Abstract
Resonance Raman spectra are measured for Tt H-NOX WT and three other Tt H-NOX proteins containing mutations at key conserved residues to determine the heme conformation in solution. The most dramatic changes in heme conformation occurred in the O2-bound forms, and the single Tt H-NOX P115A mutation was sufficient to generate a significant relaxation of the chromophore. Clear evidence of heme relaxation in the Tt H-NOX I5L, P115A, and I5L/P115A mutants in solution is demonstrated by the observation of reduced resonance Raman intensities for several out-of-plane low frequency modes (e.g. γ11, γ12, γ13, and γ15) in the 400–750 cm−1 region known to be sensitive to ruffling and saddling deformations, as well as increased vibrational frequencies for the core heme skeletal stretching modes, ν3, ν2, and ν10. In addition, all three mutants exhibited some degree of heme conformational heterogeneity based on several broad skeletal markers (e.g. ν10) in the high frequency region. These results are comparable to those observed by Olea et al. for Tt H-NOX P115A in crystal form, where four different heme structures were determined from a single unit cell. Based on the resonance Raman spectra, it is clear that the actual heme conformation for Tt H-NOX P115A in solution is considerably more relaxed than WT, with increased flexibility within the protein pocket, allowing for rapid sampling of alternate conformations.
Keywords: Resonance Raman spectroscopy, H-NOX, Thermoanaerobacter tengcongensis, porphyrin distortion, vibrational analysis, normal coordinate structural decomposition (NSD), O2 binding
Nonplanar porphyrin conformations are of great interest and widely investigated due to their potential role in regulating biochemical properties and activity in proteins containing either heme or chlorophyll cofactors (1–7). Over the last three decades, several methods have been utilized to study these heme deformations in proteins. These include theoretical approaches using DFT and QM/MM methodologies (2, 6, 8, 9), vibrational spectroscopy of model compounds isolating specific distortions (3, 5, 10–12), and the normal coordinate structural decomposition (NSD) method developed by Shelnutt et al. (4, 7, 13) to quantify specific heme deformations observed in x-ray crystal structures. In nature, some of the largest heme deformations typically occur in the c-type cytochromes and peroxidases with out-of-plane displacements greater than 1 Å (7). However, recent crystallographic work by Pellicena et al. shows that the Heme-Nitric oxide and/or OXygen binding (H-NOX) domain from Thermoanaerobacter tengcongensis also contains a highly distorted heme structure (14). Subsequent H-NOX crystal structures indicate that the heme chromophore can sample a range of nonplanar conformations (15, 16).
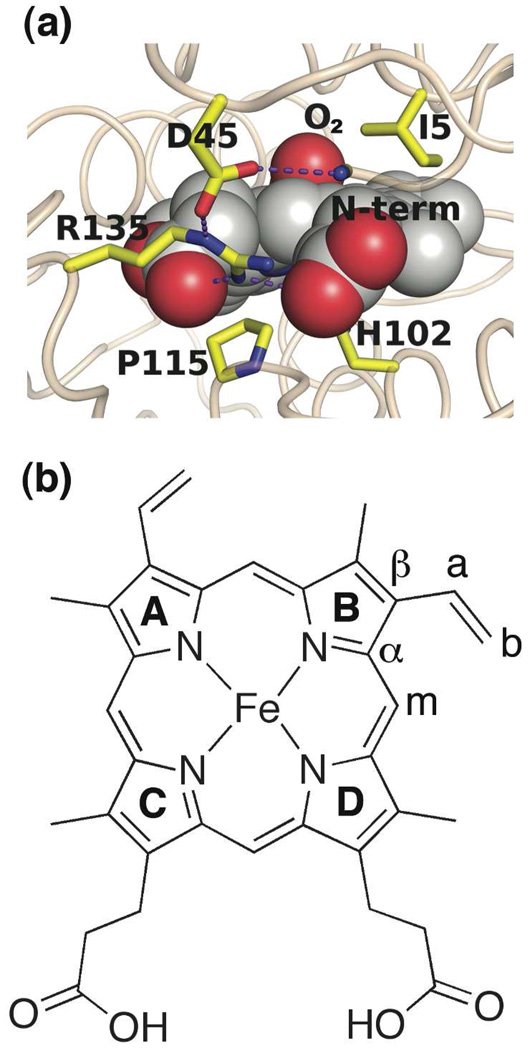
The H-NOX family of heme proteins has the unique property that some proteins bind only NO and CO, whereas others additionally bind O2 (17, 18). The first crystal structure obtained within this family was the O2-bound H-NOX domain from the bacterial obligate anaerobe, Thermoanaerobacter tengcongensis (Tt H-NOX) (14). One of the most distinctive features about this O2-bound structure is the significant heme deformation (Figure 1a). Energy minimization calculations by Pellicena et al. predicted that Ile-5, Pro-115, and Leu-144 maintained nonbonded contacts with the heme, inducing this observed deformation in Tt H-NOX (14, 19). Of those three residues, Pro-115 is of particular interest since it is conserved within this family. Thus, a key focus in investigating these H-NOX domains is to better understand how these conserved residues may contribute towards regulating heme distortion, and thereby possibly controlling ligand specificity.
Figure 1.
(a) Structure of the heme pocket of the Tt H-NOX WT FeII-O2 complex, indicating the distal Ile-5 and proximal Pro-115 thought to be responsible for the distorted heme structure (PDB 1U55). (b) Heme molecular structure and labeling scheme.
Recently, Olea et al. have shown via x-ray crystallography that mutating Pro-115 to alanine in Tt H-NOX alters the protein pocket such that the heme relaxes and samples several conformations, including more planar structures than that observed in the WT protein (20). However, the presence of four different heme structures in the unit cell complicated the determination of which conformation is actually dominant in solution. To address this question, a different technique that can probe the heme structure in solution is required.
Because of its selectivity and sensitivity to molecular structure, resonance Raman spectroscopy is an invaluable tool for specifically probing protein-bound heme conformations in solution (Figure 1b). Detailed information on the ground state geometry and electronic structure can be obtained from the vibrational frequencies; furthermore, the RR intensities provide insight into the symmetry and equilibrium geometry, and may be used to probe excited state dynamics (21, 22). In addition, RR spectroscopy is useful for investigating heme deformations because the distortions within the chromophore cause frequency shifts in the skeletal stretching modes (denoted as “ν”) and activate the intensity of low frequency out-of-plane (oop) modes (denoted as “γ”) (5, 7, 21, 23).
In this study, we use resonance Raman spectroscopy to investigate the effects of mutating the distal Ile-5 and proximal Pro-115 residues in Tt H-NOX on the heme structure. The spectra show that these residues are important in Tt H-NOX for maintaining a specific protein environment, and that mutating Ile-5 and Pro-115 relaxes the solution chromophore conformation for the O2 complexes. Specifically, our RR spectra suggest that the single P115A mutation leads to the largest change in the heme structure, whereas the Tt H-NOX I5L and I5L/P115A double mutants maintain conformations that are more similar to Tt H-NOX WT. These conformational changes likely occur as a consequence of heme pocket rearrangements to accommodate the substituted residues. We compare these relaxations to the quantified changes observed in crystal form using the normal coordinate structural decomposition (NSD) analysis developed by Shelnutt et al. (4, 7, 11, 13).
MATERIALS AND METHODS
Protein expression and purification
Expression and purification of the Tt H-NOX domain were performed as previously described (19) with the following modifications. Thawed cell pellets resuspended in buffer A [50 mM TEA, 20 mM NaCl, 5% glycerol, 1 mM Pefabloc (Pentapharm), pH 7.5] were lysed at 4 °C with an Emulsiflex-C5 high-pressure homogenizer at 15000 psi (Avestin, Inc.) upon addition of DNAse I and lysozyme (Sigma). Lysed cells were centrifuged for 1 h at 42000 rpm at 4 °C, and the supernatant was heat-denatured at 75 °C for 45 min. The denatured protein was centrifuged again for 1 h at 42000 rpm, and the supernatant was concentrated to < 10 mL using 10 K MWCO spin concentrators (Vivaspin). Concentrated protein was then loaded onto a prepacked Superdex S75 Hiload 26/60 gel filtration column (Pharmacia) equilibrated with buffer A, and fractions containing Tt H-NOX were pooled and applied to a POROS HQ 7.9 mL (1 × 10 cm, 10 µm) anion-exchange column (Applied Biosystems) at 5–10 mL/min. Flow-through containing the Tt H-NOX domain was collected and stored at −80 °C. Site-directed mutagenesis was carried out using the QuikChange® protocol (Stratagene), and verified by sequencing (UC Berkeley sequencing core).
Sample preparation
The purified Tt H-NOX protein was brought into an anaerobic glovebag, and oxidized using ~5–10 mM potassium ferricyanide to remove the bound O2. The ferricyanide was removed using a PD10 desalting column (Amersham Biosciences) equilibrated with buffer B (50 mM TEA, 50 mM NaCl, pH 7.5). Following oxidation and desalting, the protein was reduced with ~5–20 mM sodium dithionite that was removed using a PD10 desalting column upon complete reduction of the heme. The 16O2 complexes were generated by opening the reduced protein to air. To make the 18O2 (95% 18O2; Cambridge Isotopes) complexes, gas was added to a sealed Reacti-Vial (Pierce) containing FeII-unligated protein. Commercially available horse heart myoglobin and horse muscle hemoglobin (Sigma) samples were prepared similarly. Final sample concentrations for the Raman experiments were typically 15 to 50 µM. All UV/Vis absorption samples were prepared and measured as previously described (19, 24).
Resonance Raman spectroscopy
All spectra were collected using the 413.1 nm line from a Kr+ laser (Spectra-Physics model 2025) focused to a beam diameter of ~60 µm with a 50 mm focal-length excitation lens. Raman scattering was detected with a cooled, back-illuminated CCD (LN/CCD-1100/PB; Roper Scientific) controlled by an ST-133 controller coupled to a subtractive dispersion double spectrograph (25). The laser power at the sample was ~2 mW, and a microspinning sample cell was used to minimize photoinduced degradation. Typical data acquisition times were 30 to 60 min. Electronic absorption spectra were obtained before and after the Raman experiments to verify that no photoinduced degradation occurred. Raman spectra were corrected for wavelength dependence of the spectrometer efficiency with a white lamp, and the instrument was calibrated using the Raman frequencies from cyclohexane, CCl4, and toluene. The reported frequencies are accurate to ±1 cm−1, and the spectral bandpass was set to 8 cm−1. For each Raman spectrum, the raw data were baseline-corrected, and the buffer background signal was subtracted. Spectral analysis and decomposition were performed using Igor Pro (WaveMetrics). Isotopic shifts were approximated on the basis of a simple harmonic oscillator (HO) model.
Structural deformation analysis
To deconvolute the distortions from Tt H-NOX and other heme proteins, we used the web-based version of the normal-coordinate structural decomposition method (NSD) developed by Jentzen et al. to input our heme coordinates and quantify the deformations for the porphyrin macrocycle (4, 7, 11, 13). This program uses a porphyrin reference of D4h-symmetry to describe the heme distortions in terms of displacements along the lowest frequency out-of-plane normal coordinates within the molecule. The generated output quantifies the amount of each deformation type necessary to model the overall observed distortions within a given heme structure. Coordinates from previous crystal structures were obtained from the RCSB Protein Data Bank (14, 20, 26–29). For all proteins used in our NSD calculations, the heme is oriented such that the vinyl groups remain in quadrants I and II as defined by Jentzen et al. (4) in order to maintain the directionality in the absolute signs of the deformation types. In our analysis, the minimal basis set was used to quantify the different types of heme distortion. This basis set only includes the lowest frequency mode from each of the 6 in-plane and 6 out-of-plane normal deformations, and has been shown to adequately describe heme distortions in proteins (13). The overall magnitude of out-of-plane distortions (Δoop) for the heme is defined as the square root of the squared sum of observed z axis displacements for the 24-atom porphyrin macrocycle (C20N4). This total out-of-plane distortion was determined from the complete basis set, which gives the total deformation of each symmetry type (13).
RESULTS
Electronic absorption characterization
As an initial step toward characterizing the effects of mutating Ile-5 and Pro-115 in Tt H-NOX, we obtained electronic absorption spectra of the proteins with different ligands. Table 1 summarizes the Soret and α/β bands measured for Tt H-NOX in the unligated, CO, NO, and O2-bound forms, and includes the globins for comparison (30).
Table 1.
Electronic absorption properties for Tt H-NOX, Hb, and Mb in the reduced and CO, NO, and O2-bound forms.a
| Protein | Ligand | Soret | α/β | Ref. |
|---|---|---|---|---|
| Tt WT | reduced | 430 | 563 | (19) |
| Tt P115A | 432 | 563 | this work | |
| Tt I5L | 431 | 569 | this work | |
| Tt P115A/I5L | 431 | 566 | this work | |
| Hb | 430 | 555 | (30) | |
| Mb | 434 | 556 | (30) | |
| Tt WT | CO | 423 | 567/541 | (19) |
| Tt P115A | 424 | 568/538 | this work | |
| Tt I5L | 424 | 565/541 | this work | |
| Tt P115A/I5L | 424 | 570/542 | this work | |
| Hb | 419 | 569/540 | (30) | |
| Mb | 423 | 579/542 | (30) | |
| Tt WT | NO | 420 | 575/547 | (19) |
| Tt P115A | 418 | 573/543 | this work | |
| Tt I5L | 421 | 572/539 | this work | |
| Tt P115A/I5L | 421 | 568/535 | this work | |
| Hb | 418 | 575/545 | (30) | |
| Mb | 421 | 575/543 | (30) | |
| Tt WT | O2 | 416 | 591/556 | (19) |
| Tt P115A | 416 | 584/548 | this work | |
| Tt I5L | 418 | 587/550 | this work | |
| Tt P115A/I5L | 417 | 588/552 | this work | |
| Hb | 415 | 576/541 | (30) | |
| Mb | 418 | 580/542 | (30) | |
All peak positions are reported in nm.
Both the unligated and CO complexes show small changes in absorption upon mutating Ile-5 and Pro-115. The unligated Tt H-NOX WT spectrum displays a characteristic Soret at ~430 nm and a broad α/β band at ~563 nm, indicative of a 5-coordinate, high-spin species. Compared to Tt H-NOX WT, the P115A, I5L, and I5L/P115A mutants produce minor red shifts between 1–6 nm in the Soret and α/β bands. Addition of CO to the reduced, unligated Tt H-NOX WT protein shifts the electronic absorption features to display the Soret at ~423 nm and discernible α/β bands at ~567 nm and ~541 nm, confirming the presence of a 6-coordinate, low-spin CO complex. All three Tt H-NOX mutants display similar Soret values at 424 nm, but deviate by 1–3 nm in the split α/β band region compared to Tt H-NOX WT.
Addition of NO to the reduced, unligated Tt H-NOX WT protein results in a 6-coordinate, low-spin NO complex (Soret: 420 nm, α/β bands: 575 nm and 547 nm). A slight blue shift of 2–4 nm is observed in the Soret and α/β bands for the Tt H-NOX P115A NO complex. Although the Soret only shifts to 421 nm for the Tt H-NOX I5L mutant, its α/β bands decrease by 3 and 8 nm to 572 and 539 nm, respectively. The largest blue shift is observed in the α/β region for the Tt H-NOX I5L/P115A mutant (7 and 12 nm, respectively).
As previously shown, Tt H-NOX produces a stable 6-coordinate, low-spin O2 complex upon exposure of a reduced sample to air (19). The mutants shift by 6–7 nm to shorter wavelengths in the α/β region compared to Tt H-NOX WT (Soret: 416 nm, α/β bands: 591 nm and 556 nm). The largest shift is observed for the Tt H-NOX P115A mutant with α/β bands at ~584 and ~548 nm.
In summary, the NO and O2 complexes show consistent shifts in the α/β bands toward shorter wavelengths upon mutating Ile-5 and Pro-115 in Tt H-NOX, whereas the unligated and CO complexes exhibit both shorter and longer wavelength changes for the mutants. The overall changes observed in the electronic absorption spectra are relatively minor, but consistently reproducible. These observed shifts are plausibly introduced from structural rearrangements within the heme pocket, which then change the internal electric field. However, interpreting the changes in the α/β region is more complicated due to the different factors that influence its shape and splitting pattern, including contributions from electronic and vibronic perturbations, heme-protein interactions, and vibronic coupling strength redistributions (31).
Resonance Raman spectroscopy
To better characterize the effects of mutating Ile-5 and Pro-115 on the Tt H-NOX heme solution structure, we performed resonance Raman measurements on the unligated and oxygenated forms (Figure 2 and Figure 3). The main heme skeletal marker bands, ν4, ν3, ν2, and ν10, are summarized in Table 2 and compared to other proteins (32). The π-electron density marker (ν4), assigned as the pyrrole breathing mode, varies between 1350–1380 cm−1 depending on the oxidation state of the heme. The spin and coordination state markers, ν3, ν2, and ν10, correspond to Cα-Cm and Cβ-Cβ stretching vibrations in the macrocycle (21, 23). Together, these skeletal markers in the 1350–1650 cm−1 region can be used to detect different conformations as a result of their sensitivity to the porphyrin core size and π-conjugation. In addition, the Fe-His stretching vibration at ~220 cm−1 is a sensitive probe of the bound histidine, providing unique structural details about the heme environment in the proximal pocket (33).
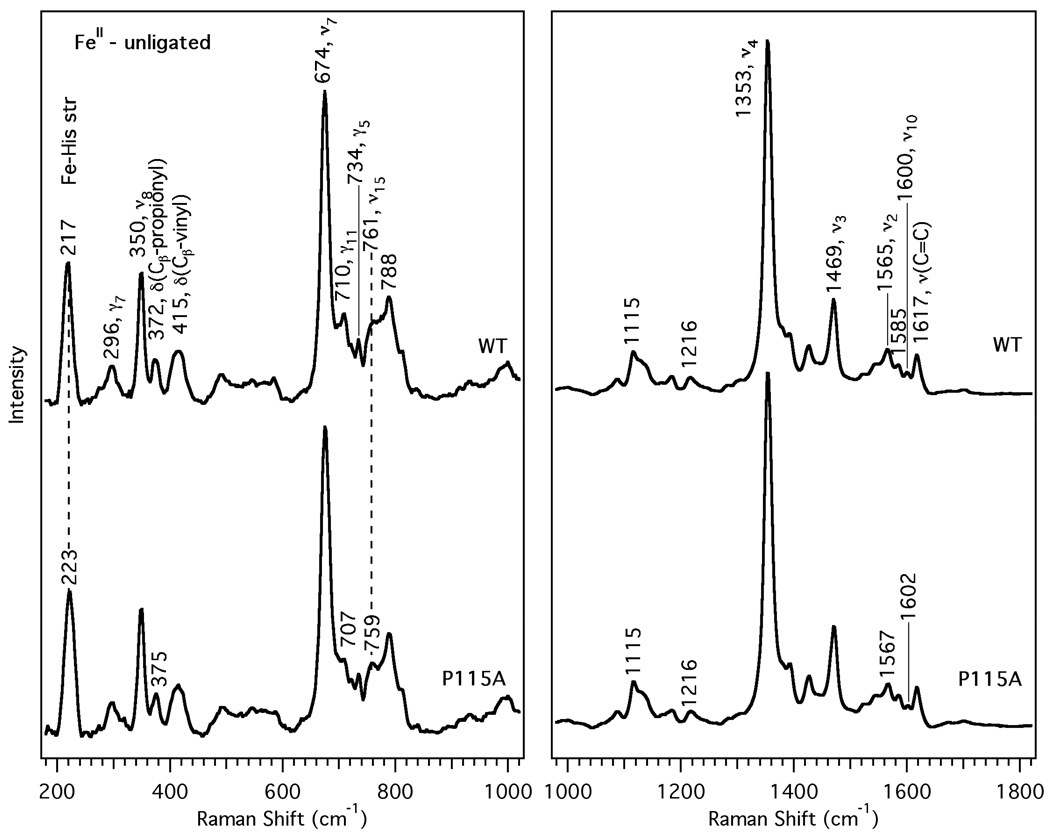
Figure 2.
Resonance Raman spectra of the FeII-unligated form of Tt H-NOX WT (upper trace) and P115A (lower trace). Spectral intensities in the low and high frequency regions were normalized to ν7 and ν4, respectively.
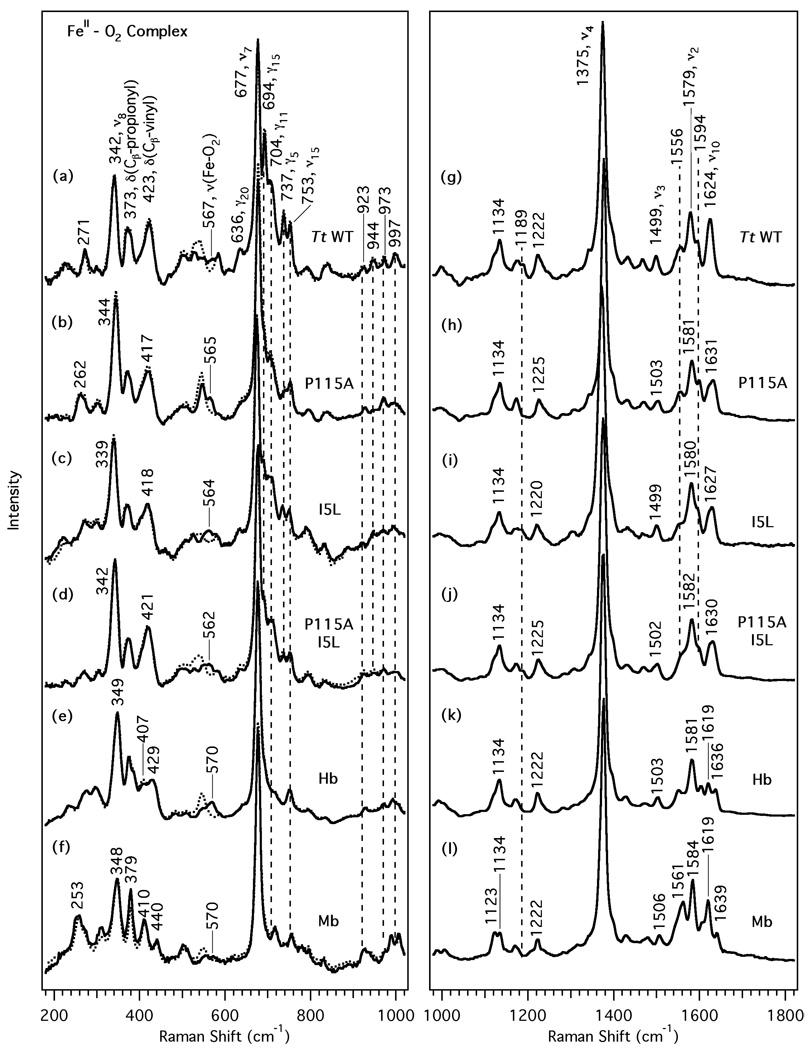
Figure 3.
Resonance Raman spectra of the O2 complexes of the Tt H-NOX domains and globins. The left panel shows the lower frequency region for Tt H-NOX WT (a), P115A (b), I5L (c), P115A/I5L (d), Hb (e), and Mb (f). 18O2 spectra (dotted line) are overlapped over the 16O2 spectra to indicate the frequency shifts upon isotopic substitution. The right panel shows the high frequency region (traces g-l).
Table 2.
Heme skeletal modes for the FeII-unligated and FeII-O2 complexes of Tt H-NOX, hemoglobin, myoglobin, and FixL.a
| Protein | Ligand | ν10 | ν2 | ν3 | ν4 | ν(Fe-X) | Ref. |
|---|---|---|---|---|---|---|---|
| Tt WT | unligated | 1600 | 1565 | 1469 | 1353 | 217 | (19) |
| Tt P115A | 1602 | 1567 | 1469 | 1353 | 223 | this work | |
| Tt I5L | 1599 | 1564 | 1469 | 1352 | 217 | this work | |
| Tt P115A/I5L | 1601 | 1566 | 1471 | 1354 | 220 | this work | |
| Hb | nrb | 1564 | 1470 | 1356 | 217 | (35,36) | |
| Mb | nr | 1563 | 1471 | 1357 | 220 | (35,36) | |
| Bj FixLHc | 1602 | 1555 | 1469 | 1353 | 218 | (31) | |
| Tt WT | O2 | 1624 | 1579 | 1499 | 1375 | 567 | (19) |
| Tt P115A | 1631 | 1581 | 1503 | 1377 | 565 | this work | |
| Tt I5L | 1627 | 1580 | 1499 | 1372 | 564 | this work | |
| Tt P115A/I5L | 1630 | 1582 | 1502 | 1375 | 562 | this work | |
| Hb | nr | 1581 | 1503 | 1376 | 570 | this work | |
| Mb | nr | 1584 | 1506 | 1377 | 570 | this work | |
| Bj FixLH | 1638 | 1579 | 1504 | 1377 | 569 | (31) | |
All vibrational frequencies are reported in cm−1.
nr = not reported.
B. japonicum FixL heme-PAS.
The resonance Raman spectra of the fully reduced, 5-coordinate Tt H-NOX WT and P115A mutant were obtained to investigate the effect of the proximal Pro-115 mutation on the Fe-His bond strength. The vibrational frequencies and intensities are similar, with the exception of the 6 cm−1 increase for the Fe-His stretch to 223 cm−1 for P115A and a ~5 cm−1 increase in bandwidth. In the high frequency region, the skeletal markers at 1353, 1469, 1565, and 1600 cm−1 correspond to typical ν4, ν3, ν2, and ν10 values for 5-coordinate, histidyl-ligated, high-spin heme proteins (32, 34, 35). The striking spectral similarity between the Tt H-NOX WT spectrum and that of P115A in Figure 2 suggests that the heme conformation is minimally perturbed by this mutation, although the Fe-His bond length may have decreased slightly.
The resonance Raman spectra of the O2 complexes are compared in Figure 3 and the vibrational frequencies are also summarized in Table 2. The 18O2-isotopically substituted RR spectra are indicated with dotted lines and overlaid upon the natural abundance O2 complex spectra in the lower frequency region to assign the Fe-O2 stretch (Figure 3, traces a-f). Based on the simple harmonic oscillator model, a downshift of 21 cm−1 is predicted for ν(Fe-O2) upon 16O2 → 18O2 substitution. As previously shown (19), Tt H-NOX WT displays a ν(Fe-O2) band at 567 cm−1 which shifts to 540 cm−1 for the 18O2 complex. Mutating Pro-115 and Ile-5 causes small downshifts of 2 to 5 cm−1 relative to Tt H-NOX WT. The Tt H-NOX P115A mutant exhibits an isotope-sensitive band at 565 cm−1 which decreases by 20 cm−1 for the 18O2 complex, corresponding very well with the expected downshift. Similarly, ν(Fe-O2) for the Tt H-NOX I5L/P115A double mutant is observed at 562 cm−1 and it decreases by 25 cm−1 upon 18O2 substitution. A cumulative downshift is observed for ν(Fe-O2) upon addition of the second mutation. This further decrease in frequency for the double mutant can plausibly be explained by a greater disruption of critical nonbonded contacts within the heme pocket that work to stabilize the O2 complex.
In the high frequency region of the O2 complexes (Figure 3, traces g-l), the skeletal markers at 1375, 1499, 1579, and 1624 cm−1 for Tt H-NOX WT are indicative of a 6-coordinate, histidyl-ligated, low-spin O2 complex (34, 36). Compared to Tt H-NOX WT, the three mutants show upshifts of 1–7 cm−1 for the skeletal markers (Table 2). Similar to the globins, the Tt H-NOX P115A mutant completely lacks a shoulder band at 1189 cm−1, whereas WT and the other mutants exhibit two overlapping bands at 1176 cm−1 and 1189 cm−1. In Tt H-NOX, ν3 remains constant at 1499 cm−1 for WT and the I5L mutant, but shifts to 1503 and 1502 cm−1 for the P115A and I5L/P115A mutants, respectively. The 1556 cm−1 peak is most pronounced in Tt H-NOX WT and P115A, but appears more as a broad shoulder in the I5L and I5L/P115A mutants. Furthermore, ν2 (1579 cm−1) and ν19 (1594 cm−1) in Tt H-NOX WT upshift by 2–3 cm−1 and 5 cm−1, respectively, upon mutating Ile-5 and Pro-115; these two bands correspond to Cβ-Cβ and Cα-Cm stretching vibrations. The breadth and slight asymmetry of ν10 (Cα-Cm stretch) in Tt H-NOX WT is likely due to overlap with the C=C stretching mode at 1619 cm−1 (assignment based on polarization and Q-band RR). Compared to Tt H-NOX WT (1624 cm−1), ν10 upshifts to 1631, 1627, and 1630 cm−1 for the Tt H-NOX P115A, I5L, and I5L/P115A mutants. These observed vibrational shifts in the high frequency region is reflective of the approximate shift pattern expected for changes in heme ruffling (ν10 > ν2 > ν3 > ν4). However, closer inspection of the high frequency region reveals shoulders for ν10 in Tt H-NOX P115A and I5L/P115A that correspond to the ν10 vibrational frequency for WT at 1624 cm−1. This band broadens and exhibits decreased intensity in the three mutants; the change is most striking in Tt H-NOX P115A, which has been shown by crystallography to sample a range of heme conformations (20). This observed crystallographic heterogeneity in the heme conformation for Tt H-NOX P115A is comparable to the broadened ν10 in the complementary RR spectra.
Below 1000 cm−1, several vibrational modes in the Tt H-NOX O2 complex spectra display reduced RR intensity upon mutation of Ile-5 and Pro-115. Heme out-of-plane modes (defined as γ to distinguish them from stretching modes, ν) involving bending, tilting, folding, and wagging motions occur in these lower frequency regions (21). The reduced RR intensities mainly occur in the 675–850 cm−1 and 900–1050 cm−1 regions as indicated by the vertical dashed lines in Figure 3 (traces a-d). Specifically, Tt H-NOX WT (trace a) exhibits peaks at 694 (γ15, sym. pyr fold), 704 (γ11, asym. pyr fold), 737 (γ5, sym. pyr fold), 944 [δ(Cm-H)], and 997 cm−1 [δ(Cm-H)] which decrease in relative RR intensity after mutating Ile-5 and Pro-115; the most striking change occurs at 694 cm−1. This band may be attributed to an in-plane pyrrole ring folding deformation mode (10). Subtle changes are also observed in the overlapping Tt H-NOX WT bands in the 500–640 cm−1 region, which are likely due to pyrrole swivels and folding vibrations (1, 5, 21). Of particular note are the weak bands at 526 (ν49), 589 (ν48), 605 (sym. pyr fold) and 636 cm−1 (γ20, asym. pyr fold) in Tt H-NOX WT that vanish in the Tt H-NOX P115A mutant. These peaks are still present in the Tt H-NOX I5L and I5L/P115A mutants, albeit slightly broader and lower in intensity than WT. The 271 cm−1 band in Tt H-NOX WT also broadens and downshifts by 9 cm−1 in the P115A mutant to 262 cm−1; this peak may correspond to either ν52 (porph-substituent bending) or a pyrrole tilting motion (γ16) within the heme (2, 37).
Spectral decomposition
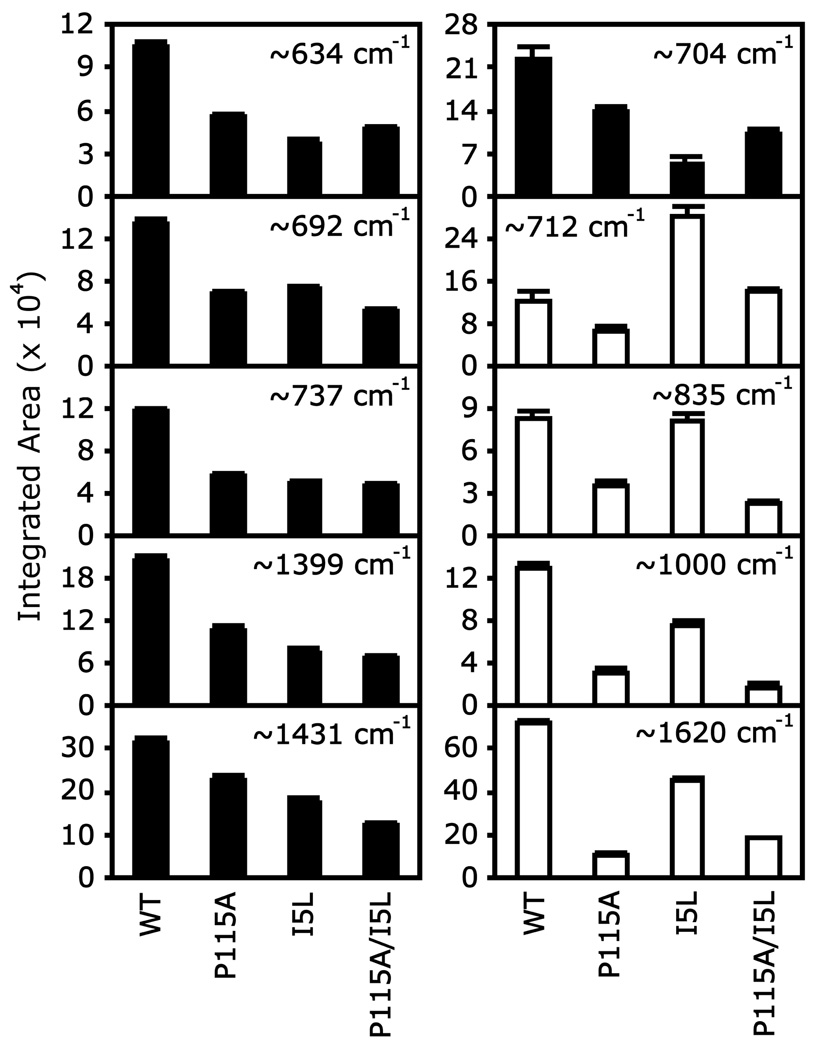
The RR intensity decreased for several low frequency modes in the O2 complexes for the Tt H-NOX mutants (Figure 3), but remained constant in the unligated (Figure 2; supplementary material, Figure 1S) and CO complexes (supplementary material, Figure 3S). These bands may show changes in intensity due to the inactivation of Raman modes upon heme relaxation. To quantify these spectral changes, the RR spectra were normalized and fit to Lorentzian peaks with fixed widths. The integrated peak areas are shown in Figure 4 to compare the RR intensity changes across Tt H-NOX WT and mutants. Intensity decreases of ~30–70% are observed upon mutating Ile-5 and Pro-115. In addition, the 704 cm−1 peak (γ11, asym. pyrrole folding) in Tt H-NOX WT broadens and develops a small shoulder at 709–712 cm−1, shifting toward the higher frequency band and decreasing in RR intensity in the mutants. Two different trends in the RR intensities were noted (the bars in Figure 4 are shaded differently to denote the two trends): one shows the intensity steadily decreasing across all mutants, whereas the other trend displays a zigzag pattern due to the minimal to moderate intensity change observed in Tt H-NOX I5L compared to the other two mutants.
Figure 4.
Integrated RR peak areas show intensity changes in Tt H-NOX upon mutation of Ile-5 and Pro-115. The black and white bars denote the two different trends in RR intensity changes.
Normal coordinate structural decomposition
Table 3 quantifies the calculated heme out-of-plane (oop) distortions for the O2 complexes of Tt H-NOX and other proteins from our NSD analysis (7). The generated output gives the amount of displacement along the normal coordinates for the six deformation types (e.g. ruffling, saddling) that properly simulate the observed distortions in the Tt H-NOX heme coordinates. NSD results obtained from other heme proteins are included for comparison. The total out-of-plane displacement (Δoop) and the amount of specific deformation types are reported according to the complete and minimal basis sets, respectively. The complete basis set includes all normal modes of the 24 atom D4h-symmetric porphyrin macrocycle (C20N4) to fully describe the observed heme nonplanarity. The minimal basis set only includes the lowest frequency normal coordinates of each symmetry type (6 in-plane and 6 out-of-plane) since these are expected to have the lowest distortion energies and should be predominant in the heme deformations (4, 13).
Table 3.
Calculated heme out-of-plane distortions from normal coordinate structural decomposition (NSD) analysis for the O2 complex of Tt H-NOX and other heme proteins.
| PDB | Protein | Δoopa | B2ub | B1u | A2u | Eg(x) | Eg(y) | A1u | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| 1U55 | Tt WT A | 1.59 | −1.07 | −1.11 | −0.1 | −0.10 | 0.24 | −0.03 | (14) |
| 1U55 | Tt WT B | 1.11 | −0.65 | −0.79 | 0.32 | −0.24 | 0.02 | −0.03 | (14) |
| 1U4H | Tt WT A | 1.59 | −1.09 | −1.09 | −0.02 | −0.30 | 0.11 | −0.06 | (14) |
| 1U4H | Tt WT B | 1.60 | −1.00 | −1.20 | 0.10 | −0.15 | 0.14 | −0.06 | (14) |
| Heme A | Tt I5L | 1.28 | −0.93 | 0.73 | −0.11 | −0.18 | 0.40 | −0.02 | c |
| Heme B | Tt I5L | 1.23 | −0.81 | −0.81 | 0.13 | −0.31 | 0.14 | −0.02 | c |
| 3EEE | Tt P115A A | 0.90 | −0.40 | −0.77 | 0.04 | −0.17 | 0.02 | 0.00 | (20) |
| 3EEE | Tt P115A B | 0.82 | −0.50 | −0.61 | 0.03 | −0.11 | 0.03 | 0.01 | (20) |
| 3EEE | Tt P115A C | 0.54 | −0.04 | −0.49 | −0.09 | −0.16 | −0.08 | 0.01 | (20) |
| 3EEE | Tt P115A D | 0.62 | 0.07 | −0.52 | −0.03 | −0.25 | −0.18 | 0.01 | (20) |
| Heme A | Tt I5L/P115A | 0.85 | −0.53 | 0.05 | −0.16 | −0.05 | 0.52 | −0.04 | c |
| Heme B | Tt I5L/P115A | 0.74 | −0.63 | 0.17 | −0.02 | 0.01 | 0.32 | −0.08 | c |
| 1DP6 | Bj FixLd | 0.60 | 0.44 | −0.32 | 0.18 | 0.01 | 0.11 | −0.03 | (26) |
| 1A6M | Sw Mbe | 0.36 | 0.19 | 0.01 | 0.22 | −0.05 | 0.19 | 0.07 | (28) |
| 1ASH | As Hbf | 0.47 | −0.11 | 0.41 | 0.08 | −0.11 | 0.07 | −0.01 | (29) |
| 1IDR | tHb Ag | 0.58 | −0.13 | 0.33 | 0.28 | −0.25 | −0.19 | 0.04 | (27) |
| 1IDR | tHb Bg | 0.55 | 0.15 | 0.45 | 0.01 | 0.05 | −0.21 | −0.04 | (27) |
Total out-of-plane displacement (Å) from the complete basis set
Reported displacements for each symmetry type are from the minimal basis set
unpublished experiments, Olea et al.
O2-sensing FixL domain of Bradyrhizobium japonicum
Sperm whale myoglobin
Ascaris suum hemoglobin domain I
Truncated hemoglobin N from Mycobacterium tuberculosis
As shown in Table 3, the largest displacements observed in the Tt H-NOX WT monoclinic (PDB 1U55) and orthorhombic (PDB 1U4H) crystal structures are from the B2u (saddling) and B1u (ruffling) deformations. The total rms out-of-plane displacement (Δoop) is ~1.6 Å for three different Tt H-NOX WT structures, but is reduced to ~1.1 Å in the monoclinic heme B structure. Pellicena et al. associate this heme relaxation with a ~11° rotation of the protein’s distal portion with respect to the proximal side and a reorientation of the distal Ile-5 residue (14). With the exception of the monoclinic heme B structure, the other three Tt H-NOX WT hemes maintain ~1.0–1.2 Å displacements in B1u and B2u deformations.
Compared to Tt H-NOX WT (Δoop ~1.6 Å), the Tt H-NOX I5L mutant showed a decrease of ~0.2–0.3 Å in overall out-of-plane displacement. The B1u deformation is more relaxed than B2u in Tt H-NOX I5L, but a small increase occurs in the Eg(x,y) (waving) displacement. In addition, the B1u directionality changes sign between the two Tt H-NOX I5L heme molecules. This also occurs for several deformations in the truncated and human hemoglobins (PDB 1IDR and 2DN1, respectively), and may result from differences in the crystal packing interactions and orientation of residues close to the heme.
The crystal structure of Tt H-NOX P115A contained four different hemes, producing a broader range of NSD displacements compared to the other proteins (PDB 3EEE). The most relaxed structure (heme C) exhibited out-of-plane distortion values (B2u, −0.04 Å; B1u, −0.49 Å; Δoop, 0.54 Å) comparable to those obtained for the crystal structures of Bj FixL (26) and the globins (27–29). The Δoop for these proteins ranged from 0.36 to 0.60 Å in contrast to Tt H-NOX WT (~1.6 Å). The total out-of-plane displacement for the least relaxed Tt H-NOX P115A structure (heme A) is 0.90 Å, which is close to the 1.11 Å Δoop observed for the monoclinic Tt H-NOX WT heme B structure.
The Tt H-NOX double mutant significantly relaxed the ruffling distortion with a B1u displacement of ~0.05–0.2 Å. Comparable to Sw Mb, these magnitudes are the lowest of the three Tt H-NOX mutants. In contrast, the B2u displacement (heme A, −0.53 Å; heme B, −0.63 Å) is greater than the < 0.1 Å observed in hemes C and D of the P115A mutant. Another unique feature exhibited by the double mutant is the ~0.3–0.5 Å Eg(y) contribution, which is similar to Tt H-NOX I5L heme A (0.4 Å). Interestingly, the Δoop for Tt H-NOX I5L/P115A (heme A, 0.85 Å; heme B, 0.74 Å) falls within the ~0.5 – 0.9 Å range obtained for the four Tt H-NOX P115A heme molecules, suggesting that the Ile-5 mutation contributes minimally to heme relaxation.
DISCUSSION
We now provide a detailed interpretation of the spectral changes exhibited by RR leading to the current model for heme distortion in Tt H-NOX. Specifically, the reduction in relative RR intensity for the low frequency modes, and the observed frequency shifts and peak broadening in the heme skeletal markers of Tt H-NOX P115A and I5L mutants provide evidence for a more relaxed chromophore within the solution form of the mutated protein. These changes are related to the heme structure and how its altered conformation, in comparison to Tt H-NOX WT, may result from heme cavity rearrangements. By comparing the spectroscopic data with the available crystallographic results, detailed information on the heme structure and insight into the possible role of these conserved residues are revealed.
RR intensity changes in Tt H-NOX O2 complex spectra
Choi and Spiro identified several out-of-plane motions of iron-porphyrin complexes, including pyrrole tilts (~260 cm−1), methine bridge deformations [γ(Cα-Cm), ~320 cm−1], pyrrole folding modes (~425–510 cm−1), and methine-hydrogen deformations [γ(Cm-H), ~840 cm−1] (10). Modes that occur below 400 cm−1 involve methine bridge wagging and other pyrrole deformations, and are expected to be heavily mixed (21). These out-of-plane modes are generally expected to be Franck-Condon and Jahn-Teller inactive for planar porphines of D4h symmetry, but may be activated upon nonplanar heme distortion and symmetry reduction (5, 10). The RR intensities of these modes are induced when their components are projected onto the in-plane electronic excitations (2, 10). Hence, only the modes that occur along specific heme distortion coordinates and have the same symmetry will be activated.
Our RR spectra of Tt H-NOX WT exhibit several bands corresponding to out-of-plane modes associated with heme deformations in the O2 complex. Specifically, the 450–600 cm−1 region displays many overlapped Raman features that are plausibly out-of-plane pyrrole tilts, swivels, and folding modes activated by the symmetry lowering effects of the protein environment on the chromophore. Other notable features include the moderately intense 692 cm−1 shoulder (γ15, sym. pyrrole folding), the 704 peak with a small shoulder at 712 cm−1 (γ11, sym. pyrrole folding), and the 737 cm−1 band (γ5/ν16, asym. pyrrole folding and deformation). These peaks are known to be sensitive to saddling (B2u) and ruffling (B1u) deformations, and are tentatively assigned by association with previous work by Spiro, Shelnutt, Schweitzer-Stenner, and others (1, 2, 5, 10, 11, 21, 38, 39). Between 900–1000 cm−1, four distinct peaks observed in the Tt H-NOX WT spectrum are tentatively assigned as hydrogen wagging motions (37). These RR features confirm that the striking heme deformations observed in the Tt H-NOX WT structure also exist in solution and are not an outcome of crystallization. Interestingly, these characteristic low frequency bands in the Tt H-NOX spectra are unique to the O2 complex, and are not prominently featured in the other Tt H-NOX complexes or bacterial H-NOXs (e.g. V. cholerae, L. pneumophila, N. punctiforme) (19, 24). In agreement with these observations, NSD analysis of the N. sp H-NOX domain crystal structures by Ma et al. indicate Δoop ~ 0.7–0.9 Å for the unligated, CO, and NO complexes; these values are lower than those observed for the Tt H-NOX O2 structures by Pellicena et al. (Δoop ~ 1.1–1.6 Å) (14, 16).
Mutation of the Ile-5 and Pro-115 residues in Tt H-NOX caused decreases in the relative RR intensities for several out-of-plane modes in the O2 complex, but did not appreciably alter any mode intensities in the unligated spectra. These observations indicate a distinct difference in how the mutations affect the heme conformation for the O2 and unligated species; although significant structural changes occur in the O2 complex, the great similarity in RR intensity for the unligated spectra strongly suggests minimal changes in the heme structure. The most important point about the loss of RR intensity is that it clearly demonstrates a relaxation of the heme upon mutating these residues. The replacement of Pro-115 in Tt H-NOX with an alanine relieves the original proximal strain on pyrrole group D by freeing up space around the random coil linking α-helix F and β-strand 1. This structural reorganization is consistent with the loss of RR intensity at 508 (γ12), 526 (ν49), 588 (ν48), and the 692–737 cm−1 region (γ15, γ11, γ5), where pyrrole swivels and folding modes that correspond to the B1u, B2u, and Eg symmetry are expected to occur (1, 2, 5, 38, 39). A similar effect, albeit to a lesser degree, can explain the changes in the Tt H-NOX I5L spectra via its distal strain over pyrrole A and influence on reorienting nearby residue contacts with the propionate groups (e.g. Asp-45).
Although the three Tt H-NOX mutants showed comparable decreases in relative RR intensity for some of the low frequency bands, the P115A mutant displayed several distinct features. These included the following observations: (i) the apparent absence of the 1189 cm−1 peak corresponding to the Cβ-substituent and Cα-N antisymmetric stretching modes, which is also exhibited in the globins spectra, (ii) the retention of the hydrogen wagging modes (850–1050 cm−1) in Tt H-NOX I5L compared to the P115A mutant, and (iii) the greater similarity in peakwidth and frequency between the Tt H-NOX I5L and WT spectra in comparison to P115A. In addition, the locations where RR intensity is lost after mutating the heme pocket residues correspond well to the less congested regions in the globin spectra, supporting our tentative assignment of these peaks to heme out-of-plane modes. These characteristics together support a more dominant effect of Pro-115 on the Tt H-NOX heme structure than Ile-5.
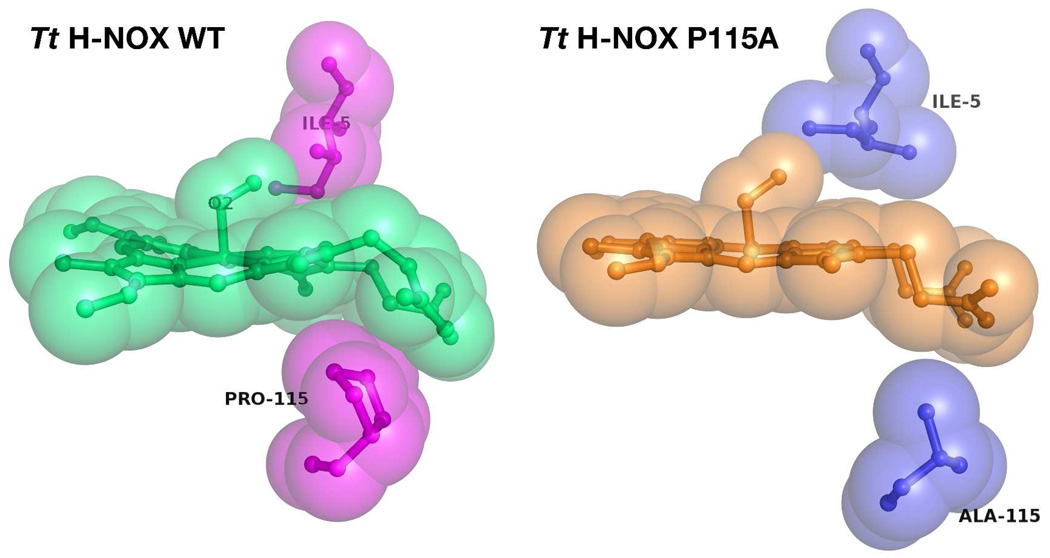
Our spectral observations also fit well with the structural data via the NSD analysis, which indicates that the lowest Δoop occurs in Tt H-NOX P115A (heme C; Δoop ~ 0.5 Å) rather than the double mutant. The Δoop range of ~0.5–0.9 Å for all four molecules in the unit cell and the differing effects on the B2u and B1u deformations further support the heme flexibility that we observed by RR in the Tt H-NOX P115A mutant. According to the NSD analysis, the Tt H-NOX I5L mutation affected the B2u displacement less than B1u; this effect may explain the two trends shown in the spectral decomposition in that some modes may be more sensitive and specific to certain heme deformations. In addition, the normal mode displacements for Tt H-NOX I5L displayed modest decreases compared to Tt H-NOX P115A, which agrees well with our conclusions from the RR spectra. Finally, the cumulative decrease in relative RR intensity exhibited by the double mutant was close to those observed for the single Tt H-NOX P115A mutant, further suggesting that the dominant perturbation of the pocket originates from the Pro-115 mutation. This reduction of the B2u and B1u deformations can be rationalized based on the nonbonded contacts between Pro-115 and pyrrole group C of the heme (Figure 1 and Figure 5). The proline ring clearly pushes up into the heme and forces the chromophore into a nonplanar conformation. Upon substitution with alanine, the ring is substituted by a more freely rotating methyl group, which allows the heme conformation to relax (Figure 5). In contrast to Pro-115, Ile-5 makes a less intrusive distal contact with the heme pyrrole group B in Tt H-NOX; thus, mutating this residue has a smaller effect on the heme conformation.
Figure 5.
Comparison of Tt H-NOX WT (PDB 1U55) with P115A (PDB 3EEE) showing reduction of steric nonbonded contacts between proximal Pro-115 and heme upon mutation to Ala-115.
Frequency shifts and peak broadening
In contrast to the pronounced changes observed in the O2 complex spectra, the great similarity in vibrational frequency and intensity between the RR spectra for the Tt H-NOX WT and P115A unligated species demonstrates the minimal impact of this mutation on the heme. The increased Fe-His stretching frequency for Tt H-NOX P115A suggests a strengthening of the Fe-His bond. In contrast, crystallographic results indicate that the bond lengths for the two proteins are within error. This discrepancy is not surprising since in resonance Raman spectroscopy, a vibrational frequency shift of 5–6 cm−1 in the most sensitive bands corresponds to bond length changes of as small as 0.01 Å (21, 33). Such a change would be below the range detectable by crystallography. Olea et al. do note, however, that the Fe-His bond tilting angle shifts from 78° in Tt H-NOX WT (heme A, monoclinic) to 87° in P115A (heme C, monoclinic), resulting in a more perpendicular orientation of His-102 with respect to the heme, and plausibly enables a better bond overlap between Fe and His-102 (20). Furthermore, the slight broadening of the peak by ~5 cm−1 suggests some heterogeneity in the proximal pocket, and supports our assertion that the Tt H-NOX P115A mutation introduces structural disorder and flexibility to the protein pocket. This heterogeneity in the RR spectra fits well with the 80–87° Fe-His tilt angle range reported for the four Tt H-NOX P115A heme molecules (20).
Kinetic studies by Olea et al. on Tt H-NOX have shown that the O2 dissociation rates decrease by an order of magnitude between WT (1.22 ± 0.09 s−1) and the P115A mutant (0.22 ± 0.01 s−1), but that the O2 association rates are similar (13.6 ± 1.0 µM−1s−1 and 10.4 ± 1.1 µM−1s−1, respectively) (20). Our RR results exhibit a small cumulative downshift in the ν(Fe-O2) frequency with the three Tt H-NOX mutants from 567–570 cm−1 (WT) to 562 cm−1 (I5L/P115A mutant). In addition, the P115A mutant exhibits a 6 cm−1 downshift to 265 cm−1 from WT (271 cm−1); this band may be a combination of in-plane Fe-Npyr stretching, pyrrole tilting, and pyrrole-substituent bending motions (ν52). Irwin et al. previously proposed that a stronger ligand field could arise from a constrained heme along the equatorial porphyrin plane (40). Since heme ruffling is thought to result in a shortening of the equatorial Fe-Npyr bonds, this may partly explain the different frequencies observed between Tt H-NOX WT and the mutants for ν(Fe-O2) and ν(Fe-Npyr), although other factors within the protein pocket (e.g. hydrogen-bonding residues, tension on the trans-Fe-imidazole bond, and steric bulk) also clearly influence the stability of the O2 complex. In particular, the ruffling deformation/Fe-Npyr bond distance dependence is consistent with our observed ν(Fe-Npyr) downshift for the Tt H-NOX P115A mutant, which also exhibits a relaxation of the ruffling deformation. In contrast, the crystal structures indicate that the Fe-Npyr distances are ~2.0 Å for both Tt H-NOX WT and P115A with only a negligible decrease for the WT Fe-Npyr distance. However, based on Badger’s Rule (41, 42), the inverse force constant/bond length relationship shows that the vibrational frequency is more sensitive to small changes (~0.01 Å) in internuclear distance. Thus, the discrepancy between the RR spectra and crystal structure can be readily explained.
Structural heterogeneity in a sample can be revealed by the broadness and asymmetry of structure sensitive lines, such as ν2 (Cβ-Cβ stretch) and ν10 (Cα-Cm stretch) (11). Our RR results exhibit several regions in which the peaks broaden upon mutating Ile-5 and Pro-115 in Tt H-NOX. The clearest example of this is ν10 in the P115A mutant; in comparison to the WT spectra, this band decreases in intensity and broadens to display a prominent shoulder band at 1631 cm−1. Broad features are also observed for the Tt H-NOX Ile-5 and double mutants in the 1550–1600 cm−1 range; the modes in this region correspond to the Cα-Cm, Cβ-Cβ, and peripheral vinyl modes. The broadening of these distinct peaks into merged bands suggests conformational flexibility in these regions, and may result from the ability of the Leu-5 side chain to rapidly flip directions within the pocket. These fluctuations likely disrupt the contacts between the protein’s YSR motif and the nearby propionate groups. Furthermore, the fact that ν2 and ν10 upshift in frequency for all three Tt H-NOX mutants supports the possibility of a better π-π overlap in the C=C, Cβ-Cβ, and Cα-Cm bonds within the porphyrin macrocycle as a result of heme relaxation.
Some peak broadening and shoulder band formation also occur below 450 cm−1, where pyrrole deformations are expected to occur. The 423 cm−1 peak [δ(Cβ-vinyl)] in Tt H-NOX WT not only downshifts to 417 cm−1 (P115A), but also broadens with a weak shoulder band and slight decrease in RR intensity. Several other bands in this region exhibit similar changes. These observations further support the possibility that the Tt H-NOX protein pocket no longer firmly holds the heme in a particular conformation.
Possible role for conserved residues and heme deformation in Tt H-NOX
The heme deformation in the H-NOX family plausibly plays a role in signal transduction from the chromophore to the protein upon ligand binding; alteration of the heme conformation may induce protein conformational changes, which then regulate the linked signaling protein (e.g. histidine kinases, diguanylate cyclases, and MCP domains). Based on our data, we conclude that the single P115A mutation is sufficient to open the Tt H-NOX distal pocket and introduce flexibility to the heme structure, enabling the chromophore to relax and sample other conformations. The possibility of the H-NOX domain alternating between closed and open conformations is reasonable based on previously observed varying rotational angles in the H-NOX distal and proximal subdomains (14, 16, 20). Similar to the protein conformation changes observed by Pellicena et al. (14) in the Tt H-NOX WT monoclinic structure (heme B), a plausible explanation for P115A’s spectral changes is that the distal pocket has more rotational motion, and may sample an open heme pocket more frequently due to the increased flexibility granted by replacing the rigidly kinked proline ring with the more freely rotating alanine methyl group. Thus, the highly conserved Pro-115 and neighboring residues can serve as a hinge point between the N- and C-terminal subdomains, which are necessary for maintaining an enclosed environment, retaining important heme-protein interactions, and conserving specific biochemical properties within the H-NOX protein pocket. A clear example of this is the dramatic changes observed in the reduction potential and O2 affinity upon decreasing the amount of heme distortion present in Tt H-NOX (20). The Tt H-NOX I5L mutation may also contribute toward opening the pocket, although to a lesser extent based on the more modest spectral changes and shifts exhibited by this single mutant in comparison to those observed by mutating Pro-115. This result is not surprising since the substitution of Ile with Leu is relatively conservative in comparison to the replacement of the proline with alanine. However, this Leu-5 side chain can also plausibly flip inward toward the distal pocket or away from it; thus, the heme may interact differently with the conserved YSR motif, depending on whether or not the chromophore is pinned down along pyrrole group A by the aliphatic chain.
In summary, we provide spectral evidence of heme relaxation in the O2-bound form of Tt H-NOX upon mutating the Ile-5 and Pro-115 residues. Our resonance Raman results exhibit the most significant changes in the Tt H-NOX P115A mutant, in contrast to the I5L mutation, which bears spectral features more similar to Tt H-NOX WT. Furthermore, the double mutant appears to behave more like an admixture of P115A with I5L, rather than a synergistic combination of the two mutations. In addition, we observe some broad shoulder features in the high frequency region that suggest the presence of other heme conformations in the three Tt H-NOX mutants. Of the different heme complex forms that we have studied with resonance Raman, the most striking changes occur in the O2-bound spectra; we see very little change in the unligated and CO complexes. These results provide a clear insight into the actual heme conformation of Tt H-NOX P115A in solution; indicating that although it is heterogeneous, the predominant form is significantly relaxed based on both changes in vibrational frequencies and RR out-of-plane mode intensities.
Supplementary Material
ACKNOWLEDGMENTS
We thank Charles Olea Jr. for providing the structures of Tt H-NOX I5L and I5L/P115A, and members of the Mathies and Marletta labs for helpful discussions. We are also grateful to Drs. Kathleen Durkin and Jamin Krinsky at the Molecular Graphics and Computation Facility for their extensive help with MD simulations and DFT calculations.
Abbreviations
- H-NOX
Heme-Nitric Oxide and/or Oxygen binding domain
- RR
resonance Raman
- Tt
Thermoanaerobacter tengcongensis
- TEA
triethanolamine
- DFT
density functional theory
- QM/MM
quantum mechanics/molecular mechanics
- NSD
normal coordinate structural decomposition
- Δoop
total out-of-plane displacement
Footnotes
This work was supported in part by NIH Grant GM070671 (M.A.M.) and the Mathies Royalty Fund.
SUPPORTING INFORMATION
Supplementary Figures have been included to show the Tt H-NOX deoxy spectra (Fig. 1S), isotopic O2 difference spectra (Fig. 2S), isotopic CO spectra (Fig. 3S) and detailed deconvolution spectra for the O2 complex (Fig. 4S, a–c). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Blackwood ME, Rush TS, Medlock A, Dailey HA, Spiro TG. Resonance Raman spectra of ferrochelatase reveal porphyrin distortion upon metal binding. J. Am. Chem. Soc. 1997;119:12170–12174. [Google Scholar]
- 2.Jarzecki AA, Spiro TG. Porphyrin distortion from resonance Raman intensities of out-of-plane modes: Computation and modeling of N-methylmesoporphlyrin, a ferrochelatase transition state analog. J. Phys. Chem. A. 2005;109:421–430. doi: 10.1021/jp0470142. [DOI] [PubMed] [Google Scholar]
- 3.Sparks LD, Anderson KK, Medforth CJ, Smith KM, Shelnutt JA. Correlations between Raman Frequencies and Structure for Planar and Nonplanar Metalloporphyrins. Inorg. Chem. 1994;33:2297–2302. [Google Scholar]
- 4.Jentzen W, Ma JG, Shelnutt JA. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys. J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Q, Medforth CJ, Schweitzer-Stenner R. Nonplanar heme deformations and excited state displacements in nickel porphyrins detected by Raman spectroscopy at Soret excitation. J. Phys. Chem. A. 2005;109:10493–10502. doi: 10.1021/jp052986a. [DOI] [PubMed] [Google Scholar]
- 6.Sigfridsson E, Ryde U. The importance of porphyrin distortions for the ferrochelatase reaction. J. Biol. Inorg. Chem. 2003;8:273–282. doi: 10.1007/s00775-002-0413-8. [DOI] [PubMed] [Google Scholar]
- 7.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Nonplanar porphyrins and their significance in proteins. Chem. Soc. Rev. 1998;27:31–41. [Google Scholar]
- 8.Kozlowski PM, Rush TS, Jarzecki AA, Zgierski MZ, Chase B, Piffat C, Ye BH, Li XY, Pulay P, Spiro TG. DFT-SQM force field for nickel porphine: Intrinsic ruffling. J. Phys. Chem. A. 1999;103:1357–1366. [Google Scholar]
- 9.Xu CL, Ibrahim M, Spiro TG. DFT analysis of axial and equatorial effects on Heme-CO vibrational modes: Applications to CooA and H-NOX heme sensor proteins. Biochemistry. 2008;47:2379–2387. doi: 10.1021/bi702254y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi SH, Spiro TG. Out-of-Plane Deformation Modes in the Resonance Raman Spectra of Metalloporphyrins and Heme Proteins. J. Am. Chem. Soc. 1983;105:3683–3692. [Google Scholar]
- 11.Jentzen W, Simpson MC, Hobbs JD, Song X, Ema T, Nelson NY, Medforth CJ, Smith KM, Veyrat M, Mazzanti M, Ramasseul R, Marchon JC, Takeuchi T, Goddard WA, Shelnutt JA. Ruffling in a Series of Nickel(II) Meso-Tetrasubstituted Porphyrins as a Model for the Conserved Ruffling of the Heme of Cytochromes-c. J. Am. Chem. Soc. 1995;117:11085–11097. doi: 10.1021/ja00150a008. [DOI] [PubMed] [Google Scholar]
- 12.Czernuszewicz RS, Li XY, Spiro TG. Nickel Octaethylporphyrin Ruffling Dynamics from Resonance Raman Spectroscopy. J. Am. Chem. Soc. 1989;111:7024–7031. [Google Scholar]
- 13.Jentzen W, Song XZ, Shelnutt JA. Structural characterization of synthetic and protein-bound porphyrins in terms of the lowest-frequency normal coordinates of the macrocycle. J. Phys. Chem. B. 1997;101:1684–1699. [Google Scholar]
- 14.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS. Femtomolar sensitivity of a NO sensor from Clostridium botulinum. Science. 2004;306:1550–1553. doi: 10.1126/science.1103596. [DOI] [PubMed] [Google Scholar]
- 16.Ma XL, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. EMBO J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boon EM, Marletta MA. Ligand specificity of H-NOX domains: from sGC to bacterial NO sensors. J. Inorg. Biochem. 2005;99:892–902. doi: 10.1016/j.jinorgbio.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 18.Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 20.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. Probing the Function of Heme Distortion in the H-NOX Family. ACS Chem. Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spiro TG, Li XY. Resonance Raman Spectroscopy of Metalloporphyrins. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy: Resonance Raman spectra of heme and metalloproteins. New York: John Wiley & Sons; 1988. pp. 1–37. [Google Scholar]
- 22.Myers AB, Mathies RA. "Resonance Raman Intensities: A Probe of Excited State Structure and Dynamics". In: Spiro TG, editor. Biological Applications of Raman Spectroscopy: Resonance Raman Spectra of Polyenes and Aromatics. New York: John Wiley & Sons; 1988. pp. 1–58. [Google Scholar]
- 23.Spiro TG, Stong JD, Stein P. Porphyrin Core Expansion and Doming in Heme Proteins - New Evidence from Resonance Raman Spectra of 6-Coordinate High-Spin Iron(III) Hemes. J. Am. Chem. Soc. 1979;101:2648–2655. [Google Scholar]
- 24.Boon EM, Davis JH, Tran R, Karow DS, Huang SH, Pan D, Miazgowicz MM, Mathies RA, Marletta MA. Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase β1 H-NOX domain. J. Biol. Chem. 2006;281:21892–21902. doi: 10.1074/jbc.M600557200. [DOI] [PubMed] [Google Scholar]
- 25.Mathies R, Yu NT. Raman Spectroscopy with Intensified Vidicon Detectors -Study of Intact Bovine Lens Proteins. J. Raman Spectrosc. 1978;7:349–352. [Google Scholar]
- 26.Gong W, Hao B, Chan MK. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry. 2000;39:3955–3962. doi: 10.1021/bi992346w. [DOI] [PubMed] [Google Scholar]
- 27.Milani M, Pesce A, Ouellet Y, Ascenzi P, Guertin M, Bolognesi M. Mycobacterium tuberculosis hemoglobin N displays a protein tunnel suited for O2 diffusion to the heme. EMBO J. 2001;20:3902–3909. doi: 10.1093/emboj/20.15.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vojtechovsky J, Chu K, Berendzen J, Sweet RM, Schlichting I. Crystal structures of myoglobin-ligand complexes at near-atomic resolution. Biophys. J. 1999;77:2153–2174. doi: 10.1016/S0006-3495(99)77056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Kloek AP, Goldberg DE, Mathews FS. The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity. Proc. Natl. Acad. Sci. U.S.A. 1995;92:4224–4228. doi: 10.1073/pnas.92.10.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonini E, Brunori M. Hemoglobin and myoglobin in their reactions with ligands. Vol. 21. Amsterdam: North-Holland Publishing Company; 1971. [Google Scholar]
- 31.Levantino M, Huang Q, Cupane A, Laberge M, Hagarman A, Schweitzer-Stenner R. The importance of vibronic perturbations in ferrocytochrome c spectra: A reevnaluation of spectral properties based on low-temperature optical absorption, resonance Raman, and molecular-dynamics simulations. J. Chem. Phys. 2005;123 doi: 10.1063/1.1961556. [DOI] [PubMed] [Google Scholar]
- 32.Tomita T, Gonzalez G, Chang AL, Ikeda-Saito M, Gilles-Gonzalez MA. A comparative resonance Raman analysis of heme-binding PAS domains: heme iron coordination structures of the BjFixL, AxPDEA1, EcDos, and MtDos proteins. Biochemistry. 2002;41:4819–4826. doi: 10.1021/bi0158831. [DOI] [PubMed] [Google Scholar]
- 33.Kitagawa T. The Heme Protein Structure and the Iron-Histidine Stretching Mode. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy: Resonance Raman Spectra of Heme and Metalloproteins. New York: John Wiley & Sons; 1988. pp. 97–131. [Google Scholar]
- 34.Tamura K, Nakamura H, Tanaka Y, Oue S, Tsukamoto K, Nomura M, Tsuchiya T, Adachi S, Takahashi S, Iizuka T, Shiro Y. Nature of endogenous ligand binding to heme iron in oxygen sensor FixL. J. Am. Chem. Soc. 1996;118:9434–9435. [Google Scholar]
- 35.Choi S, Spiro TG, Langry KC, Smith KM, Budd DL, Lamar GN. Structural Correlations and Vinyl Influences in Resonance Raman Spectra of Protoheme Complexes and Proteins. J. Am. Chem. Soc. 1982;104:4345–4351. [Google Scholar]
- 36.Takahashi S, Ishikawa K, Takeuchi N, Ikedasaito M, Yoshida T, Rousseau DL. Oxygen-Bound Heme-Heme Oxygenase Complex - Evidence for a Highly Bent Structure of the Coordinated Oxygen. J. Am. Chem. Soc. 1995;117:6002–6006. [Google Scholar]
- 37.Hu SZ, Smith KM, Spiro TG. Assignment of protoheme Resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 1996;118:12638–12646. [Google Scholar]
- 38.Rush TS, Kozlowski PM, Piffat CA, Kumble R, Zgierski MZ, Spiro TG. Computational modeling of metalloporphyrin structure and vibrational spectra: Porphyrin ruffling in NiTPP. J. Phys. Chem. B. 2000;104:5020–5034. [Google Scholar]
- 39.Huang Q, Schweitzer-Stenner R. Non-planar heme deformations and excited state displacements in horseradish peroxidase detected by Raman spectroscopy at Soret excitation. J. Raman Spectrosc. 2005;36:363–375. doi: 10.1021/jp052986a. [DOI] [PubMed] [Google Scholar]
- 40.Irwin MJ, Armstrong RS, Wright PE. Resonance Raman Studies of Soybean Leghemoglobin and Myoglobin - Origin of the Differences in O2 Dissociation Rate Constants. FEBS Lett. 1981;133:239–243. [Google Scholar]
- 41.Badger RM. The relation between the internuclear distances and force constants of molecules and its application to polyatomic molecules. J. Chem. Phys. 1935;3:710–714. [Google Scholar]
- 42.Green MT. Application of Badger's rule to heme and non-heme iron-oxygen bonds: An examination of ferryl protonation states. J. Am. Chem. Soc. 2006;128:1902–1906. doi: 10.1021/ja054074s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.