Summary
RelA/ SpoT homolog (RSH) proteins have (p)ppGpp synthetase and hydrolase activities that mediate major global responses to nutrient limitation and other stresses. RSH proteins are conserved in most bacteria and play diverse roles in bacterial pathogenesis. We report here that the RSH protein of S. pneumoniae, RelSpn, can be deleted and is the primary source of (p)ppGpp synthesis in virulent strain D39 under some conditions. A D39 ΔrelSpn mutant grew well in complex medium, but did not grow in chemically defined medium unless supplemented with the metals copper and manganese. Transcriptome analysis of D39 rel+Spn and ΔreSpn strains treated with mupirocin revealed relSpn-independent (translation stress), relSpn-dependent (stringent response), and ΔrelSpn-dependent changes suggesting that relSpn and (p)ppGpp amount play wide-ranging homeostatic roles in pneumococcal physiology, besides adjusting macromolecular synthesis and transport in response to nutrient availability. Notably, the relSpn-dependent response included significant up-regulation of the ply operon encoding pneumolysin toxin, whereas the ΔrelSpn-dependent response affected expression linked to the VicRK and CiaRH two component systems. Finally, a D39 ΔrelSpn mutant was severely attenuated and displayed a significantly altered course of disease progression in a mouse model of infection, which was restored to normal by an ectopic copy of rel+Spn.
Introduction
Streptococcus pneumoniae (pneumococcus) is a gram-positive human respiratory pathogen that causes numerous serious diseases, including pneumonia, otitis media, meningitis, and bacteremia (see (Musher, 2004)). In addition, S. pneumoniae occasionally has been associated with urinary tract, kidney, soft tissue, and peritoneal infections as a consequence of host immunosuppression, asplenia, or recent surgery (see (Dufke et al., 2004; Musher, 2004)). Pneumococcal infections can be severe in at-risk populations, such as the very young, elderly, and immunocompromised individuals without access to antibiotics, and over 1.6 million deaths are estimated to occur annually (Janoff and Rubins, 2004; WHO, 2007). Antibiotic resistance and evasion of current vaccines by serotype selection are increasing worldwide (see (Appelbaum, 2002; Dagan and Klugman, 2008; Felmingham et al., 2007)). Clearly, S. pneumoniae has the capacity to inhabit numerous niches in the human body that differ in nutrient, metal ion, and oxygen availability, and responses to these environmental cues may play roles in colonization and disease progression.
This capacity for metabolic adaptation occurs despite the fact that pneumococcal central metabolism is limited to glycolysis, pneumococcus lacks a tricarboxylic acid cycle, and is auxotrophic for several amino acids (see below; (Hoskins et al., 2001; Tettelin et al., 2001)). Several regulators contribute to pneumococcal nutrient sensing, including two component systems (TCSs) (Hendriksen et al., 2007; McKessar and Hakenbeck, 2007; Paterson et al., 2006; Ulijasz et al., 2004; Wagner et al., 2002), PsaR (Johnston et al., 2006; Kloosterman et al., 2008), SczA (Kloosterman et al., 2007), GlnR (see (Hendriksen et al., 2008b)), CodY (Hendriksen et al., 2008a), SpxR (Ramos-Montanez et al., 2008), CcpA (see (Iyer et al., 2005)), and LacI/ GalR family regulatory proteins (Chapuy-Regaud et al., 2003; Iyer and Camilli, 2007). In addition, it is likely that the alarmones guanosine-pentaphosphate and -tetraphosphate ((p)ppGpp), which are synthesized by the RelSpn protein, play critical roles in the metabolic adaptation of S. pneumoniae during infection. Recent studies have implicated (p)ppGpp-mediated responses in the adaptation of several bacterial pathogens to nutritional limitations and environmental stresses encountered during host infection (reviewed in (Braeken et al., 2006; Potrykus and Cashel, 2008); see also (Dalebroux et al., 2009; Dozot et al., 2006; Zhou et al., 2008)). A signature-tagged mutagenesis screen performed on S. pneumoniae strain TIGR4 identified the relSpn gene as a putative virulence factor in a murine pneumonia model (Hava and Camilli, 2002). However, its exact role in pathogenesis is unknown.
The role of (p)ppGpp as a regulator of growth rate and starvation responses has been studied for many years (reviewed in (Braeken et al., 2006; Cashel et al., 1996; Magnusson et al., 2005; Potrykus and Cashel, 2008; Srivatsan and Wang, 2008)). In E. coli and other gram-negative bacteria, the RelA protein synthesizes (p)ppGpp upon binding to ribosomes stalled for polypeptide synthesis. The elicited stringent response decreases the synthesis of stable RNA and ribosome proteins, increases amino acid biosynthesis, and reprograms transcription through the synthesis of alternative sigma factors and modulation of their association with core RNA polymerase (see (Potrykus and Cashel, 2008; Srivatsan and Wang, 2008)). (p)ppGpp generally shortens the half-life of RNA polymerase-promoter DNA open complexes, and its effect on decreasing or increasing transcription initiation is potentiated by the DksA protein (see (Potrykus and Cashel, 2008)). Besides RelA, E. coli and other gram-negative bacteria produce a bifunctional SpoT synthetase/ hydrolase that regulates intracellular (p)ppGpp concentrations (see (Braeken et al., 2006; Potrykus and Cashel, 2008)). SpoT-dependent (p)ppGpp accumulation occurs in response to depleted concentrations of carbon, iron, or fatty acids (see (Dalebroux et al., 2009; Potrykus and Cashel, 2008; Srivatsan and Wang, 2008)). The SpoT synthetase/ hydrolase activities are regulated by interactions with other proteins, including ribosomal proteins, ribosome-associated GTPase CgtA, and Acyl Carrier Protein (ACP), a key component of lipid biosynthesis (see (Battesti and Bouveret, 2009; Dalebroux et al., 2009; Potrykus and Cashel, 2008)).
Bacillus subtilis and other gram-positive species produce a bifunctional RSH (p)ppGpp synthetase/ hydrolase (see (Braeken et al., 2006; Potrykus and Cashel, 2008)). This enzyme has previously been called “RelSeq,” “RelBsu,” and “RelMtb” in Streptococcus equisimilis, B. subtilis, and Mycobacterium tuberculosis, respectively (Braeken et al., 2006), and we refer to the paralog in S. pneumoniae as “RelSpn” in this paper. In contrast to the gram-negative paradigm described above, (p)ppGpp synthesis by RSH proteins in gram-positive species seems to decrease rRNA transcription indirectly through depletion of the cellular pool of GTP, which is the initiating nucleotide triphosphate (iNTP) in rRNA transcripts (Kasai et al., 2006; Krasny and Gourse, 2004). This indirect mode of regulation was recently extended to other stringently regulated promoters in B. subtilis, although additional factors likely play significant roles in the expression of some up-regulated transcripts (Krasny et al., 2008; Tojo et al., 2008). Similar to gram-negative SpoT, RelMtb interacts with ribosomes (Potrykus and Cashel, 2008), but RelBsu and RelSpn do not seem to interact directly with ACP (Battesti and Bouveret, 2009), and the mechanisms that regulate gram-positive RSH protein activities are not well understood (see (Potrykus and Cashel, 2008)).
Besides RSH proteins, gram-positive species in the Firmicutes phylum encode small polypeptides homologous to the (p)ppGpp synthetase domain of gram-negative RelA that lack carboxyl terminus regulatory domains (see (Potrykus and Cashel, 2008)). These RSH fragments, which have been called “small alarmone synthetases” (SAS) (Nanamiya et al., 2008), can synthesize (p)ppGpp in a heterologous E. coli system (Battesti and Bouveret, 2009; Lemos et al., 2007; Nanamiya et al., 2008), but may not contribute significantly to the stringent response (Lemos et al., 2007; Nanamiya et al., 2008). Instead of responding to amino acid limitation, like RelA, SAS proteins seem to respond to environmental conditions such as alkaline or envelope stress, and are linked to other regulatory circuits involving alternative sigma factors and TCSs (Eiamphungporn and Helmann, 2008; Gardete et al., 2006; Lemos et al., 2007; Nanamiya et al., 2008).
We report here the first physiological characterization of the stringent response and RelSpn regulon in S. pneumoniae. We found that the pneumococcal relSpn gene was not essential under nutrient replete conditions, but unexpectedly, mediated uptake or utilization of certain metal ions in defined growth medium. Transcription profiling in response to the antibiotic mupirocin, an inhibitor of tRNAIle charging that induces (p)ppGpp synthesis (Hughes and Mellows, 1978), revealed a complex pattern of relSpn-independent, relSpn-dependent, and ΔrelSpn-dependent responses, consistent with a role for RelSpn in mediating a classic stringent response and in maintaining broader metabolic homeostasis. Finally, we showed that relSpn is a significant virulence factor that caused unanticipated alterations in the timing and route of disease progression.
Results
relSpn is not essential under nutrient replete conditions
The S. pneumoniae relSpn gene (spd1458, spr1487, and sp1645 in strains D39, R6, and TIGR4, respectively; Fig. 1A) encodes a 740 amino acid bifunctional (p)ppGpp synthetase/ hydrolase with 76% amino acid identity to the well-characterized S. equisimilis RelSeq (Hogg et al., 2004; Mechold et al., 1996) and 53% identity to B. subtilis RelBsu (Wendrich and Marahiel, 1997). relSpn is part of the pneumococcal core genome derived from 72 clinical isolates (Obert et al., 2006), and the RelSpn sequence is almost identical among the 17 S. pneumoniae strains for which sequences are available (Hiller et al., 2007). A candidate extended −10 promoter with sequence TTTTCAtattcttaaaaaagTGaTAAAAT is located 24 bp upstream of the relSpn start codon. relSpn and the downstream dtd gene (spd1457), encoding a putative D-tyrosyl-tRNATyr deacylase, are separated by 29 bp. This gene arrangement is found in other species of Streptococcus. A short hairpin structure with a 7 bp stem (ΔG= -5.3 by Mfold; http://www.bioinfo.rpi.edu/applications/mfold/) lies in the relSpn-dtd intergenic region but is not predicted to act as a transcription terminator. A predicted Rho-independent terminator lies downstream of dtdSpn (TransTermHP, http://transterm.cbcb.umd.edu/). A 100 bp long pneumococcal BOX repetitive element (see (Knutsen et al., 2006)) is located 292 bp upstream of relSpn (Fig. 1A).
Fig. 1.
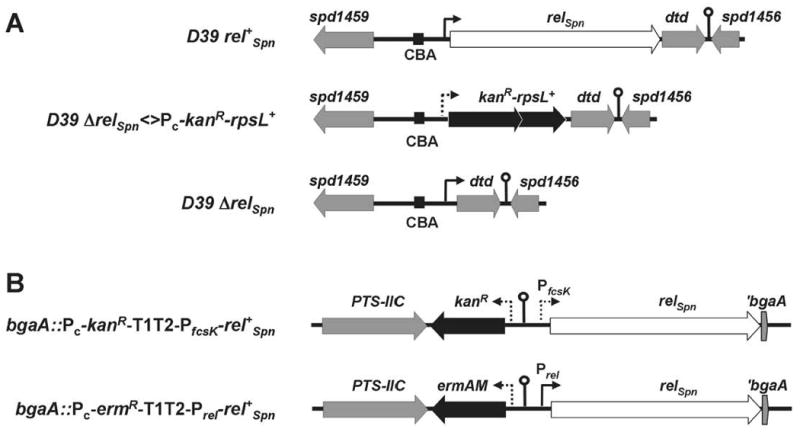
Organization of the relSpn gene region and design of constructs used in this study at the native relSpn locus (A) or in the ectopic bgaA locus (B). Reading frames are indicated by arrows; genes of interest are indicated in white, antibiotic resistance genes are in black, others are in gray. Gene names are indicated above the arrows. Promoters are indicated by arrows: solid arrows represent endogenous promoters, broken arrows represent either the synthetic constitutive Pc promoter (Claverys et al., 1995) or the L-fucose-inducible PfcsK promoter (Chan et al., 2003). The Prel-relSpn construct used for complementation studies contains the relSpn gene and 147 bp of upstream sequence. Lollipop symbols indicate putative terminator sequences. Regions are drawn to scale. CBA= BOX repetitive element; dtd encodes D-tyrosyl tRNATyr deacylase.
We deleted the entire relSpn gene in strain D39 using the two-step Janus cassette allele-replacement method (Fig. 1A, Experimental procedures; (Sung et al., 2001)). Transformants (e.g., IU1908, Table S1) were recovered at normal frequencies, formed uniform sized colonies on blood agar plates, and could be maintained without apparent size changes in rich media (BHI broth or blood agar). Because RelSpn senses perturbations in translation, strain IU1921, a streptomycin-sensitive derivative of IU1908, was used in subsequent studies (Experimental procedures). relSpn was also easily deleted in the avirulent D39-derived laboratory strain R6 (EL1513) using a similar approach. Deletion of relSpn did not change transcript levels of dtd during growth in BHI or upon mupirocin treatment (see below). Growth of the ΔrelSpn mutant (IU1921) in BHI was transiently inhibited by 2.4 mM D-tyrosine (data not shown). This effect was not due to polarity of the ΔrelSpn mutation on dtd expression (see (Soutourina et al., 2004)), because the growth of the corresponding rel+Spn parent (IU1690) and complemented ΔrelSpn bgaA∷PfcsK-rel+Spn strains (IU2024 and IU2560, Table S1) were not inhibited by D-tyrosine addition.
RelSpn is the primary source of (p)ppGpp in S. pneumoniae D39 and R6
We determined the levels of (p)ppGpp synthesized in rel+Spn (IU1690, IU1912, EL908) and ΔrelSpn (IU1921, IU1943, EL1513) strains. S. pneumoniae grew poorly or not at all in the low phosphate concentrations (Cashel, 1994) required for uniform labeling of (p)ppGpp with 32P-phosphoric acid (data not shown; (Novak et al., 1999)). Instead, we grew cultures in BHI to early exponential phase (OD620 ≈0.2), and performed non-uniform labeling of washed cells with 32Pi in MOPS-buffered CDM media lacking isoleucine, in the absence or presence of 100 ng mupirocin per mL (Experimental procedures). We obtained the same nucleotide labeling patterns for D39 strains (IU1690, IU1921, IU2024; Table S1) and R6 strains (EL908, EL1513, IU2028; Table S1) (data not shown) as for the D39 strains containing the luxABCDE operon (IU1912, IU1943, and IU2025; Fig. 2) that were used in animal infection experiments (see below).
Fig. 2.
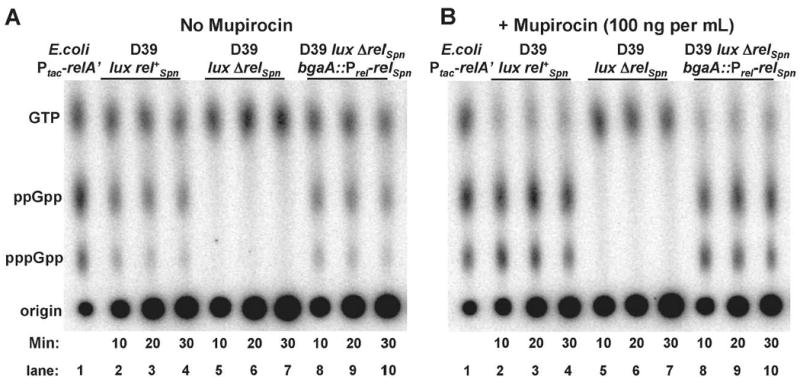
(p)ppGpp production by D39 rel+Spn, ΔrelSpn, and complemented ΔrelSpn bgaA∷Prel-rel+Spn strains in the absence (A) and presence (B) of 100 ng mupirocin per mL. Cells were non-uniformly 32Pi labeled (150 μCi per mL) in MOPS-CDM (- Ile) and extracted with formic acid, and extracts were analyzed by PEI TLC as described in Experimental procedures. The strains assayed were: IU1912 (D39 luxABCDE rel+Spn), IU1943 (D39 luxABCDE ΔrelSpn), and IU2025 (D39 luxABCDE ΔrelSpn bgaA∷Prel-rel+Spn). IU2025 contained an amino acid replacement (Glu227Gly, GAG to GGG) that matches the sequence of E. coli SpoT at that position, and a silent change at Cys607 (TGT to TGC). Labeled nucleotides were identified by comparison with standards synthesized by strain TX2737 (Table S1) expressing the constitutive E. coli RelA′ enzyme from an IPTG-inducible promoter (Svitil et al., 1993).
(p)ppGpp synthesis in the rel+Spn strains could be detected after 5 min of labeling in MOPS-CDM (-Ile) (not shown). After 10 min, approximately 55% of the 32P incorporated into guanosine nucleotides was in GTP, and 45% was in ppGpp and pppGpp in a ratio of ≈3:1 (Fig. 2A, lanes 2-4). In the presence of mupirocin, 32P incorporation into GTP dropped to 20%, and the 80% incorporated into ppGpp and pppGpp shifted to a ratio of ≈1.6:1 (Fig. 2B, lanes 2-4). No (p)ppGpp synthesis or decrease in labeled GTP levels was detected in the ΔrelSpn strain (Fig. 2A and 2B, lanes 5-7).
As controls, we introduced an ectopic copy of rel+Spn into the bgaA locus under control of the fucose-inducible PfcsK promoter (IU2024, IU2028) (Chan et al., 2003) or the native Prel promoter (IU2025) (Fig. 1B; Experimental procedures). (p)ppGpp synthesis was restored in the ΔrelSpn bgaA∷Prel-rel+Spn complementation strain (Fig. 2A and 2B, lanes 8-10). Similar complementation of rel+Spn function was detected in ΔrelSpn bgaA∷PfcsK-rel+Spn strains grown and assayed in the presence of 0.5% (wt/vol) fucose (IU2024, IU2028; data not shown). In addition, we checked whether deletion of the spr1004 gene, which encodes the sole YjbM/ RelQ-like SAS in S. pneumoniae R6 and D39, affected (p)ppGpp synthesis under these experimental conditions. Spr1004 synthesizes (p)ppGpp in a heterologous E. coli system (Battesti and Bouveret, 2009). Deletion of spr1004 in the R6 background (strain EL1516; Fig. S1A; Table S1) did not affect (p)ppGpp amounts (data not shown), whereas (p)ppGpp was not detected in mutants deleted for relSpn (EL1513 (ΔrelSpn), EL1522 (ΔrelSpn Δspr1004), and EL1576 (ΔrelSpn Δspr1004 bgaA∷PfcsK-spr1004+) (Fig. S1A; Table S1; data not shown). Addition of 100 ng mupirocin per mL for 30 min or 1 mg serine hydroxamate per mL for 20 min also did not result in (p)ppGpp synthesis by Spr1004 in a ΔrelSpn mutant (Fig. 2, lanes 5-7; data not shown), nor was (p)ppGpp synthesis by RelSpn changed in a Δspr1004 mutant (data not shown). Together, these results support the conclusion that RelSpn was the primary, and possibly only source of (p)ppGpp in S. pneumoniae D39 and R6 under these conditions.
Finally, a 32P-labeled species, which was not observed in the labeling medium, remained at the origin in one- and two-dimensional TLC of rel+Spn, ΔrelSpn, Δspr1004 and ΔrelSpn Δspr1004 samples (Fig. 2; data not shown). The origin-bound material was labile in hot acid in preliminary experiments (not shown; Experimental procedures), suggesting that it might be polyphosphate or a phosphorylated polymer. However, more definitive assays, such as digestion by exopolyphosphatase Ppx (Kornberg et al., 1999), were not performed in this study to determine whether this material was polyphosphate.
D39 ΔrelSpn mutants require copper and manganese for growth
D39 rel+Spn (IU1690) and D39 ΔrelSpn (IU1921) strains grew similarly in complex BHI broth (Fig. 3A). In contrast, D39 ΔrelSpn did not grow in a chemically defined medium (CDM) (Fig. 4A). The growth defect of the ΔrelSpn mutant was complemented to different degrees in strains IU2024 and IU2560 (D39 ΔrelSpn bgaA∷PfcsK-relSpn; Fig. 4A). D39 and IU2024 grew with similar doubling times (≈48 and 60 min, respectively), whereas IU2560, which contained a missense mutation in the ectopically expressed relSpn gene (Lys319Glu), grew with a significantly longer doubling time (≈138 min) (Fig. 4A). These complementation results support the conclusion that growth of serotype 2 strain D39 in CDM was dependent on RelSpn activity.
Fig. 3.
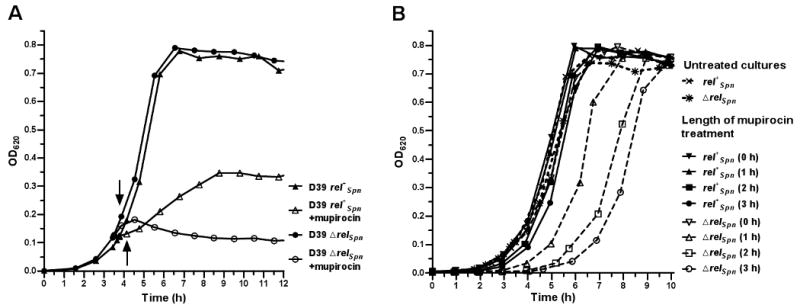
Growth of D39 rel+Spn (IU1690) and D39 ΔrelSpn (IU1921) strains in BHI broth (A) and outgrowth in BHI following mupirocin treatment (B). In (A), mupirocin was added to 100 ng per mL (open symbols) and arrows indicate when samples were harvested for RNA preparation for microarray and QPCR analyses. In (B), strains were grown in BHI and treated with 100 ng mupirocin per mL. Samples of treated cultures were removed at the indicated times and diluted into BHI containing no drug as described in Experimental procedures. Growths of untreated cultures are shown for comparison.
Fig. 4.
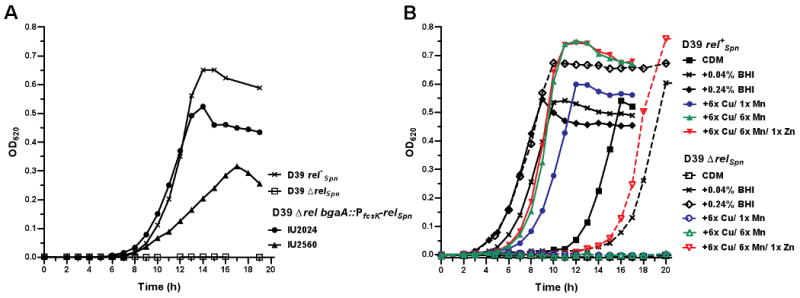
Growth of the D39 rel+Spn (IU1690) and D39 ΔrelSpn (IU1921) strains in CDM (A) and in CDM supplemented with BHI or CuSO4, MnSO4, and ZnSO4 (B). Strains were grown in BHI overnight, collected by centrifugation, washed and resuspended in CDM, and diluted into the indicated media as described in Experimental procedures. In (A), D39 ΔrelSpn bgaA∷PfcsK-relSpn complementation strains IU2024 and IU2560 contained a silent change at Asn475 (AAT to AAC) or an amino acid replacement (Lys319Glu, AAA to GAA), respectively, in RelSpn. In (B), standard CDM contained 2.8 mM MgSO4, 26.4 μM MnSO4 (1× Mn), 9.8 μM FeSO4, and 1.5 μM Fe(NO3)3. Where indicated, CuSO4 was added to 1.56 μM (6× Cu), MnSO4 was added to 158.4 μM (6× Mn), and ZnSO4 was added to 9.8 μM (1× Zn). See the text for additional details.
Some bacteria deficient in (p)ppGpp synthesis are auxotrophic for multiple amino acids (see (Tedin and Norel, 2001; Wendrich and Marahiel, 1997)); therefore, we added combinations of different amino acids to see if they supported growth of the D39 ΔrelSpn mutant in CDM. Adding high-purity amino acid preparations did not support growth of the D39 ΔrelSpn mutant. However, the ΔrelSpn strain grew when supplemented with lower purity amino acid preparations of serine, or by carryover of small volumes of BHI from overnight cultures used as inocula. D39 rel+Spn was able to grow in CDM without BHI addition, but adding BHI to a final concentration of 0.04% (vol/vol) shortened the lag time preceding exponential growth. Adding this same small amount of BHI supported growth of the D39 ΔrelSpn strain in CDM after a considerable lag time (Fig. 4B). Titration experiments showed that adding larger amounts of BHI further shortened the ΔrelSpn growth lag (data not shown), and adding a final concentration of 0.24% (vol/vol) BHI to CDM resulted in identical growth of the D39 rel+Spn and ΔrelSpn strains (Fig. 4B).
The extremely small volume of BHI required to support growth of D39 ΔrelSpn in CDM suggested a deficiency in the uptake or utilization of a trace metal or vitamin. Standard CDM contains magnesium sulfate (MgSO4, 2.8 mM), manganese sulfate (MnSO4, 26.4 μM) and iron (FeSO4, 9.8 μM and Fe(NO3)3, 1.5 μM) (van de Rijn and Kessler, 1980). Addition of high-purity (≈99%) copper sulfate (CuSO4, 0.13 μM), cobalt chloride (CoCl2, 2.9 μM), zinc sulfate (ZnSO4, 4.9 μM), and sodium molybdate (Na2MoO4, 3.2 μM) to CDM supplemented with 0.04% BHI greatly enhanced growth of the ΔrelSpn strain, and much of this enhancement was attributable to CuSO4 (data not shown). However, adding only CuSO4 to CDM over a range of concentrations did not support growth of the ΔrelSpn strain. Addition of CuSO4 and ZnSO4 and increased concentrations of MnSO4 over the amount specified in the CDM recipe were required for growth of the ΔrelSpn strain without added BHI (Fig. 4B). Zn2+ was also likely present at some level in the water and glassware used in our experiments, since sometimes ZnSO4 addition was not required for growth of the ΔrelSpn strain (data not shown). We observed some variability in the lag times of the D39 ΔrelSpn mutant grown in CDM supplemented with CuSO4, MnSO4 and ZnSO4. In addition, growth of the D39 ΔrelSpn mutant in CDM supplemented with metals still lagged more than the D39 parent in CDM lacking metals (Fig. 4B), suggesting that the ΔrelSpn mutant may be sensitive to unidentified media conditions or require additional trace nutrients for optimal growth. However, initial experiments testing vitamin requirements were difficult to interpret because some vitamins (e.g., folate, pyridine, pyridoxamine, and riboflavin) bind metals (Dawson et al., 1969).
We noted that increasing MnSO4 concentrations improved growth of the D39 rel+Spn strain in CDM supplemented with CuSO4 (Fig. 4B, compare 6× Cu/ 1× Mn and 6× Cu/ 6× Mn). Copper is a redox active metal that can potentially react with the relatively high concentration of hydrogen peroxide produced by pneumococcus (see (Ramos-Montanez et al., 2008) to generate harmful hydroxyl radicals by the Fenton reaction (Imlay, 2003; Sutton and Winterbourn, 1989). Manganese can protect bacteria from reactive oxygen species (ROS) (see (Tseng et al., 2001)) and was required for growth of the D39 ΔrelSpn mutant in CDM supplemented with CuSO4 (Fig. 4B and data not shown). However, preliminary experiments showed that the hydroxyl radical scavengers dimethyl sulfoxide (14 mM) and thiourea (10 mM) (Suthanthiran et al., 1984) could not replace increased manganese concentrations in supporting growth of the D39 ΔrelSpn mutant in CDM containing CuSO4 (data not shown).
Re-evaluation of the amino acid requirements of strain D39
As part of our characterization of the nutritional requirements of the D39 ΔrelSpn mutant and stringent response, we re-evaluated the amino acid requirements of the D39 rel+Spn parent strain (IU1690) in CDM drop-out media. These experiments led to several unanticipated results. D39 did not grow in medium lacking histidine, arginine, or leucine (Fig. 5), because it lacks histidine biosynthetic genes and encodes incomplete pathways or truncated genes for arginine and leucine biosynthesis (Hoskins et al., 2001; Lanie et al., 2007; Sonenshein et al., 2002). Unexpectedly, D39 did not grow without isoleucine or valine, omitted singly or in combination from CDM, despite the fact that D39 encodes a complete biosynthesis pathway (ilvBNC, spd0404-0406; ilvD, spd1956; ilvE, spd0749). D39 was also auxotrophic for cysteine, and methionine and inorganic sulfate could not substitute as sulfur sources (data not shown). This result is consistent with the absence of genes for uptake and utilization of inorganic sulfate in the D39 genome (Sonenshein et al., 2002) and indicates that the encoded transsulfuration pathway can synthesize methionine from cysteine but cannot synthesize cysteine from methionine. D39 grew well when cystine or near- physiological concentrations of glutathione (429 μM in airway surface liquid; (Wilson, 2005)) were provided as sole sulfur sources, but did not utilize the organosulfonate taurine (data not shown).
Fig. 5.
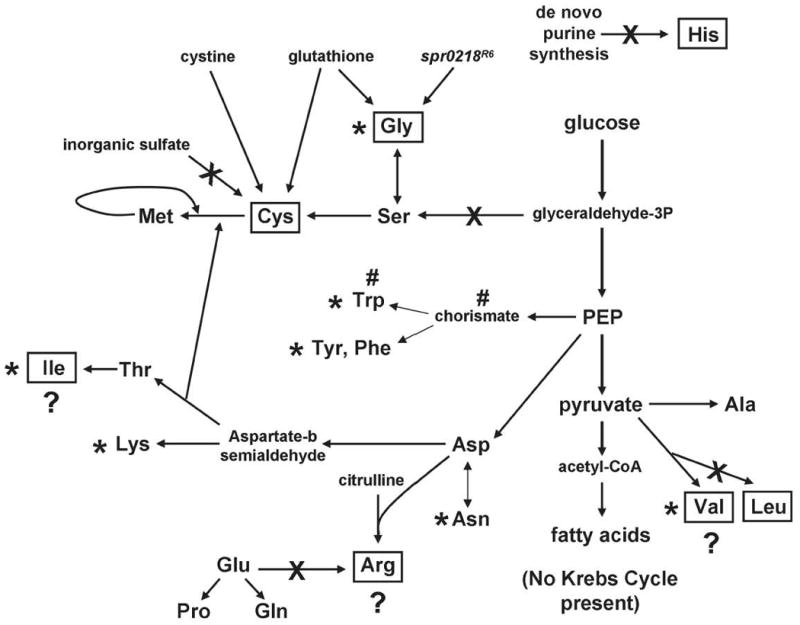
Predicted amino acid biosynthesis pathways in S. pneumoniae D39 based on genomic sequences (Hoskins et al., 2001; Lanie et al., 2007). Arrows are not scaled to reflect lengths of pathways. “X” indicates incomplete or missing pathway. “?” indicates a complete pathway that appears nonfunctional under tested conditions. D39 did not grow in CDM drop-out media lacking boxed amino acids. Relative transcript amounts for some biosynthesis genes increased (amino acids indicated with asterisks) or decreased (pound signs) upon mupirocin treatment (see text and Table 1, lines 1, 2, 9).
D39 was unable to grow in CDM lacking glycine. This result was surprising, because laboratory strain R6 (Hoskins et al., 2001; Lanie et al., 2007), which was derived from D39, is a glycine prototroph. The spd0221/ spr0218 gene, which encodes a conserved hypothetical protein of unknown function, is truncated in D39 and possibly in its progenitor, but not in R6 (Lanie et al., 2007). We found that replacing the truncated spd0221 allele in D39 with the full-length spr0218 gene from R6 restored growth in CDM lacking glycine (Fig. 5 and Fig. S1B; data not shown). Conversely, moving the truncated spd0221 allele from D39 into R6 resulted in glycine auxotrophy (data not shown). Another unexpected finding was that the glycine requirement of D39 was satisfied by glutathione, even though D39 lacks an obvious homologue of γ-glutamyltranspeptidase (ggt), which cleaves glutathione in E. coli and B. subtilis (see (Suzuki et al., 1993)).
Thus, D39 was auxotrophic in CDM for histidine, arginine, leucine, cysteine (cystine), isoleucine, valine, and glycine, and these amino acid requirements were not satisfied by adding 0.24% (vol/vol) BHI to CDM drop-out media (Fig. 5, data not shown). The D39 ΔrelSpn mutant showed the same amino acid requirements as the D39 rel+Spn parent, confirming that the ΔrelSpn mutation did not cause additional amino acid auxotrophy in this serotype 2 strain (data not shown). However, the D39 ΔrelSpn mutant did grow more slowly than its D39 parent in CDM containing 0.24% (vol/vol) BHI, but lacking glutamine (doubling times of ∼320 and 110 min, respectively), suggesting limitation of a precursor or cofactor in glutamine biosynthesis.
relSpn enhances recovery after mupirocin treatment
We compared the recovery of D39 rel+Spn and D39 ΔrelSpn strains after 0 to 3 h of exposure to 100 ng mupirocin per mL. We did not detect significant changes in pneumococcal chain length or cell shape by phase microscopy upon mupirocin treatment of up to 3 h (data not shown). Following mupirocin addition, equal CFUs (≈1.2 × 106) were diluted into fresh BHI broth and allowed to outgrow in the absence of the drug. Mupirocin treatment did not significantly affect the lag and doubling time of the D39 parent strain (Fig. 3B). In contrast, recovery of the D39 ΔrelSpn mutant was correlated with the length of mupirocin treatment; the greater the length of treatment, the longer the lag of the ΔrelSpn mutant before exponential growth resumed (Fig. 3B). Finally, we examined whether relSpn function and presumably (p)ppGpp amount affected the comparatively short stationary phase and spontaneous autolysis of these pneumococcal strains grown in BHI. We did not detect a difference in the timing of autolysis or a decrease in cell survival, determined by optical density (A620) and direct plating of viable cells, between the D39 rel+Spn and D39 ΔrelSpn strains (data not shown).
Complex responses of D39 and D39 ΔrelSpn to limitation of tRNAIle charging
We performed microarray analyses on rel+Spn and ΔrelSpn strains growing exponentially (OD620 ≈0.16 - 0.2) in BHI broth and following addition of mupirocin to limit Ile-tRNAIle synthesis and induce the stringent response. D39 and D39 ΔrelSpn grew similarly in BHI broth (Fig. 3A), and a 16-fold decrease in the relative amount of the relSpn transcript in the ΔrelSpn mutant was the only significant change (p=0.0002) detected between the two strains. Preliminary experiments showed that the addition of several compounds commonly used to induce stringent response in other bacteria (see (Cashel et al., 1996; Eymann et al., 2002; Wendrich and Marahiel, 1997)), including amino acid analogues (serine hydroxamate, 1 mg per mL; DL-norleucine or DL-norvaline, 0.5 mg per mL) or α-methylglucoside (up to 1.3% (vol/vol)) did not significantly reduce growth of R6 strains in BHI or CDM broth, respectively (data not shown). Mupirocin was the only compound tested that strongly inhibited the growth of the D39 and R6 strains. Titration experiments showed that 100 ng mupirocin per mL was a sublethal concentration that slowed growth of D39 for about 1 h (Fig. 3A), after which growth resumed with an average doubling time 4-fold longer than that of untreated cultures (177 and 45 min, respectively) before reaching a lower growth yield. By comparison, the D39 ΔrelSpn mutant continued growing at a slightly reduced rate for 20 min and then stopped within ≈1 h, after which culture densities and CFUs began to drop slowly (Fig. 3A; data not shown). RNA samples were collected 20 min after mupirocin addition (Experimental procedures) to determine relatively early transcriptional responses to translation stalling and to avoid secondary stress responses caused by completely halting growth.
The transcript amounts from a large number of genes changed in both the D39 (IU1690) and D39 ΔrelSpn (IU1921) strains treated with mupirocin (351 and 357, respectively, with p ≤ 0.003) relative to the untreated strains. This number corresponded to 17% of the pneumococcal open reading frames included on this array. Of these, relative transcript amounts of 276 or 205 genes changed at least 2-fold in D39 or D39 ΔrelSpn, respectively, after mupirocin treatment. These changes fell into the following three classes: relSpn-independent (changed in the same direction in both D39 and D39 ΔrelSpn), relSpn-dependent (changed in D39, but not in D39 ΔrelSpn), and ΔrelSpn-dependent (changed in D39 ΔrelSpn, but not in D39) (Tables 1, S3-S6).
Table 1.
Relative transcript changes for specific genes upon mupirocin treatment of D39 rel+Spn and D39 ΔrelSpn strainsa,b
| relSpn-independent | relSpn-dependent | ΔrelSpn-dependent | ||
|---|---|---|---|---|
| Increased | ||||
| 1 | Ile-tRNAIle synthetase (ileS, 4.0/ 3.1) | Biosynthesis of lysine (dapA, lysA; 2.6 ±0.7), aromatic amino acids (spd1151, trpFCDGE; 2.8 ±0.6), serine (glyA, 3.4); asparagine (asnA, 3.7) and isoleucine/ valine (ilvN, 4.8) | Ribosomal proteins (2.1 ±0.2, n=15), ribosome maturation and translation initiation (spd1413, infB; 2.2 ±0) rpsD, rplP, rpsJ | |
| 2 | Synthesis of isoleucine/ valine (ilvBC-spd0407, 4.2 ±0.4/ 1.9 ±0.1) and lysine (ykuRQ, 5.2 ±0.2/ 1.9 ±0) | Predicted amino acid transporters (spd0530-0531, spd0954-0955, spd1289-1290, spd1356-1357; 2.8 ±1.0) | Iron transport (pit2BCD, 2.7 ±0.2) | |
| 3 | Oligopeptide uptake (spd1167,1169; 2.6 ±0.1/2.1 ±0.4) and amino acid transport (spd1328-1330, 6.1 ±0.5/ 2.3 ±0.4) | Purine synthesis and interconversion (guaB, 2.2; guaC, 2.8) | adenylate kinase (adk, 2.4) | |
| 4 | β-glucoside uptake and utilization (PTS IIABC, bglA-2; 3.1 ±0.8/ 2.0 ±0) | Stress responses (yfiA, 3.9; gsp24, 2.1; uspA, 4.5; thioredoxin (trx, 2.4; trxB, 2.3); metal efflux (czcD, 3.3)) | Balancing reducing equivalents (lpdA-acoLCBA, 3.0 ±0.4; pyrKDb, 2.5 ±0.1) | |
| 5 | Drug efflux (spd1900, 3.4/ 2.4) | Pneumolysin production (ply, spd1728-1729; 3.4 ±0.2) | Sugar binding transcription factor regR (2.1) | |
| 6 | Replication initiation and gene expression (dnaA, dnaN; 3.0 ±0.6/2.1 ±0.1) | Competence stimulating peptide CSP (comC, 5.4); CSP transporter comA, 2.5; histidine kinase comD, 4.4c | VicRK regulon (spd0104, spd0703, lytB, spd1870-1874, pcsB; 4.5 ±1.8) | |
| 7 | Ribosome recycling machinery (rnr, smpB; 2.1 ±0/ 4.2 ±0.4), RelB (spd1082, 2.0/ 2.2) | Putative virulence factors (by STM: spd0249, bglG, spd0909, glgB, spd1528, spd1614, spd1643; 2.7 ±0.7) | Endoribonuclease RelE (spd1081, 2.3) | |
| Decreased | ||||
| 8 | T-box controlled genes: spd0491-0493 (tRNAVal, -7.3 ±0.6/ -8.0 ±0.9); glySQ-spd1306-1307 (tRNAGly, -3.8 ±0.2/ -6.5 ±0.8); spd1442-1443-thrS (tRNAThr, -2.9 ±0.4/ -5.2 ±1.0); alaS-spd1217 (tRNAAla, -5.3 ±2.5/ -5.2 ±2.3); pheS (tRNAPhe, -1.8/ -6.0) | Ribosomal proteins (-3.1 ±0.7, n=28), translation initiation, elongation, and release factors (-2.5 ±0.3) rpsD, rplP | Ribonuclease J1 (-2.1), ribosome dependent GTPase (rsgA, -2.0), elongation factors (-2.2 ±0.2) | |
| 9 | Cell division (ftsE, -2.0/ -2.0), chromosome structure and segregation (smc, -3.7/ -2.4; parC, -5.2/ -2.7) | DNA replication and repair (polC, dnaX, dnaQ, dnaG, ligA, ssb, mfd, rexB, recN; -2.9 ±0.8); septum formation (divIC, -2.9) | Synthesis of aromatic amino acid (trpBCDE, aroCED; -2.7 ±0.9); Trp-tRNATrp synthetase (trpS, -2.3) | |
| 10 | Glycolysis (eno, fba, tpi, pfkA, pgk; -2.2 ±0.5/ -3.7 ±1.8), alcohol dehydrogenase (adhE, -8/ -11.7) | RNA polymerase (rpoA, -2.8; rpoD, -2.3) | Glycolysis and pyruvate metabolism (gap, pyk, ldh; -2.2 ±0.2) | |
| 11 | Fructose uptake (fruA, fruR; -8.4 ±0.4/ -5.7 ±1.2) | Carbohydrate transport (spd0661, spd0740-0742; -2.6 ±0.5 | PTS proteins EI and HPr (ptsIH, -2.4 ±0) | |
| 12 | Glutamine and glutamate uptake and metabolism (glnR, -2.2/ -3.0) | Cation and iron transport (spd1436, pit2CD; -2.2 ±0.4) | Cation binding (adcA, -2.1) | |
| 13 | Heat-shock proteins and chaperones (hrcA-grpE-dnaK, groES; -5.2 ±1.3/ -7.2 ±1.5); (hslO, -2.0/ -2.0) | Pyrimidine biosynthesis (pyrFE, carAB, pyrR; -3.1 ±0.4); adenylate kinase (adk, -2.0) | Pyrimidine salvage (upp, cdd; -4.0 ±0.8); GMP synthetase (guaA, -3.6); F1F0 ATP synthase subunits (atpDGH, -2.0 ±0.1) | |
| 14 | Protein maturation and proteolysis (ppmA, -4.7/ -2.2; htrA, -2.2/ -2.1) | Fatty acid synthesis and saturation (accA, -2.2; fabM, -2.3) | Transcription antitermination factor NusB (-2.1); response regulator ciaR, -2.4; histidine kinase ciaH, -4.0c | |
| 15 | Synthesis of carrier lipid (mvaK2, mvaS; -2.8 ±0.2) and peptidoglycan (murF, -2.5) | Lipoteichoic acid modification (dltABCD, -2.5 ±0.2); protective antigen (phtA, -2.3) | ||
| 16 | Capsule biosynthesis (cps2IJKPMO; -2 ±0.1) | SpxR regulon (strH, -2.4; spxB, -3.8); ROS protection (sodA, psaD, spd0695; -2.5 ±0.2) | ||
| 17 | Redox sensing (spd0976, -3.0) |
Microarray analyses were performed using cutoffs of 2 fold and p ≤ 0.003 for relSpn-dependent and ΔrelSpn-dependent changes, and a cutoff of p ≤ 0.003 for relSpn-independent changes. All genes whose relative transcript levels changed at least 2 fold in one or both strains treated with mupirocin are listed as relSpn-independent changes as described in Experimental procedures. relSpn-independent: changed in the same direction in both D39 rel+Spn + mupirocin versus rel+Spn and D39 ΔrelSpn + mupirocin versus ΔrelSpn comparisons; relSpn-dependent: changed in the D39 rel+Spn + mupirocin versus rel+Spn, but not in the D39 ΔrelSpn + mupirocin versus ΔrelSpn comparison; and ΔrelSpn-dependent: changed in D39 ΔrelSpn + mupirocin versus ΔrelSpn, but not in the D39 rel+Spn + mupirocin versus rel+Spn comparison. For the relSpn-independent changes, the average fold changes observed for the D39 rel+Spn + mupirocin versus rel+Spn and D39 ΔrelSpn + mupirocin versus ΔrelSpn comparisons are indicated as ratios (rel+Spn/ ΔrelSpn). For the other categories, the average fold change and standard deviation is given for the mupirocin treated strain compared to the untreated strain. Individual fold changes are listed in Tables S4-S6.
Transcript changes confirmed by QPCR are indicated in bold type.
Relative transcript changes for genes of interest with p values larger than the reporting cutoff: comD, p=0.004; ciaH, p=0.01.
relSpn-independent changes
Relative transcript amounts of 77 genes increased or decreased with p ≤ 0.003 in both D39 and D39 ΔrelSpn after mupirocin treatment (Tables 1, S3, and S4). These genes function in carbohydrate and amino acid transport and metabolism, translation, and protein folding (Table S3). Some of the strongest relative transcript amount changes were for genes regulated in response to translation levels, confirming that the mupirocin treatment of D39 and D39 ΔrelSpn was successful. These included T-box regulated genes (spd0491-0493; spd0504; spd1216-1217; spd1304-1307; spd1442-1444) (Table 1, line 8), whose transcription decreases in response to increasing ratios of charged to uncharged tRNA (Grundy and Henkin, 2003), and genes encoding chaperones that catalyze protein folding (grpE-dnaK, spd0459-0460; groES, spd1710) and their transcriptional regulator (hrcA, spd0458) (Table 1, line 13). Other relSpn-independent genes are potentially regulated directly or indirectly by levels of signal or substrate molecules, including dnaA (spd0001) (Table 1, line 6); fructose (fruA, spd0773) and β-glucoside (spd0502) transporters (Table 1, lines 4, 11); the GlnR transcriptional repressor (spd0447) (Table 1, line 12); glycolytic enzymes (eno, spd1012; fba, spd0526; pfkA, spd0789; pgk, spd0445; tpi, spd1404) (Table 1, line 10); a putative amino acid transporter (spd1328-1330), and enzymes of lysine (ykuRQ, spd1922-1923) and branched chain amino acid biosynthesis (ilvB, spd0404; ilvC, spd0406; spd0407) (Table 1, lines 2, 3); and an efflux pump (spd1900) (Table 1, line 5). Possible regulatory mechanisms underlying some of these general relSpn-independent changes in response to reduced translation level are considered in the Discussion and Table S4.
relSpn-dependent changes
The majority of relSpn-dependent genes with predicted functions (87 of 213) play roles in translation and ribosome structure, amino acid metabolism and transport, and DNA replication and repair (Tables 1, S3, and S5). This set of genes matched classical stringent responses defined previously for other bacteria (see (Durfee et al., 2008)). Relative transcript amounts of genes encoding ribosomal proteins, translation initiation and elongation factors, and RNA modification enzymes decreased, as did those encoding subunits of the DNA replication machinery and proteins involved in repair of double-stranded DNA ends (Table 1, lines 8, 9). In addition, relative transcript amounts from genes encoding RNA polymerase subunits α (rpoA) and σ (rpoD) decreased; these genes appear to be co-regulated with rplQ (ribosomal protein L17) and dnaG (DNA primase), respectively (Table 1, line 10).
The relative transcript amounts of several amino acid biosynthesis genes changed in a relSpn-dependent manner. Relative transcript amounts increased for genes encoding enzymes for lysine biosynthesis (dapA, spd0901; lysA, spd1775), asparagine biosynthesis (asnA, spd1768) and glycine biosynthesis (glyA, spd0910; also produces 5,10-methylene-tetrahydrofolate) (Table 1, line 1). Curiously, relative transcript amounts for tryptophan import and biosynthesis genes (spd0954-0955; trpFCDGE, spd1598-1602 (Gutierrez-Preciado et al., 2007; Panina et al., 2003)) also increased (Table 1, lines 1, 2). These two operons are preceded by T-box elements and were expected to decrease with the majority of other T-box regulated genes (Table 1, line 8). Relative transcript amounts for numerous putative transporters, which are abundant in S. pneumoniae, also changed in a relSpn-dependent manner (Table S5).
(p)ppGpp synthesis directs the transcription of virulence factor genes in several bacteria (see (Aberg et al., 2006; Anderson et al., 2006; Braeken et al., 2006; Dozot et al., 2006; Malke et al., 2006; Nakanishi et al., 2006)). In S. pneumoniae, the operon encoding pneumolysin (ply, spd1726), which is the major cholesterol-dependent membrane toxin (see (Kadioglu et al., 2008)), and adjacent co-regulated genes encoding hypothetical proteins found only in pneumococcus (spd1728-1729) were strongly up-regulated in a relSpn-dependent manner (Table 1, line 5). Changes in the relative transcript amounts for other major virulence factors (reviewed in (Kadioglu et al., 2008)) were not detected (Table S5). However, relative transcript amounts were strongly up-regulated for several genes identified as important for virulence in signature-tagged mutagenesis and other screens, including glyA, trpDGE, asnA, aliB (spd1357) and others (Table 1, lines 1, 2, 7; Table S5).
Finally, transcription from several key genes involved in the biosynthesis of capsule and cell wall were stringently controlled. Relative transcript levels decreased for 6 of the 9 exopolysaccharide capsule biosynthesis genes included on this array (Table 1, line 16) (see (Lanie et al., 2007)), for murF (spd1483, UDP-N-acetylmuramoylalanyl-D-glutamyl-lysyl D-alanyl-D-alanyl ligase) that catalyzes the final step of peptidoglycan stem peptide synthesis (Sonenshein et al., 2002), and for mevalonate biosynthesis genes mvaK2 (spd0348) and mvaS (spd1537) that are involved in producing isopentenyl pyrophosphate and the C55-P carrier lipid (Table 1, line 15) (Wilding et al., 2000). Relative transcript amounts for fatty acid synthesis genes were largely unchanged, except for decreases in fabM (spd0378), which catalyzes unsaturated fatty acid synthesis (Marrakchi et al., 2002), and accA (spd0390), a subunit of the acetyl-CoA carboxylase complex that catalyzes the first committed step of long chain fatty acid synthesis (Sonenshein et al., 2002) (Table 1, line 14).
ΔrelSpn-dependent changes
The relative transcript amounts of a surprisingly large number (160) of genes changed only in the D39 ΔrelSpn mutant, but not the rel+Spn D39 parent, following mupirocin treatment (Table S3). In addition, many other transcript changes observed for the ΔrelSpn mutant were less than the 2-fold cutoff used in this study but highly reproducible (p ≤0.003) (see GSE14750). ΔrelSpn-dependent transcript changes included genes associated with translation, with roles in energy production and conversion, and carbohydrate and amino acid transport and metabolism (Tables 1, S3 and S6). The relative transcript levels from a subset of ribosomal protein genes, most of which correspond to the E. coli spc and S10 ribosomal operons (Keener and Nomura, 1996), increased only in the D39 ΔrelSpn mutant treated with mupirocin (Table 1, line 1). Other translation-related ΔrelSpn-dependent changes included decreased relative transcript levels for an RNAse J1 homologue (spd0130) implicated in 16S rRNA maturation and global mRNA stability (see (Mader et al., 2008)), for rsgA (spd1781) which is a putative ribosome dependent GTPase with roles in ribosome stability and tRNA dissociation (see (Kimura et al., 2008)), and for genes encoding elongation factors P and Tu (spd0395, spd1318) (Table 1, line 8). In contrast, relative transcript amounts increased for genes encoding initiation factor IF-2 (spd0482) and an ATP-dependent RNA helicase (spd1413) similar to the E. coli cold-shock protein DeaD that may play a role in 50S ribosome subunit maturation (Moll et al., 2002) (Table 1, line 1).
The relative transcript amounts from genes encoding many glycolytic enzymes decreased in a relSpn-independent manner (above; Tables 1, S4); however, relative transcription from genes encoding several important enzymes from glycolysis (glyceraldehyde 3-phosphate dehydrogenase (gap, spd1823); pyruvate kinase (pyk, spd0790); (Sonenshein et al., 2002)), pyruvate metabolism (L-lactate dehydrogenase (ldh, spd1078); pyruvate oxidase (spxB, spd0636); (Ramos-Montanez et al., 2008)), and PTS sugar transport (Enzyme I and phosphocarrier protein HPr, spd1039-1040; (Jahreis et al., 2008)) decreased only in the ΔrelSpn strain (Table 1, lines 10, 11, 16). Other striking ΔrelSpn-dependent decreases in relative transcript amounts were for genes encoding: (i) oxidative stress-resistance proteins (superoxide dismutase A (sodA, spd0667); thiol peroxidase (psaD, spd1464); a putative short chain oxidoreductase, spd0695; (McAllister et al., 2004; Yesilkaya et al., 2000)); (ii) a putative redox-sensing transcriptional repressor (spd0976; (Sickmier et al., 2005)); (iii) a protective pneumococcal antigen (phtA, spd1038; see (Ogunniyi et al., 2009)); and (iv) enzymes that decorate lipoteichoic acid with D-alanine, a modification that changes envelope charge and confers resistance to antimicrobial cationic peptides (dltABCD, spd2002-2005; (Kovacs et al., 2006)) (Table 1, lines 15, 16, 17). Finally, significant ΔrelSpn-dependent increases in relative transcript amounts were observed for genes encoding: (v) a dihydrolipoamide dehydrogenase complex (lpdA-acoLCBA, spd1024-1028) (Table 1, line 4); and (vi) members of the regulon (spd0104, spd0703, lytB (spd0853), spd1870-1874, and pcsB (spd2043)) positively controlled by the essential VicRK (WalRK) two-component system implicated in maintaining cell wall homeostasis (see (Dubrac et al., 2008; Winkler and Hoch, 2008)) (Table 1, line 6).
Opposite responses and those of metal transporters
Relative transcript amounts for some genes or groups of genes encoding related functions increased in one strain and decreased in the other upon mupirocin treatment. As noted above, the relative transcript levels for some ribosomal proteins decreased in D39 treated with mupirocin, but increased in D39 ΔrelSpn (Table 1, lines 1 and 8; Tables S5 and S6). As a second example, relative transcript amounts from some tryptophan biosynthetic genes increased in D39 treated with mupirocin, but decreased in D39 ΔrelSpn (trpC, spd1599; trpD, spd1600; trpE, spd1602) (Table 1, lines 1, 9). Transcription responses for metal ion transporters were also differentially regulated and complicated. Relative transcript amounts for a putative cation transporting ATPase (spd1436, E1-E2 family), and subunits of an iron transporter (pit2CD, spd0917-0918; (Brown et al., 2001)) decreased in D39 treated with mupirocin, whereas those corresponding to a Zn2+ and Co2+ cation efflux system protein (czcD, spd1638; (Kloosterman et al., 2007)) increased (Table 1, lines 4, 12). By contrast, in the ΔrelSpn mutant, relative transcript levels encoding a Zn2+ binding protein (adcA, spd1997; (Dintilhac et al., 1997)) decreased, whereas those encoding subunits of the Pit2 iron transporter (pit2BCD, spd0916-0918; (Brown et al., 2001)) increased (Table 1, lines 2, 12). Notably, relative transcription levels for the PsaBCA manganese transporter (spd1461-1463; (McAllister et al., 2004)) and genes predicted to have roles in copper uptake and homeostasis (copYA, spd0633, spd0635; (Solioz and Stoyanov, 2003)); cutC (spd1118; (Gupta et al., 1995)) did not change in either strain upon mupirocin treatment (see GSE14750).
Confirmation of microarray results by quantitative real-time PCR (QPCR)
We determined relative transcript amounts of several genes by QPCR analysis using the same RNA preparations analyzed by microarrays. Expression ratios obtained by QPCR and by microarray analyses were log2-transformed and plotted against each other for comparison (Figures 7 and S2). Transformed expression ratios for four genes (rpoB, gyrA, ply, eno) generated by QPCR and normalized to 16S rRNA levels agreed well (r2=0.99) with transformed average expression ratios (M values) obtained by microarray analysis for the D39 rel+Spn strain treated with mupirocin (Fig. 6A, dotted line). We also observed good agreement between microarray M values and log2-transformed QPCR expression ratios normalized to 16S rRNA for 27 other genes from this dataset (Fig. S2A; r2=0.94). In contrast, transformed expression ratio values for seven genes (rpoB, gyrA, ply, ileS, rpsJ, eno, spxB) generated by QPCR and normalized to 16S rRNA levels were higher than the corresponding microarray M values for the D39 ΔrelSpn strain treated with mupirocin (Fig. 6B, dotted line; r2=0.98). We observed a similar trend in QPCR-generated expression ratios normalized to 16S rRNA for 25 other genes from the ΔrelSpn dataset (Fig. S2B; r2=0.97), indicating that this discrepancy was not limited to a small number of genes. This shift suggested a decrease in the size or amplification efficiency of the stable RNA pool in the D39 ΔrelSpn strain treated with mupirocin. Indeed, QPCR analysis detected a modest, but significant, decrease in the relative amounts of 16S and 23S rRNA recovered from ΔrelSpn cultures treated with mupirocin compared to those of untreated ΔrelSpn and rel+Spn cultures or rel+Spn cultures treated with mupirocin (Experimental procedures). Therefore, 16S rRNA was not an appropriate QPCR normalization control for the ΔrelSpn + mupirocin data set. Consistent with this interpretation, expression ratio values generated by QPCR and normalized to either gyrA or rpoB mRNA levels, which do not change significantly in microarray experiments, agreed well with the microarray results for both the D39 and D39 ΔrelSpn datasets (Fig. 6A and B, solid lines).
Fig. 7.
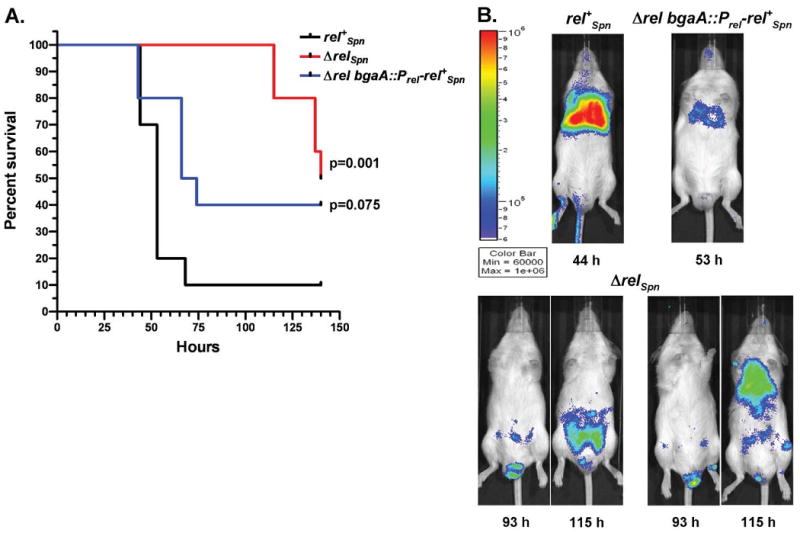
Disease progression and survival of mice infected with D39 luxABCDE rel+Spn (IU1912), D39 luxABCDE ΔrelSpn (IU1943), and complemented D39 luxABCDE ΔrelSpn bgaA∷Prel-rel+Spn (IU2025) strains. Mice were inoculated intranasally with 6 × 106 CFU and disease progression was followed in real time by survival curve analysis (A) and biophotonic imaging (B) as described in Experimental procedures. Ten animals were infected with each strain in two independent experiments, and a representative survival curve and biophotonic images are shown. Survival curves were analyzed by Kaplan-Meier statistics and log-rank tests, and P values relative to the D39 rel+Spn lux strain are given. In (B), the time at which each image was captured is indicated along with the relative color bar scale extending from blue (fewer bacteria) to red (more bacteria). All pictured mice are from the same experiment. Luminescence visible on the tail and lower foot of the rel+Spn-infected mouse (Fig. 8B, top) is light reflected from an adjacent animal photographed at the same time (not shown). Two representative ΔrelSpn-infected mice are shown at 93 h and 115 h after inoculation to illustrate altered disease progression (Fig. 8B, bottom). Additional details are in the text.
Fig. 6.
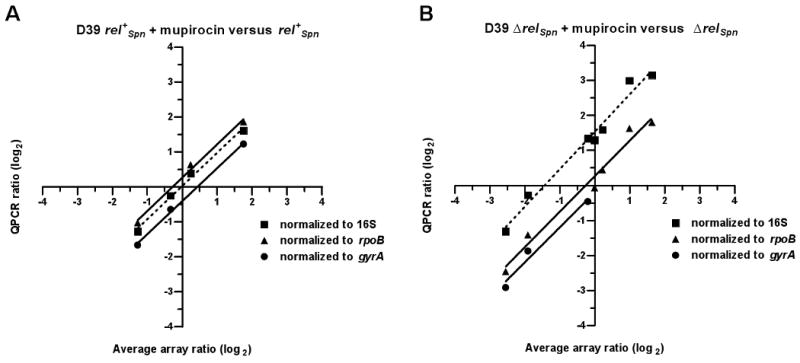
Correspondence of relative expression ratios obtained by QPCR and microarray analyses. Relative transcript amounts were determined by QPCR and normalized to 16S rRNA, rpoB or gyrA as described (Experimental procedures). Expression ratios obtained by QPCR and by microarray analyses were log2-transformed and plotted against each other. (A) Comparison of transformed expression ratios obtained for the D39 rel+Spn strain treated with mupirocin. Relative transcript levels were determined for rpoB (RNA polymerase subunit β), ply (pneumolysin), gyrA (DNA gyrase subunit), and eno (enolase). Correlation values for QPCR data normalized to the different RNAs: 16S rRNA (r2=0.99); rpoB mRNA (r2=0.99); gyrA mRNA (r2=0.99). (B) Comparison of transformed expression ratios obtained for the D39 ΔrelSpn strain treated with mupirocin. Relative transcript levels were determined for rpoB, ply, rpsJ (ribosomal protein S10), gyrA, spxB (pyruvate oxidase), eno and ileS (isoleucyl-tRNA synthetase). Correlation values for QPCR data normalized to the different RNAs: 16S rRNA (r2=0.98); rpoB mRNA (r2=0.98); gyrA mRNA (r2=0.97). Normalization to 16S rRNA resulted in a line that did not pass through the plot origin and was not appropriate for validation of the D39 ΔrelSpn data (see text).
Role of relSpn in virulence
To examine the role of RelSpn in vivo, D39 rel+Spn, D39 ΔrelSpn, and D39 ΔrelSpn bgaA∷Prel-rel+Spn complemented strains expressing the luxABCDE bacterial luminescence operon (IU1912, IU1943 and IU2025; Fig. 2; Table S1) were tested in a murine pneumonia model. ICR male mice were inoculated intranasally and disease progression was monitored by biophotonic imaging (Experimental procedures). Mice infected with the D39 rel+Spn (IU1912) or the ΔrelSpn bgaA∷Prel-relSpn (IU2025) complemented strain displayed median survival times that were not statistically different (53 and 70 h, respectively, p =0.075; Fig. 7A). In contrast, the D39 ΔrelSpn mutant was significantly attenuated in this infection model (median survival time ≈ 140 h, p =0.001; Fig. 7A). Unexpectedly, the D39 ΔrelSpn mutant did not show the same disease progression as its D39 rel+Spn parent (Fig. 7B). The majority of mice infected with D39 rel+Spn displayed lung luminescence characteristic of pneumonia as the first sign of disease (Fig. 7B, top), which progressed to bacteremia within 44-70 h after inoculation (not shown). In contrast, in two independent experiments, 71-80% of the animals inoculated with the D39 ΔrelSpn mutant that developed disease first displayed foci of bacterial luminescence near the groin or in the abdomen suggestive of lymph node involvement (Fig. 7B, bottom). There was no correlation between the presence of these foci, which were highly localized to the lower body, and bacterial luminescence in mouse nasopharynges or mouths. We also did not observe any obvious wounds on the feet or lower bodies of ΔrelSpn-infected mice. Therefore, there was no indication that ΔrelSpn bacteria were introduced into the lower body as a result of external licking or grooming. Bacterial luminescence spread from these foci in the lower body through the abdomen and in many cases upward to the lung area, although we did not confirm bacterial loads in the lungs in these experiments. Finally, introduction of an ectopic copy of the rel+Spn gene (D39 ΔrelSpn bgaA∷Prel-rel+Spn, IU2025) complemented the ΔrelSpn phenotype by restoring disease progression back to normal (Fig. 7B, top and not shown).
Discussion
Like other species of Streptococcus (see (Samen et al., 2004; Steiner and Malke, 2000)), S. pneumoniae is auxotrophic for certain amino acids. Because S. pneumoniae occupies many niches in the human body during colonization and invasive disease (Introduction), responding to changes in the supplies of required amino acids and other nutrients is likely important. In other bacterial species, RSH proteins controlling the synthesis and hydrolysis of the alarmones (p)ppGpp play major roles in responding to limitations of amino acid, carbon source, and fatty acid supplies and have pronounced effects on virulence (Introduction). We report here the first characterization of relSpn mutants and the stringent response of S. pneumoniae. As part of this study, we re-evaluated the growth requirements of serotype 2 pneumococcal strain D39 and found requirements for histidine, arginine, leucine, cysteine (cystine), isoleucine, valine, and glycine (Results). The requirements for histidine, arginine, and leucine were expected, because S. pneumoniae lacks complete pathways for these amino acids (Fig. 5). The requirements for isoleucine and valine are not understood, because these pathways are present (Fig. 5), but were not functional in the CDM used in this study. The pneumococcal genes encoding the acetolactate synthase large subunit (ilvB, spd0404) and threonine dehydratase (ilvA, spd0409) are preceded by potential anti-antiterminator/ antiterminator/ terminator structures (http://cmgm.stanford.edu/ ∼merino/) and are strongly repressed by CodY (Hendriksen et al., 2008a). A potential cre binding site for the CcpA catabolite repressor is located upstream of ilvB, suggesting complex regulation in response to multiple signals, as is observed in B. subtilis (see (Tojo et al., 2008)). In addition, we found that a mutation in a conserved hypothetical gene (spd0221; DUF711; PDB 2HA9) caused the glycine requirement of strain D39 (Results). spd0221 is adjacent to an ACT-domain protein (spd0220, Table S5) that may bind an amino acid and play a role in regulating amino acid metabolism (Grant, 2006).
We found that relSpn was not essential in S. pneumoniae grown in rich BHI broth or complete CDM supplemented with 0.24% BHI (Fig. 3A and 4B). S. pneumoniae encodes a SAS (spd0982/ spr1004), whose structure (PDB 2BE3) closely resembles the synthetase domain of RelSeq (Hogg et al., 2004) and that can synthesize (p)ppGpp in a heterologous E. coli system (Battesti and Bouveret, 2009). In B. subtilis, the SAS proteins encoded by ywaC and yjbM synthesize enough (p)ppGpp to inhibit growth of a ΔrelBsu mutant, and suppressors of the ΔrelBsu mutation inactivate ywaC and yjbM, presumably to reduce their unopposed (p)ppGpp synthesis (Srivatsan et al., 2008). In contrast to B. subtilis, we did not observe a significant difference in growth of rel+Spn, ΔrelSpn, Δspr1004, and ΔrelSpn Δspr1004 strains in rich media (Fig. 3A and data not shown). The spd0982 sequence was not mutated in ΔrelSpn mutants reported here (IU1921, IU2024), suggesting that spd0982 may not play a significant role in (p)ppGpp synthesis under normal growth conditions. Consistent with this interpretation, relSpn was the primary, and possibly only, source of (p)ppGpp synthesis under the non-uniform labeling conditions that could be used for S. pneumoniae (Results). The ΔrelSpn mutant did not synthesize (p)ppGpp in labeling medium lacking or containing mupirocin to induce stringent response (Fig. 2), nor was (p)ppGpp amount affected in a R6 rel+Spn Δspr1004 mutant (Results). Similarly, a Streptococcus mutans RSH deletion strain (ΔrelASmu) was not severely impaired for growth in complete media, despite encoding two SAS proteins, RelP and RelQ (Lemos et al., 2007). RelASmu is the major source of (p)ppGpp synthesized during stringent response, and RelQSmu, the paralog of pneumococcal Spr1004/ Spd0982, efficiently synthesized (p)ppGpp when expressed in E. coli, but displayed very low activity in S. mutans under the conditions tested (Lemos et al., 2007). Together, it seems likely that Spr1004/ Spd0982 may function under stress conditions other than stringent response, by analogy to the cases for B. subtilis and S. mutans (Introduction), or its activity may be regulated by a novel mechanism as proposed for RelQSmu (Lemos et al., 2007). In this regard, it is noteworthy that spr1004 was identified as a late competence gene in pneumococcus, whose induction corresponded with reduced transcription of many ribosomal protein genes (Peterson et al., 2004).
Surprisingly, the D39 ΔrelSpn mutant displayed an apparent defect in copper uptake or utilization during growth in CDM, instead of an amino acid auxotrophy found for many other bacteria (Results). S. pneumoniae encodes homologues of the E. coli CutC (spd1118) putative copper binding protein (Gupta et al., 1995), enterococcal CopY transcriptional repressor (spd0633) and CopA transporter implicated in copper uptake (spd0635) (Solioz and Stoyanov, 2003). It lacks recognizable homologues of the CopB copper efflux protein and the CopZ copper chaperone that are co-transcribed with CopY and CopA in many enterococcal species (Solioz and Stoyanov, 2003). The relative transcript amounts of these copper homeostasis genes did not change in the D39 ΔrelSpn mutant or its rel+Spn parent grown in BHI in the absence or presence of mupirocin (Results), nor in the R6 ΔrelSpn mutant grown in CDM supplemented with 2% BHI (data not shown). An obvious relationship between the transcription of genes that encode proteins that likely bind copper, such as oxygenases and nitrite and nitrous oxide reductases (Solioz and Stoyanov, 2003), and (p)ppGpp amount also did not emerge from microarray analyses (Results). However, relative amounts of transcripts encoding subunits of the PsaBCA Mn2+-permease increased 3-fold in the R6 ΔrelSpn mutant grown in CDM supplemented with 2% BHI (psaBC, spr1492-1493; p < 0.001) (data not shown). PsaBCA is required for resistance to hydrogen peroxide- and superoxide-mediated killing (McAllister et al., 2004). Further studies will be needed to determine why ΔrelSpn mutants require copper and whether RelSpn activity is regulated directly or indirectly in response to metal ion limitation, as was observed for iron in other bacteria (Miethke et al., 2006; Vinella et al., 2005). Likewise, further experiments are needed to determine whether the growth enhancement caused by relatively higher concentrations of manganese in the presence of copper is due to a transcriptional response or an antioxidant effect that counteracts the high concentrations of hydrogen peroxide produced by S. pneumoniae in culture (Results; see (Kloosterman et al., 2008; Ramos-Montanez et al., 2008; Tseng et al., 2001)).
Growth, microarray, and virulence studies showed that relSpn plays crucial roles in pneumococcal adaptation to conditions that are expected to limit translation, such as mupirocin addition. ΔrelSpn mutants were more sensitive to and recovered less well from sublethal concentrations of mupirocin than rel+Spn parent or complemented strains (Figs. 3A and 6; data not shown). Comparisons of global transcription responses of the ΔrelSpn mutant with that of the rel+Spn parent revealed three complex responses that were relSpn-independent, relSpn-dependent, and ΔrelSpn-dependent (Results; Tables 1, S4-S6). These responses did not reveal significant involvement of CodY, a global regulator that responds to branched-chain amino acids but not GTP levels in S. pneumoniae (see (Hendriksen et al., 2008a; Sonenshein, 2007)). The CodY regulon has been determined in pneumococcus (Hendriksen et al., 2008a), and expression of this regulon did not change within the cutoff limits used in the microarrays reported here (Tables S4-S6). Thus, unlike some other gram-positive species (see (Bennett et al., 2007)), the RelSpn and CodY regulatory responses were not coordinated or overlapping in S. pneumoniae under the conditions tested so far.
rel-independent responses have been observed previously in numerous bacteria, including B. subtilis, S. pyogenes, and S. mutans (see (Durfee et al., 2008; Eymann et al., 2002; Nascimento et al., 2008; Steiner and Malke, 2001)). There are several mechanisms that underlie aspects of this general response to translation inhibition. T-box regulation (Grundy and Henkin, 2003) is implicated in a number of changes (Tables 1, S4-S6). The relative transcript amount of ileS (spd1472) increased (Table 1, line 1), because mupirocin specifically inhibits charging of tRNAIle. In contrast, the relative transcript amounts of other genes that are likely negatively regulated by the ratio of charged-to-uncharged tRNA species (tRNAVal, tRNAPhe, tRNAAla, tRNAGly, tRNAThr) (Table 1, line 8) decreased upon translation inhibition, while relative transcript amounts of genes likely regulated by tRNATrp were unexpectedly differentially expressed (Results; Table 1, lines 1, 2, 9). Reduced translation also can account for the decreased relative transcript amounts of genes encoding protein folding chaperones (GrpE, DnaK, GroES) and their transcriptional regulator, HrcA (Table 1, line 13). Based on precedents from B. subtilis, mupirocin addition may have left sufficient GroEL available to refold HrcA, which acts as a repressor of the chaperone operons (see (Narberhaus, 1999)).
The mechanisms underlying changes in expression of other relSpn-independent genes are largely unknown. Some genes are preceded by predicted antiterminator/ terminator structures (Table S4) (see (Merino and Yanofsky, 2005)). Other genes encode glycolytic enzymes, putative amino acid transport and biosynthetic enzymes, sugar transporters, and the GlnR metabolic regulator (Table 1, lines 3, 4, 10-12). Together these expression changes suggest alterations in metabolite levels, carbon catabolite repression (see (Jahreis et al., 2008)) and elevated glutamine levels (Kloosterman et al., 2006). Other noteworthy changes in the general relSpn-independent response were increased relative transcript amounts of genes that encode the tmRNA ribosome recycling machinery (reviewed in (Keiler, 2008)) and a drug efflux pump (spd1900) known to be induced by translation inhibitors (Marrer et al., 2006) (Table 1, lines 5, 7).
The rel+Spn-dependent response resembled a classic stringent response (reviewed in (Cashel et al., 1996; Srivatsan and Wang, 2008)) with decreased relative transcription of many genes involved in translation, DNA replication, recombination and double-strand break repair, and macromolecular synthesis (Tables 1, S5). In B. subtilis and T. thermophilus, a drop in intracellular GTP amount upon (p)ppGpp synthesis likely mediates the decreasing transcription of rRNA operons (Kasai et al., 2006; Krasny and Gourse, 2004), and other promoters initiating transcription with +1G are also down-regulated during stringent response (Krasny et al., 2008). We observed a concomitant drop in labeled GTP amount during pneumococcal stringent response (Fig. 2A and 2B, lanes 2-4). In addition, the relative transcript amounts of several pyrimidine biosynthesis genes (carBA-pyrR, pyrFE) decreased in the D39 rel+Spn strain treated with mupirocin (Table 1, line 13; Table S5). S. pneumoniae PyrR is 75% similar to B. subtilis PyrR, and potential antiterminator/ terminator structures precede pyrR and pyrF (http://cmgm.stanford.edu/∼merino/), suggesting that transcription of the pyr genes may be controlled by attenuation in response to the intracellular ratio of uridine (UMP, UTP) to guanosine nucleotides (GMP, GDP, GTP) as in Bacillus (Turnbough and Switzer, 2008). According to this model, decreased pyr gene transcript levels correspond to decreased GTP levels in the D39 rel+Spn strain in response to mupirocin treatment. At the same time, the relative transcript amount of guaB (spd2055), which encodes an IMP dehydrogenase that is likely inhibited by (p)ppGpp (see (Kasai et al., 2006; Krasny and Gourse, 2004)), increased during stringent response (Table 1, line 3), possibly reflecting a mechanism to restore guanylate nucleotide levels.
S. pneumoniae encodes a vegetative sigma factor (RpoD) and only one alternative sigma factor (ComX) that is induced during competence development (see (Claverys et al., 2006)). Pneumococcus lacks a recognizable DksA homologue (Introduction). Consequently, it is not clear how S. pneumoniae re-programs transcription in response to (p)ppGpp accumulation. In B. subtilis, incorporation of an initiating ATP nucleotide seems to play a role in positive regulation of some stringently controlled genes, although the mechanism of this response is not well understood (Krasny et al., 2008; Tojo et al., 2008). The ply operon that synthesizes pneumolysin was significantly induced during stringent response (Table 1, line 5 and Table S5). Pneumolysin is a major pneumococcal toxin that lyses cholesterol-containing cell membranes at high concentrations, poisons cilia, stimulates the immune response, and plays roles in respiratory tract survival and the development of sepsis (see (Kadioglu et al., 2008)). Little is known about the regulation of the ply operon or other pneumococcal genes that are positively regulated during stringent response, including their sites of transcription initiation. Similar to the case in B. subtilis (Krasny et al., 2008; Tojo et al., 2008), many of the pneumococcal genes that are positively regulated during stringent response may be subject to multiple regulation. For example, ply operon transcription was elevated under some growth conditions in D39 mutants deleted for the rr09 (spd0574) gene encoding a TCS response regulator (Hendriksen et al., 2007). Finally, the relative transcript levels of competence genes comC (competence stimulating peptide CSP), comA (encoding a subunit of the CSP transporter), and comD (encoding a CSP-sensing histidine kinase) increased significantly during stringent response (Table 1, line 6). (p)ppGpp and guanine nucleotide pools were proposed as a stress signal that may feed into competence induction in S. pneumoniae (Claverys et al., 2006), and the induction of comC fits this model. However, we did not observe a general induction of the competence regulon, nor do we know whether DNA uptake or other functions of competence were affected under the conditions tested. Further studies of this phenomenon and the general mechanism of positive and negative stringent control in S. pneumoniae are needed.
As noted previously in S. mutans and E. coli (see (Nascimento et al., 2008)), the pneumococcal ΔrelSpn-dependent response included an aberrant increase in relative transcript levels of some ribosomal protein genes (Table 1, line 1 and Table S6). In pneumococcus, many of these aberrantly transcribed genes correspond to orthologs in the E. coli S10 operon, which is regulated through transcription attenuation mediated by NusA and ribosomal protein L4 (Keener and Nomura, 1996). In addition, the pneumococcal ΔrelSpn-dependent response included changes in relative transcript amounts of numerous genes involved in central metabolism and oxidative stress. Therefore, the synthesis of (p)ppGpp directly or indirectly plays a large homeostatic role in S. pneumoniae in maintaining metabolic balance. For example, relative transcript amounts of genes playing direct (pgk, pyk) and indirect (spxB) roles in substrate level phosphorylation reactions decreased only or decreased more strongly in the ΔrelSpn mutant upon mupirocin treatment compared to the rel+Spn parent (Table 1, lines 10, 16). As another example, the ΔrelSpn-dependent response included decreased or increased relative transcript amounts of genes encoding enzymes which utilize NADH (e.g., lactate (ldh) and alcohol (adhE) dehydrogenase) or NAD+ (lpdA-acoLCBA, spd1024-1028; pyrKDb, spd0851-0852), respectively, suggesting a re-balancing of reducing equivalents (Table 1, lines 4, 10). Consistent with this interpretation, there was a decrease in the relative transcript amount of a putative Rex homologue (spd0976), which is a redox sensor that responds to NADH: NAD+ ratio (Table 1, line 17; see (Sickmier et al., 2005)). A third example of metabolic change in the ΔrelSpn-dependent response was the increase in relative transcript levels of the adk gene (spd0214) encoding adenylate kinase. adk appeared to be co-regulated with an adjacent ribosomal protein operon and its relative transcription changed in opposite direction in the rel+Spn and ΔrelSpn strains treated with mupirocin (Table 1, lines 3, 13). Adenylate kinase phosphorylates AMP to ADP using ATP as a phosphodonor, and these divergent transcriptional responses could influence overall energy charge.
The ΔrelSpn-dependent response also contained likely changes in cell wall homeostasis that did not occur in the rel+Spn parent strain during stringent response. Relative transcription of the VicRK (WalRK) TCS regulon, which is thought to respond to changes in cell wall status (Dubrac et al., 2008; Ng et al., 2005; Winkler and Hoch, 2008), increased considerably in the ΔrelSpn, but not in the rel+Spn strain, upon mupirocin addition (Table 1, line 6). In contrast, relative transcription of the CiaRH TCS and CiaRH-regulated Dlt system decreased in a ΔrelSpn-dependent manner (Table 1, lines 14 and 15; Table S6). CiaRH is part of the gram-positive cell wall stress response that confers resistance to lysis-inducing cell wall antibiotics (see (Jordan et al., 2008)). The signals sensed by the VicRK and CiaRH TCSs are unknown. We did not observe overt morphological changes by phase contrast microscopy in rel+Spn or ΔrelSpn strains after brief mupirocin treatment, but enlarged cells began appearing after prolonged (3 h) treatment (data not shown). Lastly, the ΔrelSpn-dependent response included decreased expression of the SpxR regulon, which includes the SpxB pyruvate oxidase and StrH exoglycosidase (Ramos-Montanez et al., 2008). SpxR is thought to respond to adenylate and CoA-containing compounds, which may be reduced in amount in the mupirocin-treated ΔrelSpn mutant. SpxB is the main source of hydrogen peroxide produced by S. pneumoniae (see (Ramos-Montanez et al., 2008)), and the decreased relative transcription of genes that protect against reactive oxygen species (Table 1, line 16), may reflect decreased spxB transcription. Although the ΔrelSpn-dependent response is large and seems to reflect a disruption in metabolic homeostasis, the mechanisms underlying these gene changes and their inter-relationships are largely unknown.
Finally, we demonstrated that the S. pneumoniae relSpn gene is a major virulence factor in a murine pneumonia/ bacteremia model of infection and showed that the attenuation of infection in the ΔrelSpn mutant was complemented by an ectopically expressed rel+Spn gene (Results; Fig. 7). This finding confirmed results from a previous lung-based STM screen in which relSpn-disrupted bacteria were not recovered from mouse lungs after 48 h of infection (Hava and Camilli, 2002). Biophotonic imaging revealed that the D39 ΔrelSpn strain was initially unable to establish an infection in the mouse lungs. Bacterial luminescence of the ΔrelSpn mutant appeared first in the lower abdomen or in lower body foci that may be inguinal or lumbar lymph nodes, instead of in the lungs as observed for the rel+Spn or complemented ΔrelSpn bgaA∷Prel-rel+Spn strains (Fig. 7B). Infections of the ΔrelSpn strain only later spread upward into the thorax and lung region in some of the infected mice (Fig. 7B). Thus, the ΔrelSpn mutant was not only attenuated, but progression of the infection was dramatically altered. Several mechanisms could cause this change in infection progression, including a requirement for normal stringent response, metabolic consequences of the aberrant ΔrelSpn-dependent response described above, and the in vivo availability of metal ions, such as copper. Further physiological and genetic experiments are in progress to understand this unusual virulence response.
Experimental procedures
Chemicals and Media
Bacteria were grown in Brain Heart Infusion broth (Bacto BHI, Becton Dickinson) or chemically defined medium (CDM) prepared from individual components. CDM was formulated as described (van de Rijn and Kessler, 1980) with the following changes: L-cystine and hydroxy-L-proline were omitted from the media; 0.001 g pyridoxal hydrochloride, 0.001 g pyridoxine and 1.0 g choline chloride were added per L of media; and 0.75 g L-cysteine hydrochloride, 0.1 g L-methionine, and 0.0025 g β-NAD were added per L immediately before use. MOPS-CDM used in non-uniform (p)ppGpp labeling assays was modified to resemble the media of Neidhardt et al. (Neidhardt et al., 1974) by replacing potassium phosphate and sodium phosphate with 40 mM 3-[N-morpholino] propanesulfonic acid (MOPS) and adding 4 mM tricine, 0.28 mM potassium sulfate and 50 mM sodium chloride. Asparagine was not included in the published formulation (van de Rijn and Kessler, 1980) and was routinely omitted from all CDM media. Isoleucine was omitted from MOPS-CDM. CDM used in amino acid drop-out and metal supplementation studies was prepared without amino acids or metals, which were added to the media in the desired combinations before use. Media components were purchased from Mallinckrodt Baker, Inc. or Sigma-Aldrich (U.S.A.). Lithium mupirocin was a generous gift of Dr. Charles Jakielaszek (GlaxoSmithKline).
Strain construction
Strains and primers used in this study are listed in Supplemental Tables S1 and S2. Mutant strains were constructed by transformation with amplicon DNA generated by overlap fusion PCR using PfuTurbo (Stratagene) or rTth (Applied Biosystems) proofreading DNA polymerases and screened as described previously (see (Ng et al., 2003)). Transformants were single-colony isolated on TSAII-BA plates containing Trypticase Soy Agar II, Modified (BBL, Becton Dickinson), 5% (vol/vol) defibrinated sheep blood (Remel) and appropriate antibiotics at the following concentrations: 200 μg kanamycin (Kan) per mL, 150 μg streptomycin (Str) per mL, or 0.3 μg erythromycin (Erm) per mL. Plates were incubated at 37°C in an atmosphere of 5% CO2. Target loci and flanking regions in all constructs were sequenced using BigDye Terminator mix (Applied Biosystems) as described (Ramos-Montanez et al., 2008). Markerless deletions or replacements of target genes were generated using the kanR-rpsL+ (“Janus” cassette) allele replacement method described in (Sung et al., 2001). Target genes were disrupted by insertion of the Janus cassette in an rpsL1 (StrR) strain background, yielding KanR StrS transformants. The Janus cassette was subsequently replaced by transformation with an allele lacking antibiotic markers and selection for StrR colonies. IU1908 (D39 rpsL1 ΔrelSpn) was transformed with ∼1.2 μg of ‘pulA-rpsL+-rpsG-fusA’ amplicon and a StrS colony (IU1921, D39 ΔrelSpn) was isolated after patching ∼160 colonies. The rpsL1 allele contains one mutation conferring streptomycin resistance (Lys56Thr) and three flanking silent mutations (Table S1 and (Sung et al., 2001)). IU1921 contained the wild-type sequence at position 167 (Lys56), but retained the three silent mutations, suggesting a gene conversion event.
Determining nutritional requirements
S. pneumoniae strains were inoculated into BHI broth from frozen glycerol stocks as described (Ramos-Montanez et al., 2008) and grown statically at 37°C in an atmosphere of 5% CO2 to an optical density of OD620 ≈0.1-0.3 (16-mm tubes). Cells were collected by centrifugation at ≈3000 × g for 8 min at room temperature. Pellets were washed with 3 mL of CDM lacking nutrients being tested, resuspended in the same medium to a density of OD620 ≈0.1, and further diluted 100-fold into CDM containing the desired combinations of amino acids and metals to start final cultures. Amino acids were omitted from CDM singly (CDM drop-out media) unless otherwise indicated in the text. Amino acid requirements of the ΔrelSpn strain were determined in CDM drop-out media supplemented with 0.24% (vol/vol) BHI. Cystine, glutathione, taurine, or sodium thiosulfate were added to ≈500 μM where indicated. Growth was monitored by optical density (OD620) for ≈40 h using a Spectronic 20 spectrophotometer (Ramos-Montanez et al., 2008).
Growth in the presence of inhibitors
S. pneumoniae strains were inoculated into BHI from frozen glycerol stocks as described (Ramos-Montanez et al., 2008). Overnight cultures in exponential phase (OD620 ≈0.1-0.3) were diluted to a density of OD620 ≈0.1 with BHI and further diluted 100-fold into BHI to start final cultures. At densities of OD620 ≈0.1, cultures were divided in two and D-tyrosine (2.4 mM final concentration) or lithium mupirocin (100 ng per mL) were added to one culture. Growth was monitored by spectrometry. For outgrowth experiments, 400 μL of mupirocin-treated culture were removed after 0, 1, 2 and 3 h of drug treatment and adjusted to a density of OD620 ≈0.1 with BHI. 40 μL were diluted 100-fold into BHI containing no drug and growth was monitored by OD620. 50 μL of inocula were serially diluted in phosphate-buffered saline (PBS) and plated on TSAII-BA to determine CFUs. Pneumococcal chain length and cell morphology after mupirocin treatment were examined using a Nikon E-400 epifluorescent phase-contrast microscope equipped with a 100X oil-immersion objective and SPOT RT Monochrome camera.
Non-uniform labeling of (p)ppGpp and TLC
(p)ppGpp synthesis was monitored using a modified rapid colony screening protocol (Cashel, 1994). S. pneumoniae strains were grown in BHI broth as described above. At culture densities of OD620 ≈0.2, cells were collected by microcentrifugation at 3,300 × g for 4 min at room temperature. Pellets were washed and resuspended in MOPS-CDM (-Ile) to the same density. 200 μL of cell suspension were added to the wells of a 96-well microtiter plate and pre-warmed at 37°C for 10 min in a bench-top heating block (VWR). 25 μL of each cell suspension were mixed with 150 μL of MOPS-CDM (-Ile) containing 150 μCi per mL of H3[32P]O4 (500 μCi per mL in 0.02 N HCl; MP Biomedicals, Inc., Irvine, CA) and lithium mupirocin (100 ng per mL) where indicated. 25 μL samples were removed after 0, 5, 10, 20 and 30 min, added to an equal volume of frozen 13 M formic acid, and thawed and refrozen twice. Microtiter plates were thawed on an angle on a bed of ice and 5 μL of each sample were removed for TLC analysis without resuspending cellular debris. Samples were spotted on polyethyleneimine-cellulose (PEI) plastic-backed TLC plates (J.T. Baker) and chromatographed in 1.5 M KH2PO4 (pH 3.4). IU2024 (D39 ΔrelSpn bgaA∷PfcsK-rel+Spn) and IU2028 (R6 rspL1 ΔrelSpn bgaA∷PfcsK-rel+Spn) were grown and labeled in media containing 0.5% (wt/vol) fucose (final concentration). (p)ppGpp- producing E. coli strain TX2737 (Table S1) was streaked for single colonies on LB agar containing 100 μg per mL ampicillin. Cells from 5-6 isolated colonies were resuspended in CDM-MOPS (-Ile) containing 100 mM IPTG, labeled with H3[32P]O4 for 30 min and acid extracted as above. The E. coli sample was diluted 1:1 in CDM-MOPS: formic acid (vol/ vol) before spotting on plates. After drying, TLC plates were exposed to a phosphor screen (GE Healthcare) and radioactivity was quantitated using ImageQuant 5.2 software. Two-dimensional TLC of labeled samples was performed using solvents Fa (4 M formic acid, pH 3.5) and Pb (0.4 M K2HPO4, 0.7 M boric acid) as described (Bochner and Ames, 1982). Acid-lability of 32P-labeled material was tested by extracting PEI-cellulose scraped from the origins of TLC plates in 300 μL of 0.25 M ammonium carbonate for 1 h at room temperature (Kuroda and Kornberg, 1997). After brief centrifugation, 50 μL of supernate was acidified with 15 μL of 1 N HCl and incubated at 95°C. 10 μL samples were removed at 5 min intervals and spotted onto PEI-cellulose TLC plates, which were chromatographed and quantitated as above.
Microarray Analysis
Bacteria were grown in BHI broth as described above but diluted 50-fold into BHI to start final cultures (Lanie et al., 2007). Cultures were divided in two upon reaching a density of OD620 ≈0.1, and lithium mupirocin was added to one culture (t=0) to a final concentration of 100 ng per mL. Treated and untreated cultures were harvested after 20 min further incubation. RNA extraction and purification, cDNA synthesis, labeling and hybridization to S. pneumoniae R6 microarrays (MWG Biotech, Inc.), array washing, and data collection were performed as in (Lanie et al., 2007). Data were collected from three independent biological replicates, including one dye swap. Data analysis was performed using R packages from the Bioconductor project (Gentleman et al., 2004). Spots with background signals higher than foreground signals and spots that were flagged as bad by the investigator or GenePix Pro 5.0 software in more than one replicate were removed from further analysis. The Bioconductor marray package was used to perform loess normalization on background-subtracted data (Dudoit and Yang, 2002; Quackenbush, 2002). Ribosomal proteins were excluded from the normalization procedure, because most were on average highly expressed and were strongly differentially expressed. In contrast, few non-differentially expressed genes had similar high average expression levels so that including ribosomal proteins in the normalization procedure would have biased the resulting fold changes of all genes with high average expression levels. Statistical significance of observed expression changes was calculated using an empirical Bayes moderated t-test (limma package, (Smyth, 2004)). Multiple hypothesis correction was performed to determine statistically significant differential expression by calculating a false discovery rate (FDR; (Benjamini and Hochberg, 1995)). The cutoff for statistical significance of differential expression was set at p = 0.003, which corresponds to a Benjamini Hochberg FDR of 1.9%. In addition, we required genes to have an up or down fold change of at least 2 fold, with the following exceptions. For rel+Spn-dependent and ΔrelSpn-dependent changes, genes whose relative transcript amounts changed at least 1.8 fold (p ≤ 0.003) were included when potentially co-transcribed with genes that change at least 2 fold, as judged by transcription polarity, average microarray spot signal intensity (A value), and Operon Db prediction (http://operondb.cbcb.umd.edu/cgi-bin/operondb/operons.cgi). For relSpn-independent changes, all genes whose relative transcript levels changed at least 2 fold in one or both strains treated with mupirocin were included. Intensity and expression ratio data for ΔrelSpn versus rel+Spn, rel+Spn + mupirocin versus rel+Spn, ΔrelSpn + mupirocin versus ΔrelSpn, and ΔrelSpn + mupirocin versus rel+Spn + mupirocin comparisons have been deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE14750).
Quantitative RT-PCR (QPCR)
RNA used for microarray analyses was assayed by QPCR to confirm differential gene expression as described (Ramos-Montanez et al., 2008) with the following changes. cDNA was synthesized at 42°C using 100 ng of total RNA and random primers according to the manufacturer's recommendations (AffinityScript Multiple Temperature cDNA synthesis kit, Stratagene). RT-PCR was performed using the Brilliant SYBR Green QPCR Master Mix (Stratagene) and appropriate primers (Table S2) as described (Ramos-Montanez et al., 2008). Reactions were performed in duplicate using cDNA from at least two independent biological replicates (Fig. 6 and S2), and primers KK387 and KK388 (Table S2) were used for normalization to 16S rRNA amounts. Reactions were run in a Stratagene Mx3000P instrument. Data was collected and analyzed using the Comparative Quantitation program with calibrator and added dissociation curve (Fig. 6) or SYBR Green program with dissociation curve (Fig. S2) included in MxPro v. 3.0 software. Relative 16S and 23S rRNA amounts in all RNA preparations used for microarray and QPCR analyses were compared using primer pairs KK355 and KK356, KK387 and KK388, and KK391 and KK392 (Table S2) using the Comparative Quantitation program with calibrator and added dissociation curve program.
Mouse Models of Infection
All procedures were approved in advance by the Bloomington Institutional Animal Care and Use Committee and were performed according to recommendations of the National Research Council. Experiments were performed as described (Ramos-Montanez et al., 2008) with the following changes. Male ICR mice (21-24 grams; Harlan) were anesthetized by inhaling 4% isoflurane (Butler Animal Health Supply) for 8 min. In two independent experiments, ten mice were inoculated intranasally (IN) with each bacterial strain to be tested. To prepare inocula, bacteria were grown in BHI to mid-exponential phase (OD620 ≈0.17-0.2). 1 mL of culture was microcentrifuged for 2 min at 13,500 × g, resuspended in 1 mL of PBS, and adjusted with PBS to a bioluminescence total flux reading of ≈4.0 ×108 photons per sec, which corresponded to ≈1.3×108 CFU per mL. CFUs were confirmed by serial dilution and plating. 50 μl of suspension were administered as described (Ramos-Montanez et al., 2008). Mice were monitored at ≈8 h intervals and moribund mice were euthanized by CO2 asphyxiation, which was used as the ‘time of death’ in statistical analyses. Kaplan-Meir survival curves and log-rank tests were generated using GraphPad Prism 5 software. Images of mice infected with bioluminescent bacteria containing the luxABCDE operon were collected using Living Image v. 2.6 software provided with an IVIS Lumina imaging system (Xenogen; Alameda, CA) equipped with an isoflurane anesthesia manifold and CCD camera.
Supplementary Material
Acknowledgments
We thank Dr. Charles Jakielaszek (GlaxoSmithKline) for the gift of lithium mupirocin, Dr. Donald Morrison (University of Illinois, Chicago) for providing strains and synthetic CSP, Dr. Michael Cashel (the National Institutes of Health) for plasmid pALS13, and Dr. Tiffany Tsui for discussions regarding QPCR and microarray analyses. We thank the Center for Genomics and Bioinformatics (CGB) at Indiana University Bloomington (IUB) for access to microarray analysis equipment and programs, and members of the IUB Microarray Users group for suggestions on data analysis. The project described was supported by Grant Number AI060744 from the National Institute of Allergy and Infectious Diseases (U.S.A.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID. This work was also supported by funds from the Indiana University METACyt Initiative, funded in part through a major grant from the Lilly Endowment, Inc. to M.E.W.
References
- Aberg A, Shingler V, Balsalobre C. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol Microbiol. 2006;60:1520–1533. doi: 10.1111/j.1365-2958.2006.05191.x. [DOI] [PubMed] [Google Scholar]
- Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum PC. Resistance among Streptococcus pneumoniae: Implications for drug selection. Clin Infect Dis. 2002;34:1613–1620. doi: 10.1086/340400. [DOI] [PubMed] [Google Scholar]
- Battesti A, Bouveret E. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J Bacteriol. 2009;191:616–624. doi: 10.1128/JB.01195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B. 1995;57:289–300. [Google Scholar]
- Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63:1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006;14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Brown JS, Gilliland SM, Holden DW. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol Microbiol. 2001;40:572–585. doi: 10.1046/j.1365-2958.2001.02414.x. [DOI] [PubMed] [Google Scholar]
- Cashel M. Detection of (p)ppGpp Acccumulation Patterns in Escherichia coli Mutants. In: Adolph KW, editor. Molecular Microbiology Techniques, Part A. Vol. 3. San Diego, CA: Academic Press, Inc.; 1994. pp. 341–356. [Google Scholar]
- Cashel M, Gentry D, Hernandez VJ, Vinella D. The Stringent Response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 1. Washington, D C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Chan PF, O'Dwyer KM, Palmer LM, Ambrad JD, Ingraham KA, So C, Lonetto MA, Biswas S, Rosenberg M, Holmes DJ, Zalacain M. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J Bacteriol. 2003;185:2051–2058. doi: 10.1128/JB.185.6.2051-2058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy-Regaud S, Ogunniyi AD, Diallo N, Huet Y, Desnottes JF, Paton JC, Escaich S, Trombe MC. RegR, a global LacI/GalR family regulator, modulates virulence and competence in Streptococcus pneumoniae. Infect Immun. 2003;71:2615–2625. doi: 10.1128/IAI.71.5.2615-2625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- Dagan R, Klugman KP. Impact of conjugate pneumococcal vaccines on antibiotic resistance. Lancet Infect Dis. 2008;8:785–795. doi: 10.1016/S1473-3099(08)70281-0. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for Biochemical Research. 2nd. Oxford: Clarendon Press; 1969. 17B. Metal Binding Properties of Compounds Used in Biochemistry. [Google Scholar]
- Dintilhac A, Alloing G, Granadel C, Claverys JP. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- Dozot M, Boigegrain RA, Delrue RM, Hallez R, Ouahrani-Bettache S, Danese I, Letesson JJ, De Bolle X, Kohler S. The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol. 2006;8:1791–1802. doi: 10.1111/j.1462-5822.2006.00749.x. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Yang YH. Bioconductor R packages for exploratory analysis and normalization of cDNA microarray data. In: Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. New York: Springer; 2002. pp. 73–101. [Google Scholar]
- Dufke S, Kunze-Kronawitter H, Schubert S. Pyelonephritis and urosepsis caused by Streptococcus pneumoniae. J Clin Microbiol. 2004;42:4383–4385. doi: 10.1128/JCM.42.9.4383-4385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W, Helmann JD. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymann C, Homuth G, Scharf C, Hecker M. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J Bacteriol. 2002;184:2500–2520. doi: 10.1128/JB.184.9.2500-2520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmingham D, Canton R, Jenkins SG. Regional trends in β-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J Infect. 2007;55:111–118. doi: 10.1016/j.jinf.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Gardete S, Wu SW, Gill S, Tomasz A. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:3424–3434. doi: 10.1128/AAC.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GA. The ACT domain: a small molecule binding domain and its role as a common regulatory element. J Biol Chem. 2006;281:33825–33829. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. The T box and S box transcription termination control systems. Front Biosci. 2003;8:d20–31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- Gupta SD, Lee BT, Camakaris J, Wu HC. Identification of cutC and cutF (nlpE) genes involved in copper tolerance in Escherichia coli. J Bacteriol. 1995;177:4207–4215. doi: 10.1128/jb.177.15.4207-4215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Preciado A, Yanofsky C, Merino E. Comparison of tryptophan biosynthetic operon regulation in different Gram-positive bacterial species. Trends Genet. 2007;23:422–426. doi: 10.1016/j.tig.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Hava DL, Camilli A. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol. 2002;45:1389–1406. [PMC free article] [PubMed] [Google Scholar]
- Hendriksen WT, Silva N, Bootsma HJ, Blue CE, Paterson GK, Kerr AR, de Jong A, Kuipers OP, Hermans PW, Mitchell TJ. Regulation of gene expression in Streptococcus pneumoniae by response regulator 09 is strain dependent. J Bacteriol. 2007;189:1382–1389. doi: 10.1128/JB.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen WT, Bootsma HJ, Estevao S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PW. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J Bacteriol. 2008a;190:590–601. doi: 10.1128/JB.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen WT, Kloosterman TG, Bootsma HJ, Estevao S, de Groot R, Kuipers OP, Hermans PW. Site-specific contributions of glutamine-dependent regulator GlnR and GlnR-regulated genes to virulence of Streptococcus pneumoniae. Infect Immun. 2008b;76:1230–1238. doi: 10.1128/IAI.01004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol. 2007;189:8186–8195. doi: 10.1128/JB.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- Hoskins J, Alborn WE, Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR, Jr, Skatrud PL, Glass JI. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreis K, Pimentel-Schmitt EF, Bruckner R, Titgemeyer F. Ins and outs of glucose transport systems in eubacteria. FEMS Microbiol Rev. 2008;32:891–907. doi: 10.1111/j.1574-6976.2008.00125.x. [DOI] [PubMed] [Google Scholar]
- Janoff EN, Rubins JB. Immunodeficiency and invasive pneumococcal disease. In: Tuomanen EI, Mitchell TJ, Morrison DA, Spratt BG, editors. The Pneumococcus. Washington, D C.: ASM Press; 2004. pp. 252–280. [Google Scholar]
- Johnston JW, Briles DE, Myers LE, Hollingshead SK. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect Immun. 2006;74:1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- Kadioglu A, Weiser JN, Paton JC, Andrew PW. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 2008;6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- Kasai K, Nishizawa T, Takahashi K, Hosaka T, Aoki H, Ochi K. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J Bacteriol. 2006;188:7111–7122. doi: 10.1128/JB.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Nomura M. Regulation of Ribosome Synthesis. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 1. Washington, D C.: ASM Press; 1996. pp. 1417–1431. [Google Scholar]
- Keiler KC. Biology of trans-Translation. Annual Review of Microbiology. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Kimura T, Takagi K, Hirata Y, Hase Y, Muto A, Himeno H. Ribosome-small-subunit-dependent GTPase interacts with tRNA-binding sites on the ribosome. J Mol Biol. 2008;381:467–477. doi: 10.1016/j.jmb.2008.06.023. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, Hendriksen WT, Bijlsma JJ, Bootsma HJ, van Hijum SA, Kok J, Hermans PW, Kuipers OP. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J Biol Chem. 2006;281:25097–25109. doi: 10.1074/jbc.M601661200. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol Microbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- Kloosterman TG, Witwicki RM, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. Opposite effects of Mn2+ and Zn2+ on PsaR-mediated expression of the virulence genes pcpA, prtA, and psaBCA of Streptococcus pneumoniae. J Bacteriol. 2008;190:5382–5393. doi: 10.1128/JB.00307-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen E, Johnsborg O, Quentin Y, Claverys JP, Havarstein LS. BOX elements modulate gene expression in Streptococcus pneumoniae: impact on the fine-tuning of competence development. J Bacteriol. 2006;188:8307–8312. doi: 10.1128/JB.00850-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R. A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol. 2006;188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasny L, Tiserova H, Jonak J, Rejman D, Sanderova H. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol Microbiol. 2008;69:42–54. doi: 10.1111/j.1365-2958.2008.06256.x. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Kornberg A. Polyphosphate kinase as a nucleoside diphosphate kinase in Escherichia coli and Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1997;94:439–442. doi: 10.1073/pnas.94.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol. 2007;65:1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- Mader U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Malke H, Steiner K, McShan WM, Ferretti JJ. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int J Med Microbiol. 2006;296:259–275. doi: 10.1016/j.ijmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Marrakchi H, Choi KH, Rock CO. A new mechanism for anaerobic unsaturated fatty acid formation in Streptococcus pneumoniae. J Biol Chem. 2002;277:44809–44816. doi: 10.1074/jbc.M208920200. [DOI] [PubMed] [Google Scholar]
- Marrer E, Schad K, Satoh AT, Page MG, Johnson MM, Piddock LJ. Involvement of the putative ATP-dependent efflux proteins PatA and PatB in fluoroquinolone resistance of a multidrug-resistant mutant of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2006;50:685–693. doi: 10.1128/AAC.50.2.685-693.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister LJ, Tseng HJ, Ogunniyi AD, Jennings MP, McEwan AG, Paton JC. Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol Microbiol. 2004;53:889–901. doi: 10.1111/j.1365-2958.2004.04164.x. [DOI] [PubMed] [Google Scholar]
- McKessar SJ, Hakenbeck R. The two-component regulatory system TCS08 is involved in cellobiose metabolism of Streptococcus pneumoniae R6. J Bacteriol. 2007;189:1342–1350. doi: 10.1128/JB.01170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E, Yanofsky C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet. 2005;21:260–264. doi: 10.1016/j.tig.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Miethke M, Westers H, Blom EJ, Kuipers OP, Marahiel MA. Iron starvation triggers the stringent response and induces amino acid biosynthesis for bacillibactin production in Bacillus subtilis. J Bacteriol. 2006;188:8655–8657. doi: 10.1128/JB.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Grill S, Grundling A, Blasi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol. 2002;44:1387–1396. doi: 10.1046/j.1365-2958.2002.02971.x. [DOI] [PubMed] [Google Scholar]
- Musher DM. A pathogenetic categorization of clinical syndromes caused by Streptococcus pneumoniae. In: Tuomanen EI, Mitchell TJ, Morrison DA, Spratt BG, editors. The Pneumococcus. Washington, D C.: ASM Press; 2004. pp. 211–220. [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Lemos JA, Abranches J, Lin VK, Burne RA. Role of RelA of Streptococcus mutans in global control of gene expression. J Bacteriol. 2008;190:28–36. doi: 10.1128/JB.01395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Robertson GT, Kazmierczak KM, Zhao J, Gilmour R, Winkler ME. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol. 2003;50:1647–1663. doi: 10.1046/j.1365-2958.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- Ng WL, Tsui HC, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J Bacteriol. 2005;187:7444–7459. doi: 10.1128/JB.187.21.7444-7459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak R, Cauwels A, Charpentier E, Tuomanen E. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J Bacteriol. 1999;181:1126–1133. doi: 10.1128/jb.181.4.1126-1133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, Tuomanen EI, Orihuela CJ. Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun. 2006;74:4766–4777. doi: 10.1128/IAI.00316-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, Paton JC. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2009;23:731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- Panina EM, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of biosynthesis and transport of aromatic amino acids in low-GC Gram-positive bacteria. FEMS Microbiol Lett. 2003;222:211–220. doi: 10.1016/S0378-1097(03)00303-3. [DOI] [PubMed] [Google Scholar]
- Paterson GK, Blue CE, Mitchell TJ. Role of two-component systems in the virulence of Streptococcus pneumoniae. J Med Microbiol. 2006;55:355–363. doi: 10.1099/jmm.0.46423-0. [DOI] [PubMed] [Google Scholar]
- Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol. 2004;51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- Ramos-Montanez S, Tsui HC, Wayne KJ, Morris JL, Peters LE, Zhang F, Kazmierczak KM, Sham LT, Winkler ME. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol. 2008;67:729–746. doi: 10.1111/j.1365-2958.2007.06082.x. [DOI] [PubMed] [Google Scholar]
- Samen U, Gottschalk B, Eikmanns BJ, Reinscheid DJ. Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J Bacteriol. 2004;186:1398–1408. doi: 10.1128/JB.186.5.1398-1408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier EA, Brekasis D, Paranawithana S, Bonanno JB, Paget MS, Burley SK, Kielkopf CL. X-ray structure of a Rex-family repressor/NADH complex: insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Solioz M, Stoyanov JV. Copper homeostasis in Enterococcus hirae. FEMS Microbiol Rev. 2003;27:183–195. doi: 10.1016/S0168-6445(03)00053-6. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Its Closest Relatives: From Genes to Cells. Washington, D C.: ASM Press; 2002. [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Soutourina O, Soutourina J, Blanquet S, Plateau P. Formation of D-tyrosyl-tRNATyr accounts for the toxicity of D-tyrosine toward Escherichia coli. J Biol Chem. 2004;279:42560–42565. doi: 10.1074/jbc.M402931200. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Steiner K, Malke H. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol Microbiol. 2000;38:1004–1016. doi: 10.1046/j.1365-2958.2000.02203.x. [DOI] [PubMed] [Google Scholar]
- Steiner K, Malke H. relA-Independent amino acid starvation response network of Streptococcus pyogenes. J Bacteriol. 2001;183:7354–7364. doi: 10.1128/JB.183.24.7354-7364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CK, Li H, Claverys JP, Morrison DA. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol. 2001;67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthanthiran M, Solomon SD, Williams PS, Rubin AL, Novogrodsky A, Stenzel KH. Hydroxyl radical scavengers inhibit human natural killer cell activity. Nature. 1984;307:276–278. doi: 10.1038/307276a0. [DOI] [PubMed] [Google Scholar]
- Sutton HC, Winterbourn CC. On the participation of higher oxidation states of iron and copper in Fenton reactions. Free Radic Biol Med. 1989;6:53–60. doi: 10.1016/0891-5849(89)90160-3. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Hashimoto W, Kumagai H. Escherichia coli K-12 can utilize an exogenous γ-glutamyl peptide as an amino acid source, for which γ-glutamyltranspeptidase is essential. J Bacteriol. 1993;175:6038–6040. doi: 10.1128/jb.175.18.6038-6040.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitil AL, Cashel M, Zyskind JW. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- Tedin K, Norel F. Comparison of ΔrelA strains of Escherichia coli and Salmonella enterica serovar Typhimurium suggests a role for ppGpp in attenuation regulation of branched-chain amino acid biosynthesis. J Bacteriol. 2001;183:6184–6196. doi: 10.1128/JB.183.21.6184-6196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Tojo S, Satomura T, Kumamoto K, Hirooka K, Fujita Y. Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids. J Bacteriol. 2008;190:6134–6147. doi: 10.1128/JB.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol Microbiol. 2001;40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- Turnbough CL, Jr, Switzer RL. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol Mol Biol Rev. 2008;72:266–300. doi: 10.1128/MMBR.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulijasz AT, Andes DR, Glasner JD, Weisblum B. Regulation of iron transport in Streptococcus pneumoniae by RitR, an orphan response regulator. J Bacteriol. 2004;186:8123–8136. doi: 10.1128/JB.186.23.8123-8136.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Rijn I, Kessler RE. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 1980;27:444–448. doi: 10.1128/iai.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Wagner C, Saizieu Ad A, Schonfeld HJ, Kamber M, Lange R, Thompson CJ, Page MG. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect Immun. 2002;70:6121–6128. doi: 10.1128/IAI.70.11.6121-6128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich TM, Marahiel MA. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- WHO. Pneumococcal conjugate vaccine for childhood immunization - World Health Organization position paper. Weekly Epidemiological Record. 2007;82:93–104. [Google Scholar]
- Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J Bacteriol. 2000;182:4319–4327. doi: 10.1128/jb.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease. New York: Cambridge University Press; 2005. [Google Scholar]
- Winkler ME, Hoch JA. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J Bacteriol. 2008;190:2645–2648. doi: 10.1128/JB.01682-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect Immun. 2000;68:2819–2826. doi: 10.1128/iai.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YN, Coleman WG, Jr, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. Regulation of cell growth during serum starvation and bacterial survival in macrophages by the bifunctional enzyme SpoT in Helicobacter pylori. J Bacteriol. 2008;190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


