Abstract
Forkhead factors are important regulators of animal development and homeostasis. They are among the earliest to bind quiescent genes, which they activate in conjunction with other transcription factors. Many liver-specific genes are under the control of FoxA2, a liver-enriched forkhead protein. Here we confirmed by chromatin immunoprecipitation that FoxA2 is one of the factors bound to the promoter-proximal enhancer of the gene encoding apolipoprotein AI (a component of high density lipoprotein) and that it functions in synergy with the nuclear receptor hepatocyte nuclear factor-4α. Furthermore, toward identifying additional cofactors that could potentially regulate FoxA2 activity, we identified DNA-de pend ent protein kinase (DNA-PK) as a FoxA2-associated factor upon affinity purification of epitope-tagged FoxA2. We show that FoxA2, found to be a phosphoprotein in vivo, is also an efficient substrate for DNA-PK, which targets serine 283. This residue is contained within a conserved serine-glutamine phos pho ryl a tion signal for DNA-PK, located within the C-terminal third of the polypeptide, just distal to its winged-helix DNA binding domain. We establish that this residue is critical for FoxA2 function because FoxA2 bearing a mutation at this site is severely compromised in its ability to activate a reporter gene under the control of its cognate DNA-binding site (apoAI site B). Complementary experiments rule out that this mutation compromises the ability of FoxA2 to either translocate to the nucleus or to bind site B. We therefore conclude that DNA-PK-de pend ent phos pho ryl a tion of FoxA2 plays a critical role in its transcriptional activation function per se.
Transcriptional control of mRNA genes is achieved through multiprotein complexes that are targeted to promoter and enhancer regions of these genes by regulatory factors that recognize specific DNA sequences. Typically, several such factors, or transcriptional activators, each respond to a particular signal but act together in a synergistic fashion to control the expression of the target gene. Thus, the unique temporal and spatial expression program of a given gene is hard-wired into the regulatory sequences contained within its enhancer and promoter regions (1, 2).
Transcriptional activators recruit different classes of coactivators, which function broadly on at least two levels (3, 4). Some coactivators function at the earliest stage of gene activation by acting on the chromatin template to make it more amenable to the action of factors that enter later in the process. Many of these coactivators possess enzymatic activities whose effects range from ATP-dependent nucleosome remodeling to covalent modifications of histones that include acetylation, methylation, and ubiquitylation (2, 5). The role of the other class of coactivators, exemplified by the Mediator complex (6), is to nucleate the preinitiation complex, the RNA polymerase II-containing multiprotein entity that is ultimately responsible for the actual RNA synthesis (7).
Apolipoprotein AI (apoAI)3 is the major protein component of high density lipoprotein, blood plasma levels of which are inversely correlated with incidence of atherosclerosis. The gene encoding this protein is expressed predominantly in liver and intestine (8, 9). Its expression is primarily driven by the orphan nuclear receptor HNF-4α, which, like its target genes is also expressed in liver, intestine, and related endoderm-derived tissues (10). However, HNF-4α functions maximally in the context of other activators that also bind to the apoAI gene enhancer. Among these, FoxA2 (previously known as HNF-3β) appears to play a major role (11, 12). Interestingly, consistent with the determinative role of HNF-4α and FoxA2 in shaping the liver- and intestine-specific gene expression program, numerous genes expressed in these tissues display this joint dependence on the two activators (13). Thus, elucidating the underlying mechanisms by which each of these factors works is critical toward an understanding of tissue-specific gene expression. However, although the mechanisms by which nuclear receptors, including HNF-4α, have been extensively studied, and a fairly detailed picture of how they nucleate coactivator cascades has begun to emerge (14, 15), the corresponding mechanisms for FoxA2 remain relatively unclear.
FoxA2 and the closely related FoxA1 are members of the large family of “forkhead” factors that have been shown to regulate a wide range of genes (16–18). The forkhead factors are characterized by the presence of a “winged-helix” DNA binding domain that shows a high degree of homology with the linker histone H5 (19, 20). Consistent with this link to compacted chromatin, FoxA1 has been shown to occupy a linker histone site on the nucleosomes surrounding the transcriptional regulatory region of a target gene (21). Furthermore, binding of FoxA1 to compacted chromatin can in fact result in a somewhat more “open” nucleosome configuration of the targeted region (22). Most recently, it has also been realized that proximity of FoxA1-binding sites is a major determinant in regulating which of its numerous target sites a given nuclear receptor (estrogen receptor) would interact (23).
Taken together, and consistent with the role of forkhead proteins as “pioneer” factors in development pathways, these observations lead to a hypothetical framework for describing FoxA factor function in which a key role of the FoxA factors is to penetrate compacted chromatin, which is a hallmark of quiescent genes. Entry of other factors (which in turn recruit various coactivators) is thus predicted to be facilitated following this breach in the chromatin barrier.
Although a role for additional coactivator-like factors that work with FoxA2 in the initial decompaction of chromatin in the above pathway can be envisioned, none have been described to date. As part of our ongoing effort to understand how HNF-4α activates the apoAI gene in conjunction with other factors, our strategy has been to focus on cataloguing and biochemically characterizing the relevant coactivators. Given the close association with FoxA2, and a long term view toward understanding the basis of this synergy, we have focused on identifying factors that associate with FoxA2 and potentially contribute to its transcriptional activity. Here we identify DNA-dependent protein kinase (DNA-PK) as a FoxA2-associating protein and demonstrate that it utilizes FoxA2 as a substrate. Furthermore, we show that phosphorylation of FoxA2 by DNA-PK modulates its transcriptional activation potential. Our results thus implicate DNA-PK as a FoxA2 cofactor.
EXPERIMENTAL PROCEDURES
Epitope Tagging and Protein Purification from Mammalian Cells
Rat FoxA2 cDNA was subcloned into VP5 (24), a pIRESneo-1-derived (Clontech) vector, which allows expression as a FLAG-fused protein (f:FoxA2). Following transfection into HeLaS cells, G418-resistant clones were selected and amplified. Nuclear extract from a f:FoxA2-expressing clone was subjected to affinity purification over M2-agarose, as described previously (24). For identification of FoxA2-associated polypeptides, eluates were resolved by SDS-PAGE, and the gel was stained with colloidal Coomassie, and bands of interest were excised and subjected to mass spectrometric analysis.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed essentially as described previously (15, 25) except that quantitative (real time) PCR was used for the final readout. Briefly, upon reaching confluence in 150-mm plates, cells (HeLa, 293T, and HepG2), which were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, were fixed with formaldehyde. Following cell lysis, and chromatin shearing by sonication, the extracts were subjected to immunoprecipitation. The cross-links in the precipitated material were reversed by heating, and after deproteinization, samples were analyzed by real time PCR amplification using SYBR green probes (Applied Biosystems) and primer sets that either spanned the apoAI promoter region from −336 to −149 relative to the transcription start site (26) or a more distal region from +2645 to +2708. Raw data from each immunoprecipitate (done in triplicate) were first expressed as a fraction of the input and then normalized to the signal from the corresponding control IgG to obtain relative enrichment.
ChIP assays were also performed on extracts of 293T cells that had been cross-linked with formaldehyde 48 h after transfection with the B3XAI.LUC reporter and vectors expressing FoxA2 derivatives (see below). Immunoprecipitate cross-links were reversed, and the promoter region on the reporter template was amplified by standard PCR as described previously (15).
Transient Transfection Reporter Assay
Cells were transfected with the indicated plasmids using FuGENE 6 (Roche Applied Science). Luciferase activity was measured after 24 h using the Dual-Luciferase Reporter® assay kit from Promega. A Renilla-expressing plasmid was used to normalize for transfection efficiency.
The reporter plasmid B3XAI.LUC was constructed by subcloning three copies of the apoAI site B sequence (synthetic oligonucleotides) upstream of the AI.LUC plasmid that contains core promoter elements (TATA and initiator) from the apoAI gene upstream of the luciferase sequence in a pGL3basic backbone (Promega).
Electrophoretic Mobility Shift Assay
Reaction and electrophoresis conditions were essentially as described before (15) except that an end-labeled probe containing site B (corresponding to positions -148 to -178) from the promoter-proximal enhancer of the apoAI gene was used. All components were typically mixed on ice at initial times and incubated for 30 min at 30 °C prior to electrophoresis.
Expression and Purification of Recombinant Proteins
Recombinant FoxA2 and derivatives were expressed as histidine-tagged proteins (subcloned into pET11d-6His) in Escherichia coli BL21 pLysS (24). Following induction, lysis, and chromatography over nickel-nitrilotriacetic acid-agarose, the full-length proteins (FoxA2 WT and FoxA2(T280A,S283A) (see below)) were additionally purified over SP-Sepharose. Truncated FoxA2 derivatives FoxA2-N, FoxA2-WH, FoxA2-C, corresponding to amino acid residues 1–128, 129–275, and 276–458, respectively, were also expressed in bacteria following subcloning of their corresponding cDNAs into the pET11d-6His vector. The deletion mutant FoxA2-CΔ (in which residues 276–286 were deleted) and the point mutants (FoxA2-C S283A, FoxA2-C T280A, and FoxA2-C(T280A,S283A), in which the indicated residues were converted to alanine) were derived from the FoxA2-C domain using standard PCR-based techniques. The full-length FoxA2 mutant (FoxA2(T280A,S283A)) was also similarly constructed using a mutagenic primer set that introduced the appropriate changes.
For GST-pulldown assays (below), full-length FoxA2 and derivatives were subcloned into pGEX vectors and expressed in E. coli.
Protein-Protein Interactions
Standard GST pulldown assays were performed with 10 μg of the various FoxA2 derivatives that were immobilized onto glutathione-Sepharose beads. The beads were incubated for 4 h at 25 °C with HeLa nuclear extract (Dignam et al. (27); 5 mg of total protein) or a derived chromatographic fraction, in buffer (BC100) containing 20 mm Tris-HCl, pH 7.4, 100 mm KCl, 0.1 mm EDTA, 20% glycerol supplemented with 0.1% Nonidet P-40. The beads were washed three times in the same buffer, once with the same buffer containing 0.5 μg/ml ethidium bromide, plus three additional washes in the original buffer. Bound proteins were eluted in Laemmli sample buffer and following SDS-PAGE were analyzed by Western blotting with specified antibodies.
HeLa cell nuclear extract was chromatographed over phosphocellulose P11 (Whatman), as described (24). DNA-PK was immunologically detected in high salt-eluting fractions. The 0.5 m step eluate was spectrophotometrically analyzed for nucleic acid content (A260/A280 <1.0) prior to use in the GST-pulldown assay.
For the coimmunoprecipitation assays, whole cell extracts from HepG2 and 293T were prepared in lysis buffer containing 0.5 m NaCl and 1% Nonidet P-40 as described previously (24). Extracts were diluted to 0.15 m salt and 0.1% Nonidet P-40 and incubated overnight (4 °C) with 5 μg of either anti-FoxA2 (Santa Cruz Biotechnology, sc-9187) or a control goat IgG antibody. Protein-G-Sepharose beads were added and incubated for 4 h at 4 °C. Beads were washed extensively in BC150 buffer (plus 0.1% Nonidet P-40), and proteins were eluted in Laemmli sample buffer. Samples were analyzed by SDS-PAGE, followed by Western blotting.
Mass Spectrometric Identification of Phosphorylated Residues
The VP5-FoxA2 plasmid was transfected into 293T cells using FuGENE. After 48 h, whole cell extracts were made as above, and f:FoxA2 was purified by chromatography on M2-agarose. The samples were then processed for nano-liquid chromatography/tandem mass spectrometry as described (28). Briefly, the sample was reduced and digested with Lys-C followed by trypsin or, separately, with Glu-C. Nano-liquid chromatography/tandem mass spectrometry analyses were performed on an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific) using a slightly modified version of a previously described setup (29). Peptides and proteins were identified by means of automated data base searching using Mascot version 2.2 (Matrix Science, London, UK) against UniProt/SwissProt release 57.0 with a precursor mass tolerance of 3 ppm, a fragment ion mass tolerance of 0.8 Da, and strict trypsin specificity or combined trypsin/Glu-C specificity allowing for up to two missed cleavages. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation and phosphorylation of serine, threonine, and tyrosine were allowed as variable modifications. Peptides were considered identified if the Mascot score was over the 99% confidence limit based on the “identity” score of each peptide and at least three successive y or b ions with two and more y, b, and/or precursor origin neutral loss ions were observed. Phosphorylated sites were unambiguously determined when the post-translational modification site scores were more than 0.75 (30).
In Vitro Kinase Assays
Typical 100-μl kinase reactions contained 300 ng of the substrate polypeptide, 12.5 units of DNA-PKcs (catalytic subunit; Promega), and 2.5 μCi of [γ-32P]ATP in reaction buffer consisting of 20 mm HEPES-KOH, pH 8.2, 4 mm MgCl2, 60 mm KCl, 0.12 mm EDTA, and 12% glycerol. Reactions were assembled on ice and then incubated for 1 h at 30 °C. Samples were precipitated by trichloroacetic acid and resolved by SDS-PAGE. Gels were fixed, stained with Coomassie Blue, and autoradiographed.
Immunofluorescence
293T cells were transiently transfected with either an empty vector control plasmid or plasmids containing the cDNAs of wild type and the phosphorylation mutant of FoxA2 (FoxA2(T280A,S283A)) using Trans-IT transfection reagent (Mirus) according to the manufacturer's protocols. Twenty four hours post-transfection, cells were plated on 4-well chamber slides (Nunc) at a density of 8 × 104 cells per well. On the following day, cells were fixed in 4% paraformaldehyde. Standard protocols for indirect immunofluorescence were performed with a goat anti-FoxA2 antibody (Santa Cruz Biotechnology) and an anti-goat Alexa 488 secondary antibody (Invitrogen). Coverslips were mounted with Vectashield containing 4′,6-diamidino-2-phenylindole (Vector Laboratories). Images were collected using a ×40 objective on an Olympus BX51 epifluorescent microscope and DP-BSW Olympus image acquisition software.
RESULTS
Synergistic Function of FoxA2 and HNF-4α in apoAI Gene Transcription
Prominent binding elements within the promoter-proximal enhancer that in large part controls the liver-specific expression of the apoAI gene include site A and site B, which have been shown to bind HNF-4α and FoxA2, respectively (as well as other factors (8, 9)). Furthermore, a minimal enhancer construct containing just these two sites in tandem was demonstrated to recapitulate at least partial enhancer function (12). Before undertaking our biochemical analyses, we began by authenticating a role for HNF-4α and FoxA2 in synergistically activating the apoAI gene. We first assessed by chromatin immunoprecipitation (ChIP) experiments using antibodies against HNF-4α and FoxA2 and whether these factors are localized to the apoAI enhancer in cell types that express apoAI. As determined by reverse transcription-PCR (Fig. 1A), the apoAI mRNA is expressed in hepatocarcinoma-derived HepG2 cells (lane 3) but not in 293T (lane 1) or HeLa cells (lane 2). Consistent with this expression pattern, our ChIP analysis (Fig. 1B) clearly showed that, as compared with a distal location, HNF-4α and FoxA2 preferentially occupy the apoAI promoter-proximal region in HepG2 cells but not in 293T or HeLa cells. These data thus independently confirm and extend the conclusions of a recent genome-wide study (13).
FIGURE 1.
Involvement of HNF-4α and FoxA2 in apoAI gene transcription. A, apoAI RNA expression in 293T (lane 1), HeLa (lane 2), and HepG2 (lane 3) was analyzed by reverse transcription-PCR. β-Actin RNA is shown for normalization (lanes 4–6). B, ChIP analyses to assess occupancy of the apoAI promoter-proximal enhancer by HNF-4α and FoxA2. Cells (293T, HeLa, or HepG2) were processed for a standard ChIP assay, and immunoprecipitates from the indicated antibodies were subjected to real time PCR using amplicon primers that specifically amplify either the apoAI promoter or a distal (3′) region as indicated. For each set, the data are plotted as fold-enrichment of the specific signal relative to the corresponding IgG control. C, synergistic activation of a luciferase reporter by HNF-4α and FoxA2. For the transient transfection assay using a luciferase reporter containing cognate sites for HNF-4α and FoxA2 in close juxtaposition, 293T cells were transfected with 100 ng of a FoxA2-expressing plasmid or 50 ng of an HNF-4α-expressing plasmid or a combination of the two, as indicated.
We also confirmed that following transient transfection into 293T cells (Fig. 1C), FoxA2 and HNF-4α could synergistically activate a luciferase reporter construct containing cognate sites for these activators. Thus, whereas FoxA2 and HNF-4α independently activate this reporter 2.7- and 3.9-fold, respectively, together they activate it more than 13.4-fold. Similar results were also obtained when these constructs were transfected into HeLa cells (data not shown). These data are in accord with the previously demonstrated synergism between FoxA2 and HNF-4α (12). Importantly, our data show further that despite the fact that under their natural circumstances they activate target genes in a restricted repertoire of cell types (which in all likelihood is a direct outcome of where they are actually expressed), these factors retain the intrinsic ability to function as transcriptional activators in heterotypic cells as well.
Generation of a Stable Cell Line Expressing Epitope-tagged FoxA2 and Isolation of Polypeptides Interacting with FoxA2
To further dissect FoxA2 mechanisms, we focused on the identification of proteins that interact with it and potentially function as coactivators. For this purpose, we generated a HeLa cell line that stably expresses a FLAG epitope-tagged version of FoxA2 (f:FoxA2). The derived nuclear extracts from these cells were subjected to affinity chromatography on M2-agarose, and bound proteins were analyzed by SDS-PAGE and silver staining (Fig. 2A). Comparison of the eluted proteins against a comparable isolate from a control HeLa nuclear extract revealed the presence of several polypeptides that were preferentially associated with f:FoxA2 (Fig. 2A, lane 2 versus lane 1). Among these, a polypeptide exhibiting an apparent molecular mass exceeding 350 kDa was the most prominent.
FIGURE 2.
Copurification and specific association of FoxA2 and DNA-PK. A, SDS-PAGE analysis (silver-stained gel is shown) of a FLAG peptide-eluted preparation obtained after M2-agarose affinity purification of nuclear extract derived from a cell line that stably expresses f:FoxA2 (lane 2). A corresponding eluate from a control cell extract that does not express f:FoxA2 was also run on the gel (lane 1). Molecular mass markers (in kDa) are shown just left of the column. The f:FoxA2 polypeptide as well as the apparently specifically associated polypeptides with molecular masses estimated to be ∼100 kDa and more than 350 kDa are indicated. B, SDS-PAGE analysis (visualized by silver staining) of the preparation shown in A, lane 2, that was additionally resolved on a Superose 6 gel filtration column. Every other fraction in the main protein peak was loaded on the gel. Bands identified as f:FoxA2 and p > 350 are marked (see also D). C, specificity of association between FoxA2 and DNA-PK. The eluates shown in A (control, lane 3; f:FoxA2, lane 4) were analyzed by Western blot using antibodies against DNA-PKcs (upper panel) and FoxA2 (lower panel). Standard unfractionated nuclear extracts from HepG2 (lane 1) and HeLa (lane 2) were also included. Note that HepG2 cells express FoxA2, but at the sensitivity of the Western shown, the corresponding band is not visible in lane 1. D, Superose 6 fractions shown in B were analyzed by Western blot using antibodies against DNA-PKcs (upper panel) and FoxA2 (lower panel).
To further ascertain that the association of the >350-kDa polypeptide is specific (and not, for example, a result of a nonspecific association with the resin), we subjected the eluate to gel filtration chromatography on Superose 6 (Fig. 2B). Upon fractionation, many polypeptides in the eluate segregated away from f:FoxA2 (e.g. compare fraction 12 containing the f:FoxA2 peak (see also Fig. 2D) with fraction 14 containing a 66-kDa polypeptide) indicating that they are not bona fide FoxA2-associated polypeptides. By contrast, the elution profile of the >350-kDa polypeptide was coincident with that of f:Fox2A (both peaking at fraction 12), consistent with their being bona fide interacting proteins.
Identification of p350 as DNA-PKcs
The >350-kDa polypeptide (Fig. 2A) was subjected to mass spectrometry and identified as the catalytic subunit (cs) of DNA-dependent protein kinase (DNA-PK). An additional polypeptide (p100), which also exhibited some degree of specificity in its association with FoxA2, was identified as kinase anchor protein 8 (also known as AKAP95). Although a few additional bands appeared to correspond to specific interacting polypeptides, we were unable to unambiguously identify them because of their very low stoichiometry in the preparation. Only the interaction of DNA-PK with FoxA2 has been examined further in this study.
Specificity of Association between DNA-PK and FoxA2
Having identified the >350 kDa as DNA-PKcs, we used Western blotting to further authenticate the specificity of its interaction with FoxA2 both in the one-step f:FoxA2 affinity-selected preparation and in the fractions collected following gel filtration chromatography. In the former situation, we confirmed that DNA-PKcs is present only in eluates derived from the f:FoxA2-expressing extract and not from a control extract (Fig. 2C, lane 4 versus lane 3). In the latter case, mirroring the original silver staining (Fig. 2A), FoxA2 and DNA-PKcs elution profiles were found to coincide perfectly (Fig. 2D).
Mapping of FoxA2 Domain(s) That Interact with DNA-PK
To map the FoxA2 domain responsible for mediating interaction with DNA-PK, we first established that the intracellular association of f:FoxA2 and DNA-PK, which became evident from the combined epitope tagging and affinity purification approach described above, also takes place in a cell-free context. For this purpose, HeLa cell nuclear extract was incubated with beads containing a GST-FoxA2 fusion protein, and following extensive washing, the bound material was probed for various polypeptides (Fig. 3A). As expected, DNA-PKcs was specifically retained on GST-FoxA2 beads but not on control GST beads (Fig. 3A, lane 3 versus lane 2). By contrast, an unrelated polypeptide (MED13) showed no detectable binding either to GST-FoxA2 or to control GST. Given that DNA-PK exists as a trimeric complex that contains the Ku70 and Ku80 polypeptides, we also probed for the Ku70 subunit and found that it is specifically retained on the GST-FoxA2 beads (Fig. 3A, lane 3 versus lane 2), indicating that even though its association with FoxA2 was not evident from gels that were processed for mass spectroscopy, it is the trimeric form of DNA-PK that interacts with FoxA2. We also utilized a chromatographic fraction (see under “Experimental Procedures”) derived from HeLa nuclear extract that was devoid of nucleic acids as a source of DNA-PK in similar pulldown assays. The results again showed efficient specific retention of DNA-PK by GST-FoxA2 (Fig. 3B).
FIGURE 3.
Physical interactions between FoxA2 and DNA-PK. A, for the GST pulldown assay, GST-FoxA2 (lane 3; fl, full-length) or control GST (lane 2) was immobilized on glutathione-Sepharose beads, incubated with HeLa nuclear extract, washed, and analyzed for retention of the indicated polypeptides by Western blotting. Input HeLa extract (2%) was also loaded on the gel. B, GST pulldown assay was done as in A except that phosphocellulose P11 0.5 m fraction was used as the input (lane 1). Eluates from control GST (lane 2) or GST-FoxA2(fl) (lane 3) were probed with anti-DNA-PKcs antibodies. C, Coomassie Blue staining of the GST fusion proteins used in the pulldown assay shown in D (control GST, lane 1; FoxA2-N, lane 2; FoxA2-WH, lane 3; and FoxA2-C, lane 4). The proteins were expressed in E. coli, purified over glutathione-Sepharose beads, and resolved by SDS-PAGE prior to staining. Note that because the FoxA2-C derivative tended to degrade, normalization was based on the full-length products (arrow) rather than total protein amounts. For exact coordinates of the constructs, see also the schematic of Fig. 5B. D, GST pulldown assay was performed as described for A, except that truncated FoxA2 derivatives were bound to the beads, as indicated. GST alone (lane 2) served as a control, and 2% of the HeLa extract input was also loaded on the gel.
Apart from its centrally located winged-helix DNA binding domain, FoxA2 also contains multipartite N- and C-terminal activation domains (18). Therefore, to map the DNA-PK interaction domain of FoxA2, we generated appropriate GST derivatives and tested them for their ability to interact with FoxA2 (Fig. 3C). The results (Fig. 3D) clearly show that the DNA-PK interaction is mediated almost exclusively by the winged-helix DNA binding domain (lane 4 versus lanes 2, 3, and 5).
Intracellular Association of FoxA2 and DNA-PK
To confirm that the interactions between FoxA2 and DNA-PK that we deduced through use of epitope-tagged derivatives of FoxA2 faithfully reflect the intracellular interactions of the endogenous proteins, we also carried out a coimmunoprecipitation assay (Fig. 4A). Whole cell extracts from HepG2 cells, which constitutively express both FoxA2 and DNA-PK (Fig. 4A, lane 2), and from 293T cells, which express DNA-PK but not FoxA2 (Fig. 4A, lane 1), were incubated with antibodies against either FoxA2 or with corresponding control antibodies. The resulting immunoprecipitates were probed for both DNA-PKcs and FoxA2. Consistent with the absence of FoxA2 in 293T cells, no specific FoxA2 signal was seen in immunoprecipitates from these cells (Fig. 4A, lane 4); furthermore, no signal for DNA-PKcs was detectable. By contrast, immunoprecipitates from HepG2 extracts were, as expected, greatly enriched in FoxA2 (Fig. 4A, lane 6 versus lanes 5 and 4). Most importantly, significant amounts of DNA-PKcs were specifically brought down by the FoxA2 antibody from these extracts (lane 6 versus lanes 5 and 4). These data establish that native (untagged) FoxA2 and DNA-PK indeed physically interact with each other in liver-derived cells.
FIGURE 4.
Intracellular interactions between FoxA2 and DNA-PK. A, coimmunoprecipitation assays utilized whole cell extracts (WCE) from 293T (lane 1) and HepG2 (lane 2) cells. The extracts were incubated with control antibodies (goat IgG, lanes 3 and 5) or with anti-FoxA2 (lanes 4 and 6) antibodies. After washing, the precipitates were analyzed by Western blotting with the indicated antibody (top panel, anti-DNA-PKcs; bottom panel, anti-FoxA2). Nonspecific bands in the immunoprecipitates (IP), which likely result from cross-reactivity to immunoglobin heavy chains, are marked with asterisks. Note that whereas immunoblots shown for lanes 3–6 are from the same experiment, those showing the inputs used for this experiment (lanes 1 and 2) are from a separate run. B, ChIP analysis to assess occupancy of the apoAI promoter-proximal enhancer by DNA-PK. HepG2 cells were processed for a standard ChIP assay and immunoprecipitated with either control IgG or an anti-Ku70 antibody. The processed samples were subjected to real time PCR using amplicon primers that specifically amplify either the apoAI promoter or a distal region as in Fig. 1B. Data are plotted as enrichment of the specific signal relative to the IgG control for each amplicon.
We also asked whether DNA-PK is localized to the promoter region as would be expected for a classical coactivator. We therefore performed a ChIP assay (Fig. 4B), in which we exposed formaldehyde cross-linked extracts from HepG2 cells to an antibody against the Ku70 subunit of DNA-PK. The assay yielded a modest, but clear, signal for Ku70 at both the promoter and distal regions. Hence, rather than a coactivator recruitment scenario, these data are more suggestive of mechanisms in which FoxA2 encounters DNA-PK, which is perhaps constitutively distributed across the chromatin, leading to functional consequences (see under “Discussion”).
Phosphorylation of FoxA2 by DNA-PK
Association of FoxA2 with a protein kinase prompted us to investigate whether FoxA2 can be phosphorylated at serine residues, in vivo. For this purpose, we transiently overexpressed FoxA2 in 293T cells. Following affinity purification, FoxA2 was processed for mass spectrometric analysis aimed at identifying distinct phosphorylated residues (see under “Experimental Procedures”). The data clearly revealed that FoxA2 exists as a phosphoprotein with residues Ser-212, Ser-303, Ser-306, Ser-307, Ser-309, Ser-437, and Ser-458 being the most prominent sites of modification (Table 1).
TABLE 1.
Serine phosphorylation of FoxA2
f:FoxA2 was transiently expressed in 293T cells. Following affinity purification from whole cell extracts, the sample was analyzed for serine phosphorylation by nano-liquid chromatography/tandem mass spectrometry. Digested peptides bearing phosphorylated residues, their coordinates on the rat sequence, and the identified modified residues are shown.
| Coordinates | Peptide sequence | Phosphorylated sites |
|---|---|---|
| 209–219 | HSLSFNDCFLK | Ser-212 |
| 290–313 | AAGSASETPAGTESPHSSASPCQE | Ser-309a |
| 290–313 | AAGSASETPAGTESPHSSASPCQE | Ser-307,a Ser-309a |
| 290–313 | AAGSASETPAGTESPHSSASPCQE | Ser-303, Ser-306 |
| 290–313 | AAGSASETPAGTESPHSSASPCQE | Ser-303,a Ser-306,a Ser-307a |
| 432–458 | AGLDASPLAADTSYYQGVYSRPIMNSS | Ser-458a |
| 432–458 | AGLDASPLAADTSYYQGVYSRPIMNSS | Ser-437 |
a Ambiguous sites were determined by probabilities from the post-translational modification scores (30).
Because DNA-PK has been shown to utilize several transcription factors as a substrate for its kinase activity, we next asked if FoxA2 is also a substrate. To this end, we incubated bacterially expressed FoxA2 with the DNA-PKcs in the presence of [γ-32P]ATP and found that FoxA2 indeed is an efficient substrate (Fig. 5A, lane 1). In this assay, we also tested p53, a previously described DNA-PK substrate (31), which thus served as a positive control, and we found it to be phosphorylated albeit at a lower efficiency relative to FoxA2 (data not shown). Other controls ensured that the observed phosphorylation in fact results from the action of DNA-PKcs on the bacterially expressed FoxA2 polypeptide because DNA-PKcs alone or the FoxA2 preparation alone did not yield the corresponding labeled band (data not shown).
FIGURE 5.
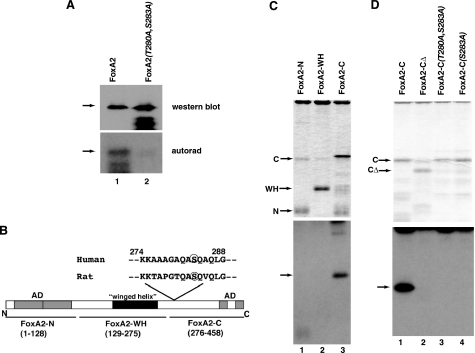
Phosphorylation of FoxA2 by DNA-PK. A, full-length bacterially expressed FoxA2 (lane 1, FoxA2 WT) and the corresponding mutant (see text for details) in the mapped DNA-PK phosphorylation site (lane 2, FoxA2(T280A,S283A)) were incubated with the catalytic subunit of DNA-PK in the presence of [γ-32P]ATP. The reaction products were visualized by autoradiography (autorad, bottom panel). Top panel shows a Western blot (WB) of the two purified proteins. Arrows indicate full-length FoxA2 in the preparations, which tend to degrade. B, schematic diagram (adapted from Ref. 18) showing the domain organization of FoxA2, including the central winged helix DNA binding domain and the N- and C-terminally located activation domains (AD). The amino acid coordinates of various derivatives used in this study are also indicated. Amino acid sequences (human and rat) of corresponding linker regions are just adjacent to the winged helix domains that appear to be targeted by DNA-PK (C and D). The serine residue identified as the DNA-PK target site within FoxA2 lies within a conserved DNA-PK signal (TQ/SQ) and is circled. Note that in our FoxA2(T280A,S283A) construct, the T280A change (in addition to the S283A) also eliminates any disparities that may be attributable to differences between the rat and human sequences. C, identification of the FoxA2 domain that is phosphorylated by DNA-PK phosphorylation. FoxA2 derivatives (FoxA2-N, lane 1; FoxA2-WH, lane 2; FoxA2-C, lane 3) were subjected to DNA-PK- mediated phosphorylation as in A. Reaction products were resolved by SDS-PAGE and visualized by both Coomassie Blue staining (to ensure that equivalent amounts of the polypeptides were loaded; upper panel) and by autoradiography (lower panel). Bands corresponding to the test proteins are identified (N, FoxA2-N; WH, FoxA2-WH; C, FoxA2-C) for the upper panel. Arrows mark the phosphorylated FoxA2-C derivative for the lower panel. D, mapping of the serine residue in the FoxA2-C domain that is phosphorylated by DNA-PK. The indicated derivatives of the FoxA2-C domain were analyzed for their ability to be phosphorylated by DNA-PK as described for C. These derivatives include the following: FoxA2-CΔ (lane 2), which contains an 11-residue N-terminal deletion that removes the putative DNA-PK sites (TQ/SQ); FoxA2-C(S283A,T280A; lane 3), a double point mutant in which both the serine and threonine are mutated; and FoxA2-C(S283A; lane 4), a point mutant in which the serine is mutated to alanine.
To identify potential amino acid residues in FoxA2 that are phosphorylated by DNA-PK, we first tested the various truncated FoxA2 derivatives in the in vitro kinase assay (Fig. 5C). Whereas the N-terminal fragment of FoxA2 also exhibited some tendency to be phosphorylated by DNA-PK (Fig. 5C, lane 1), the C-terminal fragment appeared to be the most favored domain in this regard (lane 3). Surprisingly, the central winged helix DNA binding domain, which interacts most strongly with DNA-PK, was not phosphorylated (Fig. 5C, lane 2). These results indicate that this domain could serve as a stable docking site for the kinase, which can then target relatively distal site(s) for modification (see under “Discussion”).
To further map the preferred DNA-PK phosphorylation site on FoxA2, we generated a derivative (FoxA2-CΔ) of FoxA2-C from which 11 N-terminal residues spanning putative DNA-PK consensus sites (TQ/SQ) (31, 32) were deleted (Fig. 5B). When subjected to the kinase assay, this derivative failed to serve as a DNA-PK substrate (Fig. 4D, lane 2 versus lane 1). These results thus allow us to consider the threonine and serine residues at positions 280 and 283 as the major candidates for serving as a DNA-PK target within the (rat) FoxA2. Note also that although the region in which this residue resides lies within a “linker” between the winged-helix DNA binding domain and the C-terminal activation domain that shows minor phylogenetic variation (e.g. at position 280, rat has a threonine whereas the human has an alanine), the serine residue at position 283 (Ser-283) is nonetheless conserved between rat and human (Fig. 5B).
To map more precisely the DNA-PK phosphorylation site on FoxA2, we also generated point mutants within FoxA2-C in which either Ser-283 or Thr-280 or both were altered to alanine residues, and we tested them in the in vitro kinase assay. The results (Fig. 5D, lane 3, and data not shown) definitively identified Ser-283 as the residue in FoxA2-C that is phosphorylated by DNA-PK.
The full-length FoxA2 molecule was then mutated at Thr-280/Ser-283 to assess how DNA-PK-dependent phosphorylation of the intact FoxA2 is affected. The corresponding wild-type and mutant proteins (hereinafter designated FoxA2 WT and FoxA2(T280A,S283A), respectively) were purified following expression in bacteria and tested for their ability to serve as DNA-PK substrates, as described. The results of the in vitro kinase assay showed a dramatic reduction in the ability of FoxA2 to be phosphorylated by DNA-PK (Fig. 5A, lane 2 versus lane 1). These results thus identify the Ser-283 residue as the predominant, if not the only, site that is targeted by DNA-PK within the intact FoxA2 molecule. Interestingly, our mass spectrometric methods did not identify this site as being phosphorylated at high levels (see under “Discussion”).
Effect of DNA-PK on the Transactivation Potential of FoxA2
We next assessed if the observed interaction of DNA-PK with FoxA2 and the resulting phosphorylation affects the ability of the latter to transactivate from its target sites. Therefore, we looked at the ability of FoxA2 to activate a luciferase reporter construct under the control of apoAI site B, which had been transiently transfected into a genotypically matched pair of cell lines (33), one of which carries a deletion for the DNA-PKcs gene (Fig. 6B, lane 2 versus lane 1). Although the luciferase data (Fig. 6A) trended toward a difference (4.7-fold for the DNA-PKcs+/+ cell line (SCH8) versus 2.8-fold for the DNA-PKcs−/− cell line (SCSV3)), statistical analyses suggest that this may reflect a modest effect at best. This likely is due to the fact that DNA-PK belongs to a family of ataxia telangiectasia mutated-like kinases, some with overlapping, if not identical, substrate specificities (34). Indeed, consistent with the presence of such redundant kinases, extracts from the DNA-PKcs−/− cell line (SCSV3) were able to phosphorylate the FoxA2-C polypeptide almost to an extent that was comparable with that with equivalent extracts from the DNA-PKcs+/+ cell line (Fig. 6C, lane 4 versus lane 2). Thus, a direct test of the role of DNA-PK in cellular assays is complicated by the presence of other kinases, which could subsume its role, at least in part.
FIGURE 6.
Effect of DNA-PK on transcriptional activation by FoxA2. A, B3XAI.LUC reporter plasmid (100 ng) was transfected into either SCH8 (DNA-PKcs+/+, black bars) or SCSV3 cells (DNA-PKcs−/−, gray bars) either alone or together with 250 ng of a FoxA2-expressing plasmid. For each cell type, luciferase activity was first normalized using the corresponding Renilla reading. Taking the normalized activity in the absence of ectopically expressed FoxA2 as basal (hence, 1.0), the data were expressed as fold-activation in the context of the indicated cell line. Student's t test was used to evaluate the data (p = 0.09; n = 4). B, to authenticate the phenotypes of the two cell lines, whole cell extracts from SCH8 (lane 1, DNA-PKcs+/+) or SCSV3 (lane 2, DNA-PKcs−/−) were analyzed by Western blotting using either antibodies against DNA-PKcs (upper panel) or TAF100 (lower panel). C, SCSV3 (DNA-PKcs−/−) cells contain a kinase that also can phosphorylate the FoxA2-C derivative. For the in vitro kinase assay purified FoxA2-C (lanes 2 and 4) was incubated with extracts from SCH8 (DNA-PKcs+/+, lanes 1 and 2) or from SCSV3 (DNA-PKcs−/−; lanes 3 and 4) cells in the presence of [γ-32P]ATP. Following the reaction, the samples were purified over nickel-nitrilotriacetic acid-agarose beads, concentrated by trichloroacetic acid precipitation, and resolved by SDS-PAGE. Labeled proteins (arrow) were visualized by autoradiography.
We therefore directly evaluated whether ablation of the target sites of DNA-PK (and of any related kinases present in various cell types) impinges on transcription activation function of FoxA2. For this purpose, we compared the ability of transiently transfected wild-type FoxA2 (FoxA2 WT) and the derivative (FoxA2(T280A,S283A)) that is no longer phosphorylated by DNA-PK in vitro (see Fig. 5A) to activate the apoAI site B-driven luciferase reporter in HepG2 cells (Fig. 7A). Whereas FoxA2 WT efficiently activated this template in a dose-dependent manner (up to 80-fold relative to basal), corresponding activation levels for FoxA2(T280A,S283A) were reduced by almost 8-fold. We also confirmed by Western blotting that the two FoxA2 proteins were expressed at comparable levels under these conditions (Fig. 7C). We conclude that ablation of the DNA-PK phosphorylation site on FoxA2 severely compromises its ability to activate transcription.
FIGURE 7.
Effect of ablation of the DNA-PK phosphorylation site on transcriptional activation by FoxA2. A, B3XAI.LUC reporter plasmid (100 ng) was transfected into HepG2 cells together with the indicated amounts (in nanograms) of vectors expressing FoxA2 WT or FoxA2(T280A,S283A). Luciferase activity was normalized as described for Fig. 6, and the data are expressed as fold-activation relative to control transfections with no ectopic FoxA2. B, luciferase reporter containing binding sites for both HNF-4α and FoxA2 was transfected into HepG2 cells together with indicated amounts (in nanograms) of vectors expressing HNF-4α and either FoxA2 WT or FoxA2(T280A,S283A). The results were analyzed as described for A. C, Western blot analysis of transfected FoxA2 proteins. HepG2 cells were transfected with the control vector alone (lane 1) or vectors expressing FLAG-tagged FoxA2 WT (lane 2) or FLAG-tagged FoxA2(T280A,S283A) (lane 3). After 48 h whole cell lysates from the cells were analyzed by Western blotting using an anti-FLAG antibody.
Given that FoxA2 rarely functions in a solitary mode, it was also important to ascertain whether this newly uncovered DNA-PK-dependent activity of FoxA2 remains relevant when FoxA2 is working synergistically with other factors. We therefore evaluated the ability of FoxA2(T280A,S283A) to synergize with HNF-4α by transfecting our reporter construct that is responsive to both FoxA2 and HNF-4α into HepG2 cells (Fig. 7B). Because HepG2 cells express rather high levels of HNF-4α, we note first that “basal” expression of this template is quite high (but is here normalized to 1). The additional effect of ectopic FoxA2 WT and thus of the apparent synergy with ectopic HNF-4α is relatively modest but is nonetheless significant. Importantly, this additional level of activity is not observed with FoxA2(T280A,S283A) suggesting that requirement for DNA-PK-dependent phosphorylation of FoxA2 remains in effect even when functioning in the synergy mode.
Conserved DNA-PK Site in FoxA2 Is Not Required for Its Translocation to the Nucleus
It has been shown previously that FoxA factors can be functionally regulated by AKT family kinases via mechanisms that control their subcellular localization (35, 36). Toward establishing whether or not such a mechanism underlies the above-described effect of DNA-PK-dependent phosphorylation of FoxA2 on its transcriptional activation function, we determined, by indirect immunofluorescence, whether the subcellular localization of the FoxA2(T280A,S283A) was similar to that of FoxA2 WT. For this purpose, we transiently transfected 293T cells with the appropriate expression vectors. Both FoxA2 WT and the FoxA2(T280A,S283A) were almost exclusively localized to the nuclei, as demarcated by counterstaining with 4′,6-diamidino-2-phenylindole (Fig. 8, compare upper panels with lower panels). Control cells, which were transfected with just the vector, showed no detectable staining for FoxA2, which is not expressed in 293T cells. These data indicate that phosphorylation by DNA-PK does not affect the subcellular localization of FoxA2.
FIGURE 8.
Ablation of the DNA-PK phosphorylation site on FoxA2 does not alter its subcellular localization. Representative images of immunostained 293T cells transfected with either vector control, FoxA2 WT, or FoxA2(T280A,S283A) are shown. FoxA2 protein (green) is localized to the nuclei, which were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). There was no detectable staining of FoxA2 in vector control cells.
Mutation of the Conserved DNA-PK Site in FoxA2 Does Not Impair Its Ability to Bind apoAI Site B
Toward understanding the basis of the transcriptional defect of FoxA2(T280A,S283A), we also ruled out that the mutation directed at precluding DNA-PK-dependent phosphorylation somehow affects the ability of FoxA2 to bind to its cognate DNA sites. For this purpose we focused on recombinant FoxA2 WT and FoxA2(T280A,S283A), each purified from bacteria, and we evaluated the ability of each to bind site B (Fig. 9). Using a probe that spans site B from the apoAI promoter-proximal enhancer, we first established conditions for an electrophoretic mobility shift assay (EMSA) to generate a specific FoxA2-DNA complex (Fig. 9A). Specificity of this complex was ascertained by the following criteria. First, we demonstrated that the complex could be competed away by a molar excess of unlabeled site B oligonucleotide but not by an equivalent amount of a DNA fragment spanning site A, which does not bind FoxA2 (Fig. 9A, compare lane 1 with lanes 5–7). Second, we showed that only with a specific antibody against FoxA2, but not control IgG (Fig. 9B, lane 3 versus lane 1), an additional band corresponding to a supershifted FoxA2-DNA complex could be generated. Next we incubated the probe with increasing amounts of each of FoxA2 WT and FoxA2(T280A,S283A) (Fig. 9C). The EMSA confirmed that both proteins could bind to the probe in a dose-dependent manner and with comparable efficiencies.
FIGURE 9.
DNA binding analysis of FoxA2 to its target site. A, EMSA showing binding of recombinant FoxA2 to a site B probe (lane 1) and its competition by increasing amount of unlabeled site B oligonucleotide (lanes 2–4) but not equivalent amount of site A oligonucleotide (lanes 5–7). The molar ratios of cold competitor over labeled probe were as follows: lanes 2 and 5, 2.5; lanes 3 and 6, 10; lanes 4 and 7, 50. B, further validation of the FoxA2 EMSA using an anti-FoxA2 antibody (lane 2) to supershift the specific complex (identified by the arrow). As control, total goat IgG was used (lane 1). C, increasing amounts of FoxA2 WT (lane 1, 1.6 ng; lane 2, 4 ng; lane 3, 8 ng) or FoxA2(T280A,S283A) (lane 4, 2.8 ng; lane 5, 7 ng; lane 6, 14 ng) were incubated with the site B probe and analyzed by EMSA. D, using the indicated bodies, ChIP assays were performed on extracts of 293T cells that had been cross-linked with formaldehyde 48 h after transfection with the B3XAI.LUC reporter and either the empty vector (pVP5; top panel), the vector expressing wild-type FoxA2 (middle panel), or the vector expressing the mutant FoxA2 protein (lower panel). Immunoprecipitate cross-links were reversed, and the promoter region on the reporter template was amplified by PCR.
Finally, we also asked if FoxA2(T280A,S283A) similarly retains the ability to bind to its target sites in the context of the cell. We transfected wild-type FoxA2 and FoxA2(T280A,S283A) via expression vectors into 293T cells together with a reporter plasmid that carries apoAI site B. ChIP analysis (Fig. 9D) of the extracts from these cells using antibodies against FoxA2 clearly demonstrated that the mutant protein, despite its inability to be phosphorylated at Ser-283, is efficiently capable of binding to its target site. Indeed, if anything, our results suggest that, under the conditions of the experiment, the mutant protein binds more efficiently than the wild type. We further conclude that the transcriptional defect exhibited by FoxA2(T280A,S283A) is unlikely to arise from impairment in its ability to bind to its target sites. Taken together, the data presented here allow us to conclude that DNA-PK-dependent phosphorylation of FoxA2 at Ser-283 contributes to its transcriptional activation function at a step following its recruitment to the target gene.
DISCUSSION
The main conclusion from this study is that DNA-PK can interact with FoxA2 and modulate its transcription activation potential by phosphorylating a critical serine residue. Given an emerging role of the superfamily of forkhead transcription factors as pioneer factors in developmental pathways through alterations in chromatin configuration, these results have important implications for how the activity of these factors might in turn be regulated.
Role for DNA-PK in Transcription
DNA-PK appears to be a multifunctional protein. It has been most extensively characterized as a key participant in DNA repair pathways in which, intriguingly, it initiates a repair cascade by phosphorylating the variant histone H2AX (32, 34, 37). At the same time, numerous reports have implicated it directly in various aspects of transcription, including via mechanisms such as double-stranded DNA breaks that may be common to both processes. Thus, beginning with its initial identification as a kinase for the C-terminal domain of the largest subunit of RNA polymerase II (38), DNA-PK was shown to phosphorylate other components of the preinitiation complex (39) as well as to stimulate multiple rounds of transcription in in vitro transcription systems (40). DNA-PKcs was also detected as one of the polypeptides associated with the putative nuclear receptor coactivator TRBP (41). It has also been implicated as a regulator of many transcriptional activators such as Sp1 (42) and of the antitumor p53 protein, a transcriptional activator in its own right (31). DNA-PK was also shown to be a component of the multiprotein complex nucleated by the estrogen receptor (43). Most recently, and perhaps of immediate relevance to this study, DNA-PK was implicated in regulating transcription factor upstream stimulatory factor-dependent expression of metabolic genes in response to feeding-induced signals (44). Thus, our demonstration here that DNA-PK binds to and phosphorylates FoxA2, with significant effects on its ability to activate its target genes, highlights yet another transcription-related role of DNA-PK.
Potential Mechanisms for DNA-PK Vis à Vis FoxA Factors as Pioneer Factors
Our results allow us to conclude that phosphorylation of FoxA2 by DNA-PK at Ser-283 greatly stimulates its transcriptional activation function. In contrast to what has been reported for AKT-mediated phosphorylation of FoxA2 (35, 36), our data suggest that the effects mediated through modification of Ser-283 are unlikely to be realized from control of subcellular localization of FoxA2. Furthermore, although we have not yet analyzed effects of the modification on interactions with chromatin (see below), our comparative analysis of the specific DNA binding ability of wild-type FoxA2 and FoxA2 bearing a mutation in the DNA-PK phosphorylation site (on which our conclusions regarding the functional consequences of the modification are based) suggests that the mutation does not have any gross effects on the ability of FoxA2 to bind to its target site. Thus the evidence presented here is most consistent with the notion that a main functional consequence of this modification is realized at a step following the recruitment of FoxA2 to its target genes.
Recent mechanistic dissection of how forkhead factors exert their transcriptional activation functions has focused on their ability to bind higher order chromatin and render it more amenable for additional transcriptional factors to bind in the immediate vicinity (22, 23). As attractive as this idea is with respect to the function of forkhead factors as pioneer factors and the similarity of their DNA binding domains to linker histones, it remains likely that these factors also function as conventional transcriptional activators that act in part by recruiting additional coactivators. Indeed, forkhead factors have long been known to contain autonomous “activation domains” (18) that function independently of the winged-helix DNA binding domain. Thus, these transcription factors can reasonably be expected to act at multiple levels.
Therefore, a most straightforward mechanism that can be envisioned on the basis of our data is that DNA-PK-mediated phosphorylation of the C-terminal domain of FoxA2 facilitates interactions of downstream factors necessary for full activation. In the simplest case these could include factors responsible for chromatin transactions or interactions with the general transcription machinery. More intriguingly, as is emerging for several other transcriptional factors (45–47), it may be that efficient activation by FoxA2 is coupled to proteasome-dependent degradation, which serves to “reset” the transcription complex at each round of transcription to maintain elevated expression levels. Phosphorylation by DNA-PK could contribute to the turnover, and apparent coactivation effects, by marking FoxA2 for degradation. In this regard, it is worth mentioning that steady-state intracellular concentrations of FoxA2(T280A,S283A), which could not be phosphorylated, were consistently found to build up to significantly higher levels as compared with wild-type FoxA2 (data not shown). Also potentially consistent with the relatively fleeting lifetime of wild-type FoxA2 is the observation that mass spectrometry analyses failed to detect FoxA2 species bearing this modification. Regardless of the precise downstream mechanism, phosphorylation induced by DNA-PK would initiate a cascade of events that culminate in the efficient establishment and activity of the preinitiation complex, the hallmark of an active gene.
Unexpectedly for a coactivator, our ChIP data indicate that DNA-PK (Ku70) is distributed across the entire apoAI gene albeit at low levels. This could imply that FoxA2 encounters DNA-PK, a chromatin-associated protein upon interaction with its cognate sites within chromatin. Based on the above discussion, and although this is not simply a passive interaction, important functional outcomes could nevertheless ensue at the local sites of interaction. Alternatively, given the recent report that DNA-PK recruitment to upstream stimulatory factor is dependent on extracellular signals (insulin) (44), its full mobilization to the apoAI promoter may also be dependent on some as-yet-unidentified signal. It also remains unclear as to what role, if any, the Ku70 and Ku80 subunits, which in the context of DNA repair are the primary determinants in guiding DNA-PK to the various lesions (48), play in the context of transcription.
Indeed, it cannot be ruled out that FoxA2 interaction with DNA-PK is additionally a means to deliver the latter to the transcription machinery as a conventional coactivator. The linker histone H1 has been identified previously as a substrate for DNA-PK (49) and further that phosphorylation of linker histones has been linked to gene activation (50, 51). Thus, DNA-PK, recruited by FoxA2, could also potentially contribute in this context by phosphorylating the linker histone H1 in tandem with its effects on FoxA2. In the end, although FoxA2 binding to H1-compacted chromatin, per se, might reflect a substantial local relief of compaction, additional unfolding may depend on DNA-PK-mediated phosphorylation of H1. How the phosphorylation at Ser-283 would tie in with this mode is not immediately apparent, but scenarios can be imagined in which this modification is coupled to interactions with the remodeled chromatin. In this regard, it is interesting to note that whereas the winged helix domain of FoxA2 serves as the main docking site for DNA-PK, Ser-283 is located somewhat distally in the C-terminal region of the molecule suggesting a physical basis for such a coupled mechanism.
Ongoing studies, which include establishing an in vitro transcription system for FoxA2, along the lines that we have previously implemented for HNF-4α (15, 24), will allow us to test the effects of DNA-PK on FoxA2 whether as a bona fide coactivator or, more generally, as a cofactor.
Acknowledgments
We thank Dr. R. G. Roeder for continued support and encouragement and for critical reading of the manuscript. We also thank Drs. A. Bharti and Don Kufe for DNA-PK cell lines, Joe Fernandez of the Proteomics Core Facility at Rockefeller University for mass spectrometric analysis, and Dr. Miki Jishage for logistical help. We thank Drs. Manuel Ascano and T. Tuschl for both guidance and use of their fluorescence microscope.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 DK060764 (to S. M.).
- apoAI
- apolipoprotein AI
- HNF
- hepatocyte nuclear factor
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- cs
- catalytic subunit
- DNA-PK
- DNA-dependent protein kinase
- GST
- glutathione S-transferase.
REFERENCES
- 1.Brivanlou A. H., Darnell J. E., Jr. (2002) Science 295, 813–818 [DOI] [PubMed] [Google Scholar]
- 2.Roeder R. G. (2005) FEBS Lett. 579, 909–915 [DOI] [PubMed] [Google Scholar]
- 3.Roeder R. G. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 201–218 [DOI] [PubMed] [Google Scholar]
- 4.Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 5.Lemon B., Tjian R. (2000) Genes Dev. 14, 2551–2569 [DOI] [PubMed] [Google Scholar]
- 6.Malik S., Roeder R. G. (2005) Trends Biochem. Sci. 30, 256–263 [DOI] [PubMed] [Google Scholar]
- 7.Malik S., Roeder R. G. (2003) in Handbook of Cell Signaling (Bradshaw R. A., Dennis E. A. eds) Vol. 3, pp. 11–19, Elsevier Academic Press, San Diego, CA [Google Scholar]
- 8.Zannis V. I., Kan H. Y., Kritis A., Zanni E., Kardassis D. (2001) Front. Biosci. 6, D456–504 [DOI] [PubMed] [Google Scholar]
- 9.Malik S. (2003) Front. Biosci. 8, d360–368 [DOI] [PubMed] [Google Scholar]
- 10.Sladek F. M. (1993) Receptor 3, 223–232 [PubMed] [Google Scholar]
- 11.Harnish D. C., Malik S., Karathanasis S. K. (1994) J. Biol. Chem. 269, 28220–28226 [PubMed] [Google Scholar]
- 12.Harnish D. C., Malik S., Kilbourne E., Costa R., Karathanasis S. K. (1996) J. Biol. Chem. 271, 13621–13628 [DOI] [PubMed] [Google Scholar]
- 13.Rada-Iglesias A., Wallerman O., Koch C., Ameur A., Enroth S., Clelland G., Wester K., Wilcox S., Dovey O. M., Ellis P. D., Wraight V. L., James K., Andrews R., Langford C., Dhami P., Carter N., Vetrie D., Pontén F., Komorowski J., Dunham I., Wadelius C. (2005) Hum. Mol. Genet. 14, 3435–3447 [DOI] [PubMed] [Google Scholar]
- 14.Rochette-Egly C. (2005) J. Biol. Chem. 280, 32565–32568 [DOI] [PubMed] [Google Scholar]
- 15.Barrero M. J., Malik S. (2006) Mol. Cell 24, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuteja G., Kaestner K. H. (2007) Cell 131, 192. [DOI] [PubMed] [Google Scholar]
- 17.Tuteja G., Kaestner K. H. (2007) Cell 130, 1160. [DOI] [PubMed] [Google Scholar]
- 18.Friedman J. R., Kaestner K. H. (2006) Cell. Mol. Life Sci. 63, 2317–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark K. L., Halay E. D., Lai E., Burley S. K. (1993) Nature 364, 412–420 [DOI] [PubMed] [Google Scholar]
- 20.Lai E., Clark K. L., Burley S. K., Darnell J. E., Jr. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10421–10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirillo L. A., Zaret K. S. (1999) Mol. Cell 4, 961–969 [DOI] [PubMed] [Google Scholar]
- 22.Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., Zaret K. S. (2002) Mol. Cell 9, 279–289 [DOI] [PubMed] [Google Scholar]
- 23.Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 24.Malik S., Roeder R. G. (2003) Methods Enzymol. 364, 257–284 [DOI] [PubMed] [Google Scholar]
- 25.Kuo M. H., Allis C. D. (1999) Methods 19, 425–433 [DOI] [PubMed] [Google Scholar]
- 26.Sastry K. N., Seedorf U., Karathanasis S. K. (1988) Mol. Cell. Biol. 8, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. (1983) Methods Enzymol. 101, 582–598 [DOI] [PubMed] [Google Scholar]
- 28.Kyono Y., Sugiyama N., Imami K., Tomita M., Ishihama Y. (2008) J. Proteome Res. 7, 4585–4593 [DOI] [PubMed] [Google Scholar]
- 29.Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 30.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 31.Lees-Miller S. P., Sakaguchi K., Ullrich S. J., Appella E., Anderson C. W. (1992) Mol. Cell. Biol. 12, 5041–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson C. W., Lees-Miller S. P. (1992) Crit. Rev. Eukaryot. Gene Expr. 2, 283–314 [PubMed] [Google Scholar]
- 33.Bharti A., Kraeft S. K., Gounder M., Pandey P., Jin S., Yuan Z. M., Lees-Miller S. P., Weichselbaum R., Weaver D., Chen L. B., Kufe D., Kharbanda S. (1998) Mol. Cell. Biol. 18, 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durocher D., Jackson S. P. (2001) Curr. Opin. Cell Biol. 13, 225–231 [DOI] [PubMed] [Google Scholar]
- 35.Wolfrum C., Besser D., Luca E., Stoffel M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11624–11629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfrum C., Asilmaz E., Luca E., Friedman J. M., Stoffel M. (2004) Nature 432, 1027–1032 [DOI] [PubMed] [Google Scholar]
- 37.Anderson C. W. (1993) Trends Biochem. Sci. 18, 433–437 [DOI] [PubMed] [Google Scholar]
- 38.Dvir A., Stein L. Y., Calore B. L., Dynan W. S. (1993) J. Biol. Chem. 268, 10440–10447 [PubMed] [Google Scholar]
- 39.Chibazakura T., Watanabe F., Kitajima S., Tsukada K., Yasukochi Y., Teraoka H. (1997) Eur. J. Biochem. 247, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 40.Woodard R. L., Anderson M. G., Dynan W. S. (1999) J. Biol. Chem. 274, 478–485 [DOI] [PubMed] [Google Scholar]
- 41.Ko L., Chin W. W. (2003) J. Biol. Chem. 278, 11471–11479 [DOI] [PubMed] [Google Scholar]
- 42.Jackson S. P., MacDonald J. J., Lees-Miller S., Tjian R. (1990) Cell 63, 155–165 [DOI] [PubMed] [Google Scholar]
- 43.Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. (2006) Science 312, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 44.Wong R. H., Chang I., Hudak C. S., Hyun S., Kwan H. Y., Sul H. S. (2009) Cell 136, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipford J. R., Deshaies R. J. (2003) Nat. Cell Biol. 5, 845–850 [DOI] [PubMed] [Google Scholar]
- 46.Collins G. A., Tansey W. P. (2006) Curr. Opin. Genet. Dev. 16, 197–202 [DOI] [PubMed] [Google Scholar]
- 47.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 48.Downs J. A., Jackson S. P. (2004) Nat. Rev. Mol. Cell Biol. 5, 367–378 [DOI] [PubMed] [Google Scholar]
- 49.Kysela B., Chovanec M., Jeggo P. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dou Y., Song X., Liu Y., Gorovsky M. A. (2005) Mol. Cell. Biol. 25, 3914–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koop R., Di Croce L., Beato M. (2003) EMBO J. 22, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]