Abstract
Magnetic resonance imaging (MRI) and MR spectroscopy can probe a variety of physiological (e.g. blood vessel permeability) and metabolic characteristics of prostate cancer. However, little is known about the changes in gene expression that underlie the spectral and imaging features observed in prostate cancer. Tumor induced changes in vascular permeability and angiogenesis are thought to contribute to patterns of dynamic contrast enhanced (DCE) MRI images of prostate cancer even though the genetic basis of tumor vasculogenesis is complex and the specific mechanisms underlying these DCEMRI features have not yet been determined. In order to identify the changes in gene expression that correspond to MRS and DCEMRI patterns in human prostate cancers, we have utilized tissue print micropeel techniques to generate “whole mount” molecular maps of radical prostatectomy specimens that correspond to pre-surgical MRI/MRS studies. These molecular maps include RNA expression profiles from both Affymetrix GeneChip microarrays and quantitative reverse transcriptase PCR (qrt-PCR) analysis, as well as immunohistochemical studies. Using these methods on patients with prostate cancer, we found robust over-expression of choline kinase a in the majority of primary tumors. We also observed overexpression of neuropeptide Y (NPY), a newly identified angiogenic factor, in a subset of DCEMRI positive prostate cancers. These studies set the stage for establishing MRI/MRS parameters as validated biomarkers for human prostate cancer.
Keywords: MRI, prostate, gene expression
The wide range of biologic activity exhibited by prostate cancer creates dilemmas for the management of individuals with known or suspected disease. Prostate cancer is one of the few cancers that can grow so slowly that it never threatens the lives of many patients. Yet, in some patients, the disease spreads beyond the gland, and is incurable. This characteristic of prostate cancer is reflected in the fact that, although one out of six men will develop prostate cancer, only one out of thirty five will die of the disease [1].
Prostate cancer patients are being identified at an earlier and potentially treatable stage because of the adoption of screening using serum prostate-specific antigen (PSA) and transrectal ultrasound (TRUS) guided biopsy [2–6]. A major challenge to the medical community is to characterize the behavior of an individual patient’s prostate cancer and to select the appropriate level of management while preserving the patient’s quality of life. Current pre-treatment paradigms have significant limitations, both individually and collectively. One of the most widely used pretreatment paradigms is based on the Partin tables [7, 8], which are derived from a combination of demographic factors, as well as the Gleason score from biopsy samples, PSA assessments and DRE results. These tables are computed using population based probabilities and, as such, have limitations when making decisions about individual patients. Thus, a more comprehensive and objective means to characterize prostate cancer after its detection and prior to therapy would be very useful in clinical practice.
In general, apart from TRUS guided biopsy, imaging has had little impact on the detection, and subsequent characterization, of prostate cancer. This may seem somewhat surprising in light of the impact imaging has had on other cancers. Of all of the modalities available, magnetic resonance imaging (MRI) holds the most promise for providing both high-resolution anatomical images [9], functional [10, 11], and metabolic images of prostate cancer [9, 12]. These advantages have already been exploited by many groups who have evaluated MRI alone, or in combination with MRS, to stage and characterize prostate cancer. The development and application of dynamic contrast enhanced MRI has provided a potential tool to image the manifestations of angiogenesis in tumors, in general [10, 11, 13, 14], and in prostate cancer, specifically [10, 11]. The close connection between DCEMRI and angiogenesis and the current consensus that more aggressive tumors exhibit greater angiogenesis form the basis for suggesting that DCEMRI is an “imaging” biomarker. As an example, a recent review by Rehman and Jayson [15] discusses the role of DCEMRI as a molecular imaging tool in assessing the efficacy of antiangiogenic agents, both in clinical and preclinical studies. More broadly, Oehr, in his recent article [16], reviews the use of non-invasive imaging modalities to probe genomics, proteomics and metabolomics. The probing of metabolomics is particularly relevant to MRS where there is good evidence to suggest that the choline resonance can serve as a biomarker for malignancy [9, 12, 17]. While most of Oehr’s discussion revolves around the development and use of targeted contrast agents, Segal et al. [18] have shown that it is already possible to relate morphological features found in CT images of liver cancer in humans with underlying gene expression profiles.
All of these considerations prompted our group to undertake systematic studies in prostate cancer that attempt to correlate gene expression profiles with the different imaging features found in DCEMRI and MRS. Aside from the clinical importance of these studies, prostate cancer is a good system for these kinds of investigations since tissue from the entire gland is available from men who have undergone radical prostatectomies. Carefully step-sectioning of the gland in the same slice orientation employed for the MR studies provides the potential capability of spatially registering the results of “omic” kinds of studies with non-invasive in-vivo MRI. We have described (Gaston et al [19]) a “tissue printing” method that preserves the prostate tissue for detailed histopathology, while providing sufficient material to carry out a variety of biochemical and molecular profiling studies. In this report we describe, by example, how it is technically possible to obtain genomic, proteomic, and metabolimic data that can be spatially registered to the in-vivo MRI data and discuss the implications of our first results.
Materials and Methods
MR Protocols
At our institution all prostate cancer patients, whether they are evaluated for pre- or post-therapeutic assessment, undergo the same standardized comprehensive MR protocol:
Patient Preparation
Patients are asked to refrain from ejaculation for three (3) days preceding the exam to ensure optimal distension of the seminal vesicles. A sodium phosphate enema is administered on the day of the study in order to minimize fecal residue in the rectum. A 1 mg glucagon i.m. injection is administered to reduce peristaltic motion.
The endo-rectal coil (MRInnervu, Medrad, Pittsburgh, PA) is inserted into the rectum and connected to a pelvic phased-array coil with a coupling device to combine the surface phased array coil with the endorectal coil. A barium suspension is used to fill the balloon in the endo-rectal coil with a typical volume of 80 cc’s, however, adjusted for patient tolerance. The use of Barium reduces susceptibility effects when compared to the use of air and should improve MRS and DWI results [20] .
MRI and MRS are performed on a 3.0-T MR imaging system (Genesis Signa LX Excite, General Electric, Milwaukee, WI). The endo-rectal coil position is verified with sagittal T2-weighted localizer images and adjusted, if needed, so that the prostate gland is optimally situated with respect to the coil.
T2W images
transverse and coronal fast spin-echo T2-weighted images are obtained from below the prostatic apex to above the seminal vesicles using the following parameters: repetition time msec/echo time msec (effective) 4,500–7600/165, BW = 31.25 kHz, 2.0–2.2mm section thickness and no intersection gap, 3 averages, 14-cm field of view, 256 × 192 matrix, and no phase wrap. A unique attribute of this protocol is the use of thinner sections than have been the previous routine. This results in 30% – 50% more sampling of the gland compared to the traditional 3–4 mm section approach. The phase encoding direction is left-right.
DCEMRI. Three-dimensional (3D) T1-weighted (T1W) gradient echo (GRE) images
These are acquired prior to, during, and after contrast injection. Dynamic contrast enhanced (DCE) images are obtained after bolus injection of gadopentetate dimeglumine (Magnevist®; Berlex Laboratories, Wayne, NJ) at a dose of 0.1 mmol / kg of body weight administered with a mechanical injection system (Spectris® MR Injection System, MEDRAD, Pittsburgh, PA) at a flow rate of 4ml/sec. The imaging parameters of the 3D GRE sequence are: TR/TE 7.2/2.1 msec, BW = 31.25 kHz, flip angle 18°, FOV 14 cm, matrix 256 × 224, ST: 2.0–2.6mm, no phase wrap, which can be obtained with a temporal resolution of approximately 1 minute, 35 seconds. Two pre-contrast and five post-contrast sequential acquisitions are obtained.
For the pre-contrast scans, the first is used to ensure relevant anatomic coverage; the second is used as part of a continuous series of pre- and post-acquisitions in which the instrument settings (gain and attenuation values) are identical. Contrast injection is initiated during the last 10 seconds of the 2nd pre-contrast acquisition. See [21] for details.
MRSI Acquisitions
These studies were performed using the endo-rectal coil as the only receiver using the pulse sequence described by Cunningham et al. [22]. The 3D volume of interest was prescribed graphically from the axial T2W images. Spatial saturation pulses were also graphically prescribed around this volume of interest to avoid areas with high susceptibility gradients and areas that contained high lipid signals. The homogeneity of the selected region was automatically optimized using the automated linear shimming procedure provided by the manufacturer. Typical acquisition parameters were 8×8×8 phase encodes, TR 1300 msec, TE 144 mec. The resulting MRSI data were transferred to an offline workstation and processed using the SAGE spectral analysis package provided by GE.
Analysis of DCEMRI Images
The time series of 3D gradient echo images were processed at pixel resolution, using the three-time-point (3TP) model [23] to analyze the time evolution of contrast enhancement. The results were translated into a color-coded scheme as published previously [23–25].
Gene Expression Profiles
Tissue print micropeel mapping of molecular biomarkers
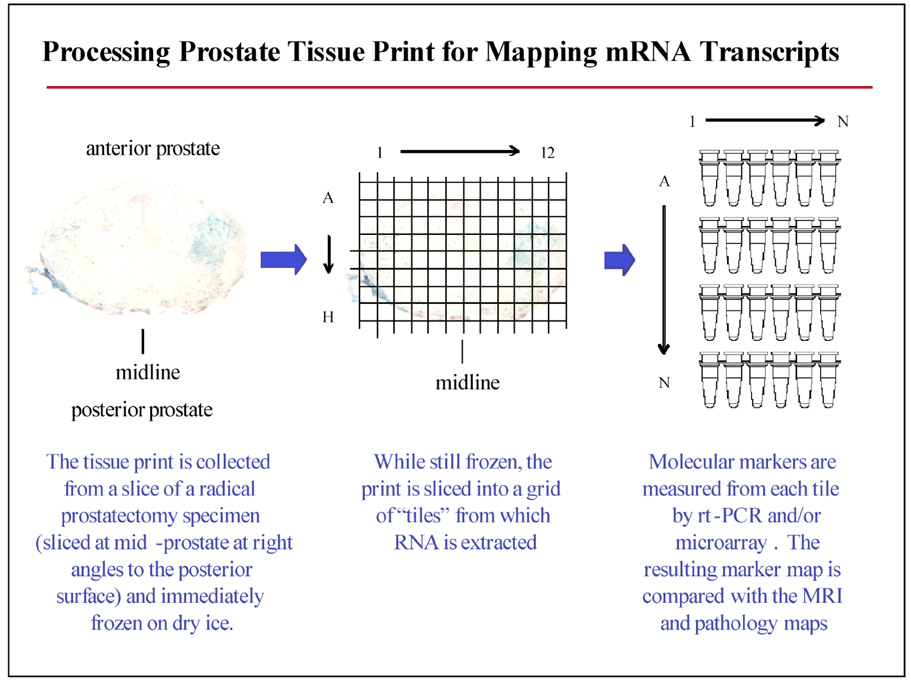
In order to identify the changes in gene expression that correspond to MRI-MRS and DCE-MRI visible changes in human prostate cancers, we have utilized tissue print micro-peel techniques [19] to generate “whole mount” molecular maps of radical prostatectomy specimens that correspond to pre-surgical MRI imaging studies. An example of this methodology is shown in Figure 1. Within one hour of surgical excision, the radical prostatectomy specimen was sliced in parallel to the presurgical MRI imaging planes. Tissue print micropeels were collected in duplicate from each specimen slice of the prostate specimen and immediately snap-frozen on dry ice. Registration between the print and tissue specimens is maintained by fiduciary markings and detailed photographic record. For molecular biomarker mapping, the print was marked and cut while still frozen in order to generate a grid of “tiles” from which RNA, DNA, and protein fractions were extracted. Using this approach, it is possible to efficiently extract DNA, RNA and protein from the prostate tissue prints in such a way that the molecular markers can be mapped back to a specific tile from the print, with the print in register with the formalin fixed, paraffin embedded (FFPE), tissue. Using this approach, we routinely obtain 200–400 ng of high quality RNA (RNA Integrity Index of 6 or better; Schroeder et al.,2006 [26]) from each 5×5 mm tile for the RNA analysis. Technical validation studies using Affymetrix focus arrays (over 8,500 verified human sequences) show that the global gene expression concordance between a tissue print and its corresponding 1 mm diameter tissue core sample is excellent, in the range of Pearson correlation coefficient of 0.96 – 0.98. (R2 approximately, 0.92 – 0.96) (Gaston et al. Manuscript in preparation)
Figure 1.
Generating “whole mount” gene expression maps using the tissue print micropeel technique.
RNA analysis
In our global gene expression analysis of RNA purified from tissue print tiles, we employed the Affymetrix U133 Plus 2.0 array platform (U133 Plus 2.0 represents 47,000 human gene transcripts and variants). RNA was extracted and purified from a 5×5 mm tile centered in the tumor and another 5×5 mm tile centered in an anatomically matched benign region on the opposite side (see Figure 2). Prior to labeling, a 100 ng aliquot of total RNA purified from each tissue print tile was amplified using a modified Eberwine-based protocol designed to maintain the representation of mRNA expression in samples with minimal starting material, and the array metrics were consistent with excellent biological representation of the microarray data (present calls >40%). Array results obtained by our academic core genomics facility were confirmed by a commercial laboratory that serves as a fully licensed service provider for Affymetrix. We employed qrt-PCR RNA analysis to validate the results of our microarray differential gene expression profiles in the original samples. Validated genes of interest were then assessed using qrt-PCR in the RNA samples that had been isolated from each of the tissue print tiles in the map, as described in [19].
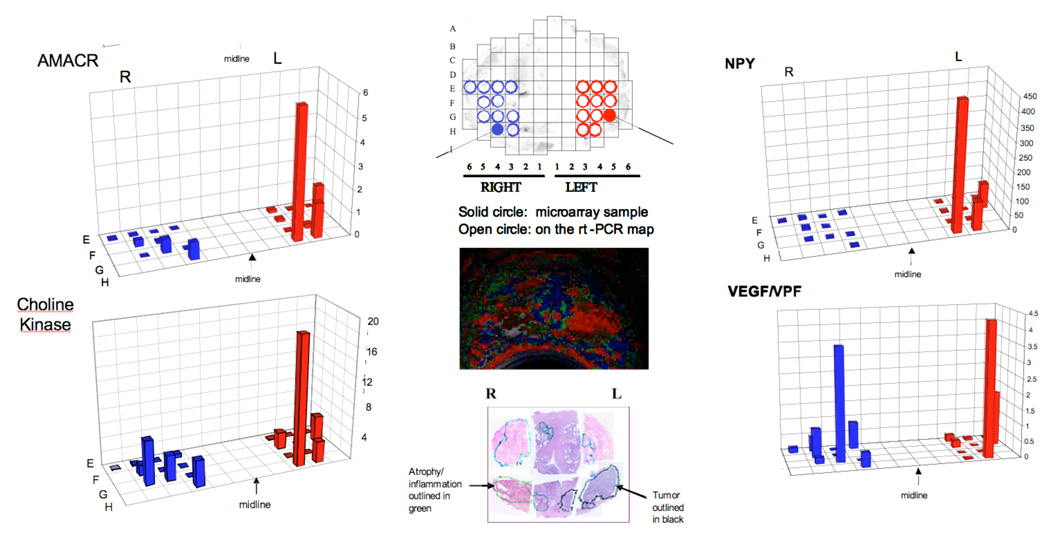
Figure 2.
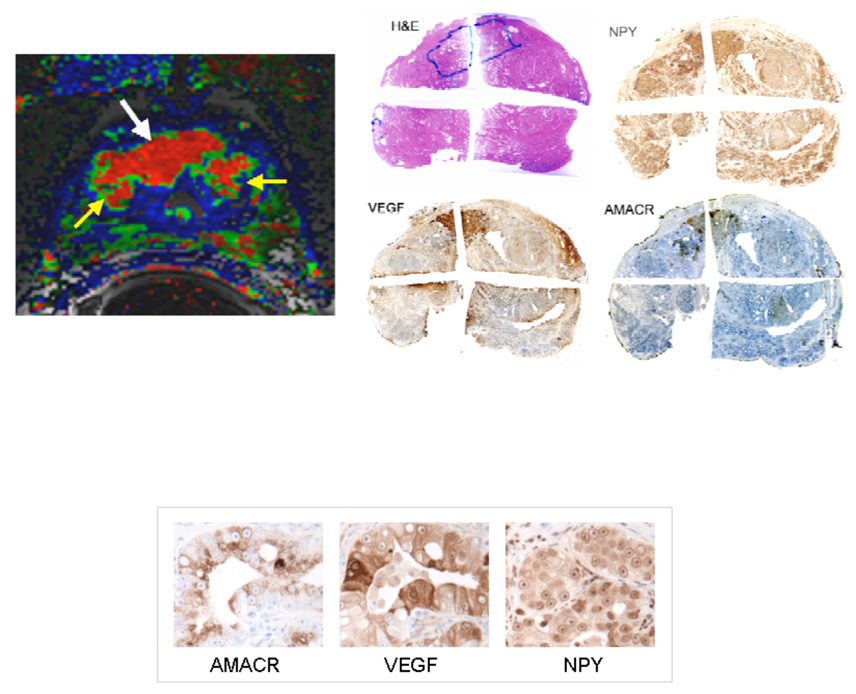
Spatial overlay maps showing results of RNA microarray and rt-PCR analysis for AMACR, Choline kinase, NPY and VEGF. The color-coded maps derived from the analysis of DCEMRI as well as the histopathology (H and E stain) are shown for spatial reference. Note the two solid circles indicated in the tissue print grid that show the regions used for microarray analysis.
Immunohistochemical analysis
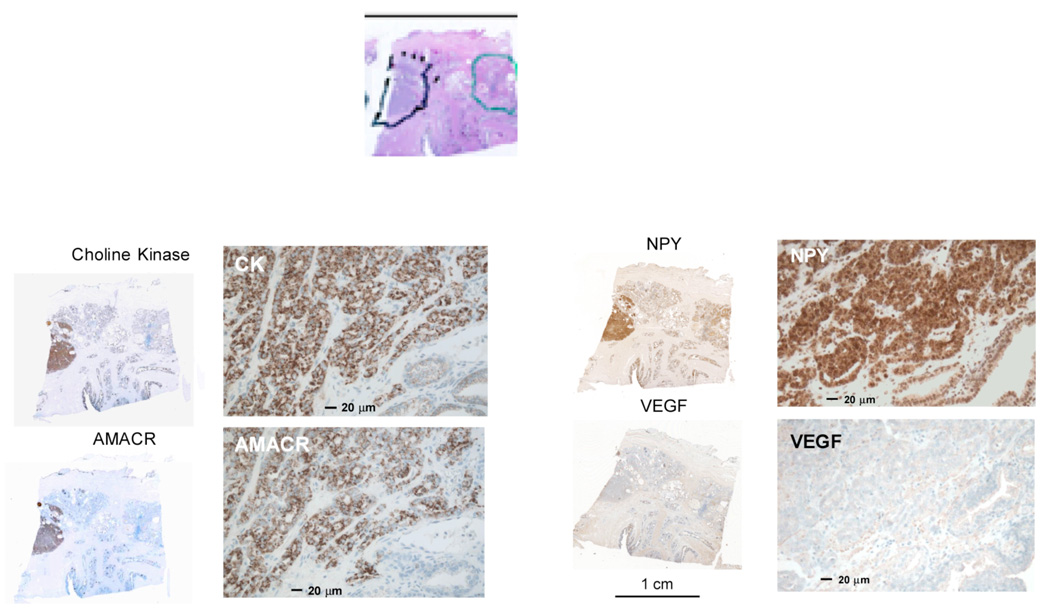
4 micron serial sections were prepared from FFPE blocks prepared from the tissue region shown in Figure 3. The first and tenth sections were stained using a standard H&E and reviewed and marked by a GU pathologist in order to identify regions of histological cancer, inflammatory changes, and tissue atrophy. For immunohistochemistry, successive sections from each block were stained individually using primary antibodies (rat, human) for a-aylmethyl co-enzyme A racemase (AMACR; Zeta Corp., Sierra Madre, CA), hepsin (#100022, Cayman Chemical Company, Ann Arbor MI), choline kinase (SC-23382, Santa Cruz Biotechnology, Santa Cruz CA), vascular endothelial growth factor (VEGF) (SC7269, Santa Cruz Biotechnology, Santa Cruz CA), and neuropeptide Y (NPY) (#RDI-PRO16054, Research Diagnostics, Inc, Concord MA). These were then stained using horseradish peroxidase methods optimized for prostate tissue [27]. The slides were then examined and digitally photographed under a light microscope. Staining intensity was evaluated relative to a tissue microarray with a reference set of prostate cancers that was processed in parallel with the slides of interest and analyzed with the an Automated Cellular Imaging System (ASIC II, Chromavision Medical Systems Inc.). Where available, slides were also prepared using the appropriate blocking peptide or the secondary antibody as controls.
Figure 3.
Immunohistochemical stains obtained using horseradish peroxidase readout of sections taken from the bottom-middle section of the gland shown in Figure 2 as indicated above in the stained sections. The tumor is outlined in black, a region showing chronic inflammatory changes is shown in green. Note that these images are shown in the pathologist’s convention (real left, right), whereas the images shown in Figure 2 are shown in the radiology convention, (patient left, right). The subjective intensity of the stain is greatest for NPY and least for VEGF; these visual impressions were confirmed by automated image analysis.
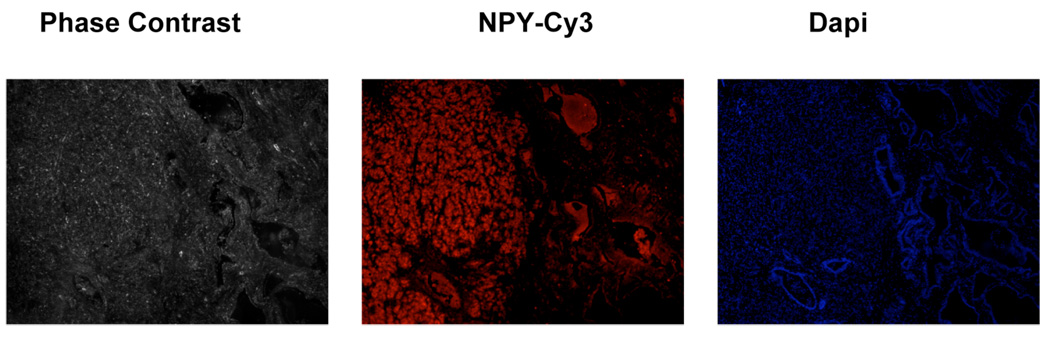
The tissue slices were prepared as above for the immunofluorescence studies. A slice of tissue was labeled with the primary antibody for NPY, followed by staining with Jackson Cy3-conjugated Donkey anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, Inc, West Grove PA). An additional slice was
stained using 4',6-diamidino-2-phenylindole (DAPI), (Invitrogen Corporation, Carlsbad, CA) which was used to stain for nuclear DNA. These slices were examined using a NIKON Eclipse TE 300 Microscope, in the phase contrast and fluorescence mode.
Results
The color-coded map derived from a pixel-by-pixel analysis of the axial DCEMRI images is shown in the middle of Figure 2. Note the presence of the red nodular region in the left side of the gland. This region corresponds to the tumor outlined in the histopathology shown below in Figure 2. The global gene expression profile carried out on the two tiles indicated in Figure 2 showed that there were 380 genes that showed significant up-regulation, as judged by a signal log ratio of tumor/contra-lateral side that was greater than 1.0 (greater than 2 fold change). Of these, eleven genes known to play a role in angiogenesis showed 3 fold or greater over-expression. The most highly over-expressed of these genes was neuropeptide Y (NPY) which showed a 49 fold ratio of tumor to benign, whereas VEGF showed a 3.8 fold ratio of tumor to benign. The distribution of hepsin, choline kinase, VEGF and NPY as determined using rt-PCR analysis for the RNA from the 5×5 mm tiles is shown in Figure 2. AMACR is over-expressed in almost all localized prostate cancers; AMACR staining is used in Figure 3 to visualize the area of tumor. Note that in this case of localized prostate cancer, a high level of NPY mRNA over-expression is evident both by Affymetrix array (49 fold over-expression) and rt-PCR (400 fold over-expression comparing highest tiles, tumor/benign) and confirmed by immunohistochemistry shown in Figure 3 (staining intensity in the highest 20% of reference primary prostate cancers, with intense cytoplasmic staining, ACIS IHC quantitation). In contrast, VEGF mRNA over-expression is relatively low in this tumor, with 3.8 fold over expression by Affymetrix and 1.2 fold over-expression (highest tiles, tumor/benign) by rt-PCR. By immunohistochemistry, VEGF staining intensity for this tumor is in the lowest 20% of reference primary prostate cancers (ACIS IHC quantitation). The levels of choline-kinase over-expression, shown in Figure 2A by rt-PCR and in Figure 3 by immunohistochemistry, are greater than those found for VEGF but considerably below those found for NPY. Figure 4 confirms that there is robust overexpression of NPY in this tumor; immunofluroescent detection of NPY is shown in comparison to both a phase-contrast and DAPI stained image of the same section. In principle, this method, (immunofluorescence), should lend itself to more precise and accurate quantitation than the horseradish peroxide staining [27, 28].
Figure 4.
An immunofluorescence stained section showing NPY (middle) taken from the bottom-middle section of the gland shown in Figure 2. The phase-contrast image is shown on the left. The DAPI stain is shown on the right.
Another example of the results of immunohistochemical staining is shown in Figure 5. This tumor (marked in blue) is located predominantly in the anterior portion of the gland. The color-coded 3TP map shows good concordance with the H and E stain, the NPY stain and the AMACR stain. The VEGF stain shows that most but not all of this tumor over-expresses VEGF. The VEGF positive areas of this cancer show greater staining intensity than observed in the tumor in figure 3; corresponding to the third (middle) quintile of reference primary prostate cancers. In contrast to the very high level of NPY overexpression seen in the figure 3 tumor, NPY staining in the figure 5 tumor corresponded to the second lowest quintile in the same reference series (ACIS IHC quantitation), with minimal cytoplasmic staining.
Figure 5.
Immunohistochemical stains obtained using horseradish peroxidase readout of the prostate of a patient with proven prostate cancer. The tumor is outlined in blue of the H and E stain in the series of low magnification images showing an entire slice of the gland. The color-coded image obtained from an analysis of the non-invasive DCEMRI sequence is shown on the left. The red region indicated by the white arrow is the tumor. The yellow arrows are showing two BPH nodules adjacent to the tumor. The intensity of VEGF and NPY expression shown in the high magnification images reflects the same conditions of IHC staining as shown in the high magnification images in Figure 3.
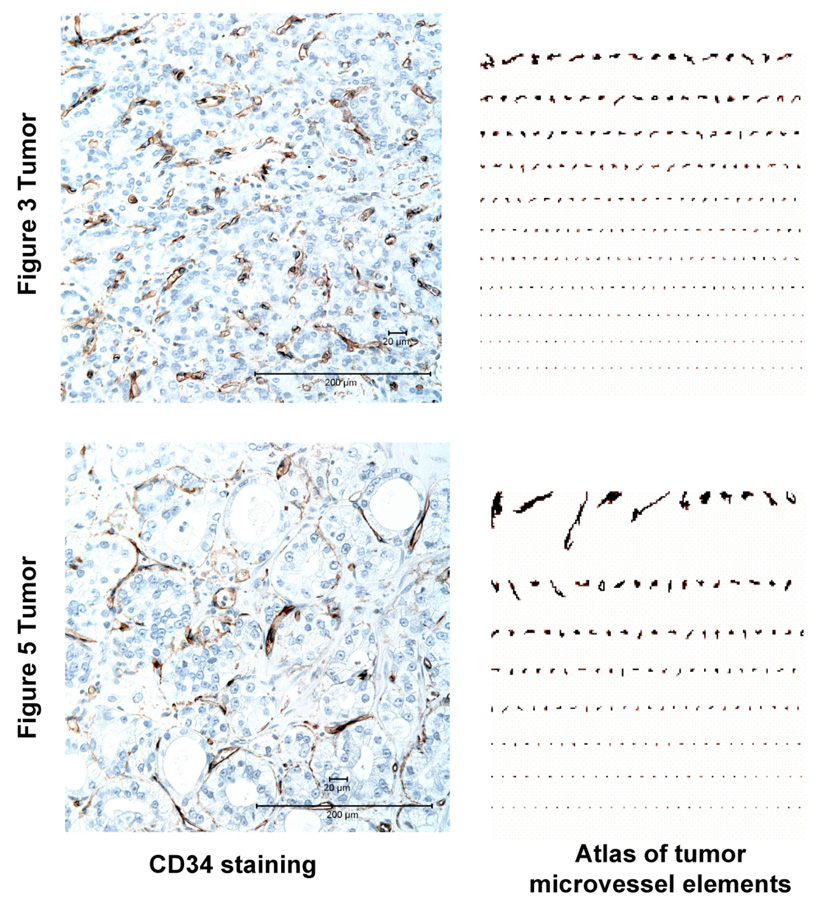
We compared the pattern of tumor microvessels in the Figure 3 and Figure 5 prostate cancers using CD34 staining (Figure 6). In the Figure 3 tumor, CD34 shows a microvasculature that is relatively uniform in vessel diameter and spacing; in contrast, the Figure 5 tumor microvasculature is highly heterogeneous in structure. The difference in the pattern of tumor microvasculature is particularly evident in an image analysis of the vascular elements, presented in Figure 6 as a microvascular atlas [29].
Figure 6.
Contrasting Patterns of Tumor Microvasculature in the Prostate Cancers Shown in Figure 3 and Figure 5. CD34 IHC staining was used to visualize tumor microvessels. An image analysis “atlas” of the microvessel elements highlight the difference between these two cancers, with the more uniform pattern evident in the Figure 3 tumor being characteristic of a more “mature” tumor vasculature.
Discussion
The general ability to map genomic and proteomic expressions to in-vivo imaging information opens the potential for new insights into disease characterization and a better understanding of disease mechanisms. In turn, such new insights seem well suited for diseases like prostate cancer that show heterogeneous clinical expression and that demand more specific information upon which to base management decisions for an afflicted individual’s personalized management. MRI and MRS offer a variety of ways to probe the in-vivo features of biologic processes, including cancer.
One of the almost universal findings in MRS of tumors is the elevation of the composite resonance assigned to the trimethyl group of choline containing compounds (for a review see [17]). The spectral patterns observed on MRS of the prostate and their interpretations have recently been reviewed by Rajesh et al. [30] . The relative level of the choline peak has been shown to correlate with the Gleason grade of the tumor [31, 32]. In spite of the importance of this peak, it is still unclear as to what alterations in genetic expression leads to choline elevation in prostate cancer. The lifetime of choline in the blood and its distribution in the body, as well as its transport into the cells, are critical steps in the metabolic fate of choline. The transport is the rate-limiting step for the synthesis of acetylcholine [33] and of PCho [34]. The transporters of choline, as well as choline kinase isozymes, were recently characterized by Eliyahu et al. [35] in normal and malignant mammary cells. In addition to two specific transporters, (CHT1 and OCT2), choline kinase a was shown to be up-regulated in the malignant transformation of breast tissue. Choline kinase a has been shown to be up-regulated in prostate cancer [36], and the implications of this up-regulation for potential targeted therapies has been discussed by Glunde and Bhujwalla [37]. In this light, it is interesting to point out that both F-18 [38, 39] and C-11 [40] labeled choline have been used in PET studies to visualize primary prostate tumors. Since choline kinase a is one of the major enzymes that converts choline to phosphocholine, it is reasonable to hypothesize that these radiotracers may be probing its over-expression. The methods shown here provide a means for spatially correlating the levels of over-expression of choline kinase a with the level of the choline resonance observed on in vivo MRSI studies.
The induction of new blood vessels is a key event in the progression of a solid tumor as it grows beyond 1–2 mm in diameter and can no longer depend on simple passive diffusion to provide nutrients and clear toxic wastes (for a review see Folkman [41]). Both the outgrowth of new blood vessels from established vessels (angiogenesis) and the formation of new microvessels from circulating endothelial cells (vasculogenesis) can be co-opted by a cancer [42]. Tumor induced blood vessel formation has proven to be a complex pathological event, and the early hopes for anti-angiogenesis therapies have been tempered by the realization that cancers utilize multiple molecular pathways to induce and remodel a vascular network. Model systems can be used to selectively manipulate the formation, regression and maturation of tumor blood vessels in order to identify the genetic pathways involved in these processes. However, it is unclear how faithfully these models replicate the pathologies of human tumors. Assessments of angiogenic phenotypes using a variety of biomarkers are only beginning to be included into the clinical assessments of a patient’s malignancy. There is great interest in developing, validating and utilizing, non-invasive imaging technologies to replace these more invasive tests.
Vascular endothelial growth factor (VEGF) is one of the most intensively studied angiogenic factors in human cancers, and VEGF has generally been reported to be over-expressed in prostate cancer relative to benign prostate tissue. Thus, it is reasonable to expect that most, if not all, of the tumors that exhibit abnormal DCEMRI patterns would show over-expression of VEGF. However, it has been reported that the levels of VEGF over-expression in primary human prostate cancers are highly variable, both at the RNA and protein levels [43–46]. The tissue print mapping techniques described in this paper have allowed us to undertake a detailed analyses of gene expression patterns in low-VEGF, DCE-MRI positive prostate cancers to determine which pathways were involved in the angiogenesis of these tumors. Interestingly, one of the genes most highly over-expressed in the low-VEGF cancer shown in Figure 2 is neuropeptide Y (NPY), which showed a 49-fold ratio tumor/benign on Gene Array. This high level of NPY over-expression was confirmed by rt-PCR and immunohistochemistry. In our ACIS IHC analysis of tissue microarrays that contain cores from progressively higher grades of radical prostatectomy cancers, we observed NPY overexpression early in development of this malignancy, with the most intense staining in a subset of cases that include approximately 15% of the Gleason 7 tumors. Rasiah et al (47) saw a similar pattern of expression in their patient population, and noted that NPY overexpression could also be detected in high grade PIN.
Although other groups have recongized NPY as one of many neuroendocrine markers that are over-expressed in a subset of prostate cancers, the ramifications of its over-expression has not yet been established [47, 48]. In a number of experimental systems, NPY has been shown to be a potent factor for stimulating angiogenesis in benign tissues [49–53]. Moreover, NPY can act as a proangiogenesis factor in experimental models of two different neuroendocrine cancers: neuroblastoma and Ewing’s sarcoma [54]. In addition to its direct proangiogenic activity, it has been suggested that NPY can also modify the angiogenic response to VEGF [55]. However, very few published studies have attempted a direct evaluation of NPY as an angiogenic factor in naturally occurring human cancers. The significance of NPY over-expression is that in model systems NPY produces micro vessels that appear to be more mature and less “leaky” than those induced by VEGF [49, 55]. In principle, DCEMRI should be able to distinguish tumors with these more mature vessels from tumors with more leaky vessels. For example, in the models commonly employed in the pharmacokinetic analyses of DCEMRI results, e.g. the so called Tofts Model [56–58], the values of the parameter, Ktrans, should be higher in tumors with leaky vessels. If this hypothesis were correct, there would be a non-invasive way to identify high VEGF expressing tumors from high NPY expressing tumors. DCEMRI could be used to identify patients who would respond to anti-angiogenic therapy based on anti-VEGF approaches and also monitor changes in response to therapy.
In summary, the examples shown here indicate that it may be possible to map expression profiles of proteins and enzymes that are upregulated in tumors with MRI/MRS related parameters on a pixel-by-pixel basis. MR imaging data can be employed to direct tissue sampling for genetic micro-array analysis to identify candidate genes for further immunohistochemical studies. The advantage of the immunohistochemical studies using fluorescence readouts is that they can provide quantitative spatial maps of protein expression that can be directly related to the MRI/MRS results. This approach can be exploited to determine whether the overexpression of a particular protein or enzyme results in measurable change in a MRI/MRS parameter. For example, it is possible to determine whether the level of total choline seen in the prostate cancer by MRS is correlated with the level of overexpression of choline kinase a or the levels of choline transporters, or both. Using this kind of approach, it should be possible to determine which, if any, of the many MRI/MRS parameters that are currently available are true biomarkers for characterization of the disease processes being investigated.
Acknowledgments
This work was supported, in part, by a grant from the Ellison Foundation and grants from the National Institutes of Health, CA116465-02 (NMR), CA112220 and CA116866 (SMG). BNB was supported, in part, by Bayer Healthcare Pharmaceuticals. The authors would like to thank Dr. Melissa Upton, Dr. Jihad Hayek (pathology), and Dr. Fabio Grizzi (microvessel image analysis) for their guidance and assistance.
References
- 1.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 2.Andriole G, Djavan B, Fleshner N, Schroder F. The case for prostate cancer screening with prostate-specific antigen. European Urology Supplements. 2006;5:737–745. [Google Scholar]
- 3.Catalona WJ, Smith DS, Ratliff TL, Basler JW. Detection of Organ-Confined Prostate-Cancer Is Increased through Prostate-Specific Antigen-Based Screening. Jama-Journal of the American Medical Association. 1993;270:948–954. [PubMed] [Google Scholar]
- 4.Efstathiou JA, Chen MH, Catalona WJ, McLeod DG, Carroll PR, Moul JW, Roehl KA, D’Amico AV. Prostate-specific antigen-based serial screening may decrease prostate cancer-specific mortality. Urology. 2006;68:342–347. doi: 10.1016/j.urology.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 5.Han M, Partin AW, Piantadosi S, Epstein JI, Walsh PC. Era specific biochemical recurrence-free survival following radical prostatectomy for clinically localized prostate cancer. Journal of Urology. 2001;166:416–419. [PubMed] [Google Scholar]
- 6.van der Cruijsen-Koeter IW, van der Kwast TH, Schroder FH. Interval carcinomas in the European Randomized Study of Screening for Prostate Cancer (ERSPC)-Rotterdam. Journal of the National Cancer Institute. 2003;95:1462–1466. doi: 10.1093/jnci/djg057. [DOI] [PubMed] [Google Scholar]
- 7.Partin AW, Kattan MW, Subong ENP, Walsh PC, Wojno KJ, Oesterling JE, Scardino PT, Pearson JD. Combination of prostate-specific antigen, clinical stage, and gleason score to predict pathological stage of localized prostate cancer - A multi-institutional update. Jama-Journal of the American Medical Association. 1997;277:1445–1451. [PubMed] [Google Scholar]
- 8.Partin AW, Mangold LA, Lamm DM, Walsh PC, Epstein JI, Pearson JD. Contemporary update of prostate cancer staging nomograms (Partin Tables) for the new millennium. Urology. 2001;58:843–848. doi: 10.1016/s0090-4295(01)01441-8. [DOI] [PubMed] [Google Scholar]
- 9.Huzjan R, Sala E, Hricak H. Magnetic resonance imaging and magnetic resonance spectroscopic imaging of prostate cancer. Nature Clinical Practice Urology. 2005;2:434–442. doi: 10.1038/ncpuro0296. [DOI] [PubMed] [Google Scholar]
- 10.Alonzi R, Padhani AR, Allen C. Dynamic contrast enhanced MRI in prostate cancer. European Journal of Radiology. 2007;63:335–350. doi: 10.1016/j.ejrad.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 11.Padhani AR, Harvey CJ, Cosgrove DO. Angiogenesis imaging in the management of prostate cancer. Nature Clinical Practice Urology. 2005;2:596–607. doi: 10.1038/ncpuro0356. [DOI] [PubMed] [Google Scholar]
- 12.Casciani E, Gualdi GF. Prostate cancer: value of magnetic resonance spectroscopy 3D chemical shift imaging. Abdom Imaging. 2006 doi: 10.1007/s00261-006-9029-8. [DOI] [PubMed] [Google Scholar]
- 13.Dadiani M, Furman-Haran E, Degani H. The application of NMR in tumor angiogenesis research. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;49:27–44. [Google Scholar]
- 14.Hylton N. Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. Journal of Clinical Oncology. 2006;24:3293–3298. doi: 10.1200/JCO.2006.06.8080. [DOI] [PubMed] [Google Scholar]
- 15.Rehman S, Jayson GC. Molecular imaging of antiangiogenic agents. Oncologist. 2005;10:92–103. doi: 10.1634/theoncologist.10-2-92. [DOI] [PubMed] [Google Scholar]
- 16.Oehr P. 'Omics'-based imaging in cancer detection and therapy. Personalized Medicine. 2006;3:19–32. doi: 10.2217/17410541.3.1.19. [DOI] [PubMed] [Google Scholar]
- 17.Negendank W. Studies of human tumors by MRS: a review. NMR Biomed. 1992;5:303–324. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- 18.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 19.Gaston SM, Soares MA, Siddiqui MM, Vu D, Lee JM, Goldner DL, Brice MJ, Shih JC, Upton MP, Perides G, Baptista J, Lavin PT, Bloch BN, Genega EM, Rubin MA, Lenkinski RE. Tissue-print and print-phoresis as platform technologies for the molecular analysis of human surgical specimens: mapping tumor invasion of the prostate capsule. Nat Med. 2005;11:95–101. doi: 10.1038/nm1169. [DOI] [PubMed] [Google Scholar]
- 20.Rosen Y, Bloch BN, Lenkinski RE, Greenman RL, Marquis RP, Rofsky NM. 3T MR of the prostate: reducing susceptibility gradients by inflating the endorectal coil with a barium sulfate suspension. Magn Reson Med. 2007;57:898–904. doi: 10.1002/mrm.21166. [DOI] [PubMed] [Google Scholar]
- 21.Bloch BN, Rofsky NM, Baroni RH, Marquis RP, Pedrosa I, Lenkinski RE. 3 Tesla magnetic resonance imaging of the prostate with combined pelvic phased-array and endorectal coils; Initial experience(1) Acad Radiol. 2004;11:863–867. doi: 10.1016/j.acra.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham CH, Vigneron DB, Marjanska M, Chen AP, Xu D, Hurd RE, Kurhanewicz J, Garwood M, Pauly JM. Sequence design for magnetic resonance spectroscopic imaging of prostate cancer at 3 T. Magn Reson Med. 2005;53:1033–1039. doi: 10.1002/mrm.20478. [DOI] [PubMed] [Google Scholar]
- 23.Degani H, Gusis V, Weinstein D, Fields S, Strano S. Mapping pathophysiological features of breast tumors by MRI at high spatial resolution. Nat Med. 1997;3:780–782. doi: 10.1038/nm0797-780. [DOI] [PubMed] [Google Scholar]
- 24.Bloch BN, Furman-Haran E, Helbich TH, Lenkinski RE, Degani H, Kratzik C, Susani M, Haitel A, Jaromi S, Ngo L, Rofsky NM. Prostate cancer: accurate determination of extracapsular extension with high-spatial-resolution dynamic contrast-enhanced and T2-weighted MR imaging--initial results. Radiology. 2007;245:176–185. doi: 10.1148/radiol.2451061502. [DOI] [PubMed] [Google Scholar]
- 25.Furman-Haran E, Degani H. Parametric analysis of breast MRI. J Comput Assist Tomogr. 2002;26:376–386. doi: 10.1097/00004728-200205000-00012. [DOI] [PubMed] [Google Scholar]
- 26.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubin MA, Zerkowski MP, Camp RL, Kuefer R, Hofer MD, Chinnaiyan AM, Rimm DL. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164:831–840. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 29.Grizzi F, Russo C, Colombo P, Franceschini B, Frezza EE, Cobos E, Chiriva-Internati M. Quantitative evaluation and modeling of twodimensional neovascular network complexity: the surface fractal dimension. BMC Cancer. 2005;5:14. doi: 10.1186/1471-2407-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajesh A, Coakley FV, Kurhanewicz J. 3D MR spectroscopic imaging in the evaluation of prostate cancer. Clin Radiol. 2007;62:921–929. doi: 10.1016/j.crad.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Coakley FV, Qayyum A, Kurhanewicz J. Magnetic resonance imaging and spectroscopic imaging of prostate cancer. J Urol. 2003;170:S69–S75. doi: 10.1097/01.ju.0000094958.23276.c4. discussion S75-6. [DOI] [PubMed] [Google Scholar]
- 32.Zakian KL, Sircar K, Hricak H, Chen HN, Shukla-Dave A, Eberhardt S, Muruganandham M, Ebora L, Kattan MW, Reuter VE, Scardino PT, Koutcher JA. Correlation of proton MR spectroscopic imaging with gleason score based on step-section pathologic analysis after radical prostatectomy. Radiology. 2005;234:804–814. doi: 10.1148/radiol.2343040363. [DOI] [PubMed] [Google Scholar]
- 33.Haga T. Synthesis and release of ( 14 C)acetylcholine in synaptosomes. J Neurochem. 1971;18:781–798. doi: 10.1111/j.1471-4159.1971.tb12008.x. [DOI] [PubMed] [Google Scholar]
- 34.Katz-Brull R, Degani H. Kinetics of choline transport and phosphorylation in human breast cancer cells; NMR application of the zero trans method. Anticancer Res. 1996;16:1375–1380. [PubMed] [Google Scholar]
- 35.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: Molecular and biochemical studies. Int J Cancer. 2007 doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 36.de Molina AR, Rodriguez-Gonzalez A, Gutierrez R, Martinez-Pineiro L, Sanchez JJ, Bonilla F, Rosell R, Lacal JC. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochemical and Biophysical Research Communications. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 37.Glunde K, Jacobs MA, Bhujwalla ZM. Choline metabolism in cancer: implications for diagnosis and therapy. Expert Review of Molecular Diagnostics. 2006;6:821–829. doi: 10.1586/14737159.6.6.821. [DOI] [PubMed] [Google Scholar]
- 38.Kwee SA, Coel MN, Lim J, Ko JHP. Prostate cancer localization with (18)fluorine fluorocholine positron emission tomography. Journal of Urology. 2005;173:252–255. doi: 10.1097/01.ju.0000142099.80156.85. [DOI] [PubMed] [Google Scholar]
- 39.Kwee SA, Wei H, Sesterhenn I, Yun D, Coel MN. Localization of primary prostate cancer with dual-phase F-18-fluorocholine PET. Journal of Nuclear Medicine. 2006;47:262–269. [PubMed] [Google Scholar]
- 40.Reske SN, Blumstein NM, Neumaier B, Gottfried HW, Finsterbusch F, Kocot D, Moller P, Glatting G, Perner S. Imaging prostate cancer with C-11-choline PET/CT. Journal of Nuclear Medicine. 2006;47:1249–1254. [PubMed] [Google Scholar]
- 41.Folkman J. Angiogenesis. Annu Rev Med. 2006;57:1–18. doi: 10.1146/annurev.med.57.121304.131306. [DOI] [PubMed] [Google Scholar]
- 42.Dome B, Hendrix MJ, Paku S, Tovari J, Timar J. Alternative vascularization mechanisms in cancer: Pathology and therapeutic implications. Am J Pathol. 2007;170:1–15. doi: 10.2353/ajpath.2007.060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaib H, Cockrell EK, Rubin MA, Macoska JA. Profiling and verification of gene expression patterns in normal and malignant human prostate tissues by cDNA microarray analysis. Neoplasia. 2001;3:43–52. doi: 10.1038/sj.neo.7900126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Gohary YM, Silverman JF, Olson PR, Liu YL, Cohen JK, Miller R, Saad RS. Endoglin (CD105) and vascular endothelial growth factor as prognostic markers in prostatic adenocarcinoma. Am J Clin Pathol. 2007;127:572–579. doi: 10.1309/X6NXYE57DLUE2NQ8. [DOI] [PubMed] [Google Scholar]
- 45.Soulitzis N, Karyotis I, Delakas D, Spandidos DA. Expression analysis of peptide growth factors VEGF, FGF2, TGFB1, EGF and IGF1 in prostate cancer and benign prostatic hyperplasia. Int J Oncol. 2006;29:305–314. [PubMed] [Google Scholar]
- 46.Walsh K, Sriprasad S, Hopster D, Codd J, Mulvin D. Distribution of vascular endothelial growth factor (VEGF) in prostate disease. Prostate Cancer Prostatic Dis. 2002;5:119–122. doi: 10.1038/sj.pcan.4500575. [DOI] [PubMed] [Google Scholar]
- 47.Rasiah KK, Kench JG, Gardiner-Garden M, Biankin AV, Golovsky D, Brenner PC, Kooner R, O’Neill GF, Turner JJ, Delprado W, Lee CS, Brown DA, Breit SN, Grygiel JJ, Horvath LG, Stricker PD, Sutherland RL, Henshall SM. Aberrant neuropeptide Y and macrophage inhibitory cytokine-1 expression are early events in prostate cancer development and are associated with poor prognosis. Cancer Epidemiol Biomarkers Prev. 2006;15:711–716. doi: 10.1158/1055-9965.EPI-05-0752. [DOI] [PubMed] [Google Scholar]
- 48.Ruscica M, Dozio E, Motta M, Magni P. Modulatory actions of neuropeptide Y on prostate cancer growth: role of MAP kinase/ERK 1/2 activation. Adv Exp Med Biol. 2007;604:96–100. doi: 10.1007/978-0-387-69116-9_7. [DOI] [PubMed] [Google Scholar]
- 49.Ekstrand AJ, Cao R, Bjorndahl M, Nystrom S, Jonsson-Rylander AC, Hassani H, Hallberg B, Nordlander M, Cao Y. Deletion of neuropeptide Y (NPY) 2 receptor in mice results in blockage of NPY-induced angiogenesis and delayed wound healing. Proc Natl Acad Sci U S A. 2003;100:6033–6038. doi: 10.1073/pnas.1135965100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee EW, Grant DS, Movafagh S, Zukowska Z. Impaired angiogenesis in neuropeptide Y (NPY)-Y2 receptor knockout mice. Peptides. 2003;24:99–106. doi: 10.1016/s0196-9781(02)00281-4. [DOI] [PubMed] [Google Scholar]
- 51.Lee EW, Michalkiewicz M, Kitlinska J, Kalezic I, Switalska H, Yoo P, Sangkharat A, Ji H, Li L, Michalkiewicz T, Ljubisavljevic M, Johansson H, Grant DS, Zukowska Z. Neuropeptide Y induces ischemic angiogenesis and restores function of ischemic skeletal muscles. J Clin Invest. 2003;111:1853–1862. doi: 10.1172/JCI16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zukowska Z, Grant DS, Lee EW. Neuropeptide Y: a novel mechanism for ischemic angiogenesis. Trends Cardiovasc Med. 2003;13:86–92. doi: 10.1016/s1050-1738(02)00232-3. [DOI] [PubMed] [Google Scholar]
- 53.Zukowska-Grojec Z, Karwatowska-Prokopczuk E, Rose W, Rone J, Movafagh S, Ji H, Yeh Y, Chen WT, Kleinman HK, Grouzmann E, Grant DS. Neuropeptide Y: a novel angiogenic factor from the sympathetic nerves and endothelium. Circ Res. 1998;83:187–195. doi: 10.1161/01.res.83.2.187. [DOI] [PubMed] [Google Scholar]
- 54.Kitlinska J. Neuropeptide Y (NPY) in neuroblastoma: effect on growth and vascularization. Peptides. 2007;28:405–412. doi: 10.1016/j.peptides.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 55.Kitlinska J, Abe K, Kuo L, Pons J, Yu M, Li L, Tilan J, Everhart L, Lee EW, Zukowska Z, Toretsky JA. Differential effects of neuropeptide Y on the growth and vascularization of neural crest-derived tumors. Cancer Res. 2005;65:1719–1728. doi: 10.1158/0008-5472.CAN-04-2192. [DOI] [PubMed] [Google Scholar]
- 56.Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, Jayson G, Judson IR, Knopp MV, Maxwell RJ, McIntyre D, Padhani AR, Price P, Rathbone R, Rustin G, Tofts PS, Tozer GM, Vennart W, Waterton JC, Williams SR, Workman P. Assessment of antiangiogenic and antivascular therapeutics using MRI: recommendations for appropriate methodology for clinical trials. British Journal of Radiology. 2003;76:S87–S91. doi: 10.1259/bjr/15917261. [DOI] [PubMed] [Google Scholar]
- 57.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. Jmri-Journal of Magnetic Resonance Imaging. 1997;7:91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 58.Tofts PS, Brix G, Buckley DL, Evelhoch JL, Henderson E, Knopp M, Larsson HBW, Lee TY, Mayr NA, Parker GJM, Port RE, Taylor J, Weisskoff RM. Estimating kinetic parameters from dynamic contrastenhanced T-1-weighted MRI of a diffusable tracer: Standardized quantities and symbols. Jmri-Journal of Magnetic Resonance Imaging. 1999;10:223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]