Summary
Fibrotic rigidification following a myocardial infarct is known to impair cardiac output, and it is also known that cardiomyocytes on rigid culture substrates show a progressive loss of rhythmic beating. Here, isolated embryonic cardiomyocytes cultured on a series of flexible substrates show that matrices that mimic the elasticity of the developing myocardial microenvironment are optimal for transmitting contractile work to the matrix and for promoting actomyosin striation and 1-Hz beating. On hard matrices that mechanically mimic a post-infarct fibrotic scar, cells overstrain themselves, lack striated myofibrils and stop beating; on very soft matrices, cells preserve contractile beating for days in culture but do very little work. Optimal matrix leads to a strain match between cell and matrix, and suggests dynamic differences in intracellular protein structures. A ‘cysteine shotgun’ method of labeling the in situ proteome reveals differences in assembly or conformation of several abundant cytoskeletal proteins, including vimentin, filamin and myosin. Combined with recent results, which show that stem cell differentiation is also highly sensitive to matrix elasticity, the methods and analyses might be useful in the culture and assessment of cardiogenesis of both embryonic stem cells and induced pluripotent stem cells. The results described here also highlight the need for greater attention to fibrosis and mechanical microenvironments in cell therapy and development.
Keywords: Cardiomyocyte, Elasticity, Matrix, Myocardium, Myosin
Introduction
In healthy myocardium, cardiomyocytes attach to a collagen-based extracellular matrix (ECM) that must be sufficiently compliant for actomyosin forces to pump the heart. The elasticity (E) of ECM is an ‘insoluble cue’ for many cells, influencing cell shape, protein expression and organization, as well as differentiation (Discher et al., 2005; Pelham and Wang, 1997; Peyton and Putnam, 2005). E varies considerably between tissues: the lateral elasticity of striated muscle (with E ∼10 kPa) is intermediate between that of brain tissue, which is soft (with E ∼1 kPa), and the elasticity of osteoid, which is the heavily crosslinked collagen that initiates bone growth (with E ∼25−50 kPa) (Engler et al., 2007). Elasticity is also frequently altered in disease: sclerosis (as in atherosclerosis, osteosclerosis, etc.) literally means ‘hardening’ in Greek. Indeed, following a myocardial infarction, beating cardiomyocytes are replaced by a fibrotic scar (Ehard≈35−70 kPa) that is several-fold stiffer than normal myocardium and mechanically more similar to osteoid (Berry et al., 2006; Dean et al., 2005). Stem cells injected into such a scar can lead to myocardial calcifications (Breitbach et al., 2007). The effects of such mechanical changes have yet to be explored, and understanding their influence seems likely to provide new insight into processes in development, disease and stem-cell-based regeneration.
Myofibril assembly and striation of cardiomyocytes occurs after mitosis both in situ (Du et al., 2008; Ehler et al., 1999) and in tissue explants (Du et al., 2003): cells first disassemble this specialized contractile apparatus and subsequently reassemble it. Likewise, when cardiomyocytes are isolated from embryos and grown in vitro, they first attach to a substrate and then their pre-myofibrils, which contain non-muscle α-actinin and myosin IIB, are replaced by muscle isoforms that develop A-bands and I-Z-I bands of mature myofibrils (Du et al., 2003; Rhee et al., 1994; Sanger et al., 2005). Rhythmic contraction can then be sustained for day(s) when cardiomyocytes are grown on hard, ligand-coated culture materials (e.g. glass, polystyrene), but cells ultimately lose their rhythmic contractions, their myofibrils and their ability to divide (Dabiri et al., 1999). In comparison, skeletal muscle cells that fuse from myoblasts must synthesize and assemble all myofibril components de novo – but myofibrillogenesis is inhibited in cells on rigid, ligand-coated matrices where isometric contractions appear sustained in cells that are adhesively pinned ‘at fixed length’ (Engler et al., 2004b). Embryonic cardiomyocytes already possess the necessary components as well as some pre-assembled myofibrils (Sanger et al., 2005) and, so, rigid environments are perhaps less inhibitory to myofibril formation. Nonetheless, during excessive force production, such as contraction against a rigid substrate or a hard fibrotic scar (Berry et al., 2006), myosin is expected to shift its force-velocity curve to a less efficient duty cycle (Piazzesi et al., 2002; Takagi et al., 2006). We, therefore, hypothesized that a decreased efficiency affects rhythmic beating and sought to clarify molecular mechanism(s) for how microenvironment mechanics induces dysfunction in striated muscle.
Cardiomyocytes cultured on thick collagen (Bird et al., 2003; Sanger et al., 2005) or synthetic gels (Shapira-Schweitzer and Seliktar, 2007) can contract their matrix, but if the matrices are too ‘soft,’ the power stroke of myosin is expected to be inefficient (against minimal resistance). Since myocardium falls within an intermediate range of tissue elasticity (Berry et al., 2006) [similar to other types of muscle (Collinsworth et al., 2002; Engler et al., 2004b; Pasternak et al., 1995)], a normal shortening-contraction cycle could in principle optimize force development. To evaluate the impact of in vitro microenvironment compliance on cardiomyocyte strain, structure and function, we used a well-established, elastically tunable polyacrylamide gel system (Pelham and Wang, 1997) with collagen I covalently attached to promote adhesion. Microenvironments that mimic those found in vivo (when E is ∼10−15 kPa) (Berry et al., 2006) maintain a contractile phenotype that beats at 1 Hz, whereas cells on harder matrices can sustain beating only for a short time. During this period, the largest difference between intra- and extracellular deformations was found in cells on hard matrices, suggesting enhanced intracellular strains in protein structure as examined by a proteomic-scale, ‘cysteine shotgun’ method for in situ labeling and identification of protein structures (Johnson et al., 2007).
Results
Cell contraction versus matrix contraction
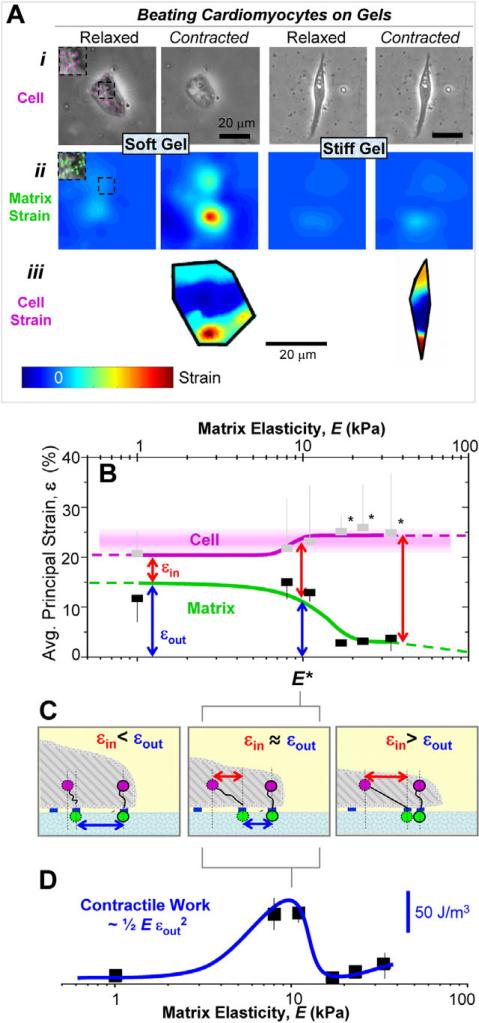
The matrix elasticity E was systematically varied to measure maximum cell contraction versus matrix contraction as cardiomytocytes beat. Embryonic cardiomyocytes were plated onto collagen-I-coated polyacrylamide gels for 24 hours with pre-embedded fluorescent beads (Fig. 1Aii, inset) prior to measuring bead displacements within the matrix. Particulates near the cell periphery 3−5 μm above the cell-gel interface were also tracked to determine intracellular displacements (Fig. 1Ai, inset). Average principal strains observed in the gel and the cell during rhythmic beating were estimated and mapped as εout and εcell, respectively (Fig. 1A; supplementary material Movies 1 and 2 show cells beating on 1 kPa or 34 kPa matrices). Note that the matrix and cellular strains tend to be maximal near the cell periphery (see Fig. 1Aii,iii). Cell and gel strains in or under the cell during contraction were averaged and plotted as a function of matrix E (Fig. 1B). E* is the point at which the intracellular strain (εin), the difference between matrix (Fig. 1Aii) and cell strain (Fig. 1Aiii), is statistically different (by paired t-tests) when compared with softer matrices.
Fig. 1.
Cell and matrix contraction. (A) Cells were imaged in brightfield mode 3−5 μm above the cell-matrix interface (i) and in fluorescence at the cell-matrix interface to observe particle motion during contraction (see inset pseudo-colored beads) to determine cellular and matrix deformations, respectively, that were then used to generate matrix (ii) and cellular (iii) principal strain maps. Note the difference in matrix strain between soft and stiff gels reflected by decreased deformation of the contracted cell. (B) The maximum cellular strain εcell is similar to the maximum matrix strain for soft matrices εout, resulting in small εin values. Hard substrates only allow the cell to deform itself during contraction. The transition between these regimes occurs at E*, where internal and external strain appear equivalent. Curve fits to guide the eye are of the form y=a+bEm×n/(Em+E1/2m) with m=6 and n=0 (matrix), n=1 (cell). Error bars, ± s.d. for at least five cells in duplicate studies. Paired t-tests were used to identify significant differences between εin of cells on softer versus stiffer gels (*P<0.05) and between cell strains on different gels. In the latter case, no two cell strains were significantly different. (C) Schematic of strained states for soft gels (E* gels) and stiff gels that respectively yield εin<εout, εin=εout, and εin>εout. (D) The contractile ‘work’ done by cardiomyocytes on the substrate peaks at E*.
As expected, cell strain always exceeded matrix strain, i.e. εcell≥εout, because the cells are the source of contraction. The strain sustained within the cell is the difference that is ‘felt’ intracellularly during contraction: εin=εcell–εout. On soft matrices (E<8 kPa), cell strains transmit almost completely to the matrix (Fig. 1C, left) and, so, intracellular strain εin is small. On the stiffest matrices, cells are highly strained but cannot significantly strain the matrix (εout→0), yielding εin≈εcell. Myosin-based contraction on this hard-matrix limit is thus fully sustained by the cell (Fig. 1C, right), perhaps hyperactivating stretch-sensitive proteins, with the eventual result being – as shown below – an inhibition of cell beating.
For an intermediate stiffness that we estimate is at the low end of the range E*≈11−17 kPa, both the matrix and the cell are about equally strained with εin≈εout. The contractile work (as an energy density) that is generated by the cardiomyocyte in the matrix is estimated as work done per volume (w) ∼½E×εout2 and is also maximized near E* (Fig. 1D) with a value w∼100 J/m3. On softer gels, strains are large but the resisting E is small, whereas the opposite scenario applies for stiffer matrices.
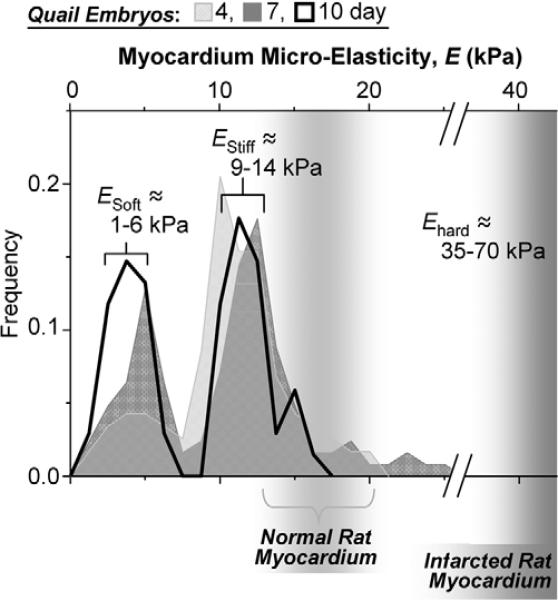
Micro-elasticity of the developing myocardium
During development, many cellular and matrix-based changes occur in the outermost myocardium layers (Gittenberger-de Groot et al., 1998), which could be mimicked with these PA gels to give it a physiological context. To assess the mechanical state of a microenvironment at a scale that cells would ‘feel’ this developing layer, the apical layers of embryonic myocardium were investigated using atomic force microscopy (AFM).
Resulting histograms of tissue micro-elasticity appear bimodal with approximately Gaussian distributions for E. The softer peak at ESoft ≈3−4 kPa (Fig. 2) develops over the time to ∼40−50% of the total number of force curves at day 10 [and is similar to the elasticity of fibroblasts in culture (Mahaffy et al., 2000)]. The stiffer peak of EStiff≈11−12 kPa remains constant over time and accounted for 30−50% of the data. EStiff is similar to previously measured elasticity of cultured and isolated myotubes (Collinsworth et al., 2002; Engler et al., 2004b) but is softer than infarcted myocardium, Ehard ≈35−70 kPa (Berry et al., 2006). Importantly, the range of EStiff overlaps with the E* value (see Fig. 1).
Fig. 2.
Myocardial elasticity during embryogenesis. Histograms of elastic moduli (∼150 locations per sample) determined from quail embryos show two peaks, one indicating passive elasticity of contractile myocardium (EStiff) and a softer, second peak (ESoft) which surrounds the myocardium and increases in frequency with development. Results for normal and infarcted rat myocardium are indicated for comparison (Berry et al., 2006).
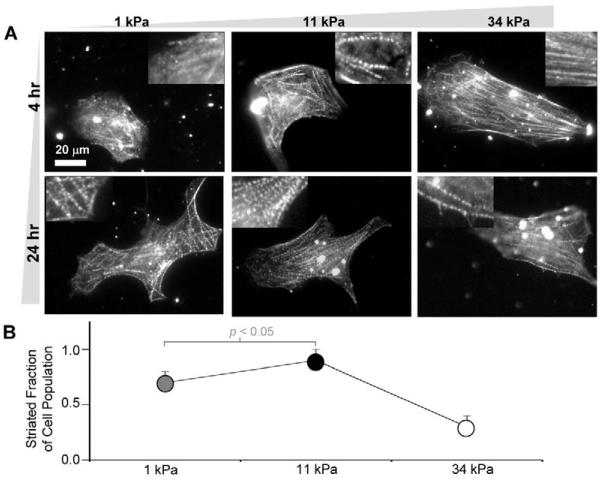
Striation and cell spreading are functions of matrix elasticity
Rhythmic beating of isolated cardiomyocytes is dependent first on successful spreading of the cells and then on myofibril reassembly – which appears similar to the reassembly that follows cell division in embryos and explants (Du et al., 2003; Ehler et al., 1999). To assess the assembly as a function of both matrix elasticity and time in culture, purified cardiomyocytes were plated on soft (1 kPa), optimally stiff (11 kPa ∼E*), or hard gels (34 kPa). Cells were immunostained using striated-muscle-specific anti-α-actinin at 4 or 24 hours post plating (Fig. 3A) to assess myofibril reassembly. Cells were selected in brightfield mode and imaged to quantify the fraction of cells on each gel that exhibited the typical striated myofibrils throughout the cytoplasm (and not just the cell edges per cell on the hard 34-kPa matrix). After 24 hours in culture, a larger fraction of striated cardiomyocytes was counted on the two softer matrices of 1 kPa and 11 kPa (Fig. 3B). For cells on the hard substrate, striation was present but observed in very few cells, despite the cells appearing well spread. Thus, the ability of cells to reassemble their striations reveals an optimal gel stiffness of E∼E*, consistent with both the maximum in contractile work transmitted to the matrix (Fig. 1D) and the tissue micro-elasticity measurements (Fig. 2).
Fig. 3.
In vitro striation of cardiomyocytes. (A) Purified cardiomyocytes from 10-day-old embryonic myocardium were plated onto substrates of varying elasticity to observe striated cytoskeletal organization with skeletal α-actinin. Many cells on both soft gels and intermediate E* gels reassembled myofibrils whereas cells on hard matrices exhibited less myofibril reassembly. Inset images show magnified views of the larger images. (B) Fraction of cardiomyocytes that exhibit striation throughout the cytoplasm (± s.d. for >15 cells in triplicate studies). Organization appeared greatest on E* gels.
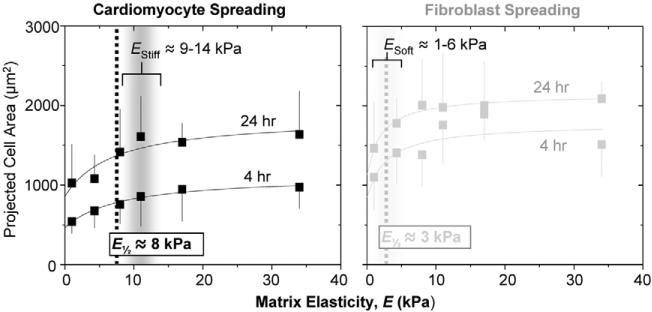
Cardiomyocytes and co-isolated fibroblasts spread more on stiffer substrates as do many other soft tissue cells that often exhibit a hyperbolic form in spreading area: Projected area =a+b×E ÷ (E½+E). In our experiments cardiomyocytes and fibroblasts (Fig. 4) spread with a similar functional dependence, with E½ (EStiff or ESoft) being the half-maximum ‘set-point’ where the cell is neither too round nor too spread. For cardiomyocytes EStiff ≈8 kPa, and for fibroblasts ESoft ≈2−3 kPa, independent of time; such set-points are again remarkably close to the respective micro-elasticities of tissue (Fig. 2B), and suggest a mechanical match between cells and matrix.
Fig. 4.
Cardiomyocyte and fibroblast spreading. Cardiomyocytes and pericardial fibroblasts were plated from 10-day old embryonic myocardium without purification and spread areas were measured (± s.d. for >15 cells in triplicate studies) after 4 or 24 hours. Fibroblasts were detected as α-actinin-negative cells and were the larger fraction of cells, consistent with in vivo proliferation (Fig. 2B). Half maximal ‘set points’ for projected cell area correspond closely to the peaks in passive tissue elasticity in Fig. 2B.
Cardiomyocyte beating depends on matrix elasticity
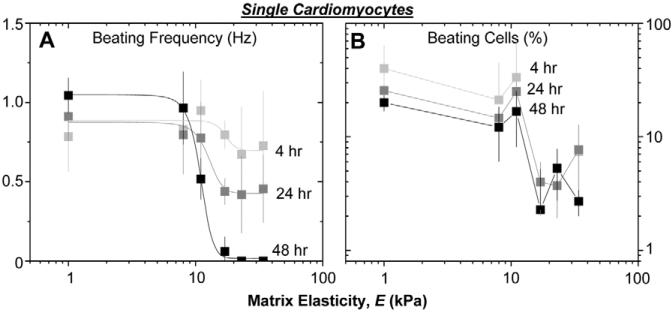
To address longer-term cardiomyocyte function, single cells were observed over several minutes in order to quantify both beating frequency and the beating fraction of the population (Fig. 5). Cell cultures previously seen to have assembled myofibrils (E≤11 kPa) were also observed to beat at ∼1 Hz with 20−40% of the cell population beating. However, on the hardest matrices and under otherwise identical conditions, the contraction frequency decays from ∼1 Hz to sporadic contraction with a half-life of ∼24 hours, and only 2−8% of a population of such isolated cells exhibited pulsatile beating. Pre-incubation of the gel substrates in serum for 24 hours does not alter these trends, suggesting matrix elasticity is the important factor.
Fig. 5.
Cardiomyocyte beating is sensitive to matrix elasticity. Purified cardiomyocytes were plated at low density on matrices of varied stiffness, and beating was observed in phase contrast. (A) Beat frequency is elasticity-dependent, with cells on hard matrices slowing their beat frequency over days. (B) The percentage of cells beating was also quantified after 4, 24 and 48 hours post isolation. Beating cells persisted only on matrices with E≤E*. Error bars are ± s.d. for >5 cells, 10 seconds each in triplicate.
Cardiomyocytes in vivo maintain cell-cell contacts; therefore, to assess the contribution of such a compliant coupling, cell clustering was promoted by seeding cells at high density. A greater number of clustered cells were seen at 4 hours to beat at 1 Hz on the hard matrices (supplementary material Fig. S2A), but protection proved temporary as both beating frequency and the fraction of beating cells decayed with time for E>E* (supplementary material Fig. S2). By 48 hours, less than 10% of cells on hard substrates were beating, with cells beating only sporadically.
Protein structural changes on hard versus compliant substrates
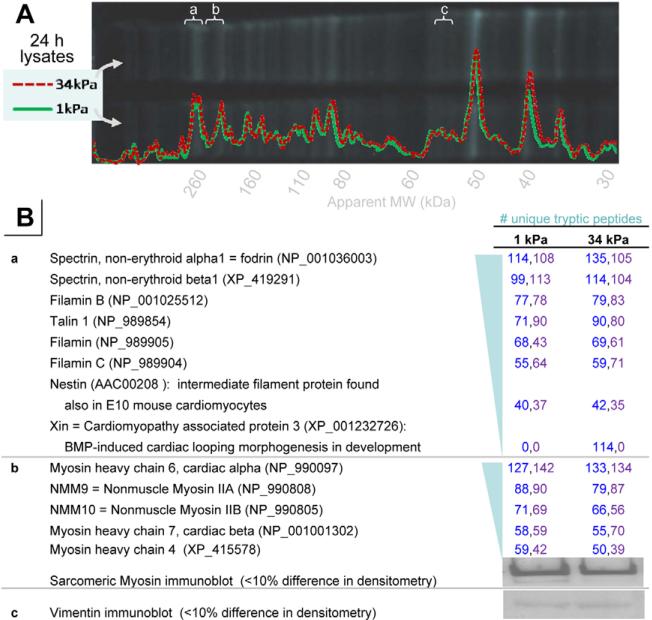
To begin investigating molecular differences between cells on the hard versus soft matrices, cardiomyocyte lysates were generated from 24-hour cultures, and then separated by SDS-PAGE and analyzed by densitometry (Fig. 6A). To maximize resolution, the Cys-reactive dye monobromobimane (mBBr) – which was put to further use as described below – was added with the denatured proteins before electrophoresis. Mass spectrometry of some of the major bands near where one would expect to find key structural proteins, such as filamin, myosin or vimentin, did indeed identify these proteins, their isoforms and several other proteins consistent with a cardiac lineage (Fig. 6B). However, densitometry analyses and peptide abundance from mass spectrometry as well as immunoblotting – for vimentin and also sarcomeric myosin – revealed no major difference in mean abundance (<0.15) of the identified proteins – including nonmuscle and cardiac myosin isoforms – between cells on hard versus soft substrates despite major functional differences (Figs 1-5).
Fig. 6.
Protein abundance assessments in embryonic chicken cardiomyocytes grown on soft or hard matrices. (A) SDS-PAGE separation and densitometry analyses of lysates from 7-day embryonic chicken cardiomyocytes grown on 1 kPa or 34 kPa grown for 24 hours and then lysed, denatured and labeled by mBBr. (B) Mass spectrometry analyses of tryptic peptides or immunoblot results from the indicated gel bands (a-c). These bands were chosen as candidate cytoskeletal proteins using differential labeling in in-situ Cys-shotgun experiments (Table 1). The numbers of unique peptides are indicated from two different experiments. Immunoblotting for sarcomeric myosin and vimentin also indicated no significant differences in abundance.
Mechanical strains within cells were estimated to be much larger in cells on hard matrices than on soft matrices (Fig. 1) and, given the above result for similar abundance of at least some major cytoskeletal proteins, we hypothesized that differences in protein structure rather than abundance differences underlie the elasticity-dependent behavior of cells (Figs 2-5). To assess changes in protein conformation or assembly within living cardiomyocytes, a recently developed cysteine-shotgun method (Johnson et al., 2007) was applied. Cys is hydrophobic and often buried in tertiary or quaternary structure; mBBr is membrane-permeable and can label exposed Cys. Changes in the accessibility of buried Cys can occur with differences in contractile activities, conformational states or shielding due to structural assembly – all leading to differential in-cell labeling by mBBr (Fig. 6). Differences in the redox states could interfere in principle and, so, we first verified that there were no significant differences in cytoplasmic levels of glutathione – which is a main redox stabilizing polypeptide in cells that can also be assayed with mBBr labeling and HPLC separation (supplementary material Fig. S3).
For Cys-shotgun studies of intact cells, cardiomyocytes were again separated from fibroblasts using a pre-plating method, as verified by immunostaining parallel cultures for skeletal muscle α-actinin. Cells were grown on 1-kPa or 34-kPa gels for 24 hours, labeled live with mBBr, lysed and then analyzed similar to those shown in Fig. 6. Fluorescence per protein mass was determined by normalizing for protein amount by Coommassie staining (which is adequate because of abundance determinations shown in Fig. 6B). When comparing cells grown on hard versus soft gels, four prominent bands (Table 1, bold entries) showed significant differential labeling (>0.l5): positive or negative changes indicate greater labeling on hard or soft substrates, respectively. The four prominent mBBr-labeled bands were removed from the gel, trypsinized and identified by mass spectrometry as myosin heavy chains (IIB and cardiac), filamin, vimentin and the muscle-specific pyruvate kinase M1. The latter is a glycolytic phospho-enzyme that transfers phosphate phosphoenolpyruvate to ADP yielding ATP (Mela-Riker and Bukoski, 1985).
Table 1.
Results of Cys-shotgun experiments of cardiomyocyte proteome
| Estimated molecular mass (kD) | Differential labeling (34 kPa and 1 kPa) | Proteins indentified by mass spectrometry |
|---|---|---|
| ∼285 | +0.21 | Filamin A and B |
| ∼217 | +0.36 | Non-muscle myosin IIB |
| ∼125 | 0.03 | |
| ∼90 | 0.05 | |
| ∼70 | −0.08 | |
| ∼58 | +0.23 | Pyruvate kinase M1 |
| ∼51 | −0.37 | Vimentin |
| ∼40 | 0.04 | |
| ∼35 | −0.09 | |
| ∼20 | −0.08 |
Analysis of proteins from SDS-PAGE and the difference of fluorescent signal between proteins labeled in cells on hard versus compliant substrates. Proteins showing a difference of >20% in triplicate studies (bold) were subsequently identified by mass spectrometry.
Mass spectrometry also identified specific protein residues that were modified dependent on matrix elasticity. Filamin, a homodimeric crosslinker of F-actin, has 38 Cys residues that are distributed mostly among its 24 immunoglobulin-like domains. Cys59 in filamin A showed substantial labeling and is located in the actin-binding domain (supplementary material Fig. S4A) (Barry et al., 1993). Vimentin has just one Cys residue (Cys319) in the middle of its rod domain, and has been predicted to be shielded in fibrils (Strelkov et al., 2002). Decreased fluorescent labeling has indeed been found with polymerization of purified vimentin and also with mesenchymal stem cells that are tensed on a hard matrix versus relaxed by myosin inhibition (Johnson et al., 2007).
Nonmuscle myosin IIB (NMM IIB) and cardiac myosin heavy chain (cardiac MHC) are the major myosin isoforms in cardiac premyofibrils and myofibrils, respectively (Du et al., 2003; Sanger et al., 2005). Enhanced mBBr labeling of these on hard matrices (Table 1; supplementary material Table S1) suggest protein changes that are induced by the larger net strain (Fig. 1). Based on peptide location, myosin structure (Houdusse et al., 1999; Rayment et al., 1993) and homology modeling (Schwede et al., 2003), structural changes in the myosin head and lever arm were assessed. Two Cys residues (Cys95 and Cys701 in both NMM IIB, and what could be cardiac MHC) were mBBr-labeled out of the seven Cys residues detected in the >40%-sequence coverage (total of Cys 16 residues). Cys95 is on the protein surface (Houdusse et al., 1999; Rayment et al., 1993), and so facile labeling is understandable. Cys701 is located close to the ATP-binding pocket on an SH1-SH2 helix (Nitao et al., 2002) (supplementary material Fig. S4B), and the proximal Cys701 appears to be completely buried within the myosin head. Surprisingly, Cys701 is labeled in cells on hard matrices with an efficiency similar to a surface-exposed Cys residue, which implies a change in myosin conformation.
Discussion
Although myofibril formation in cardiomyocytes has been extensively studied in culture, the difficulty of sustaining a rhythmically contractile phenotype on hard substrates is made clear in the results here (Figs 1,3,5). Optimal matrix elasticity in vitro shows a close correspondence with tissue elasticity (Fig. 2) and supports cardiomyocyte function for longer times (Fig. 4). Since myosin II contractility is the essential motor activity here, a simple analogy with skeletal muscle can perhaps provide some initial intuition: cells doing work on 1-kPa matrices might be equated – at least in principle – to ‘lifting’ a 1-kg dumbbell, cells on a 10-kPa matrix thus lift a proportionally larger 10-kg dumbbell, whereas cells on a scar-like 50-kPa matrix must try to repeatedly lift a 50-kg barbell. For many of us, the latter would be an isometric effort with little to no motion.
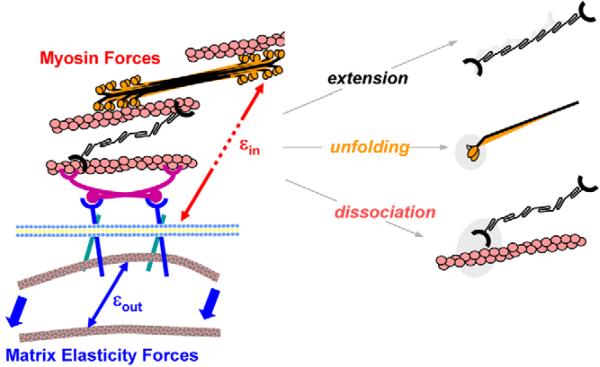
A simplistic analysis of the maximal matrix-strain energy W (in J), might also provide some insight. W=w×A×R with w being the contractile work done by the cell (Fig. 1D) (measured in J/m3), A being the cell area (1500 μm2) (Fig. 4A), and an R value of ∼10 μm being an estimate of the radial penetration of the strain into the gel. Since w is the work done in a volume, its multiplication by the volume of the strained gel (i.e. A×R) yields W, which is maximal at E* and results in a W value of ∼1.5 pJ. Given that the work by a single myosin in one cycle is 15 zJ (Takagi et al., 2006), the estimated W value implies 105 myosin cycles per contraction. Considering the average myofibril density (450/μm2) (Hernandez-Nicaise et al., 1982), and sarcomere longitudinal spacing and cell size (i.e. A× height) of 2 μm, our estimate of 105 myosin cycles per contraction is surprisingly close to the maximum number of myofibrils possible, ∼106 myofibrils/cell. For E>E*, the matrix is too stiff and myosin is likely to be overloaded – similar perhaps to the rollover in stroke volume with pressure in the Starling curve. Because myofibrils often form at the cell periphery (Du et al., 2003; Rhee et al., 1994; Sanger et al., 2005), our finding that cell strains appear largest at the periphery of cells on the stiffest matrices (Fig. 1Aiii) provides a possible physical mechanism for limited myofibril reassembly (Fig. 3B) that would only be compounded if disassembly is also accelerated by cell strain and stress. Initial molecular insights as to why cardiomyocytes stop beating on stiff matrices is provided here by Cys-shotgun analysis (Table 1, Figs 6,7; supplementary material Table S1 and Fig. S4;) that indeed implicate overextension in cytoskeletal proteins or complexes that often localize to the cell cortex (NMM IIB, filamin-actin) and also suggest stabilization of a static cytoskeleton with increased vimentin assembly.
Fig. 7.
Cell-driven, matrix-coupled remodeling through a combination of forced extension, unfolding or dissociation of proteins. On hard matrices, the intracellular strains εin exceed the extracellular strains εout and intracellular remodeling is promoted.
Mechanical influences on myocardial development and cell contraction
The spatiotemporal expression of both ECM and specific cell types seems almost certain to influence myocardial development (Tsuda et al., 1996). As in other vertebrates, avian myocardium develops from pre-epicardial cells that form a collagenous, U-shaped tubular heart (Hamburger and Hamilton, 1951). At day 4, embryonic proepicardium can be expected to be mechanically homogeneous – consistent with the data shown in Fig. 2 – because of the relatively few pericardial-like fibroblasts, but over time the tissue matures into a visceral epicardium and becomes more heterogeneous and stratified (Gittenberger-de Groot et al., 1998). The layers consist of an endomysial collagen network containing cardiac fibroblasts that surround distinct sheets of cardiac myocytes (Goldsmith et al., 2004) to establish specific mechanical layers (Novak et al., 1994). Although such changes almost certainly contribute to the differentiation program, they also are likely to affect the mechanical microenvironment: Fig. 2 indeed reveals the ‘mechanical development’ with emergence of an ESoft-microtissue that supplements the EStiff-microtissue. Cardiac disorders can adversely change the myocardium by accumulating collagen fibers and increasing heart-chamber stiffness (Yamazaki et al., 1995). Such changes are measurable by AFM – as can be seen in Fig. 2 with EHard – and, based on the data here, rigidification seems likely to limit cell-based myocardial therapeutics by limiting contractile capacity (Berry et al., 2006). Limited contractile capacity was also noticed in observations of cell clusters (supplementary material Fig. S2); despite a permissive environment, with cell-cell contacts and a potential for signaling and paracrine effects that are not present in isolated cardiomyocytes, gels with an elasticity value of ∼EHard suppress contractility even after 4 hours. The results may be relevant to differences in the beating of embryoid bodies generated by embryonic stem cells (ESC) versus induced pluripotent stem (iPS) cells, with the latter appearing to beat less (Mauritz et al., 2008).
As with any cell culture system intended to mimic some aspect(s) of a tissue, there are many caveats. Cardiomyocytes in vivo interact with other cell types as well as with each other through receptor and transport proteins – but all of these would require further study to document their possible protective roles in the sparse cultures and cell clusters studied here. The 3D nature of most mesenchymal tissue would, in the context described in this study, suggest that cardiomyocytes adhere and contract tissue all around them, perhaps doing even more work than that reported in this study, with polarized cells that strain only their basal matrix. Strains in the cells are also likely to change and perhaps shift owing to interactions with other cells in a more-3D microenvironment, and although important effects could also be mediated by membrane receptors and second messengers among other mechanisms, the Cys-shotgun method might still hit some of the same strain-remodeled proteins as here.
The observation that matricellular composition and mechanics can direct cell function is not specific to heart (see Engler et al., 2004b; Paszek et al., 2005; Yeung et al., 2005). Unique to our study, however, is the estimation of periodic cell strains in order to compare matrix strains, with the finding that the latter vanish on hard gels, whereas cell strains do not equal zero. The overall elliptical or elongated shapes of cells described here also tend to align the myofibrils (Fig. 3), which probably explains why bead displacements are small near the cell center and why the strains appear anisotropic and bipolar in both the matrix and the cell (supplementary material Movies 1 and 2; Fig. 1A, bottom panels). Further investigation of this contractile anisotropy could benefit from the use of anisotropic materials, which have also been used to show that cells adjust their contractility in response to the substrate stiffness (Saez et al., 2005).
Our results suggest that, at an optimal matrix elasticity E*, matrix strains and cell strains are similar in magnitude (Fig. 1A, middle panels). However, cell strains are generally difficult to quantify, and the fiduciary particles used in this study might not seem ideal but are typical for such measurements (Hoffman et al., 2006). Particles that undergo persistent transport are eliminated, and only particles located within 5 μm of the cell edge and 3−5 μm above the cell-matrix interface are analyzed. Even with such a defined locus, differences in z-position and out-of-plane motion are likely to be greater in the cell than for the small beads embedded in the matrix interface. Regardless, the large difference between the cell- and matrix strains for cells on hard gels is at least qualitatively descriptive, and suggestive of overstretching in the cell (Fig. 1C). Cytoskeletal changes associate with myofibril disorganization Whereas most of the cytoskeletal proteins examined show much less than a 20% change in abundance, using either densitometry and/or tryptic peptide abundance, one protein the cardiomyopathy-associated protein 3 (also known as Xin actin-binding repeat-containing protein 2, XIRP2, hereafter referred to as Xin) was found, by using mass spectrometry, to be expressed only in cells on hard matrices (Fig. 6B). The relation of Xin to BMP-induced looping morphogenesis (Wang et al., 1999) or cardiomyopathy requires further study, but some confidence in these results is provided by the detection of nestin. This intermediate filament protein is usually considered as a marker of neurogenesis, but it is also reported to be the first marker for mid-embryonic (E10) heart development (Kachinsky et al., 1995). Despite being a marker of cardiomyocyte differentiation state, nestin is found here to be equally abundant in cells on soft versus hard matrices. However, the Cys-shotgun method applied to intact cells suggested structural differences in a few of the otherwise equally abundant proteins.
Contractile dysfunction of cells on hard gels might be due to unfavorable assembly of protein and/or unfavorable conformations, and the Cys-shotgun results (Table 1; supplementary material Table S1) combined with analyses of protein structures (Figs 6,7) can add some clarity. In filamin, Cys59 is in the actin-binding domain (Fig. 7A) and enhanced labeling of this site (indicative of increased Cys exposure) therefore suggests stretch-induced rupture of the filamin-actin interaction under isometric contraction. Since filamin is required for heart development (Feng et al., 2006), is mutated in some cardiomyopathies (Unger et al., 2007) and is upregulated during the recovery from cardiac injury (Birks et al., 2005), disruption of actin binding in cells on hard matrices probably undermines sustained beating (Fig. 5). Vimentin labeling, however, is decreased in cells on hard gels, and it therefore appears to reflect a shift to the assembled state (Johnson et al., 2007), which would tend to mechanically resist rhythmic contraction and stabilize the cell.
Force-generating myosin is perhaps more complicated. A prominent model of muscle development posits myofibril assembly through membrane-localized assembly of NMM IIB, actin and α-actinin (Rhee et al., 1994; Sanger et al., 2005). The cell periphery is where the largest intracellular strains appear (Fig. 1Aiii) and, whereas NMM II probably has an additional role in cytokinesis during heart formation (Conrad et al., 1991; Du et al., 2003), its assembly into striated premyofibrils that link to the ends of more-mature cardiac-myosin-containing myofibrils means that NMM II physically couples the contractile structures to the matrix (Dabiri et al., 1997; Sanger et al., 1986). The SH1-SH2 helix of myosin is capable to unwind to a loop and to function as one of three key ATP-driven joints in the head (Nitao et al., 2002); therefore, facile labeling of Cys701 in this structure implies a localized unfolding of myosin in cells on hard matrices with possible implications for contractile activity. Forced unfolding seems consistent with greater εin values in cells on hard matrices (Fig. 1), and it could inhibit the assembly of pre-myofibrils, providing a molecular explanation for the loss of striation and the decreases in beating. The general scheme that seems plausible on the basis of our results is that cell-driven, matrix-coupled remodeling pathways (Fig. 7) include both forced unfolding and forced dissociation, affecting what structures assemble or what structures persist in an actively contracting cell.
Elasticity of ECM is emerging as a key cue of the insoluble microenvironment around cells in development and is also likely to contribute to dysfunction in disease. Beating cardiomyocytes facilitate estimations of strain, both in the matrix and in the cell, and the two appear almost equal at an optimal matrix elasticity (E*≈11−17 kPa) that maximizes cardiac work. In vivo, such work would ultimately be the pressure-volume work performed by pumped blood. In diseased states, such as the hardened post-infarct state, where E≥3E* (Berry et al., 2006), the our results predict dramatic losses in contractile function. Taken together, our work highlights the need for greater attention to matrix rigidity and matrix remodeling in cell therapies.
Materials and Methods
Myocardium and cell isolation, and mechanical probing
Embryonic heart tissue was isolated by approved protocols from 4-day-, 7-day- and 10-day-old Japanese quail (Coturnix japonica; CBT Farms; Chestertown, MD) at key developmental stages (Hamburger and Hamilton, 1951), and the tissue was placed in α-MEM medium containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cardiomyocytes were isolated from 9-day quail or 7-day old White Leghorn chicken (Gallus gallus) embryos and pre-plated to remove fibroblasts (Du et al., 2003; Sanger et al., 2005); supernatant was then plated onto matrices at 103 cells/cm2. Medium lacking L-glutamine was used and is crucial for controlling fibroblast contamination. To obtain cell clusters, 105 cells/cm2 were plated. Molecular mass markers were ‘Magic Mark’ for Cys Shotgun studies and ‘Novex Sharp’ for measurements of protein abundance (Invitrogen; Carlsbad, CA).
Atomic force microscopy probing of cardiac tissue
Tissues were bisected and mounted on coverslips with epoxy to expose the apical surface of the myocardium (supplementary material Fig. S1A). Samples were then placed on a microscope-mounted Asylum 1-D atomic force microscopy (AFM) (Asylum Research; Santa Barbara, CA) and probed with a pyramid-tipped cantilever (Veeco; Santa Barbara, CA) having a spring constant (∼60 pN/nm nominal) as determined by thermal calibration (Engler et al., 2004a). Samples were randomly indented hundreds of times over their surface, generating probe force versus indentation plots (supplementary material Fig. S1B) and all within ∼30 minutes of tissue isolation. Data were fit with a Hertz cone model over a range of cantilever deflections of ∼10−100 nm (Domke and Radmacher, 1998; Rotsch et al., 1999) that corresponds to an indentation range of ∼100−1000 nm (supplementary material Fig. S1B). The fitted elastic modulus E represents the collective mechanical response of cell and matrix on a local cellular scale.
Live cell and immunofluorescence microscopy
To assess contractile properties of cardiomyocytes, cells were plated on polyacrylamide gels and allowed to adhere. Cell area was observed for both cardiomyocytes and fibroblasts, and cell types were differentiated by staining for skeletal muscle α-actinin or non-muscle myosin IIB (Sigma; St Louis, MO) to identify the cardiomyocytes. For live cell imaging, coverslips were attached to the hole cut into the bottom of a custom chamber slide using vacuum grease to ensure a seal and allow media to be added directly on top of the coverslip. This system was then placed on a 37°C heated stage where cells were observed with a 60× objective in brightfield or fluorescence mode at low magnification for up to 30 minutes. A Photometric Cascade CCD camera capturing at 20 frames/second was used to image cell contractions (supplementary material Movies 1 and 2). For analyses of contraction, images in which the average displacement vector appeared maximal were considered ‘maximally contracted’.
Matrix preparation, particle tracking, and strain and energy estimations
Cells were plated on polyacrylamide gels (Pelham and Wang, 1997) with collagen I (BD Biosciences; San Jose, CA) attached covalently at an optimal density of 0.25−1 μg/cm2 (Engler et al., 2004a). Acrylamide and bis-acrylamide crosslinker were mixed at concentrations suitable to obtain elastic moduli as measured by AFM (Engler et al., 2007); the moduli do not change with collagen attachment or serum incubation. Cell-generated substrate principal strains were measured by tracking the displacement of fluorescent polystyrene beads embedded in the gel during polymerization (Dembo and Wang, 1999; Oliver et al., 1995; Wang et al., 2002) (Fig. 1Aii, inset). The positions were of fluorescent beads determined by thresholding images and then using the analyze particle function in Image-J (NIH; Bethesda, MD). Bead position information was tracked for both cardiomyocyte contraction and resting phases where particles were matched between images. Bead positions were also determined after the cell was removed (by scraping or aspirating with a micropipette), to establish the bead locations in gels unstressed by cells. Comparisons were then made between the first contracted and reference states and the second ‘relaxed’ and reference states to generate the displacement vector of each bead with respect to its undisturbed position. This comparison neglects a non-zero matrix strain that is present even when the cell is not beating due to the resting traction forces in the cell (Fig. 1Aii, relaxed cells). Regardless, displacements were determined by the difference in bead positions using the cell centroid for a local reference coordinate system. Displacements computed at each location were then converted to Cauchy strains, and the maximum principal strain was used for further analysis. Cells pull inward and so this strain tended to be compressive (positive). Next, these discrete points were mapped to a surface in Matlab using the grid-data function and a cubic spline-like smooth routine previously established (Sandwell, 1987), the colormaps of which are displayed in Fig. 1A with the indicated strain values (lower panels). Lastly, this strain due to active contraction was averaged across the entire cell, and the difference between contracted and relaxed states was plotted as εout (Fig. 1B, black data points).
The cell-average principal strain in the matrix was then compared with a principal strain in the cell (εcell), which is the averaged principal strain in the cell itself due to contraction (Fig. 1Ai). This was computed from the motion of fiduciary particles within the cell (e.g. vesicles, etc.) (Fig. 1Ai inset) as per particle tracking methods (Hoffman et al., 2006). Particles were identified simply by their phase contrast, and only those located within 5 μm of the cell periphery within the focal plain and 3−5 μm above the cell-matrix interface were included in the estimations. Motion was again compared between the contracted and relaxed states, the latter of which was used as a reference state. Only rhythmic particle motions due to contraction were analyzed to obtain εcell; particles with large motions – typical of active transport – were excluded. Analyses similar to those for εout were used with strains computed from discrete particle motion between contracted and relaxed states. After smoothing and mapping in Matlab, the average strain across the cell, εcell, was estimated.
The difference between the total principal strain εcell and the matrix principal strain εout represents the principal strain dissipated internally (εin). For εout>εin, εout=εin, and εout<εin, the substrate sustains greater, equal, or less strain, respectively, than the internal cytoskeleton, (Fig. 1C). The last case implies large cytoskeletal stretching.
Cysteine-shotgun labeling and mass spectrometry
On the basis of recent studies (Johnson et al., 2007), a membrane-permeable cysteine-reactive dye, monobromobimane (mBBr; Molecular Probes–Invitrogen), was added at 125 μM to 1-day quail cardiomyocyte cultures for 20 min prior to trypsinization. Importantly, mBBr labeling at 125 μM did not significantly alter cell beating or morphology but was sufficient to label proteins. Cells were then lysed in 200 μL buffer solution containing 0.1% Triton X-100, 6 M Urea, 5 mM ETDA, and 10 μM Protease Inhibitor Cocktail (Sigma) diluted in PBS. Labeling was quenched by addition of 10 μL of 282 mM β-mercaptoethanol, and lysate was separated by SDS-PAGE. Total protein levels were measured using the Bradford assay and equal loads were run; actin immunostaining was used to verify transferred loads in immunoblots. Fluorescent intensities of labeled proteins were measured by densitometry and normalized to protein level by using Coomassie Blue, (i.e. blue stain intensity). Bands with a large normalized difference between 1 and 34 kPa samples were excised, trypsinized, analyzed by mass spectrometry and compared with available proteomes. To determine whether a peptide was mBBr labeled or not, sequence searches included looking for the mass of the peptide alone, the peptide in an oxidized state and the peptide plus label. For studies of protein abundance, chicken cardiomyocytes (separated from pre-plated fibroblasts) were cultured as above for 24 hours, washed with ice-cold PBS, and then scraped with matrix and some gel substrate into a tube. Cells were lysed as above, and subsequently labeled using 400 M mBBr at pH 8. Proteins were separated and analyzed per SDS-PAGE and mass spectrometry methods above except that peptide sequences were compared with the Gallus gallus proteome. Immunoblotting for vimentin or sarcomeric myosin respectively used monoclonals V9 (Sigma) or MF20 (DSHB, University of Iowa).
Glutathione assay
Adapting previously described methods (Fernandez-Checa and Kaplowitz, 1990), we determined the levels of free glutathione (GSH). Briefly, cells were treated with 1 mM buthionine sulfoximine (Sigma) in serum-free, L-glutamine-free MEM for 1 hour to inhibit glutathione synthesis. Cells were then rinsed twice with serum-free medium and incubated with labeling solution (400 μM mBBr in medium) for 20 minutes. Cells were lysed with 5% trifluoroacetic acid and scraped. Total protein was verified to be the same by Bradford assay. The resultant GSH extract was cleared by centrifugation and separated on an LC-20 Prominence Liquid Chromatograph with an SPD-M20A Prominence Diode Array Detector (Shimadzu; Kyoto, Japan). Samples were eluted with 0.1% TFA in acetonitrile, and the presence of peptide in a putative peak of mBBr-thiol adduct was verified by the 220 nm : 396 nm intensity ratio.
Supplementary Material
Supplemental Movie 1: Isolated Cardiomyocyte Beating on Soft Matrix
Depicted here is the principal strain field (left) created by the cell (right) as it deforms the soft gel with a basal level of traction forces. As the cardiomyocyte contracts the gel, however, strain rapidly increases as a result of increased traction forces, which also decay rapidly again. The magnitude of the strain field is determined from bead displacements and uses the same strain scale as in Figure 1.
Supplemental Movie 2: Isolated Cardiomyocyte Beating on Stiff Matrix
Shown here is the principal strain field (left) created by the cell (right) as it attempts to deform the stiff gel. Traction forces are small, but non-zero. As the cardiomyocyte contracts, the resulting traction forces do not greatly change, although the cell significantly contracts. Strain magnitude uses the same scale as in Figure 1.
Table S1: Summary of In Situ Cysteine Labeling in the Non-muscle Myosin IIB Head and Lever Arm for Cardiomyocytes on Stiff Matrices
Figure S1: AFM Measurement of Myocardial Elasticity
(A) Brightfield image of freshly isolated day 4 myocardium. Sample is then mounted on a AFM stage as depicted schematically with the apical surface exposed. An AFM tip with r ∼ 50 nm and experimentally determined spring constant, ksp, is then indented into the myocardium to yield tip deflection, d, versus tip position, z (B). Elasticity was determined by fitting a hertz cone model from the contact to the maximal indentation points, with the former determined by a range of analysis method from z1, d1 to z2, d2.
Figure S2: Cardiomyocytes at High Density also show Beating depends on Matrix Elasticity Purified cardiomyocytes were plated at high density on matrices of varied stiffness, and beating was observed in phase contrast. (A) Beat frequency is elasticity-dependent, but decreases in beat frequency are smaller compared to low density cultures. (B) The percentage of cells beating at high density was higher compared to low density cultures, though stiff matrices above E* still induced loss of beating over time. Error bars are SD for >5 cells, 10 sec each in triplicate studies.
Figure S3: Assay for Glutathione shows no difference between Cardiomyocytes on Soft and Stiff Matrices (A) Representative chromatogram from Reverse Phase HPLC separation of hydrophilic glutathione (GSH; MW = 307 Da) derivatized with excess of hydrophobic mBBr (MW 271 Da). The two wavelengths were used to identify where the peptide absorbs (220 nm) and where mBBr alone has an absorption peak (396 nm). Free GSH (dotted line) elutes early (∼2.5 min), whereas free mBBr elutes at 18 min. mBBr-GSH shows two peaks that absorb significantly at both 220 and 396 nm. (B) Total levels of reacted GSH-mBBr product, determined by integrating the appropriate peaks, are similar for cardiomyocytes grown on soft or hard matrices for 24 h.
Figure S4: Labeled Cysteine Sites in Structures of Filamin-like Actin Binding Domain and Myosin II head. The sites were identified in Cys Shotgun labeling with mBBr of live cardiomyocytes on hard matrix. (A) The actin binding domain structure of human α-actinin-3 (1TJT), combined with sequence alignment to filamin A and B, identifies the location of C59 (in filamin A) at the end of helix-A near a conserved, actin-binding Asn. (B) The structure of the head of chicken smooth muscle myosin II (1BR2), combined with sequence alignment to human nonmuscle myosin IIB and cardiac myosin, identifies the location of the labeled and partially buried thiol (SH1) at one end of the canonical SH1-SH2 helix.
Acknowledgments
The authors thank Jushuo Wang and Aiping Du for assistance with cardiomyocyte isolation and cell culture. Funding is gratefully acknowledged from NIH (to D.W.S., J.W.S., J.M.S. and D.E.D.), the Ashton Foundation Pre-doctoral Fellowship fund (to A.J.E.) and an NIH-NHLBI Training Grant fellowship (to C.C.-K. and C.P.J.).
Footnotes
Present address: Department of Molecular Biology, Princeton University, Princeton, NJ 08544, USA
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/121/22/3794/DC1
References
- Barry CP, Xie J, Lemmon V, Young AP. Molecular characterization of a multi-promoter gene encoding a chicken filamin protein. J. Biol. Chem. 1993;268:25577–25586. [PubMed] [Google Scholar]
- Berry MF, Engler AJ, Woo YJ, Pirolli TJ, Bish LT, Jayasankar V, Morine KJ, Gardner TJ, Discher DE, Sweeney HL. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H2196–H2203. doi: 10.1152/ajpheart.01017.2005. [DOI] [PubMed] [Google Scholar]
- Bird SD, Doevendans PA, van Rooijen MA, Brutel de la Riviere A, Hassink RJ, Passier R, Mummery CL. The human adult cardiomyocyte phenotype. Cardiovasc. Res. 2003;58:423–434. doi: 10.1016/s0008-6363(03)00253-0. [DOI] [PubMed] [Google Scholar]
- Birks E, Hall J, Barton P, Grindle S, Latif N, Hardy J, Rider J, Banner N, Khaghani A, Miller L, et al. Gene profiling changes in cytoskeletal proteins during clinical recovery after left ventricular-assist device support. Circulation. 2005;112:157–164. doi: 10.1161/CIRCULATIONAHA.104.526137. [DOI] [PubMed] [Google Scholar]
- Breitbach M, Bostani T, Roell W, Xia Y, Dewald O, Nygren JM, Fries JW, Tiemann K, Bohlen H, Hescheler J, et al. Potential risks of bone marrow cell transplantation into infarcted hearts. Blood. 2007;110:1362–1369. doi: 10.1182/blood-2006-12-063412. [DOI] [PubMed] [Google Scholar]
- Collinsworth AM, Zhang S, Kraus WE, Truskey GA. Apparent elastic modulus and hysteresis of skeletal muscle cells throughout differentiation. Am. J. Physiol. Cell Physiol. 2002;283:C1219–C1227. doi: 10.1152/ajpcell.00502.2001. [DOI] [PubMed] [Google Scholar]
- Conrad AH, Clark WA, Conrad GW. Subcellular compartmentalization of myosin isoforms in embryonic chick heart ventricle myocytes during cytokinesis. Cell Motil. Cytoskeleton. 1991;19:189–206. doi: 10.1002/cm.970190307. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc. Natl. Acad. Sci. USA. 1997;94:9493–9498. doi: 10.1073/pnas.94.17.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri GA, Ayoob JC, Turnacioglu KK, Sanger JM, Sanger JW. Use of green fluorescent proteins linked to cytoskeletal proteins to analyze myofibrillogenesis in living cells. Meth. Enzymol. 1999;302:171–186. doi: 10.1016/s0076-6879(99)02017-0. [DOI] [PubMed] [Google Scholar]
- Dean RG, Balding LC, Candido R, Burns WC, Cao Z, Twigg SM, Burrell LM. Connective tissue growth factor and cardiac fibrosis after myocardial infarction. J. Histochem. Cytochem. 2005;53:1245–1256. doi: 10.1369/jhc.4A6560.2005. [DOI] [PubMed] [Google Scholar]
- Dembo M, Wang Y-L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999;76:2307–2316. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher DE, Janmey PA, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Domke J, Radmacher M. Measuring the elastic properties of thin polymer films with the atomic force microscope. Langmuir. 1998;14:3320–3325. [Google Scholar]
- Du A, Sanger JM, Linask KK, Sanger JW. Myofibrillogenesis in the first cardiomyocytes formed from isolated quail precardiac mesoderm. Dev. Biol. 2003;257:382–394. doi: 10.1016/s0012-1606(03)00104-0. [DOI] [PubMed] [Google Scholar]
- Du A, Sanger JM, Sanger JW. Cardiac myofibrillogenesis inside intact embryonic hearts. Dev. Biol. 2008;318:236–246. doi: 10.1016/j.ydbio.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, Rothen BM, Hammerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J. Cell Sci. 1999;112:1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Engler A, Bacakova L, Newman C, Hategan A, Griffin M, Discher D. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 2004a;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stiffness: pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004b;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Rehfeldt F, Sen S, Discher DE. Micro-tissue elasticity: measurements by atomic force microscopy and its influence on cell differentiation. In: Discher DE, Wang Y-L, editors. Methods in Cell Biology. Vol. 83. Academic Press; New York: 2007. pp. 521–545. [DOI] [PubMed] [Google Scholar]
- Feng Y, Chen M, Moskowitz I, Mendonza A, Vidali L, Nakamura F, Kwiatkowski D, Walsh C. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. USA. 2006;103:19836–19841. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Checa JC, Kaplowitz N. The use of monochlorobimane to determine hepatic GSH levels and synthesis. Anal. Biochem. 1990;190:212–219. doi: 10.1016/0003-2697(90)90183-a. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ. Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Goldsmith EC, Hoffman A, Morales MO, Potts JD, Price RL, McFadden A, Rice M, Borg TK. Organization of fibroblasts in the heart. Dev. Dyn. 2004;230:787–794. doi: 10.1002/dvdy.20095. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryos. J. Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hernandez-Nicaise ML, Bilbaut A, Malaval L, Nicaise G. Isolation of functional giant smooth muscle cells from an invertebrate: structural features of relaxed and contracted fibers. Proc. Natl. Acad. Sci. USA. 1982;79:1884–1888. doi: 10.1073/pnas.79.6.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BD, Massiera G, Van Citters KM, Crocker JC. The consensus mechanics of cultured mammalian cells. Proc. Natl. Acad. Sci. USA. 2006;103:10259–10264. doi: 10.1073/pnas.0510348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdusse A, Kalabokis VN, Himmel D, Szent-Gyorgyi AG, Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachinsky AM, Dominov JA, Miller JB. Intermediate filaments in cardiac myogenesis: nestin in the developing mouse heart. J. Histochem. Cytochem. 1995;43:843–847. doi: 10.1177/43.8.7542682. [DOI] [PubMed] [Google Scholar]
- Mahaffy RE, Shih CK, MacKintosh FC, Kas J. Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys. Rev. Lett. 2000;85:880–883. doi: 10.1103/PhysRevLett.85.880. [DOI] [PubMed] [Google Scholar]
- Mauritz C, Schwanke K, Reppel M, Neef S, Katsirntaki K, Maier LS, Nguemo F, Menke S, Haustein M, Hescheler J, et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118:507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- Mela-Riker LM, Bukoski RD. Regulation of mitochondrial activity in cardiac cells. Annu. Rev. Physiol. 1985;47:645–663. doi: 10.1146/annurev.ph.47.030185.003241. [DOI] [PubMed] [Google Scholar]
- Nitao L, Yeates T, Reisler E. Conformational dynamics of the SH1-SH2 helix in the transition states of myosin subfragment-1. Biophys. J. 2002;83:2733–2741. doi: 10.1016/S0006-3495(02)75283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak VP, Yin FC, Humphrey JD. Regional mechanical properties of passive myocardium. J. Biomech. 1994;27:403–412. doi: 10.1016/0021-9290(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Oliver T, Dembo M, Jacobson K. Traction forces in locomoting cells. Cell Motil. Cytoskeleton. 1995;31:225–240. doi: 10.1002/cm.970310306. [DOI] [PubMed] [Google Scholar]
- Pasternak C, Wong S, Elson EL. Mechanical function of dystrophin in muscle cells. J. Cell Biol. 1995;128:355–361. doi: 10.1083/jcb.128.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Pelham RJ, Wang Y-L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- Piazzesi G, Lucii L, Lombardi V. The size and the speed of the working stroke of muscle myosin and its dependence on the force. J. Physiol. 2002;545:145–151. doi: 10.1113/jphysiol.2002.028969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G, Holden HM. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil. Cytoskeleton. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- Rotsch C, Jacobson K, Radmacher M. Dimensional and mechanical dynamics of active and stable edges in motile fibroblasts investigated by using atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1999;96:921–926. doi: 10.1073/pnas.96.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Buguin A, Silberzan P, Ladoux B. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys. J. 2005;89:L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandwell DT. Biharmonic spline interpolation of GEOS-3 and SEASAT altimeter data. Geophys. Res. Lett. 1987;2:139–142. [Google Scholar]
- Sanger JM, Mittal B, Pochapin MB, Sanger JW. Myofibrillogenesis in living cells microinjected with fluorescently labeled alpha-actinin. J. Cell Biol. 1986;102:2053–2066. doi: 10.1083/jcb.102.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J. Muscle Res. Cell. Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira-Schweitzer K, Seliktar D. Matrix stiffness affects spontaneous contraction of cardiomyocytes cultured within a PEGylated fibrinogen biomaterial. Acta Biomater. 2007;3:33–41. doi: 10.1016/j.actbio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J. 2002;21:1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys. J. 2006;90:1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda T, Philp N, Zile MH, Linask KK. Left-right asymmetric localization of flectin in the extracellular matrix during heart looping. Dev. Biol. 1996;173:39–50. doi: 10.1006/dbio.1996.0005. [DOI] [PubMed] [Google Scholar]
- Unger S, Mainberger A, Spitz C, Bähr A, Zeschnigk C, Zabel B, Superti-Furga A, Morris-Rosendahl D. Filamin A mutation is one cause of FG syndrome. Am. J. Med. Genet. A. 2007;143:1876–1879. doi: 10.1002/ajmg.a.31751. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Reiter RS, Lin JL, Wang Q, Williams HS, Krob SL, Schultheiss TM, Evans S, Lin JJ. Requirement of a novel gene, Xin, in cardiac morphogenesis. Development. 1999;126:1281–1294. doi: 10.1242/dev.126.6.1281. [DOI] [PubMed] [Google Scholar]
- Wang N, Tolic-Norrelykke IM, Chen J, Mijailovich SM, Butler JP, Fredberg JJ, Stamenovic D. Cell prestress. I. Stiffness and prestress are closely associated in adherent contractile cells. Am. J. Physiol. Cell Physiol. 2002;282:C606–C616. doi: 10.1152/ajpcell.00269.2001. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Shiojima I, Komuro I, Nagai R, Yazaki Y. Interaction of cardiac myocytes and non-myocytes in mechanical stress-induced hypertrophy. Herz. 1995;20:109–117. [PubMed] [Google Scholar]
- Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1: Isolated Cardiomyocyte Beating on Soft Matrix
Depicted here is the principal strain field (left) created by the cell (right) as it deforms the soft gel with a basal level of traction forces. As the cardiomyocyte contracts the gel, however, strain rapidly increases as a result of increased traction forces, which also decay rapidly again. The magnitude of the strain field is determined from bead displacements and uses the same strain scale as in Figure 1.
Supplemental Movie 2: Isolated Cardiomyocyte Beating on Stiff Matrix
Shown here is the principal strain field (left) created by the cell (right) as it attempts to deform the stiff gel. Traction forces are small, but non-zero. As the cardiomyocyte contracts, the resulting traction forces do not greatly change, although the cell significantly contracts. Strain magnitude uses the same scale as in Figure 1.
Table S1: Summary of In Situ Cysteine Labeling in the Non-muscle Myosin IIB Head and Lever Arm for Cardiomyocytes on Stiff Matrices
Figure S1: AFM Measurement of Myocardial Elasticity
(A) Brightfield image of freshly isolated day 4 myocardium. Sample is then mounted on a AFM stage as depicted schematically with the apical surface exposed. An AFM tip with r ∼ 50 nm and experimentally determined spring constant, ksp, is then indented into the myocardium to yield tip deflection, d, versus tip position, z (B). Elasticity was determined by fitting a hertz cone model from the contact to the maximal indentation points, with the former determined by a range of analysis method from z1, d1 to z2, d2.
Figure S2: Cardiomyocytes at High Density also show Beating depends on Matrix Elasticity Purified cardiomyocytes were plated at high density on matrices of varied stiffness, and beating was observed in phase contrast. (A) Beat frequency is elasticity-dependent, but decreases in beat frequency are smaller compared to low density cultures. (B) The percentage of cells beating at high density was higher compared to low density cultures, though stiff matrices above E* still induced loss of beating over time. Error bars are SD for >5 cells, 10 sec each in triplicate studies.
Figure S3: Assay for Glutathione shows no difference between Cardiomyocytes on Soft and Stiff Matrices (A) Representative chromatogram from Reverse Phase HPLC separation of hydrophilic glutathione (GSH; MW = 307 Da) derivatized with excess of hydrophobic mBBr (MW 271 Da). The two wavelengths were used to identify where the peptide absorbs (220 nm) and where mBBr alone has an absorption peak (396 nm). Free GSH (dotted line) elutes early (∼2.5 min), whereas free mBBr elutes at 18 min. mBBr-GSH shows two peaks that absorb significantly at both 220 and 396 nm. (B) Total levels of reacted GSH-mBBr product, determined by integrating the appropriate peaks, are similar for cardiomyocytes grown on soft or hard matrices for 24 h.
Figure S4: Labeled Cysteine Sites in Structures of Filamin-like Actin Binding Domain and Myosin II head. The sites were identified in Cys Shotgun labeling with mBBr of live cardiomyocytes on hard matrix. (A) The actin binding domain structure of human α-actinin-3 (1TJT), combined with sequence alignment to filamin A and B, identifies the location of C59 (in filamin A) at the end of helix-A near a conserved, actin-binding Asn. (B) The structure of the head of chicken smooth muscle myosin II (1BR2), combined with sequence alignment to human nonmuscle myosin IIB and cardiac myosin, identifies the location of the labeled and partially buried thiol (SH1) at one end of the canonical SH1-SH2 helix.