Abstract
Y family DNA polymerases have proven to be remarkably diverse in their functions and in strategies for replicating through DNA lesions. The structure of yeast Rev1 ternary complex has revealed the most radical replication strategy, where the polymerase itself dictates the identity of the incoming nucleotide, as well as the identity of the templating base. We show here that many of the key elements of this highly unusual strategy are conserved between yeast and human Rev1, including the eviction of template G from the DNA helix and the pairing of incoming dCTP with a surrogate arginine residue. We also show that the catalytic core of human Rev1 is uniquely augmented by two large inserts, I1 and I2, wherein I1 extends >20Å away from the active site and may serve as a platform for protein-protein interactions specific for Rev1’s role in translesion DNA synthesis (TLS) in human cells, and I2 acts as a “flap” on the hydrophobic pocket accommodating the template G. We suggest that these novel structural features are important for providing human Rev1 greater latitude in promoting efficient and error-free TLS through the diverse array of bulky and potentially carcinogenic N2-dG DNA adducts in human cells.
Introduction
The integrity of genomic DNA is continually threatened by external and internal DNA damaging agents. Although organisms have evolved a variety of repair mechanisms to remove the resulting lesions, some lesions escape repair and block the replication machinery. The recently discovered Y-family DNA polymerases permit the continuity of the replication fork by allowing replication through such DNA lesions1. Humans have four Y-family polymerases – Polη, Polι, Polκ, and Rev1 – each with a unique DNA damage bypass profile. Polη, for example, is unique in its ability to replicate through an ultraviolet (UV)-induced cis-syn thymine-thymine (T–T) dimer by inserting two As opposite the two Ts of the dimer with the same efficiency and accuracy as opposite undamaged Ts 2; 3; 4; 5. Because of the involvement of Polη in promoting error free replication through cyclobutane pyrimidine dimers, its inactivation in humans causes the variant form of xeroderma pigmentosum, a genetic disorder characterized by a greatly enhanced predisposition to sun induced skin cancers6; 7. Polι, on the other hand, is unable to replicate through a cis-syn T–T dimer8; 9, but it can proficiently incorporate nucleotides opposite adenine and guanine lesions that are impaired in their ability to make Watson-Crick (WC) base pairs10; 11; 12; 13; 14; 15. Polκ is specialized for the extension step of lesion bypass16; 17; 18, carrying out, for example, efficient extension from a C incorporated opposite bulky N2-adducted guanine lesions by another polymerase, such as Polι10; 11.
Rev1 is the most extreme member of the eukaryotic Y-family Pols. It is highly specific for incorporating a C opposite template G 19; 20. In this respect, Rev1 differs not only from replicative and repair polymerases, which incorporate the correct nucleotide opposite all four template bases with nearly equivalent catalytic efficiencies, but it differs also from the other three eukaryotic Y-family Pols, η, ι, and κ. Of these, Pols η and κ form the four Watson-Crick base pairs with similar catalytic efficiencies, whereas Polι incorporates the correct nucleotide opposite template purines with a much higher efficiency than opposite template pyrimidines1.
The crystal structure of the ternary complex of yeast Rev1 bound to template G and incoming dCTP has provided a basis for the extreme specificity of this polymerase21. Strikingly, in the active site of yeast Rev1, template G and dCTP do not pair with each other. Instead, template G is evicted from the DNA helix and it makes two hydrogen bonds via its Hoogsteen edge (N7 and O6) with amino acids in a segment of Rev1 called the G-loop. Moreover, the incoming dCTP pairs with an arginine in another segment of Rev1 called the N-digit; ensuring the incorporation of dCTP over other incoming nucleotides. Thus, unlike any other DNA polymerase, in yeast Rev1, specificity for both the templating and the incoming nucleotide is provided by the protein itself rather than the DNA. Because of the novelty of this mechanism a question arises as to whether human Rev1 also shares the same protein-template-directed strategy of DNA synthesis? The catalytic cores of human and yeast Rev1 share only ~27% sequence identity. The human Rev1 catalytic core is also substantially larger and characterized by a number of large inserts, the structural significance of which remains unclear. To gain insights into the mechanism of DNA synthesis by human Rev1, we have solved the structure of the polymerase in ternary complex with a template-primer and an incoming nucleotide. We show here that the central elements of protein-template-directed mechanism of DNA synthesis are conserved in the human enzyme, including the pairing of template G and incoming dCTP with segments of the polymerase. The structures differ, however, in the palm and fingers domains. The palm in human Rev1 is supplemented by an ~40 residue insert (I1), which extends away from the active site and maybe involved in protein-protein interactions. The fingers domain in human Rev1 is augmented by an insert (I2) that acts as a “flap” on the hydrophobic pocket accommodating template G.
Results
Overall arrangement
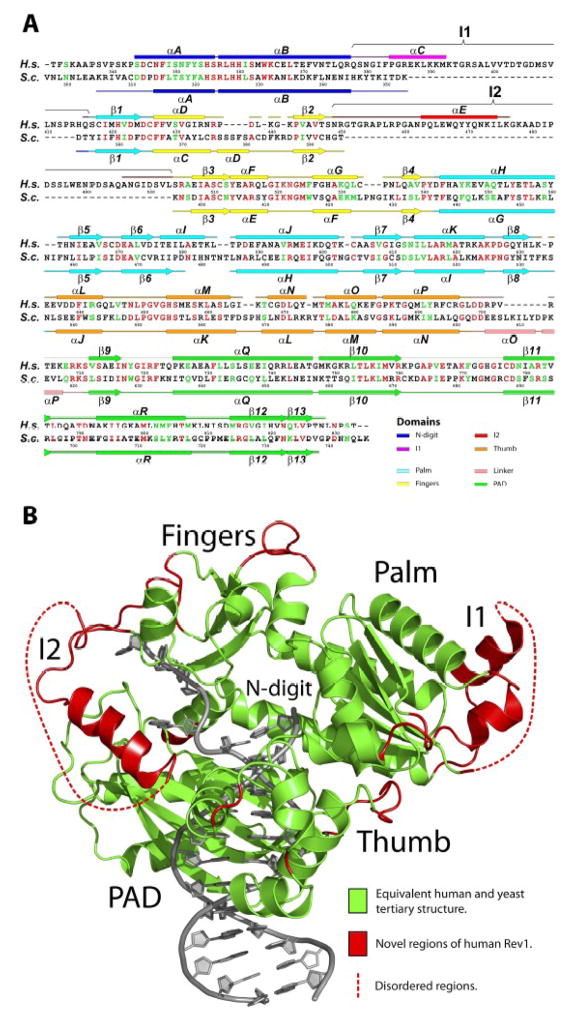
As in the yeast Rev1 ternary complex21, the human enzyme grasps the template-primer with palm (residues 417 to 426 and 542 to 632; αH-αK, β1, β5-β8), fingers (residues 427 to 448 and 504 to 541;αD, αF, αG, β2-β4) and thumb domains (residues 633 to 698; αL-αP) that are common to all DNA polymerases, and the PAD (polymerase associated domain; residues 711 to 828; αQ, αR, β9-β13) that is specific to Y-family polymerases (Figs. 1 and 2). The palm carries the active site residues (Asp423, Asp570 and Glu571) that catalyze the nucleotidyl transfer reaction. The fingers domain lies over the replicative end of the template-primer and interacts primarily with incoming dCTP and the unpaired nucleotides at the 5′ end of the template (Fig. 1). The thumb and the PAD approach the template-primer from opposite sides, connected by a long linker. The thumb makes contacts with DNA in the minor groove, while the PAD makes contacts in the major groove. The right-handed grip of palm, fingers, thumb and the PAD on the template-primer is augmented by a loose α-loop substructure at the N-terminus, an N-digit (residues 344-377; αA and αB), which interacts with incoming dCTP, and a loop in the PAD, a G-loop, which interacts directly with the templating G (Figs. 1 and 2A).
Fig. 1.
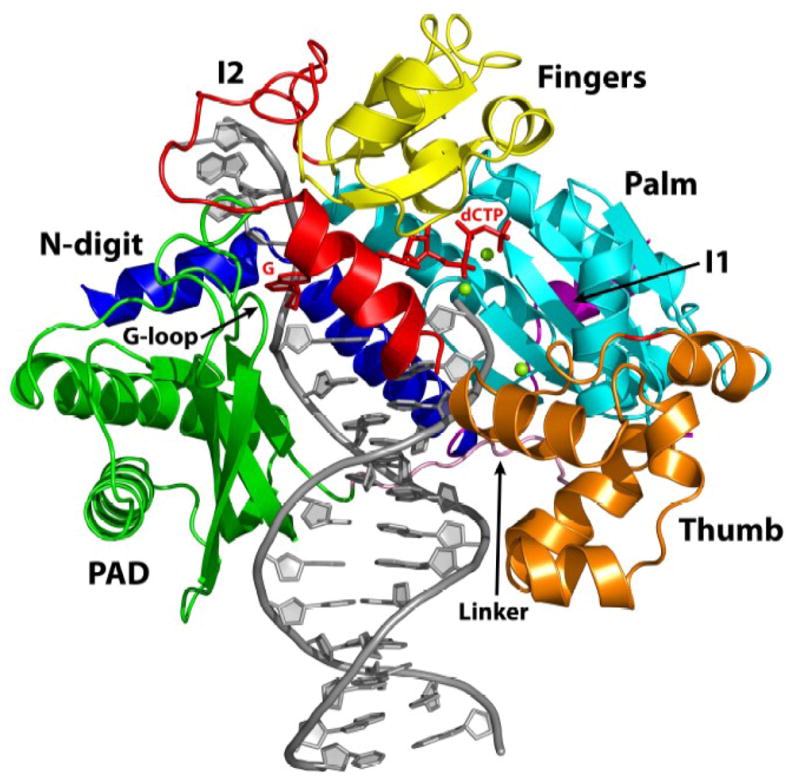
Human Rev1-DNA-dCTP ternary complex. The palm, fingers, thumb and PAD domains are colored cyan, yellow, orange and green respectively. The two large insertions in human Rev1, I1 and I2, are colored magenta and red, respectively. The linker joining the thumb to the PAD domains is shown in pink. The N-digit is shown in dark blue. DNA is gray, template G and the incoming dCTP are red, and the putative Mg2+ ions are green spheres.
Fig. 2.
Comparison of human and yeast Rev1. (A) Sequence and secondary structure comparison between the catalytic cores of human (H.s) and yeast (S.c) Rev1. Identical residues are highlighted in red and semi-conserved residues are shown in green. (B) Human Rev1 catalytic core displayed in two colors: green, for elements that are conserved between the human and yeast Rev1; red, for elements (including, inserts I1 and I2) that are unique to human Rev1. The DNA is shown in gray.
Protein-template directed mechanism of DNA synthesis
Template G and incoming dCTP do not pair with each other (Fig. 3). Template G is evicted from the active site by Leu358 (from the N-digit), which inserts into the DNA helix. The ejected G slips into a hydrophobic pocket delineated by His774 and Lys681 from the G-loop, and Phe525 from the fingers domain (Fig. 3A). Lys681 is conserved in yeast Rev1, whereas His774 and Phe525 are substituted by methionine and tryptophan residues, respectively. In yeast Rev1, the minor groove edge of template G is solvent exposed in the space between the PAD and the fingers domain (Fig. 4). In human Rev1, the fingers domain lacks a helix (αD) and two short loops observed in yeast Rev1, but it contains a large 54 residue insert (I2) that packs against the N-digit (Fig. 2B). Part of I2 folds into an α-helix (αE) that acts as a “flap” on the hydrophobic pocket holding the template G (Figs. 3A and 4A). Tyr470 and Trp467 on αE protrude into the pocket and may help to stabilize the binding of large N2-dG adducted DNAs (see Discussion). The Hoogsteen edge of template G (atoms N7 and O6) is engaged in hydrogen bonds with the main-chain amides of His774 and Gly775 on the G-loop (Fig. 3A). The O6 of G is further linked to a water molecule bonded to Glu466, and the phosphate groups on either side of G are anchored in place by direct hydrogen bonds to Ser356, Arg357, and Asn717 (Fig. 3A). Together, these backbone interactions help to maintain the extrahelical path of template G outside of the active site cleft.
Fig. 3.
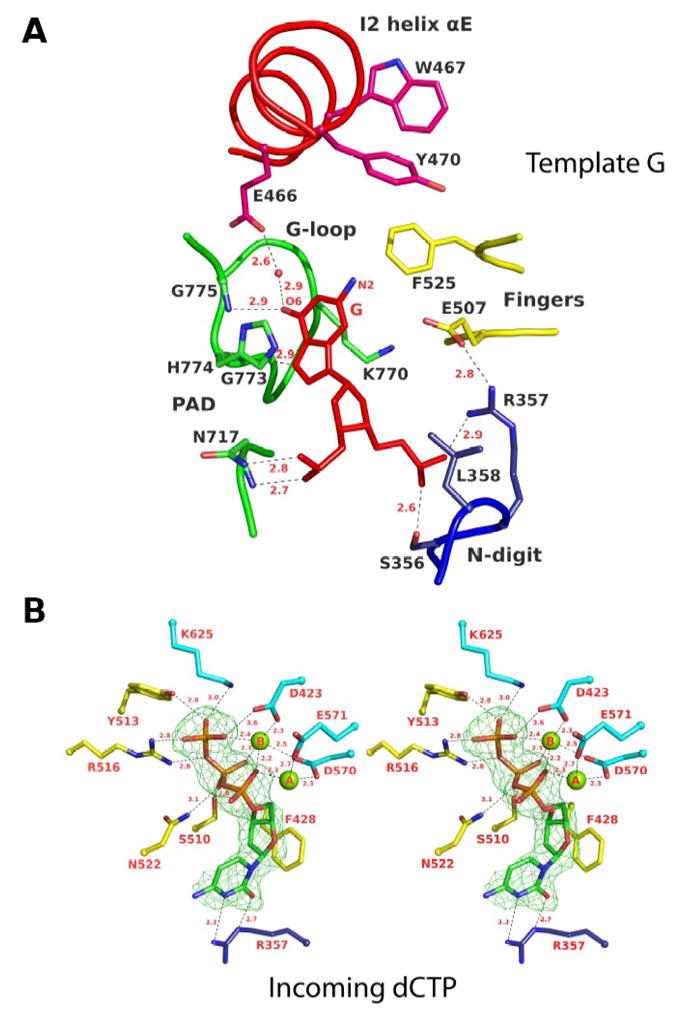
Rev1 interactions with template G and incoming dCTP. (A) A close-up view of the interaction between human Rev1 and template G. Residues are colored according to the domain or subdomain/insert they belong to (c.f. Figure 1): yellow, fingers; green, PAD; dark blue, N-digit; red, insert I2. Template G is shown in red. Hydrogen bonds are depicted as dotted lines and the distances are in Angstroms. (B) A close-up stereo view of the human Rev1 active site showing interactions with incoming dCTP. Residues are colored according to the domain they belong to: yellow, fingers; cyan, palm; dark blue, N-digit. dCTP is shown with Fo-Fc difference density (green) at a contour level of 3s. The two catalytic magnesium ions (A and B) are shown as green spheres. Distances are given in Angstroms.
Fig. 4.
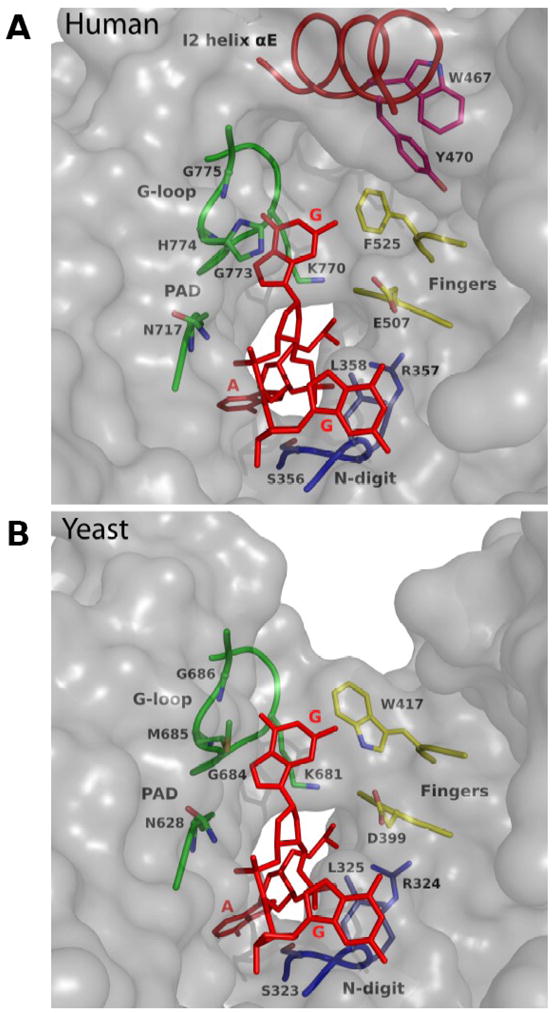
Surface near template G in human and yeast Rev1. A) Human Rev1 surface. The DNA is in red, and the key protein residues are colored according to the domain they belong to: yellow, fingers; green, PAD; dark blue, N-digit; red; insert I2. Note that helix αE in insert I2 acts as a “flap” on the hydrophobic pocket ccommodating template G. B) Yeast Rev1 surface near template G. Note that N2 of template G is fully exposed to solvent.
In the active site cleft incoming dCTP pairs with Arg357 that emanates from the N-digit (Fig. 3B). Two hydrogen bonds are established between N3 and O2 at the Watson-Crick edge of dCTP and the Nη2 and Nε donor groups of Arg357, respectively (Fig. 3B). The pattern of hydrogen bonding is such that substitution of dCTP by any other incoming nucleotide would result in the loss of hydrogen bonds and/or electrostatic and steric intrusion (on Arg357). The triphosphate moiety of dCTP weaves a contorted path between the fingers and palm domains, with Ser 510, Tyr 513, Arg 516, Asn522 from the fingers and Lys 625 from the palm making hydrogen bonds along its length (Fig. 3B). The catalytic residues, Asp423, Asp570 and Glu571 are arrayed between the triphosphate moiety and the primer terminus, and two octahedrally coordinated Mg2+ ions – analogous to metals “A” and “B” in replicative DNA polymerases 22; 23; 24 – complete the human Rev1 active site (Fig. 3B). Intriguingly, the palm in human Rev1 is augmented by a large 38 residue insert (I1) that extends away from the active site (Fig. 2B). Thus, whereas the N-digit in yeast Rev1 is connected to the first β-strand of the palm domain, in human Rev1 the connection is interceded by I1, which is composed of an α-helix (αC) and an unstructured region (not visible in the electron density) (Fig. 2).
A third metal ion
An intriguing feature of the present structure is a putative Mg2+ ion within a loop between helices αL and αM of the thumb domain (Fig. 5). We assign it as a Mg2+ ion (rather than as a water molecule) based on its coordination pattern and relatively short ligation distances. The “ion” is coordinated by a set of main chain carbonyls and the phosphate group of the penultimate base at the primer terminus (Fig. 5). There are analogous interactions in yeast Rev1, human Polι and human Polκ ternary complexes 12; 13; 15; 18; 21, though in these cases the ion was assigned as a water molecule. These bridging interactions appear to be a conserved feature in these polymerases that may help to fix the primer terminus for the nucleophilic attack on incoming dNTP. Interestingly, an ion at a nearby position has been observed in ternary complexes of an archaeal homolog of Polκ, namely Dpo4 25,26.
Fig. 5.
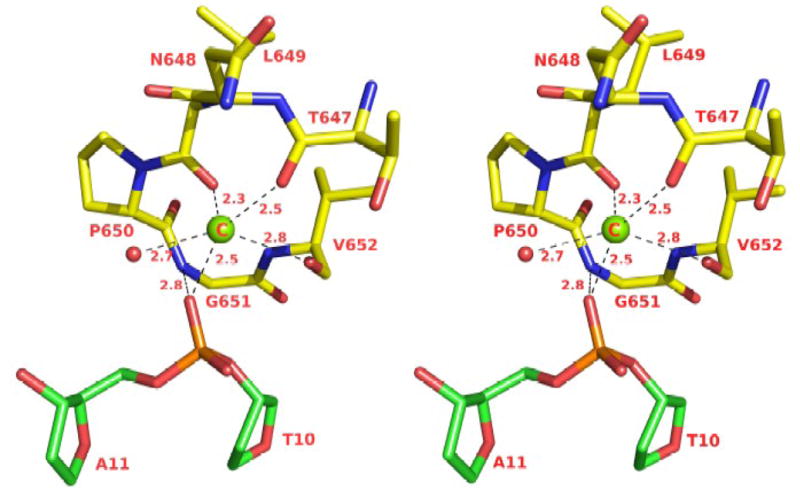
Stereo illustration of the coordination geometry of a putative 3rd magnesium ion. The putative magnesium ion is shown as a green sphere (C). The DNA is green and the protein (a loop between helices αL and αM in the thumb domain) is yellow. The single water molecule is colored red. The distances are in Angstroms.
Discussion
Human Rev1 conforms to the domain arrangement in yeast Rev1 with the exception of two unique regions (Fig. 2B). First, a large insertion (I1) in the palm that is solvent exposed and extends >20Å away from the active site. Curiously, only the first 14 residues of this ~38 residue insert are visible in the electron density. The remaining residues may only become ordered upon association with another protein. Perhaps more than any other Y-family polymerase, Rev1 has emerged as a “hub” for protein-protein interactions in translesion DNA synthesis (TLS) 1; 27; 28; 29; 30. Independent of its polymerase activity, Rev1 has been shown to play a key scaffolding role in the recruitment of polymerases opposite certain DNA lesions. Human and yeast Rev1 differ, however, in their interactions with other polymerases. For example, whereas yeast Rev1 has been shown to associate with Polη via the PAD in the catalytic core 31, human Rev1 binds Polη through a C-terminal ~100 residue segment outside of the catalytic core 32; 33. Similarly, whereas yeast Rev1 interacts with Rev7 via the PAD (and motifs outside of the catalytic core) 34; 35, in human Rev1 the interaction has been again mapped to the C-terminal ~100 residue segment 36; 37. The insert I1 in human Rev1 may provide an additional site for the binding of as yet unidentified protein cofactors needed specifically for Rev1-mediated TLS in human cells.
The most pronounced difference between yeast and human Rev1 catalytic cores is a 54 residue insert in the human enzyme that acts as a “flap” on the hydrophobic pocket accommodating the template G. In both structures, the Hoogsteen edge of template G is paired to the G-loop, and the N2 amino group lies in the space between the PAD and the fingers domain. The N2 of G is highly susceptible to modification by various potential carcinogens. For example, in cells, the N2 of G can react with various aldehydes generated during the peroxidation of lipids, including acrolein, an α,β-saturated aldehyde, to produce a cyclic γ–HOPdG adduct that presents a strong block to normal replication 38; 39; 40. Rev1 can promote replication through such γ– HOPdG adducts 41, which have been detected in mammalian tissues at considerable levels and appear to play a role in the onset of cancer42. A recently determined structure of yeast Rev1 bound to a permanently ring-closed form of γ– HOPdG shows the adduct accommodated “comfortably” in the space between the PAD and the fingers domain and stabilized in part by van der Waals contacts with Trp417 (from the fingers domain) 43. From the structure of human Rev1 presented here, the enzyme would be equally capable of accommodating a γ– HOPdG adduct, with the exocyclic ring of the adduct stabilized by van der Waals contacts with Phe525 (which substitutes for Trp417 in the yeast Rev1) (Fig. 4). More bulkier and potentially more interfering adducts derive from reactions of N2 of G with aldehydes such as trans-4-hydroxy-2-noenal (from the peroxidation of lipids) and polycyclic aromatic hydrocarbons (PAHs) such as benzo[a]pyrene (BP) that is present in automobile and factory emissions and cigarette smoke44; 45; 46. In yeast Rev1, the larger N2-dG adducts may protrude from the three sided pocket accommodating the extrahelical G (Fig. 4B). By contrast, in human Rev1, the bulky N2-dG adducts may be stabilized in part by contacts with insert I2 (Fig. 4A). That is, Tyr470 and Trp467 emanating from helix αE on I2 present a ‘wall” of potential hydrophobic contacts at the outer edge of the pocket accommodating the extrahelical base. Consistent with this idea, human Rev1 has recently been shown to bind N2-dG-BP adducted DNA 3-fold more tightly than unmodified G-containing DNA47. Interestingly, only 35 of the 54 residues in the I2 insert are visible in the electron density. The remaining residues (many of which ensue αE) may become ordered when human Rev1 binds to large N2-dG adducted DNAs. The entire I2 insert is relatively mobile with an average B factor of 52.0Å2, as compared to 41.7Å2 for the rest of the protein. Thus, it is possible that I2 (as a “flap”) assumes different conformations when bound to different N2-dG adducted DNAs.
In conclusion, Y family polymerases have proven to be remarkable in their functions and in strategies for replicating through DNA lesions. Rev1 adopts the most radical strategy, where the polymerase itself dictates the identity of the incoming nucleotide, as well as the identity of the templating base. We show here that many of the key elements of this novel strategy are conserved between yeast and human Rev1, including the eviction of template G from the DNA helix and the pairing of incoming dCTP with a surrogate arginine residue. We also show that the catalytic core of human Rev1 is augmented by two large inserts, I1 and I2, wherein I1 may serve as a platform for protein-protein interactions specific for Rev1’s TLS role in human cells and I2 may aid in the binding of bulky N2-dG adducted DNAs. Taken together, these new structural features could be important for providing human Rev1 greater latitude in promoting error-free TLS through a diverse array of bulky N2-dG adducts than that possible for yeast Rev1.
Materials and Methods
Protein and DNA preparation
The human GST-Rev1 (residues 330–833) fusion protein was expressed in E. coli from plasmid pBJ1237. The Rev1 (330–833) protein is as catalytically active as the full length protein and has the same nucleotide incorporation specificity. The fusion protein was purified by (NH4)2SO4 precipitation followed by affinity purification using a glutathione-Sepharose column. Rev1 (330–833) lacking the GST tag was eluted from the column, following cleavage on the resin with PreScission protease (Amersham Pharmacia). The protein was further purified over a gel filtration SD200 column, concentrated, and used for co-crystallization. The primer was synthesized as a 12-mer, with a dideoxy cytosine at its 3′ end (5′-ATCCTCCCCTACdd-3′). This was annealed with a 16-nt template (5′-TAAGGTAGGGGAGGAT-3′) to yield a 12/16 primer-template. Prior to annealing, the oligonucleotides were purified by reverse phase over a C18 column, desalted, and lyophilized.
Crystallization and structure determination
The native complex was prepared by incubating Rev1 with the 12/16 primer-template in a molar ratio of 1:1.5 with 10 mM dCTP and 20mM MgCl2, and then crystallized from solutions containing 20% PEG 5000 MME, 0.1 M (NH4)2SO4, 0.2M NaCl, and 0.4M sodium malonate (pH 4.6). For data collection, the cocrystals were cryoprotected by soaks in mother liquor solutions containing a gradient of 0–24% glycerol and then flash frozen in liquid nitrogen. X-ray diffraction data on the cocrystals (2.5Å) were measured at the Advanced Photon Source (APS; beamline 17-ID) (Table 1). All data were indexed and integrated using DENZO and reduced using SCALEPACK 48. The data reduced most accurately to space group P21 with cell dimensions of a=56.4 Å, b=172.8 Å, c=129.2Å, and β=90.6. The data could also be reduced to space group P212121 albeit with a much higher number of rejected reflections. For refinement in space group P21 the test reflections for Rfree calculations were selected from data reduced in space group P212121 (to omit Bijvoet pairs related by pseudo-orthorhombic symmetry). The structure was solved by molecular replacement (MR) with the program PHASER 49- using a simplified construct of the yeast Rev1 ternary complex model based on sequence alignment and secondary structure matching. The MR solution revealed four Rev1 ternary complexes in the crystallographic asymmetric unit (A-D), and electron density maps calculated after a round of refinement with CNS 50 showed densities for several segments not included in the search model. Refinement was continued with REFMAC 51, which allows for the anisotropic motion of rigid bodies, described as TLS parameters 52. Each Rev1 protein molecule and DNA strand was treated as a separate group. Iterative refinement, building with COOT 53, and water picking using ARP/wARP 54, lowered the Rfree to 26.7%. The final model includes residues 344–377, 383–391, 416–477 and 499–828 of molecule A; residues 344–392, 416–473 and 496–828 of molecule B; residues 344–378, 383–385, 416–472 and 500–827 of molecule C; residues 346–389, 416–472 and 496–827 of molecule D; nucleotides 1–16 for each template strand; nucleotides 1–12 for each primer strand; 4 molecules of dCTP; 11 Mg2+ ions; and 98 water molecules. The model has excellent stereochemistry, as shown by MOLPROBITY 55, with over 97% of the residues in the most favored regions of the Ramachandran plot and only 0.2% in the disallowed regions.
Table 1.
Data Collection and Refinement Statistics
| Native Data collection: | |
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 50.0–2.5 (2.6–2.5) |
| No. of measured reflections | 266877 |
| No. of unique reflections | 80609 |
| Completeness (%) | 94.7 (91.6) |
| Mean I > σI | 27.4 (2.9) |
| Rmerge1 (%) | 5.9 (46.2) |
| Refinement: | |
| Resolution range (Å) | 50.0–2.5 |
| R factor (%) | 21.4 |
| R free (%) | 26.7 |
| Number of atoms | |
| Protein | 13025 |
| DNA | 2276 |
| dCTP | 112 |
| Mg2+ ions | 11 |
| Water | 98 |
| RMS deviations from ideal stereochemistry:- | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.34 |
| rms deviation in main chain B factor (Å2) | 0.43 |
| rms deviation in side chain B factors (Å2) | 1.14 |
| Mean B factors: (Å2) | |
| Protein (overall) | 42.3 |
| Protein (main chain) | 42.0 |
| Protein (side chains) | 42.3 |
| DNA | 40.2 |
| dCTP | 35.8 |
| Mg2+ ion A | 64.3 |
| Mg2+ ion B | 30.7 |
| Mg2+ ion C | 49.8 |
| Water | 39.1 |
| Ramachandran plot: | |
| % residues in most favored region | 97.8 |
| % residues in allowed regions | 2.0 |
| % residues in disallowed regions | 0.2 |
Rmerge =ΣhklΣi |Ii(h k l) – <I(h k l)>|/ΣhklΣiIi(h k l)]
Numbers in parentheses are for the outer shell of data.
Accession Number
The coordinates have been deposited in the PDB with code 3GQC.
Acknowledgments
We thank the staff at the Advanced Photon Source (beamline 17-ID) and at the Brookhaven National Laboratory (beamline X29) for facilitating X-ray data collection. We also thank Axel Brunger and Deepak Nair for helpful discussions. This work was supported by grant ES016666 from the U. S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 2.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–4. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J Biol Chem. 2000;275:7447–50. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 4.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta [see comments] Proc Natl Acad Sci U S A. 2000;97:3094–9. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Washington MT, Prakash L, Prakash S. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase eta. Proc Natl Acad Sci U S A. 2003;100:12093–8. doi: 10.1073/pnas.2134223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum [see comments] Science. 1999;285:263–5. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 7.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta [see comments] Nature. 1999;399:700–4. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–9. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 9.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc Natl Acad Sci U S A. 2001;98:14256–61. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2004;24:5687–93. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases iota and kappa. Mol Cell Biol. 2006;26:381–6. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–80. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 13.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA Polymerase iota Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair. Structure (Camb) 2005;13:1569–77. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006;13:619–25. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 15.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14:749–55. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta [see comments] Proc Natl Acad Sci U S A. 2000;97:3838–43. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washington MT, Johnson RE, Prakash L, Prakash S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc Natl Acad Sci U S A. 2002;99:1910–4. doi: 10.1073/pnas.032594399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–14. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277:15546–51. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 20.Nelson JR, Lawrence CW, Hinkle DC. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–31. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 21.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–22. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 22.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution [see comments] Nature. 1998;391:251–8. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–25. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–8. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 25.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 26.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acharya N, Johnson RE, Prakash S, Prakash L. Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol Cell Biol. 2006;26:9555–63. doi: 10.1128/MCB.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baynton K, Bresson-Roy A, Fuchs RP. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol Microbiol. 1999;34:124–33. doi: 10.1046/j.1365-2958.1999.01583.x. [DOI] [PubMed] [Google Scholar]
- 29.Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–54. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–54. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 31.Acharya N, Haracska L, Prakash S, Prakash L. Complex formation of yeast Rev1 with DNA polymerase eta. Mol Cell Biol. 2007;27:8401–8. doi: 10.1128/MCB.01478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohashi E, Murakumo Y, Kanjo N, Akagi J, Masutani C, Hanaoka F, Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–31. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 33.Tissier A, Kannouche P, Reck MP, Lehmann AR, Fuchs RP, Cordonnier A. Co-localization in replication foci and interaction of human Y-family members, DNA polymerase pol eta and REVl protein. DNA Repair (Amst) 2004;3:1503–14. doi: 10.1016/j.dnarep.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 34.Acharya N, Haracska L, Johnson RE, Unk I, Prakash S, Prakash L. Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol Cell Biol. 2005;25:9734–40. doi: 10.1128/MCB.25.21.9734-9740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D’Souza S, Waters LS, Walker GC. Novel conserved motifs in Rev1 C-terminus are required for mutagenic DNA damage tolerance. DNA Repair (Amst) 2008;7:1455–70. doi: 10.1016/j.dnarep.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278:12356–60. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- 37.Murakumo Y, Ogura Y, Ishii H, Numata S, Ichihara M, Croce CM, Fishel R, Takahashi M. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–51. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 38.Chung FL, Zhang L, Ocando JE, Nath RG. Role of 1,N2-propanodeoxyguanosine adducts as endogenous DNA lesions in rodents and humans. IARC Sci Publ. 1999:45–54. [PubMed] [Google Scholar]
- 39.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 40.Chung FL, Chen HJ, Nath RG. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 1996;17:2105–11. doi: 10.1093/carcin/17.10.2105. [DOI] [PubMed] [Google Scholar]
- 41.Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase zeta. Mol Cell Biol. 2004;24:6900–6. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nath RG, Ocando JE, Chung FL. Detection of 1, N2-propanodeoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56:452–6. [PubMed] [Google Scholar]
- 43.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure. 2008;16:239–45. doi: 10.1016/j.str.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 44.Meehan T, Straub K. Double-stranded DNA steroselectively binds benzo(a)pyrene diol epoxides. Nature. 1979;277:410–2. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 45.Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. 1999;443:139–47. doi: 10.1016/s1383-5742(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 46.Phillips DH. Fifty years of benzo(a)pyrene. Nature. 1983;303:468–72. doi: 10.1038/303468a0. [DOI] [PubMed] [Google Scholar]
- 47.Choi JY, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J Biol Chem. 2008;283:23645–55. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 50.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve R, Jiang W, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]