Abstract
Directional transport of specific mRNAs is of primary biological relevance. In Xenopus oocytes, mRNA localization to the vegetal pole is important for germ layer formation and germ cell development. Using a biochemical approach, we identified Xenopus Elr-type proteins, homologs of the Hu/ELAV proteins, as novel components of the vegetal mRNA localization machinery. They bind specifically to the localization elements of several different vegetally localizing Xenopus mRNAs, and they are part of one RNP together with other localization proteins, such as Vg1RBP and XStaufen 1. Blocking Elr-type protein binding by either localization element mutagenesis or antisense morpholino oligonucleotide-mediated masking of their target RNA structures, as well as overexpression of wild type and mutant ElrB proteins, interferes with vegetal localization in Xenopus oocytes.
mRNA localization to the vegetal pole of Xenopus oocytes establishes a primary axis of asymmetry that is crucial for early embryonic development. Two major transport pathways that guide specific mRNAs to the vegetal cortex can be distinguished from each other. The early or METRO pathway operates via the mitochondrial cloud during earliest stages of oogenesis. Several early localizing mRNAs have been found to be involved in germ cell development (1). Although early localizing RNAs like Xcat2 or Xdazl become first enriched in the mitochondrial cloud by a microtubule-independent diffusion/entrapment mechanism and relocate to the vegetal cortex during stage II along with components of the fragmented mitochondrial cloud (2–5), late pathway RNAs like Vg1, VegT, and Velo1 are initially homogenously dispersed throughout the cytoplasm (3, 6–11). The late transport pathway is activated at mid-oogenesis (stages III and IV) and is mediated by a motor-driven, microtubule-dependent mechanism (12–15). Several of the late localizing mRNAs are critical for germ layer formation (16). A small population of RNAs exhibits localization features of both pathways and is therefore referred to as intermediate pathway RNAs (17–19).
Both early and late localization pathways are under the control of regulatory RNA elements, usually residing in the 3′-UTR3 of localized mRNAs, referred to as localization elements (LEs) or mitochondrial cloud localization element (reviewed in Refs. 20 and 21). LEs recruit proteins to form a localization complex. Although proteins that exclusively interact with LEs from early localizing RNAs and that could mediate the entrapment in the MC have not been identified to date, a number of proteins that interact with the localization element of the late localizing Vg1 mRNA have been identified; they include Vg1RBP, hnRNP I, Prrp, VgRBP71/KSRP, XStaufen 1, and 40LoVe (15, 22–27). Interestingly, mitochondrial cloud localization elements of all early pathway RNAs tested to date can enter the late localization pathway if injected into stage III/IV oocytes, suggesting that they are able to recruit late transport proteins (17, 28–31). This may serve as a fail-proof mechanism to ensure vegetal cortex localization of early pathway RNAs that are transcribed late, after mitochondrial cloud breakdown.
A core transport RNP containing hnRNP I and Vg1RBP is formed in the nucleus and exported to the cytoplasm. Although Vg1RBP and hnRNP I form direct protein-protein interactions in the nucleus, complex formation becomes RNase-sensitive in the cytoplasm, suggesting that a remodeling step occurs after export to the cytoplasm (32, 33). VgRBP71/KSRP and 40LoVe can also be detected in the nucleus, but whether they are indeed part of a nuclear transport RNP remains to be determined (26, 27). The reassembly step in the cytoplasm includes the recruitment of additional proteins; whereas hnRNP I, Vg1RBP, Prrp, XStaufen 1, and 40LoVe accompany the localizing RNA in the vegetal cytoplasm and get enriched at the cortex (15, 25, 27, 32, 34, 35), VgRBP71/KSRP is found throughout the cytoplasm with a slight enrichment at the animal cortex (26). Rather than directly participating in the vegetal transport, VgRBP71/KSRP has been suggested to translationally activate cortical Vg1 mRNA by stimulating a nuclease that cleaves off the Vg1 translational control element (TCE) (36). Because of its interaction with profilin, a regulator of actin dynamics, Prrp has been proposed to function in the microfilament-dependent anchoring of localized RNA at the cortex (25). The recruitment of Staufen 1 into the transport particle might be mediated by hnRNP I, because dominant negative Staufen 1 mutants not only affect vegetal localization of injected RNAs but also lose interaction with hnRNP I (15).
The active particle transport along microtubule filaments is mediated by overlapping functions of kinesin I and II plus-end directed motor proteins (12, 13, 15). Further remodeling of the localization RNP is likely to occur at the vegetal cortex, where late localizing RNAs become anchored. Cytokeratin intermediate filaments, in addition to microfilaments, seem to be required for anchoring (14, 37). Interestingly, vegetally localized RNAs themselves may function as structural components of the cortical cytokeratin meshwork, because antisense oligodeoxynucleotide-mediated depletion of localized RNAs like VegT or xlsirts disrupts the cytokeratin network and leads to a release of specific RNAs from the vegetal cortex (38–40).
Proteins of the ELAV/Hu protein family are RNA-binding proteins that contain three RRM domains, with RRM2 and 3 being separated by a linker region (41). Vertebrates express four members of ELAV/Hu-type proteins, namely the ubiquitous HuR (also called HuA or ElrA in Xenopus), HuB (Hel-N1, ElrB), which is expressed in neurons, testes and ovaries, as well as the strictly neuronal proteins HuC (ElrC) and HuD (ElrD). ELAV/Hu-type proteins bind to AU-rich regions and function in a wide variety of posttranscriptional processes, such as control of RNA stability, translational regulation, splicing, and polyadenylation, as well as nucleocytoplasmic transport (reviewed in Ref. 42). As part of its role in translational regulation, HuR has recently been described to mediate the stress-induced release of microRNA-silenced RNAs from P-bodies for active translation (43). Xenopus ElrA/B proteins have been shown to interact with the translational control element of Vg1 mRNA (VTE) and anti-HuR antibody injections support a role for ElrB in the translational repression of Vg1 mRNA during early to mid-oogenesis (44).
We have previously characterized the function of the late localizing XDead end (XDE) mRNA in Xenopus. As revealed by UV cross-linking experiments, its localization element (XDE-LE) binds to a set of unknown proteins with molecular masses between 35 and 45 kDa (45). Here, we report on the identification of these proteins as Xenopus ElrA/B proteins. We further show that Elr-type proteins are part of one RNP complex together with other localization proteins from Xenopus oocytes. ElrA/B bind to a number of additional LEs from both early and late localizing mRNAs. Interfering with Elr-LE interaction, as well as overexpression of wild type or mutant versions of ElrB, results in a loss of vegetal mRNA localization, arguing for a critical role of Elr-type proteins during vegetal RNA localization.
EXPERIMENTAL PROCEDURES
Cloning Procedures and Protein Expression
Full-length cDNA clones of ElrA (accession number BC086269) and ElrB (accession number NM_001087566) in pSPORT vector were obtained from the RZPD genome resource center. The corresponding IMAGE clone numbers for ElrA and ElrB are IMAGp998E0114218Q and IRBHp990C0179D, respectively. The open reading frames of ElrA and B were amplified by PCR from plasmids and subcloned into StuI and XbaI sites of the pCS2+MT (46) and pCS2+ FLAG (47) using the following primers: ElrB_F, AATAGGCCTATGGCAGTCAGACTGTGTGATGTGGCT; ElrB_R, GCCTCTAGATTAAGCTTTGTGTGTTTTGCTCGTTTTGAAC; ElrA_F, TCAAGGCCTATGTCTAACGGTTATGAAGATCACA; and ElrA_R, ATATCTAGATTATTTGTGTGACTTGCTGGTTTTG. For recombinant protein expression, ElrB open reading frame excised from the pCS2+ FLAG construct and subcloned into NcoI and NotI sites of pET21d (Novagen) was employed. The Vg1RBP expression construct was as described (22). His-tagged ElrB and Vg1RBP proteins were purified as described previously (48).
For point mutagenesis of the XDE-LE, the XDE-LE was amplified with primers reported in (45) and cloned into the pGEM-T easy vector (Promega). Mutant versions of XDE-LE were generated by site-directed mutagenesis using a two-step overlap extension PCR as originally described in Ref. 49. PCRs were done using a high fidelity DNA polymerase kit (Fermentas), and DNA fragments were purified from agarose gels by use of a PCR purification kit (Qiagen). The mutant XDE-LE was cleaved with BamHI and XhoI for cloning into the pBK-CMV-lacZ tagged vector (11, 45). Sequences of mutagenesis primers are available in the supplemental materials.
ElrB mutants defective in RNA binding were created by introducing point mutations in the RRM domains by site directed mutagenesis. Mutation sites were chosen according to (50). Sequences of mutagenesis primers are available in the supplemental materials. Most likely because of inaccuracies of the DNA polymerase used for the mutagenesis PCR, ElrBmut1/2 and ElrBmut1/3 contain additional amino acid changes in the first RRM at amino acid position 100 (D100G, ElrBmut1/2) and amino acid position 73 (L73F, ElrBmut1/3).
Isolation of Oocytes and Extract Preparation
Oocytes were obtained as described (11). Blendzyme (Sigma) treatment was performed to release the oocytes from the ovary sac. The oocytes were homogenized using a syringe mounted with needles (27 and 24 gauge) in S100 buffer (50 mm Tris, pH 7.5, 50 mm KCl, 0.1 mm EDTA, protease inhibitor mixture (Roche Applied Science) and 5% glycerol); after three passages crude extracts were centrifuged for 20 min at 13,000 rpm, 4 °C in a table top centrifuge (S16 extract). The yolk material was routinely removed by Freon (Merck) extraction (51).
Glycerol Gradient Centrifugation and Biochemical Enrichment of Localization Proteins
This procedure was adapted from Ref. 52; the gradient was formed using S100 buffer containing 5 and 60% glycerol (50 mm Tris, pH 7.5, 50 mm KCl, 0.1 mm EDTA, protease inhibitor mixture (Roche Applied Science), 5 or 60% glycerol). A polyallomer centrifuge tube (2.2 ml) was filled with 1 ml of the 5% glycerol S100 buffer, and then 1 ml of the 60% glycerol S100 buffer was gently loaded underneath. The tubes were sealed with parafilm and gently placed sideways on a flat plate and incubated at 4 °C for at least 2 h. An approximately linear gradient is formed after the tubes are returned to the upright position. Protein samples (usually a total of 200 μg of S16 in 200 μl of volume) were carefully loaded on top of the preformed gradient. High speed centrifugation was performed in the TLS 55 rotor at a speed of 50,000 rpm (214,200 × g) for 4 h at 4 °C in an Optima TL Ultracentrifuge (Beckmann). Fractions of 200 μl were carefully taken from the top of the gradient and processed for Western blot analysis. Protein fractions enriched for localization proteins were pooled. The glycerol concentration of these fractions was reduced to 5% by ultrafiltration using Amicon Ultra (Millipore) or Vivaspin (Vivascience) tubes in a swinging bucket rotor (Sorvall) at 4 °C (4000 rpm centrifugation until volume is reduced to desired levels). RNPs were disrupted either by RNase A treatment (10 μg/ml) or by increasing the salt concentration to 400 mm NaCl. After 2 h at 4 °C, the samples were again subjected to ultracentrifugation at ∼200,000 × g (68,000 rpm) in a TLA100.2 rotor (Beckmann). The high speed supernatants were concentrated to the desired levels by using ultrafiltration tubes.
Two-dimensional Gel Electrophoresis and Protein Identification
The Bio-Rad two-dimensional gel system was used according to the manufacturers instructions; the sample buffer recipe was modified, replacing dithiothreitol with hydroxyethyl-disulfide, DeStreak reagent (GE Healthcare; see Ref. 53). Isoelectric focusing was done with immobilized pH gradients of 3–10 (Bio-Rad) and 7–11 (GE Healthcare), with the second dimension on 11–12% SDS-PAGE. Manually excised gel plugs were subjected to an automated platform for the identification of gel-separated proteins (54) as described in the framework of recent large scale proteome studies (55, 56). An Ultraflex matrix-assisted laser desorption ionization time-of-flight-mass spectrometer (Bruker Daltonics) was used to acquire both peptide mass fingerprint and fragment ion spectra, resulting in confident protein identifications based on peptide mass and sequence information. Data base searches in the NCBI nonredundant primary sequence data base restricted to the taxonomy Xenopus laevis were performed using Mascot software 2.0 (Matrix Science) with parameter settings described earlier (55, 56). All of the data sets were searched again without taxonomy restriction to account for potential matches to sequences from Xenopus tropicalis. The minimal requirement for accepting a protein as identified was at least one peptide sequence match above homology threshold in coincidence with at least four peptide masses assigned in the peptide mass fingerprint.
Western Blotting
Protein samples separated by SDS-PAGE were blotted onto nitrocellulose membrane and blocked with 5% nonfat milk powder in Tris buffered saline with Tween 20 (20 mm Tris-Cl, pH 6.8, 150 mm NaCl, 0.05% Tween 20) overnight at 4 °C. Primary antibodies used were diluted as follows: α-HuR (sc-5261; Santa Cruz Biotechnology, Inc.) 1/4000, α-Vg1RBP (35) 1/10,000, α-XStaufen 1 (34) 1/10,000, and anti-40LoVe (27) 1/5000. Horseradish peroxidase-coupled α-mouse (Santa Cruz Biotechnology, Inc.) and α-rabbit (Dianova) secondary antibodies were used at 1/10,000 dilutions, and detection was performed using the ECL system (GE Healthcare).
Immunostaining on Oocyte Sections
Stage II/IV oocytes were embedded in O.C.T. Compound (Tissue-Tek) for cryotome sectioning. The sections were fixed in paraformaldehyde for 10 min, washed in PBS containing 0,5% Triton X-100 (PBS-T) and blocked for 30 min in PBS-T containing 1% bovine serum albumin. Primary antibodies (α-HuR (sc-5261; Santa Cruz Biotechnology, Inc.), α-XStaufen 1 (1% bovine serum albumin), and α-40LoVe (27), as well as secondary α-mouse Alexa594 (Invitrogen) and α-rabbit fluorescein isothiocyanate (Sigma) antibodies were diluted 1:400 in PBS-T (1% bovine serum albumin) and incubated for 1 h in a humid chamber. The specimens were embedded in Dako fluorescent mounting medium and analyzed with a Axioimager M1 microscope (Zeiss).
Co-immunoprecipitations
Co-immunoprecipitation reactions were performed in principle as described (44) using NET buffer containing 10% glycerol instead of 0.25% gelatin. To identify ElrA/B-interacting proteins, preparative scale IPs were performed with 50 mg of total oocyte lysate in 50 ml of total IP volume in 50 aliquots, whereas an equivalent of two aliquots was used for silver stained SDS-PAGE analysis. Co-immunoprecipitation of Alexa-labeled XDE-LE RNA were performed essentially as described (11). 1 pmol of Alexa-labeled XDE-LE RNA was added to the in vitro translated proteins. Binding reactions were performed in the absence or presence of 2.5 pmol of morpholino oligonucleotides added 30 min prior to the Alexa-XDE-LE. Sequences of morpholino oligonucleotides are available in the supplemental materials.
Quantitative RNA-Co-IP
To assess for the enrichment of localizing RNAs in ElrA/B-containing RNPs, RNA co-immunoprecipitation from stage II–IV oocyte S16 extracts was performed using either α-HuR (sc-5261; Santa Cruz Biotechnology, Inc.) or α-Myc (Sigma) (control) antibodies immobilized on GammaBind Plus Sepharose (GE Healthcare). Co-IPs were performed using either YSS buffer (32) or IPP buffer (10 mm Tris, pH 8,0, 145 mm NaCl, 0.1% Nonidet P-40), supplemented with both RNase inhibitors (RNaseOut, Invitrogen) and protease inhibitors (Complete; Roche Applied Science). Co-precipitated RNAs were released by addition of 1% SDS, phenol chloroform-extracted, and precipitated with ethanol/ammonium acetate. The amount of individual mRNAs in the ElrA/B-RNPs, the control IP or 2% of the input material was determined by quantitative reverse transcription-PCR using the iQ SYBR Green Supermix and iCycler-system (Bio-Rad). Primer sequences are available in the supplemental materials.
The relative enrichment of individual mRNAs in the ElrA/B-RNPs versus the input (E1 = 2−(CT value (α-HuR-IP)-CT value (input))) or versus the control-IP (E2 = 2−(CT value (α-HuR-IP)-CT value (control-IP))), as well as control-IP versus input (E3 = 2−(CT value (control-IP)-CT-value (input))) were calculated according to the 2−ΔCT method (57). mRNA enrichment in the ElrA/B-RNPs was scored by the enrichment factor F = (E1 × E2)/E3. To compare the results of independent Co-IP experiments, the enrichment factors of the different mRNAs tested were normalized against the enrichment factor for glyceraldehyde-3-phosphate dehydrogenase, respectively.
In Vitro Transcription
Capped RNAs for oocyte injection were transcribed using the T3 mESSAGE mACHINE kit (Ambion), and pBK-CMV-lacZ-tagged XDE LE and mutant version templates were linearized with XhoI. Labeled RNAs or unlabeled competitor RNAs were synthesized using the T7 MEGAshortscript kit (Ambion). For radiolabeling, the unlabeled UTP provided with the kit was replaced by 5 μl of 20 μCi/μl [α-32P]UTP (GE Healthcare); for Alexa- or Cy3-labeled RNA, the reaction was supplemented with 1 μl of 1 mm ChromaTide Alexa Fluor 546–14-UTP (Molecular Probes/Invitrogen) or Cy3-UTP (PerkinElmer Life Sciences). Templates of XDE LE and its mutant versions were prepared by cutting the pGEMT plasmids constructs with XhoI. In vitro transcribed RNAs were purified using the RNeasy mini kit (Qiagen). T7 promoter-containing primers were used to generate templates for in vitro transcription of LE-RNAs by PCR amplification as listed in the supplemental materials.
UV Cross-linking Assay
This assay was done as described previously (11, 58). Cross-linked proteins were separated by SDS-PAGE and analyzed by phosphorimaging (Typhoon 9400; Amersham Biosciences).
Electrophoretic Mobility Shift Assay
Mobility shift analysis was carried out essentially as described (59). The reaction mixtures in 1× UV cross-linking buffer (5× cross-link buffer, 5 mg/ml heparin, 1% glycerol, 50 mm KCl, 10 mm dithiothreitol, 5 mm HEPES, pH 7.8, 1 mm MgCl2, 0.1 mm EDTA, 40 μg/ml yeast tRNA), contained recombinant protein and Alexa-labeled RNA, as well as various amounts of competitor RNA. After incubation at room temperature for 30 min, an equal volume of loading buffer (50% glycerol, 0.1 mm EDTA, and 0.01% bromphenol blue) was added and loaded on a 1% agarose/Tris/acetic acid/EDTA buffer gel. The gel was run in 1× Tris/acetic acid/EDTA buffer at 40 V for 1.5 h and analyzed by phosphorimaging.
Vegetal Localization Assay
Localization assays were performed essentially as described (11). In brief, capped RNAs (5 nl of 10 ng/μl) were injected into stage III and IV oocytes and maintained in culture for 4 or 5 days in a vitellogenin-containing L15 medium as described in Refs. 60 and 61. For the morpholino co-injection experiments mixtures of 0.3 fmol of lacZ-tagged RNAs and 50 fmol of morpholino oligonucleotides were injected into stage III and IV oocytes and cultured for 3 or 4 days. RNA distribution was visualized by whole mount in situ hybridization as described (62, 63), using digoxygenin-labeled antisense lacZ RNA.
Nucleocytoplasmic Transport
Nucleocytoplasmic transport assays were carried out as described (64). In brief, 35S-labeled Myc-tagged ElrA and ElrB proteins were in vitro translated using the TnT system (Promega) and [35S]methionine (Hartmann Analytic), with pCS2+MT-ElrA and ElrB serving as templates. The 35S-labeled proteins were injected in either nucleus or cytoplasm of stage VI oocytes. MT-ElrA and B were immunoprecipitated from manually dissected oocytes, separated by SDS-PAGE, and analyzed by phosphorimaging.
RESULTS
Identification Xenopus ElrA and ElrB as Vegetal Localization Element-binding Proteins
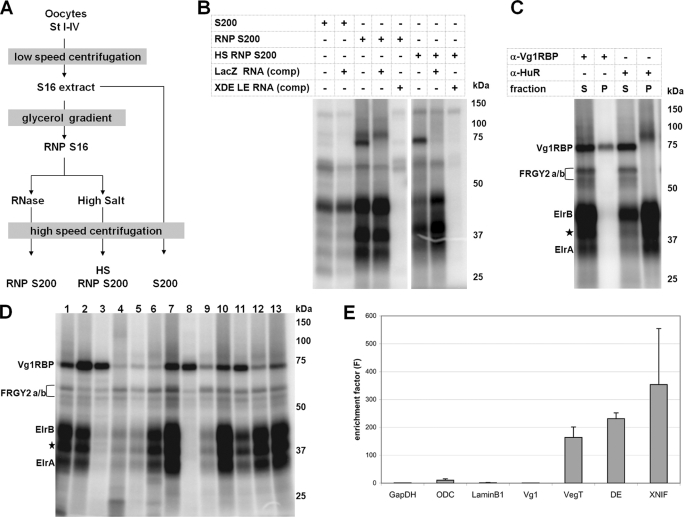
In an effort to isolate novel proteins that function in vegetal mRNA localization in Xenopus oocytes, we established a biochemical purification strategy to generate protein fractions enriched in components of the localization complex (Fig. 1 A). In brief, total extracts from early/mid-stage (stage I–IV) Xenopus oocytes were fractionated by use of a glycerol gradient, and the fractions containing known localization proteins, such as Vg1RBP and XStaufen 1, were collected (supplemental Fig. S1). After disruption of the RNPs by either RNase digestion or high salt treatment, high speed centrifugation yielded preparations highly enriched in RNA-binding proteins (RNP S200 and HS RNP S200), as revealed by UV cross-linking to the XDead end-LE (XDE-LE) (Fig. 1B). In particular, these experiments reveal enrichment of a group of low molecular mass proteins in the range of 35–45 kDa. Individual proteins from these enriched extracts were identified by mass spectrometry analysis after two-dimensional gel electrophoretic separation (supplemental Fig. S2) and found to contain the known vegetal localization proteins Vg1RBP, XStaufen 1, and 40LoVe (supplemental Table S1). Candidate LE-interacting proteins in the range of 35–45 kDa include the Xenopus homolog of the human RNA-binding protein HuB, ElrB (41) (supplemental Fig. S2 and Table S1). To test for a direct interaction of Elr-type proteins with the XDead end-LE, immunoprecipitation experiments from cross-linking reactions using a monoclonal anti-HuR antibody, which detects both forms of oocyte Elr-type proteins (44), were performed. Radiolabeled proteins corresponding to ElrA and ElrB were specifically precipitated (Fig. 1C). Cross-linking of Vg1RBP to the XDE-LE was verified using the same type of assay.
FIGURE 1.
Identification of Xenopus Elr-type proteins as vegetal localization element-binding proteins. A, oocyte fractions enriched for proteins of the vegetal localization complex were prepared as illustrated. B, UV cross-linking using 32P-labeled XDE-LE RNA was used to monitor the enrichment of LE-binding proteins in the different protein preparations. Unlabeled XDE-LE and LacZ RNAs served as specific and nonspecific competitors, respectively. C, immunoprecipitation of radiolabeled Vg1RBP and Elr-type proteins from UV cross-linking reactions. The proteins were precipitated from the RNP S200/[32P]XDE-LE cross-linking reaction by using α-Vg1RBP and α-HuR antibodies. Supernatant (S) and bound (P) fractions were analyzed by SDS-PAGE. Proteins that might correspond to putative ElrB isoforms are marked by stars. D, comparative UV cross-linking of RNP S200 and 32P-labeled XDEADSouth-LE (lane 1), XGrip2-LE (lane 2), XNIF-LE (lane 3), Velo40-LE (lane 4), Velo45-LE (lane 5), XDE-LE (lane 6), Velo76-LE (lane 7), XVelo1-LE (lane 8), XVelo1–3′ UTR (lane 9), Velo7-LE (lane 10), Vg1-LE (lane 11), Vg1-TE (lane 12), and VegT-LE (lane 13) RNAs. E, enrichment of localizing RNAs in ElrA/B-containing RNPs. Immunoprecipitations from stage II-IV oocyte extracts were performed by using either α-HuR or α-Myc control antibodies. Co-precipitated RNAs were detected by quantitative reverse transcription-PCR; relative RNA enrichment in the IP-fractions versus the input and control IP-fractions is depicted. Average enrichment factors of three independent Co-IP experiments are shown. The error bars indicate standard deviation.
Binding of Elr-type proteins could be a specific property of the XDE localization element but also a more general property of LEs in vegetally localizing mRNAs. To address this possibility, a comparative UV cross-linking assay making use of a collection of different LEs was performed (Fig. 1D). These LEs contained previously known as well as a set of novel LEs that were mapped in our laboratory.4 Of 11 different LEs belonging to either the early, the intermediate or late localizing RNAs, seven were found to cross-link with high efficiency and two additional ones with low efficiency to the Elr-type proteins, whereas only two (XNIF and Velo1 LEs) exhibit no or hardly any such binding activity in this assay (Fig. 1D, lanes 3 and 8). We notice that these latter two LEs cross-link efficiently with Vg1RBP. Furthermore, when tagged versions of ElrA and ElrB proteins were overexpressed in stage VI oocytes, they were found to exhibit similar binding specificities (data not shown). In previous studies, Xenopus ElrB has been described to act as translational repressor, specifically interacting with the Vg1 translational control element (VTE), whereas no or only weak interaction with the Vg1 LE was observed (44). Our experiments reveal relatively weak but significant binding of Elr-type proteins to the Vg1-LE upon use of extracts heavily enriched in ElrA/B proteins (Fig. 1D, lanes 11 and 12).
To assess whether ElrA/B proteins associate preferentially with localized mRNAs, RNA co-immunoprecipitation experiments with stage II-IV oocyte extracts using an α-HuR antibody were performed. The enrichment of certain mRNAs in such ElrA/B-RNPs relative to the overall abundance of the RNA was determined as enrichment factor (see “Experimental Procedures” for details). Late localizing mRNAs, such as VegT and XDead end, as well as the early localizing XNIF mRNA, are indeed found to be heavily enriched in ElrA/B-RNPs, as compared with nonlocalizing mRNAs such as glyceraldehyde-3-phosphate dehydrogenase, ornithine decarboxylase, and Lamin B1 (Fig. 1E). However, although robust interactions of Vg1-LE with ElrA/B proteins were detected in the UV cross-linking assay, we were not able to detect a particular enrichment of Vg1 mRNA in ElrA/B RNPs. In this particular case, RNPs forming in vivo on the endogenous Vg1 mRNA could differ from those forming in vitro with ElrA/B-enriched protein fractions.
ElrA/B Proteins Are Part of One RNP Together with Vg1RBP
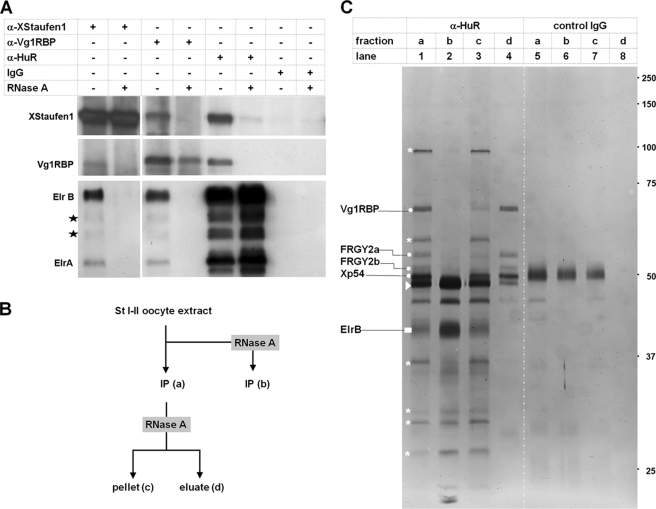
Co-purification of ElrA/B with Vg1RBP and XStaufen 1 by use of the biochemical strategy outlined above and the ability of such proteins to bind directly to different LEs does not necessarily imply that these proteins are part of one and the same RNP. To address this question, co-immunoprecipitation experiments were carried out (Fig. 2A). In full agreement with earlier studies (32), antibodies directed against XStaufen 1 co-precipitate Vg1RBP and vice versa. The same antibodies also pull down ElrA/B proteins, indicating that Vg1RBP, XStaufen 1, and Elr-type proteins are part of one RNP. Similarly, antibodies directed against Elr-type proteins also precipitate XStaufen 1 and Vg1RBP, providing further support for the same notion. Formation of these complexes is lost upon RNase treatment prior to protein isolation, indicating that these three different proteins are held together via an RNA scaffold (Fig. 2A).
FIGURE 2.
ElrA and B proteins interact with Vg1RBP and XStaufen 1 in an RNA-dependent manner. A, co-immunoprecipitations using α-XStaufen 1, α-Vg1RBP, α-HuR, and control IgGs were performed in the absence or presence of RNase A. Co-precipitated proteins were detected by Western blot. Proteins that might correspond to putative ElrB isoforms are marked by stars. B, work flow for the identification of proteins co-precipitating with Elr-type proteins (α-HuR) from stage I and II S16 extracts. C, protein fractions generated as depicted in B were separated by SDS-PAGE and detected by silver staining. The dots indicate proteins that co-precipitate in an RNA-dependent manner and were identified by mass spectrometry (see supplemental Fig. S3 and Table S2 in the supplemental material). Proteins that co-precipitate in an RNA-independent manner (fraction c) are marked by stars. Proteins corresponding to IgG heavy chains are marked by arrowheads.
In an attempt to identify other proteins that might be part of the ElrA/B containing RNPs, co-immunoprecipitation experiments were performed on a preparative scale, followed by release of ElrA/B-associated proteins via RNase treatment and identification by mass spectrometry (Fig. 2, B and C, as well as supplemental Fig. S3 and Table S2). Again, Vg1RBP was found to co-purify specifically with ElrB (which is the major Elr-type protein in the oocyte extracts used; see also Fig. 2A), as well as with the unspecific FRGY-type RNA-binding proteins. However, XStaufen 1, as demonstrated above by Western blot analysis to co-precipitate with Elr-type proteins (Fig. 2A), was not among the proteins identified in this second approach, indicating that it is either only present in substoichiometric amounts in ElrA/B containing RNPs or, alternatively, that there exist different classes of ElrA/B-RNPs, those with and those without XStaufen 1. Smaller amounts of other proteins eluted by RNase treatment from the Elr protein containing RNPs included the RNA-helicase Xp54 (65), as well as two previously unknown proteins related to the Lsm domain protein RAP55 (66), which were therefore termed RAP42 and RAP46 (see supplemental Table S2 and protein alignment in supplemental Fig. S4). Several additional proteins were not released by RNase treatment and therefore found in the pellet fraction together with significant amounts of ElrB (Fig. 2C, lane 3); the identity of these proteins remains to be established.
Inhibition of ElrA/B Correlates with a Loss of Vegetal mRNA Transport
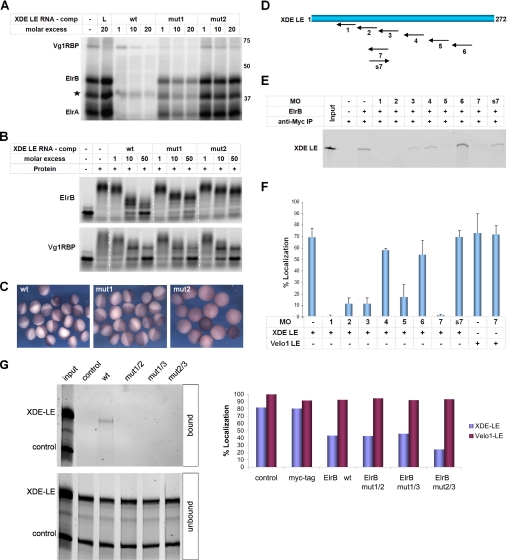
The experiments described above establish that the ElrA/B proteins bind specifically to the XDE-LE as well as to several other LEs and that they are part of one RNP together with other proteins known to be involved in vegetal transport. To address the functional role of Elr-type proteins in this latter process more directly, mutant versions of the XDE-LE were generated that are impaired in their ability to bind to these proteins. Because Elr-type proteins are known to bind A/U-rich regions, two mutant versions of XDE-LE, containing different sets of point mutations in several of its A/U-rich elements, were generated (supplemental Fig. S5). These mutants were first analyzed in respect to their ability to compete for binding of ElrA/B to the wild type version of XDE-LE in a UV cross-linking assay (Fig. 3A), as well as in an electrophoretic mobility shift assay making use of bacterially expressed versions of ElrB and Vg1RBP (Fig. 3B). Although mut1 is only moderately reduced in its ability to compete for ElrA/B binding, mut2 is severely impaired to do so. Interestingly, binding to Vg1RBP seems to be equally competed by all of the competitor RNAs tested, indicating that the mutations introduced do not perturb binding to this localization protein (Fig. 3, A and B).
FIGURE 3.
Interference with ElrA/B-binding blocks vegetal localization of the XDE-LE. A, UV cross-linking was performed with RNP S200 and 32P-labeled XDE-LE wt in the absence or presence of either nonspecific lacZ (L) or increasing amounts of XDE-LE wt, XDE-LE mut1 and XDE-LE mut2 competitor RNAs. A putative ElrB isoform is marked by an star. B, electrophoretic mobility shift assay using 1 pmol of Alexa-labeled XDE-LE wt and 10 or 25 pmol of recombinant ElrB and Vg1RBP proteins, respectively. RNA-protein complexes were allowed to assemble either in the absence or presence of increasing amounts of XDE-LE wt, XDE-LE mut1, or XDE-LE mut2 competitor RNAs. C, LacZ-tagged XDE-LE wt and mutant RNAs were injected into stage IV oocytes and analyzed for subcellular localization by in situ-hybridization after 4 or 5 days. The localization efficiencies, standard deviations, and numbers of oocytes analyzed were as follows: wt (92% ± 8.7; n = 177), mut1 (95% ± 8.6; n = 119), and mut2 (0%; n = 170). D, schematic representation of the target region for the different morpholinos in the XDE-LE. E, Alexa-labeled XDE-LE RNA was co-immunoprecipitated along with Myc-tagged, in vitro translated ElrB protein in the absence or presence of the different morpholino oligonucleotides. F, average localization efficiencies of XDE-LE RNA alone (0.3 fmol) and co-injected with different MOs (50 fmol) of three independent injection experiments are indicated (see also supplemental Fig. S6). XVelo1-LE does not contain a MO7 target site and served as a specificity control. G, Cy3-labeled lacZ-tag-XDE-LE RNA and lacZ tag (control) RNAs were co-immunoprecipitated along with Myc-tagged, in vitro translated wt ElrB protein as well as ElrB RRM domain point mutants. Average localization efficiencies of XDE-LE and Velo1-LE RNAs in control oocytes and oocytes preinjected with RNAs encoding for Myc tag, wt, and mutant versions of ElrB from two independent experiments are indicated.
The same XDE-LE mutants were then employed in oocyte microinjection assays testing for vegetal transport activity (Fig. 3C). Although transport efficiency of a reporter RNA fused to mut1 was comparable with wt, mut2 was not able to mediate localization to the vegetal cortex of the oocytes.
In an alternative experimental approach, a set of antisense morpholino oligonucleotides was designed that target different regions of the XDE-LE (Fig. 3D) and tested for their ability to compete for ElrB binding to XDE-LE in co-precipitation experiments on the one hand (Fig. 3E) and to interfere with vegetal RNA localization in the oocyte on the other (Fig. 3F and supplemental Fig. S6). As above, inhibition of ElrB binding correlated well with an inhibition of vegetal localization.
Finally, we have also assayed for the effects of overexpressing mutant versions of ElrB that are deficient in RNA binding because of point mutations in RRM1 and 2 (mut1/2), RRM1 and 3 (mut1/3), or RRM2 and 3 (mut2/3) (Fig. 3G and supplemental Fig. S7). All three mutants tested had a dominant negative effect on vegetal localization of an ElrB-binding RNA substrate (XDE-LE), whereas localization via the Velo1-LE, which does not bind to Elr-type proteins, was not affected. Interestingly, overexpression of an epitope-tagged wt version of ElrB also exerted an inhibitory effect, indicating that excess ElrB might sequester other localization factors by protein-protein interactions without forming a complex on a target RNA. Interestingly, ElrB has been reported to oligomerize in a concentration- and RNA-dependent manner (67); although the in vivo relevance of this effect is not understood, it may counteract formation of transport-competent complexes in the oocyte. Taken together, these different experiments provide strong indications for Elr-type proteins to be actively involved in the events that allow for RNA localization to the vegetal cortex in Xenopus oocytes.
Elr-type Proteins Are Enriched at the Vegetal Cortex of Xenopus Oocytes
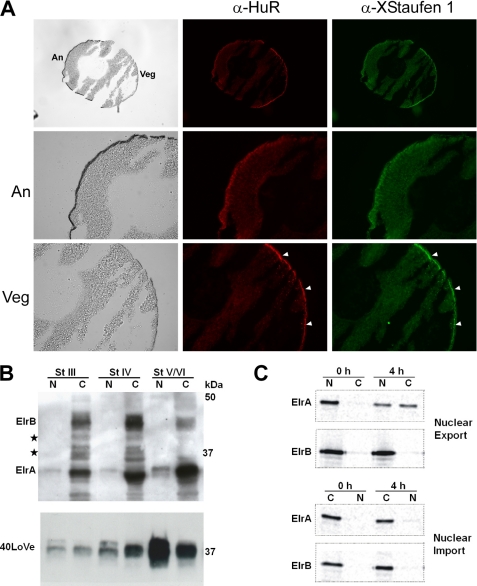
It has been shown that proteins with a function in vegetal mRNA localization, such as Vg1RBP (35), XStaufen 1 (15), hnRNP I (32), and 40LoVe (27), exhibit vegetal cortex enrichment reminiscent of the distribution of their target mRNAs. Assuming a functional role for ElrA and B in vegetal mRNA localization, we performed co-immunostaining experiments on cryosections of stage III and IV oocytes to examine the subcellular distribution of these proteins (Fig. 4A). ElrA/B staining is detected in the cytoplasm with vegetal cortex enrichment in sections from both stage IV and stage III oocytes (Fig. 4A and data not shown). Staining for XStaufen 1 served as a positive control and revealed co-localization for ElrA/B and XStaufen 1 at the vegetal cortex (Fig. 4A, arrowheads mark vegetal cortex co-localization of ElrA/B and XStaufen 1). Similar results were observed for co-immunostaining using a α40LoVe antibody (data not shown). In addition, co-localization was also observed in discrete particle-like structures in the vegetal cytoplasm, which may correspond to RNA transport particles in the process of localization.
FIGURE 4.
Subcellular distribution of ElrA and B in oocytes. A, co-immunostaining on cryo sections of a stage IV oocyte using α-HuR and α-XStaufen 1 antibodies. Animal (An) and vegetal (Veg) hemispheres are marked in the brightfield image (upper left). α-HuR and α-XStaufen 1 stainings are shown in red and green, respectively. Magnifications of animal and vegetal halves of the oocyte are shown below. Arrowheads mark vegetal cortex enrichment of ElrA/B and XStaufen 1, respectively. B, nuclei and cytoplasmatic fractions of stage III–VI oocytes were manually dissected and ElrA/B and 40LoVe proteins detected by Western blotting. Proteins that might correspond to putative ElrB isoforms are marked by stars. C, nucleocytoplasmic transport assay with ElrA and B. 35S-Labeled, Myc-tagged versions of the proteins were injected into the cytoplasms or nuclei of stage VI oocytes. These were dissected either directly or 4 h after injection. Myc-tagged proteins were immunoprecipitated from these fractions, separated by SDS-PAGE, and detected by phosphorimaging.
Analysis of manually dissected oocyte nuclei and cytoplasm reveals a predominantly cytoplasmic localization of ElrA/B proteins with very low levels of ElrA, but not ElrB, in the nuclear fractions (Fig. 4B). Anti-40LoVe immunoblots served as a loading control and detect the protein in the nucleus and cytoplasm. However, there could be dynamic ElrA/B protein exchange between the oocyte nucleus and cytoplasm, as shown for HuR in somatic cells (68). To test for a possible shuttling of ElrA and B between nucleus and cytoplasm, microinjection experiments were performed using Myc-tagged versions of ElrA and B (Fig. 4C). Myc-ElrA was found to be readily exported into the cytoplasm upon injection into the nucleus, whereas ElrB remained almost entirely in the nucleus under the same conditions. Conversely, when both proteins were injected into the cytoplasm, neither protein was found to accumulate the nucleus.
Taken together, these results indicate that Elr-type proteins co-localize with the known vegetal localization proteins XStaufen 1 and 40LoVe at the vegetal cortex. Furthermore, the rapid export of ElrA from nucleus to cytoplasm is compatible with the idea of an early function for this protein in the transport pathway that leads from the nucleus to the vegetal cortex in Xenopus oocytes.
DISCUSSION
Data reported in this communication reveal that Xenopus Elr-type proteins, homologs of Hu/ELAV proteins, bind specifically to the XDead end-LE, as well as to a number of other Xenopus localization elements. ElrA/B proteins co-fractionate in an RNA-dependent manner along with known localization proteins such as Vg1RBP and XStaufen 1 on glycerol gradients, and co-precipitation analysis indicates that that they assemble into the same RNP. Interference with ElrA/B binding by either LE-mutagenesis or co-injection of antisense morpholino oligonucleotides, as well as results obtained from overexpression of wt or mutant versions of ElrB, provide strong evidence that ElrA/B binding is indeed crucial for the vegetal localization of the XDE-LE RNA. UV cross-linking as well as co-immunoprecipitation analysis indicates that ElrA/B proteins might also function in the localization of other vegetally localized RNAs. However, ElrA/B binding does not seem to reflect a general requirement for vegetal localization, because the LE of the late localizing RNA Xvelo1 does not exhibit binding activities for these proteins and is not affected by overexpression of dominant negative ElrB mutants. It thus seems possible that localizing RNAs might assemble into different localization RNPs, either containing or lacking ElrA/B proteins.
It is of interest to define how exactly ElrA/B proteins function during vegetal RNA localization; because a small quantity of ElrA can be detected in the oocyte nuclei, it might assemble with the localization RNP already in the nucleus. Nucleocytoplasmic transport studies indicate that Xenopus ElrA is rapidly exported to the cytoplasm after nuclear injection, which is probably mediated by the conserved HNS (HuR nuclear shuttling sequence) that is located in the hinge region separating RRM2 and 3 and which has previously been shown to mediate nuclear shuttling of HuR (69). Because ElrB, similar to the strictly neuronal Hu family members HuC and HuD, exhibits an insertion in the hinge region interfering with the HNS function, it seems likely that ElrB enters the localization RNP after export to the cytoplasm. Whether ElrA and B differ in respect to their functional role during vegetal transport remains to be determined. Co-immunoprecipitation data revealed an RNA-dependent interaction with Vg1RBP, 40LoVe, and XStaufen 1, indicating that ElrA/B are part of localization particles during their migration in the cytoplasm. Xenopus ElrB has been reported to oligomerize on its target RNAs (67), suggesting that ElrB might function as an assembly factor for the formation of large RNA transport granules that may also contain multiple localized transcripts. Immunostaining for ElrA/B, XStaufen 1, and 40LoVe revealed co-localization in large particles in the vegetal cytoplasm, which may correspond to such transport granules. In addition, these transport granules might also mediate translational repression of the localizing RNA, either by sequestering the RNAs in silencing complexes that are not accessible for the translation machinery and/or by recruitment of translational repressors into the transport granule, as has been shown for the Bruno-mediated translational repression of oskar mRNA particles e.g. (70). Co-immunoprecipitation of the RNA-helicase Xp54 and Rap55-related Lsm domain proteins RAP42 and RAP46 along with ElrA/B indicates that ElrA/B containing localization RNPs might indeed be transported in a translationally repressed state. Xenopus Xp54 has been described as a component of stored mRNPs that also represses translation of reporter RNAs in the MS2-tethered function assay (65, 71), and the Drosophila Xp54 homolog Me31b has been shown to participate in translational repression of the posterior localizing RNA oskar (72). Similarly, the Lsm domain protein Rap55 has been reported to be part of translationally repressed mRNP complexes in Xenopus oocytes and acts a translational repressor in vitro as well as in oocytes if tethered to a reporter RNA (Ref. 66; reviewed in Ref. 73). In addition to a role during the transport process, co-localization with XStaufen 1 and 40LoVe at the vegetal cortex suggests that ElrA/B might also function in anchoring the RNP complex after transport has been completed.
Hu/ELAV RNA-binding proteins function in various aspects of RNA metabolism, including splicing and nuclear export, as well as regulation of mRNA stability and translation (reviewed in Refs. 42, 74, and 75). Hu/ELAV proteins are known to specifically target localized RNAs in other systems, although no indications for a direct function of Hu/ELAV proteins in RNA localization have been described to date. In particular, HuR was reported to bind to the 3′-UTR of β-actin mRNA, which localizes to the leading edge of migrating cells, a process that also involves the Vg1RBP homolog ZBP1 (76, 77). It was demonstrated that depletion of HuR results in a reduced migratory capacity of such cells, perhaps as a consequence of reduced β-actin mRNA stability (76). In neurons, the Vg1RBP homolog IMP1, together with HuD, is found to exert a repressing effect on the translation of Tau mRNA 3′-UTR reporters (78). Tau 3′-UTR not only regulates translation but also contains the axonal localization signal that mediates mRNA localization to the axon in neuronal cells (79). Although, in these cases, Hu/ELAV proteins are reported to control the stability and translation of localized RNAs, our study provides evidence for a direct function of Xenopus ElrA/B proteins during vegetal transport in oocytes.
Binding of Elr-type proteins to the AU-rich translational control element in the 3′-UTR of the vegetally localizing mRNA Vg1 (VTE) was reported to exert a repressing effect on translation (44). However, although we could reproduce the translational repressor function of the ElrA/B-binding VTE, we did not observe a strict correlation between ElrA/B binding activity and translational repression or transcript stability in the context of diverse vegetal localization elements,. In particular, luciferase reporter assays revealed that wild type XDE-LE compared with mut2, lacking ElrA/B-binding sites, did not mediate a repressive effect but rather a slight stimulation of translation (data not shown). Thus, it seems likely, that the diverse functions of Elr-type proteins in Xenopus and those of their homologs in other biological systems are context-dependent and are modulated by additional factors, such as Vg1RBP, hnRNP I, and XStaufen 1 in the process of RNA localization, and so-far unknown factors mediating the translational repression of VTE-containing RNAs. This dependence on co-factors could also explain why Xenopus ElrB protein alone is not able to mediate translational repression if tethered to a MS2 stem-loop containing luciferase reporter RNA by the viral MS2 coat protein.5 To unravel the diverse functions of Elr-type/Hu proteins in RNA metabolism in more detail, it will be of crucial future interest to define the composition and structural arrangement of the different functional RNPs that contain these proteins.
Supplementary Material
Acknowledgments
We thank J. Yisraeli for the α-Vg1RBP antibody, N. Standart for the α-XStaufen 1 antibody, K. Czaplinski and I. W. Mattaj for the α-40LoVe antibody, S. Hüttelmaier and N. Stöhr for help with the calculation of RNA enrichment factors, C. Viebahn and colleagues for cryosectioning of oocytes, and I. Eckhardt for excellent technical assistance.
This work was supported by funds from the Deutsche Forsch ungs ge mein schaft Grant SFB523 (to T. P.) and by the research program of the Faculty of Medicine, Georg-August-University Göttingen (to M. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
S. Koch and T. Pieler, unpublished observations.
M. Claussen and T. Pieler, unpublished results.
- UTR
- untranslated region
- LE
- localization element
- PBS
- phosphate-buffered saline
- IP
- immunoprecipitation
- wt
- wild type
- RRM
- RNA recognition motif
- VTE
- Vg1 translational element.
REFERENCES
- 1.Zhou Y., King M. L. (2004) IUBMB Life 56, 19–27 [DOI] [PubMed] [Google Scholar]
- 2.Forristall C., Pondel M., Chen L., King M. L. (1995) Development 121, 201–208 [DOI] [PubMed] [Google Scholar]
- 3.Kloc M., Etkin L. D. (1995) Development 121, 287–297 [DOI] [PubMed] [Google Scholar]
- 4.Mosquera L., Forristall C., Zhou Y., King M. L. (1993) Development 117, 377–386 [DOI] [PubMed] [Google Scholar]
- 5.Chang P., Torres J., Lewis R. A., Mowry K. L., Houliston E., King M. L. (2004) Mol. Biol. Cell 15, 4669–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebagliati M. R., Weeks D. L., Harvey R. P., Melton D. A. (1985) Cell 42, 769–777 [DOI] [PubMed] [Google Scholar]
- 7.Weeks D. L., Melton D. A. (1987) Cell 51, 861–867 [DOI] [PubMed] [Google Scholar]
- 8.Lustig K. D., Kroll K. L., Sun E. E., Kirschner M. W. (1996) Development 122, 4001–4012 [DOI] [PubMed] [Google Scholar]
- 9.Stennard F., Carnac G., Gurdon J. B. (1996) Development 122, 4179–4188 [DOI] [PubMed] [Google Scholar]
- 10.Zhang J., King M. L. (1996) Development 122, 4119–4129 [DOI] [PubMed] [Google Scholar]
- 11.Claussen M., Pieler T. (2004) Dev. Biol. 266, 270–284 [DOI] [PubMed] [Google Scholar]
- 12.Betley J. N., Heinrich B., Vernos I., Sardet C., Prodon F., Deshler J. O. (2004) Curr. Biol. 14, 219–224 [DOI] [PubMed] [Google Scholar]
- 13.Messitt T. J., Gagnon J. A., Kreiling J. A., Pratt C. A., Yoon Y. J., Mowry K. L. (2008) Dev. Cell 15, 426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yisraeli J. K., Sokol S., Melton D. A. (1990) Development 108, 289–298 [DOI] [PubMed] [Google Scholar]
- 15.Yoon Y. J., Mowry K. L. (2004) Development 131, 3035–3045 [DOI] [PubMed] [Google Scholar]
- 16.White J. A., Heasman J. (2008) J. Exp. Zoolog. B. Mol. Dev. Evol. 310, 73–84 [DOI] [PubMed] [Google Scholar]
- 17.Chan A. P., Kloc M., Etkin L. D. (1999) Development 126, 4943–4953 [DOI] [PubMed] [Google Scholar]
- 18.Pannese M., Cagliani R., Pardini C. L., Boncinelli E. (2000) Mech. Dev. 90, 111–114 [DOI] [PubMed] [Google Scholar]
- 19.Zearfoss N. R., Chan A. P., Wu C. F., Kloc M., Etkin L. D. (2004) Dev. Biol. 267, 60–71 [DOI] [PubMed] [Google Scholar]
- 20.Jambhekar A., Derisi J. L. (2007) RNA 13, 625–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King M. L., Messitt T. J., Mowry K. L. (2005) Biol. Cell 97, 19–33 [DOI] [PubMed] [Google Scholar]
- 22.Havin L., Git A., Elisha Z., Oberman F., Yaniv K., Schwartz S. P., Standart N., Yisraeli J. K. (1998) Genes Dev. 12, 1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshler J. O., Highett M. I., Abramson T., Schnapp B. J. (1998) Curr. Biol. 8, 489–496 [DOI] [PubMed] [Google Scholar]
- 24.Cote C. A., Gautreau D., Denegre J. M., Kress T. L., Terry N. A., Mowry K. L. (1999) Mol. Cell 4, 431–437 [DOI] [PubMed] [Google Scholar]
- 25.Zhao W. M., Jiang C., Kroll T. T., Huber P. W. (2001) EMBO J. 20, 2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroll T. T., Zhao W. M., Jiang C., Huber P. W. (2002) Development 129, 5609–5619 [DOI] [PubMed] [Google Scholar]
- 27.Czaplinski K., Köcher T., Schelder M., Segref A., Wilm M., Mattaj I. W. (2005) Dev. Cell 8, 505–515 [DOI] [PubMed] [Google Scholar]
- 28.Claussen M., Horvay K., Pieler T. (2004) Development 131, 4263–4273 [DOI] [PubMed] [Google Scholar]
- 29.Hudson C., Woodland H. R. (1998) Mech. Dev. 73, 159–168 [DOI] [PubMed] [Google Scholar]
- 30.Allen L., Kloc M., Etkin L. D. (2003) Differentiation 71, 311–321 [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y., King M. L. (1996) Dev. Biol. 179, 173–183 [DOI] [PubMed] [Google Scholar]
- 32.Kress T. L., Yoon Y. J., Mowry K. L. (2004) J. Cell Biol. 165, 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis R. A., Gagnon J. A., Mowry K. L. (2008) Mol. Cell. Biol. 28, 678–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allison R., Czaplinski K., Git A., Adegbenro E., Stennard F., Houliston E., Standart N. (2004) RNA 10, 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q., Yaniv K., Oberman F., Wolke U., Git A., Fromer M., Taylor W. L., Meyer D., Standart N., Raz E., Yisraeli J. K. (1999) Mech. Dev. 88, 101–106 [DOI] [PubMed] [Google Scholar]
- 36.Kolev N. G., Huber P. W. (2003) Mol. Cell 11, 745–755 [DOI] [PubMed] [Google Scholar]
- 37.Alarcón V. B., Elinson R. P. (2001) J. Cell Sci. 114, 1731–1741 [DOI] [PubMed] [Google Scholar]
- 38.Heasman J., Wessely O., Langland R., Craig E. J., Kessler D. S. (2001) Dev. Biol. 240, 377–386 [DOI] [PubMed] [Google Scholar]
- 39.Kloc M., Etkin L. D. (1994) Science 265, 1101–1103 [DOI] [PubMed] [Google Scholar]
- 40.Kloc M., Wilk K., Vargas D., Shirato Y., Bilinski S., Etkin L. D. (2005) Development 132, 3445–3457 [DOI] [PubMed] [Google Scholar]
- 41.Good P. J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4557–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinman M. N., Lou H. (2008) Cell Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006) Cell 125, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 44.Colegrove-Otero L. J., Devaux A., Standart N. (2005) Mol. Cell. Biol. 25, 9028–9039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvay K., Claussen M., Katzer M., Landgrebe J., Pieler T. (2006) Dev. Biol. 291, 1–11 [DOI] [PubMed] [Google Scholar]
- 46.Rupp R. A., Snider L., Weintraub H. (1994) Genes Dev. 8, 1311–1323 [DOI] [PubMed] [Google Scholar]
- 47.Koebernick K., Hollemann T., Pieler T. (2003) Dev. Biol. 260, 325–338 [DOI] [PubMed] [Google Scholar]
- 48.Git A., Standart N. (2002) RNA 8, 1319–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 50.Lisbin M. J., Gordon M., Yannoni Y. M., White K. (2000) Genetics 155, 1789–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans J. P., Kay B. K. (1991) Methods Cell Biol. 36, 133–148 [DOI] [PubMed] [Google Scholar]
- 52.Abe S., Davies E. (1986) in Memoirs of the College of Agriculture, Vol. 31, pp. 187–199, Ehime University, Ehime, Japan [Google Scholar]
- 53.Görg A., Weiss W., Dunn M. J. (2004) Proteomics 4, 3665–3685 [DOI] [PubMed] [Google Scholar]
- 54.Jahn O., Hesse D., Reinelt M., Kratzin H. D. (2006) Anal. Bioanal. Chem. 386, 92–103 [DOI] [PubMed] [Google Scholar]
- 55.Werner H. B., Kuhlmann K., Shen S., Uecker M., Schardt A., Dimova K., Orfaniotou F., Dhaunchak A., Brinkmann B. G., Möbius W., Guarente L., Casaccia-Bonnefil P., Jahn O., Nave K. A. (2007) J. Neurosci. 27, 7717–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reumann S., Babujee L., Ma C., Wienkoop S., Siemsen T., Antonicelli G. E., Rasche N., Lüder F., Weckwerth W., Jahn O. (2007) Plant Cell 19, 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 58.Mowry K. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14608–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung S., Jiang L., Cheng S., Furneaux H. (1996) J. Biol. Chem. 271, 11518–11524 [DOI] [PubMed] [Google Scholar]
- 60.Yisraeli J. K., Melton D. A. (1988) Nature 336, 592–595 [DOI] [PubMed] [Google Scholar]
- 61.Kloc M., Etkin L. D. (1999) in A Comparative Methods Approach to the Study of Oocytes and Embryos (Richter J. D. ed) pp. 256–278, Oxford University Press Inc., Oxford, UK [Google Scholar]
- 62.Harland R. M. (1991) Methods Cell Biol. 36, 685–695 [DOI] [PubMed] [Google Scholar]
- 63.Hollemann T., Panitz F., Pieler T. (1999) in A Comparative Methods Approach to the Study of Oocytes and Embryos (Richter J. D. ed) pp. 279–290, Oxford University Press Inc., Oxford, UK [Google Scholar]
- 64.Rudt F., Pieler T. (1996) EMBO J. 15, 1383–1391 [PMC free article] [PubMed] [Google Scholar]
- 65.Ladomery M., Wade E., Sommerville J. (1997) Nucleic Acids Res. 25, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka K. J., Ogawa K., Takagi M., Imamoto N., Matsumoto K., Tsujimoto M. (2006) J. Biol. Chem. 281, 40096–40106 [DOI] [PubMed] [Google Scholar]
- 67.Devaux A., Colegrove-Otero L. J., Standart N. (2006) FEBS Lett. 580, 4947–4952 [DOI] [PubMed] [Google Scholar]
- 68.Atasoy U., Watson J., Patel D., Keene J. D. (1998) J. Cell Sci. 111, 3145–3156 [DOI] [PubMed] [Google Scholar]
- 69.Fan X. C., Steitz J. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chekulaeva M., Hentze M. W., Ephrussi A. (2006) Cell 124, 521–533 [DOI] [PubMed] [Google Scholar]
- 71.Minshall N., Standart N. (2004) Nucleic Acids Res. 32, 1325–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakamura A., Amikura R., Hanyu K., Kobayashi S. (2001) Development 128, 3233–3242 [DOI] [PubMed] [Google Scholar]
- 73.Marnef A., Sommerville J., Ladomery M. R. (2009) Int. J. Biochem. Cell Biol. 41, 977–981 [DOI] [PubMed] [Google Scholar]
- 74.Brennan C. M., Steitz J. A. (2001) Cell Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorospe M. (2003) Cell Cycle 2, 412–414 [PubMed] [Google Scholar]
- 76.Dormoy-Raclet V., Ménard I., Clair E., Kurban G., Mazroui R., Di Marco S., von Roretz C., Pause A., Gallouzi I. E. (2007) Mol. Cell. Biol. 27, 5365–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Condeelis J., Singer R. H. (2005) Biol. Cell 97, 97–110 [DOI] [PubMed] [Google Scholar]
- 78.Atlas R., Behar L., Sapoznik S., Ginzburg I. (2007) J. Neurosci. Res. 85, 173–183 [DOI] [PubMed] [Google Scholar]
- 79.Aronov S., Aranda G., Behar L., Ginzburg I. (2001) J. Neurosci. 21, 6577–6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.