Abstract
We have sequenced the genome and identified the structural proteins and lipids of the novel membrane-containing, icosahedral virus P23-77 of Thermus thermophilus. P23-77 has an ∼17-kb circular double-stranded DNA genome, which was annotated to contain 37 putative genes. Virions were subjected to dissociation analysis, and five protein species were shown to associate with the internal viral membrane, while three were constituents of the protein capsid. Analysis of the bacteriophage genome revealed it to be evolutionarily related to another Thermus phage (IN93), archaeal Halobacterium plasmid (pHH205), a genetic element integrated into Haloarcula genome (designated here as IHP for integrated Haloarcula provirus), and the Haloarcula virus SH1. These genetic elements share two major capsid proteins and a putative packaging ATPase. The ATPase is similar with the ATPases found in the PRD1-type viruses, thus providing an evolutionary link to these viruses and furthering our knowledge on the origin of viruses.
Three-dimensional structures of the major capsid proteins, as well as the architecture of the virion and the sequence similarity of putative genome packaging ATPases, have revealed unexpected evolutionary connection between virus families. Viruses infecting hosts residing in different domains of life (Bacteria, Archaea, and Eukarya) share common structural elements and possibly also ways to package the viral genome (8, 13, 41). It has been proposed that the set of genes responsible for virion assembly is a hallmark of the virus and is designated as the innate viral “self,” which may retain its identity through evolutionary times (5). Based on this, it is proposed that viruses can be classified into lineages that span the different domains of life. Therefore, the studies of new virus isolates might provide insights into the events that led to the origin of viruses and maybe even the origin of life itself (34, 40). However, viruses are known to be genetic mosaics (28), and these structural lineages therefore do not reflect the evolutionary history of all genes in a given virus. For example, the genome replication strategies vary significantly even in the currently established lineages (41) and, consequently, a structural approach does not point out to a specific form of replication in the ancestor. Nevertheless, as the proposal for a viral self is driven from information on viral structures and pathways of genome encapsidation, the ancestral form of the self was likely to be composed of a protective coat and the necessary mechanisms to incorporate the genetic material within the coat.
Viruses structurally related to bacteriophage PRD1, a phage infecting gram-negative bacteria, have been identified in all three domains of life, and the lineage hypothesis was first proposed based on structural information on such viruses. Initially, PRD1 and human adenovirus were proposed to originate from a common ancestor mainly due to the same capsid organization (T=25) and the major coat protein topology, the trimeric double β-barrel fold (12). In addition, these viruses share a common vertex organization and replication mechanism (20, 31, 53, 63). PRD1 is an icosahedral virus with an inner membrane, whereas adenovirus lacks the membrane. Later, many viruses with similar double β-barrel fold in the major coat protein have been discovered and included to this viral lineage. For example, the fold is present in Paramecium bursaria Chlorella virus 1 (56) of algae, Bam35 (45) of gram-positive bacteria, PM2 (2) of gram-negative marine bacteria, and Sulfolobus turreted icosahedral virus (STIV) (38) of an archaeal host. Moreover, genomic analyses have revealed a common set of genes in a number of nucleocytoplasmic large DNA viruses. Chilo iridescent virus and African swine fever virus 1 are related to Paramecium bursaria Chlorella virus 1 and most probably share structural similarity to PRD1-type viruses (13, 30, 31, 68). The largest known viruses, represented by mimivirus and poxvirus, may also belong to this lineage (29, 77). Two euryarchaeal proviruses, TKV4 and MVV, are also proposed to belong to this lineage based on bioinformatic searches (42). The proposed PRD1-related viruses share the same basic architectural principles despite major differences in the host organisms and particle and genome sizes (1, 2, 38, 56). PM2, for example, has a genome of only 10 kbp, whereas mimivirus (infecting Acanthamoeba polyphaga) double-stranded DNA (dsDNA) genome is 1.2 Mbp in size (59).
How many virion structure-based lineages might there be? This obviously relates to the number of protein folds that have the properties needed to make viral capsids. It has been noted that, in addition to PRD1-type viruses, at least tailed bacterial and archaeal viruses, as well as herpesviruses, share the same coat protein fold. Also, certain dsRNA viruses seem to have structural and functional similarities, although their hosts include bacteria and yeasts, as well as plants and animals (6, 18, 19, 27, 55, 60, 74). Obviously, many structural principles to build a virus capsid exist, and it has been suggested that especially geothermally heated environments have preserved many of the anciently formed virus morphotypes (35).
Thermophilic dsDNA bacteriophage P23-77 was isolated from an alkaline hot spring in New Zealand on Thermus thermophilus (17) ATCC 33923 (deposited as Thermus flavus). P23-77 was shown to have an icosahedral capsid and possibly an internal membrane but no tail (81). Previously, another Thermus virus, IN93, with a similar morphology has been described (50). IN93 was inducible from a lysogenic strain of Thermus aquaticus TZ2, which was isolated from hot spring soil in Japan. Recently, P23-77 was characterized in more detail (33). It has an icosahedral protein coat, organized in a T=28 capsid lattice (21). The presence of an internal membrane was confirmed, and lipids were shown to be constituents of the virion. Ten structural proteins were identified, with apparent molecular masses ranging from 8 to 35 kDa. Two major protein species with molecular masses of 20 and 35 kDa were proposed to make the capsomers, one forming the hexagonal building blocks and the other the two towers that decorate the capsomer bases (33). Surprisingly, P23-77 is structurally closest to the haloarchaeal virus SH1, which is the only other example of a T=28 virion architecture (32, 33). In both cases it was proposed that the capsomers are made of six single β-barrels opposing the situation with the other structurally related viruses where the hexagonal capsomers are made of three double β-barrel coat protein monomers (8).
In the present study we analyze the dsDNA genome of P23-77. Viral membrane proteins and those associated with the capsid were identified by virion dissociation studies. The protein chemistry data and genome annotation are consistent with the results of the disruption studies. A detailed analysis of the lipid composition of P23-77 and its T. thermophilus host was carried out. The data collected here reveal additional challenges in attempts to generate viral lineages based on the structural and architectural properties of the virion.
MATERIALS AND METHODS
Biological material.
T. thermophilus ATCC 33923 was used as a host for P23-77 propagation. The virus was grown and purified as described earlier (33). Viruses purified by rate zonal centrifugation in sucrose (once purified) were used for dissociation studies. Twice-purified (rate zonal and equilibrium centrifugation in sucrose; twice purified) virus particles were used for DNA extraction as well as for protein and lipid analyses.
Sequencing of the virus genome.
Viruses (0.3 mg of protein/ml) were disrupted by sodium dodecyl sulfate (SDS) protease treatment, followed by multiple extractions with phenol (9). The genome was double digested with BamHI and HindIII restriction enzymes, resulting in fragments of ∼500, 1,500, 2,000, 6,000, and 7,000 nucleotides. The fragments were cloned in pSU18 cloning vector (10) between the BamHI and HindIII restriction sites. The genome sequences were determined first by using universal and reverse sequencing primers hybridizing to the vector sequence, followed by primer walking the insert with custom made 19- to 22-nucleotide long primers. Both strands were sequenced approximately four times. Sequencing was performed by using a BigDye Terminator v3.1 sequencing kit on an ABI Prism 3130 automated sequencer (both from Applied Biosystems). The sequence assembly was done by using Vector NTI 10 (Invitrogen, Inc.).
To verify the sequence at the ends of the restriction fragments, regions covering the joints were amplified by PCR using Herculase II Fusion DNA polymerase, specific primers, and the virus DNA as a template. DNA sequences of the products were determined as described above using oligonucleotide nucleotides hybridizing to the ends of the fragments.
Dissociation of virus particles.
Viruses in TV buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 150 mM NaCl, and ∼6.5 mg of protein/ml [14]) were stored maximally 5 days at 28°C (optimal storage temperature [33]). The following final conditions were used for virus dissociation: (i) 0.1 M sodium acetate (Merck) (pH 6.0), (ii) 3 M urea (MP Biomedicals), (iii) 3 M guanidine hydrochloride (GuHCl; Fluka BioChemicals), and (iv) 0.1% SDS (Serva) in TV buffer (except for condition i), using 250 μg (protein) of purified viruses for each reaction. The reactions were incubated for 30 min at 22°C (300 μl). Aggregates formed were removed by centrifugation (Eppendorf microcentrifuge; 13,000 rpm, 22°C, 1 min) or, alternatively, left in the reaction mixture, which leads to the formation of a smear appearing in the first gradient fraction (not shown in Fig. 3). The reaction mixtures were loaded either on top of a 5 to 20% (wt/vol) (SDS and low-pH treatments) or a 10 to 40% (wt/vol) (urea and GuHCl treatments) linear sucrose gradient, followed by centrifugation (Beckman SW41 rotor; 23,000 rpm, 25°C) for 45 min or 1 h 20 min, for the SDS-pH treatments or for the urea-GuHCl treatments, respectively. After centrifugation, the gradients were fractionated in 10 fractions, and the proteins were precipitated with trichloroacetic acid and analyzed by Tricine SDS-polyacrylamide gel electrophoresis (PAGE) containing 17% acrylamide (66).
FIG. 3.
Analysis of proteins released from P23-77 particles after different chemical treatments. (A) Dissociation products of P23-77 particles treated with 3 M GuHCl and separated by rate zonal centrifugation were analyzed by Tricine SDS-PAGE. Twice-purified P23-77 virus is shown at the left (with marked virion proteins) and gradient fractions from top to pellet at right. (B) Summary of the results of the other dissociation experiments. (C) Schematic illustration of the P23-77 virion showing the major structural proteins in the capsid and membrane moieties.
Protein analyses.
SDS-PAGE-separated proteins were analyzed by mass spectrometry. For this the gel was stained with Coomassie brilliant blue or silver. Bands were cut from the gels and “in gel” digested as described by Shevchenko et al. (67). Proteins were reduced with dithiothreitol and alkylated with iodoacetamide before digestion with trypsin (sequencing grade modified trypsin, V5111; Promega). Peptides generated by enzymatic cleavage were analyzed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry for mass fingerprinting using an Ultraflex TOF/TOF instrument (Bruker-Daltonik GmbH, Bremen, Germany) as described previously (58). The obtained peptide masses were compared to the sequences of the putative proteins translated from the nucleotide sequence. N-terminal sequence analysis was carried out as described by Bamford et al. (8).
Lipid analyses.
Lipids from the freshly purified P23-77 and its host membranes were extracted as described by Folch et al. (25), redissolved in chloroform-methanol (9:1 [vol/vol]) and stored at −20°C until analyzed. Thin-layer chromatography (TLC) of the total lipids was performed on heat-activated silica gel 60 plates (Merck) using chloroform-methanol-acetic acid-water (80:25:15:4 [vol/vol/vol/vol]) as the eluting solvent for polar lipids and hexane-diethyl ether-acetic acid (80:20:1 [vol/vol/vol]) for neutral lipids (37). Total lipids were visualized by iodine vapor, whereas phospho- and glycolipids were detected by spraying the plate with the specific molybdate (23) and orcinol reagents (69), respectively. After visualization, the lipid-containing spots were scraped from the plate, and their phosphorus content was determined (36). Mass spectrometric analyses of the lipids in each spot of the TLC plate and in total lipid extracts were carried out as described previously (8). The lipids were identified by their staining with specific dyes and comparing their m/z values, as well as the product and precursor ion profiles, to those of previously published lipid spectra (46, 78).
Computational methods.
DNA and the putative protein sequences were analyzed by using Vector NTI 10. Transmembrane region identifications and domain predictions were conducted by SMART (47; http://smart.embl-heidelberg.de/). Similarities to known proteins were searched with PSI-BLAST (3; http://www.ncbi.nlm.nih.gov/BLAST/). PSI-BLAST was executed for each open reading frame (ORF) until no new hits were reported. Protein fold recognition was performed by using the PHYRE server (11; http://www.sbg.bio.ic.ac.uk/phyre/). Multiple alignments of virus proteins were done by Vector NTI using the identity matrix Blossum62.
The evolutionary history of major capsid proteins was inferred by using the minimum-evolution method (61). The bootstrap consensus tree inferred from 500 replicates is taken to represent the evolutionary history of the taxa analyzed (24). Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The phylogenetic tree was linearized assuming equal evolutionary rates in all lineages (71). The evolutionary distances were computed by using the Poisson correction method (83). The tree was searched by using the close-neighbor-interchange algorithm (57) at a search level of 3. The neighbor-joining algorithm (64) was used to generate the initial tree. All positions containing gaps and missing data were eliminated from the data set. There were a total of 101 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (72).
RESULTS
Lipid composition of P23-77 differs from that of the host.
Several different lipid species were observed when the total lipid extract from the T. thermophilus were examined by TLC analysis (Fig. 1A, right lane). Staining of the TLC plate with specific dyes indicated the presence of at least one phospholipid (spot 4) and glycolipid (spot 5) in the membrane of this organism (data not shown). In addition to polar lipids, T. thermophilus also contains neutral lipids of which the majority are fatty acids, diacylglycerols, and some unknown lipids (data not shown). Mass spectrometric analysis of the lipid extracts of each individual spot was carried out in order to identify the lipids. By this approach, lipids in the spots 1, 2, 6, and 7 (Fig. 1) could not be identified. Spectra from the spots 3 to 5, on the other hand, contained clear lipid peaks that were identified based on their m/z masses and (in some cases) on the profile of their fragmentation products. The spectrum of spot 5 contained multiple peaks in the m/z range of 1,400 to 1,570, of which the major ones with m/z values of 1,409.5 and 1,467.5 were in a good agreement with the masses of glycolipids GL1 and GL2 from T. thermophilus Samu-SA1 (46). In spot 4, a minor peak at m/z value 1,118.2 probably represents phosphoglycolipid 1 (PGL1) of the Thermus and Meothermus thermophiles (77), whereas two major ones at m/z values 1,176.3 and 1204.3 most likely are the major molecular species of PGL2 of Thermus and Meothermus species (PGL2) (46, 77). Phosphate analysis suggested that more than 90% of total phosphorus was contained within spot 4 (data not shown). The rest of the total phosphate (<1% in the host and ∼5% in the virus) was found in spot 3. Mass spectrometric analyses showed that one peak has the same m/z value (1,204.3) as in spot 4, suggesting that this lipid is also present in spot 3. Another major peak in this spot was at m/z value 1,232.3, which corresponds to the mass PGL2 containing longer acyl chains than that at m/z value 1,204.3. The reason why PGL2 with longer acyl chains migrate as a separate spot on the TLC plate is not clear.
FIG. 1.
Lipids of P23-77 and its Thermus sp. 33923 host. (A) TLC analysis of polar lipids from P23-77 (virus lane) and Thermus sp. (host lane). All spots of polar lipids are numbered (1-7). NL, neutral lipids. (B) Negative-ion mode mass spectra of the lipid extracts from (from top to bottom) virus, spot 3, spot 4, spot 5, and total host membranes. See the text for details.
Genome of P23-77 is a 17,036-bp circular dsDNA molecule.
For determination of the genome sequence, P23-77 DNA was cloned in a plasmid vector as restriction fragments (see Materials and Methods). The approach was taken since high-quality sequence was not obtained using the full-length phage DNA as a template. This was probably due to the high GC content (68%) of the genome. The phage genome was shown to be circular (by agarose gel electrophoresis), and it was linearized by NotI restriction enzyme yielding one ∼17-kb molecule (data not shown). In order to assemble the restriction fragment sequences, the joint regions were amplified by PCR, and the products were sequenced. This confirmed the circularity of the P23-77 genome and revealed its exact length (17,036 nucleotides). As shown in Fig. 2, the genome of P23-77 was annotated to contain 37 putative protein coding ORFs, 10 of which were confirmed here to be true genes (see below).
FIG. 2.
Genome of P23-77 is a 17,036-bp circular dsDNA molecule and encodes 37 putative proteins (GenBank accession no. GQ403789). The genes encoding the 10 determined structural protein species are marked with the adopted gene nomenclature (see Table 1). Some of the ORFs gave hits to known genes in a BLAST search, and these are indicated in the figure.
Identification of the genes encoding structural proteins of the P23-77 virion.
It has previously been shown that lipid-containing P23-77 virion consists of about 10 structural protein species (33). The genes responsible for encoding the structural proteins (denoted as VPs for virion proteins) were identified by peptide mass fingerprint spectrometric analysis. Masses of trypsin-generated peptides were compared to the theoretical tryptic peptide masses. The identified ORFs and the theoretical molecular mass for each identified virion protein are shown in Table 1. In addition, the N-terminal sequence was determined for the 35-kDa protein VP17 to determine its ORF as the mass analysis gave two signals. A sequence GVFDRIRGA… was obtained, confirming that ORF 17 encodes VP17. The gene and protein nomenclature is presented in Table 1, and this replaces the previous protein nomenclature used in the study by Jaatinen et al. (33).
TABLE 1.
Determination of ORFs encoding virion-associated proteins in P23-77a
| ORF | Coding capacity (kDa)b | Protein | Gene | No. of peptides matched | Peptide sequence coverage (%) |
|---|---|---|---|---|---|
| 6 | 19.5 | VP6 | 6 | 6 | 33 |
| 11 | 22.1 | VP11 | 11 | 8 | 33 |
| 15 | 14.7 | VP15 | 15 | 9 | 55 |
| 16 | 19.1 | VP16 | 16 | 13 | 86 |
| 17 | 31.9 | VP17 | 17 | 5 | 24 |
| 19 | 8.9 | VP19 | 19 | 1 | 20 |
| 20 | 24.6 | VP20 | 20 | 6 | 26 |
| 22 | 9.3 | VP22 | 22 | 2 | 50 |
| 23 | 7.4 | VP23 | 23 | 2 | 57 |
| 29 | 40.7 | VP29 | 29 | 9 | 37 |
The number of peptides matched and the peptide sequence coverage indicate the number of theoretical tryptic fragments matched and their percent sequence coverage, respectively, in the search result determined by MASCOT for the identified protein. The numerical values shown are based on MALDI-TOF peptide mass fingerprint analyses.
That is, the theoretical molecular mass of the protein translated from the corresponding ORF.
In order to determine which structural proteins are associated with the protein coat or with the internal membrane, purified virions were subjected to dissociation studies. P23-77 particles were dissociated with 3 M urea or 3 M GuHCl, low-pH, or SDS treatments. The resulting subviral particles were separated by rate zonal sucrose gradient centrifugation, and the protein content in the gradient fractions and pellet was analyzed by Tricine SDS-PAGE (i.e., the gradient for GuHCl treatment in is shown Fig. 3A and is summarized for the other treatments in Fig. 3B).
In 3 M urea or 3 M GuHCl most of VP16 and VP17 were released as soluble proteins residing at the top of the gradient. By lowering the pH to 6.0 VP11 was also released. The viral membrane with the DNA aggregated and was found in the pellet fraction. The pellet fractions contained proteins VP11, VP15, VP20, VP22, and VP23. Also, a fraction of VP16 and VP15 proteins were present in the pellet. After a mild (0.1%) SDS treatment the membrane-associated proteins VP15, VP19, VP20, VP22, and VP23 also appeared at the top, soluble, fractions. Figure 3C schematically summarizes the dissection of the coat and membrane-associated virion proteins. Transmembrane helix predictions indicated that there may be 13 putative integral membrane proteins encoded by the virus (Table 2), including proteins VP15, VP19, VP20, and VP22. There was no transmembrane helix predicted for VP23, even though dissociation analyses propose its membrane association.
TABLE 2.
P23-77 ORFs or genes examined in this study
| P23-77 ORF or gene | No. of residues (position) | ORF IN93 (position in IN93 genome) | Identity (%) vs IN93 | Virion-associated protein and/or predicted function (BLAST) | No. of transmembrane region(s) and significant structure prediction hits (PHYRE hit %) |
|---|---|---|---|---|---|
| 1 | 425 (241-1515) | CD2+CD3 (23-1260) | 65.0 | DNA replication protein | |
| 2 | 125 (1515-1889) | CD4 (1250-1594) | 47.2 | Topoisomerase (50) | |
| 3 | 255 (1889-2653) | Phosphoadenosine phosphosulfate reductase | Adenine nucleotide alpha hydrolase (100) | ||
| 4 | 129 (2653-3039) | CD5 (1614-2003) | 40.0 | One transmembrane | |
| 5 | 97 (3070-3360) | CD6 (2020-2313) | 61.0 | ||
| Gene 6 | 187 (3456-4016) | CD7 (2415-2975) | 60.1 | VP6 | Two transmembranes |
| 7 | 131 (4068-4460) | CD8 (3026-3340) | 37.8 | One transmembrane | |
| 8 | 173 (4378-4896) | CD9 (3341-3862) | 61.4 | ||
| 9 | 203 (4829-5437) | Endolysin | l-Alanyl-d-glutamate peptidase (100) | ||
| 10 | 178 (5443-5976) | Amidase endolysin | N-Acetylmuramoyl-l-alanine amidase (100) | ||
| Gene 11 | 187 (5996-6556) | VP11 | |||
| 12 | 64 (6556-6747) | ||||
| 13 | 224 (6740-7411) | ORF 5 (4829-5506) | 79.2 | ATPase | AAA-ATPase (95) |
| 14 | 66 (7414-7611) | ||||
| Gene 15 | 138 (7556-7969) | CD13 (5646-6062) | 82.6 | VP15 | Three transmembranes |
| Gene 16 | 173 (7983-8501) | CD14 (6073-6588) | 79.2 | 20-kDa major capsid protein (VP16) | |
| Gene 17 | 291 (8514-9386) | CD15 (6597-7472) | 73.8 | 35-kDa major capsid protein (VP17) | STIV-MCP (45) |
| 18 | 116 (9407-9754) | CD16 (7485-7898) | 43.1 | One transmembrane | |
| Gene 19 | 87 (9761-10021) | CD17 (7901-8155) | 65.5 | VP19 | One transmembrane |
| Gene 20 | 227 (10021-10701) | CD18 (8152-8805) | 71.4 | VP20 | One transmembrane |
| 21 | 72 (10704-10919) | CD19 (8829-9029) | 39.2 | One transmembrane | |
| Gene 22 | 92 (10912-11187) | CD20 (9019-9321) | 48.0 | VP22 | One transmembrane |
| Gene 23 | 75 (11199-11423) | CD21 (9330-9563) | 49.4 | VP23 | |
| 24 | 140 (11423-11842) | ORF 6 (9560-9985) | 69.0 | Two transmembranes | |
| 25 | 41 (11842-11964) | ORF 13 (9985-10101) | 54.8 | One transmembrane | |
| 26 | 62 (11960-12145) | ORF 8 (10106-10291) | 68.3 | One transmembrane | |
| 27 | 150 (12127-12576) | CD22 (10273-10725) | 67.3 | ||
| 28 | 341 (12651-13673) | CD23 (10803-11765) | 51.3 | ||
| Gene 29 | 369 (13686-14792) | CD24 (11924-12841) | 17.4 | Lysozyme (VP29) | |
| 30 | 88 (14795-15058) | CD26 (13130-13396) | 54.5 | ||
| 31 | 233 (15039-15737) | CD27 (13362-14081) | 41.9 | Transglycosylase | Lysozyme (100%) |
| 32 | 72 (15997-16212) | ORF 2 (17959-18159) | 37.0 | ||
| 33 | 62 (16208-16393) | ORF 11 (18159-18332) | 56.11 | One transmembrane | |
| 34 | 35 (16393-16497) | ||||
| 35 | 90 (16503-16772) | CD34 (18766-18990) | 35.2 | ||
| 36 | 145 (16772-170) | ||||
| 37 | 78 (8-241) | CD35 (19145-19429) | 40.61 |
It was not possible to quantitatively remove and separate the different proteins, and the dissociation resulted in the aggregation of the membrane. This is similar to what has been previously observed with lipid-containing bacteriophage PRD1 (7).
Other P23-77 genes.
PSI-BLAST analysis of the putative proteins revealed possible functions for some of the proteins. ORF 13 encodes a putative ATPase (SMART domain prediction result: AAA ATPase, E-value 5.72e−03) containing conserved Walker A-, Walker B-, and PRD1-specific motifs (70) (Fig. 4). The PRD1 ATPase mediates the genome packaging into the capsid (70, 82); consequently, ORF 13 may also code for a packaging ATPase. The gene product of ORF 1 is similar to the replication initiation protein of Thermus sp. (ATCC 27737) plasmid pMY1 (22; GenBank no. CAA71700, E-value 0.0258). Since no polymerase genes were identified in the genome of P23-77, we assume that a host polymerase is used for the viral DNA synthesis. Predictions for conserved protein domains suggest that ORFs 9, 10, and 31 could encode a peptidase (E-value 1.10e−02), an amidase (4.50e−24), and a transglycosylase (8.10e−03), respectively, all catalyzing bacterial cell wall degradation that is needed in virus exit and sometimes in entry (26, 49, 80). For the product of gene 29 no conserved lysozyme domain was recognized, but a highly similar protein for which lysozyme activity has been demonstrated (51) was identified by BLAST (GenBank no. BAC55303, E-value 8.2e-81). The putative protein encoded by ORF 3 showed similarity to phosphoadenosine phosphosulfate reductases (for example, to such an enzyme from Streptomyces clavuligerus, GenBank no. ZP_03181578, E-value 5.29e-008) and protein structure prediction produced a significant hit to adenine nucleotide alpha hydrolase (E-value 8.60e-02). Other putative proteins of P23-77 showed no detectable similarities to any proteins with known functions.
FIG. 4.
Comparison of putative ATPase sequences from membrane-containing dsDNA viruses, plasmids, or genome integrated genetic elements of bacterial, archaeal, and eukaryotic origin. Walker A and B regions, as well as the phage PRD1 packaging ATPase P9 region that is conserved in all known PRD1-like viruses. Putative or experimentally demonstrated ATPase sequences from bacterial (P23-77, IN93, PRD1, AP50, Bam35, Gil16c, and PM2), archaeal (SH1, pHH205, IHP, and STIV), and eukaryotic viruses (mimivirus), plasmids, or genome integrated sequences are aligned (the acronyms IHP and STIV are defined in the text). Sequences were chosen on the basis of previously proposed evolutionary relationships of the viruses, structural and genetic comparisons, or sequence similarity to P23-77, as detected by BLAST. Amino acid residues identical in all sequences are depicted in red, conservative sequences are depicted in blue, and blocks of similar amino acids are depicted with a green background. The GenBank or RefSeq numbers of putative ATPase sequences are indicated in parentheses as follows: IN93 (BAC55291), SH1 (AY950802), pHH205 (YP_01687808), IHP (AAV45617), PRD1 (P27381), AP50 (ACB54903), Bam35 (NP_943760), STIV (AAS89100), PM2 (AF155037), and mimivirus (AAV50705).
P23-77 is homologous to T. aquaticus phage IN93.
A database search revealed that the DNA sequence of P23-77 is similar to that of the phage IN93 (GenBank no. AB063393) infecting T. aquaticus, a close relative to T. thermophilus. The IN93 genome has been submitted to GenBank, but its sequence has not been analyzed previously. The overall DNA-level identity between P23-77 and IN93 is 47.1%. Of the 37 ORFs of P23-77, 29 could also be recognized in IN93. A comparison of the genomes is presented in Table 2. The small and large major capsid proteins and the putative genome packaging ATPase were among the most conserved proteins since their amino acid level identities were 79, 74, and 79%, respectively.
IN93 genome is 2,567 bp longer than the genome of P23-77. IN93 contains a region (nucleotides 14481 to 17932) that includes genes on the opposite DNA strand with respect to all other genes of the genome. The lack of this region in P23-77 genome explains the differences in the genome sizes. A phage integrase (62), a restriction endonuclease, and a prophage repressor were predicted to be the gene products of the opposite-strand ORFs of IN93. P23-77 has no such an integration cassette. These observations are in accordance with the notion that IN93 is a prophage in strain T. aquaticus TZ2 (50) and that P23-77 is lytic.
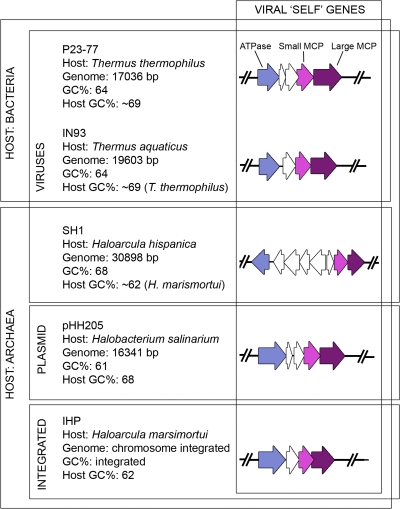
P23-77 virus elements are present in plasmids and viruses of halophilic archaea.
P23-77 has a predicted packaging ATPase and two major capsid proteins (see above). Interestingly, we discovered similar gene products in Haloarcula hispanica virus SH1 (GenBank no. AY950802), in the genome of Haloarcula marismortui ATCC 43049 (AY596297), and in Halobacterium salinarium plasmid pHH205 (AY048850 [79]). Despite its name, Halobacterium is an archaeal organism. The putative integrated plasmid or virus in Haloarcula marismortui (ATCC 43049) is situated within ca. 540 to 560 kb of the 3.13-Mb genome. This element is designated here as IHP (for integrated Haloarcula provirus). Most other putative proteins encoded by IHP are related to halophilic archaea (data not shown). However, the putative IHP protein rrnAC0597 (AAV45608) is 35% identical to Halorubrum phage HF2 putative protein CAOIfh, and one IHP gene is annotated as putative phage integrase (AAV45601). Alignments of the putative major capsid proteins encoded by these genetic elements are presented in Fig. 5. Although there are only few truly conserved amino acids, multiple amino acids are shared by three or four proteins. The arrangements of these similar sets of genes are presented in Fig. 6. The evolutionary history of major capsid genes was inferred by using the minimum-evolution method, and the obtained tree is represented in Fig. 7. Structure prediction of P23-77 large major capsid protein revealed a possible similarity to the major capsid protein of STIV (38) with a probability of 45% (PHYRE), thus suggesting a β-barrel fold for the protein. Comparison of the structure of P23-77 to that of SH1 (32, 33) suggests that P23-77 capsomers may be composed of hexamers of VP16 that are decorated with two or three copies of VP17 (depending on the capsomer type). Halobacterium salinarium plasmid pHH205 has previously been shown to carry a homologous recombination function (52). It is related to IHP since they share six putative proteins with identity values higher than 20%. The putative large major capsid protein, the putative small major capsid protein, and the putative ATPase share identities of 34.7, 27.1, and 33.3%, respectively, and are the three most conserved proteins. This strongly suggests that pHH205 is a provirus or a defective one.
FIG. 5.
Alignment of the large major capsid proteins (A) and small major capsid proteins (B) of the P23-77-like viruses.
FIG. 6.
Organization of the viral “self” genes of P23-77, IN93, SH1, pHH205, and IHP. These virus core genes demonstrate a homologous virus lineage with members infecting archaeal or bacterial hosts.
FIG. 7.
Minimum-evolution tree of large and major capsid proteins. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The units on the bar represent of the number of amino acid substitutions per site.
DISCUSSION
P23-77 was found to be related to T. aquaticus bacteriophage IN93. Most of the structural proteins of the phages have ca. 75% identity at the amino acid level, but for the 40.7-kDa protein encoded by gene 29 the identity was only 17.4%. Low identity might indicate a recent acquisition of the gene as a moron. Morons are foreign DNA fragments that have recombined into virus genome and are usually detectable by abnormal GC content (28). However, the GC content of gene 29 is the same as in the rest of the genome, suggesting alternative causes behind the low identity. Viral genes that encode receptor-binding proteins are known to evolve fast (6, 65); thus, one possibility is that VP29 has a role in receptor recognition. On the other hand, the product of a corresponding gene of IN93 genome (CD24) has been shown to be an active, thermostable lysozyme (51). Thus, VP29 could be a capsid-associated protein that (among other possible functions) facilitates the intrusion of the viral genome into cytoplasm by mediating the degradation of the protective host cell wall. Similar putative proteins also exist in two other Thermus-specific phages, P74-26 (GenBank no. ABU97052, BLAST E-value 2.2e−12) and P23-45 (GenBank no. ABU96936, E-value 8.1e−12), both assigned to the Siphoviridae family (54). A wide distribution of a potential capsid associated lysozyme among nonhomologous Thermus phages suggests that the activity of VP29-like proteins is crucial for the survival of Thermus viruses.
Based on genome and protein analyses of P23-77, 10 structural proteins encoding genes were identified (Fig. 2 and Table 1). Dissociation analyses revealed five structural proteins (VP15, VP19, VP20, VP22, and VP23) to associate with the viral membrane aggregate and three proteins (VP11, VP16, and VP17) with the external protein capsid. Dissociation studies of structurally related SH1 revealed a similar lipid core but containing only two major proteins: VP10 and VP12. At least eight of the SH1 proteins were capsid associated (VP1, VP2, VP3, VP4, VP5, VP6, VP7, and VP9) (39). Three of these (VP2, VP3, and VP6) were associated with the vertex spike structure (32). Regardless of the fact that P23-77 and SH1 have a similar capsid organization, the number of membrane-associated protein species differ. In the case of PRD1, most of the membrane-associated proteins are linked to the cell entry (26); consequently, the differences in the numbers of membrane-associated proteins may reflect differences in the entry mechanisms of these viruses. Such adaptations in entry strategies seem only logical given that the hosts of these viruses diverged a long time ago and thus differ in many ways.
Analysis of P23-77 and its host T. thermophilus lipid compositions (Fig. 1) indicates that some host lipids are selected to the viral membrane (spots 1, 3, and 7), whereas some others are almost completely excluded (spots 2 and 6 and glycolipids). The exact mechanisms of lipid selection during membrane assembly are not clear, but similar selective incorporation of lipids has been shown for archaeal virus SH1 (8) and for bacterial viruses PM2 and PRD1, both containing an internal lipid membrane (15, 16, 43). In all of these viruses, the lipid selection is thought to be driven, despite the structural differences between lipids of bacteria and archaea, by the physicochemical properties of lipids, such as the shape of the lipid molecule and the charge of the lipid head groups, as well as by their interaction with membrane proteins (44). Therefore, it is possible that the same factors are responsible for the selective incorporation of lipids into the membrane of P23-77. Recently, we isolated new types of archaeal lipid-containing viruses. They deviate structurally from icosahedral internal membrane containing viruses such as P23-77, and their lipids reflect the host lipids (unpublished data). Obviously, lipid composition allows grouping of viruses, thus linking P23-77 to PRD1-type viruses in this respect.
We found here that, in addition to SH1, the haloarchaeal plasmid pHH205, the T. aquaticus virus IN93, and a possible Haloarcula marismortui genome integrated (pro)virus (IHP) share similarity with P23-77. Previously, it has been shown that even if there is no detectable sequence similarity between viruses infecting hosts in the different domains of life, they may still have conserved coat protein fold and virion architecture. These observations have led to the proposal that viruses may be ancient, predating the separation of the three cellular domains of life (8, 41). The homologous relationship between P23-77, IN93, SH1, pHH205, and IHP was traceable by aligning the primary protein sequences (Fig. 4 and 5). This indicates that the proteins of these viral genetic elements were already conserved when the ancestor of P23-77 and SH1 diverged. If we assume that viruses have followed the phylogenetic branching of their hosts, then the time point for this was close to the existence of the last universal common ancestor. However, the other possibility is that bacterial and archaeal viruses could cross the domain barrier, making the separation of these viruses more recent. Consequently, these viruses could have mediated the observed horizontal gene transfer between the prokaryotic domains (48, 73). Nevertheless, genes that are not part of the self have most likely evolved and accumulated into the current genomes of P23-77 and SH1 only after their divergence (if we observe the genome from the “self” point of view). Genes responsible for the replication of genetic material and the machinery of genome entry, among others, may represent local adaptations that were essential for the viruses to survive within their current host organisms. In line with this is the notion that all viruses might not survive as true, virion-encoding parasites. Viruses may evolve into stable plasmids (as might have happened in the case of pHH205) or become permanently a part of the host chromosome (which might be the situation for IHP).
Finally, P23-77 type virions might be related to PRD1- and STIV-like viruses due to the similar putative genome packaging ATPases and the possible similar folding of the major capsid proteins. We envision an ancestor that contained a coating formed of a single β-barrel protein and which used an ATPase to include the genome within the coating structure. This ancestor then diverged into the double β-barrel lineage (PRD1) and into the lineage with a (putative) single β-barrel coat protein and a capsomer decorating protein (P23-77). In other words, while the packaging machinery remained fundamentally similar, these viruses evolved to stabilize their virions differently. Given this line of reasoning, P23-77 and SH1 form the earliest separating branch of the DNA viruses in the previously demonstrated β-barrel viral lineage (41). The presented notion suggests an interesting pathway for broadening our knowledge on the origin of viruses within the hypothesized communally evolving, predomain era of life (34, 75, 76).
Acknowledgments
We thank Elina Laanto for skillful technical assistance and Maija P. Jalasvuori for assistance with the bioinformatic analyses.
This study was supported by Finnish Centre of Excellence Program of the Academy of Finland (2006-2011) grant 1129648 (J.K.H.B. and D.H.B.) and grant 1210253 (D.H.B.).
Footnotes
Published ahead of print on 8 July 2009.
REFERENCES
- 1.Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. [DOI] [PubMed] [Google Scholar]
- 2.Abrescia, N. G., J. M. Grimes, H. M. Kivela, R. Assenberg, G. C. Sutton, S. J. Butcher, J. K. Bamford, D. H. Bamford, and D. I. Stuart. 2008. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Mol. Cell 31:749-761. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akita, F., K. T. Chong, H. Tanaka, E. Yamashita, N. Miyazaki, Y. Nakaishi, M. Suzuki, K. Namba, Y. Ono, T. Tsukihara, and A. Nakagawa. 2007. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J. Mol. Biol. 368:1469-1483. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, D. H. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231-236. [DOI] [PubMed] [Google Scholar]
- 6.Bamford, D. H., R. M. Burnett, and D. I. Stuart. 2002. Evolution of viral structure. Theor. Popul. Biol. 61:461-470. [DOI] [PubMed] [Google Scholar]
- 7.Bamford, D., and L. Mindich. 1982. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bamford, D. H., J. J. Ravantti, G. Rönnholm, S. Laurinavicius, P. Kukkaro, M. Dyall-Smith, P. Somerharju, N. Kalkkinen, and J. K. Bamford. 2005. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 79:9097-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamford, J. K., and D. H. Bamford. 1991. Large-scale purification of membrane-containing bacteriophage PRD1 and its subviral particles. Virology 181:348-352. [DOI] [PubMed] [Google Scholar]
- 10.Bartholomé, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 11.Bennett-Lovsey, R. M., A. D. Herbert, M. J. Sternberg, and L. A. Kelley. 2008. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program PHYRE. Proteins 70:611-625. [DOI] [PubMed] [Google Scholar]
- 12.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 13.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 14.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 15.Braunstein, S. N., and R. M. Franklin. 1971. Structure and synthesis of a lipid-containing bacteriophage. V. Phospholipids of the host BAL-31 and of the bacteriophage PM2. Virology 43:685-695. [DOI] [PubMed] [Google Scholar]
- 16.Brewer, G. J., and R. M. Goto. 1983. Accessibility of phosphatidylethanolamine in bacteriophage PM2 and in its gram-negative host. J. Virol. 48:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brock, T. D. 2005. Genus Thermus Brock and Freeze 1969, 295AL, p. 333-337. In J. G. Holt (ed.), Bergey's manual of systematic bacteriology. The Williams & Wilkins Co., Philadelphia, PA.
- 18.Butcher, S. J., T. Dokland, P. Ojala, D. H. Bamford, and S. D. Fuller. 1997. Intermediates in the assembly pathway of the double-stranded RNA virus phi6. EMBO J. 16:4477-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Böttcher, B., N. A. Kiselef, V. Y. Mashchuk, N. A. Perevozchikova, A. V. Borissov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldentey, J., L. Blanco, D. H. Bamford, and M. Salas. 1993. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 21:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caspar, D. L., and A. Klung. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1-24. [DOI] [PubMed] [Google Scholar]
- 22.de Grado, M., I. Lasa, and J. Berenguer. 1998. Characterization of a plasmid replicative origin from an extreme thermophile. FEMS Microbiol. Lett. 165:51-57. [DOI] [PubMed] [Google Scholar]
- 23.Dittmer, J. C., and R. L. Lester. 1964. A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 15:126-127. [PubMed] [Google Scholar]
- 24.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 25.Folch, J. M., M. Lees, and G. H. Sloane-Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissue. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 26.Grahn, A. M., R. Daugelavičius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46:1199-1209. [DOI] [PubMed] [Google Scholar]
- 27.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 28.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 29.Hyun, J. K., F. Coulibaly, A. P. Turner, E. N. Baker, A. A. Mercer, and A. K. Mitra. 2007. The structure of a putative scaffolding protein of immature poxvirus particles as determined by electron microscopy suggests similarity with capsid proteins of large icosahedral DNA viruses. J. Virol. 81:11075-11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer, L. M., S. Balaji, E. V. Koonin, and L. Aravind. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156-184. [DOI] [PubMed] [Google Scholar]
- 32.Jaalinoja, H. T., E. Roine, P. Laurinmaki, H. M. Kivela, D. H. Bamford, and S. J. Butcher. 2008. Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc. Natl. Acad. Sci. USA 105:8008-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaatinen, S. T., L. J. Happonen, P. Laurinmaki, S. J. Butcher, and D. H. Bamford. 2008. Biochemical and structural characterisation of membrane-containing icosahedral dsDNA bacteriophages infecting thermophilic Thermus thermophilus. Virology 379:10-19. [DOI] [PubMed] [Google Scholar]
- 34.Jalasvuori, M., and J. K. Bamford. 2008. Structural co-evolution of viruses and cells in the primordial world. Orig. Life Evol. Biosph. 38:165-181. [DOI] [PubMed] [Google Scholar]
- 35.Jalasvuori, M., and J. K. Bamford. 2009. Did the ancient crenarchaeal viruses from the dawn of life survive exceptionally well the eons of meteorite bombardment? Astrobiology 9:131-137. [DOI] [PubMed] [Google Scholar]
- 36.Kahma, K., J. Brotherus, M. Haltia, and O. Renkonen. 1976. Low and moderate concentrations of lysobisphosphatidic acid in brain and liver of patients affected by some storage diseases. Lipids 11:539-544. [DOI] [PubMed] [Google Scholar]
- 37.Kates, M. 1972. Techniques of lipidology: isolation, analysis, and identification of lipids, p. 344. In T. S. Work and E. Work (ed.), Laboratory techniques in biochemistry and molecular biology, vol. 3. North-Holland Publishing Company, Amsterdam, The Netherlands. [Google Scholar]
- 38.Khayat, R., L. Tang, E. T. Larson, C. M. Lawrence, M. Young, and J. E. Johnson. 2005. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc. Natl. Acad. Sci. USA 102:18944-18949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kivelä, H. M., E. Roine, P. Kukkaro, S. Laurinavicius, P. Somerharju, and D. H. Bamford. 2006. Quantitative dissociation of archaeal virus SH1 reveals distinct capsid proteins and a lipid core. Virology 356:4-11. [DOI] [PubMed] [Google Scholar]
- 40.Koonin, E. V., T. G. Senkevich, and V. V. Dolja. 2006. The ancient virus world and evolution of cells. Biol. Direct. 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krupovic, M., and D. H. Bamford. 2008. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat. Rev. Microbiol. 6:941-948. [DOI] [PubMed] [Google Scholar]
- 42.Krupovic, M., and D. H. Bamford. 2008. Archaeal proviruses TKV4 and MVV extend the PRD1-adenovirus lineage to the phylum Euryarchaeota. Virology 375:292-300. [DOI] [PubMed] [Google Scholar]
- 43.Laurinavicius, S., R. Käkelä, P. Somerharju, and D. H. Bamford. 2004. Phospholipid molecular species profiles of tectiviruses infecting gram-negative and gram-positive hosts. Virology 322:328-336. [DOI] [PubMed] [Google Scholar]
- 44.Laurinavicius, S., D. H. Bamford, and P. Somerharju. 2007. Transbilayer distribution of phospholipids in bacteriophage membranes. Biochim. Biophys. Acta 1768:2568-2577. [DOI] [PubMed] [Google Scholar]
- 45.Laurinmäki, P. A., J. T. Huiskonen, D. H. Bamford, and S. J. Butcher. 2005. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 13:1819-1828. [DOI] [PubMed] [Google Scholar]
- 46.Leone, S., A. Molinaro, B. Lindner, I. Romano, B. Nicolaus, M. Parrilli, R. Lanzetta, and O. Holst. 2006. The structures of glycolipids isolated from the highly thermophilic bacterium Thermus thermophilus Samu-SA1. Glycobiology 16:766-775. [DOI] [PubMed] [Google Scholar]
- 47.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin, Z., M. Nei, and H. Ma. 2007. The origins and early evolution of DNA mismatch repair genes: multiple horizontal gene transfers and co-evolution. Nucleic Acids Res. 35:7591-7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loessner, M. J. 2005. Bacteriophage endolysins: current state of research and applications. Curr. Opin. Microbiol. 8:480-487. [DOI] [PubMed] [Google Scholar]
- 50.Matsushita, I., N. Yamashita, and A. Yokota. 1995. Isolation and characterization of bacteriophage induced from a new isolate of Thermus aquaticus. Microbiol. Cult. Coll. 11:133-138. [Google Scholar]
- 51.Matsushita, I., and H. Yanase. 2008. A novel thermophilic lysozyme from bacteriophage phiIN93. Biochem. Biophys. Res. Commun. 377:89-92. [DOI] [PubMed] [Google Scholar]
- 52.Mei, Y., D. Chen, D. Sun, X. Wang, Y. Huang, X. Chen, and P. Shen. 2007. Identification homologous recombination function from haloarchaea plasmid pHH205. Curr. Microbiol. 55:76-80. [DOI] [PubMed] [Google Scholar]
- 53.Merckel, M. C., J. T. Huiskonen, D. H. Bamford, A. Goldman, and R. Tuma. 2005. The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol. Cell 18:161-170. [DOI] [PubMed] [Google Scholar]
- 54.Minakhin, L., M. Goel, Z. Berdygulova, E. Ramanculov, L. Florens, G. Glazko, V. N. Karamychev, A. I. Slesarev, S. A. Kozyavkin, I. Khromov, H. W. Ackermann, M. Washburn, A. Mushegian, and K. Severinov. 2008. Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23-45 and P74-26: siphoviruses with triplex-forming sequences and the longest known tails. J. Mol. Biol. 378:468-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naitow, H., J. Tang, M. Canady, R. B. Wickner, and J. E. Johnson. 2002. L-A virus at 3.4 Å resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9:725-728. [DOI] [PubMed] [Google Scholar]
- 56.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 58.Poutanen, M., L. Salusjarvi, L. Ruohonen, M. Penttila, and N. Kalkkinen. 2001. Use of matrix-assisted laser desorption/ionization time-of-flight mass mapping and nanospray liquid chromatography/electrospray ionization tandem mass spectrometry sequence tag analysis for high sensitivity identification of yeast proteins separated by two-dimensional gel electrophoresis. Rapid Commun. Mass Spectrom. 15:1685-1692. [DOI] [PubMed] [Google Scholar]
- 59.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 60.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 61.Rzhetsky, A., and M. Nei. 1992. A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 9:945-967. [Google Scholar]
- 62.Ruan, L., and X. Xu. 2007. Sequence analysis and characterizations of two novel plasmids isolated from Thermus sp. 4C. Plasmid 58:84-87. [DOI] [PubMed] [Google Scholar]
- 63.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 64.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 65.Saren, A. M., J. J. Ravantti, S. D. Benson, R. M. Burnett, L. Paulin, D. H. Bamford, and J. K. Bamford. 2005. A snapshot of viral evolution from genome analysis of the Tectiviridae family. J. Mol. Biol. 350:427-440. [DOI] [PubMed] [Google Scholar]
- 66.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 67.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, A. Shevchenko, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simpson, A. A., N. Nandhagopal, J. L. Van Etten, and M. G. Rossmann. 2003. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr. D Biol. Crystallogr. 59:2053-2059. [DOI] [PubMed] [Google Scholar]
- 69.Skipski, V. P., and M. Barclay. 1969. Thin-layer chromatography of lipids. Methods Enzymol. 14:530-598. [Google Scholar]
- 70.Stromsten, N. J., D. H. Bamford, and J. K. Bamford. 2005. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J. Mol. Biol. 348:617-629. [DOI] [PubMed] [Google Scholar]
- 71.Takezaki, N., A. Rzhetsky, and M. Nei. 2004. Phylogenetic test of the molecular clock and linearized trees. Mol. Biol. Evol. 12:823-833. [DOI] [PubMed] [Google Scholar]
- 72.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 73.Urbonavicius, J., S. Auxilien, H. Walbott, K. Trachana, B. Golinelli-Pimpaneau, C. Brochier-Armanet, and H. Grosjean. 2008. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol. Microbiol. 67:323-335. [DOI] [PubMed] [Google Scholar]
- 74.Wickner, R. B. 1996. Double-stranded RNA viruses of yeast. Microbiol. Rev. 60:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woese, C. R. 1998. The universal ancestor. Proc. Natl. Acad. Sci. USA 95:6854-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woese, C. R. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. USA 99:8742-8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan, X., P. R. Chipman, T. Castberg, G. Bratbak, and T. S. Baker. 2005. The marine algal virus PpV01 has an icosahedral capsid with T=219 quasisymmetry. J. Virol. 79:9236-9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang, Y. L., F. L. Yang, S. C. Jao, M. Y. Chen, S. S. Tsay, W. Zou, and S. H. Wu. 2006. Structural elucidation of phosphoglycolipids from strains of the bacterial thermophiles Thermus and Meiothermus. J. Lipid Res. 47:1823-1832. [DOI] [PubMed] [Google Scholar]
- 79.Ye, X., J. Ou, L. Ni, W. Shi, and P. Shen. 2003. Characterization of a novel plasmid from extremely halophilic Archaea: nucleotide sequence and function analysis. FEMS Microbiol. Lett. 221:53-57. [DOI] [PubMed] [Google Scholar]
- 80.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu, M. X., M. R. Slater, and H. W. Ackermann. 2006. Isolation and characterization of Thermus bacteriophages. Arch. Virol. 151:663-679. [DOI] [PubMed] [Google Scholar]
- 82.Žiedaitė, G., H. M. Kivelä, J. K. H. Bamford, and D. H. Bamford. 2009. Purified membrane-containing procapsids of bacteriophage PRD1 package the viral genome. J. Mol. Biol. 386:637-647. [DOI] [PubMed] [Google Scholar]
- 83.Zuckerkandl, E., and L. Pauling. 1965. Evolutionary divergence and convergence in proteins, p. 97-166. In V. Bryson and H. J. Vogel (ed.), Evolving genes and proteins. Academic Press, Inc., New York, NY.