Abstract
We have cloned two new triadin isoforms from rat skeletal muscle, Trisk 49 and Trisk 32, named according to their theoretical molecular weights, 49 kDa and 32 kDa respectively. Specific antibodies directed against each protein were produced to characterize both new triadins. Both are expressed in adult rat skeletal muscle, and their expression in slow twitch muscle is lower than in fast twitch muscle. Using double immunofluorescent labeling, the localization of these two triadins was studied in comparison to well characterized proteins such as ryanodine receptor, calsequestrin, desmin, Ca2+-ATPase, titin. None of these two triadins are localized within the rat skeletal muscle triad. Both are instead found in different parts of the longitudinal sarcoplasmic reticulum. We attempted to identify partners for each isoform: neither are associated with ryanodine receptor, Trisk 49 could be associated with titin or another sarcomeric protein, and Trisk 32 with IP3-receptor. These results open further fields of research concerning the functions of these two proteins, in particular they could be involved in the setting up and the maintenance of a precise sarcoplasmic reticulum structure.
Keywords: Animals; Calcium-Transporting ATPases; metabolism; Carrier Proteins; analysis; metabolism; Molecular Sequence Data; Muscle Proteins; analysis; metabolism; Muscle, Skeletal; metabolism; Protein Isoforms; genetics; metabolism; Protein Kinases; metabolism; Rats; Ryanodine Receptor Calcium Release Channel; metabolism; Sarcoplasmic Reticulum; metabolism; ultrastructure
Skeletal muscle cells have highly organized structures. Sarcomeres, the contractile units of striated muscles, are assembled from thousands of proteins to produce the largest and most regular macromolecular complex known. In addition, this organized structure is designed to undergo strong mechanical stress during muscle contraction. Muscle contraction is activated by Ca2+ release from the sarcoplasmic reticulum in response to plasma membrane depolarization. This process is referred to as excitation-contraction coupling (E-C coupling), and takes place at the skeletal muscle triad junction, where T-tubules and the sarcoplasmic reticulum terminal cisternae are in close contact [1]. To perform its function, the sarcoplasmic reticulum is built as a sleeve-like structure around the myofibrils, and is compartmentalized in highly specialized structures (terminal cisternae, longitudinal reticulum) with specialized functions (calcium release and calcium uptake, respectively) [2].
Calcium release occurs via a macromolecular complex, the calcium release complex, specifically localized in the skeletal muscle triad [3]. Major components of this calcium release complex are the two calcium channels, the ryanodine receptor (RyR) and the dihydropyridine receptor (DHPR) [4]. Triadin is an integral membrane protein of the sarcoplasmic reticulum, first identified in rabbit skeletal muscle in 1990 [5, 6] as a 95-kDa glycoprotein specifically located in the triads. Because of its co-localization with RyR in the triads, involvement of triadin in excitation-contraction coupling has been presumed [7, 8]. Protein interaction studies have shown that molecular partners of triadin are RyR [9, 10, 11], calsequestrin (CSQ), the protein which traps calcium inside the sarcoplasmic reticulum [12–14], junctin [15] and histidine-rich Ca2+-binding protein [16]. Functional studies have shown that triadin by itself is able to regulate the activity of the RyR calcium channel in vitro [11, 17].
Triadin is expressed in both skeletal and cardiac muscle. Several isoforms of triadin have been identified in cardiac muscle. Three isoforms called CT1, CT2 and CT3 (Cardiac Triadin 1, 2, 3), with molecular weights of 35, 40 and 92-kDa respectively, have been cloned in rabbit heart [18]. Of these, CT1 (35-kDa) is the major triadin isoform expressed in canine heart muscle whereas CT2 (40-kDa) is not detectable as a protein, and CT3 (92-kDa) is expressed at very low levels in this species [19]. More recently, three triadin isoforms have been cloned from mouse heart muscle with molecular weights of 35, 35.5 and 40-kDa [20]. While the 35-kDa and 40-kDa isoforms presumably correspond to CT1 and CT2 isoforms of rabbit heart muscle, the 35.5-kDa protein presumably represents a new isoform.
We have previously shown that multiple isoforms of triadin are also expressed in rat skeletal muscle [21], and we identified a new skeletal muscle triadin isoform with an apparent molecular weight of 51-kDa. This new isoform was cloned from rat skeletal muscle [21] and from human skeletal muscle [22]. The skeletal muscle triadin isoforms were named according to their apparent molecular weights: Trisk (for TRIadin SKeletal) 95 for the 95-kDa isoform, and Trisk 51 for the 51-kDa isoform. We have also shown that Trisk 95 and Trisk 51 are expressed in equivalent amounts in rabbit and rat skeletal muscles.
In the present study, two new shorter rat skeletal muscle triadins were cloned, Trisk 49 and Trisk 32. Specific antibodies were developed and used to characterize both proteins more precisely. The triadins’ expression patterns in fast and slow twitch muscles were studied, as well as during differentiation. The localization of these two triadins was studied with respect to other well characterized proteins localized in known regions of the sarcomere. This study demonstrates that both 49 kDa and 32 kDa triadins are not located within the triad, like Trisk 95 and Trisk 51, but are rather found in the longitudinal sarcoplasmic reticulum. Through double immunofluorescent labeling, this study precisely specifies their localization within the longitudinal sarcoplasmic reticulum, and identifies possible partners for each protein. This raises new questions concerning their possible function: Trisk 49 and Trisk 32 could be involved in the maintenance of sarcomere structure during contraction, and Trisk 32 could also be involved in the regulation of non triadic calcium release complex.
Experimental procedures
cDNA Cloning
Total RNA was extracted from adult rat skeletal muscle using RNA-Plus (Q Biogene). mRNA were then purified twice using the Oligotex mRNA purification system (Qiagen). The first cDNA strand was synthetized by Superscript reverse transcriptase (Invitrogen) using the “Smart Race PCR cDNA Amplification” kit (Clontech, BD Biosciences), during 1h30 at 65°C in presence of 0.6 M trehalose (Sigma-Aldrich) with the 3′-CDS primer (AAGCAGTGGTAACAACGCAGAGTAC(T)30 - 3′) and under all other conditions/products supplied in the kit. On the structural basis of triadin clone search (common 5′-end, and divergent 3′-end), a 3′-RACE PCR was performed with a common 5′-end primer, starting in the non-coding sequence of triadin at −19 (5′-ATTGATTTCTGCACCCACCATGACTGAG-3′), and extended toward the 3′ divergent extremity up to the CDS primer used for reverse transcription (universal primer supplied in the kit: 5′-CTAATACGACTCACTATAGGGCAAGCAGT GGTAACAACGCAGAGT-3′). PCR products were then subcloned using TA-cloning PGEM-T easy (Promega). Vectors containing inserts sized >1kb were then sequenced, resulting in the occurrence of two clones named 8F and 10D.
Antibodies
Anti-calsequestrin monoclonal antibody (clone VIIID12) was obtained from Affinity BioReagents. Mouse anti-IP3R-type III antibody was from Transduction laboratories (BD Biosciences). Monoclonal anti-desmin antibody (clone DE-R-11) was from DakoCytomation. Monoclonal anti-titin antibody (T11), specific to the A-I junction, originated from Abcam (Abcam limited, UK). Guinea pig anti Ca2+-ATPase was a gift from Dr. A.M. Lompré [23]. Rabbit anti-bovine mitochondrial F1-ATPase antibody was previously characterized [24]. Anti-ryanodine receptor antibodies (A-RyR), obtained by rabbit immunization with ryanodine receptor purified from pig skeletal muscle, have been described previously [4], as well as antibodies against Trisk 95 and Trisk 51 [21], and antibodies against the N-terminal end of triadins [25]. Using the same experimental techniques, anti-Trisk 49 and anti-Trisk 32 have been produced. Anti-Trisk 49 results from rabbit immunization with the C-terminal peptide of Trisk 49, with an extra tyrosine residue added to its N-terminal end to allow coupling with ovalbumin (YSTTGKHS). Anti-Trisk 32 results from the immunization of a rabbit or guinea pig with the C-terminal peptide of Trisk 32, with an extra tyrosine residue added to its N-terminal end to allow coupling with ovalbumin (YGGPKRILDKKQI). Except for antibodies used against Trisk 49 and Trisk 32, all other antibodies used in this study have previously been characterized and used.
Microsome preparation
Two types of skeletal muscle were collected from adult rat hind legs: extensor digitorum longus (EDL) and soleus. Crude microsomes were prepared from each muscle with slight modifications to the procedures previously described [21]. Indeed, after muscle homogenization in a 30 ml solution made of 200 mM sucrose, 20 mM HEPES (pH = 7.4), 0.4 mM CaCl2, 2 mM leupeptin, 100 mM PMSF, 1 mM DFP, the low speed (10 min at 1500 g) pellet was discarded, and the microsomes obtained after 50 min centrifugation at 41 000 g. They were homogenized at 8–10 mg/ml in 0.1 M NaCl, 30 mM Imidazole (pH 6.8), 8% sucrose, proteases inhibitors, and stored in liquid nitrogen.
Satellite cell cultures
Rat skeletal muscle cell primary cultures resulted from satellite cells obtained by trypsinization of muscle fragments extracted from 20 day old rat embryos. They were induced through differentiation and formed myotubes, as previously described [21].
L6 cell cultures and transfection
Rat myogenic L6 cells (clone C5) were cultured in DMEM supplemented with 10% FBS (Invitrogen) and 1% PS. They were transfected with Lipofectamine and Plus Reagent (Invitrogen) as reported [21]. The expression plasmid used for transfection was plasmid pcDNA3.1 (Invitrogen), with cDNA coding for Trisk 49 (full length sequence of clone 10D) or Trisk 32 (full length sequence of clone 8F).
Western blot analysis
The presence of Trisk 49 or Trisk 32 in samples was assayed by Western blot analysis, using a chemiluminescent reagent (Western lightning Chemiluminescence reagent plus, Perkin Elmer) after electrophoretic separation of the protein on a 5–15% acrylamide gel, and electrotransfer on Immobilon P (Millipore) [21]. The secondary antibodies were labeled with HRP and supplied by Jackson ImmunoResearch Laboratories.
Immunolabelling
Adult rat skeletal muscle (EDL) was embedded in Tissue-Tek O.C.T. compound (Miles Inc.), frozen and stored at −20°C. Cryosections (10 μm thickness) collected on microscope slides were fixed 10 min with 4% paraformaldehyde in PBS at RT, then washed 2 × 10 min with PBS at 4°C. After 5 min permeabilization with 0.5% Triton X100 in PBS at RT, and 30 min saturation with PBS supplemented with Triton X100 0.1%, 0.5% BSA and 2% goat serum, the sections were incubated for 2 h at RT with the primary antibodies (dilution 1/100 to 1/800) in PBS supplemented with 0.1% Triton X100, 0.5% BSA and 2% goat serum, and washed 3 × 10 min at 4°C with PBS-0.1% Triton X100. The sections were then incubated with secondary antibodies for 30 min at RT. After 3 × 10 min washes in PBS-0.1% Triton X100 at RT, the nuclei were stained with TO-PRO-3 (Molecular Probes), and the samples were mounted with an anti-fading solution (DakoCytomation fluorescent mounting medium, DakoCytomation). For double labeling, the two primary antibodies (from different species) and the two secondary antibodies were added at the same time. The secondary antibodies were either labeled with Alexa-488, Alexa-546 (both from Molecular Probes), or Cy3 (from Jackson ImmunoResearch Laboratories). The muscle sections were analyzed by confocal laser scanning microscopy using a Leica TCS-SP2 operating system. Alexa-488, Alexa-546, or Cy3 fluorescence were sequentially excited and collected (400 Hz line by line) by using a 488 nm argon laser line for Alexa-488, and a 543 nm helium-neon laser line for Alexa-546 and Cy3 excitation. Fluorescence emission was collected from 498 to 541 nm for Alexa-488, and from 554 to 625 nm for Alexa-546 and Cy3. To assess the specificity of some antibodies, the antibody was pre-incubated with the corresponding antigenic peptide for 1 h at RT, and a control was performed after incubation of the antibody with the corresponding antigenic peptide for 1 h at RT. For myotubes (differentiated satellite cells), the immunolabeling was performed directly in the culture plate. After fixation of the cells with a 4% paraformaldehyde solution for 15 min, and 2 × 5 min washes with PBS, the cells were permeabilized 5 min with PBS - 0.25% Triton X100 and the immunolabeling was achieved similarly to that muscle sections.
Immunoprecipitation
Rat skeletal muscle microsomes or Trisk 32-transfected cells were solubilized for 30 min at 4°C at 3mg/ml in a buffer composed of 0.9M NaCl, 1.6% CHAPS, 0.1% phospholipids (egg total phosphatide extract, Avanti Polar Lipids), 20mM Pipes (pH 7.1), PMSF 200μM, DFP 1mM. After solubilization, the solubilized proteins were diluted with 1 volume PBS and 1 volume H2O. The immunoprecipitation was then performed with the chosen antibody and protein-A sepharose as described previously [4]. The immunoprecipitated proteins were then analyzed by Western blotting.
RESULTS
Cloning of Trisk 49 and Trisk 32
We have previously shown that multiple triadin mRNA are produced in rat skeletal muscle [21], and we have cloned two major triadin isoforms, Trisk 95 and Trisk 51, which correspond to 4.5 kb transcripts. We then went on with cloning the remaining smaller transcripts.
The classic reverse transcription technique using oligo-dT18 failed to provide full length clones because of the presence of A-rich regions in the sequence of the cDNA, and only 3′-truncated clones were obtained. A more stringent technique was thus used to eliminate hybridization of the oligo-dT primer within the internal stretches of A in the region spanning triadin nucleotides 900 to 1200. The reverse transcription step was performed with a 30-mer oligo-dT, at a temperature of 65°C, and in presence of trehalose to stabilize the reverse transcriptase at such a high temperature [26, 27]. In these conditions, most of the cDNAs produced by Superscript reverse transcriptase were full length clones, resulting from an oligo-dT hybridization only on the correct poly-A stretches. Two clones, named 10D and 8F, were further analyzed. Clone 8F contains an open reading frame of 864 bp followed by a non-coding sequence of 127 bp with a polyadenylation signal. It encodes a 287 amino acid protein with a molecular weight of 32.1 kDa, and was thus named Trisk 32 according to its molecular weight (TRIadin SKeletal muscle 32 kDa), as proposed for the two other skeletal muscle triadins Trisk 95 and Trisk 51. Trisk 32 results from an alternative splicing after the 8 first exons of human triadin [22], and possesses a unique C-terminal end made of 23 amino acids (figure 1). The size of this triadin isoform is similar to CT1, the major cardiac triadin [19]. CT1 is nevertheless distinct from Trisk 32, even though both result from alternative splicing after exon 8, as Trisk 32 has a different specific C-terminal end (figure 1).
Figure 1.

Sequence alignment of the C-terminal specific sequences of Trisk 32 and Trisk 49 with Trisk 95, Trisk 51 and mouse CT1 (cardiac isoform). Amino acids within the box represent the conserved sequences between the different isoforms. Upper numbers correspond to amino acid positions in the Trisk 95 sequence.
Clone 10D, containing an open reading frame of 1326 bp and a short non-coding sequence of 24 bp with a polyadenylation signal, encodes a protein of 442 amino acids, and results from an alternative splicing after the 20 first exons of human triadin. It possesses a unique C-terminal end of 4 amino acids (figure 1). The corresponding protein formed of 442 amino acids has a theoretical molecular weight of 49.5 kDa, and was thus named Trisk 49.
Evidence for the existence of Trisk 49 and Trisk 32
In order to check that the cloned cDNA are physiologically translated in the corresponding proteins, specific antibodies were produced for each isoform. Taking advantage of their C-terminal specific sequences, synthetic peptides were produced and used to immunize either rabbits or guinea pigs. Reactivity of these antibodies was first checked on cells transfected with the corresponding cloned cDNA (figure 2A), and then on rat skeletal muscle (figure 2B).
Figure 2. Western blot analysis of Trisk 32 and Trisk 49.
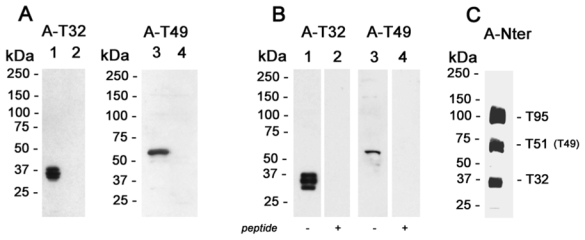
Panel A: L6 cells transfected with Trisk 32 or Trisk 49 cDNA. Fifty micrograms of cell lysates were loaded in each lane. Lane 1: L6 transfected with Trisk 32. Lanes 2 and 4: non transfected L6 cells. Lane 3: L6 transfected with Trisk 49. Western blot analysis was performed with anti-Trisk 32 (lanes 1 and 2) or anti-Trisk 49 (lanes 3 and 4).
Panel B: Identification of Trisk 32 and Trisk 49 in rat skeletal muscle. Forty micrograms of rat skeletal muscle microsomes were loaded in each lane. Western blot analysis was performed with anti-Trisk 32, without (lane 1) and with (lane 2) pre-incubation of the antibody with the corresponding peptide (10 μg/ml). Western blot analysis in lanes 3 and 4 was performed with anti-Trisk 49, without (lane 3) and with (lane 4) pre-incubation with the corresponding peptide (10 μg/ml).
Panel C: Western blot analysis of rat skeletal muscle with an antibody directed against the N-terminal end of triadin, common to all triadin isoforms. Five micrograms of rat skeletal microsomes were loaded. The three major bands correspond to Trisk 95, Trisk 51 and Trisk 32.
Transfection of L6 cells with Trisk 32 cDNA gave rise to a triplet of approx. 37 kDa, specifically recognized by anti-T32 antibodies (figure 2A, lane 1). Transfection of L6 cells with Trisk 49 cDNA gave rise to a unique band of approx. 60 kDa, specifically recognized by anti-T49 antibodies (figure 2A, lane 3). The multiple bands observed for Trisk 32 are thought to arise from post-transcriptional modifications (such as glycosylation), as a single cDNA leads to the expression of two/three proteins, a situation that has previously been observed for Trisk 51 expressed in L6 cells [21], and for cardiac triadin CT1 expressed in Sf21 insect cells [19].
These antibodies were then tested by Western blot analysis on rat skeletal muscle, in order to identify the corresponding protein within skeletal muscle. Anti-Trisk 32 specifically recognizes a triplet within rat skeletal muscle (figure 2B, lane 1), which was abolished by pre-incubation with the corresponding peptide (figure 2B, lane 2). Anti-Trisk 49 antibodies recognized a faint but specific band in rat skeletal muscle (figure 2B, lane 3), as shown by the abolition of the signal upon antibody-peptide pre-incubation (figure 2B, lane 4). Using anti-Nterminal end antibodies, common to all triadin isoforms and which thus allow relative quantification, this study showed that Trisk 32 is present in about the same quantities as Trisk 95 and Trisk 51 (figure 2C). Since Trisk 49 migrates at the same molecular weight as Trisk 51, it is impossible to tell the respective contributions of Trisk 51 and Trisk 49 in the observed band. Nevertheless, it is likely that Trisk 49 is a less abundant isoform compared to the other three, as observed from anti-Trisk 49 reactivity on rat microsomes (figure 2B), and from RT-PCR amplification (data not shown).
Are these isoforms specific to muscle type ?
Since at least four triadin isoforms are expressed in rat skeletal muscle (Trisk 95, Trisk 51, Trisk 49 and Trisk 32), this study further investigated whether some of these isoforms are muscle type specific. Indeed, myosin and many other skeletal muscle proteins (for review, see [28]) are expressed under different isoform (“fast” or “slow”) when they are localized in fast twitch or slow twitch muscles. Microsomes from two types of rat muscle were prepared, in order to test the reactivity of the different triadin antibodies: extensor digitorum longus (EDL), a fast twitch muscle, and soleus, a slow twitch muscle. As shown on figure 3, reactivity to all four isoforms is lower in rat slow twitch muscle compared to fast twitch muscle, with Trisk 49 being almost undetectable. Nevertheless, except for Trisk 49, none of these isoforms seem to be slow twitch or fast twitch specific proteins. Trisk 49 is present in low amounts in fast twitch muscle, and either its expression in slow twitch muscle decreases to reach detection limit of Western blot, or it is not expressed in slow twitch muscle.
Figure 3. Expression of triadin isoforms in slow twitch and fast twitch muscle.
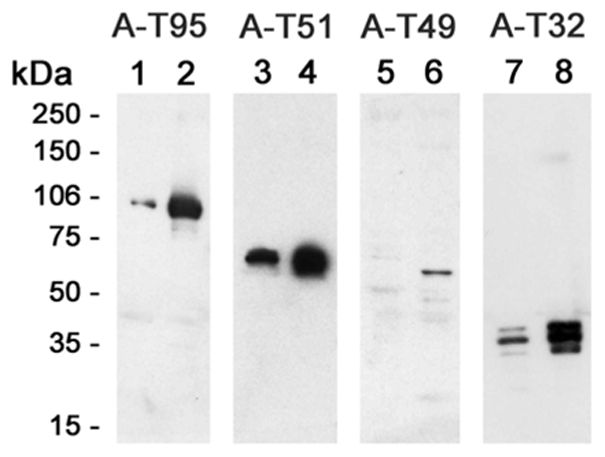
The relative amount of each triadin isoform was assessed by Western blot analysis on slow twitch muscle (soleus, lanes 1, 3, 5, 7) and on fast twitch muscle (EDL, lanes 2, 4, 6, 8). Twenty micrograms microsomes from both muscle types were loaded in each lane, except for lane 5, where 40 μg of soleus microsomes were loaded. Analysis was performed with antibodies against Trisk 95 (A-T95, lanes 1 and 2), against Trisk 51 (A-T51, lanes 3 and 4), against Trisk 49 (A-T49, lanes 5 and 6), and against Trisk 32 (A-T32, lanes 7 and 8).
Localization of Trisk 49 and Trisk 32 in adult skeletal muscle
In order to identify the localization of the two new isoforms in rat skeletal muscle, immunohistolabeling of rat EDL longitudinal section was performed with different antibodies: RyR, CSQ, Trisk 95, Trisk 51, Trisk 49 and Trisk 32. As previously observed [21], Trisk 95 and Trisk 51 perfectly co-localize with RyR and CSQ. Figure 4 presents images of the Trisk 95/RyR and Trisk 51/CSQ double labeling, indicating that these two isoforms are localized within the adult rat skeletal muscle triad. The same experiment was performed with anti-Trisk 49 and anti-Trisk 32. These two triadin isoforms presented a striated pattern perpendicular to the fiber axis when observed at low magnification similarly to RyR, CSQ, Trisk 95, Trisk 51 (data not shown). Nevertheless, at higher magnification, results are completely different for the two isoforms: neither are co-localized with RyR or CSQ (figure 5), nor with Trisk 95, Trisk 51 or DHPR (data not shown), these 5 proteins being all co-localized. The staining for Trisk 49 appears as a double row of rods situated in the middle of the double row of dots observed for CSQ (figure 5 panels a, b, c). This staining is very intense and contrasts with the faint labeling observed in Western blot, probably because Trisk 49 is present in low amount but it is very concentrated in a precise part of the muscle. The staining for Trisk 32 shows a diffused band of dots in the middle of RyR staining (figure 5, panels d, e, f). In addition, localization of both isoforms is not similar, Trisk 32 being inside the labeling of Trisk 49 (figure 5, panels g, h, i). In order to map their localization more precisely, double immunolabelings were performed with anti-Trisk 49 and anti-Ca2+-ATPase antibodies, anti-desmin or anti-titin (figure 6). Ca2+-ATPase is a protein specifically localized in the sarcoplasmic reticulum responsible for the uptake of calcium by the sarcoplasmic reticulum [29], and mostly abundant in the longitudinal part of sarcoplasmic reticulum. Desmin is considered as a marker of the Z-disc [30]. Titin is a giant protein (each molecule, extending from the Z-disc to the M-line, is more than 1 μm long) which controls the layout of the sarcomere, and by acting like a spring provides elasticity to the sarcomere (reviewed in [31]). The antibody used to label titin is specific to the A-I junction part of the molecule [32]. Trisk 49 is localized within the longitudinal sarcoplasmic reticulum, but it is not present in all the longitudinal reticulum as observed from overlay with Ca2+-ATPase labeling (figure 6, panels a, b, c). Comparison with desmin’s localization showed that it is centered on the Z disc, the double row of rods surrounding the Z line (figure 6, panels d, e, f). Trisk 49 seems more precisely co-localized with titin at the A-I junction (figure 6, panels g, h, i).
Figure 4. Co-localization of RyR and T95, and of CSQ and T51 in rat skeletal muscle sections.
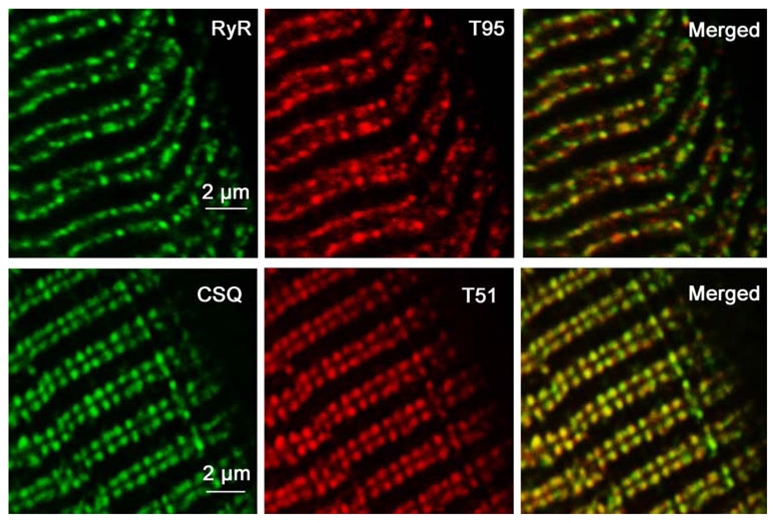
Longitudinal sections of adult rat EDL were immunolabeled with specific antibodies against RyR and Trisk 95 (T95), and CSQ and Trisk 51 (T51). They presented the typical triad protein labeling pattern: double rows of fluorescent dots corresponding to triads pairs on either side of the Z-line.
Figure 5. Trisk 49 and Trisk 32 are not in rat skeletal muscle triad.
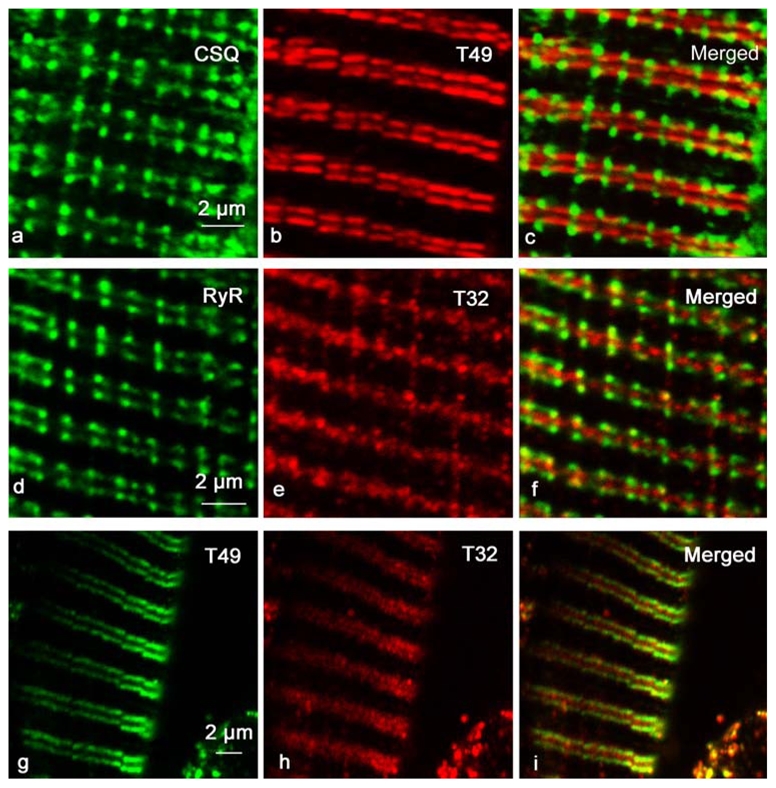
Longitudinal sections of adult rat EDL were immunolabeled with specific antibodies against CSQ and Trisk 49 (T49) (panels a, b, c), RyR and Trisk 32 (T32) (panels d, e, f) or Trisk 49 and Trisk 32 (panels g, h, i). Trisk 49 labeling (panel b) is presented as a double row of rods, closer to the Z-line than the triads are (represented by CSQ labeling, panel a). Trisk 32 labeling (panel e) appears as a row of dots in the middle of the triad double rows.
Figure 6. Trisk 49 is localized within the longitudinal sarcoplasmic reticulum, at the interface of the A and I bands.
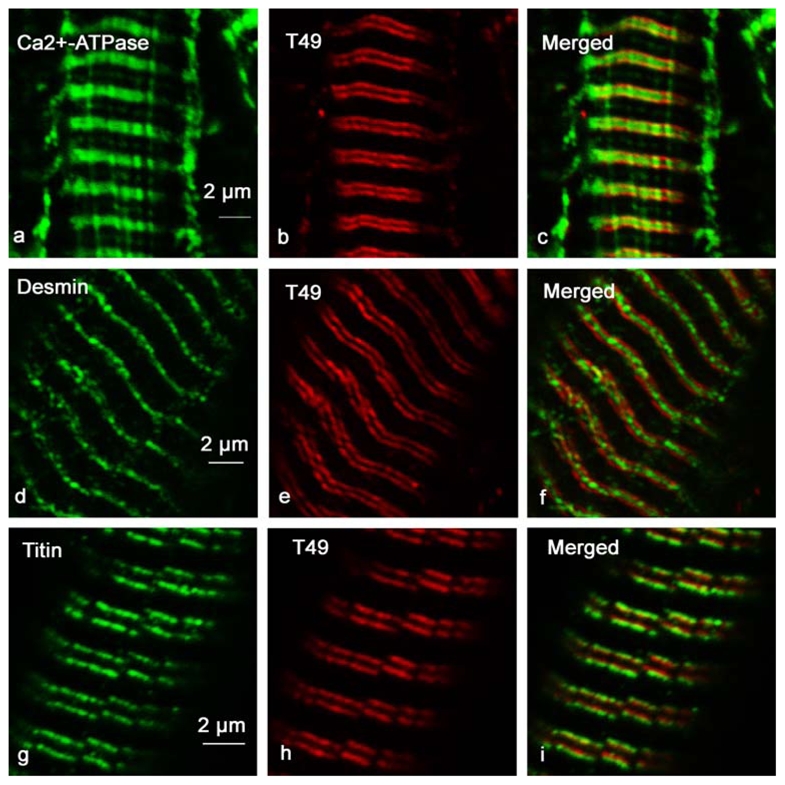
Longitudinal sections of adult rat EDL were immunolabeled with specific antibodies against Ca2+-ATPase and Trisk 49 (panels a, b and c), desmin and Trisk 49 (panels d, e and f), and titin and Trisk 49 (panels g, h and i). Ca2+-ATPase is a longitudinal sarcoplasmic reticulum marker, desmin labels the Z-line, and antibodies used against titin are specific to the A-I interface.
In order to localize Trisk 32 more precisely, double labeling was performed with anti-Trisk 32 and anti-Ca2+-ATPase, anti-desmin, anti-IP3R-type III or anti-mitochondrial FoF1-ATPase. It has been demonstrated that IP3R is found within the longitudinal sarcoplasmic reticulum, at the Z-line [33], and FoF1-ATPase, a specific mitochondrial protein responsible for ATP synthesis, is localized in the inner membrane of mitochondria [34]. Trisk 32 is localized within the longitudinal sarcoplasmic reticulum (figure 7, panels a, b, c), where it co-localizes with Ca2+-ATPase, and is specifically centered on the Z-disc (co-localization with desmin, figure 7, panels d, e, f). Trisk 32 is also present in the same part of the reticulum as IP3R (figure 7, panels g, h, i), and as the mitochondria (figure 7, panels j, k, l).
Figure 7. Trisk 32 is located within the longitudinal sarcoplasmic reticulum, co-localized with IP3R and mitochondria.
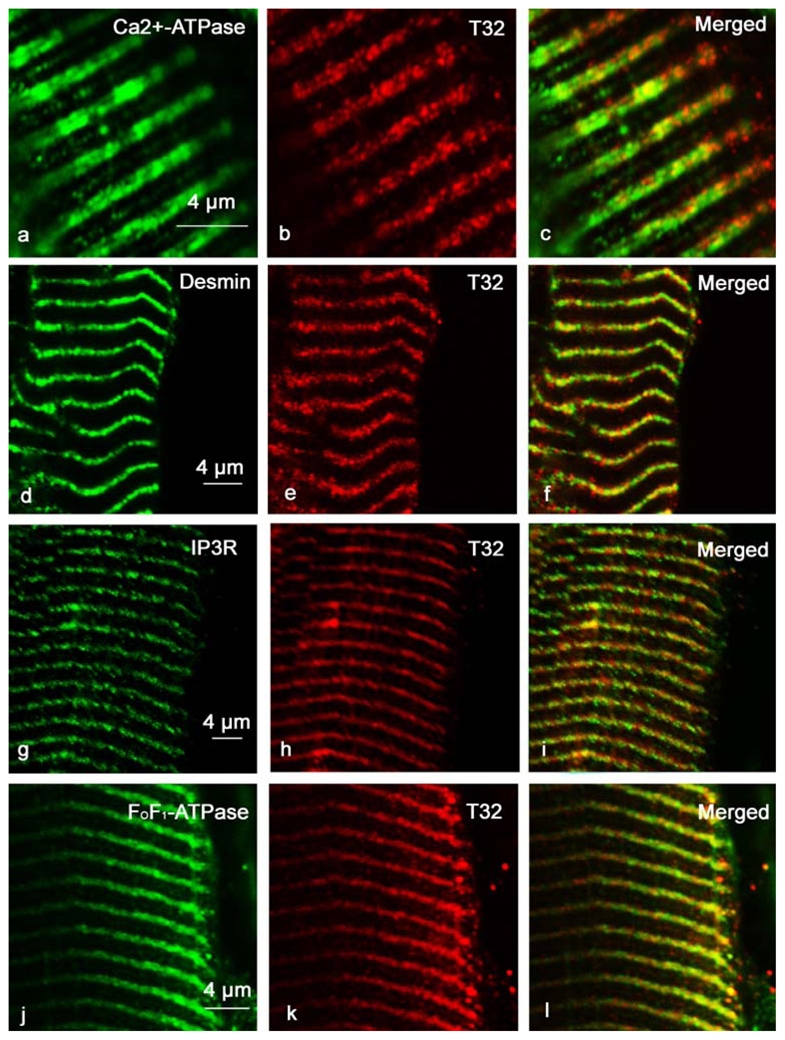
Longitudinal sections of adult rat EDL were immunolabeled with specific antibodies against Ca2+-ATPase and Trisk 32 (panels a, b and c), desmin and Trisk 32 (panels d, e and f), IP3R and Trisk 32 (panels g, h and i), and FoF1-ATPase and Trisk 32 (panels j, k and l). Ca2+-ATPase is a longitudinal sarcoplasmic reticulum marker, desmin labels the Z-line, IP3R is located in the longitudinal sarcoplasmic reticulum, and FoF1-ATPase is a mitochondria marker.
Identification of Trisk 32 and Trisk 49 partners
We attempted to identify the specific partners of Trisk 32 and Trisk 49 using immunoprecipitation with specific antibodies, and their association with RyR was first investigated. As expected, RyR was neither associated with Trisk 49 nor with Trisk 32 (or only in very minor amounts), in contrast with Trisk 95 and Trisk 51 which are both associated with RyR (figure 8A). Based on the possible co-localization observed in immunolabeling, association of Trisk 32 with IP3R was also examined. Since IP3R is a protein that shares homologous domains with RyR, and since Trisk 32 shares common parts with Trisk 95, which is associated with RyR, one could assume that Trisk 32 and IP3R are not only partially co-localized, but also associated. Nevertheless, it was not possible to demonstrate IP3R’s association with Trisk 32 by immunoprecipitation on rat skeletal muscle, probably because of the low expression of IP3R in skeletal muscle [35, 36]. Thus, in order to test this hypothesis, skeletal muscle culture cells (L6 cells) were used, as they are known to express high amounts of IP3R-type III [36, 37]. These cells were transfected with Trisk 32 cDNA, or with Trisk 49, Trisk 51 or Trisk 95 cDNAs. Then the immunoprecipitation was performed with antibodies directed against the corresponding triadin isoform (A-Trisk 32, A-Trisk 49, A-Trisk 51 or A-Trisk 95). The presence of IP3R-III among the immunoprecipitated proteins was assayed by Western blot analysis. Results are presented in figure 8B. IP3R-type III is expressed in the L6 cells, as observed in lane 1. Immunoprecipitation was performed on Trisk 32 transfected (lanes 3 and 5) or non-transfected (lanes 2 and 4) cells, with two different antibodies directed against Trisk 32 (guinea pig anti-T32, lanes 2 and 3, or rabbit anti- T32, lanes 4 and 5). Both anti-T32 antibodies induced co-immunoprecipitation of IP3R-III only in cells expressing Trisk 32. In the cells transfected with another triadin isoform, the immunoprecipitation of this triadin didn’t lead to the co-immunoprecipitation of IP3R-III (figure 8B, lanes 6, 7 and 8). This indicates that Trisk 32 and IP3R-III are associated within the cells, and that the other triadin isoforms (Trisk 49, Trisk 51 and Trisk 95) are unable to interact with IP3R-III. The association with IP3R-III is therefore specific of Trisk 32. It is thus plausible that in skeletal muscle, not only are Trisk 32 and IP3R co-localized, but also associated.
Figure 8.
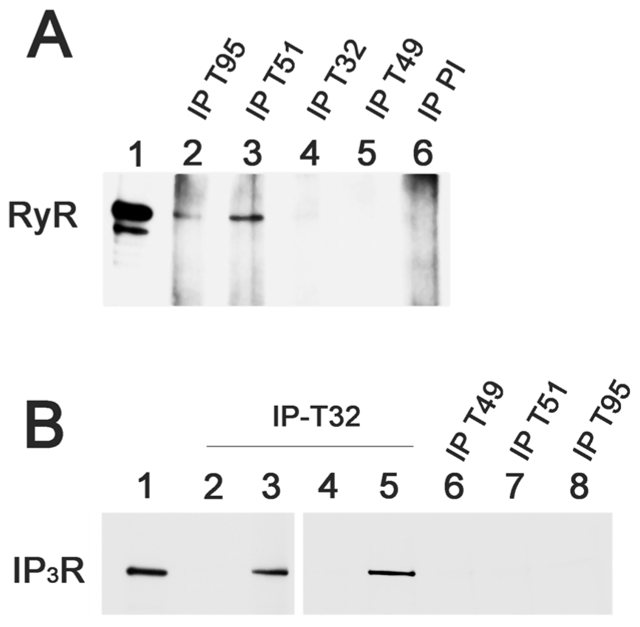
A -Trisk 95 and Trisk 51, and not Trisk 32 nor Trisk 49, are associated with RyR. Immunoprecipitations were performed on rat skeletal muscle with antibodies against Trisk 95 (IP-T95, lane 2), against Trisk 51 (IP-T51, lane 3), against Trisk 32 (IP-T32), against Trisk 49 (IP-T49), and with non-immune serum (pre-immune: IP-PI). The presence of RyR in the immunoprecipitated proteins was analyzed by western blot. Lane 1: 5 μg of rat skeletal muscle microsomes. The two bands with high molecular weights are characteristic of RyR, the lower one, present in smaller amount, being a proteolytic degradation of RyR.
B- Trisk 32, but not Trisk 49, Trisk 51 or Trisk 95, is associated with IP3R in L6 cells. L6 cells were transfected (lanes 3 and 5) or not (lanes 2 and 4) with Trisk 32 cDNA, or transfected with the cDNA of Trisk 49 (lane 6), Trisk 51 (lane 7) or Trisk 95 (lane 8). Immunoprecipitations were then performed with the corresponding anti-Trisk antibody (anti-Trisk 32 developed in guinea pig, lanes 2–3; anti-Trisk 32 developed in rabbit, lanes 4–5; anti-Trisk 49, lane 6; anti-Trisk 51, lane 7 and anti-Trisk 95, lane 8). The immunoprecipitated proteins were analyzed by western blot using anti-IP3R antibodies. Lane 1: 60 μg of non-transfected L6 cells, for control of IP3R-type III expression levels
As titin and Trisk 49 appear co-localized on muscle sections, co-immunoprecipitation on rat micorosomes of titin and Trisk 49 was also attempted, using either anti-titin or anti-Trisk 49 antibodies, but without success. This is probably due either to the low expression of Trisk 49 in skeletal muscle (figure 3), or to the microsome preparation which could lead to dissociation of the complex, as both proteins are expressed in different compartments.
Trisk 49 and Trisk 32 expression during in vitro myotube differentiation
We have previously shown that Trisk 95 and Trisk 51 are progressively expressed in primary cultures of rat skeletal muscle as differentiation and myotube formation takes place. Expression of Trisk 49 and Trisk 32 was examined during in vitro differentiation of rat myoblast primary cultures. The cells were identified by nuclei staining (represented in blue on figure 9, panels c and f). The myotubes were identified through nuclei alignment, while other cells remained as myoblasts or fibroblasts. Similarly to Trisk 95 and Trisk 51, Trisk 49 is not expressed within myoblasts, but within myotubes (figure 9, panel b). As soon as myotubes are formed, Trisk 49 is present under a striated pattern (figure 9, panel b), identical to the pattern observed for titin (figure 9, panel a). This striated pattern is specific to the protein and abolished by pre-incubation of antibodies with the peptide (data not shown). Trisk 49’s early striated pattern is another indication towards a possible interaction with titin, and probably reflects a specific role and interaction at this early developmental stage.
Figure 9. Trisk 49 and Trisk 32 expression during differentiation of myoblasts.
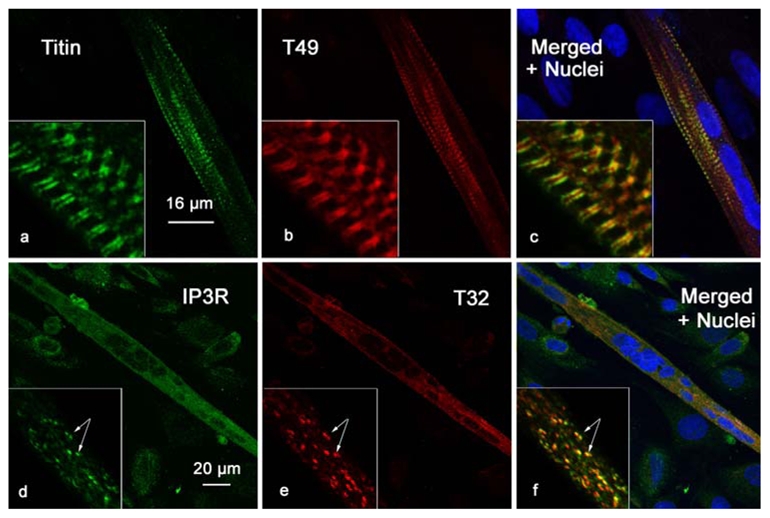
Primary cultures of satellite cells were induced in differentiation. When myotubes formed, the cells were fixed and labeled with antibodies against titin and Trisk 49 (panels a, b and c), or IP3R and Trisk 32 (panels d, e and f). The nuclei were stained with TO-PRO 3 (blue labeling in panels c and f). Titin and Trisk 49 antibodies labeled only myotubes (aligned nuclei) and not myoblasts (isolated nuclei), and both presented a striated pattern. IP3R and Trisk 32 antibodies label the myotubes (multinucleated cells) and the myoblasts (isolated nuclei), with a diffuse punctuated pattern.
On the contrary, Trisk 32 shows punctuate labeling in myotubes (figure 9 panel e), similar to observations for Trisk 95 and Trisk 51, and it is expressed at a low level in myoblasts. This punctuated staining in myotubes is already co-localized with IP3R-type III (figure 9, panels d, e and f inserts).
DISCUSSION
In this study, two new triadin isoforms, Trisk 49 and Trisk 32, were cloned from rat skeletal muscle, and the presence of the corresponding protein was demonstrated using specific antibodies. While Trisk 32 is present in comparable amounts with the two previously described isoforms, Trisk 95 and Trisk 51, Trisk 49 is apparently less abundant. No isoform switch was observed between slow or fast twitch muscles. All triadin isoforms seemed less abundant in slow twitch muscle, probably because the relative amount of sarcoplasmic reticulum is lower in slow twitch muscle than in fast twitch muscle [38].
Analysis of the cellular distribution of these two triadin isoforms in rat skeletal muscle showed that, in contrast with Trisk 95 and Trisk 51, Trisk 49 and Trisk 32 are not located within the skeletal muscle triad. Since all triadins share common sequences, the localization of the triadins inside the muscle cell is somehow connected to the specific C-terminal end of each isoform. While it could be concluded that the name “triadin”, in its original meaning “located in the triad”, is not fully appropriate for these two triadin isoforms, it nevertheless should be kept because all triadin proteins result from alternative splicings of the same gene. These results were confirmed by immunoprecipitation, and once again it was observed that Trisk 95 and Trisk 51 are associated with RyR, and thus members of the calcium release complex, whereas no association was shown for Trisk 49 and Trisk 32. Their involvement in the calcium release complex or excitation-contraction coupling is still unclear. In order to find clues regarding their function, their precise localization was investigated in skeletal muscle. Using double immunofluorescent labeling on skeletal muscle sections, it was shown that Trisk 32 is localized within the longitudinal part of the sarcoplasmic reticulum, close to the Z-line. This part of the SR which has already been shown to contain IP3R, the other intracellular calcium channel [33, 39], and we indeed observed that Trisk 32 and IP3R are located within the same region of the sarcoplasmic reticulum. The similar localization of these two proteins enabled speculation concerning the existence of a calcium release complex centered around IP3R and involving Trisk 32, identical to the one centered around RyR and involving Trisk 95 and Trisk 51. This hypothesis was reinforced by sequence homologies identified between RyR and IP3R in the channel domains of both proteins [40, 41]. This domain of RyR has recently been demonstrated to be an interaction site between RyR and triadin [42]. Thus it is tempting to conclude towards a Trisk 32 - IP3R interaction. In order to test this hypothesis, immunoprecipitation was performed on a rat skeletal muscle cell line, known to express higher amount of IP3R than differentiated cells or adult muscle, transfected with Trisk 32 cDNA. Within the transfected cells, results showed that immunoprecipitation of Trisk 32 induced co-immunoprecipitation of IP3R-type III. Thus association of Trisk 32 and IP3R-type III can be observed if both partners are expressed in reasonable amounts. This interaction is specific of Trisk 32 and can not be reproduced with the other triadin isoforms, Trisk 49, Trisk 51 or Trisk 95. We conclude that this association could probably also exist in skeletal muscle, and part of Trisk 32 could be associated with IP3R-III. By double labeling a muscle section with anti-Trisk 32 antibodies and antibodies directed against a specific mitochondrial protein (FoF1-ATP synthase), it was observed that the part of the SR which contains Trisk 32 also co-localized with the mitochondria. The function of Trisk 32 could then be connected to calcium release via IP3R and/or calcium storage in the mitochondria, which is today a well established fact [43–46].
Double immunofluorescence labeling was also used to study the precise localization of Trisk 49. As early as the first developmental stages, Trisk 49 differs in its localization from other triadin isoforms. Exterior to the triad, it is organized in a double row of rods localized between the double row of triads and the Z line. This localization corresponds to the A-I part of titin, a giant protein involved in the organization and elasticity of the sarcomere. Because of Trisk 49’s low expression in skeletal muscle, identification of its molecular partners has not yet been achieved, but will be the focus of future research.
As previously observed for Trisk 95 and Trisk 51, the two new triadins Trisk 49 and Trisk 32 are strongly expressed in myotubes. Whereas Trisk 95, Trisk 51 and Trisk 49 are absent from myoblasts, Trisk 32 seems to have an earlier expression and is weakly expressed before differentiation. Trisk 32 presents a punctuated pattern in myotubes, co-localized with IP3R-III, whereas Trisk 49 presents a striated pattern, identical to the one observed for titin. Once again, this could be an indication that Trisk 32 is associated with IP3R, and Trisk 49 is associated with titin, ever since the early differentiation stages. Trisk 49 has already reached an organized localization compared to the Z-line, while the triads have not. Trisk 49 could be directly or not connected to the A-I part of titin, or connected to another sarcomeric protein localized in this region. It could then be involved in sarcomeric structures positioning and in organization of the triads.
Figure 10 summarizes data concerning the localization of Trisk 32 and Trisk 49 in rat skeletal muscle. The next step will involve a more precise identification of their function, which in contrast to Trisk 95 and Trisk 51, will probably not be connected to excitation-contraction coupling. Because of its similar localization and possible interaction with IP3R, Trisk 32 could be involved in the regulation of a calcium release complex centered on IP3R, in a similar fashion as the regulation of the calcium release complex centered on RyR by Trisk 95. Trisk 32 is already expressed in myoblasts, as well as IP3R, in contrast to Trisk 95 or Trisk 51 which, as RyR, are expressed only in myotubes. One possible role for these two triadin isoforms could involve an anchoring function. During contraction, elasticity of muscle fibers is such that, in order to remain organized, some parts have to be anchored while others are mobile. Position of the triads is fixed compared to the Z-line [47]. It is likely that all sarcoplasmic reticulum around the Z-line is fixed, including the triads, and mitochondria. As opposed, the sarcoplasmic reticulum centered around the myosin undergoes muscle contraction and elongation. In this context, the function of Trisk 32 in a “fixed” sarcoplasmic reticulum could be related to the anchoring of the reticulum and IP3R to adjacent mitochondria. Similarly, Trisk 49 could be involved in connecting titin or another sarcomeric protein to the reticulum close to the triad. Thus, both triadins Trisk 49 and Trisk 32 could contribute to maintaining this part of the reticulum stable during contraction. The existence of a cytoskeleton spatially separated from the contractile apparatus is increasingly recognized today [48]. A great number of proteins are involved in connecting the plasma membrane with the sarcomere Z-line at costamere [49]. Others are involved in connecting the sarcoplasmic reticulum to the myofibrils (see [50] for review), like ankyrin [51], or in connecting the plasma membrane to the sarcoplasmic reticulum membrane, like junctophilin [52]. Disruption to any of these systems has dramatic consequences on muscle and triad structure or function [52, 53], similarly to conditions observed in some myopathies. Thus the putative function of Trisk 32 and Trisk 49 in maintaining sarcoplasmic reticulum positioning could be of major importance and needs to be researched thoroughly. In addition, the presence of these two triadin in human skeletal muscle has to be assayed.
Figure 10. Schematic organization of the triad area and localization of Trisk 49 and Trisk 32.
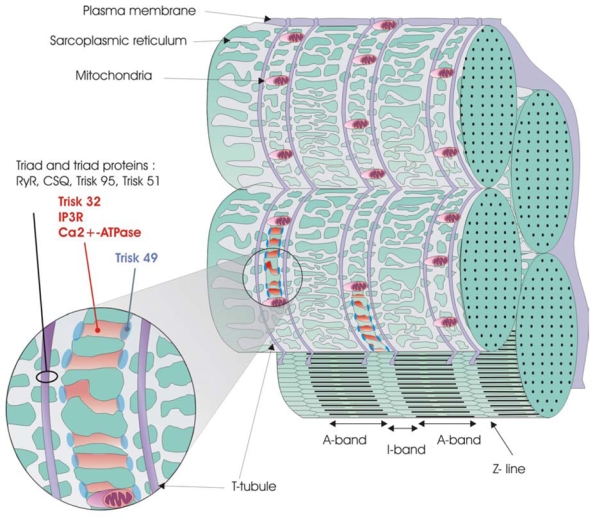
Possible organizations of the sarcoplasmic reticulum and triad, and localization of Trisk 49 and Trisk 32 compared to other known proteins are represented on this figure. Both Trisk 49 and Trisk 32 are located at slightly different places within the non-junctional sarcoplasmic reticulum surrounding the Z-line, while triad proteins are found around the T-tubules.
Another muscle protein, tropomyosin, has recently been discovered under different isoforms in various localizations of the skeletal muscle, either associated with thin actin filaments, or with the Z-line [54]. Thus, triadins are likely to be part of a large family of skeletal muscle proteins which can have multiple localizations and functions within the same cell. Different types of reticulum exist within skeletal muscle, and it would be worth identifying which type of reticulum (endoplamic, sarcoplasmic,…) these triadins are located in, in order to understand their sorting and function [55].
Acknowledgments
We thank Dr A. M. Lompré for antibodies against Ca2+-ATPase. This work was partly supported by grants from the Association Française contre les Myopathies (AFM), as well as by grants from the GIS-Institut des Maladies Rares. We acknowledge financial support from INSERM, CEA and the French ministry of research.
Abbreviations list
- CSQ
calsequestrin
- Cy3
Indocarbocyanine 3
- DHPR
dihydropyridine receptor
- EDL
Extensor digitorum longus
- E-C coupling
excitation-contraction coupling
- IP3R
IP3 receptor
- RACE-PCR
Rapid amplification of cDNA End by Polymerase Chain Reaction
- RyR
ryanodine receptor
- T95
Trisk 95
- T51
Trisk 51
- T49
Trisk 49
- T32
Trisk 32
Footnotes
References
- 1.Flucher BE. Dev Biol. 1992;154:245–260. doi: 10.1016/0012-1606(92)90065-o. [DOI] [PubMed] [Google Scholar]
- 2.Franzini-Armstrong C. In: Myology. Engel AE, Franzini-Armstrong C, editors. McGraw-Hill Inc; New York: 1994. pp. 176–199. [Google Scholar]
- 3.Flucher BE, Franzini-Armstrong C. Proc Natl Acad Sci USA. 1996;93:8101–8106. doi: 10.1073/pnas.93.15.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marty I, Robert M, Villaz M, Lai Y, De Jongh KS, Catterall WA, Ronjat M. Proc Natl Acad Sci USA. 1994;91:2270–2274. doi: 10.1073/pnas.91.6.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt NR, Caswell AH, Wen SR, Talvenheimo JA. J Membr Biol. 1990;113:237–251. doi: 10.1007/BF01870075. [DOI] [PubMed] [Google Scholar]
- 6.Kim KC, Caswell AH, Talvenheimo JA, Brandt NR. Biochemistry. 1990;29:9281–9289. doi: 10.1021/bi00491a025. [DOI] [PubMed] [Google Scholar]
- 7.Caswell AH, Brandt NR, Brunschwig JP, Purkerson S. Biochemistry. 1991;30:7507–7513. doi: 10.1021/bi00244a020. [DOI] [PubMed] [Google Scholar]
- 8.Knudson CM, Stang KK, Moomaw CR, Slaughter CA, Campbell KP. J Biol Chem. 1993;268:12646–12654. [PubMed] [Google Scholar]
- 9.Liu G, Pessah IN. J Biol Chem. 1994;269:33028–33034. [PubMed] [Google Scholar]
- 10.Caswell AH, Motoike HK, Fan H, Brandt NR. Biochemistry. 1999;38:90–97. doi: 10.1021/bi981306+. [DOI] [PubMed] [Google Scholar]
- 11.Groh S, Marty I, Ottolia M, Prestipino G, Chapel A, Villaz M, Ronjat M. J Biol Chem. 1999;274:12278–12283. doi: 10.1074/jbc.274.18.12278. [DOI] [PubMed] [Google Scholar]
- 12.Guo W, Campbell KP. J Biol Chem. 1995;270:9027–9030. doi: 10.1074/jbc.270.16.9027. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi YM, Alseikhan BA, Jones LR. J Biol Chem. 2000;275:17639–17646. doi: 10.1074/jbc.M002091200. [DOI] [PubMed] [Google Scholar]
- 14.Shin DW, Ma J, Kim DH. FEBS Lett. 2000;486:178–182. doi: 10.1016/s0014-5793(00)02246-8. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Kelley J, Schmeisser G, Kobayashi YM, Jones LR. J Biol Chem. 1997;272:23389–23397. doi: 10.1074/jbc.272.37.23389. [DOI] [PubMed] [Google Scholar]
- 16.Lee HG, Kang H, Kim DH, Park WJ. J Biol Chem. 2001;276:39533–39538. doi: 10.1074/jbc.M010664200. [DOI] [PubMed] [Google Scholar]
- 17.Ohkura M, Furukawa K, Fujimori H, Kuruma A, Kawano S, Hiraoka M, Kuniyasu A, Nakayama H, Ohizumi Y. Biochemistry. 1998;37:12987–12993. doi: 10.1021/bi972803d. [DOI] [PubMed] [Google Scholar]
- 18.Guo W, Jorgensen AO, Jones LR, Campbell KP. J Biol Chem. 1996;271:458–465. doi: 10.1074/jbc.271.1.458. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi YM, Jones LR. J Biol Chem. 1999;274:28660–28668. doi: 10.1074/jbc.274.40.28660. [DOI] [PubMed] [Google Scholar]
- 20.Hong CS, Ji JH, Kim JP, Jung DH, Kim DH. Gene. 2001;278:193–199. doi: 10.1016/s0378-1119(01)00718-1. [DOI] [PubMed] [Google Scholar]
- 21.Marty I, Thevenon D, Scotto C, Groh S, Sainnier S, Robert M, Grunwald D, Villaz M. J Biol Chem. 2000;275:8206–8212. doi: 10.1074/jbc.275.11.8206. [DOI] [PubMed] [Google Scholar]
- 22.Thevenon D, Smida-Rezgui S, Chevessier F, Groh S, Henry-Berger J, Romero NB, Villaz V, De Waard M, Marty I. Biochem Biophys Research Com. 2003;303:669–675. doi: 10.1016/s0006-291x(03)00406-6. [DOI] [PubMed] [Google Scholar]
- 23.Enouf J, Lompre AM, Bredoux R, Bourdeau N, de La Bastie D, Levy-Toledano S. J Biol Chem. 1988;263:13922–13929. [PubMed] [Google Scholar]
- 24.Lunardi J, Dupuis A, Frobert Y, Grassi J, Vignais PV. FEBS Lett. 1989;245:223–228. doi: 10.1016/0014-5793(89)80226-1. [DOI] [PubMed] [Google Scholar]
- 25.Marty I, Robert M, Ronjat M, Bally I, Arlaud G, Villaz M. Biochem J. 1995;307:769–774. doi: 10.1042/bj3070769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carninci P, Nishiyama Y, Westover A, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Hayashizaki Y. Proc Natl Acad Sci USA. 1998;95:520–524. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuno Y, Carninci P, Okazaki Y, Tateno M, Kawai J, Amanuma H, Muramatsu M, Hayashizaki Y. Nucleic Acids Res. 1999;27:1345–1349. doi: 10.1093/nar/27.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottinelli R, Reggiani C. Prog Biophys Mol Biol. 2000;73:195–262. doi: 10.1016/s0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen AO, Shen AC, MacLennan DH, Tokuyasu KT. J Cell Biol. 1982;92:409–416. doi: 10.1083/jcb.92.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokuyasu KT, Dutton AH, Singer SJ. J Cell Biol. 1983;96:1727–1735. doi: 10.1083/jcb.96.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skeie GO. Cell Mol Life Sci. 2000;57:1570–1576. doi: 10.1007/PL00000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furst DO, Osborn M, Nave R, Weber K. J Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salanova M, Priori G, Barone V, Intravaia E, Flucher B, Ciruela F, McIlhinney RA, Parys JB, Mikoshiba K, Sorrentino V. Cell Calcium. 2002;32:193–200. doi: 10.1016/s0143416002001549. [DOI] [PubMed] [Google Scholar]
- 34.Kopaczyk K, Asai J, Allmann DW, Oda T, Green DE. Arch Biochem Biophys. 1968;123:602–621. doi: 10.1016/0003-9861(68)90181-1. [DOI] [PubMed] [Google Scholar]
- 35.Furuichi T, Shiota C, Mikoshiba K. FEBS Lett. 1990;267:85–8. doi: 10.1016/0014-5793(90)80294-s. [DOI] [PubMed] [Google Scholar]
- 36.Vermassen E, Parys JB, Mauger JP. Biol Cell. 2004;96:3–17. doi: 10.1016/j.biolcel.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 37.De Smedt H, Missiaen L, Parys JB, Henning RH, Sienaert I, Vanlingen S, Gijsens A, Himpens B, Casteels R. Biochem J. 1997;322:575–83. doi: 10.1042/bj3220575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisenberg BR, Kuda AM. J Ultrastruct Res. 1976;54:76–88. doi: 10.1016/s0022-5320(76)80010-x. [DOI] [PubMed] [Google Scholar]
- 39.Powell JA, Carrasco MA, Adams DS, Drouet B, Rios J, Muller M, Estrada M, Jaimovich E. J Cell Sci. 2001;114:3673–3683. doi: 10.1242/jcs.114.20.3673. [DOI] [PubMed] [Google Scholar]
- 40.Furuichi T, Yoshikawa S, Miyawaki A, Wada K, Maeda N, Mikoshiba K. Nature. 1989;342:32–38. doi: 10.1038/342032a0. [DOI] [PubMed] [Google Scholar]
- 41.Mignery GA, Sudhof TC, Takei K, De Camilli P. Nature. 1989;342:192–195. doi: 10.1038/342192a0. [DOI] [PubMed] [Google Scholar]
- 42.Lee JM, Rho SH, Shin DW, Cho C, Park WJ, Eom SH, Ma J, Kim DH. J Biol Chem. 2004;279:6994–7000. doi: 10.1074/jbc.M312446200. [DOI] [PubMed] [Google Scholar]
- 43.Brini M. Cell Calcium. 2003;34:399–405. doi: 10.1016/s0143-4160(03)00145-3. [DOI] [PubMed] [Google Scholar]
- 44.Malli R, Frieden M, Osibow K, Zoratti C, Mayer M, Demaurex N, Graier WF. J Biol Chem. 2003;278:44769–44779. doi: 10.1074/jbc.M302511200. [DOI] [PubMed] [Google Scholar]
- 45.Filippin L, Magalhaes PJ, Di Benedetto G, Colella M, Pozzan T. J Biol Chem. 2003;278:39224–39234. doi: 10.1074/jbc.M302301200. [DOI] [PubMed] [Google Scholar]
- 46.Rudolf R, Mongillo M, Magalhaes PJ, Pozzan T. J Cell Biol. 2004;166:527–536. doi: 10.1083/jcb.200403102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown IE, Kim DH, Loeb GE. J Muscle Res Cell Motil. 1998;19:473–477. doi: 10.1023/a:1005309107903. [DOI] [PubMed] [Google Scholar]
- 48.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 49.Ervasti JM. Costameres: the Achilles’ heel of Herculean muscle. J Biol Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 50.Sorrentino V. Biochim Biophys Acta. 2004;1742:113–118. doi: 10.1016/j.bbamcr.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 51.Bagnato P, Barone V, Giacomello E, Rossi D, Sorrentino V. J Cell Biol. 2003;160:245–253. doi: 10.1083/jcb.200208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito K, Komazaki S, Sasamoto K, Yoshida M, Nishi M, Kitamura K, Takeshima H. J Cell Biol. 2001;154:1059–1067. doi: 10.1083/jcb.200105040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuvia S, Buhusi M, Davis L, Reedy M, Bennett V. J Cell Biol. 1999;147:995–1008. doi: 10.1083/jcb.147.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kee AJ, Schevzov G, Nair-Shalliker V, Robinson CS, Vrhovski B, Ghoddusi M, Qiu MR, Lin JJ, Weinberger R, Gunning PW, Hardeman EC. J Cell Biol. 2004;166:685–696. doi: 10.1083/jcb.200406181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaisto T, Metsikko K. Exp Cell Res. 2003;289:47–57. doi: 10.1016/s0014-4827(03)00231-3. [DOI] [PubMed] [Google Scholar]


