Abstract
Selected vulnerability of neurons in Huntington’s disease (HD) suggests alterations in a cellular process particularly critical for neuronal function. Supporting this idea, pathogenic Htt (polyQ-Htt) inhibits fast axonal transport (FAT) in various cellular and animal HD models (mouse and squid), but the molecular basis of this effect remains unknown. Here we show that polyQ-Htt inhibits FAT through a mechanism involving activation of axonal JNK. Accordingly, increased activation of JNK was observed in vivo in cellular and animal HD models. Additional experiments indicate that polyQ-Htt effects on FAT are mediated by the neuron-specific JNK3, and not ubiquitously expressed JNK1, providing a molecular basis for neuron-specific pathology in HD. Mass spectrometry identified a residue in the kinesin-1 motor domain phosphorylated by JNK3, and this modification reduces kinesin-1 binding to microtubules. These data identify JNK3 as a critical mediator of polyQ-Htt toxicity and provides a molecular basis for polyQ-Htt-induced inhibition of FAT.
Keywords: Huntingtin, Huntington’s disease, conventional kinesin, kinesin-1, JNK, JNK3, axonal transport, neurodegeneration
Huntington’s disease (HD) is an autosomal dominant, adult-onset neurodegenerative disease1. Patients with a single mutant huntingtin (Htt) gene develop symptoms in midlife with 100% penetrance. The mutation is a toxic gain of function, because loss of a single gene has no phenotype and the Htt-null is embryonic lethal2. The disease-causing mutation is expansion of a CAG trinucleotide repeat encoding a polyglutamine (polyQ) tract and polyQ-expansions in other genes may also lead to adult-onset neurodegeneration, but pathogenic mechanisms remained uncertain2. Several lines of evidence indicated that pathogenesis in Huntington’s disease include inhibition of fast axonal transport (FAT)3. Although consequences of FAT inhibition in neurons are well established, mechanisms underlying inhibition of FAT by pathogenic Htt (polyQ-Htt) were unknown. Proposed mechanisms included wild-type Htt (WT-Htt) loss of function4, physical blockade by polyQ-Htt aggregates5,6, sequestration of motor molecules4, 6, and misregulation of FAT3, 7, 8.
We found no evidence for direct interactions of conventional kinesin and cytoplasmic dynein (CDyn) with WT-Htt or polyQ-Htt in vivo. However, analysis of HD models showed that polyQ-Htt, but not WT-Htt increased cJun N-terminal kinase (JNK) activity. Pharmacological and peptide inhibitors of JNK prevented inhibition of FAT by polyQ-Htt. Surprisingly, only a subset of JNK isoforms inhibited FAT, with neuron-specific JNK3 selectively mimicking effects of polyQ-Htt. In vitro phosphorylation and mass spectrometry studies showed that JNK3, but not ubiquitously expressed JNK1, phosphorylated Ser176 in the kinesin heavy chain (kinesin-1, KHC) motor domain. Consistent with this location, phosphorylation of kinesin-1 by JNK3 inhibited kinesin binding to microtubules and translocation along axons. Our data indicate that polyQ-Htt inhibits FAT by a mechanism involving axonal JNK3 activation and phosphorylation of kinesin-1.
RESULTS
Htt does not interact with microtubule-based motors
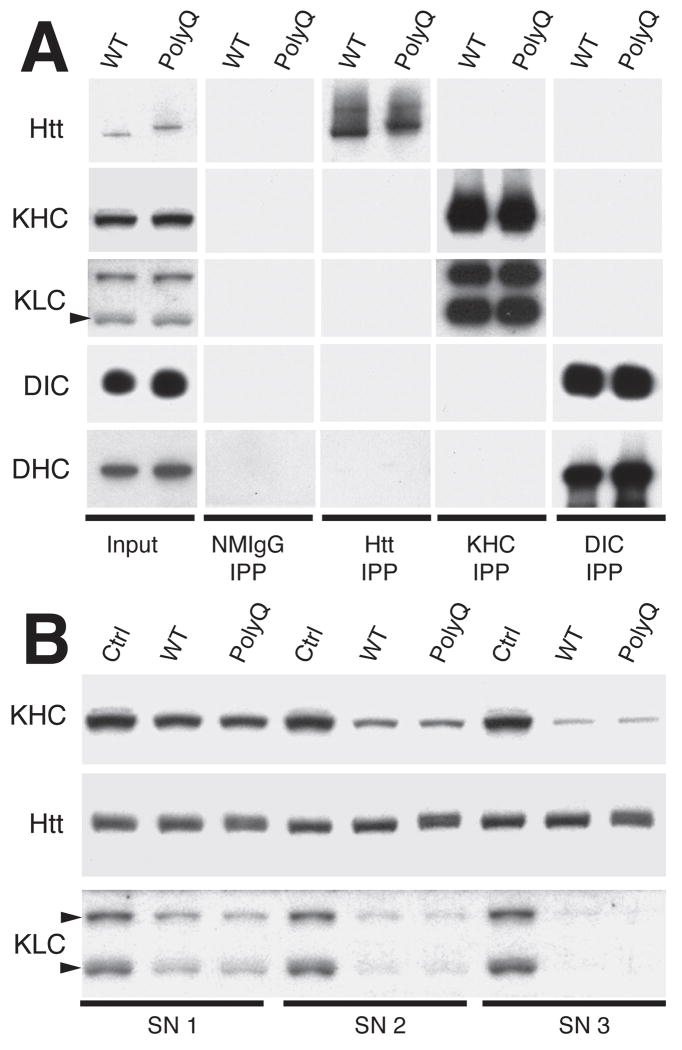
PolyQ-Htt inhibits FAT in various experimental systems, including Drosophila4, 5, neuroblastoma cells9, 10, and isolated squid axoplasm8, but the molecular basis of inhibition was undetermined3. Interactions have been reported between exogenously overexpressed Htt and conventional kinesin4 or various subunits of cytoplasmic dynein (CDyn)9-11. Alternatively, polyQ-expansion was proposed to affect Htt function as a scaffolding protein for molecular motors9, 12. However, interactions between endogenous WT-Htt and molecular motors were not evaluated. We tested interactions by immunoprecipitation and subcellular fractionation (Fig. 1), as previously described13, 14. To avoid overexpression-related artifacts, we used brain tissue from 14 month-old homozygous HdhQ109 knock-in mice and age-matched controls, which express polyQ-Htt or WT-Htt at endogenous levels15. At this age, both polyQ-Htt-derived nuclear inclusions and insoluble aggregates are found in the brains of HdhQ109 knock-in mice15. Conventional kinesin is a heterotetramer composed of two heavy chains (kinesin-1, KHC) and two light chains (KLC)13. Antibodies recognizing kinesin-1 (H2) effectively immunoprecipitated kinesin-1 from detergent-soluble brain lysates independent of genotype (Fig. 1A). Kinesin-1 antibodies co-immunoprecipitated KLCs13, 14, but failed to co-immunoprecipitate either WT-Htt or polyQ-Htt. Similarly, antibodies against DIC co-immunoprecipitated DHC16, but did not immunoprecipitate WT-Htt or polyQ-Htt (Fig. 1A). Conversely, anti-Htt antibodies immunoprecipitated Htt from both wild type and homozygous HdhQ109 knock-in mouse brain lysates, but no kinesin-1, KLC, DIC nor DHC could be detected in Htt immunoprecipitates. To detect substoichiometric amounts of Htt associated with conventional kinesin or CDyn, we performed three rounds of immunoprecipitation, sufficient to nearly deplete mouse brain lysates of kinesin-1 (Fig. 1B) or DIC (Supplemental Fig. 1B). As in previous studies, marked reductions in kinesin-1 and KLC levels occurred with each immunoprecipitation cycle13, 14. However, Htt levels remained unchanged after three immunoprecipitation cycles, regardless of genotype.
Figure 1. Endogenous Htt does not interact with molecular motors.
A) Detergent-soluble brain lysates obtained from wild type (WT) and 14 month-old HdhQ109 knock-in (polyQ) mice were immunoprecipitated with antibodies against Htt, kinesin-1 (KHC), or dynein intermediate chains (DIC). Immunoblots of resulting immunoprecipates (IPP) showed that anti-kinesin-1 antibodies (H213) effectively precipitated both kinesin-1 heavy (KHC) and light chains (KLC), but failed to precipitate Htt, dynein heavy chain (DHC) or dynein intermediate chain (DIC). Similarly, anti-DIC antibodies (74.116) immunoprecipitated DIC and DHC, but not Htt. Conversely, anti-Htt antibodies effectively immunoprecipitated Htt, but not KHC, KLC, DIC or DHC. Immunoprecipitation with non-immune mouse IgG (NMIgG) provided a control for non-specific immunoprecipitation. An aliquot of each brain lysate before immunoprecipitation (Input) was used as a positive control. B) Detergent-soluble brain lysates from wild type (WT-Htt) and HdhQ109 knock-in (polyQ-Htt) mice were subjected to three cycles of immunoprecipitation with antibodies against kinesin-1. Aliquots of each supernatant (SN1-3) were analyzed by immunoblot. Note reductions in both KHC and KLC immunoreactivity with each cycle. In contrast, no change in Htt levels was detected, regardless of mouse genotype. Immunoprecipitations with a non-immune mouse IgG (Ctrl) served as a control for non-specific precipitation of proteins.
PolyQ-Htt was reported to sequester molecular motors in detergent-insoluble aggregates when overexpressed4, 6. To evaluate this at endogenous levels, brain lysates from wild type and homozygous HdhQ109 knock-in mice were fractionated into detergent-soluble and insoluble fractions, and partitioning of Htt and molecular motors analyzed by immunoblot (Supplemental Fig. 1). WT-Htt and polyQ-Htt levels were comparable in detergent soluble and insoluble fractions, but the bulk of kinesin-1, DHC and DIC were detergent-soluble. Molecular motor levels were similar for wild type and homozygous HdhQ109 knock-in mice. Thus, WT-Htt and polyQ-Htt expressed at endogenous levels do not interact with molecular motors; so FAT inhibition associated with polyQ-Htt must result from a different mechanism.
JNK activity mediates FAT defects induced by polyQ-Htt
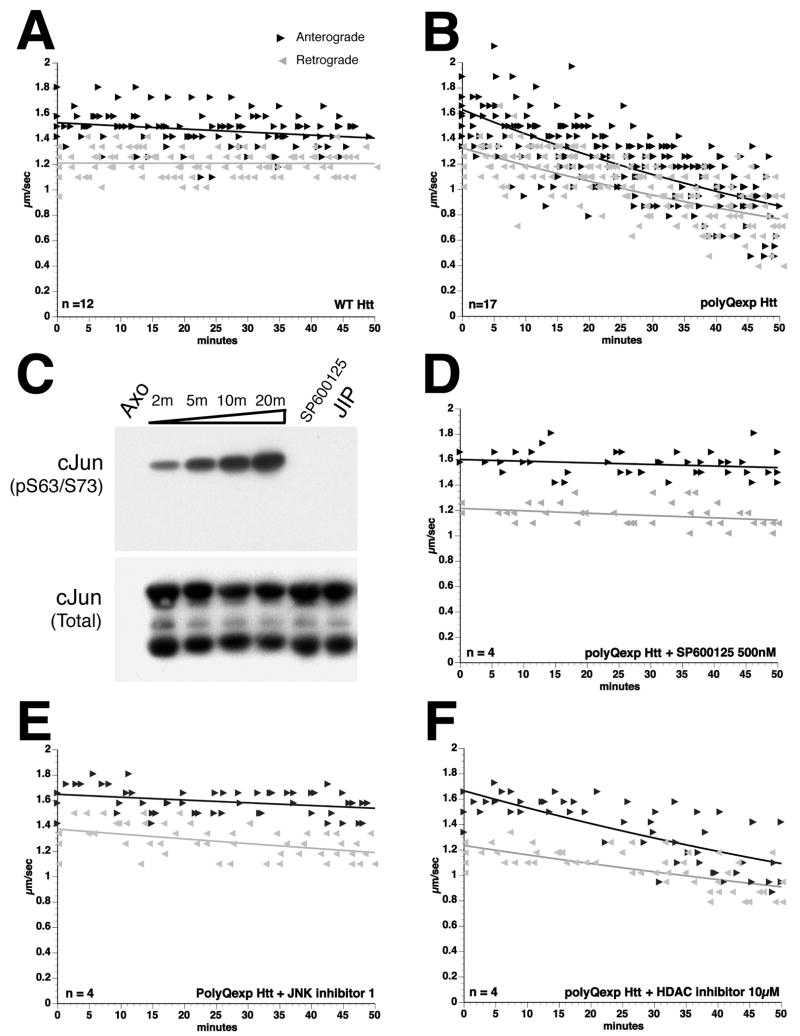
Effects of WT-Htt and polyQ-Htt on FAT were assayed in isolated squid axoplasm8. Perfusion of WT-Htt showed no effect (Fig. 2A), but perfusion of polyQ-Htt at a concentration 100-1000 times lower than kinesin-13, 8 dramatically inhibited both anterograde (kinesin-dependent) and retrograde (CDyn-dependent) FAT (Fig. 2B). In axoplasm, anterograde FAT of membrane-bounded organelles (MBOs) depends primarily upon conventional kinesin17, which is regulated by phosphorylation7, 18, 19. Multiple kinase activities may be deregulated in Huntington’s disease20-22 and inhibition of FAT by pathogenic androgen receptor (polyQ-AR) depends on JNK activation in axoplasm7. This led us to evaluate the role of JNK in polyQ-Htt-induced inhibition of FAT. Axoplasmic JNK activity was assessed in vitro using recombinant GST-cJun, a well-characterized JNK substrate23 (Fig. 2C). Incubation with axoplasm lysates produced a time-dependent increase in cJun phosphorylation at JNK sites (serines 63/73), reflecting basal JNK activity in axoplasm. Pharmacological properties of endogenous axoplasmic JNK were validated with two JNK inhibitors (SP600125 and JIP peptide)23. SP600125 is a pharmacological inhibitor of JNK, showing >20-fold selectivity for JNK over a range of kinases24. JIP peptide (JNK inhibitor I) is a 20-amino acid peptide derived from a JNK-binding protein, which selectively inhibits JNK, but not p38 activity25. Both SP600125 and JIP peptide dramatically inhibited cJun phosphorylation, suggesting common pharmacological properties between squid and mammalian JNKs (Fig. 2C).
Figure 2. JNK inhibitors prevent polyQ-Htt-induced FAT inhibition.
Vesicle motility assays in isolated axoplasm show individual velocity measurements (arrowheads) as a function of time. Dark arrowheads and line show anterograde kinesin-dependent FAT rates and reverse grey arrowheads and line show retrograde, cytoplasmic dynein-dependent FAT rates. In vitro translated Htt constructs were perfused as before8. A) Perfusion of WT-Htt does not affect FAT8 B) Perfusion of polyQ-Htt inhibited both anterograde and retrograde FAT8. C) Endogenous JNK activity in squid axoplasm is revealed by in vitro phosphorylation assays. A time-dependent increase in cJun phosphorylation at JNK sites (cJun S63/73) is seen when GST-cJun is incubated with squid axoplasm lysates (Axo). Phosphorylation of GST-cJun is completely inhibited by either SP600125 or JIP peptide (JIP), two well-characterized JNK inhibitors. Anti-GST antibody shows similar GST-cJun levels for each reaction (cJun Total). Co-perfusion of polyQ-Htt with either D) SP600125 (500nM) or E) JIP peptide (100 μM) completely prevented inhibition of FAT elicited by polyQ-Htt (compare D-E with B). E) In contrast, co-perfusion of polyQ-Htt with the histone deacetylase inhibitor BC-6-2527 (10μM) failed to prevent polyQ-Htt effects on FAT. These results indicate that polyQ-Htt-mediated inhibition of FAT involves activation of endogenous axoplasmic JNK.
As with polyQ-AR7, co-perfusion of axoplasm with polyQ-Htt and either SP600125 (500nM, Fig. 2D) or JIP peptide (100 nM, Fig. 2E) blocked effects of polyQ-Htt on FAT, suggesting that polyQ-Htt inhibition of FAT requires JNK activity. In cultured cells, pharmacological inhibitors of histone deacetylase (HDAC) were reported to rescue defects in FAT induced by polyQ-Htt by increasing tubulin acetylation26. However, coperfusion of polyQ-Htt with the HDAC6 inhibitor BC-6-25 (10μM) (Compound 227) did not prevent inhibition of FAT by polyQ-Htt in axoplasm (Fig. 2F). Thus, inhibition of FAT by polyQ-Htt involves activation of endogenous axonal JNK.
PolyQ-Htt expression increases JNK activity
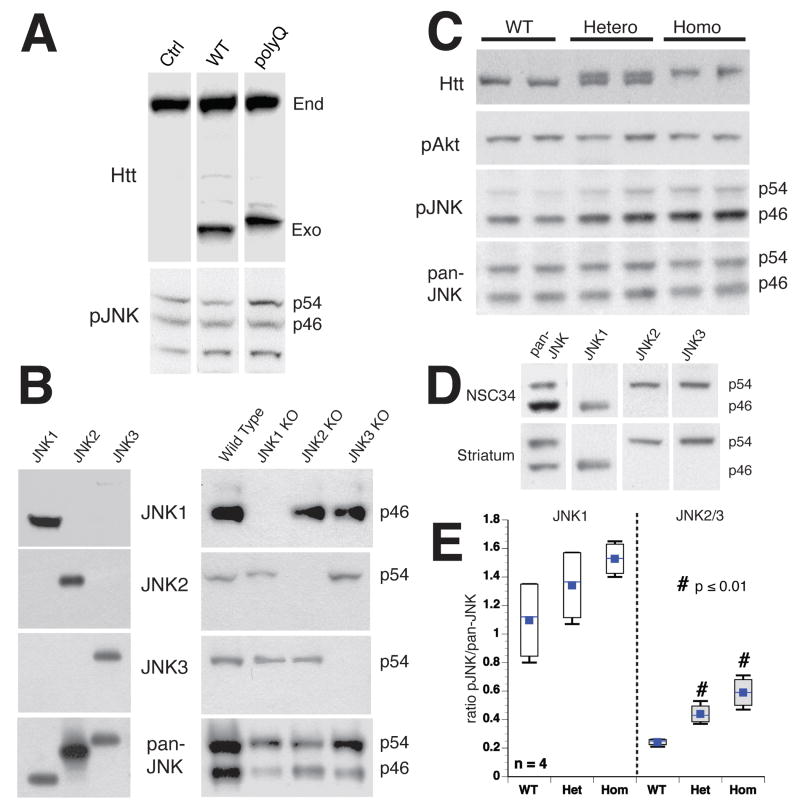
Vesicle motility and biochemical assays indicated that JNK activity mediates polyQ-Htt inhibition of FAT. JNK activation involves phosphorylation by upstream mitogen-activated protein kinase kinases (MKKs, typically MKK4/MKK7)28. Effects of polyQ-Htt on JNK phosphorylation were evaluated in cellular and animal Huntington’s models, using antibodies against total JNK (pan-JNK, phosphorylation-independent) or against phosphorylated, catalytically active JNKs (pJNK). Effects of polyQ-Htt expression on JNK activation were evaluated qualitatively after transient transfection (Fig. 3A). NSC34 neuroblastoma cells were transfected with WT-Htt (Q18) or polyQ-Htt (Q56)29. Immunoblots show exogenous Htt constructs were expressed at levels similar to endogenous Htt. Pan-JNK and pJNK antibodies recognized two major immunoreactive bands (p46 and p54) corresponding to mammalian JNKs in NSC34 cell lysates and striatal mouse tissue (Fig. 3A, C)30. Expression levels for both pan-JNK immunoreactive bands were comparable in cells expressing WT-Htt and polyQ-Htt. However, pJNK antibodies showed a clear increase in p54 immunoreactivity in cells expressing polyQ-Htt (Fig. 3A), consistent with previous reports20, 22, 31.
Figure 3. PolyQ-Htt increases JNK activity in Huntington’s disease models.
JNK activation was evaluated by immunoblots with active JNK (pJNK) and total JNK (pan-JNK) antibodies, which mainly recognized bands at p54 and p46. A) NSC34 cells transfected with WT-Htt (18Q) or polyQ-Htt (56Q) constructs were analyzed after 24h. Both endogenous (End) and exogenous (Exo) Htt were detectable. Total JNK (Pan-JNK) was similar for untransfected (Ctrl), WT-Htt (WT) and polyQ-Htt (polyQ) transfected cells, but phosphorylated p54 JNK (pJNK) increased with polyQ-Htt. B) JNK1, JNK2 and JNK3 antibody specificity were validated with GST-JNKs (left), and striatal lysates with individual JNK genes ablated (right). JNK1, JNK2 and JNK3 antibodies showed no immunoreactivity in brains from corresponding knockout samples. C) Striata from wild type (WT), heterozygous (Hetero) and homozygous (Homo) HdhQ109 mice showed similar Total JNK (pan-JNK) regardless of genotype, whereas active JNK (pJNK) increased in HdhQ109 mice. Akt activation (p-Akt) was similar for all mice. D) Immunoblots using NSC34 (top) and mouse brain (bottom) showed JNK1 co-migrating with p46, whereas JNK2 and JNK3 co-migrated with p54. E) Quantitative analysis of immunoblots in C showed activation of both p54 and p46 JNK in HdhQ109 mice. pJNK/JNK ratios for JNK1 (p46) or JNK2/3 (p54) indicated that both JNK2/3 and JNK1 activity were higher in homozygous than heterozygous HdhQ109 mice. JNK2/3 was activated to a greater extent than JNK1, suggesting differential activation of JNKs by polyQ-Htt. Differences between JNK2/3 activity in WT and HdhQ109 were significant (p ≤ 0.01 in a pooled t-test).
Differential activation of p54 and p46 JNKs led us to evaluate their identity using antibodies selective for JNK1, JNK2 and JNK3. Specificity was validated by immunoblotting against recombinant GST-tagged JNKs, as well as brain lysates from mice with ablation of individual JNK genes30 (Fig. 3B). Anti-JNK1, 2 and 3 antibodies selectively recognized their target JNK (Fig. 3B), showing no immunoreactivity against brain lysates from mice deleted for that JNK gene (Fig. 3B). We examined JNK expression in striatal tissue from 8 month-old wild-type, heterozygous and homozygous HdhQ109 mice (Fig. 3C). HdhQ109 knock-in mice are asymptomatic at this age15. Phosphorylation of Akt at Ser473 was not affected (Fig. 3C) and pan-JNK immunoreactivity was comparable, regardless of genotype. However, pJNK immunoreactivity was significantly greater in lysates from heterozygous and homozygous HdhQ109 knock-in mice, relative to wild-type (Fig. 3C). Cortical lysates gave similar results (data not shown). P54 and p46 were differentially activated as observed in transfected cells. P54 showed greater increases in pJNK immunoreactivity than p46 in HdhQ109 mice relative to wild type in both striatum (Fig. 3E) and cortex (not shown). Consistent with autosomal dominance of HD, JNK activation increased in both heterozygous and homozygous HdhQ109 mice, but activation was greater in homozygotes than heterozygotes.
Immunoreactivity corresponding to p54 and p46 in NSC34 and striatal lysates mainly corresponded to JNK2/3 and JNK1, respectively (Fig. 3D). An activation ratio (Fig. 3E) was defined as the ratio of pJNK/JNK immunoreactivities for JNK1 (p46) or JNK2/3 (p54). Both JNK1 and JNK2/3 activation ratios were elevated in heterozygotes and homozygotes relative to wild-type brain. JNK1 activity in wild-type mice was 1.097±0.146 (mean ± SEM), whereas JNK1 activity was 1.527±0.061 in homozygotes and 1.342±0.133 in heterozygotes HdhQ109 mice (n = 4 in all cases). Differences in JNK1 activity between wild-type and HdhQ109 homozygotes was significant at p≤0.05 in a pooled t-test, but failed to reach significance between wild-type and HdhQ109 heterozygotes. JNK2/3 activity for wild-type mice was 0.242±0.012 (mean ± SEM), whereas JNK2/3 activity was 0.590±0.055 for homozygotes and 0.440±0.035 for heterozygotes (n = 4 in all cases). JNK2/3 activity increased in both homozygote and heterozygote HdhQ109 mice relative to WT mice and differences were significant at p≤0.01 in a pooled t-test. Both JNK1 and JNK2/3 activities were increased by polyQ-Htt, but polyQ-Htt had a greater effect on JNK2/3 than JNK1.
JNK3 mediates effects of polyQ-Htt on FAT
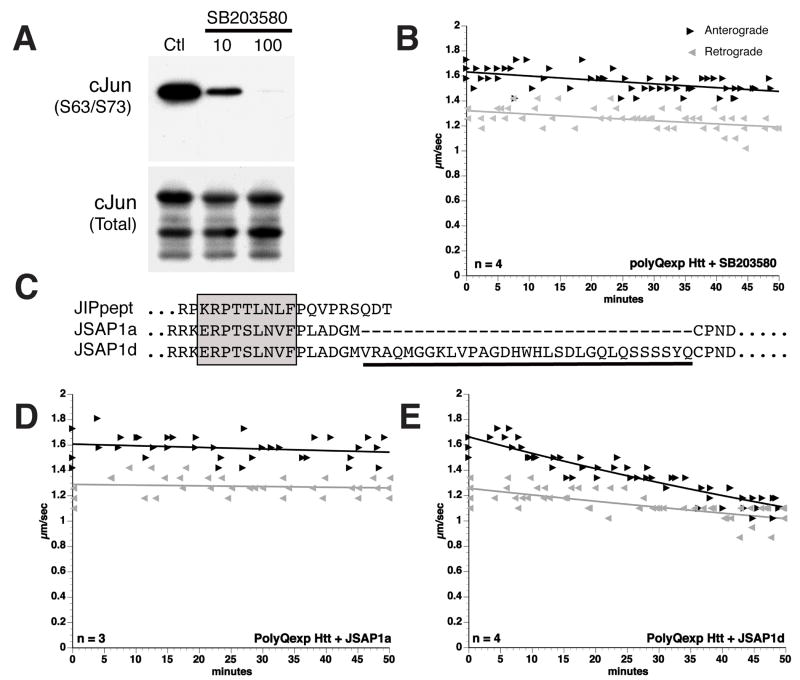
JNK2/3 is selectively activated in neurons by stress in the presence of constitutive JNK1 activity32, 33. Based on differential activation of JNKs (Fig. 3A–B), effects of specific JNK isoforms on FAT were evaluated. SB203580 differentially affects JNK1 and JNK2/3 in vitro24. At 10μM concentration, SB203580 preferentially inhibits JNK2/3; JNK1 inhibition requires a 10-fold higher concentration23. Axoplasmic JNK activity was almost eliminated by 100μM SB203580, but significant JNK activity remained with 10μM SB203580 (Fig. 4A). However, 10μM SB203580 completely prevented inhibition of FAT by polyQ-Htt (Fig. 4B), suggesting that polyQ-Htt effects involved JNK2 or JNK3. Significantly, 10μM SB203580 also prevented inhibition of FAT by polyQ-AR7. JNK isoforms were distinguished using biochemical approaches. JSAP1 is a scaffold protein regulating JNK activation34. Four JSAP1 isoforms were identified (a,b,c and d), all containing a 17-amino acid JNK binding domain (JBD) homologous to the JIP peptide (Fig. 4C). A 31 amino acid insert in JSAP1c/JSAP1d selectively reduces binding to JNK334. As a result, JSAP1d binds JNK3 with lower affinity than JSAP1a, and inhibits JNK3 less efficiently34. Recombinant JSAP1a (amino acids 115–233) and JSAP1d (amino acids 115–264) polypeptides were expressed and purified, then co-perfused at 5μM with polyQ-Htt (Fig. 4D). Perfusion of JSAP1a and JSAP1d alone had no effect on FAT (not shown). As with JIP peptide (Fig. 2E), co-perfusion of polyQ-Htt and JSAP1a completely prevented inhibition of FAT by polyQ-Htt (Fig. 4D). In contrast, JSAP1d reduced, but did not eliminate polyQ-Htt effects on FAT (Fig. 4E), suggesting JNK3 mediates polyQ-Htt effects in FAT.
Figure 4. The effects of polyQ-Htt on FAT are mediated by JNK3.
SB203580 blocks the action of pathogenic androgen receptor on FAT8, but SB203580 is a poor inhibitor of JNK1 (IC50≥100 μM) and a good inhibitor of JNK2/3 (IC50≈10μM)23. A)In vitro phosphorylation assays in axoplasm showed a dose-dependent inhibition of axoplasmic JNK activity by SB203580. Notably, significant JNK activity remained in the presence of 10μM SB203580, presumably representing endogenous JNK1 activity. B) In contrast, vesicle motility assays in isolated squid axoplasm show that effects of polyQ-Htt on FAT were blocked by SB203580 at 10μM, which is consistent with an action on JNK2/3. C) Sequence alignment of a JIP peptide inhibitor (Calbiochem) with portions of JSAP1a and JSAP1d polypeptides34. The boxed region indicates the highly conserved JNK binding domain (JBD). A 31-aminoacid peptide sequence unique to JSAP1d (underlined) selectively interferes with binding to JNK334. D) Co-perfusion of polyQ-Htt with JSAP1a (5 μM) which blocks both JNK1 and JNK3 activity, effectively prevents effects of polyQ-Htt on FAT. E) Co-perfusion of polyQ-Htt with JSAP1d (5μM), which preferentially inhibits JNK1, only partially blocks polyQ-Htt effects. These results suggest that JNK3 mediates the effects of polyQ-Htt on FAT.
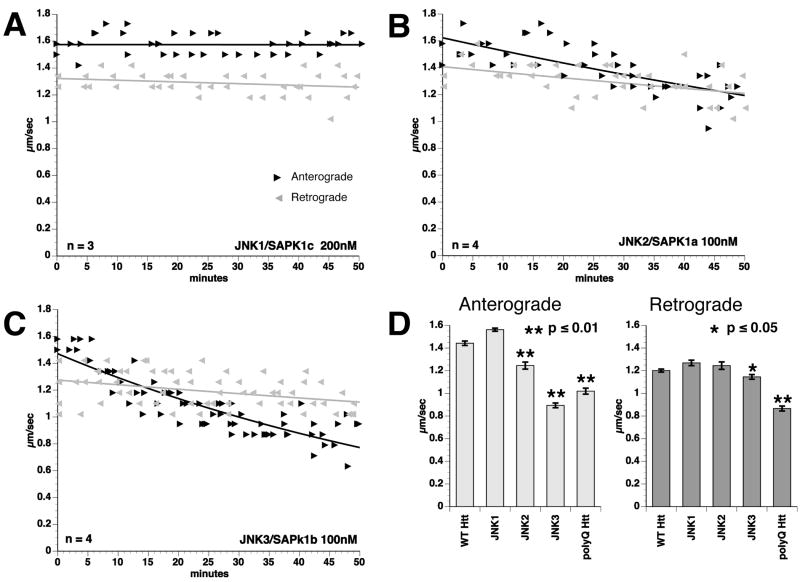
Effects of specific JNK isoforms on FAT were evaluated in isolated axoplasm (Fig. 5). The activity of recombinant JNKs was normalized in vitro using c-Jun as substrate (data not shown). JNK1 (200nM) did not affect FAT (Fig. 5A), consistent with reports of constitutive JNK1 activity in neurons32, 33. JNK2 (100nM) decreased anterograde FAT rates slightly (1.25 μm/sec, compared to 1.6 μm/sec for control buffer; P≤ 0.01, two-sample t-test) (Fig. 5B), but did not affect retrograde FAT (Fig. 5D). As with polyQ-Htt perfusion (Fig. 5D), JNK3 (100nM) inhibited both directions of FAT (Fig. 5C). Quantitative analysis showed that JNK3 inhibited both anterograde (0.9 μm/sec; compared to 1.6 μM/sec in control buffer) and retrograde (1.25 μm/sec, compared to 1.4 μm/sec in control buffer) FAT at p≤ 0.01, two-sample t-test, suggesting that JNK3 is principally responsible for polyQ-Htt effects on FAT.
Figure 5. Active JNK3 mimics the effect of polyQ-Htt on FAT.
Effects of active, recombinant JNK1, JNK2 and JNK3 were evaluated by vesicle motility assays in isolated axoplasm. A) Perfusion of active JNK1 kinase (200nM) in axoplasm had no effect on FAT. B) Perfusion of active JNK2 (100nM) slightly inhibited anterograde FAT, but had no significant effect on retrograde FAT. C) Perfusion of JNK3 (100nM) significantly inhibited FAT in both directions, as observed with polyQ-Htt (compare to Fig. 2B) D) Bar graphs showing mean FAT rates in axoplasm perfused with active JNKs and polyQ-Htt. Error bars show SEM. Data represent pooled FAT rate measurements between 30-50 min of observation. Anterograde FAT rates after polyQ-Htt perfusion were comparable to those elicited by perfusion of active JNK3 and both JNK3 and polyQ-Htt significantly inhibit retrograde FAT.
JNK3 phosphorylates kinesin-1 at Ser176
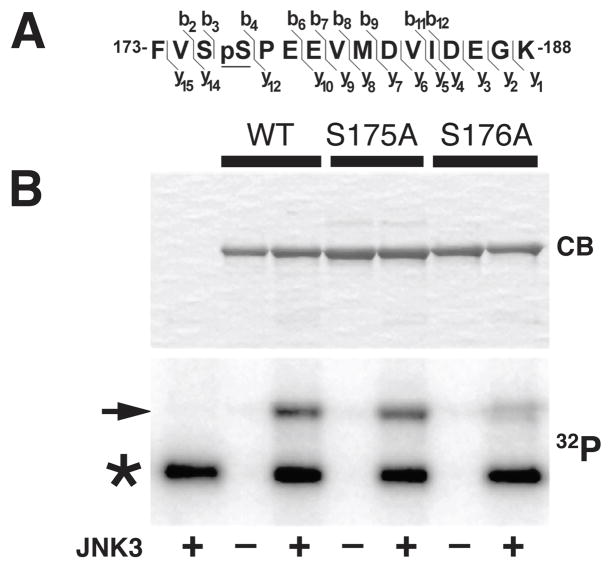
JNK3 phosphorylated kinesin-1, but not KLCs7. Using liquid chromatography tandem mass spectrometry (LC/MS/MS) we mapped residues in kinesin-1 phosphorylated by JNK3 (Fig. 6 and Supplemental Fig. 2). Kinesin-1 constructs encompassing the first 584 amino acids of kinesin-1C (KHC584) were phosphorylated in vitro with JNK3 (Fig. 6B)7 and compared to non-phosphorylated KHC584. Phosphorylated polypeptides were digested with trypsin directly for LC/MS/MS analysis or for phosphopeptide enrichment by IMAC35 and LC/MS/MS analysis (Supplemental Fig. 2). Identification of mass spectrometry-generated peptides used the SEQUEST algorithm. KHC584 identity was confirmed by identification of multiple kinesin-1C peptides (coverage of 72%) (Supplemental Fig. 2). Residual phosphorylation of several peptides was detected (not shown), but using analysis of protease cleavage sites, MS spectra, cross correlation (X-corr) as well as delta correlation (dCn) values, a unique peptide corresponding to amino acids 173-188, was identified with unequivocal evidence of phosphorylation by JNK3 (Fig. 6A, and Supplemental Fig. 2). Phosphatase treatment reduced the phosphorylated peak intensity compared to neighboring peaks (not shown). This JNK3-phosphorylated peptide was within the kinesin-1c motor domain and contained two serines (Fig. 6A). LC/MS/MS allowed precise mapping of the specific serine residue carrying the phosphorylation. Tandem mass spectrometry analysis by collision-induced dissociation (CID) indicated that phosphorylation occurs on Ser176, but not Ser175 (Fig. 6 and Supplemental Fig. 2). Presence of a proline residue adjacent to Ser176 (a hallmark of JNK substrates) supported identification of Ser176 as the JNK3 phosphorylation site. Mass spectrometric analysis of JNK1-phosphorylated KHC584 did not reveal phosphorylation at this site (Supplemental Fig. 3), consistent with differential effects of JNK1 and JNK3 on FAT (Fig. 5A, C). To confirm ESI-LC/MS/MS analysis, site-directed mutagenesis substituted alanine for either Ser175 or Ser176 of KHC584 (KHC584-S175A and KHC584-S176A). Bacterially-expressed mutant proteins were purified (Fig. 6B), then phosphorylated in vitro with JNK3, and 32P incorporation analyzed by Phosphorimaging. Phosphorylation of KHC584-S176A, but not KHC584-S175A, was reduced relative to wild-type KHC584 by≈30% (KHC584-WT, Fig. 6B). Ser176 is conserved among squid, mouse and human kinesin-1s (Supplemental Fig. 4A). Levels of 32P incorporated in KHC584-S176A were consistent with mass spectrometry findings showing partial phosphorylation of KHC584in vitro at non-conserved residues outside the motor domain (not shown). Ser176 phosphorylation was also detected in kinesin-1 immunoprecipitated from mouse brain (not shown). Together, these data identified Ser176 as a major kinesin-1 residue phosphorylated by JNK3.
Figure 6. JNK3 phosphorylates kinesin-1 heavy chains at Ser176.
A) Mass spectrometry studies identified a tryptic peptide within the motor domain of kinesin-1 (amino acids 173-188) showing unequivocal evidence of phosphorylation by JNK3 (see also Supplemental Fig. 2). B) Recombinant KHC584-WT protein was incubated in the presence (+) or absence (-) of active JNK3 and 32P-ATP, and analyzed by SDS-PAGE. The ability of JNK3 to phosphorylate KHC584 at S175 and S176 was also evaluated using KHC584-S175 and KHC584-S176A mutants. An autoradiogram (32P) showed a dramatic reduction in the phosphorylation of a KHC584-S176A (S176A) construct relative to wild type (WT) and KHC584-S175A (S175A) constructs (arrow). Coomassie blue (CB) staining showed comparable amounts of KHC584 in all lanes. Asterisk indicates autophosphorylated JNK3. These results indicated that S176 is the main residue phosphorylated by JNK3.
Ser176 phosphorylation affects kinesin-1 function
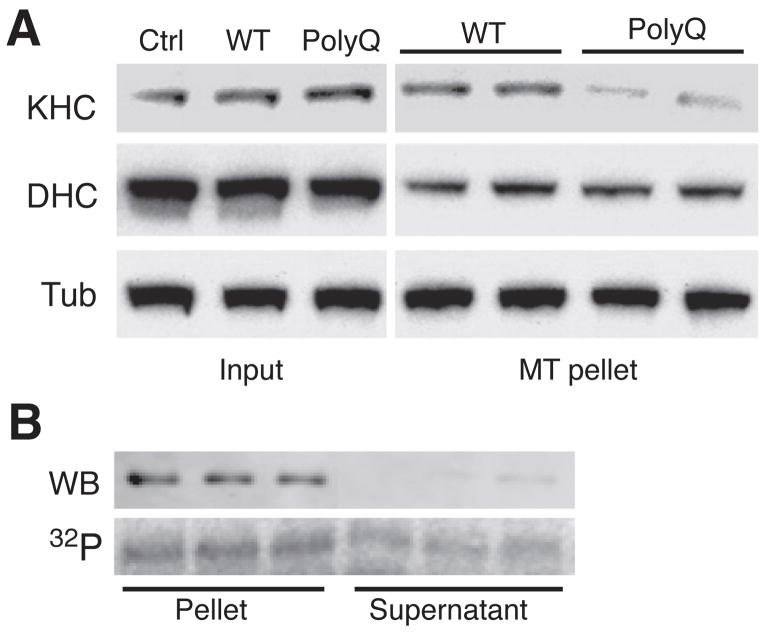
Effects of polyQ-AR on FAT involved JNK activation, increased kinesin-1 phosphorylation and reduced microtubule binding of kinesin-17. Ser176 localizes to loop 8, a sequence within the motor domain of kinesin-1 implicated in kinesin-1 interaction with microtubules (Supplemental Fig. 4B)36, suggesting its phosphorylation might affect kinesin-1 binding to microtubules. NSC34 cells were transiently transfected with WT-Htt, or polyQ-Htt (see Fig. 3A). Total kinesin-1 levels were comparable for untransfected, WT-Htt, and polyQ-Htt-expressing cells (Fig. 7A). However, microtubule-binding assays using the non-hydrolysable ATP analog AMP-PNP7 revealed a reduction in kinesin-1 binding to microtubules in polyQ-Htt-expressing cells relative to WT-Htt-expressing cells (Fig. 7A). Similar effects were observed when using immortalized striatal cell lines derived from wild type and HdhQ109 mice (not shown). This was consistent with reduced kinesin-1 binding to microtubules in cells expressing polyQ-AR7.
Figure 7. PolyQ-Htt expression inhibits kinesin-1 binding to microtubules.
A) Lysates of NSC34 cells transfected with WT-Htt or polyQ-Htt as in Fig. 3A were analyzed by immunoblot. Total levels (Input) of kinesin-1 (KHC), dynein heavy chain (DHC) and tubulin (Tub) were comparable for untransfected (Ctrl), WT-Htt and polyQ-Htt-expressing cells. However, the fraction of kinesin-1 recovered in association with microtubules was reduced for lysates from polyQ-Htt expressing cells, when compared to untransfected and WT-Htt-expressing cells. B) Microtubule-binding assays using recombinant kinesin-1 (KHC584). Immunoblot (WB) shows that unphosphorylated KHC584 is mainly recovered in association with microtubules (Pellet). An autoradiogram (32P) reveals a significant fraction of JNK3-phosphorylated KHC584 remains in the supernatant.
Recombinant KHC584 was incubated with 100μM radiolabeled 32P-γATP in the presence or absence of active JNK3 (Fig. 7B), then incubated with purified microtubules in the presence of AMP-PNP (2 mM) or 2 mM ATP (to control for non-specific pelleting, not shown). Microtubule pellets (P) and corresponding supernatants (SN) were analyzed by immunoblot to visualize non-phosphorylated KHC584 and autoradiography to visualize JNK3-phosphorylated KHC584. In the presence of AMP-PNP, kinesin-1 forms a rigor structure with microtubules13. Virtually all non-phosphorylated KHC584 partitioned with microtubules by immunoblot (Fig. 7B), but a large fraction of JNK3-phosphorylated KHC584 remained in the SN (Fig. 7B), suggesting that phosphorylation of kinesin-1 by JNK3 inhibits kinesin-1 binding to microtubules. The presence of JNK3-phosphorylated KHC584 in P fractions was consistent with residual phosphorylation at residues outside the motor domain. JNK-3 phosphorylation of kinesin-1 reduced binding to microtubules with AMP-PNP by ≈50% (Supplemental Fig. 5).
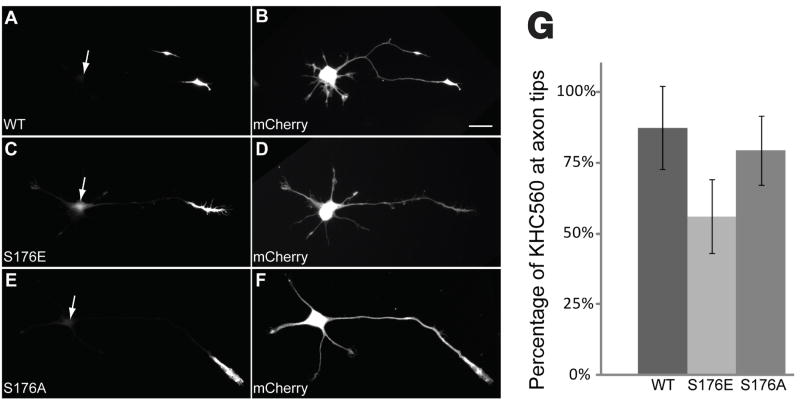
Effects of S176 phosphorylation on kinesin-1 motility in vivo were assessed by expressing GFP-tagged, truncated kinesin-1 constructs in cultured hippocampal neurons (Fig. 8). When expressed in hippocampal cells, a GFP-tagged kinesin-1 construct (KHC560-GFP -WT) selectively translocates to axon distal ends and accumulates, but not in dendrites, indicating that this motor translocates preferentially along axonal microtubules37. KHC560-GFP-WT translocation is so efficient that little or no fluorescence can be detected within cell bodies or along axons37. To evaluate whether S176 phosphorylation affects kinesin translocation efficiency, we compared localization of phosphorylation-mimicking construct KHC560-GFP-S176E and its unphosphorylatable counterpart KHC560-GFP-S176A to that of KHC560- GFP-WT by quantitative fluorescence microscopy (Fig. 8A–F). Much less phosphomimetic KHC560-GFP-S176E construct accumulated at axonal tips than the KHC560-GFP-WT construct (56 ± 2% vs. 87 ±3%, respectively, mean ± SEM; T-test, p < 0.0001) (Fig. 8G), with more KHC560- GFP- S176E fluorescence in cell bodies and faint staining of neurites (Fig. 8C), the expected pattern if a significant fraction of phosphomimetic kinesin-1 were distributed like a soluble protein. Slightly less KHC560-GFP-S176A accumulated at axon tips (79% ± 2%) compared to KHC560- GFP-WT, but accumulations were still significantly higher than KHC560-GFP-S176E (t-test, p<0.0001) (Fig. 8G). Thus, a mutation of Ser176 mimicking phosphorylation reduces efficiency of processive kinesin-1 translocation in cultured neurons.
Figure 8. A mutation mimicking Ser176 phosphorylation reduces the translocation efficiency of constitutively active kinesin-1 in cultured hippocampal neurons.
A-F: Representative stage 3 neurons were co-transfected with GFP-tagged constructs encompassing the first 560 amino acids of kinesin-1 (KHC560, A, C, and E) and mCherry, a soluble marker protein that diffuses throughout the cell (B, D, and F, scale bar= 20μm). Cells were fixed with formaldehyde and imaged 5 hours after transfection. Wild type (WT, A) and non-phosphorylatable S176A mutant (S176A, E) constructs efficiently translocated so that they were localized almost exclusively at the axon tip. In contrast, a significant fraction of the pseudophosphorylated S176 mutant (S176E, C) was found throughout the cell body. Arrowheads in A, C and E indicate the location of the neuronal cell bodies as seen in B, D, and F. G: The percentage of total KHC560-GFP fluorescence localized at the axon tip was measured in cells expressing KHC560 WT (N=22), KHC560 S176E (N=33) or KHC560 S176A (N=30). Significantly less KHC560 S176E accumulated at axon tips with the wild-type construct (t-test, p<0.0001); only marginally less KHC560 S176A accumulated at the axon tip than the wild-type construct (t-test, p~0.05). Bars show mean and standard deviation.
DISCUSSION
Despite ubiquitous expression of Htt, neuronal cells are uniquely vulnerable to the toxic gain-of-function in polyQ-expansion, suggesting that processes critical for proper neuronal function and survival are selectively altered in HD3. Axonal transport is essential for neurons, due to their large size and complex functional architecture 3. Significantly, alterations in kinesin-1 function and regulation are associated with various human neuropathologies3, 7, 38-40.
Several reports link FAT inhibition to pathogenesis in HD4, 5, 8 10 and other polyQ-expansion diseases3, 7. For example, overexpression of polyQ-Htt in cultured cells10 and Drosophila4, 5 leads to reduced transport and accumulation of vesicular cargos within axons. PolyQ-Htt and polyQ-AR, two otherwise unrelated proteins, similarly inhibit FAT vesicle motility in isolated axoplasm independently of changes in gene transcription or aggregate formation7, 8. Such observations implicate FAT defects in HD pathogenesis, which would partially explain the selective vulnerability of neurons in HD and other polyQ-expanded diseases3.
Several models were proposed to explain the inhibition of FAT induced by polyQ-Htt. Some require physical interactions between Htt and conventional kinesin41 or CDyn subunits9, 10, 12. PolyQ-Htt aggregates were proposed to sequester molecular motors4 6, or physically block FAT5, 42. Alternatively, Htt mutations were suggested to interfere with normal functions of Htt as a scaffold linking molecular motors to transported cargoes9, 10 11, or to affect the subunit composition and function of molecular motors12. However, axoplasm experiments indicated that polyQ-Htt inhibited FAT at concentrations 100-1000 fold lower than molecular motors, suggesting activation of enzymatic activities3, 7, 8.
Cell fractionation and immunoprecipitation evaluated interactions between Htt and molecular motors in brain tissue from wild-type and homozygous HdhQ109 mice. Regardless of genotype, similar levels of full-length Htt were found in detergent-soluble (S) and detergent-insoluble (I) fractions. In contrast, KHCs, KLCs, DIC and DHC levels were much higher in soluble fractions, suggesting that most molecular motors are not sequestered by polyQ-Htt. Unlike experiments involving Htt overexpression, immunoprecipitation failed to detect interactions between endogenous Htt and kinesin41 or CDyn12 (Fig. 1). Detection of KLCs in KHC- and DHC in DIC-immunoprecipitates confirmed that physiologically relevant protein complexes were preserved13, 14. Such observations are inconsistent with models in which polyQ-Htt inhibits FAT through direct interactions with molecular motors.
Another model proposed that pathogenic polyQ-Htt activates neuronal regulatory pathways that affect FAT3. Kinesin-1 and CDyn are regulated by phosphorylation of specific motor subunits7, 18. Kinase activities that regulate FAT, including GSK-343; Akt44, and JNK20, 22, 45 are altered in Huntington’s and polyQ-AR activates JNK7. JNK inhibitors blocked polyQ-Htt effects on FAT and JNK activation was documented in polyQ-Htt-expressing cells and HdhQ109 mice (Fig. 3). JNK activation was more pronounced in homozygous HdhQ109 mice than heterozygous littermates, but increased in both, consistent with autosomal dominant inheritance of HD. JNK activation in presymptomatic HdhQ109 mice indicated that this change represents an early event in HD.
Mammals have three JNK genes (JNK1, JNK2 and JNK3)46. Analysis of JNK in HdhQ109 mice and polyQ-Htt-transfected cells indicated that polyQ-Htt increased JNK2/3 activity more than JNK1 (Fig. 3), consistent with reports showing constitutive JNK1 activity and selective JNK2/3 activation in neurons with stress32, 33. This prompted us to examine roles for specific JNK isoforms mediating polyQ-Htt effects on FAT. SB203580 inhibits JNK2/3 at 10μM, but JNK1 inhibition requires ≥100μM23. Significant JNK activity (presumably JNK1) remains in axoplasms treated with 10μM SB203580 (Fig. 4A), but 10μM SB203580 prevented polyQ-Htt effects on FAT (Fig. 4B), suggesting that polyQ-Htt effects on FAT involve JNK2/3. JSAP1 polypeptides further supported a role for JNK3, because JSAP1a, which inhibits all JNKs, preserved FAT effectively, but JSAP1d, with a 31- amino acid insert that selectively interferes with JNK3 binding34 did not (Fig. 4D–E). To evaluate the role of individual JNKs on FAT, effects of active recombinant JNK1, JNK2 and JNK3 on FAT were evaluated in axoplasm. Active JNK1 had no effect on FAT (Fig. 5A), consistent with constitutive JNK1 activity in axons32, 33 (Fig. 4A). JNK2 produced a selective, moderate effect on anterograde FAT, but no effect on retrograde FAT. In contrast, JNK3 effects on FAT paralleled those of polyQ-Htt by inhibiting both anterograde and retrograde FAT. Although JNK2 may contribute, JNK3 appears to primarily mediate polyQ-Htt effects on FAT. Notably, JNK3 is preferentially expressed in nervous tissues47 and polyQ-Htt pathology is restricted to neurons.
To provide a molecular basis for FAT inhibition by JNK3, relevant phosphorylation sites were identified. Mass spectrometry mapped JNK3 phosphorylation of kinesin-1 to S176, a conserved residue in the motor domain (Fig. 6A, Supplemental Fig. 2–3). S176 was also modified in kinesin-1 immunoprecipitated from mouse brain (not shown). Mutagenesis studies confirmed that JNK3 phosphorylated Ser176 (Fig. 6B). JNK1 did not phosphorylate kinesin-1 at Ser176 (Supplemental Fig. 3), consistent with unique substrate preferences for JNK348. Ser176 was conserved among squid, mouse and human kinesin-1s (Supplemental Fig. 4A), validating the squid axoplasm model. Collectively, these data demonstrate that JNK3 activity is elevated in brains of HdhQ109 mice and axoplasm perfused with polyQ-Htt (Fig. 3–4); that JNK3 phosphorylates kinesin-1 at Ser176 (Fig.6 and Supplemental Fig. 2–3), and that Ser176 modification is sufficient to inhibit trafficking of kinesin-1 (Fig. 8). Antibodies selective for this epitope are under development and should allow us to quantify the fraction of neuronal kinesin-1 modified by JNK3 in HD brains.
Based on the Ser176 position within the motor domain36, we evaluated effects of JNK3 phosphorylation on kinesin-1 binding to microtubules. JNK3-treated recombinant kinesin-1 and kinesin-1 in polyQ-Htt-transfected cells exhibited reduced binding to microtubules (Fig. 7). Further, pseudophosphorylation at Ser176 dramatically decreased kinesin-1 translocation in vivo (Fig. 8), indicating that Ser176 phosphorylation interferes with kinesin-1 function.
Our results suggest that polyQ-Htt inhibits FAT by activation of axonal JNKs. Additionally, JNK activation by polyQ-Htt is also consistent with changes in transcription49, and JNKs can be pro-apoptotic30 (Supplemental Fig. 6). Significantly, JNK3, and not JNK1, activity mediated polyQ-Htt effects on FAT, providing a partial explanation for selective neuronal vulnerability in HD. JNK1 and JNK2 are ubiquitously expressed, whereas JNK3 is selectively expressed in brain and testes47. Identification of kinesin-1 as a novel JNK3 substrate provided a molecular basis for changes in FAT consistent with neuron-selective pathology of polyQ-Htt.
In sum, neuronal JNK3 is activated by polyQ-Htt. JNK3 in turn phosphorylates kinesin-1, decreasing its ability to bind microtubules and move cargoes in FAT (Fig. 8 and Supplemental Fig. 6). Identification of Ser176-phosphorylated kinesin-1 in normal mouse brain suggests a role of JNK3 in a normal pathway for regulating kinesin and delivering specific MBO cargoes to particular neuronal domains. Increased activation of this pathway by polyQ-Htt would lead to reductions in kinesin-based transport, deficits in synaptic and axonal function, and eventual dying-back neuronal degeneration3. Inhibition of JNK3 sufficed to prevent inhibition of FAT due to polyQ-Htt. These results suggest that elevation of JNK activity and consequent inhibition of FAT may represent primary pathogenic events in Huntington’s disease. Further studies should illuminate molecular components mediating JNK activation by polyQ-Htt. Regardless; inhibition of JNK activity to protect FAT represents a promising therapeutic target for treatment of Huntington’s disease.
MATERIAL AND METHODS
Antibodies and Reagents
The following antibodies were used: anti-KHC (H2 clone, Chemicon)13, anti-pan KLCs (63-90 clone, Chemicon)13, anti-dynein intermediate chain (Santa Cruz #13524), anti-dynein heavy chain (Santa Cruz #9115), anti-phospho Akt (44-622G, Biosource), anti-Htt (2166, Chemicon), anti-pan-JNK (Upstate #06-748), anti-JNK1 (Pharmingen #554268), anti-JNK2 (Cell Signaling #4672), anti-JNK3 (Cell Signaling #2305), anti-phospho JNK (Cell Signaling #9251), anti-GST (Sigma #G7781), anti-phospho c-Jun (Santa Cruz #16312), anti-tubulin (DM1A, Sigma). All antibodies were characterized for specificity against the tissues used (see Supplemental Figure 7). The following secondary antibodies were used: Jackson 111-035-045 HRP-conjugated goat anti-rabbit, Jackson 115-035-146 HRP-conjugated goat anti-mouse IgG, and Jackson 805-035-180 HRP-conjugated bovine anti-goat IgG.
SB203580, SP600125 (JNK inhibitor II), okadaic acid, staurosporine, K252a, 50nM okadaic acid, microcystin and c-Jun were obtained from Calbiochem. The HDAC inhibitor BC-6-2527 was a generous gift from Alan Kozikowski (UIC). Inhibitor stocks were prepared in DMSO. Active JNK kinases were from Upstate (JNK1 #14-327, JNK2 #14-329, and JNK3 #14-501). His-tagged KHC584 protein constructs were expressed in BL21-Codon Plus (DE3) E. coli (Stratagene), and purified using Talon beads (Clontech). Trx-tagged JSAP1 polypeptides were expressed and purified as described before34.
Immunoblots
Proteins were separated by SDS-PAGE on 4-12% Bis-Tris gels (NuPage minigels, Invitrogen), using MOPS Running Buffer (Invitrogen) and transferred to PVDF using Towbin buffer supplemented with 10% (v/v) methanol (90 minutes at 400mA using Hoeffer TE22 apparatus). Immunoblots were blocked with 1% (w/v) non-fat dried milk in TBS (25 mM Tris pH 7.2, 2.68 mM KCl, 136.8 mM NaCl). Membranes were incubated with primary antibodies overnight at 4° C in 1% IgG free-BSA (Jackson Immunoresearch), and washed four times with 0.1% Tween-20 in TBS. Primary antibody binding was detected with HRP-conjugated anti-mouse, anti-rabbit or anti-goat antibodies (Jackson Immunoresearch), and visualized by chemiluminescence (ECL, Amersham). Quantitative immunoblotting was performed as before39.
Immunoprecipitation
Mouse brains from wild-type and HdhQ109 knock-in mice were homogenized in lysis buffer (LB; 25 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, and mammalian protease inhibitor cocktail (Sigma, 1/100 dilution)]. Lysates were centrifuged twice for 5 minutes at 55,000 rpm (163,640 gmax) using a TLA 100.3 rotor (Beckman Instruments, Palo Alto, CA). The resulting supernatant fractions were precleared using a mixture of Protein G agarose beads (Pierce), and non-immune mouse IgG-conjugated Sepharose beads (Jackson Immunoresearch) for 1 hour at RT. 400 μg of each precleared brain lysate were brought to 1 ml with LB, and incubated with 5μg of the appropriate antibody plus 10μl of Protein G agarose beads at 4° C for 3hs. Immunocomplexes were recovered by centrifugation (3000 gmax for 30 seconds), washed four times with 1 ml LB, once with 50mM HEPES pH 7.4, and resuspended in Laemmli buffer. For immunodepletion experiments (Fig. 1B and supplemental Fig. 1B), precleared mouse brain lysates were subject to three cycles of immunoprecipitation with H2 or DIC antibodies as above. After each immunoprecipitation cycle, a 50μl aliquot of each supernatant was saved for immunoblot analysis, as before13, 14.
Vesicle Motility assays in isolated axoplasm
Axoplasm was extruded from giant axons of the squid Loligo pealii (Marine Biological Laboratory) as described previously7, 8. Recombinant proteins, peptides or inhibitors were diluted into X/2 buffer (175 mM potassium aspartate, 65 mM taurine, 35 mM betaine, 25 mM glycine, 10 mM HEPES, 6.5 mM MgCl2, 5 mM EGTA, 1.5 mM CaCl2, 0.5 mM glucose, pH 7.2) supplemented with 2-5 mM ATP and 20 μl added to perfusion chambers. Preparations were analyzed on a Zeiss Axiomat with a 100X, 1.3 n.a. objective, and DIC optics. Hamamatsu Argus 20 and Model 2400 CCD camera were used for image processing and analysis. Organelle velocities were measured with a Photonics Microscopy C2117 video manipulator (Hamamatsu).
Recombinant Constructs
Expression vectors encoding amino acids 1–548 of human Htt with an N-terminal FLAG epitope tag were translated in vitro as described previously8, and used for vesicle motility assays. Expression vectors encoding amino acids 1-969 of human Htt (wild type (Q16) and pathogenic (Q46)) with an N-terminal FLAG tag were used for transfection studies (Fig. 3A and 7A)29. His-tagged KHC584-S176A, KHC584-S176A, and GFP-tagged KHC560 mutant constructs were generated using the Quick-site mutagenesis kit (Stratagene).
Axoplasmic JNK activity assays
For experiments on kinase activity in axoplasm (Figs. 2C and 4A), 3- 4 axoplasms were triturated in 280μl of KB buffer (KB: 10mM Hepes pH 7.4, 10mM MgCl2, 1mM DTT) minus ATP. Reactions (50μl) were incubated at RT and started by adding ATP (50μM). Aliquots were collected at various time points up to 20 minutes and analyzed by immunoblotting.
Brain Tissue/Cell lysate preparation
Mouse brain tissue and cultured cells were homogenized in ROLB buffer [10mM Hepes pH7.4, 0,5% Triton X-100, 80mM β-glycerophosphate, 50mM Sodium Fluoride, 2mM Sodium Orthovanadate, 100nM Staurosporine, 100nM K252a, 50nM Okadaic acid, 50nM Microcystin, 100mM Potassium Phosphate and mammalian protease inhibitor cocktail (Sigma)]. Lysates were clarified by centrifugation, and protein concentration determined using BCA kit (Pierce). All mouse use was under protocols approved by the Institutional Animal Care and Use Committee at UIC and followed NIH guidelines for vertebrate animal use.
NSC34 Cell Culture and Transfection
NSC34 cells were plated at 10,000 cells/cm2 on 10 cm tissue culture dishes for biochemical studies and grown on DMEM containing 1mM glutamine and 10% Fetal Bovine Serum (Hyclone). When cells reached 60-70% confluency, media was replaced by DMEM plus 1mM glutamine, and cells transfected with WT-Htt and polyQ-Htt constructs (1-969)29 using Lipofectamine 2000 (Invitrogen), as previously described39. JNK activation was evaluated 24h after transfection.
Microtubule-binding Assays
NSC34 cells were transfected with WT-Htt and polyQ-Htt constructs as described above. After 24 hours, cells were scraped in BRB80 buffer [80 mM PIPES pH 7, 1 mM MgCl2, 1 mM EGTA, 1 μM staurosporine, 1 μM K252a, 50 nM okadaic acid, 200 nM microcystin, 1/100 dilution of phosphatase inhibitor cocktail II (Calbiochem), and1/100 dilution of mammalian protease inhibitor cocktail (Sigma)] at 4°C, and homogenized using a 30g syringe (500μl of BRB80/10 cm dish). Lysates were centrifuged at 14,000 rpm in a tabletop centrifuge for 5 minutes, and centrifuged at 55,000 rpm in TLA100.3 rotor for additional 5 minutes. 700 μg of total protein from clarified supernatants were incubated with 0.2 mg of taxol-stabilized microtubules and 2.5 mM AMP-PNP (Sigma) for 30 min at 37 °C. The mixture above was loaded on top of a cushion made of 20% sucrose in BRB80 buffer, and centrifuged for 15 minutes at 50,000 rpm using a Beckman TLA-100.3 rotor at room temperature. Microtubule-enriched pellets were resuspended in Laemmli buffer and analyzed by immunoblotting. For microtubule binding experiments (Fig. 7B), 4μl aliquots of in vitro-phosphorylated KHC584 (prepared as above) were incubated with 64μg of taxol-stabilized microtubules in BRB80 buffer and either 2mM AMP-PNP or 2mM ATP (100μl total volume). After 30 minutes incubation, these mixtures were centrifuged for 15 minutes at 50,000 rpm over a cushion of 20% in BRB80 in a Beckman TLA-100.3 rotor at 37° C. Pellets and supernatants were collected, and the levels of kinesin in each fraction quantified by phosphorimager scanning of radioactivity or with fluorescently-labeled secondary antibodies.
Kinesin-1 Translocation Assays
Accumulation of KHC560-GFP-WT and KHC560-GFP-S176E constructs in neurites of cultured hippocampal neurons was evaluated as before37.
Statistical Analysis
All experiments were repeated at least 3 times. Unless otherwise stated, the data was analyzed by ANOVA followed by post-hoc Student-Newman-Keul’s test in order to make all possible comparisons. Data was expressed as mean ± SEM and significance was assessed at p<0.05 or 0.01 as noted.
Mouse Brain Fractionation
Brains from wild-type (12 months old) and HdhQ109 knock-in (14 months old) were homogenized in 3mls of HB buffer (50mM Hepes pH 7.4, 150 mM NaCl, 1% Triton X-100) and centrifuged 2X 2,500g for 10 minutes, and 1X at 7,000g for 10 minutes to pellet debris and nuclei. Clarified lysates were spun at 150,000g for 20 minutes, in a Beckman TLA 100.3 rotor at 55,000 RPM. The pellets and supernatant represented detergent insoluble and detergent-soluble fractions, respectively. Detergent insoluble pellets were resuspended in HB. Equal amounts of protein from each fraction were analyzed by immunoblotting.
In vitro Phosphorylation
In vitro phosphorylation experiments (40 μl volume) were performed by incubating KHC584 protein constructs (3 μM) with 0.1μM JNK3 (Upstate) or 0.2μM JNK1 (Upstate) in HEM buffer (50 mM HEPES, 12 mM MgSO4, 100 μM ATP pH 7.4), as previously described7
Mass Spectrometry Studies
Phosphorylated KHC584 protein was subjected to in solution trypsin digestion. Briefly, dried samples were resuspended in 30 mM Hepes, 30 mM NaF in the presence of 1 μg of trypsin (Sigma, proteomics grade) and incubated at 37° C overnight (16-18h). The resulting tryptic peptides were later resuspended in buffer A (5% acetonitrile, 0.4% acetic acid, 0.005% of HFBA in water) for mass spectrometry analysis or prepared for IMAC35. Samples were analyzed by liquid chromatography on line with a LTQ (a two-dimensional ion trap) instrument equipped with a commercial nanospray source (Thermo Finnigan, San Jose, CA). Samples were automatically loaded by a microautosampler (Famos, LC Packings, Sunnyvale, CA) onto an 11-cm × 100μm fused silica capillary column packed with reverse C18 material (5-μm, 100-Å Magic beads, Michrom Bioresources, Auburn, CA). The solvent system was delivered by an HP1100 pump (Agilent Technologies, Palo Alto, CA). Samples were analyzed by performing full scan followed by tandem MS or MS/MS or MS2 of the 5 most intense ions (top 5) by collision induced dissociation (CID). Sample loading, solvent delivery, and scan function were controlled with Xcalibur software (Thermo Finnigan). Liquid chromatography tandem mass spectrometry files were searched using SEQUEST algorithm against a generated database containing KIF5C_Rat and KIF5C_Mouse, among others proteins. SEQUEST search parameters included mass tolerance of ±1.5 Da, trypsin specificity, and differential search for serine, threonine and tyrosine phosphorylation. The generated data set was filtered using INTERACT50 based on the following criteria: delta correlation, dCn: 0.1, X correlation value for +1 peptides ≥ 1.8, +2 ≥ 2.2 and +3 ≥ 3.2.
Acknowledgments
The authors would like to thank Drs Marcy MacDonald and Marian DiFiglia for knock-in mice and huntingtin constructs, respectively, and Bin Wang for excellent technical assistance. This work was supported by 2007/2008 MBL summer fellowship to GM; an HDSA grant to GM; NIH grants MH066179 to GB; and ALSA, Muscular Dystrophy Association, and NIH (NS23868, NS23320, NS41170) grants to STB.
Footnotes
Author Contributions: GAM and STB performed transport and biochemistry experiments and wrote the manuscript; YY, SLP, AK, KL performed transport and biochemistry experiments; KY provided and characterized recombinant JIP; BB, ETC, CB and DH performed Mass Spectrometry studies; and CFH and GB did GFP-kinesin experiments. All authors reviewed and edited the manuscript
Supplementary Material
Bibliography
- 1.Okun MS. Huntington’s disease: what we learned from the original essay. Neurologist. 2003;9:175–179. doi: 10.1097/01.nrl.0000080952.78533.1f. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 3.Morfini G, Pigino G, Brady ST. Polyglutamine Expansion Diseases: Failing to Deliver. Trends Molec Med. 2005;11:64–70. doi: 10.1016/j.molmed.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Gunawardena S, et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- 5.Lee WC, Yoshihara M, Littleton JT. Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a Drosophila model of Huntington’s disease. Proc Natl Acad Sci U S A. 2004;101:3224–3229. doi: 10.1073/pnas.0400243101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morfini G, et al. JNK Mediates Pathogenic Effects of Polyglutamine-expanded Androgen Receptor on Fast Axonal Transport. Nat Neurosci. 2006;9:907–916. doi: 10.1038/nn1717. [DOI] [PubMed] [Google Scholar]
- 8.Szebenyi G, et al. Neuropathogenic Forms of Huntingtin and Androgen Receptor Inhibit Fast Axonal Transport. Neuron. 2003;40:41–52. doi: 10.1016/s0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 9.Colin E, et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. Embo J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauthier LR, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 11.Engelender S, et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 12.Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deboer SR, et al. Conventional Kinesin Holoenzymes Are Composed of Heavy and Light Chain Homodimers. Biochemistry. 2008;47:4535–4543. doi: 10.1021/bi702445j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarov O, et al. Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: revisited. Journal of Neuroscience. 2005;25:2386–2395. doi: 10.1523/JNEUROSCI.3089-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wheeler VC, et al. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum Molec Genet. 2000;9:503–513. doi: 10.1093/hmg/9.4.503. [DOI] [PubMed] [Google Scholar]
- 16.Brill LB, 2nd, Pfister KK. Biochemical and molecular analysis of the mammalian cytoplasmic dynein intermediate chain. Methods. 2000;22:307–316. doi: 10.1006/meth.2000.1083. [DOI] [PubMed] [Google Scholar]
- 17.Brady ST, Pfister KK, Bloom GS. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Nat Acad Sci USA. 1990;87:1061–1065. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morfini G, Szebenyi G, Elluru R, Ratner N, Brady ST. Glycogen Synthase Kinase 3 Phosphorylates Kinesin Light Chains and Negatively Regulates Kinesin-based Motility. EMBO Journal. 2002;23:281–293. doi: 10.1093/emboj/21.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donelan MJ, et al. Ca2+-dependent dephosphorylation of kinesin heavy chain on beta-granules in pancreatic beta-cells. Implications for regulated beta-granule transport and insulin exocytosis. J Biol Chem. 2002;277:24232–24242. doi: 10.1074/jbc.M203345200. [DOI] [PubMed] [Google Scholar]
- 20.Apostol BL, et al. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- 21.Merienne K, Helmlinger D, Perkin GR, Devys D, Trottier Y. Polyglutamine expansion induces a protein-damaging stress connecting heat shock protein 70 to the JNK pathway. J Biol Chem. 2003;278:16957–16967. doi: 10.1074/jbc.M212049200. [DOI] [PubMed] [Google Scholar]
- 22.Liu YF. Expression of polyglutamine-expanded Huntingtin activates the SEK1-JNK pathway and induces apoptosis in a hippocampal neuronal cell line. J Biol Chem. 1998;273:28873–28877. doi: 10.1074/jbc.273.44.28873. [DOI] [PubMed] [Google Scholar]
- 23.Coffey ET, et al. c-Jun N-terminal protein kinase (JNK) 2/3 is specifically activated by stress, mediating c-Jun activation, in the presence of constitutive JNK1 activity in cerebellar neurons. J Neurosci. 2002;22:4335–4345. doi: 10.1523/JNEUROSCI.22-11-04335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 25.Barr RK, Kendrick TS, Bogoyevitch MA. Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem. 2002;277:10987–10997. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- 26.Dompierre JP, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozikowski AP, et al. Functional Differences in Epigenetic Modulators-Superiority of Mercaptoacetamide-Based Histone Deacetylase Inhibitors Relative to Hydroxamates in Cortical Neuron Neuroprotection Studies. J Med Chem. 2007;50:3054–3061. doi: 10.1021/jm070178x. [DOI] [PubMed] [Google Scholar]
- 28.Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- 29.Qin ZH, et al. Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J Neurosci. 2004;24:269–281. doi: 10.1523/JNEUROSCI.1409-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjorkblom B, et al. All JNKs Can Kill, but Nuclear Localization Is Critical for Neuronal Death. J Biol Chem. 2008;283:19704–19713. doi: 10.1074/jbc.M707744200. [DOI] [PubMed] [Google Scholar]
- 31.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coffey ET, Hongisto V, Dickens M, Davis RJ, Courtney MJ. Dual roles for c-Jun N-terminal kinase in developmental and stress responses in cerebellar granule neurons. J Neurosci. 2000;20:7602–7613. doi: 10.1523/JNEUROSCI.20-20-07602.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas GM, Lin DT, Nuriya M, Huganir RL. Rapid and bi-directional regulation of AMPA receptor phosphorylation and trafficking by JNK. EMBO J. 2008;27:361–372. doi: 10.1038/sj.emboj.7601969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito M, et al. Isoforms of JSAP1 scaffold protein generated through alternative splicing. Gene. 2000;255:229–234. doi: 10.1016/s0378-1119(00)00335-8. [DOI] [PubMed] [Google Scholar]
- 35.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Sack S, et al. X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry. 1997;36:16155–16165. doi: 10.1021/bi9722498. [DOI] [PubMed] [Google Scholar]
- 37.Jacobson C, Schnapp B, Banker GA. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 2006;49:797–804. doi: 10.1016/j.neuron.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Lapointe NE, et al. The amino terminus of tau inhibits kinesin-dependent axonal transport: Implications for filament toxicity. J Neurosci Res. 2009;87:440–451. doi: 10.1002/jnr.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pigino G, Morfini G, Mattson MP, Brady ST, Busciglio J. Alzheimer’s Presenilin 1 Mutations Impair Kinesin-Based Axonal Transport. J Neurosci. 2003;23:4499–4508. doi: 10.1523/JNEUROSCI.23-11-04499.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy S, Zhang B, Lee VM, Trojanowski JQ. Axonal transport defects: a common theme in neurodegenerative diseases. Acta Neuropathol (Berl) 2005;109:5–13. doi: 10.1007/s00401-004-0952-x. [DOI] [PubMed] [Google Scholar]
- 41.McGuire JR, Rong J, Li SH, Li XJ. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carmichael J, Sugars KL, Bao YP, Rubinsztein DC. Glycogen synthase kinase-3beta inhibitors prevent cellular polyglutamine toxicity caused by the Huntington’s disease mutation. J Biol Chem. 2002;277:33791–33798. doi: 10.1074/jbc.M204861200. [DOI] [PubMed] [Google Scholar]
- 44.Colin E, et al. Akt is altered in an animal model of Huntington’s disease and in patients. Eur J Neurosci. 2005;21:1478–1488. doi: 10.1111/j.1460-9568.2005.03985.x. [DOI] [PubMed] [Google Scholar]
- 45.Garcia M, Charvin D, Caboche J. Expanded huntingtin activates the c-Jun terminal kinase/c-Jun pathway prior to aggregate formation in striatal neurons in culture. Neuroscience. 2004;127:859–870. doi: 10.1016/j.neuroscience.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 46.Gupta S, et al. Selective interaction of JNK protein kinase isoforms with transcription factors. Embo J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 47.Yang DD, et al. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 48.Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cha JH. Transcriptional signatures in Huntington’s disease. Prog Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.