Abstract
The mechanism of action of platinum-based anticancer drugs such as cis-diamminedichloro-platinum( II), or cisplatin, involves three sequential steps: cell entry, drug activation, and target binding. A major target in the cell, responsible for the anticancer activity, is nuclear DNA, which is packaged in nucleosomes that comprise chromatin. It is therefore important to understand the nature of platinum-DNA interactions at the level of the nucleosome. The cis-{Pt(NH3)2}2+ 1,2-d(GpG) intrastrand cross-link is the DNA lesion most commonly encountered following cisplatin treatment. We therefore assembled two site-specifically platinated nucleosomes using synthetic DNA containing defined cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-links and core histones from HeLa-S3 cancer cells. The structures of these complexes were investigated by hydroxyl radical footprinting and exonuclease III mapping. Our experiments demonstrate that the 1,2-d(GpG) cross-link alters the rotational setting of the DNA on the histone octamer core such that the lesion faces toward inward, with disposition angles of the major grove relative to the core of ξ ≈ −20° and ξ ≈ 40°. We observe increased solvent-accessibility of the platinated DNA strand, which may be caused by a structural perturbation in proximity of the 1,2-d(GpG) cisplatin lesion. The effect of the 1,2-d(GpG) cisplatin adduct on the translational setting of the nucleosomal DNA depends strongly on the position of the adduct within the sequence. If the cross-link is located at a site that is in phase with the preferred rotational setting of the unplatinated nucleosomal DNA, the effect on the translational position is negligible. Minor exo III digestion products in this substrate indicate that the cisplatin adduct permits only those translational settings that differ from one another by integral numbers of DNA helical turns. If the lesion is located out of phase with the preferred rotational setting, the translational position of the main conformation was shifted by 5 bp. Additionally, a fraction of platinated nucleosomes with widely distributed translational positions was observed, suggesting increased nucleosome sliding relative to platinated nucleosomes containing the 1,3-intrastrand d(GpTpG) cross-link investigated previously.
Introduction
Platinum-based drugs such as cisplatin, cis-diamminedichloroplatinum(II), and related agents such as carboplatin or oxaliplatin are successfully used to treat various types of cancer, including testicular, ovarian, head, neck, certain types of lung, and colorectal cancer.1–4 The drugs enter the cell by diffusion or with the aid of a transporter, become activated by forming aqua complexes, and bind to nuclear DNA, which in eukaryotes is packaged in highly condensed chromatin structures comprising nucleosome building blocks. Cisplatin reacts well with both free and nucleosomal DNA,5 showing a slight preference for the linker region of the latter.6 Because 75–90% of genomic DNA is wrapped in nucleosomes,7 intact nucleosomes are the more likely targets in vivo. The most common adduct formed by reaction of activated cisplatin with DNA is the 1,2-d(GpG) cis-diammineplatinum(II) intrastrand cross-link, with 1,2-d(ApG) and 1,3-d(GpNpG) intrastrand cross-links being formed to a lesser extent.8
By employing hydroxyl radical footprinting, we recently discovered that the 1,3-d(GpTpG) platinum adduct in nucleosomal DNA induces a well defined rotational setting of the DNA with respect to the histone core.9 The structural effect of the platinum cross-link is strong enough to override the rotational preference as defined by a strong nucleosome positioning sequence.10 Whereas the properties of nucleosomes containing the d(GpTpG) platinum adduct and other DNA lesions including N-methylpurines11 and the TT cyclobutane pyrimidine dimer12–16 have been previously investigated, the structural influence of the most abundant 1,2-d(GpG) cisplatin cross-link within a nucleosome remained to be determined.
A key question is whether the 1,2-d(GpG) cisplatin cross-link, like the 1,3-d(GpTpG) intrastrand adduct, would be able to influence the position of the DNA relative to the histone core. Recent studies suggest that such positioning of the nucleosomes on the genome may control important regulatory properties within the cell, both structural and functional.7,17–19 Determining where cisplatin cross-links are located within the nucleosome structure represents an important step in understanding their cellular recognition and processing and may contribute to the working mechanism of the drug.
In order to answer the question of how a site-specific cisplatin intrastrand 1,2-(dGpG) cross-link would affect the properties of the nucleosome, we constructed a substrate containing the adduct in a DNA duplex wrapped around core histones obtained from HeLa-S3 cancer cell chromatin. We studied these sample nucleosomes by hydroxyl radical footprinting20 and exonuclease III mapping21 to determine the rotational and translational setting of the DNA in these reconstituted particles.
Results and Discussion
Design and Synthesis of DNA Duplexes
For our experiments we chose to study DNA that has a moderate affinity for histone octamers and forms weakly positioned nucleosomes. We modified a nucleosome sequence used previously by our laboratory9 in which a single d(GpG) dinucleotide is position in the central region to allow for site-specific platination.
In order to construct radioactively labeled duplex samples we employed a two-step protocol. In the first step, we ligated six oligonucleotides of which one contained the site-specific cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link to generate a 171-/173-mer DNA duplex R1-Pt. The DNA sequence was designed with orthogonal blunt-end restriction sites close to the 3’ (EcoRV) and 5’ (AfeI) ends of the duplex. The 32P label was introduced in the second step. In order to mark exclusively the platinated coding strand of the duplex, the 3’ end was first truncated with EcoRV and dephosphorylated. Next, the coding and template strand were 5’-end-labeled with γ-32P-ATP. A second digestion with AfeI finally truncated the duplex at the 5’ end, thereby removing the label from the template strand. The remaining, single-end-labeled duplex D1-Ptc had a length of 146 bp (Scheme S1, Supporting Information) and contained a single, site-specific cis-{Pt(NH3)2}2+ 1,2-d(GpG) intrastrand cross-link within the coding strand. In order to label exclusively the template strands of the duplex for separate experiments, the sequence of the EcoRV and AfeI digestion steps was interchanged to afford D1-Ptt as the product.
Six additional labeled duplexes were constructed in the same fashion. D2-Ptc and D2-Ptt were obtained from R2-Pt, which has an identical sequence to that in D1-Ptc and D2-Ptt except that the platinated d(GpG)-dinucleotide was shifted by six base pairs from position 71.72 to position 77.78. Duplexes D1c, D1t, D2c and D2t were unplatinated reference probes. The sequences of the oligonucleotide components and final strands are provided in Scheme S2 (Supporting Information).
Construction of Platinated Nucleosomes
We prepared the coding or template strand labeled nucleosomes nD1c/t, nD1-Ptc/t, nD2c/t, and nD2-Ptc/t from the respective DNA duplexes D1c/t, D1-Ptc/t, D2c/t, and D2-Ptc/t by using a histone transfer protocol,22 followed by heat equilibration. The donor chromatin used in the histone transfer reactions was prepared from HeLa-S3 cancer cells10 and had an average length of 450 base pairs.
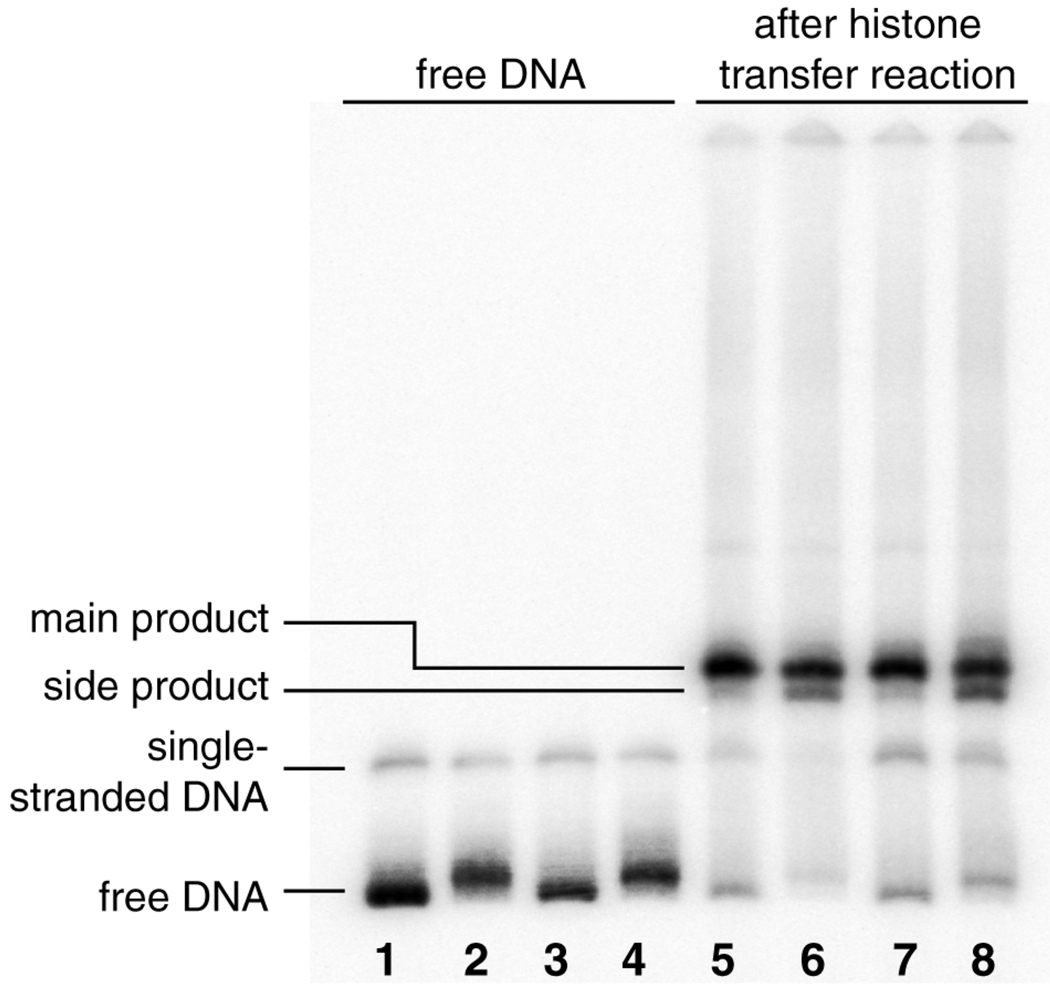
Analysis of the reactions by non-denaturing polyacrylamide electrophoresis showed a single band for nucleosomes containing unplatinated DNA. The platinated nucleosomes reproducibly contained, in addition to this main band, a faster migrating side product with an intensity of approximately 20–25% of the main band (Fig. 1). Although we have not been able conclusively to confirm the identity of this side product, there are several factors that may result in multiple bands in reconstituted nucleosome samples. First, differences in the translational setting of the DNA wrapped around the nucleosome octamer core could contribute to measurable differences in mobility during gel electrophoresis. Nucleosomes with translational settings that are asymmetric exhibit higher mobility then those with symmetrical settings.23 By comparing data from exonuclease III footprinting (see below) of unplatinated nucleosome samples nD1c/t with those for the platinated samples nD1-Ptc/t, however, we were unable to detect significant differences in the distribution of translational settings that would support this possibility.
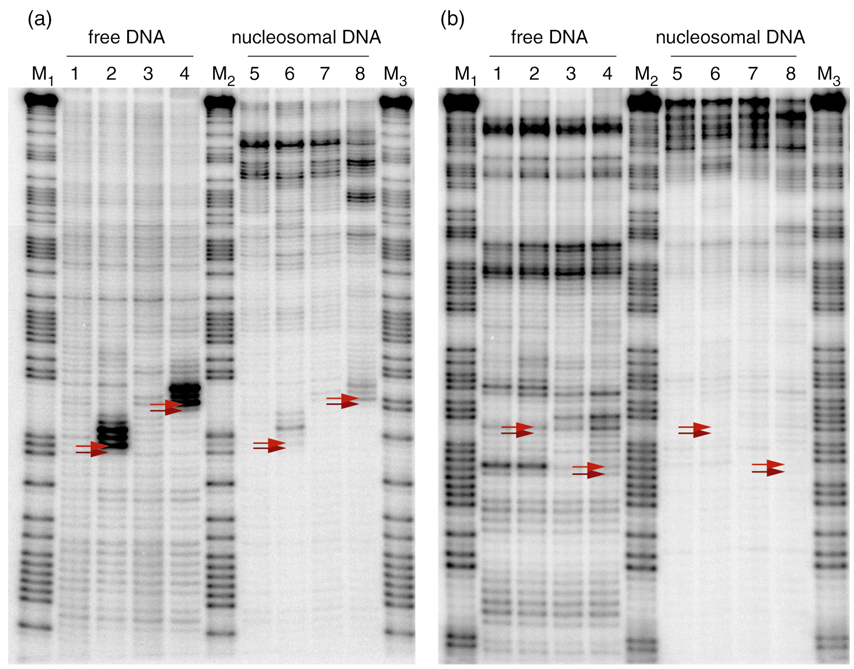
Figure 1.
Product distribution after histone transfer reaction onto the sample DNA duplexes. Lanes 1–4: free DNA. The platinated DNA samples migrate more slowly than the corresponding unplatinated samples. Lanes 5–6: after completion of transfer reaction at 4°C and heat equilibration at 45 °C, 2h. The platinated samples show a side product that migrates more rapidly than the main product. Lane 1: D1, lane 2: D1-Pt, lane 3: D2, lane 4: D2-Pt, lane 5: nD1, lane 6: nD1-Pt, lane 7: nD2, lane 8: nD2-Pt.
Second, it is possible that the lower band contains particles with incomplete histone cores. The interactions of H2a/H2b histone heterodimers with DNA is weaker then the interaction of the H32H42 tetramer. Association of the H2a/H2b heterodimers during salt-gradient dialysis, which is used to reconstitute the nucleosomes, occurs at a lower ionic strength than association of the H32H42 tetramer with DNA.24 Dissociation of H2a/H2b heterodimers from the core tetramer can also be induced by various cellular mechanisms. Incomplete particles lacking one or two H2a/H2b histone heterodimers, hexasomes or tetrasomes, also migrate more rapidly during electrophoresis.25 It is possible that the structural distortions induced by the cisplatin 1,2-intrastrand cross-link obstructs the formation of complete histone octamers. Further experiments are needed to verify this hypothesis, however.
Hydroxyl Radical Footprinting of Platinated Nucleosomes
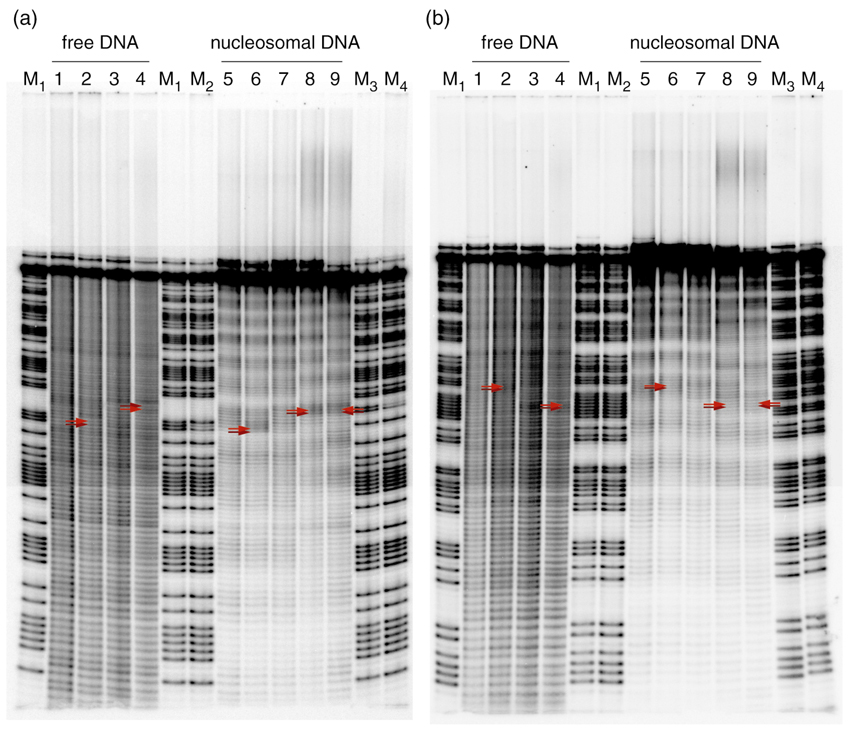
We used hydroxyl radical footprinting of the substrates described above to reveal the rotational setting of both the coding and the template strand of all nucleosomal DNA samples relative to the histone core.26,27 After reaction with hydroxyl radicals, the nucleosome samples were subsequently purified by native polyacrylamide gel electrophoresis. The resolution of this purification step was sufficiently high to separate the faster migrating side band from the main product of the platinated nucleosome samples nD2-Ptc/t. Consequently, we were able to examine the structures of the DNA from particles isolated from two bands individually (Fig. S1, Supporting Information). For nucleosome samples nD1-Ptc/t there was insufficient material and we were only able to isolate and study of material from the main band. Apart from analyzing the nucleosome samples, we examined as a point of reference the labeled coding and template strands of the free DNA duplexes D1c/t, D1-Ptc/t, D2c/t, and D2-Ptc/t. Autoradiographs from these experiments are presented in Fig. 2 and enlargements of the areas of interest are provided in Fig. S2 (Supporting Information).
Figure 2.
Autoradiographs of hydroxyl radical footprinting experiments. (a) The coding strands of the duplexes were 5’-labeled with 32P. The positions of the platinum adduct are highlighted by red arrows. Lane 1: D1c, lane 2: D1-Ptc, lane 3: D2c, lane 4: D2-Ptc, lane 5: nD1c, lane 6: nD1-Ptc, lane 7: nD2c, lane 8: nD2-Ptc (main product), lane 9: nD2-Ptc (side product), lanes M1: Maxam-Gilbert A/G reaction of D1c, lane M2: Maxam-Gilbert A/G reaction of D1-Ptc, lane M3: Maxam-Gilbert A/G reaction of D2c, lane M4: Maxam-Gilbert A/G reaction of D2-Ptc; (b) the template strands of the duplexes were 5’-labeled. The positions of the d(CpC) dinucleotide opposite the platinum adduct are highlighted by red arrows. Lane 1: D1t, lane 2: D1-Ptt, lane 3: D2t, lane 4: D2-Ptt, lane 5: nD1t, lane 6: nD1-Ptt, lane 7: nD2t, lane 8: nD2-Ptt (main product), lane 9: nD2-Ptt (side product), lanes M1: Maxam-Gilbert A/G reaction of D1t, lane M2: Maxam-Gilbert A/G reaction of D1-Ptt, lane M3: Maxam-Gilbert A/G reaction of D2t, lane M4: Maxam-Gilbert A/G reaction of D2-Ptt.
We aligned each band with the corresponding DNA sequence with the aid of A/G reference lanes from a Maxam-Gilbert gel analysis. Aligned densitometric traces of every lane together with sequence assignments are presented in Figs. S3 and S4 (Supporting Information). The positions of the cisplatin adduct were revealed by diminished double bands in the Maxam-Gilbert density traces upon comparison of the traces of D1c with D1-Ptc and D2c with D2-Ptc.
Lanes 1–4 in Figs. 2a and 2b represent the footprinting patterns of the platinated DNA duplexes in the absence of a histone core. As expected, smooth fragmentation patterns were produced because the backbones of these strands were isotropically exposed to hydroxyl radicals. Lanes 5–8 show the fragmentation patterns of the reconstituted nucleosomes that were isolated as the main product (major band, see above). Lane 9 in both autoradiographs portrays the hydroxyl radical footprint of the side products of nD2-Ptc and nD2-Ptt that were isolated from the lower band (see above).
The pattern in lanes 5–9 exhibits a periodic modulation of intensity that reflects the helical structure of the DNA packed against the histone octamer core. Hydroxyl radicals react preferentially with the C5’- and, to a lesser extent, with the C4’-hydrogen atoms of the sugarphosphate backbone.20 The hydroxyl radical footprinting pattern correlates fairly well with the solvent-accessibility of these backbone atoms. Preferential cleavage and darker bands are observed in areas where the backbone faces away from the histone core. Reduced cleavage and lighter bands are produced in areas where the backbone is packed against the histone octamer, which reduces the accessibility to hydroxyl radical cleavage.
By plotting the deconvoluted intensity of each band against its position in the sequence, we obtained hydroxyl radical cleavage curves revealing the solvent accessibility of the backbone at each nucleotide. Detailed structural information about the site-specifically platinated nucleosomal DNA was derived by overlaying the cleavage curves of both strands in a duplex. We were able to identify the disposition of the major and minor grooves of the nucleosomal DNA relative to the histone core in every sample. This disposition is described by the angle ξ defined previously.10 It approaches 0° in areas where the major groove faces toward the histone core and 180° where it points directly toward the solvent, away from the surface of the core octamer.
Influence of the Cisplatin Cross-Link on the Rotational Setting
We obtained sinusoidal hydroxyl radical cleavage curves for all samples. The position of the platinum lesion strongly influences the rotational setting of the nucleosomal DNA and overrides the slight rotational preference induced by the DNA sequence.
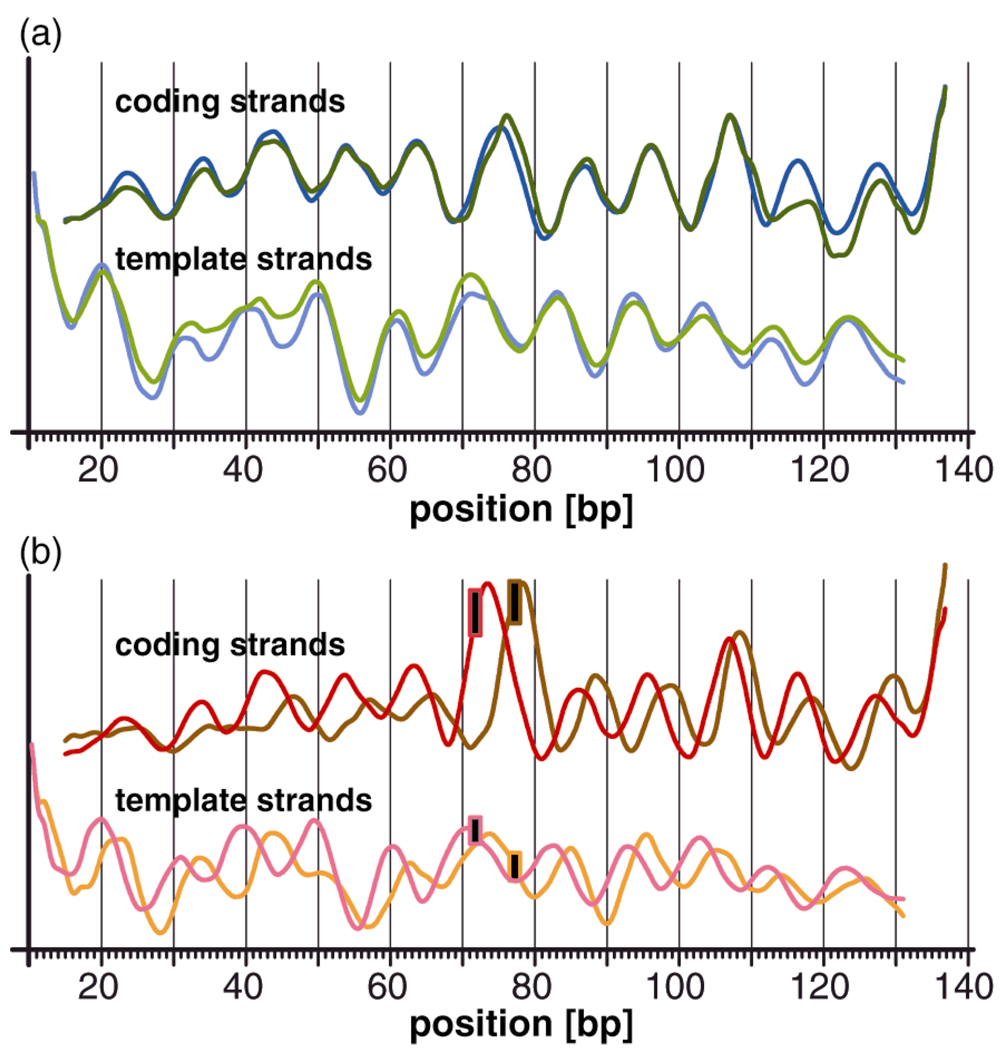
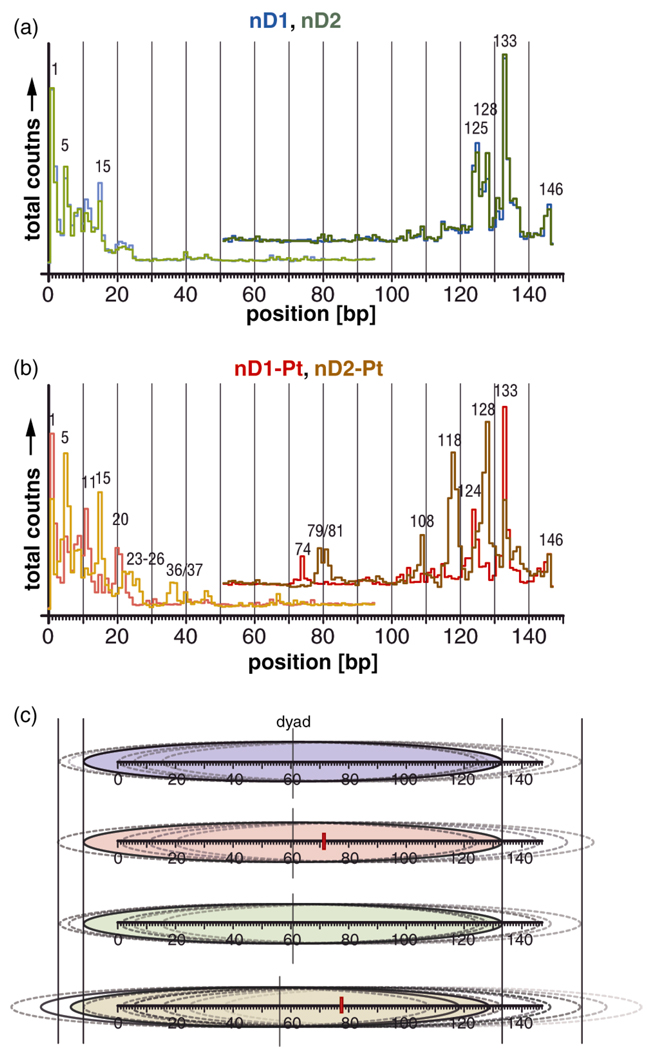
In order to investigate the extent to which the existence and position of the cisplatin adduct, rather than the position of the d(GpG) dinucleotide itself, is responsible for this effect, we superimposed the hydroxyl radical cleavage curves of the reference nucleosomes nD1 and nD2 (Fig. 3a). Shifting the unplatinated d(GpG) dinucleotide by six base pairs within our DNA sequence has almost no effect on the rotational setting of the nucleosomal DNA. The cleavage curves exhibit a phase difference of < 1 base pair (30° ± 6°) in the vicinity of the d(GpG) dinucleotide, between positions 71.72 and 77.78. The phase difference becomes negligible in the adjacent helix turns and the curves superimpose almost perfectly at sites further remote from the dinucleotide.
Figure 3.
Superposition of the cleavage curves of (a) unplatinated and (b) platinated nucleosomes. (a) The DNA sequences of the unplatinated nucleosomes nD1c/t (blue) and nD2c/t (green) are identical except for the fact that a d(GpG) dinucleotide within the bottom strand was shifted by 6 bp from position 71.72 to position 77.78. The corresponding cleavage curves are nearly congruent. (b) The platinated nucleosomes nD1-Ptc/t (red) and nD2-Ptc/t (orange/brown) have identical sequences except for the fact that the 1,2-d(GpG) cisplatin adduct, symbolized by a black rectangle, was shifted by 6 bp. The corresponding cleavage curves are out of phase by 2–4 bp. The phasing difference is approximately 125° near the cisplatin adduct.
Next, we superimposed the cleavage curves of the platinated nucleosomes nD1-Pt and nD2-Pt (Fig. 3b). Shifting the position of the cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link by six base pairs to the 3’-direction within our nucleosomal DNA sequence has a profound effect on its rotational setting. We observed a phase difference in the cleavage curves of approximately 3–4 base pairs (125 ± 20°) in immediate proximity of the lesion that gradually decreases to a phase difference of less then a quarter turn (71 ± 6°) at regions further remote from the adduct.
This finding represents the first example in which it has been possible to demonstrate that a structural difference between platinated nucleosome particles, nD1-Pt and nD2-Pt in the present case, is exclusively caused by the position of the cisplatin adduct within the sequence. It is not a consequence of the slight sequence difference between nD1-Pt and nD2-Pt that was necessary in order to accommodate the lesion at different locations along the duplex.
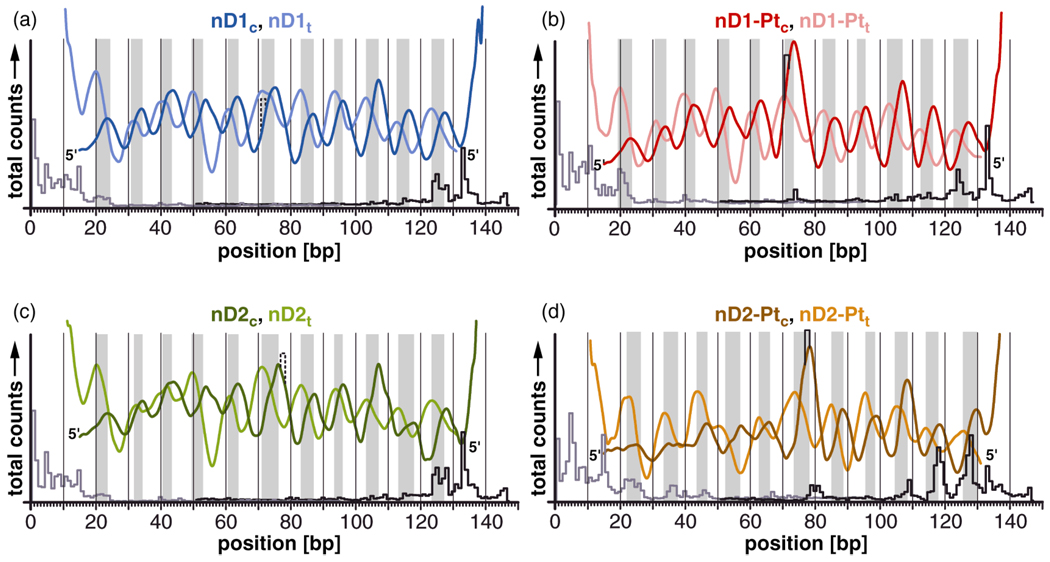
In order to develop a structural model to account for the rotational setting of DNA in our nucleosomes, we superimposed the cleavage curves of the coding and template strands for all nucleosome samples. The cis-{Pt(NH3)2}2+ 1,2-d(GpG) adduct, which is located in the major groove, rotates the duplex in both samples in such a manner that it faces more toward than away from the histone core proteins.
Within the unplatinated nucleosome nD1, the major groove faces approximately toward the histone core with ξ ≈ −50° at the position of the d(GpG) cisplatin target dinucleotide (Fig. 4a). After platination to form nD1-Pt, the structure changes slightly such that the cisplatin adduct points directly toward the histone core (ξ ≈ −20°, Fig 4b). A structural model of this conformational change is presented in Fig. S5a (Supporting Information).
Figure 4.
The cleavage curves reveal the rotational setting of the nucleosomal DNA duplexes nD1c/t, nD1-Ptc/t, nD2c/t, and nD2-Ptc/t in the nucleosome. Gray bars indicate areas in which the minor groove is exposed to the solvent and the major groove faces inward toward the nucleosome. The position of the unplatinated and platinated d(GpG) dinucleotide is indicated. Exonuclease III stop sites of each sample are presented underneath the respective hydroxyl radical cleavage curve. (a) The major groove of the unplatinated d(GpG) trinucleotide points approximately toward the histone core. (b) After platination, the major groove and the platinum adduct faces completely toward the histone core. (c) The major groove of the d(GpG) dinucleotide points sideways toward the solvent with ξ ≈ 120°. (d) After platination, the major groove faces approximately toward the histone core with ξ ≈ 40°. Cisplatin, which binds to guanine in the major groove of DNA, is in both cases buried inside the nucleosome, inaccessible to the solvent.
The major groove of the d(GpG) dinucleotide in nD2 faces approximately toward the solvent with ξ ≈ 120°. Introduction of the cisplatin cross-link in nD2-Pt changes the orientation of the major groove in such a manner that it faces approximately toward the histone core (ξ ≈ 40°). A model is again presented, in Fig. S5b (Supporting Information). The structural flexibility of the cis-{Pt(NH3)2}2+ 1,2-d(GpG) cross-link with respect to possible rotational settings seems to be greater than that of the cis-{Pt(NH3)2}2+ 1,3-d(GpTpG) cross-link described previously.10 The deviation of the rotational settings of nD1-Pt and nD2-Pt is approximately 60°, measured from the mid-point of the cross-link. By comparison, for the cis-{Pt(NH3)2}2+ 1,3-d(GpTpG) we typically observed values very close to ξ ≈ 0° with a deviation of < 15°, even in the presence of a strong nucleosome positioning sequence.9,10
The footprinting patterns of the coding strands nD1-Ptc and nD2-Ptc display significantly darker bands in direct proximity to the 1,2 cisplatin adduct compared to the unplatinated coding strands nD1c and nD2c at the same positions (Figs. S2a and S6, Supporting Information). This preferred cleavage of the coding strand is probably a consequence of the local structural perturbation induced by the cisplatin adduct, resulting in increased solvent accessibility. A chemical effect, such as cisplatin-induced DNA nicking, can be ruled out, because no such increased cleavage is observed in the proximity of the adduct for the free DNA strands D1-Ptc and D2-Ptc. Densitometric scans of the respective free DNA strands are presented in Fig. S3, rows 1–4 (Supporting Information).
We also investigated the structure of the DNA in the faster migrating side product that was isolated during the construction of nucleosome samples nD2-Ptc/t. This nucleosomal DNA also displays a sinusoidal cleavage pattern that persists throughout the entire DNA sequence. The side product contains DNA that is therefore most likely wrapped around histone core proteins – the footprinting patterns of the product isolated from the main and side bands are quite similar. The intensity modulation of the footprinting pattern has a global phase difference of 120–180° (3–6 bp) compared to that of the unplatinated samples nD2c/t (Fig. S7a, Supporting Information). The cisplatin adduct assumes a position of ξ ≈ 15° with the major groove, again pointing away from the solvent. (Fig. S7b–d, Supporting Information).
As we show below, the DNA in samples nD1c/t, nD2c/t and nD1-Ptc/t adopts a preferred single translational position with respect to the histone core. Because of the weak nucleosome positioning capability of the underlying DNA sequence, we observe additional particle conformations with other translational settings. It is not possible to separate these constructs from those having the major conformation. We wish to stress that these differently positioned nucleosomes will influence the intensity modulation pattern of our footprinting data only to a small extent. The most affected are the nD2-Ptc/t samples, which contain an increased fraction of particles with less defined translational positions.
Exonuclease III Footprinting of Damaged Nucleosomes
We studied the translational positions of the nucleosomal DNA by partial digestion of the nucleosome with exonuclease III.28 This enzyme digests double stranded DNA in a stepwise manner from the 3’ termini. Digestion of DNA shielded by the histone core occurs significantly more slowly, resulting in darker bands. This method reveals the approximate translational boundaries of the DNA that interacts with the histone core. As a reference, the coding or template strand-labeled free DNA duplexes D1c/t, D1-Ptc/t, D2c/t, and D2-Ptc/t were similarly analyzed. Autoradiographs of these experiments are presented in Fig. 5. Again, it was possible to assign each individual band to the corresponding nucleotides in the DNA sequence with the help of Maxam-Gilbert A/G reference lanes.
Figure 5.
Autoradiographs of exonuclease III mapping experiments. (a) The coding strands of the duplexes were 5’-labeled with 32P. The positions of the platinum adduct are highlighted by red arrows. Lane 1: D1c, lane 2: D1-Ptc, lane 3: D2c, lane 4: D2-Ptc, lane 5: nD1c, lane 6: nD1-Ptc, lane 7: nD2c, lane 8: nD2-Ptc, lane M1: Maxam-Gilbert A/G reaction of D1c, lane M2: Maxam-Gilbert A/G reaction of D1-Ptc, lane M3: Maxam-Gilbert A/G reaction of D2c. (b) The template strands of the duplexes were 5’-labeled. The positions of the d(CpC) dinucleotide opposite the platinum adduct are highlighted by red arrows. Lane 1: D1t, lane 2: D1-Ptt, lane 3: D2t, lane 4: D2-Ptt, lane 5: nD1t, lane 6: nD1-Ptt, lane 7: nD2t, lane 8: nD2-Ptt, lane M1: Maxam-Gilbert A/G reaction of D1t, lane M2: Maxam-Gilbert A/G reaction of D1-Ptt, lane M3: Maxam-Gilbert A/G reaction of D2t.
Lanes 1–4 in Fig. 5a and 5b show the digestion patterns of the unbound DNA duplexes. The cisplatin cross-link significantly decelerates digestion of the coding strand by exonuclease III, as revealed by a dark triple band toward the 3’-direction of the adduct (lanes 2 and 4, Fig 5a). This behavior is consistent with previous work from our laboratory.21 Cisplatin does not interfere with digestion of the template strand opposite the adduct (lanes 2 and 4, Fig 5b). Unlike the results for the coding strand, the template strands exhibits several sequence-dependent stop sites for exonuclease III, appearing as dark bands in lanes 1–4, Fig 5b.
Lanes 5–8 display the digestion pattern of the reconstituted nucleosomes. The DNA-histone interaction causes the exonuclease III to pause, an effect that results in dark bands toward the top of the gel indicating indicate the translational position of the histone core relative to the DNA duplex. These bands occur at locations different from any of the sequence-dependent stop sites. The cisplatin cross-link-induced exonuclease III pause sites are significantly diminished in the reconstituted nucleosomes because only a small fraction of the enzyme progresses that far along the DNA.
Influence of the Cisplatin Cross-Link on the Translational Setting
We used the data from the exonuclease III mapping experiments to reconstruct the translational position of the nucleosomal DNA. All nucleosome samples were positioned asymmetrically with respect to the histone core.
Exonuclear digestion of unplatinated samples nD1c and nD2c with a labeled coding strand generated a main band at position 133 (Fig. 6a), indicating that DNA in the majority of the nucleosome particles is bound to the histone core as indicated by the blue and green ovals in Fig. 6c. Digestion of the template strand is largely blocked by the histone core, as revealed when the experiment was repeated for samples nD1t and nD2t. This asymmetric translational setting protects the 3’-ends of the template strands. In addition to the main stop sites, additional bands were generated at positions 146, 128 and 125 in the coding strands as well as 5 and 15 in the template strand of both unplatinated nucleosome samples. The translational positions corresponding to these additional stop sites are demarcated by dotted ovals in Fig. 6c. The minor difference in sequence for the nucleosome samples nD1c/t and nD2c/t has no influence on the respective distribution of their translational settings. The platinated samples nD1-Ptc/t display a digestion pattern similar to those of the unplatinated nucleosomes. The major stop site in the coding strand again corresponds to position 133, and in the majority of the particles the template strand is protected from digestion. Additional stop sites occur at positions 11 and 20, whereas the bands at sites 5 and 128 are significantly reduced in intensity (Fig. 6b, red trace). Overall, the effect of the cisplatin adduct on the translational setting of the DNA in nD1-Pt is minor. The major fraction of the nucleosome core particles has the same position as in the unplatinated nucleosomes (Fig. 6c). The distances between the stop sites in the samples nD1-Ptc/t are roughly multiples of 10. The unplatinated nucleosomes show a less regular pattern in comparison. The cisplatin adduct thus appears to hold the nucleosomal DNA tightly, in a well defined rotational position, thereby permitting only translational settings that differ from one another by integral numbers of complete DNA helical turns.
Figure 6.
Exonuclease III stop sites and derived translational setting of the nucleosomal DNA. Superposition of the stop sites of (a) unplatinated and (b) platinated nucleosomes. (a) The slight sequence difference of the nucleosome samples nD1c/t (blue) and nD2c/t (green) has no effect on the exonuclease III digestion pattern. (b) The platinated nucleosomes nD1-Ptc/t (red) and nD2-Ptc/t (orange/brown) have significantly different stop positions. (c) Reconstructed translational setting of DNA in the nucleosomes nD1 (blue, top), nD1-Pt (red), nD2 (green), and nD2-Pt (brown, bottom). The DNA is represented as graduated horizontal bars. The major conformations in each sample are indicated by colored ovals and minor conformations, by non colored ovals. The positions of the cisplatin adducts are indicated by red rectangles. In addition, the approximate pseudodyad position of the major conformation in each sample is indicated. The translational setting of the main conformation in nD2-Pt is shifted by 5 bp from that in all other nucleosome samples. An increased fraction of particles with widely distributed translational positions is evident.
The data obtained for samples nD2-Ptc/t are different. The main stop site in the coding strand was shifted by five bp to position 128, and a second strong band appeared at position 118, one complete helical turn away. Correspondingly, in a significant fraction of the particles, the template strand is protected against digestion (Fig. 6b, orange/brown trace). The related translational conformations are shown as solid-bordered ovals in Fig 6c. Additional minor bands appeared at positions 146, 133 and 108 in the coding strands and at 5, 15, 23–26 and 36–37 in the template strands. Overall, there is a greater distribution of translational settings than observed in the other nucleosome samples.
The following factors may contribute to this effect. First, the position of the cisplatin adduct in nD2-Pt at position 77.78 may abrogate to some degree a sequence inherent preference for specific translational settings, resulting in the increased structural variability. Second, the cisplatin adduct may reduce the strength of the DNA-histone interaction and facilitate “nucleosome sliding,”29 increasing the possibility that the histone core may be translocated by the action of exonuclease III. Third, reduced interaction of the oligonucleotide duplex ends with the histone core may alter the association/dissociation dynamics of DNA and the histone core.30 This behavior could increase the probability that exonuclease III would digest DNA into the particle, rather than dissociating at the border of the nucleosome.28 Experiments beyond the scope of the present study, such as site-directed hydroxyl radical footprinting,31 might provide further information about these observations.
In summary, depending on its position within the nucleosome, a cisplatin adduct can have the following effects on the distribution of translational settings. When located in phase, that is at a site that does not require any adjustment of the rotational setting of the DNA, the cis-{Pt(NH3)2}2+ 1,2-intrastrand d(GpG) cross-link locks the DNA tightly into that rotational position. As a consequence, there is only a narrow cluster of possible translational settings that are separated from each other by multiples of 10 base pairs. Alternatively, if positioned out of phase with the natural rotational setting of the DNA, we observe a shift in the translational setting by half a helical turn and a strongly increased variability of possible translational settings.
Structural Differences Between 1,2- and 1,3-Intrastrand Cisplatin Cross-Links and Their Implications
We previously investigated in detail the effect on the nucleosome structure of cis-{Pt(NH3)2}2+ 1,3-d(GpTpG) intrastrand cross-links and learned that it defines the rotational setting and strongly affects the translational setting of DNA in a nucleosome particle. The present results clearly demonstrate that the more abundant cis-{Pt(NH3)2}2+ 1,2-d(GpG) intrastrand cross-link also influences the DNA conformation within a nucleosome, but in a manner that depends on its position within the DNA sequence.
Both the 1,2- and 1,3-intrastrand cross-links enforce a DNA rotational setting such that the lesion, positioned in the major groove, faces toward the histone core. The structural flexibility of nucleosomes having such a site-specific 1,3-intrastrand cross-link is less than that observed for the 1,2-intrastrand cross-link, however. Nucleosomes with a 1,3-adduct have rotational settings very close to ξ ≈ 0° irrespective of where it is located within the sequence. In these nucleosomes, the deviation in rotational orientations of the cisplatin lesion are < 15°, even when a strong, counteracting positioning sequence was employed. In the present study, the rotational settings of nD1-Pt and nD2-Pt were determined to differ by approximately 60° at the respective platinum adduct locations.
Two synergistic effects were responsible for the overall structural effect of the 1,3-d(GpTpG) intrastrand cross-link. The position of the unplatinated d(GpTpG) trinucleotide in the oligonucletide sequence itself had an influence on the rotational setting of the DNA. The 1,3-d(GpTpG) cisplatin cross-link significantly enhanced this effect. In the present case, however, 1,2-d(GpG) cisplatin adduct is exclusively responsible for the observed structural changes. A positional shift of the unplatinated dinucleotide has virtually no effect on the nucleosome structure.
The 1,2-intrastrand d(GpG) lesion unwinds the nucleosomal DNA only very slightly in the proximity of the lesion. This small effect is compensated in the adjacent helical turns and does not induce global unwinding of nucleosomal DNA, as we observed for the 1,3-intrastrand d(GpTpG) cisplatin adduct.10 This finding parallels that in previous studies with free DNA, in which we determined an unwinding angle of just 13° for the 1,2-intrastrand d(GpG) cisplatin cross-link compared to a value of approximately 23° for a 1,3-intrastrand lesion.32
The accessibility of nucleosomes to hydroxyl radical cleavage on the coding strand near the 1,2-intrastrand d(GpG) cisplatin cross-link is significantly enhanced by comparison to results for the unplatinated reference nucleosomes at the same position. This increased accessibility most likely reflects a local structural perturbation of the nucleosomal DNA induced by the cisplatin adduct. Possibly, the 1,2-intrastrand cisplatin cross-link cannot be as well accommodated within the nucleosome structure as the 1,3-intrastrand cross-link. Moreover, the structural distortion induced by the 1,2 cisplatin lesion appears to obstruct the assembly of complete histone octamers, resulting in formation of a side product with a greater gel electrophoretic mobility than that of the main product. We did not observe such an effect either with unplatinated DNA or with DNA carrying a site-specific 1,3-intrastrand cisplatin cross-link.
The 1,2-d(GpG) cisplatin intrastrand cross-link has a less profound effect on the translational setting at position 71.72 than the 1,3-d(GpTpG) adduct at position 71.73 in an otherwise identical context.9 Whereas the former alters the distribution of translational positions only slightly, the latter induces formation of two main nucleosome structures with translational settings that differ from those of the respective unplatinated nucleosomes. On the other hand, the 1,2-d(GpG) intrastrand cisplatin cross-link at position 77.78 and the 1,3-d(GpTpG) adduct at position 77.79 both strongly affect the translational settings and position the DNA in a highly asymmetric fashion relative to the histone octamer core. The 1,2-cross-link at site 77.78 generates two main nucleosome settings with DNA overhangs, starting from either position, of 118 or 128 base pair, in addition to a variety of minor conformations, suggesting increased “nucleosome sliding” and reduced histone-DNA interactions. The 1,3-adduct, by comparison, locks the DNA mainly into a single asymmetric translational setting.
The 1,2-d(GpG) and 1,3-d(GpNpG) cisplatin instrastrand cross-links are recognized and repaired via the nucleotide excision repair (NER) pathway. The nucleosome inhibits NER of both DNA lesions significantly by comparison to free DNA bearing the same platinated sequence. Whereas the nucleosome reduces the repair rate of a 1,3-d(GpTpG) cisplatin adduct by about a factor of 10, a nucleosome sample containing a 1,2-d(GpG) cross-link is repaired only approximately three times less efficiently then the corresponding free DNA.33
The details by which the NER machinery restores nucleosomal DNA that has been damaged, including the sequence of binding by the various proteins of the repair complex as well as their specific functions, are not completely clear at present. According to a current model, first step involves recognition of a structural distortion at the undamaged strand, probably by XPC. This initial binding apparently aids in location of the actual site of damage.34 Damage recognition induces then chromatin remodeling, dissociation of the DNA from the histone core and excision of the damaged DNA.35
In light of our current results, there are at least three possible factors that might account for the significant disparity in the ability of a nucleosome to inhibit the repair of a 1,2-d(GpG) cisplatin cross-link compared to the 1,3-d(GpTpG) cisplatin adduct. First, the greater structural flexibility observed for the nucleosomal 1,2-cisplatin adduct could improve access to the first response protein of the NER pathway, presumably XPC. Second, the local structural perturbation of the nucleosomal DNA induced only by the 1,2-cisplatin adduct, could enable preferred recognition by the NER machinery. Finally, a possible reduction in the stability of the histone octamer core triggered by the 1,2-lesion may further assist nucleosome remodeling needed for NER.
According to recent studies, the majority of the genomic nucleosomes are well-positioned, 7,19,36 only a subset of which are directly determined by specific nucleosome positioning sequences.37 Such sequences are encountered in promoter regions and at translation start sites.7,17,38 These strongly positioned nucleosomes indirectly determine the translational location of other nucleosomes by effects like steric hindrance.7,17,38 Positioned nucleosomes perform many important regulatory functions, such as attenuating or increasing gene expression,17 and they fulfill structural tasks, such as protecting the centromere.7 The strong positioning effects of the major cisplatin 1,2-d(GpG) and 1,3-d(GpTpG) lesions that we observe at the single nucleosome level may influence regulatory mechanisms in the cell that are linked to chromatin structure may even disrupt the local geometry in proximity to the lesion.
The ability of DNA to position itself on the histones is achieved by curvature of the double helix that predefines the binding face that interacts with octamer core. The intrinsic DNA sequence can promote DNA bending and preferred rotational and translational settings of the nucleosome by subtle additive effects of dinucleotide steps at specific positions.7,39 The 1,2-d(GpG) cisplatin adduct induces, in a manner similar to that previously encountered for the 1,3-d(GpTpG) adduct, a kink toward the major groove with an angle of approximately 30°.32 This structural motif has a dramatic effect on the overall curvature of the DNA and explains why these platinum adducts have such a strong influence on the nucleosome structure.
Summary and Outlook
In this work we investigated the effect of a 1,2-d(GpG)-cisplatin intrastrand cross-link on the rotational and translational position of nucleosomal DNA by hydroxyl radical footprinting and exonuclease III mapping. In comparison to structural studies of nucleosomes containing the related 1,3-d(GpTpG)-cisplatin cross-link, we identified some similarities but also major differences. Most interesting is the hyper-reactivity of the DNA toward hydroxyl radicals near the d(GpG) cross-link which, together with greater structural flexibility, indicates a distortion of the DNA that was not evident for the 1,3-d(GpTpG) cross-link. The two different cisplatin adducts also influence the translational setting in a different fashion.
Our nucleosome studies to date have been focused on cis-{Pt(NH3)2}2+ cross-links that are induced by the drugs cisplatin or carboplatin. The more recently approved platinum-based anticancer drugs oxaliplatin and tetraplatin, as well as compounds currently under development, react with DNA to form adducts that carry pendant ligands different from ammonia. The effect of these so-called “carrier ligands,” which are present in cis-{Pt(R,R-DACH)}2+ cross-links, where R,R-DACH is the R,R-stereoisomer of 1,2-diaminocyclhexane present in oxaliplatin, or in DNA adducts of monofunctional complexes such as {Ptpy(NH3)2)}2+, on the nucleosome structure is unknown and will be subject of future research. Additional studies will also endeavor to elucidate the biological consequences of nucleosomal platination, including damage recognition, repair, and interference with transcription.
Experimental Section
Materials and Methods
Chemical reagents and solvents were purchased from commercial sources. Cisplatin was synthesized from K2[PtCl4] according to a published procedure.40 γ-32PATP having a specific activity of at least 6000 Ci/mmol was obtained from Perkin Elmer (Boston, MA) and used on the day of arrival. DNA synthesis was conducted with an Applied Biosystems 392 DNA/RNA synthesizer on a 1 µmol scale by using standard phosphoramidite chemistry. The oligonucleotides were deprotected overnight with a saturated ammonium hydroxide solution at 60°C and dried with an Eppendorf Vacufuge. Chromatographic analysis and preparative HPLC purification of oligonucleotides was performed on a Varian Star HPLC system with a Hydopore-5-AX 10 × 100 mm SAX column. Buffer A contained 5 M urea and 20 mM aqueous sodium phosphate, pH 6.5, and buffer B contained 5 M urea, 1 M (NH4)2SO4, in 20 mM aqueous sodium phosphate buffer. The rb values, mol-Pt bound per nucleotide strand, were determined by quantification of platinum using flameless atomic absorption spectrophotometry (Perkin-Elmer AAnalyst 300 system) and of DNA by UV/vis spectroscopy (Varian Carey 1E-spectrometer). The specific extinction coefficient of each oligonucleotide was estimated as the sum of the individual extinction coefficients of the nucleotides in the sequence.41,42 MALDI measurements were conducted on a Bruker Omniflex mass spectrometer at the Department of Chemistry Instrumentation Facility (DCIF), Massachusetts Institute of Technology (MIT) or on an Applied Biosystems Model Voyager DE-STR mass spectrometer at the MIT CCR Biopolymers Laboratory. Enzymes were obtained from New England Biolabs (NEB, Ipswich, MA) unless otherwise specified. Enzymatic reactions were carried out in reaction buffers provided by the supplier unless otherwise noted. Pelleted HeLa-S3 cells were obtained from the National Cell Culture Center (NCCC; Minneapolis, MN). Beckmann-Coulter centrifuges Avanti J-25 or Optima L were used for chromatin preparation. For chromatin sonication, a Branson Digital Sonifier was utilized. All steps involving proteins, such as chromatin isolation, purification, and histone transfer, were carried out at 4 °C or on ice if not otherwise noted. Dialysis membranes were obtained from Spectra/Por and treated with hot 50 mM EDTA-solution followed by treatment with 10 mM mercaptoethanol and several washes with high purity water with a conductivity of > 12 MΩ before use to remove trace divalent cations. Denaturing DNA polyacrylamide gel electrophoresis was performed on a Life Technologies S2 sequencing gel electrophoresis apparatus. For native DNA and nucleosome gel electrophoresis, a Protean II xi cell from Biorad was used. Fluorescent agarose electrophoresis gels were documented on a BioRad Fluor-S MultiImager. Electrophoresis gels with radioactive samples were dried and documented on a Storm 840 Phosphorimager system from Amersham or exposed wet to a Kodak Biomax MS film. Radioactive samples were quantified on a Beckman LS 6500 scintillation counter. FPLC was performed with asystem from Amersham (P1 peristaltic pump and a programmable Frac-100 fraction collector).
Synthesis of Platinated Oligonucleotides
First, eight oligonucleotides a, b1, b2, c, d, e1, e2 and f, the sequences of which are provided in Scheme S2 (Supporting Information) were chemically synthesized and purified by preparative PAGE in the presence of 7.5 M urea. The oligonucleotides were ethanol precipitated and desalted with SepPak C18 cartridges. The purified strands were characterized by MALDI-ToF mass spectrometry and quantified by UV/vis spectroscopy. The two 21-mer oligonucleotides b1 and b2 were converted into b1-Pt and b2-Pt by introduction of specific cis-{Pt(NH3)2}2+ 1,2-d(GpG) intrastrand cross-links according to published procedures.43 The platinated strands were purified by semi-preparative HPLC and the quality of the fractions was monitored by analytical HPLC. The pure fractions were pooled and concentrated by ethanol precipitation. The rb values of the 21mers b1-Pt and b2-Pt were determined to be 0.98/21 and 1.05/21, respectively, corresponding to one cisplatin adduct per oligonucleotide.
Preparation of DNA Duplexes
The oligonucleotides b1, b1-Pt, b2, b2-Pt, c, d, e1, and e2 were 5’-phosphorylated with T4 polynucleotide kinase under standard conditions. A 500 pmol quantity of each appropriate phosphorylated strand, 1000 pmol of either 21-mer b1, b1-Pt, b2 or b2-Pt, and oligonucleotides a (500 pmol) and e (1000 pmol) were annealed in buffer (100 mM NaCl, 70 mM Tris/HCl, pH 7.5, 10 mM MgCl2), by applying a temperature gradient from 90 °C to 4 °C over 3 h and ligated in situ (50 mM NaCl, 60 mM Tris/HCl, 10 mM MgCl2, 10 mM DTT, 1.5 mM ATP, 25 µg/mL BSA, 10 U/µL T4 DNA ligase, 48 h, 16 °C) to give the 171/173mer duplex DNA strands R1, R1-Pt, R2 and R2-Pt detailed in Scheme S1 (Supporting Information). The sample strands were then phenol extracted, ether extracted, ethanol precipitated, and purified on a 6% denaturing polyacrylamide gel. The gel pieces were extracted with 50 mM NaCl, 10 mM Tris/HCl, 1mM EDTA, ethanol precipitated and further purified by gel filtration chromatography (Probequant G-50 cartridges, GE Healthcare). The eluted DNA was brought to 150 mM NaCl, annealed by applying a temperature gradient from 90 °C to 4 °C over 3 h and concentrated with Nanosep MWCO 10kD cartridges (Pall Corporation).
In order to label exclusively the coding strands of the duplexes, 100 pmol of all duplexes were overdigested with EcoRV (20 U) overnight in separate reactions. The strands were dephosphorylated with Antarctic phosphatase (10 U, 1 h), and the enzymes were removed by phenol extraction and ethanol precipitation. Next, 20 pmol of the truncated strands were phosphorylated with 250 µCi γ-32P-ATP per reaction by using T4 polynucleotide kinase (20 U, 37 °C, 2 h). The reactions were brought to 20 mM EDTA, the enzyme was heat deactivated and the EDTA removed by gel filtration (Microspin G-25 cartridges, GE Healthcare). Finally, the strands were overdigested with AfeI (20 U, 4 h); the reactions brought to 20 mM EDTA and purified Micropure EZ cartridges (Millipore), ethanol precipitation and subsequent denaturing PAGE (8%, 20:1 acrylamide/bisacrylamide). Single-end radiolabeled (2.8 – 4.2 × 106 cpm) 146-mer duplexes (D1c, D1-Ptc, D2c and D2-Ptc, sequences in Scheme S2, Supporting Information) were obtained, which were stored frozen at concentrations no higher than 2 × 104 cpm/µL to reduce nicking by autoradiolysis. In order to label exclusively the primer strands of the duplexes, the sequence of the EcoRV and AfeI digestions in the protocol above was swapped to yield the sequence-identical duplexes D1t, D1-Ptt, D2t and D2-Ptt in similar quantities.
Characterization of DNA Duplexes
The resulting duplexes were analyzed by Maxam- Gilbert A/G-reactions44 to confirm the sequence and position of the cisplatin adducts. Before electrophoretic analysis of the Maxam-Gilbert fragments, the samples were treated with 0.2 M KCN at 50 °C overnight to remove the platinum adducts. Residual KCN was separated by ethanol precipitation and redissolution of the DNA. This procedure removed DNA-bound cis-diammineplatinum(II) moiety completely, resulting in significantly increased resolution of the sequencing reactions. Aligned profiles of the Maxam-Gilbert traces were generated with the program SAFA V1.1,45 which is available free of charge from https://simtk.org/home/safa/. Approximately 120 bands were resolved that corresponded to the designed sequence. The position of the cisplatin lesion was identified by the missing double bands in the platinated samples when compared to the corresponding unplatinated reference strands.
Nucleosome Reconstitution
A 7 × 105 – 1 106 cpm quantity of D1c, D1-Ptc, D2c and D2-Ptc or D1t, D1-Ptt, D2t and D2-Ptt was combined with donor chromatin from HeLa-S3 cells10,22 having an average length of 2–3 nucleosomes. The reaction was brought to 50 µL volume in 10 mM Tris, 2 M NaCl, 5 mM DTT, 0.5 mM PMSF, 0.2 mM EDTA, 0.01% (v/v) Nonidet-P40, pH 7.5 and dialyzed for two days in microdialysis buttons that were constructed from the cap of a 500 µL PCR reaction tube and a 1 cm2 Spectra/Por MWCO 6–8 kDa dialysis membrane.46 The membrane was treated with EDTA and soaked in a 2 M NaCl solution prior to use. The buttons were first dialyzed at 4 °C for 1 h against 20 mL of a high-salt buffer TB-1 containing 10 mM Tris, 2 M NaCl, 1 mM DTT, 0.5 mM PMSF, and 0.01 % (v/v) Nonidet-P40 at pH 7.5. The buffer TB-1 was transferred together with the buttons into a secondary dialysis tube (Spectra/Por MWCO 6–8 kDa). The secondary tube was dialyzed against 1.5 L of a low salt buffer TB-2 (10 mM Tris, 0.2 mM PMSF, pH 7.5, 4 °C) overnight. The coding strand labeled nucleosome samples nD1c, nD1-Ptc, nD2c and nD2-Ptc or template strand labeled nucleosome samples nD1t, nD1-Ptt, nD2t and nD2-Ptt were recovered from the dialysis buttons and equilibrated at 45 °C for 2 h.
Hydroxyl Radical Footprinting of Nucleosomes
Hydroxyl radical footprinting27 and nucleosome purification were performed as follows. A 7.5 µL aliquot of 10 mM sodium ascorbate, 7.5 µL of a solution containing 1 mM of Fe(NH4)2(SO4)2·6H2O and 2 mM of EDTA, and 7.5 µL of a 0.12 % (w/w) H2O2 solution were pre-mixed and added within 5 s to 52.5 µL of the nucleosome sample (approx. 5 × 105 cpm). The reaction was incubated for 120 s at room temperature and stopped by addition of 8 µL 50 % (v/v) glycerol and 2 µL of a 500 mM EDTA solution. The samples were immediately loaded onto a cooled 4.5 % native gel (37.5:1 acrylamide/bisacrylamide, thickness 1.6 mm, 0.3 × TBE) and run for 3 h at 200 V. The gel was wrapped in plastic and exposed to a Biomax MS film for 25 min. The bands were cut out using the exposed film as a template. An additional band with an intensity of ca. 25% of the main band that migrated faster than the main band was visible for samples nD2-Ptc and nD2-Ptt. It was separated from the main band and worked up individually (Fig. S1, Supporting Information)
The nucleosomes were isolated from the gel by electroelution with a Millipore Centrilutor Microeluter system into Centricon 3 K microfiltration cartridges (0.3 × TBE, 200 V, 2 h). The sample volumes were reduced and the buffer was exchanged against 300 µL TE buffer by centrifugation. The histones were digested afterward by treatment with 20 ng/mL of Proteinase K and 0.1 % (w/v) SDS for 3 h at 50 °C and removed by two subsequent phenol extractions. Residual phenol was then removed by ether extraction followed by ethanol precipitation. The pellets were dissolved in 40 µL of 0.2 M KCN and shaken at 50 °C overnight to remove the platinum adduct. The samples were then ethanol precipitated twice and the pellet was rinsed with 70% ethanol. The purified DNA was finally taken up in TE-Buffer and stored at 5 °C. Typically, 8 × 104 – 2 105 cpm of radiolabeled DNA was recovered. As a reference, we conducted hydroxyl radical footprinting with all unbound sample duplexes D1c, D1-Ptc, D2c, D2-Ptc and D1t, D1-Ptt, D2t, and D2-Ptt as previously described.10,27
A 3.5 × 104 cpm aliquot of each footprinting sample was pipetted into new reaction tubes and completely dried by vacuum centrifugation. The dried samples were taken up in 2.5 µL of freshly deionized formamide containing 10 mM EDTA and heated to 90 °C for 120 s. Immediately after heating, the samples were cooled on ice and loaded onto an 8% denaturing sequencing gel (0.4 mm thickness, 25:1 acrylamide/bisacrylamide, 7.5 M urea, 1 × TBE). The electrophoresis was run for 2–3 h at 65 W until the bromphenol blue marker (run in an extra lane) was about 4 cm above the bottom edge of the gel. Gels were blotted onto Whatman paper, dried, and exposed for three days to a phosphor screen.
Exonuclease III Digestion Analysis of Nucleosomes
A 5 µL aliquot of each nucleosome sample (approx. 4 × 104 cpm) was diluted to 50 µL and brought to 10 mM 1,3-bis (tris(hydroxyl-methyl) methylamino)propane/HCl , pH 7 0 ,10 mM MgCl2 and 1 mM dithiothreithol. Exonucleoase III (1 U) was added and the mixture was incubated for 300 s at 25°C. The reaction was quenched by addition of 50 µL of a solution containing 40 mM EDTA-Na2, 0.8 % (w/w) SDS, 50 µg/mL salmon sperm DNA, 5.5 mg/mL salmon sperm DNA. The samples were heated to 50 °C and ethanol precipitated. The pellets were dissolved in 45 µL of 0.2 M KCN, shaken at 50 °C overnight to remove the platinum adduct, ethanol precipitated twice, washed with 70 % (v/v) ethanol, finally taken up in TE-Buffer and stored at 5 °C. Analysis by 8% denaturing sequencing gel electrophoresis was performed as described above.
Data Collection and Processing
The gel images were processed by the footprinting analysis software SAFA V1.1.45 First, the gel was horizontally and vertically aligned. Next, each band of the Maxam-Gilbert reference lane was aligned with the known DNA sequence. The program uses this information as a starting guess to conduct a least squares fit of a sum of Lorentzians to the intensity profiles derived from the footprinting data. Each identified band was assigned a relative value derived by integration of the fitted Lorentzian. The individual values for hydroxyl- and exonuclease footprinting experiments are shown in Tabs. S1 and S2 (Supporting Information).
Hydroxyl radical cleavage curves were calculated from these values by applying a moving average with a window of five bp and normalized. The areas between nearer maxima of the hydroxyl radical cleavage curves of the complementary strands of a DNA duplex indicate where the minor groove was exposed to solvent on the nucleosome surface. At the remaining locations,the major groove is solvent-exposed. The resulting exonuclease cleave curves were normalized. The aligned densitometric profiles Figs. S3 and S4, Supporting Information, were also determined with SAFA. All traces were consistently color-coded according their sequence as follows: (n)D1, blue; (n)D1-Pt, red; (n)D2, green; (n)D2-Pt, orange/brown.
Acknowledgements
This work was generously supported by NIH grant CA34992. Part of the experimental work was performed at the University of Michigan, Dept. of Chemistry. M. Ober thanks Prof. Nils G. Walter for the opportunity to work in his laboratory as a visiting scholar and the Alexander von Humboldt Foundation, Germany, for support through a Feodor-Lynen Fellowship.
Footnotes
Supporting Information Available: Sequences and assembly of sample duplexes, autoradiographs of preparative nucleosome purification gels, enlargement of autoradiographs, densitometric scans of footprinting reactions and sequence alignment, structural models of the nucleosome samples, deconvoluted 32P signals of hydroxyl radical and exonuclease footprinting (tables).
References
- 1.Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 2.Wong E, Giandomenico CM. Chem. Rev. 1999;99:2451–2466. doi: 10.1021/cr980420v. [DOI] [PubMed] [Google Scholar]
- 3.Misset JL, Bleiberg H, Sutherland W, Bekradda M, Cvitkovic E. Crit. Rev. Oncol. Hematol. 2000;35:75–93. doi: 10.1016/s1040-8428(00)00070-6. [DOI] [PubMed] [Google Scholar]
- 4.Wang D, Lippard SJ. Nat. Rev. Drug Discovery. 2005;4:307–319. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 5.Lippard SJ, Hoeschele JD. Proc. Natl. Acad. Sci. U. S. A. 1979;76:6091–6095. doi: 10.1073/pnas.76.12.6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galea AM, Murray V. Biochem. Biophys. Res. Commun. 2002;1578:142–152. [Google Scholar]
- 7.Segal E, Fondufe-Mittendorf Y, Chen LY, Thåström A, Field Y, Moore IK, Wang JPZ, Widom J. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blommaert FA, van Dijk-Knijnenburg HCM, Dijt FJ, den Engelse L, Baan RA, Berends F, Fichtinger-Schepman AMJ. Biochemistry. 1995;34:8474–8480. doi: 10.1021/bi00026a031. [DOI] [PubMed] [Google Scholar]
- 9.Danford AJ, Wang D, Wang Q, Tullius TD, Lippard SJ. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12311–12316. doi: 10.1073/pnas.0506025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ober M, Lippard SJ. J. Am. Chem. Soc. 2007;129:6278–6286. doi: 10.1021/ja0706145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SS, Smerdon MJ. J. Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 12.Kosmoski JV, Ackerman EJ, Smerdon MJ. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10113–10118. doi: 10.1073/pnas.181073398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Svedružić žM;, Wang C, Kosmoski JV, Smerdon MJ. J. Biol. Chem. 2005;280:40051–40057. doi: 10.1074/jbc.M509478200. [DOI] [PubMed] [Google Scholar]
- 14.Thoma F. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schieferstein U, Thoma F. EMBO J. 1998;17:306–316. doi: 10.1093/emboj/17.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schieferstein U, Thoma F. Biochemistry. 1996;35:7705–7714. doi: 10.1021/bi953011r. [DOI] [PubMed] [Google Scholar]
- 17.Ioshikhes I, Bolshoy A, Derenshteyn K, Borodovsky M, Trifonov EN. J. Mol. Biol. 1996;262:129–139. doi: 10.1006/jmbi.1996.0503. [DOI] [PubMed] [Google Scholar]
- 18.Kornberg RD. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 20.Balasubramanian B, Pogozelski WK, Tullius TD. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tullius TD, Lippard SJ. J. Am. Chem. Soc. 1981;103:4620. [Google Scholar]
- 22.Utley RT, Owen-Hughes TA, Juan LJ, Côté J, Adams CC, Workman JL. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 23.Pennings S. Methods Enzymol. 1999;304:298–312. doi: 10.1016/s0076-6879(99)04018-5. [DOI] [PubMed] [Google Scholar]
- 24.Widom J. Q. Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 25.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM. Mol. Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 26.Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD. Methods Enzymol. 1991;208:380–413. doi: 10.1016/0076-6879(91)08021-9. [DOI] [PubMed] [Google Scholar]
- 27.Vitolo JM, Thiriet C, Hayes JJ. Current Protocols in Molecular Biology. 1999:21.24.21–21.24.29. doi: 10.1002/0471142727.mb2104s48. [DOI] [PubMed] [Google Scholar]
- 28.Prunell A. Biochemistry. 1983;22:4887–4894. doi: 10.1021/bi00290a004. [DOI] [PubMed] [Google Scholar]
- 29.Becker PB. EMBO J. 2002;21:4749–4753. doi: 10.1093/emboj/cdf486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li G, Levitus M, Bustamante C, Widom J. Nat. Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 31.Flaus A, Luger K, Tan S, Richmond TJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bellon SF, Coleman JH, Lippard SJ. Biochemistry. 1991;30:8026–8035. doi: 10.1021/bi00246a021. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Hara R, Singh G, Sancar A, Lippard SJ. Biochemistry. 2003;42:6747–6753. doi: 10.1021/bi034264k. [DOI] [PubMed] [Google Scholar]
- 34.Buterin T, Meyer C, Giese B, Naegeli H. Chem. Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 35.Thoma F. EMBO J. 1999;18:6585–6598. doi: 10.1093/emboj/18.23.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Science. 2006;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 37.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ercan S, Lieb JD. Nat. Genet. 2006;38:1104–1105. doi: 10.1038/ng1006-1104. [DOI] [PubMed] [Google Scholar]
- 39.Satchwell SC, Drew HR, Travers AA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 40.Dhara SC. Indian Journal of Chemistry. 1970;8:193–194. [Google Scholar]
- 41.Cantor CR, Warshaw MM, Shapiro H. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 42.Fasman GD. Optical Properties of Nucleic Acids, Absorption, and Circular Dichroism Spectra. 3 ed. Vol. 1. Cleveland (Ohio): CRC Press; 1975. [Google Scholar]
- 43.Wei M, Cohen SM, Silverman AP, Lippard SJ. J. Biol. Chem. 2001;276:38774–38780. doi: 10.1074/jbc.M106374200. [DOI] [PubMed] [Google Scholar]
- 44.Ambrose BJB, Pless RC. Methods Enzymol. 1987;152:522–538. doi: 10.1016/0076-6879(87)52059-6. [DOI] [PubMed] [Google Scholar]
- 45.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thåström A, Lowary PT, Widom J. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]