Abstract
Single-cell transfection of adherent cells has been accomplished using single-cell electroporation (SCEP) with a pulled capillary. HEPES-buffered physiological saline solution containing pEGFP plasmid at a low concentration (0.16 ∼ 0.78 µg/µL) filled a 15 cm long capillary with a tip opening of 2 µm. The electric field is applied to individual cells by bringing the tip close to the cell and subsequently applying one or two brief electric pulses. Many individual cells can thus be transfected with a small volume of plasmid-containing solution (∼ 1 µL). The extent of electroporation is determined by measuring the percentage loss of freely diffusing thiols (chiefly reduced glutathione) that have been derivatized with the fluorogenic ThioGlo 1. A mass transport model is used to fit the time-dependent fluorescence intensity decay in the target cells. The fits, which are excellent, yield the electroporation-induced fluorescence loss at steady state and the mass transfer rate through the electroporated cell membrane. Steady-state fluorescence loss ranged approximately from 0 to about 80% (based on the fluorescence intensity before electroporation). For the cells having a loss of thiol-ThioGlo 1 fluorescence intensity greater than 10%, and mass transfer rate greater than 0.03 s−1, EGFP fluorescence is observed after 24 hours. The EGFP fluorescence is increased at 48 hours. With a loss smaller than 10% and a mass transfer rate smaller than 0.03 s−1, no EGFP fluorescence is detected. Thus, transfection success is closely related to the small molecule mass transport dynamics as indicated by the loss of fluorescence from thiol-ThioGlo 1 conjugates. The EGFP expression is weaker than bulk lipid-mediated transfection, as indicated by the EGFP fluorescence intensities. However, the success with the single-cell approach is considerably greater than lipid-mediated transfection.
INTRODUCTION
Studies on single cells are becoming the frontier of biochemical research. Effective and reliable single-cell transfection technologies have great value to construct single-cell models, and control biochemistry at the single-cell level. Besides the traditional carrier-mediated and plasma-membrane permeabilization protocols which require that individual cells are isolated for individual cell gene delivery, microinjection techniques using ultra-small probes (glass needles 1, nanoneedles 2, femtosyringe 3 and AFM tips 4) with the naked DNA plasmid either inside or surrounding the probes have been successfully used for single-cell transfection. However, all these microinjection techniques rely on penetrating the cell membrane with a probe which will induce physical damage to the target cells. One solution to this problem is to use a focused femtosecond near-infrared laser beam to facilitate DNA intake, causing little cellular damage 5.
Although electroporation is an established method for gene delivery into cells in suspension and tissues, there is a limited number of reports on single-cell transfection by single-cell electroporation (SCEP). The first success in single-cell transfection by SCEP was to introduce the plasmid vector pRAY1 into individual COS 7 cells using carbon fiber microelectrodes 6. One significant limitation in the microelectrode approach to SCEP is the potential for the electrode to form cytotoxic products such as reactive oxygen species (ROS). Plasmid/buffer-filled glass micropipettes containing a metal electrode have been developed and applied for plasmid DNA delivery into single cells 7–11. In this case, a glass pipette connected to a voltage supply through a metal wire inside is pushed against the target cell, and an external pressure is applied to push the plasmid-containing solution in the micropipette into the target cell during the application of electrical pulses. Hass et al. reported the first use of micropipettes for SCEP. They used micropipettes with openings in the 0.6–1 µm range for single cell transfection with plasmids encoding enhanced green fluorescent protein (EGFP) and DsRed in the brain of intact Xenopus tadpoles and rat hippocampal slices 11. Their highest transfection efficiency was only 30% after optimization of the pulse protocols. The poor transfection efficiency has been improved in two different approaches which are based on electroinjection i.e. the plasmid-filled capillary is in direct physical contact with the cell membrane. First, Rae and Levis pressed a smaller (0.5 µm) tip onto the target cells while monitoring the resistance increase 10. Second, Kitamura et al., coupled the whole-cell patch-clamp technique with two-photon microscopy 9 for in vivo transfection of neurons with plasmid encoding EGFP7. This micropipette method was also successfully applied to the delivery of RNA into neurons recently by Boudes et al. 8. Despite the above advances, problems arise, as addressed by Rae and Levis, from the cell damage caused by indenting, micropipette tip clogging, and cell adhesion to the tips after pulses. Recently, microchips coupling microelectrodes have been designed for high throughput SCEP and show promising results for gene delivery into singe cells 12–14.However, this approach is not suitable for adherent cells, tissue slices, tissue cultures, or in vivo applications. Although many papers have reported the effect of the electrical pulsing protocol on plasmid transfer during SCEP or bulk electroporation 10, 13, 15, nobody has tried to correlate cell membrane permeability to small molecules with DNA delivery.
As we have shown previously 16, 17, exposing cells to the thiol-reactive fluorogenic reagent, ThioGlo 1 leads to intracellular fluorescence, chiefly from the freely diffusible reduced glutathione (γ-Glu-Cys-Gly, GSH). Thus, in this work, we follow the loss of intracellular fluorescence (from ThioGlo 1-labeled thiols) from single cells being electroporated with a pEGFP-C2 plasmid-filled capillary with fine tips. The results show that this method of gene delivery is effective, and that gene delivery into cells is related to the extent of electroporation as indicated by the extent of labeled thiol loss from cells.
MATERIALS AND METHODS
Materials
The chemicals used for buffers were all of analytical grade and purchased from Sigma (St. Louis, MO). The HEPES-buffered saline solution, which we will call ‘extracellular buffer’, consisted of NaCl (140 mM), KCl (5 mM), MgCl2 (1.5 mM), CaCl2 (2 mM), D-glucose (10 mM) and HEPES (20 mM). The pH was adjusted to 7.4 with 0.5 M NaOH and the buffer was filtered with 0.45 µm nylon filters prior to use. Plasmid pEGFP-C2 vector (donated by Dr. Michael Trakselis) was precipitated with ethanol and resuspended in extracellular buffer to a final concentration of 0.16–0.78 µg/µL (measured by UV absorbance at 260 nm). Thioglo 1 was purchased from Covalent Associates (Woburn, MA). A549 cell lines were obtained from American Type Culture Association (Manassas, VA). Basal medium Eagle (BME), 0.25% trypsin-EDTA, One Shot™ fetal bovine serum (FBS), L-glutamine, and penicillin-streptomycin solution were all obtained from Invitrogen, GIBCO (Carlsbad, CA). Milli-Q (Millipore Synthesis A10, Billerica, MA) water was used. Pt wires (diameter 0.5 mm, high purity 99.99+ %) were purchased from Goodfellow (Oakdale, PA).
Cell culture and preparation
A549 human lung carcinoma cells were cultured for transfection. The culture medium was prepared by adding 50 mL FBS, 5 mL 200 mM L-glutamine and 5 mL 10,000 units/mL penicillin-10,000 µg/mL streptomycin to 500 mL BME. Cells were grown in 75-cm2 cell culture flasks (Nunc* Sterile EasYFlask) in a CO2 incubator (HERA cell incubator, Newtown, CT) at 37 °C and 5% CO2/95% air, and were subcultured when reaching ∼ 80% confluence every 3–4 days. Before the experiments, cells were seeded on 35-mm gridded uncoated glass-bottom cell culture dishes (MatTek Corp., Ashland, MA) at a seeding density of 1× 104 cells/dish. Experiments were performed on the second and third days following the cell plating.
Thioglo 1 staining for electroporation visualization
A stock solution of ThioGlo 1 was prepared at a concentration of 0.5 mM in pure DMSO and stored in a desiccator at −20 °C in the dark. A 2 µM ThioGlo-1 loading solution was freshly prepared by diluting 2 µL ThioGlo 1 stock solution with 0.5 mL extracellular buffer before staining the cells. For staining, each cell dish was first washed 3 times with extracellular buffer. Following the washing step the cells were exposed to 0.5 mL ThioGlo 1 for 30 s at room temperature. The cell dish was washed again with extracellular buffer and finally bathed in 2 mL extracellular buffer for electroporation.
Capillary fabrication
The fabrication of electrolyte-filled capillaries (EFC) with pulled tips was done in a laminar flow hood. Fused-silica capillaries from Polymicro Technology (Phoenix, AZ) with an outer diameter of 360 µm and an inner diameter of 100 µm were used. Capillaries were pulled at one end by using a CO2 laser puller (Sutter Instruments Co. P-2000, Novato, CA). Before the capillary was pulled, a 2-cm-center section of a 35-cm-long capillary was burned with a flame to remove the protective polyimide coating, and then the capillary was flushed with pre-filtered (by 0.2 µm syringe filter) Milli-Q grade water. These capillaries were pulled to create reproducible capillaries with a short pulled tip having an opening inner diameter of ∼ 2 µm. Before the experiments the capillaries were carefully truncated with a Shortix™ fused-silica tubing cutter to get a final length of 15 cm.
Microscope imaging
The cell dish was fixed in a cell chamber (DH 35i culture dish incubator, Warner Instruments, Holliston, MA) mounted on the stage of an inverted fluorescence microscope (Olympus IX 71, Melville, NY) coupled with a CCD (ORCA-285 IEEE 1394 -Based Digital Camera, B & B Microscopes Limited, Pittsburgh, PA) for fluorescence imaging. A Cermax 175 W Xenon Arc lamp (PE175BF, PerkinElemer Optoelectronics, Fremont, CA) in the microscope was used as the excitation source. Cells were observed through a 20× 0.70 NA UPlanApo objective lens (Olympus). Image processing was performed by the image acquisition software SimplePCI from Compix Inc (Sewickley, PA). Fluorescence intensities of regions of interest were corrected by subtracting the average fluorescence intensity of four nearby background regions.
For Thioglo 1, an Omega fluorescence filter cube (especially built, exciter XF1075-387AF28, dichroic XF2004-410DRLP, emitter XF3087-480ALP, Omega Optical, Brattleboro, VT) was used for excitation at 378 nm and emission at 480 nm. For EGFP imaging, a U-MWIB2 filter set from Olympus (Exciter 460–490, emitter 510IF, dichroic 505) was used.
Experimental circuit and electronics
The experimental setup is depicted in Figure 1. It has two switchable circuits: an electroporation circuit and a test circuit. When doing experiments, the test circuit (described below) helps to examine the status of the capillary. Once the capillary is placed near a cell, the switches are thrown to activate the electroporation circuit (described below) for electroporation accompanied by current monitoring.
Figure 1.
Schematic diagram of the experimental setup. Either of two circuits, electroporation with current monitoring and test circuit may be used by throwing a switch. By switching from A to B, the active circuit converts from the test circuit to the electroporation circuit. A multipostion rotary selector switch shelled in a grounded box is used for switching. (1) Switch position at A. The test circuit is on. The function generator produces a 2 V (rms) sine wave at 100 Hz. The resulting current goes through the same path taken by the current during electroporation including the interface of Pt with the electrolyte, its container, the capillary, the pulled tip, the cell dish filled with buffer, and the other electrolyte/Pt interface and to ground through a current-to-voltage converter in a lock-in amplifier. The lock-in amplifier has its reference input connected to the signal output of the function generator, thus locks the 100 Hz frequency and gives current reading at this specific frequency. This circuit is used for capillary and connection testing. (2) Switch position at B. The electroporation circuit is on. An electroporator provides a high voltage pulse applied for electroporation by pushing a trigger button. The current goes through the same elecrode and electrolyte path as described above. It goes to ground through a resistor and an oscilloscope in parallel. The oscilloscope is used to examine the pulse shape and magnitude as well as monitoring the current going through the electroporation circuit.
Electroporation circuit
When the switches were put in position B, the test circuit was inactive. The 15-cm-long pulled capillary was positioned using a MP-285 motorized micromanipulator from Sutter (Novato, CA). The tip end was carefully placed near the target cell at a desired distance. The other end of the capillary was inserted into a vial filled with extracellular buffer. A platinum electrode placed in this vial was connected to the electroporator (BTX ECM 830, Harvard Apparatus, San Diego, CA), and the electrical circuit was completed with another platinum electrode in the cell dish connected to ground through a 100 kΩ resistor, Rm. The voltage across the resistor created by the electroporation current was measured with a digital oscilloscope (NI 5911 Digital Oscilloscope for PCI, National Instruments Corp. Austin, Texas) for current monitoring.
The added resistor was chosen to be 100 kΩ in order to avoid exceeding the maximum voltage for the oscilloscope, 10 V, while maximizing the signal/noise ratio. Since the resistance of EFC was always > 10 MΩ, the extra 100 kΩ resistor barely affected the electroporation current. The current during electroporation is thus given by,
| (1) |
where Vo′ is the reading from oscilloscope, Rm is the 100 kΩ resistance in parallel with the oscilloscope, and Ro′ is the input resistance of the oscilloscope (1 MΩ).
Test circuit
The test circuit was used to assess the status of the tip opening. The function generator (SRS Model DS 340, Stanford Research Systems, Inc., Sunnyvale, CA) gave a continuous 2 V AC signal at 100 Hz. This signal passed through the EFC and cell dish and was finally sent to a low input impedance (1 kΩ) current-to-voltage converter that is an integral part of the lock-in amplifier (SRS Model SR 830 DSP). The lock-in amplifier measured the magnitude and phase angle of the current at 100 Hz. The magnitude of the current primarily reflects the resistance of the capillary tip.
Electroporation
A cell dish containing adherent A549 cells stained with Thioglo 1 was mounted in the cell chamber on the microscope and connected to the electroporation circuit. The capillary tip was placed at an angle of 45° with respect to the dish surface with a tip-to-cell distance dm of about 3–5 µm and a distance to the dish surface of about 4 µm. The distance dm was determined with the help of a scale bar to visually estimate the distance between the projection of capillary tip in the horizontal imaging plane and the projection of the edge of the cell in the horizontal imaging. It was chosen according to the size of a particular cell in order to achieve electroporation without inducing cell death 17. To be specific, for the capillaries with 2 µm opening, the A549 cells we electroporated had a size distribution of 17 ∼ 42 µm in diameter. This cell ‘diameter’ is calculated (SimplePCI) from the area, A, in pixels as (A/π)1/2. For a cell diameter < 20 µm, dm was set to 6 µm. For diameter of 20 ∼ 27 µm, dm was 5 µm, and dm was 4 µm when the cell’s diameter went greater than 27 µm. Two types of pulses were used for electroporation: one was a single, 300-ms 500 V dc pulse which we called the single-pulse method, and the other is 2 × 150 ms 500 V dc pulses with an interval of 150 ms referred to as the two-pulse method. For each cell dish, one capillary filled with plasmid/extracellular buffer was used for SCEP of a number of cells. Fluorescence images were captured every second during the electroporation process. The grids on the dish surface helped to record the electroporated cells’ positions for tracking. After electroporation, the cell dish was allowed to rest in situ for 20 minutes. Following the twenty-minute resting period, the extracellular buffer in the cell dish was replaced with 2 mL of BME and the cell dish was transferred back to the CO2 incubator. After overnight culture the cells were washed 3 times with extracellular buffer and checked for EGFP fluorescence under the fluorescence microscope. The cells were returned to the incubator (in BME) for one more day following which fluorescence micrographs for EGFP fluorescence were recorded.
Lipofectamine-mediated bulk transfection
The Lipofectamine 2000 transfection experiment was carried out following the general protocol (Invitrogen Lipofectamine 2000 technical manual, see Supporting Information for our particular procedure).
Data analysis
A quantitative model for loss of whole-cell thiol-ThioGlo1 fluorescence intensity was fit to the data based on a nonlinear least squares routine in Mathcad (‘genfit’). The fit quality was judged by the coefficient of determination, R2. The resulting estimates of the variables were used to perform further statistical studies to reveal the relationship between the transfection efficiency and the pore kinetics.
RESULTS AND DISCUSSION
Capillary tip sizes and resistances
We measured the resistance for capillaries with different tip sizes. EFC tips with a 2-µm opening had currents of 0.12 – 0.13 µA, corresponding to a resistance of ∼ 16 MΩ, and tip openings of 4 µm, gave currents of 0.13 – 0.14 µA corresponding to a 14 MΩ resistance. These experimentally obtained resistances were very close to the calculated values (Comsol version 3.4, data not shown). If the capillary was clogged, the current decreased substantially. In this way we were able to confirm the integrity of the capillary before carrying out electroporation by simply reading from the lock-in amplifier. All of our prior work has been with 4–5 µm tips 16, 17. Preliminary experiments indicated that these were not suitable for transfection (no cells transfected out of 16). We have fortunately found that 2 µm tips are effective. We have not investigated why the diameter makes a difference.
Thioglo 1 staining for observation of electroporation in real time
To make sure electroporation occurs when attempting to deliver plasmid DNA into A549 cells, we used cell-permeable Thioglo 1 for observation of the electroporation process. Thioglo 1 is a non-fluorescent maleimide-based reagent that gives a green fluorescent product upon its reaction with active SH groups 18. As the predominant thiol in cells is GSH, the majority of the fluorescence can be ascribed to the GSH-Thioglo 1 adduct. Thus, Thioglo 1 can be used for detection and titration of GSH 19. The Thioglo 1-GSH conjugate has an absorbance λmax at 379 nm and emission λmax at 513 nm. Low concentrations of Thioglo 1 (< 10 µM) are reported to be non-cytotoxic 19. Nevertheless, the depletion of GSH may induce cell death, especially under the stress of oxidants produced accompanying fluorescence. Interestingly, it was reported that even with a 90% depletion of GSH, mammalian cancer cells remained viable 20. Differences in the fluorescence spectra of EGFP and the Thioglo 1-GSH adduct ensure that the fluorescence from the Thioglo 1-GSH adduct will not interfere the observation of EGFP fluorescence (See Supporting Information for absorbance and emission spectra of EGFP and Thioglo 1-GSH adduct). One parameter that we measure is the whole-cell loss of fluorescence of Thioglo 1 adducts following electroporation, ΔF%.
Single-cell transfection following electroporation
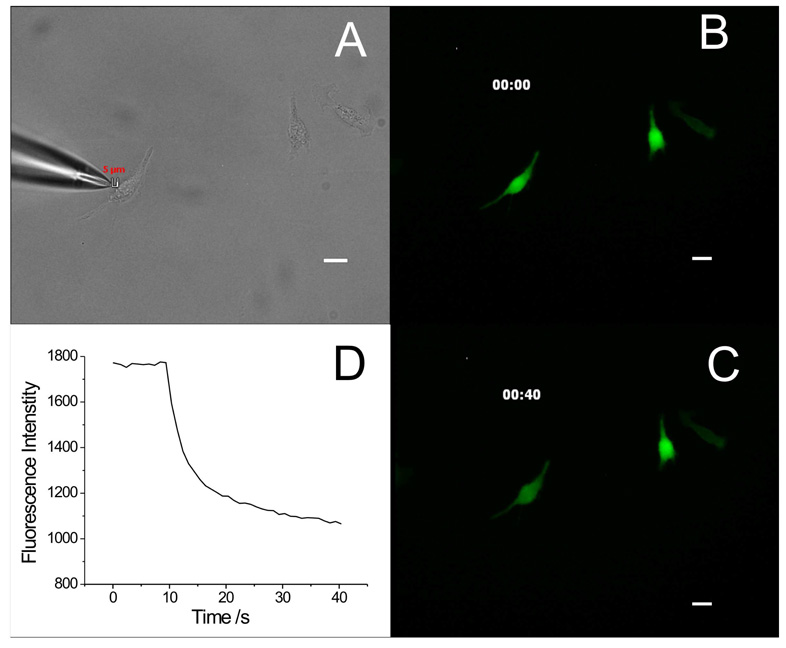
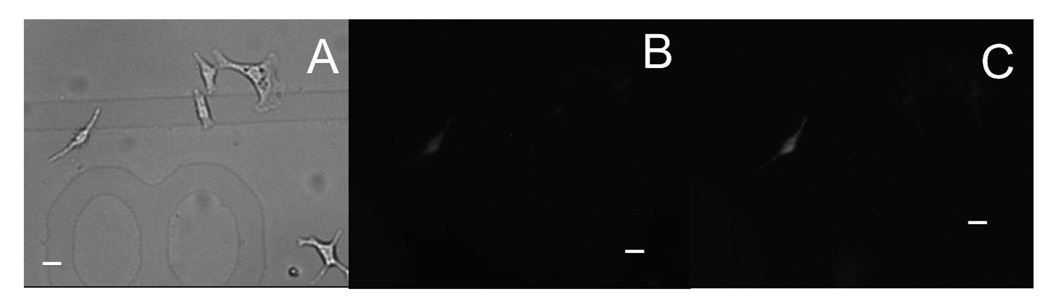
Using this SCEP method, we have successfully delivered pEGFP-C2 plasmid into A549 cells with an extracellular buffer/ plasmid-filled capillary having a pulled tip with a 2 µm opening. Figure 2 shows an example of SCEP of an individual cell and Figure 3 shows the subsequent EGFP expression inside this cell. A capillary was positioned 5 µm away from the target A549 cell with two cells nearby (Figure 2A). These cells were pre-stained with Thioglo 1, thus displaying strong green fluorescence (Figure 2B). Then a two-pulse train ( 2×150 ms pulse train with an interval of 150 ms and a voltage of 500 V) was applied to the capillary, causing electroporation of the target cell and loss of the green fluorescent Thioglo 1-GSH adduct through the transient pores in the membrane. Figure 2C shows a weaker fluorescence for the electroporated cell after electroporation, while the surrounding cells have no obvious change in fluorescence intensity. Figure 2D displays the fluorescence intensity decay of the target cell upon electroporation. At t = 9 s the pulse was applied. The fluorescence decays very rapidly in the beginning then slows down in an exponential way as the pores seal (a video file of the electroporation is included in Supporting Information). After 24 hours of incubation in BME, the cell subjected to electroporation showed fluorescence from EGFP, while the other, untreated surrounding cells showed no EGFP fluorescence (Figure 3A & 3B). The EGFP fluorescence in the target cell increased after 48 hours (Figure 3C). This demonstrated successful delivery and expression of pEGFP-C2 plasmid in the target cell through electroporation. We note that the transfected cell is in the same position as it was when it was electroporated. This is common. Occasionally, a cell expressing EGFP was found at a different location on the coverslip than the electroporated cell occupied at the time of electroporation. The electroporated cell is, in such cases, considered not to have been transfected. In our experiments, we noted that the fraction of cells that are missing from their original grid positions (following 24 h incubation) is correlated with the electroporation conditions. We believe that cells are lost primarily by necrosis and migration caused by high electric field exposure. For this study, we have assumed that all lost cells have died.
Figure 2.
Electroporation of a target A549 cells. (A) Bright field image of the capillary and A549 cells in a cell dish before electroporation. A 2-µm opening capillary tip filled with extracellular buffer/pEGFP-C2 plasmid is placed from the left of the target cell with a dm of 5 µm. (B) Thioglo 1-GSH adduct fluorescence image of the cells before the pulse. (C) Thioglo 1-GSH adduct fluorescence image of the cells 40 s after the pulse. (D) The target cell Thioglo 1-GSH adduct fluorescence decay curve. Fluorescence intensity drops suddenly when the pulse is trigged followed by a slow decay while the transient pores real. (All images taken using a 20× objective, scale bar 20 µm)
Figure 3.
Transfection following electroporation. (A) Bright field image of the target A549 cell and surrounding cells in cell dish 24 hours after electroporation. (B) EGFP fluorescence image of the target cell and surrounding cells as displayed in (E) 24 hours after electroporation. Only the electroporated cell shows EGFP fluorescence. (C) EGFP fluorescence image of the target cell and surrounding cells 48 hours after electroporation. The electroporated cell shows increased EGFP fluorescence over the image at 24 hours. (All images taken using a 20× objective, scale bar 20 µm)
Electrokinetic phenomena during electroporation
Electroosmotic flow plays an important role in plasmid delivery in our electroporation protocol. As shown in Figure 2, the ground Pt wire is the cathode so the electrode at the distal end of the capillary is the anode. This results in two opposing fluxes: electroosmotic flux goes in a direction of capillary → target cell →ground while the electrophoretic flux goes in the opposite direction because of the negative charge of plasmid DNA. The observed linear velocity of plasmid DNA is thus given by
| (2) |
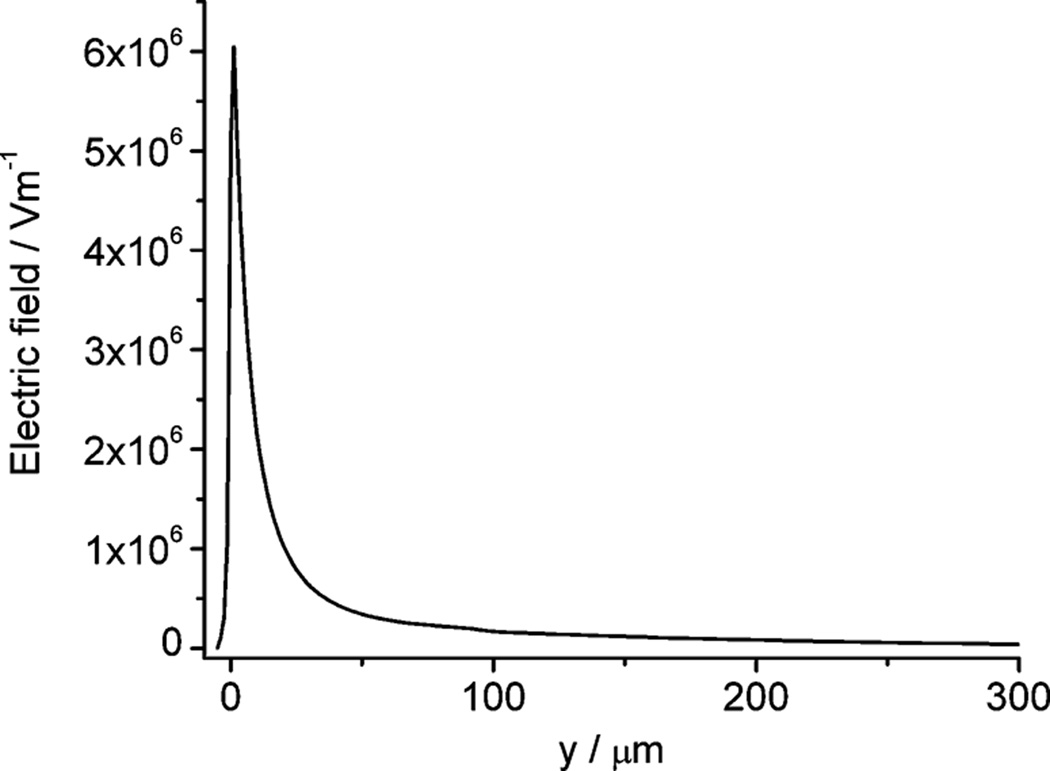
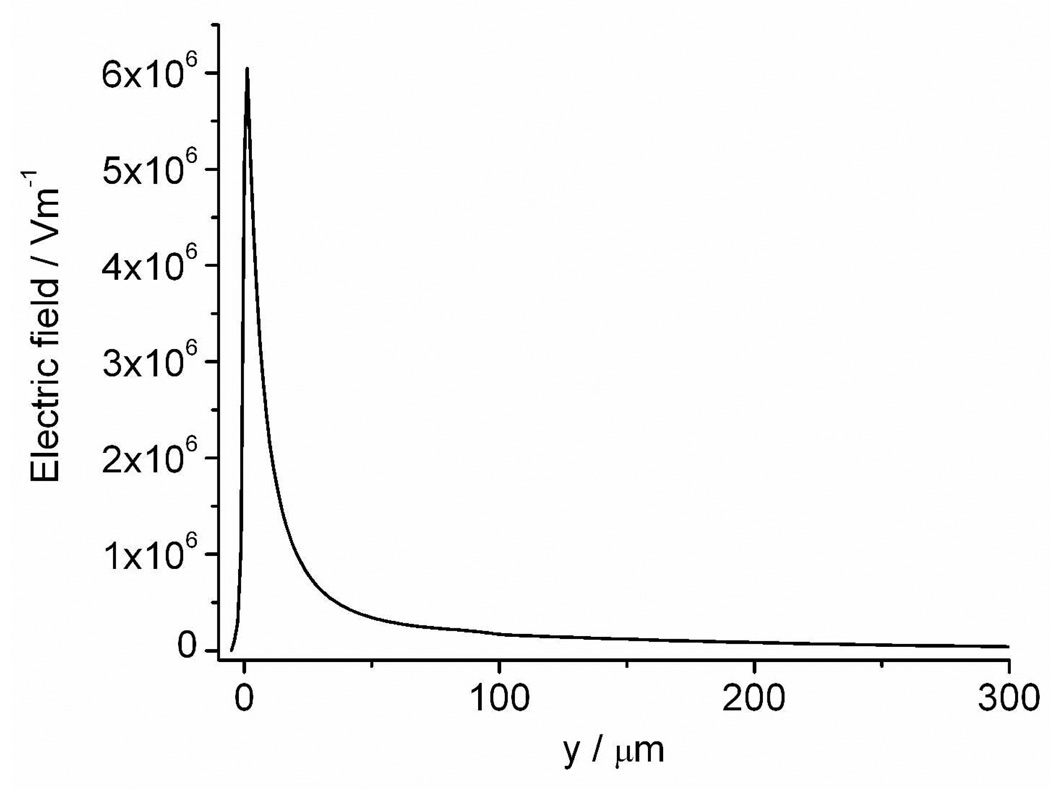
where μep is electrophoretic mobility, μeo is electroosmotic mobility, ε is the permittivity of the buffer solution, η is the dynamic viscosity of the buffer solution, ζDNA is the zeta potential of the plasmid in buffer solution, ζwall is the zeta potential of the fused silica capillary wall and E is the electric field. The sum of these two mobilities as well as the electric field strength determines the direction and the velocity of the plasmid. It has been reported that in uncoated fused silica capillaries with high ionic strength buffer, when the supercoiled plasmid is in the range of 2–16 kb, μeo > μep 21, so the plasmid moves in the direction of electroosmosis. The pEGFP-C2 plasmid has a sequence length of 4.7 kb, and our buffer is similar to the one reported, so we expect that electroosmotic flux is dominant inside the capillary. We did a simulation based on the measured capillary geometry to pursue the electric field distribution using a model similar to the one we described elsewhere 22. The simulation results reveal the electric field at the tip can be as high as ∼ 6000 kV/m, decaying rapidly in the solution outside the tip (Figure 4. The model and boundary conditions are included in the Supporting Information). Thus, near the tip we can postulate a significant net mobility pushing the pEGFP-C2 plasmid from the tip towards the cell. While it is impossible to be certain about the dynamics of the plasmid reaching the cell interior, at least we can say that it is unlikely that electrophoresis, which is in the direction away from the cell, is dominant. This is proven by the successful delivery of plasmid into the A549 cells.
Figure 4.
Simulated electric field distribution along the symmetry axis of the pulled capillary when a 500 V dc potential is applied. The capillary has a tip opening of 2 µm. The x axis labeled as ‘y’ is the distance from the tip opening, with a negative value outside the capillary and a positive value in the capillary. A cell with a diameter of 25 µm is placed 5 µm away from the tip opening at the position of y = −5 µm. Refer to reference 22 for the model set up (the model and boundary conditions are given in Supporting Information). Temperature is set to 298.15 K.
Effect of pulse types on transfection
Two types of pulse protocols were applied as explained in the experimental section: the two-pulse method, and the single-pulse method. With both types, cells were transfected. Nevertheless, the two-pulse method showed higher transfection efficiency as shown in Table 1. In Table 1, there are two sets of data, one for each pulse protocol. Each row corresponds to a range of values of ΔF%. In these experiments, there were three possible outcomes: the cells were transfected, they were not transfected but were present on the plate, or they were missing. At low ΔF%, transfection efficiency is poor. Transfection efficiency is much better for ΔF% greater than 10%. In the single-pulse protocol, there is a considerable fraction of missing cells for ΔF% greater than 10%. However, for the two-pulse protocol, the fraction of missing cells is 11%, while 89% are transfected successfully (ΔF% > 10%). The better performance of the two-pulse method may be explained by the effect of pulse duration Td and number of pulses N on electroporation. Theoretical modeling shows prolonged Td enhances the transport through the pores but does not increase the pore density, while larger N can create more pores in the cell membrane 23. Rols and Teissie reported that the transfer of molecules depends strongly on the time between pulses at a constant NTd product, and longer Td aids the transfer of macromolecules in bulk electroporation, which seems contrary to our results 15. However, they use a much shorter pulse time (1 ms), so this advantage of prolonged Td may disappear when applying 150 ms pulses in our experiments. Moreover, the two-pulse method has the advantage of accumulaing plasmid DNA near the cell membrane after the first pulse, which facilitates the transfer of plasmid DNA during the second pulse.
Table 1.
Effect of pulse types on SCEP induced pEGFP-C2 transfection
| Electroporated Cells ΔF% | Pulse type |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 300 ms |
2*150 ms with 150 ms interval |
|||||||||
| Total # | Missing€ # | Missing % | Transfected # | Transfected % | Total # | Missing€ # | Missing % | Transfected # | Transfected % | |
| 0–10% | 5 | 0 | 0.0 | 0 | 0.0 | 10 | 1 | 10.0 | 0 | 0.0 |
| 10–40% | 8 | 4 | 50.0 | 5 | 50.0 | 20 | 1 | 5.0 | 19 | 95.0 |
| 40–80% | 11 | 7 | 63.6 | 4 | 36.4 | 8 | 2 | 25.0 | 6 | 75.0 |
| Total | 24 | 11 | 45.8 | 8 | 33.3 | 38 | 4 | 10.5 | 25 | 65.8 |
Missing cells 24 hours after electroporation
Correlation of single-cell transfection success and extent of electroporation
We noticed some cells were not transfected although they were electroporated. This is because the mechanism of DNA delivery into the cells is different from that of small molecules. Rathenberg has visually demonstrated this point by tracking of plasmid delivery during SCEP with fluorescent labeled pDsRed-C1 plasmid and a small 54-bp fluoresceine-conjuated oligonucleotide. The small oligonucleotide entered cells immediately upon electroporation while the larger plasmid stayed at the electroporation point for at least 10 min 9. A further study by Golzio et al. depicted the same view. They conducted electroporation of CHO cells with propidium iodide (PI) and rhodamine labeled fluorescent GFP plasmid. As opposed to the prompt entering of PI into the cells after the pulses, the plasmid aggregated in the electropermeabilized part of the cells before it was transferred into the cytoplasm in the following minutes post the pulses 24. Although it is not known from direct experimental observation, a large number of theoretical papers claim that the pore size is about 1 nm in the electroporated region of the cell 25–27. This is obviously not sufficiently large for molecules like a DNA plasmid to diffuse through freely. Several proposals have been made to explain the different behavior of DNA when electro-transferred into the cells, including the formation of endosome-like vesicles stimulated by an electric field, transient complexation between bilayer and DNA, insertion of DNA into the bilayer and involvement of electrophoretic forces 28–32. In this work, we have tried to relate the extent of electroporation (as judged by ΔF%) with the transfection result as discussed below. The extent of electroporation is determined by the electroporation-induced loss of fluorescent Thioglo 1-GSH adduct and mass transfer rate of the small molecule across the transient pores.
We modified a quantitative model presented by Puc et al., 33 to fit the fluorescence intensity decay curves upon electroporation. This model describes the transmembrane transport of small molecules caused by electroporation. Our modifications are made based on (1) The cell volume (several pL) is much smaller than the buffer solution surrounding the cells (2 mL); (2) the pulse duration (300 ms) is negligible when compared to the whole electroporation kinetic process; (3) The fluorescence intensity has a linear relationship with the concentration of Thioglo 1-GSH inside the cells; (4) There is photobleaching that follows a first-order decay. We get an equation from the model to fit a whole-cell fluorescence decay profile upon electroporation as follows.
| (3) |
| (4) |
In equation (3), t is time from the start of the pulse, I(t) is the fluorescence intensity at time t, I0 is the fluorescence intensity at t = 0, k is the photobleaching rate (s−1 ), M is the mass transfer rate (s−1 ) through the transient pores and α is the pore resealing rate (s−1). Equation (3) has a photobleaching term and an electroporation term. From the electroporation term we can get the fluorescence intensity, Ī purely induced by electroporation. When the time t approaches infinity, the steady, photobleaching corrected fluorescence is given by
| (5) |
Thus the percentage fluorescence loss induced by electroporation is
| (6) |
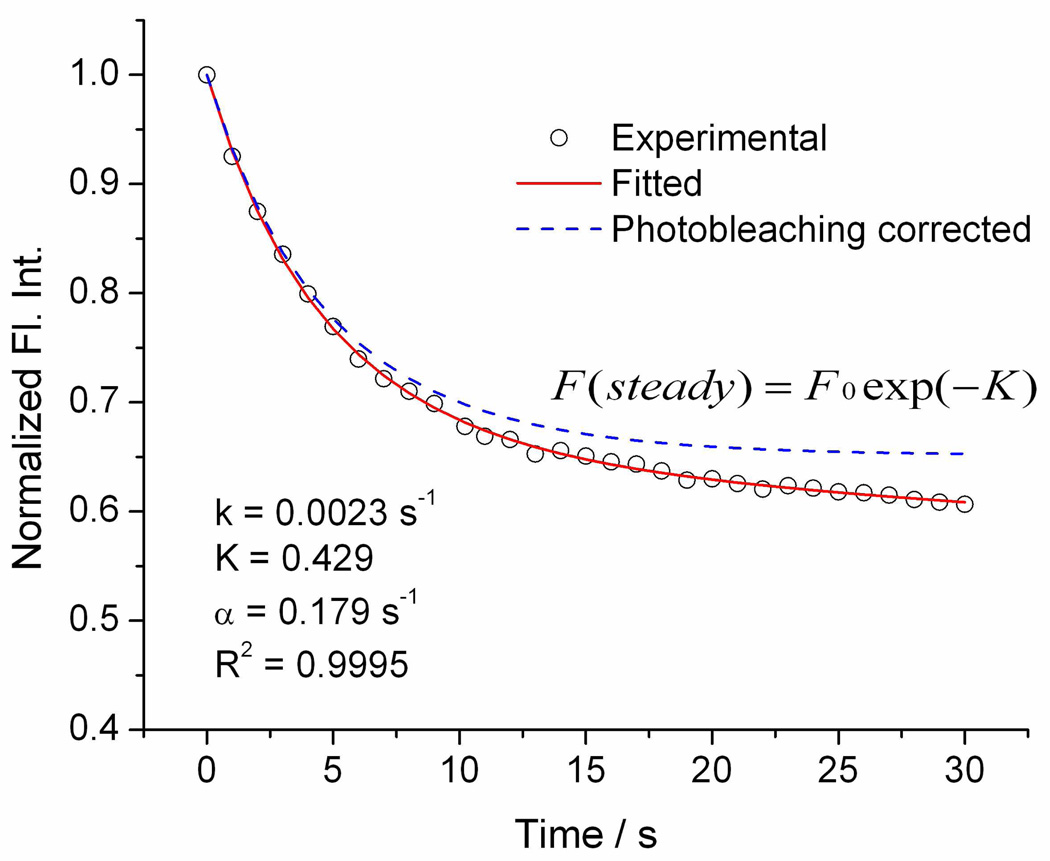
Fitting of the decay profiles of fluorescence after electroporation using Equation (3) gives good correlation coefficients. Figure 5 shows an example of fitting. Time zero is the point right before the sudden drop of whole-cell fluorescence intensity, which we consider as the time when the pores form. From the fitting we obtain k, α and K. Then M and ΔF% are calculated from equation (4) and equation (6) separately.
Figure 5.
An example of fitting Eq (1) to the normalized fluorescence intensity decay curve. Circles are experimentally observed fluorescence intensity, the red solid curve is the result from the fitting, and the blue dotted curve represents fluorescence intensity change induced purely by electroporation (i.e., photobleaching removed).
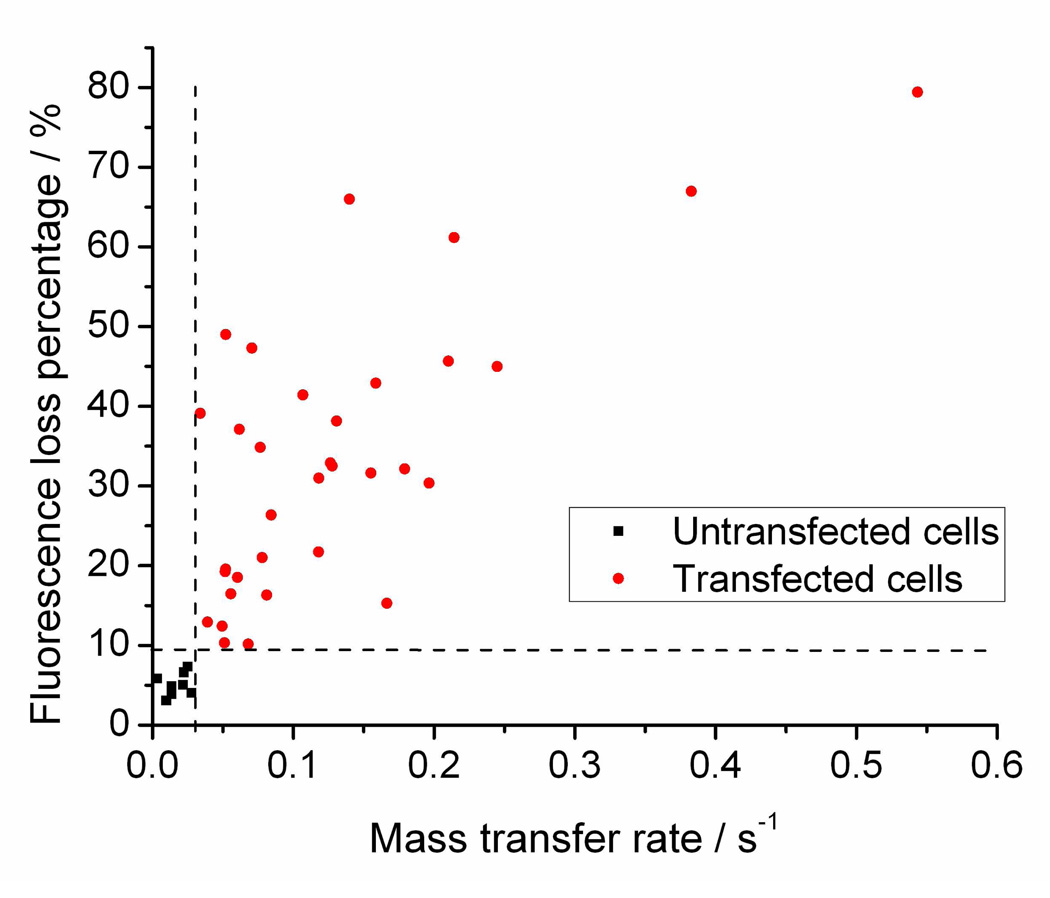
Exploring the effect of these parameters ΔF% (K), M and α on transfection gives us a clear relationship between the first two parameters and transfection. Table 2 shows results of a Student’s t-test testing the null hypothesis that the parameter in question is the same in transfected and untransfected cells. We assume that the transfected cells have larger mass transfer and longer resealing constant during electroporation, so one-tailed tests are performed here. The results give extremely small P (0.0000) values for M, K and ΔF%, indicating a strong relationship between these parameters and transfection, while α is apparently not related to transfection. Plotting of M and ΔF% on the transfected and untransfected cells gives a perfect prediction of successful transfections with M > 0.03 s−1 and ΔF% >10%, as shown in Figure 6. We have hereby for the first time established that the occurrence of transfection with SCEP using pulled capillaries is associated with a certain extent of electroporation characterized by small molecule efflux. This also clarifies that the mechanism of DNA delivery is different from small molecule diffusion. However, we have to point out that this criterion (M > 0.03 s−1 and ΔF% >10%) may change for different sized DNA. As Rae has shown, with increased DNA size, the voltage needed for transfection increases and the efficiency decreases 10. Vassanelli reported a similar trend using different plasmids 13. This is imaginable because larger DNA may need larger pore sizes for successful delivery. It would be very interesting to investigate further into how the dynamics of small molecule flux resulting from electroporation relate to the size and shape of DNA.
Table 2.
One-tailed Student’s t-test on the parameters K, M, α and ΔF% for transfected cells vs. untransfected cells.
| Parameters | Group* | Mean | SE. | [95% Conf. Interval] | P < t | |
|---|---|---|---|---|---|---|
| α | 0 | 0.339 | 0.060 | 0.196 | 0.481 | 0.5525 |
| 1 | 0.330 | 0.031 | 0.266 | 0.393 | ||
| M | 0 | 0.017 | 0.003 | 0.010 | 0.024 | 0.0000 |
| 1 | 0.129 | 0.018 | 0.093 | 0.167 | ||
| K | 0 | 0.052 | 0.005 | 0.040 | 0.065 | 0.0000 |
| 1 | 0.452 | 0.057 | 0.336 | 0.569 | ||
| ΔF% | 0 | 5.09 | 0.51 | 3.89 | 6.30 | 0.0000 |
| 1 | 33.49 | 3.05 | 27.27 | 39.70 | ||
Group 0 = untransfected cells, N=8; Group 1 = transfected cells, N=33.
Figure 6.
Each point represents a cell. Black squares stand for untransfected cells and red dots represent transfected cells. Clearly, large M corresponds to a large ΔF% and successful transfection. The two dashed lines define the zone for successful transfection: M > 0.03 s−1 and ΔF% > 10%.
Comparison with Lipofectamine 2000-mediated bulk transfection
When we compare the SCEP-based transfection to the control experiment of bulk transfection mediated by Lipofectamine 2000, we notice an average brighter EGFP fluorescence for the bulk method than the SCEP method. The dimmer fluorescence for EGFP expression after electroporation might be due to the loss of intracellular substance and resulting lag in cell growth. It is reported that depletion of GSH in A549 cells inhibited cell growth characterized by a longer lag phase than untreated cells 34. The same slow growth of the electroporated cells was also observed during our experiments, as we compare the target cell in Figure 2 to its neighboring cells. Nonetheless, our technique has much higher transfection efficiency than the bulk method. We found that 25 – 30% of cells were transfected with Lipofectamine 2000 treatment, while at least 80% of the cells electroporated to the required extent (M > 0.03 s−1 and ΔF% >10%) are transfected using the two-pulse method. Moreover, as we stated elsewhere, we can control the extent of cell electroporation by manipulating the electric field parameters (applied voltage, pulse duration, tip-cell distance, etc) to get maximized cell survivability and electroporation efficiency16, 17. Therefore, we can control SCEP-introduced transfection in cells as well.
Another advantage of our approach for single-cell transfection is that a small amount of plasmid-containing solution (< 1 µl) is needed for transfection of many cells. In our experiments, we limit the number of cells electroporated with a single capillary based on the time that the cells are on the electroporation/microscope stage. Usually the cells were handled outside the incubator for no more than 2 hours to keep them healthy. Thus, the maximum number of cells electroporated for a single cell dish was limited to about 15 using one capillary within the two-hour experiments. A further automation of the operation may increase the number. Of course, smaller capillaries can also be used to conserve plasmid.
CONCLUSIONS
We have shown successful transfection of single cells with pEGFP-C2 plasmid DNA by SCEP using a pulled capillary. This is the first time that this high-resolution electroporation approach has been used for single-cell transfection. Although the mechanism of DNA transport through the cell membrane is considered to be different from small molecules and remains unknown, our results demonstrate a strong correlation of transfection with small molecule loss. The use of the real time information provided by ΔF% is useful for predicting and also guiding transfection. Further study on other plasmid vectors with different sizes may reveal if the vector size affects this relationship and how. We took advantage of the electroosmotic flow inside the fused silica capillary and the extremely high electric field at the tip part to push plasmid into the cells through the transient pores, thus avoiding the extra effort of putting pressure on the capillary. A 2×150 ms pulse train with an interval of 150 ms worked better than a single 300 ms pulse in both transfection and in preventing cells from going missing 24 hours after electroporation.
Supplementary Material
The puller program for capillary fabrication, Lipofectamine-mediated bulk transfection prodecure, absorbance and emission spectra of EGFP and Thioglo 1-GSH adduct, numerical simulation of the electric field distribution around a 2-µm opening capillary during the pulses, and a video demonstrating electroporation of the transfected cell in Figure 2.
ACKNOWLEDGEMENTS
This research has been supported by NIH Grant R01 GM066018. We sincerely thank Dr. Michael Trakselis (Univeristy of Pittsburgh, Department of Chemistry) for providing the pEGFP-C2 vector and guidance in the plasmid solution treatment and bulk transfection.
REFERENCES
- 1.Davis BR, Brown DB, Prokopishyn NL, Yannariello-Brown J. Curr. Opin. Mol. Ther. 2000;2:412–419. [PubMed] [Google Scholar]
- 2.Han SW, Nakamura C, Obataya I, Nakamura N, Miyake J. Biochem. Biophys. Res. Commun. 2005;332:633–639. doi: 10.1016/j.bbrc.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 3.Knoblauch M, Hibberd JM, Gray JC, van Bel AJE. Nat. Biotechnol. 1999;17:906–909. doi: 10.1038/12902. [DOI] [PubMed] [Google Scholar]
- 4.Cuerrier CM, Lebel R, Grandbois M. Biochem. Biophys. Res. Commun. 2007;355:632–636. doi: 10.1016/j.bbrc.2007.01.190. [DOI] [PubMed] [Google Scholar]
- 5.Tirlapur UK, Konig K. Nature. 2002;418:290–291. doi: 10.1038/418290a. [DOI] [PubMed] [Google Scholar]
- 6.Lundqvist JA, Sahlin F, Aberg MAI, Stromberg A, Eriksson PS, Orwar O. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10356–10360. doi: 10.1073/pnas.95.18.10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Nat. Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- 8.Boudes M, Pieraut S, Valmier J, Carroll P, Scamps F. J. Neurosci. Methods. 2008;170:204–211. doi: 10.1016/j.jneumeth.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Rathenberg J, Nevian T, Witzemann V. J. Neurosci. Methods. 2003;126:91–98. doi: 10.1016/s0165-0270(03)00069-4. [DOI] [PubMed] [Google Scholar]
- 10.Rae JL, Levis RA. Pflugers Arch. 2002;443:664–670. doi: 10.1007/s00424-001-0753-1. [DOI] [PubMed] [Google Scholar]
- 11.Haas K, Sin W-C, Javaherian A, Li Z, Cline HT. Neuron. 2001;29:583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- 12.Wang HY, Lu C. Biotechnol. Bioeng. 2008;100:579–586. doi: 10.1002/bit.21784. [DOI] [PubMed] [Google Scholar]
- 13.Vassanelli S, Bandiera L, Borgo M, Cellere G, Santoni L, Bersani C, Salamon M, Zaccolo M, Lorenzelli L, Girardi S, Maschietto M, Dal Maschio M, Paccagnella A. N Biotechnol. 2008;25:55–67. doi: 10.1016/j.nbt.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Valero A, Post JN, van Nieuwkasteele JW, ter Braak PM, Kruijer W, van den Berg A. Lab Chip. 2008;8:62–67. doi: 10.1039/b713420g. [DOI] [PubMed] [Google Scholar]
- 15.Rols M-P, Teissie J. Biophys. J. 1998;75:1415–1423. doi: 10.1016/S0006-3495(98)74060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal A, Zudans I, Orwar O, Weber SG. Anal. Chem. 2007;79:161–167. doi: 10.1021/ac061270o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal A, Zudans I, Weber EA, Olofsson J, Orwar O, Weber SG. Analytical Chemistry. 2007;79:3589–3596. doi: 10.1021/ac062049e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmuir ME, Yang J-R, LeCompte KA, Durand RE. Fluorescence Microscopy and Fluorescent Probes; [Based on the Proceedings of the Conference on Fluorescence Microscopy and Fluorescent Probes]; June 25–28, 1995; Prague. 1996. pp. 229–233. [Google Scholar]
- 19.Kagan VE, Kuzmenko AI, Tyurina YY, Shvedova AA, Matsura T, Yalowich JC. Cancer Res. 2001;61:7777–7784. [PubMed] [Google Scholar]
- 20.Gaetjens EC, Chen P, Broome JD. Biochem. Biophys. Res. Commun. 1984;123:626–632. doi: 10.1016/0006-291x(84)90275-4. [DOI] [PubMed] [Google Scholar]
- 21.de Carmejane O, Schwinefus JJ, Wang S-C, Morris MD. J. Chromatogr., A. 1999;849:267–276. doi: 10.1016/s0021-9673(99)00527-0. [DOI] [PubMed] [Google Scholar]
- 22.Zudans I, Agarwal A, Orwar O, Weber SG. Biophys. J. 2007;92:3696–3705. doi: 10.1529/biophysj.106.097683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bilska AO, DeBruin KA, Krassowska W. Bioelectrochemistry. 2000;51:133–143. doi: 10.1016/s0302-4598(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 24.Golzio M, Rols MP, Teissie J. Methods. 2004;33:126–135. doi: 10.1016/j.ymeth.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Shil P, Bidaye S, Vidyasagar PB. J. Phys. D: Appl. Phys. 2008;41:055502. [Google Scholar]
- 26.Krassowska W, Filev PD. Biophys. J. 2007;92:404–417. doi: 10.1529/biophysj.106.094235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeBruin KA, Krassowska W. Biophys. J. 1999;77:1213–1224. doi: 10.1016/S0006-3495(99)76973-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mir LM. Methods Mol. Biol. 2008;423:3–17. doi: 10.1007/978-1-59745-194-9_1. [DOI] [PubMed] [Google Scholar]
- 29.Rols M-P. Biochim. Biophys. Acta, Biomembr. 2006;1758:423–428. doi: 10.1016/j.bbamem.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Spassova M, Tsoneva I, Petrov AG, Petkova JI, Neumann E. Biophys. Chem. 1994;52:267–274. doi: 10.1016/0301-4622(94)00097-4. [DOI] [PubMed] [Google Scholar]
- 31.Chernomordik LV, Sokolov AV, Budker VG. Biochim. Biophys. Acta, Biomembr. 1990;1024:179–183. doi: 10.1016/0005-2736(90)90222-a. [DOI] [PubMed] [Google Scholar]
- 32.Hristova NI, Tsoneva I, Neumann E. FEBS Lett. 1997;415:81–86. doi: 10.1016/s0014-5793(97)01097-1. [DOI] [PubMed] [Google Scholar]
- 33.Puc M, Kotnik T, Mir LM, Miklavcic D. Bioelectrochemistry. 2003;60:1–10. doi: 10.1016/s1567-5394(03)00021-5. [DOI] [PubMed] [Google Scholar]
- 34.Kang YJ, Enger MD. Exp. Cell Res. 1990;187:177–179. doi: 10.1016/0014-4827(90)90134-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The puller program for capillary fabrication, Lipofectamine-mediated bulk transfection prodecure, absorbance and emission spectra of EGFP and Thioglo 1-GSH adduct, numerical simulation of the electric field distribution around a 2-µm opening capillary during the pulses, and a video demonstrating electroporation of the transfected cell in Figure 2.