and could therefore only amplify the wild-type gene (1,071 bp) were used (F-GAACGATCATCAAGGCCGTCGTTC and R-GTCCTGGTACTTGGCCATCAGTTC). The absence of pSS4245 was confirmed by growth on BG-streptomycin plates and lack of growth on BG-kanamycin plates.
Cytotoxicity assay.
Cytotoxicity assays were carried out as previously described (34, 62). Briefly, J774 macrophages were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, 1% nonessential amino acids, and 1% sodium pyruvate to 85% confluence in 5% CO2 at 37°C. Then, warmed RPMI medium lacking phenol red and with 5% fetal bovine serum, 1% l-glutamine, 1% nonessential amino acids, and 1% sodium pyruvate was used to replace the Dulbecco's modified Eagle's medium. Bacterial infections were carried out using a multiplicity of infection (MOI) of 10, and bacterial suspensions were centrifuged onto the macrophage cells at 250 × g for 5 min and incubated in 5% CO2 at 37°C for the amount of time indicated below. The cell culture supernatants were collected, and the percent lipodehydrogenase (LDH) release was analyzed by using a Cytotox96 kit (Promega) according to the manufacturer's instructions. Statistical significance in percent cytotoxicity between strains was calculated by using a Tukey simultaneous test in Minitab (version 13.30; Minitab, Inc.) (40). A P value of ≤0.05 was taken as statistically significant.
MLST and phylogenetic tree construction.
MLST analysis was performed as previously described (5, 11). In this study, the STs of three strains (S308, S314, and 973) were determined (see Table S1 in the supplemental material). All alleles were double-strand sequenced at The Pennsylvania State University's Sequencing Center. The sequences were trimmed, and alleles and STs were designated by using the Bordetella MLST database (http://pubmlst.org/bordetella) (5, 11, 23). Using MEGA 4.0, the alleles were concatenated and aligned, and an unweighted pair group method with arithmetic mean tree with 1,000 bootstraps using the K2 model was constructed for these 3 strains and for 58 strains whose STs were previously determined (5, 11, 57) (see Table S1 in the supplemental material).
Accession numbers.
All microarray expression data have been deposited in MAIMExpress under the accession number E-MEXP-1736, and all CGH data have been deposited in MIAMExpress under the accession number E-MEXP-1737.
RESULTS
B. bronchiseptica strain 1289 is more virulent than strain RB50 in a mouse intranasal challenge model.
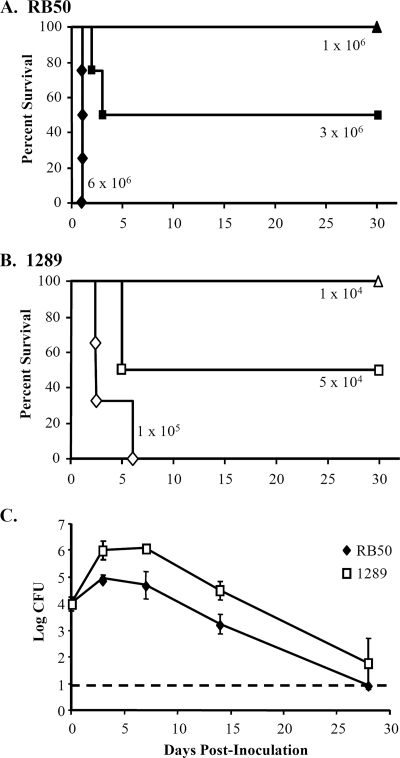
Previous studies have shown that B. bronchiseptica strains can vary widely in virulence (5, 19, 20). To establish a system in which we could measure the contribution of specific factors to the difference in virulence between strains, we compared the LD50, a measure of their virulence, of B. bronchiseptica strains in inbred, specific-pathogen-free mice. B. bronchiseptica strain RB50 was isolated from the nasal cavity of an asymptomatically infected host (8). Consistent with previous studies of this strain, inoculation with 1 × 106, 3 × 106, or 6 × 106 CFU of B. bronchiseptica strain RB50 led to 100%, 50%, and 0% survival, respectively (Fig. 1A) (5). B. bronchiseptica strain 1289 was isolated from the thoracic cavity of a host with a lethal B. bronchiseptica infection (see Table S1 in the supplemental material). When inoculated with 1 × 104, 5 × 104, or 1 × 105 CFU of strain 1289, 100%, 50%, and 0% of the mice survived the infection, respectively (Fig. 1B), which indicates that the LD50 of strain 1289 is 60-fold lower, or 2.95 million CFU fewer, than that of strain RB50. Although these two isolates did not differ in growth rate in vitro (data not shown), we examined whether the greater virulence of strain 1289 might allow it to colonize the respiratory tract to a higher level than strain RB50. Groups of 15 mice were inoculated with a sublethal dose (1 × 104 CFU) of either strain RB50 or 1289, and respiratory organs were excised to quantify bacterial loads on days 0, 3, 7, 14, and 28 postinoculation (Fig. 1C). In the lungs, strain RB50 peaked at approximately 1 × 105 CFU and was reduced to below the limit of detection (10 CFU) by day 28 postinoculation (Fig. 1C). The bacterial load of strain 1289 was approximately 10-fold higher than that of strain RB50 over the course of infection (Fig. 1C) (for bacterial strain comparisons, F1.27 = 7.03 and P = 0.013). Even 24 h postinoculation, the bacterial load of strain 1289 was 10-fold higher than that of strain RB50 (P = 0.002), indicating that the numbers of strain 1289 are higher than the numbers of strain RB50 early after infection (data not shown). The bacterial load did not differ significantly between strain RB50 and 1289 in the trachea or nasal cavity over the course of infection (see Fig. S4 in the supplemental material). Combined, these data indicate that strain 1289 is more virulent than strain RB50 and colonizes the lower respiratory tract at a higher level.
FIG. 1.
LD50s and quantification of lung bacterial loads of B. bronchiseptica strains RB50 and 1289. Groups of three or four C57BL/6 mice were inoculated intranasally with the indicated doses of strains RB50 (A) or 1289 (B). Survival curves were generated by inoculating mice with the indicated dose and determining the percent survival over a 28-day period. (C) Groups of three mice were inoculated intranasally with 1 × 104 CFU of strain RB50 or strain 1289. Bacterial loads in the lungs were quantified 0, 3, 7, 14, and 28 days postinoculation. The dashed line indicates the lower limit of detection. Bacterial numbers are expressed as the log10 means ± standard errors (error bars).
TTSS genes are upregulated in strain 1289.
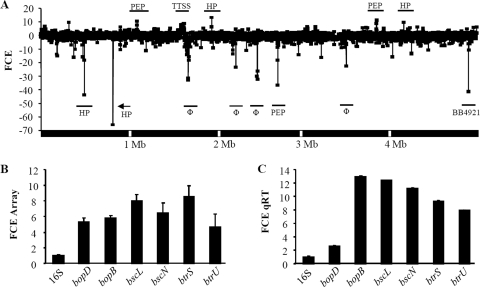
To identify candidate bacterial genes that correlated with the increased virulence of strain 1289, a comparison of whole-genome transcriptome analyses was performed between strains 1289 and RB50 (Fig. 2A). Of the 5,013 genes represented on the RB50-specific microarray, 646 were downregulated in strain 1289 relative to their expression levels in strain RB50. These included 49 transporter genes; 47 metabolism-related transcripts; 51 transcriptional regulator genes; 27 electron transporter genes; 11 two-component system genes; 7 transcriptional or translational genes; 1 protein biosynthesis-related transcript; 74 exported or membrane protein genes; 72 phage-related transcripts; and 202 hypothetical, predicted, or probable genes. Additionally, five genes classified as virulence factors were downregulated two- to eightfold in strain 1289 compared to their levels of expression in strain RB50; these were genes for filamentous hemagglutinin B (fhaB), filamentous hemagglutinin S (fhaS), Bordetella colonization factor (bcfA), Bordetella resistance to killing A (brkA), and one O-antigen-related gene (wbmS) (Fig. 2A) (see Table S2 in the supplemental material) (6, 9, 13, 24, 55). The downregulated expression of brkA and fhaB in strain 1289 was confirmed by qRT-PCR (see Table S2 in the supplemental material). When CGH analysis was completed on these strains, none of the known virulence factors were identified as divergent in strain 1289 (see Table S3 in the supplemental material), suggesting that the decreased signal of these virulence factors in strain 1289 identified in our transcriptome analysis is due to downregulation rather than sequence divergence.
FIG. 2.
Whole-transcriptome and TTSS expression analysis of B. bronchiseptica strains RB50 and 1289. (A) Comparison of whole-transcriptome analyses between strains RB50 and 1289. The x axis indicates the order of genes along the B. bronchiseptica strain RB50 5.3-megabase (Mb) chromosome. The y axis indicates the change in expression level (as fold change in expression [FCE]) of each gene. Negative FCE values indicate decreased gene expression of genes in strain 1289 compared to their levels of expression in strain RB50, and positive FCE values indicate increased gene expression in strain 1289 compared to their levels in strain RB50. Genes of interest are labeled, with corresponding underscores. HP, hypothetical protein gene; Φ, phage-related gene; PEP, putative exported protein gene; BB4921, putative ferredoxin gene. (B and C) Comparison of TTSS-related gene expression between strains RB50 and 1289 by microarray analysis (B) and qRT-PCR (C). The x axis indicates the genes analyzed. The y axis indicates the FCE in strain 1289 over the expression level in strain RB50. Error bars represent the plus-or-minus standard errors in panels A and B and the standard deviation in panel C.
We were particularly interested in determining which genes were upregulated in strain 1289, as they might contribute to the greater virulence of this strain. Six hundred seven genes were identified as upregulated in strain 1289 relative to their expression levels in strain RB50. These included 60 transporter genes; 67 metabolism-related transcripts; 23 transcriptional regulator genes; 42 electron transporter genes; 7 two-component system genes; 16 transcriptional or translational genes; 40 protein biosynthesis-related transcripts; 75 exported or membrane protein genes; 16 phage-related transcripts; and 154 hypothetical, predicted, or probable genes. Thirty-three genes associated with known virulence factors were upregulated in strain 1289 compared to their expression levels in strain RB50. Four of these, the genes for cyclolysin-activating lysine-acyltransferase (cyaC), Bordetella resistance to killing B (brkB), an O-antigen-related protein (wbmJ), and pertussis toxin subunit 4 precursor (ptxD), were upregulated by 1.8-fold or more in strain 1289 (Fig. 2A) (see Table S2 in the supplemental material). The remaining 29 virulence-associated genes are related to the TTSS and were upregulated from 1.4- to 8.5-fold in strain 1289 over their expression levels in strain RB50 (Fig. 2A and B). As expected, there was a strong correlation between expression levels analyzed by microarray and qRT-PCR results (R = 0.914) (see Table S2 in the supplemental material). Although qRT-PCR did not confirm the upregulation of cyaC, all the TTSS-related genes examined were upregulated 2.6- to 12.8-fold in strain 1289 compared to their levels of expression in strain RB50 (Fig. 2C; see also Table S2 in the supplemental material). The upregulation of these virulence factor genes in strain 1289 does not appear to be due to gene duplication, as no genes were identified as duplicated in CGH analysis (see Table S3 in the supplemental material).
The TTSS is involved in the increased cytotoxicity and virulence of strain 1289.
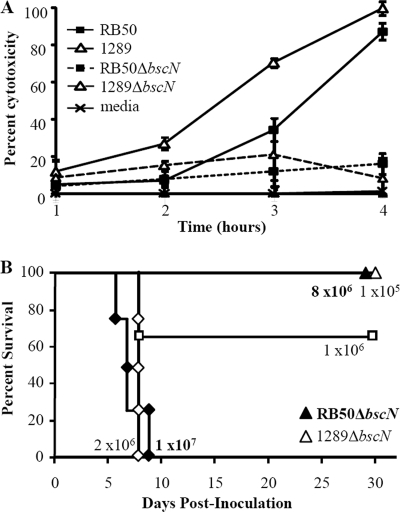
The increased expression of such a large set of genes with a known coordinated function in virulence led us to hypothesize that strain 1289 exhibited greater TTSS-mediated effects than strain RB50. One well-described function attributed to the TTSS is cytotoxicity for a variety of mammalian cells (14, 34, 62). Since nearly all TTSS-related genes are upregulated in strain 1289, we hypothesized that this strain may cause more TTSS-mediated cytotoxicity than strain RB50. Although J774 macrophages treated with medium alone did not release cytoplasmic LDH, infection of these macrophages with strain RB50 at an MOI of 10 caused 5%, 7%, 34%, and 87% of their LDH to be released after 1, 2, 3, and 4 h of infection, respectively, indicating that RB50 is cytotoxic toward macrophages (P = 0.0001) (Fig. 3A), as previously reported (34, 62). Upon infection with the same dose of strain 1289, the percent LDH release was higher than that caused by strain RB50 (P = 0.0360), which indicates that strain 1289 causes more rapid cytotoxicity of macrophages than strain RB50 (Fig. 3A). To determine if the cytotoxicity induced by these strains is caused by the TTSS, isogenic mutants each lacking the bscN gene (RB50ΔbscN and 1289ΔbscN) were used to compare their cytotoxicity for J774 macrophages (Fig. 3A). The percent LDH release caused by infection with RB50ΔbscN was lower than that caused by its parental strain, RB50, and was not significantly different from that of the medium control (P = 0.0047 and P = 0.5758, respectively), which confirms that the TTSS of strain RB50 causes cytotoxicity toward macrophages (Fig. 3A) (21, 62). Similarly, the percent LDH release caused by infection with 1289ΔbscN was lower than that caused by its wild-type counterpart and was not significantly different from that in the medium control or RB50ΔbscN (P < 0.0001, P = 0.2960, and P = 0.9857, respectively), the former of which indicates that strain 1289 does not have a measurable TTSS-independent mechanism of cytotoxicity (Fig. 3A). The greater TTSS-dependent cytotoxicity caused by strain 1289 at earlier time points suggests that strain 1289 causes more rapid TTSS-mediated cytotoxicity than strain RB50.
FIG. 3.
TTSS-mediated effect on cytotoxicity and virulence of B. bronchiseptica strains RB50 and 1289. (A) Cytotoxicity in J774 macrophages treated with medium or infected with RB50, RB50ΔbscN, 1289, or 1289ΔbscN for 1, 2, 3, and 4 h at an MOI of 10. The error bars represent the plus-or-minus standard deviations. (B) Groups of three or four C57BL/6 mice were inoculated intranasally with the indicated doses of strains RB50ΔbscN or 1289ΔbscN. Survival curves were generated by inoculating mice with the indicated dose and determining the percent survival over a 28-day period.
Since the TTSS causes cytotoxicity in vitro and increases bacterial numbers in vivo (21, 44), we hypothesized that the TTSS contributes more to the virulence of strain 1289 than to that of strain RB50. To test this, mice were inoculated with the bscN deletion strains of RB50 and 1289 and the LD50s of these isogenic mutant strains were determined. When inoculated with 8.0 × 106 or 1.0 × 107 CFU of RB50ΔbscN, 100% and 0% of the mice survived the infection, respectively (Fig. 3B), indicating that the LD50 of strain RB50ΔbscN is approximately 9.0 × 106 CFU, threefold greater than the LD50 of strain RB50 (Fig. 3B and 1A). When mice were inoculated with 1 × 105, 1.0 × 106, or 2.0 × 106 CFU of 1289ΔbscN, 100%, 66%, and 0% of the mice survived the infection, respectively (Fig. 3B), indicating that the LD50 of strain 1289ΔbscN is approximately 1.2 × 106 CFU, 24-fold greater than that of strain 1289 (Fig. 3B and 1B). Therefore, the TTSS appears to contribute more to the virulence of strain 1289 than to that of strain RB50 (Fig. 1A, 1B, and 3B). Since the LD50 of 1289ΔbscN is lower than that of RB50ΔbscN, it suggests that another factor besides the TTSS also contributes to the increased virulence of strain 1289 (Fig. 3B). Together, these data indicate that while the TTSS is not the sole factor, it is partially responsible for the increased virulence of strain 1289 compared to the virulence of strain RB50.
The TTSS is implicated in the increased virulence of ST32 strains.
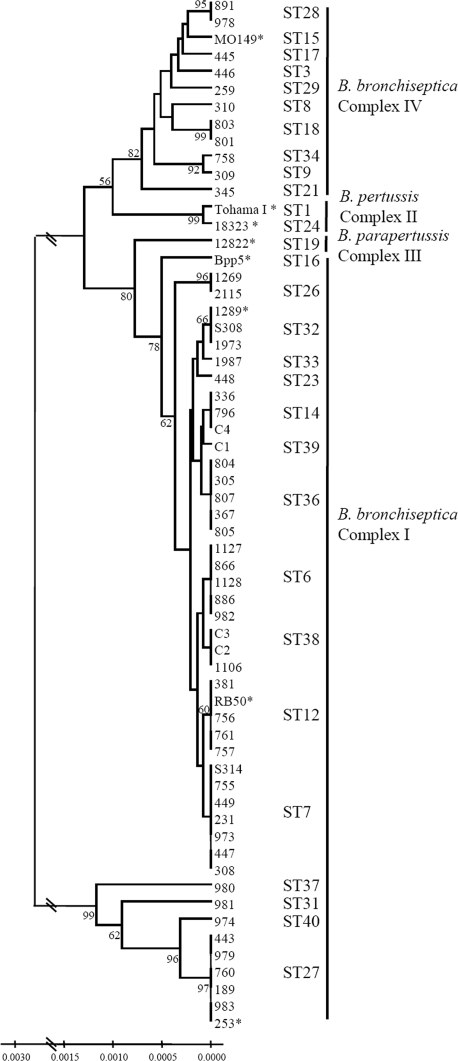
To examine whether isolates associated with B. bronchiseptica-related disease were of the same phylogenetic lineage, we completed MLST and phylogenetic analyses using two B. bronchiseptica strains from diseased hosts to determine if they fell into the same ST as strain 1289. In addition to these three disease-associated isolates, our analysis also included 58 additional Bordetella isolates, of which 55 were B. bronchiseptica strains (none known to be associated with diseased hosts and all from a broad range of locations, dates, and hosts), 2 B. pertussis strains, and 1 B. parapertussis strain (Fig. 4; see also Table S1 in the supplemental material) (5, 11). These 58 isolates served to demonstrate the genetic relatedness of strains RB50 and 1289 and to confirm the evolutionary history of the classical bordetellae. Consistent with other phylogenetic analyses, both B. pertussis and B. parapertussis appear to have evolved independently from B. bronchiseptica-like progenitors (Fig. 4) (11, 42, 60). As previously described, strains RB50 and 1289 were identified as ST12 and ST32 isolates, respectively (Fig. 4) (5, 11). Importantly, strains belonging to the same STs as strains RB50 (11) and 1289 (Fig. 4; see also Table S1 in the supplemental material) have been isolated from several continents, suggesting that these two STs exist worldwide. Two isolates, strains S308 and S314, were selected for MLST analysis because they were collected from hosts with B. bronchiseptica-induced disease (see Table S1 in the supplemental material). While one of these strains, S314, belongs to ST7, the other strain, S308, belongs to the same ST as strain 1289 (Fig. 4). These data suggest that not all strains causing B. bronchiseptica-induced disease are of the same phylogenetic lineage. However, within the ST32 lineage, both strains with an accompanying pathology report (strains 1289 and S308) (Fig. 4) were associated with B. bronchiseptica-induced disease, suggesting that ST32 constitutes a more virulent lineage.
FIG. 4.
MLST analysis of 61 Bordetella strains. Unweighted pair group method with arithmetic mean tree with 1,000 bootstraps based on concatenated MLST gene sequences of 61 Bordetella isolates (58 B. bronchiseptica, 2 B. pertussis, and 1 B. parapertussis isolates). The identification number of each strain is listed. The asterisks indicate strains that have undergone or are undergoing full-genome sequencing (42). The ST is labeled next to each strain and the complex is labeled next to each set of STs (complex I, which includes B. bronchiseptica strains, complex II, which includes B. pertussis strains, complex III, which includes B. parapertussis strains, and complex IV, which includes B. bronchiseptica strains that appear to be most closely related to B. pertussis) as previously described (5, 11). The numbers on the tree branches indicate branch strength. All branch strengths below 50 were removed. The scale indicates the relative genetic distances along the branches.
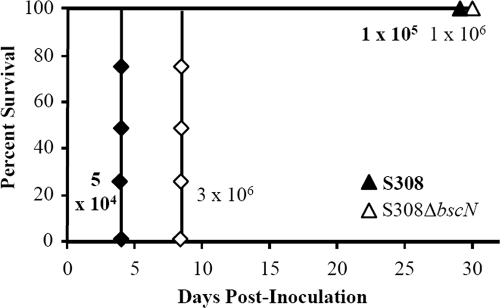
To determine if ST32 strains share increased virulence, groups of three or four mice were inoculated with different doses of strain S308 and were monitored for survival (see criteria for strain selection in Materials and Methods) (Fig. 5). While 0% of the mice survived an inoculation of 1 × 105 CFU, 100% survived an inoculation with 5 × 104 CFU, suggesting that the LD50 is approximately 7.5 × 104 CFU (Fig. 5), similar to that of strain 1289 (5 × 104 CFU) and approximately 40-fold lower than that of strain RB50 (3 × 106 CFU) (Fig. 1A, 1B, and 5). To determine if the TTSS contributed to the increased virulence of strain S308, we deleted the bscN gene from this strain and determined the LD50. When inoculated with 3 × 106 or 1 × 106 CFU of S308ΔbscN, 0% and 100% of the mice, respectively, survived the infection (Fig. 5). Therefore, the LD50 of this strain is approximately 2 × 106 CFU, which is 27-fold higher than that of its parental wild-type strain (Fig. 5). These data indicate that the TTSS contributes more to the virulence of strain S308 than to that of strain RB50 (Fig. 5 and 3B). Similar to strain 1289ΔbscN, the LD50 of S308ΔbscN is lower than that of RB50ΔbscN, which suggests that another factor besides the TTSS also contributes to the increased virulence of strain S308. Together, these data suggest that the increased virulence of ST32 strains is partially dependent on the TTSS.
FIG. 5.
TTSS-mediated effect on virulence of B. bronchiseptica strain S308, a ST32 isolate. Groups of three or four C57BL/6 mice were inoculated intranasally with the indicated doses of strain S308 or S308ΔbscN. Survival curves were generated by inoculating mice with the indicated dose and determining the percent survival over a 28-day period.
MLST analysis has been completed for approximately 260 Bordetella strains, the vast majority of which are B. bronchiseptica (S.E. Hester, K. E. Creppage, M. C. Dunagin, K. Register, and E. T. Harvill, unpublished data) (5, 11). Of these, only three ST32 strains have been identified (Fig. 4), which suggests that while these strains exist worldwide (see Table S1 in the supplemental material), this ST may not contain as many strains as other STs. Therefore, we wanted to determine if other strains closely related to ST32 are more virulent than strain RB50. We analyzed strain 448 from ST23, as it was the strain most closely related to ST32 and having Bvg+ morphology that was available at the time of this study (Fig. 4). The LD50 of strain 448 was approximately 1 × 106 CFU, threefold lower than that of strain RB50 (data not shown), and its bacterial loads in the lung were higher than those of strain RB50 (see Fig. S4 in the supplemental material) over the course of the infection. Therefore, these data suggest that strains closely related to ST32 are also more virulent than strain RB50 and may represent lineages of increased virulence.
DISCUSSION
The severity of a B. bronchiseptica infection can range from long-term asymptomatic carriage in the upper respiratory tract to fatal pneumonia (18). While previous studies have correlated differences in virulence or severity of disease to particular bacterial factors (5, 39, 47), few studies have shown that these factors actually contribute to the virulence of particular lineages (4). Here, we identify a bacterial factor that contributes to the increased virulence of a B. bronchiseptica lineage by combining comparative genomic analyses, bacterial mutagenesis, phylogenetics, and a host infection model. B. bronchiseptica strain RB50, which was isolated from an asymptomatically infected host, was less virulent than strain 1289, which was isolated from a diseased host (Fig. 1; see also Table S1 in the supplemental material) (8). Transcriptome analysis revealed that TTSS-related genes were more highly expressed in strain 1289 than in strain RB50 (Fig. 2). Using allelic exchange, we determined that the TTSS causes more-rapid cytotoxic effects in macrophages, that there was negligible cytotoxicity in its absence, and that it contributes more to the virulence of strain 1289 than to that of strain RB50 (Fig. 3). When assessing another strain that belonged to the same ST as strain 1289 and was also associated with B. bronchiseptica-induced disease, we found that the increased virulence of this strain was also partially attributable to the TTSS (Fig. 4 and 5). Combined, these data suggest that the TTSS is involved in the increased virulence of a B. bronchiseptica lineage.
The amount of genomic content shared between strains of a single microbial species can vary substantially (30). The “core genome” represents all genes shared between strains of the same species, while the “flexible genome” includes those genes that are variably present. The flexible genome is thought to confer differences in phenotypes, such as virulence, host range, and/or environmental niches, of different strains. Recently, Cummings et al. proposed an analogous distinction to describe those genes similarly expressed (the core regulon) or differentially expressed (the flexible regulon) between strains, as not all genes are similarly regulated by BvgAS among Bordetella strains (10). The TTSS of B. pertussis, which mediates cellular attachment rather than cytotoxicity, is expressed by some strains but not others (14). Therefore, the TTSS of B. pertussis appears to be part of the flexible regulon, as some B. pertussis strains express the TTSS while others do not (14, 62). The work described herein provides evidence that the TTSS is also part of the flexible regulon of B. bronchiseptica, as strains of this species express TTSS-related genes differentially. Together, these data suggest that the TTSS can have different functions and/or levels of these functions in different strains or species of Bordetella.
Since nearly all known TTSS-related genes were upregulated in strain 1289 compared to their levels of expression in strain RB50, we speculate that the underlying mechanism behind the increased TTSS-mediated virulence of strain 1289 is that increased TTSS gene expression leads to enhanced protein expression and secretion, which in turn increases TTSS-mediated cytotoxicity and virulence. While TTSS-related genes were upregulated in strain 1289, the genes encoding the master regulator of the TTSS, bvgAS, were not differentially expressed. Since the TTSS is controlled by a complex, multilayered, trans-regulatory gene network (34, 62), we propose that the increased expression of a yet-unidentified, Bvg-activated activator or decreased expression of a Bvg-activated repressor may contribute to the differential expression of the TTSS between strains. This regulator may be one of the 23 upregulated or 51 downregulated transcriptional regulators identified in strain 1289.
Since 1289ΔbscN and S308ΔbscN are more virulent than RB50ΔbscN, we conclude that the TTSS is not the only factor that contributes to the increased virulence of ST32 strains. Novel genes acquired via phage or horizontal gene transfer, loss or downregulation of hypovirulence genes, or mutations in strain 1289 may also contribute to this strain's increased virulence, although they do not appear to be sufficient for cytotoxicity in vitro (15, 16). While many phage-related genes present in strain RB50 were identified as absent in strain 1289, another study of ours showed that a B. bronchiseptica strain lacking these prophage genes is less virulent than strain RB50, making it unlikely that the lack of these genes contributes to the increased virulence of strain 1289 (5). A few known virulence-related genes and many genes with unknown function were identified as differentially expressed or divergent between these strains. These genes may also contribute to the increase in virulence of strain 1289 compared to that of strain RB50. Differences in gene expression over the course of infection, promoter mutations, or a gain of novel genes in strain 1289 would not be detected in our analyses and may also contribute to the increased virulence of strain 1289. We are currently sequencing the genome of strain 1289 and will then be able to assess promoter mutations and novel genes that may play a role in the increased virulence of this strain (A. M. Buboltz, X. Zhang, S. C. Schüster, E. T. Harvill et al., unpublished data).
The most widely accepted view of virulence evolution assumes that there is a cost-benefit trade-off to virulence, which is defined as any reduction in host fitness following infection (2, 12, 45). Under this framework, evolutionary processes that lead to the maintenance of harmful effects are thought to be characterized by the presence of other beneficial qualities (12). Thus, a fitness-decreasing change in one trait in the pathogen is accompanied by a fitness-increasing change in a different trait (12). The ST32 strains described herein appear to be quite successful, as they have been isolated from three separate continents (South America, Europe, and the United States) (see Table S1 in the supplemental material) (60). Here, we show that ST32 strains appear to be associated with respiratory disease and exhibit increased TTSS-mediated virulence, a potential cost for the bacteria because pathogen success is dependent upon host survival before transmission. Since the TTSS increases colonization and persistence of B. bronchiseptica in the lungs of mice (44, 62), this effect may benefit the bacteria in maximizing transmission, allowing for the selection and maintenance of these highly virulent ST32 strains. Thus, the fitness enhancement caused by increased TTSS-mediated effects may be accompanied by the unavoidable side effect of increased virulence (45).
A high degree of clonal diversity appears to exist among B. bronchiseptica strains (3, 17, 19, 31, 36, 47). Recently, we reported that strains belonging to ST27 and ST40, which were collected from a wide geographical area, are hypovirulent and have lost the genes encoding adenylate cyclase toxin, which was previously believed to be among the few core factors required for the success of the classical bordetellae (5). Here, we report that the TTSS contributes to the increased virulence of strains belonging to ST32, which appear to exist worldwide. Combined, these studies support the conclusion that phylogenetic lineages of B. bronchiseptica differentially regulate and utilize distinct sets of virulence factors which can affect the overall virulence of these STs. This versatility may contribute to the wide variety in severity of respiratory disease observed upon B. bronchiseptica infection.
Supplementary Material
Acknowledgments
We thank Catherine Beckwith at the Pennsylvania State University, College of Medicine; Bob Livingston at the University of Missouri Research Animal Diagnostic Laboratory; David Relman at the Department of Microbiology and Immunology, Stanford University; Frits Mooi at the Laboratory for Vaccine-Preventable Diseases, National Institute of Public Health and the Environment; and Gary Sanden at the Center for Disease Prevention and Control for B. bronchiseptica isolates. We thank all members of the Harvill laboratory for discussion and critical review of the manuscript and Gráinne Long for assistance with statistical analysis.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
This work was supported by NIH grants AI 053075, AI 065507, and GM083113 (E.T.H.). The authors declare no conflicting financial interests.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 13 July 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80611-620. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. M., and R. M. May. 1982. Coevolution of hosts and parasites. Parasitology 85411-426. [DOI] [PubMed] [Google Scholar]
- 3.Bemis, D. A., H. A. Greisen, and M. J. Appel. 1977. Bacteriological variation among Bordetella bronchiseptica isolates from dogs and other species. J. Clin. Microbiol. 5471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockmeier, S. L., K. B. Register, T. Magyar, A. J. Lax, G. D. Pullinger, and R. A. Kunkle. 2002. Role of the dermonecrotic toxin of Bordetella bronchiseptica in the pathogenesis of respiratory disease in swine. Infect. Immun. 70481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buboltz, A. M., T. L. Nicholson, M. R. Parette, S. E. Hester, J. Parkhill, and E. T. Harvill. 2008. Replacement of adenylate cyclase toxin in a lineage of Bordetella bronchiseptica. J. Bacteriol. 1905502-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, V. C., E. J. Pishko, A. Preston, D. J. Maskell, and E. T. Harvill. 2003. Role of Bordetella O antigen in respiratory tract infection. Infect. Immun. 7186-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11367-373. [DOI] [PubMed] [Google Scholar]
- 8.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 623381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 665921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings, C. A., H. J. Bootsma, D. A. Relman, and J. F. Miller. 2006. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J. Bacteriol. 1881775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diavatopoulos, D. A., C. A. Cummings, L. M. Schouls, M. M. Brinig, D. A. Relman, and F. R. Mooi. 2005. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog. 1e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebert, D., and E. A. Herre. 1996. The evolution of parasitic diseases. Parasitol. Today 1296-101. [DOI] [PubMed] [Google Scholar]
- 13.Elder, K. D., and E. T. Harvill. 2004. Strain-dependent role of BrkA during Bordetella pertussis infection of the murine respiratory tract. Infect. Immun. 725919-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fennelly, N. K., F. Sisti, S. C. Higgins, P. J. Ross, H. van der Heide, F. R. Mooi, A. Boyd, and K. H. G. Mills. 2008. Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect. Immun. 761257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald, J. R., and J. M. Musser. 2001. Evolutionary genomics of pathogenic bacteria. Trends Microbiol. 9547-553. [DOI] [PubMed] [Google Scholar]
- 16.Foreman-Wykert, A. K., and J. F. Miller. 2003. Hypervirulence and pathogen fitness. Trends Microbiol. 11105-108. [DOI] [PubMed] [Google Scholar]
- 17.Giardina, P. C., L. A. Foster, J. M. Musser, B. J. Akerley, J. F. Miller, and D. W. Dyer. 1995. bvg repression of alcaligin synthesis in Bordetella bronchiseptica is associated with phylogenetic lineage. J. Bacteriol. 1776058-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodnow, R. A. 1980. Biology of Bordetella bronchiseptica. Microbiol. Rev. 44722-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gueirard, P., and N. Guiso. 1993. Virulence of Bordetella bronchiseptica: role of adenylate cyclase-hemolysin. Infect. Immun. 614072-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gueirard, P., C. Weber, A. Le Coustumier, and N. Guiso. 1995. Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J. Clin. Microbiol. 332002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvill, E. T., P. A. Cotter, M. H. Yuk, and J. F. Miller. 1999. Probing the function of Bordetella bronchiseptica adenylate cyclase toxin by manipulating host immunity. Infect. Immun. 671493-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvill, E. T., A. Preston, P. A. Cotter, A. G. Allen, D. J. Maskell, and J. F. Miller. 2000. Multiple roles for Bordetella lipopolysaccharide molecules during respiratory tract infection. Infect. Immun. 686720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julio, S. M., and P. A. Cotter. 2005. Characterization of the filamentous hemagglutinin-like protein FhaS in Bordetella bronchiseptica. Infect. Immun. 734960-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirimanjeswara, G. S., L. M. Agosto, M. J. Kennett, O. N. Bjornstad, and E. T. Harvill. 2005. Pertussis toxin inhibits neutrophil recruitment to delay antibody-mediated clearance of Bordetella pertussis. J. Clin. Investig. 1153594-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirimanjeswara, G. S., P. B. Mann, M. Pilione, M. J. Kennett, and E. T. Harvill. 2005. The complex mechanism of antibody-mediated clearance of Bordetella from the lungs requires TLR4. J. Immunol. 1757504-7511. [DOI] [PubMed] [Google Scholar]
- 27.Kubori, T., A. Sukhan, S. I. Aizawa, and J. E. Galan. 2000. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. USA 9710225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwae, A., T. Matsuzawa, N. Ishikawa, H. Abe, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, and A. Abe. 2006. BopC is a novel type III effector secreted by Bordetella bronchiseptica and has a critical role in type III-dependent necrotic cell death. J. Biol. Chem. 2816589-6600. [DOI] [PubMed] [Google Scholar]
- 29.Kuwae, A., M. Ohishi, M. Watanabe, M. Nagai, and A. Abe. 2003. BopB is a type III secreted protein in Bordetella bronchiseptica and is required for cytotoxicity against cultured mammalian cells. Cell. Microbiol. 5973-983. [DOI] [PubMed] [Google Scholar]
- 30.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8396-401. [DOI] [PubMed] [Google Scholar]
- 31.Le Blay, K., P. Gueirard, N. Guiso, and R. Chaby. 1997. Antigenic polymorphism of the lipopolysaccharides from human and animal isolates of Bordetella bronchiseptica. Microbiology 1431433-1441. [DOI] [PubMed] [Google Scholar]
- 32.Mann, P. B., K. D. Elder, M. J. Kennett, and E. T. Harvill. 2004. Toll-like receptor 4-dependent early elicited tumor necrosis factor alpha expression is critical for innate host defense against Bordetella bronchiseptica. Infect. Immun. 726650-6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mattoo, S., and J. D. Cherry. 2005. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin. Microbiol. Rev. 18326-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattoo, S., M. H. Yuk, L. L. Huang, and J. F. Miller. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 521201-1214. [DOI] [PubMed] [Google Scholar]
- 35.Melton, A. R., and A. A. Weiss. 1989. Environmental regulation of expression of virulence determinants in Bordetella pertussis. J. Bacteriol. 1716206-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musser, J. M., D. A. Bemis, H. Ishikawa, and R. K. Selander. 1987. Clonal diversity and host distribution in Bordetella bronchiseptica. J. Bacteriol. 1692793-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musser, J. M., E. L. Hewlett, M. S. Peppler, and R. K. Selander. 1986. Genetic diversity and relationships in populations of Bordetella spp. J. Bacteriol. 166230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson, T. L. 2007. Construction and validation of a first-generation Bordetella bronchiseptica long-oligonucleotide microarray by transcriptional profiling of the Bvg regulon. BMC Genomics 8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novotny, P., A. P. Chubb, K. Cownley, and J. A. Montaraz. 1985. Adenylate cyclase activity of a 68,000-molecular-weight protein isolated from the outer membrane of Bordetella bronchiseptica. Infect. Immun. 50199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen, C. H. 2003. Review of the use of statistics in infection and immunity. Infect. Immun. 716689-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panina, E. M., S. Mattoo, N. Griffith, N. A. Kozak, M. H. Yuk, and J. F. Miller. 2005. A genome-wide screen identifies a Bordetella type III secretion effector and candidate effectors in other species. Mol. Microbiol. 58267-279. [DOI] [PubMed] [Google Scholar]
- 42.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O'Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat. Genet. 3532-40. [DOI] [PubMed] [Google Scholar]
- 43.Pilione, M. R., L. M. Agosto, M. J. Kennett, and E. T. Harvill. 2006. CD11b is required for the resolution of inflammation induced by Bordetella bronchiseptica respiratory infection. Cell. Microbiol. 8758-768. [DOI] [PubMed] [Google Scholar]
- 44.Pilione, M. R., and E. T. Harvill. 2006. The Bordetella bronchiseptica type III secretion system inhibits gamma interferon production that is required for efficient antibody-mediated bacterial clearance. Infect. Immun. 741043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Read, A. F. 1994. The evolution of virulence. Trends Microbiol. 273-76. [DOI] [PubMed] [Google Scholar]
- 46.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 40664-67. [DOI] [PubMed] [Google Scholar]
- 47.Roop, R. M., II, H. P. Veit, R. J. Sinsky, S. P. Veit, E. L. Hewlett, and E. T. Kornegay. 1987. Virulence factors of Bordetella bronchiseptica associated with the production of infectious atrophic rhinitis and pneumonia in experimentally infected neonatal swine. Infect. Immun. 55217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34374-378. [DOI] [PubMed] [Google Scholar]
- 49.Saeij, J. P. J., J. P. Boyle, S. Coller, S. Taylor, L. D. Sibley, E. T. Brooke-Powell, J. W. Ajioka, and J. C. Boothroyd. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 3141780-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schoen, C., J. Blom, H. Claus, A. Schramm-Gluck, P. Brandt, T. Muller, A. Goesmann, B. Joseph, S. Konietzny, O. Kurzai, C. Schmitt, T. Friedrich, B. Linke, U. Vogel, and M. Frosch. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 1053473-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sitkiewicz, I., M. J. Nagiec, P. Sumby, S. D. Butler, C. Cywes-Bentley, and J. M. Musser. 2006. Emergence of a bacterial clone with enhanced virulence by acquisition of a phage encoding a secreted phospholipase A2. Proc. Natl. Acad. Sci. USA 10316009-16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Stainer, D. W., and M. J. Scholte. 1970. A simple defined medium for the production of phase I Bordetella pertussis. J. Gen. Microbiol. 63211-220. [DOI] [PubMed] [Google Scholar]
- 52.Stibitz, S., W. Aaronson, D. Monack, and S. Falkow. 1989. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature 338266-269. [DOI] [PubMed] [Google Scholar]
- 53.Stibitz, S., W. Aaronson, D. Monack, and S. Falkow. 1988. The vir locus and phase-variation in Bordetella pertussis. Tokai J. Exp. Clin. Med. 13(Suppl.)223-226. [PubMed] [Google Scholar]
- 54.Stibitz, S., and N. H. Carbonetti. 1994. Hfr mapping of mutations in Bordetella pertussis that define a genetic locus involved in virulence gene regulation. J. Bacteriol. 1767260-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sukumar, N., M. Mishra, G. P. Sloan, T. Ogi, and R. Deora. 2007. Differential Bvg phase-dependent regulation and combinatorial role in pathogenesis of two Bordetella paralogs, BipA and BcfA. J. Bacteriol. 1893695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 58.Taylor, S., A. Barragan, C. Su, B. Fux, S. J. Fentress, K. Tang, W. L. Beatty, H. E. Hajj, M. Jerome, M. S. Behnke, M. White, J. C. Wootton, and L. D. Sibley. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science 3141776-1780. [DOI] [PubMed] [Google Scholar]
- 59.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 1796609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuk, M. H., E. T. Harvill, P. A. Cotter, and J. F. Miller. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-kappaB activation by the Bordetella type III secretion system. Mol. Microbiol. 35991-1004. [DOI] [PubMed] [Google Scholar]
- 62.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28945-959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.