Abstract
Clinically, amniotic membrane (AM) suppresses inflammation, scarring, and angiogenesis. AM contains abundant hyaluronan (HA) but its function in exerting these therapeutic actions remains unclear. Herein, AM was extracted sequentially with buffers A, B, and C, or separately by phosphate-buffered saline (PBS) alone. Agarose gel electrophoresis showed that high molecular weight (HMW) HA (an average of ∼3000 kDa) was predominantly extracted in isotonic Extract A (70.1 ± 6.0%) and PBS (37.7 ± 3.2%). Western blot analysis of these extracts with hyaluronidase digestion or NaOH treatment revealed that HMW HA was covalently linked with the heavy chains (HCs) of inter-α-inhibitor (IαI) via a NaOH-sensitive bond, likely transferred by the tumor necrosis factor-α stimulated gene-6 protein (TSG-6). This HC·HA complex (nHC·HA) could be purified from Extract PBS by two rounds of CsCl/guanidine HCl ultracentrifugation as well as in vitro reconstituted (rcHC·HA) by mixing HMW HA, serum IαI, and recombinant TSG-6. Consistent with previous reports, Extract PBS suppressed transforming growth factor-β1 promoter activation in corneal fibroblasts and induced mac ro phage apo pto sis. However, these effects were abolished by hyaluronidase digestion or heat treatment. More importantly, the effects were retained in the nHC·HA or rcHC·HA. These data collectively suggest that the HC·HA complex is the active component in AM responsible in part for clinically observed anti-inflammatory and anti-scarring actions.
Hyaluronan (HA)4 is widely distributed in extracellular matrices, tissues, body fluids, and even in intracellular compartments (reviewed in Refs. 1 and 2). The molecular weight of HA ranges from 200 to 10,000 kDa depending on the source (3), but can also exist as smaller fragments and oligosaccharides under certain physiological or pathological conditions (1). Investigations over the last 15 years have suggested that low Mr HA can induce the gene expression of proinflammatory mediators and proangiogenesis, whereas high molecular weight (HMW) HA inhibits these processes (4–7).
Several proteins have been shown to bind to HA (8) such as aggrecan (9), cartilage link protein (10), versican (11), CD44 (12, 13), inter-α-inhibitor (IαI) (14, 15), and tumor necrosis factor-α stimulated gene-6 protein (TSG-6) (16, 17). IαI consists of two heavy chains (HCs) (HC1 and HC2), both of which are linked through ester bonds to a chondroitin sulfate chain that is attached to the light chain, i.e. bikunin. Among all HA-binding proteins, only the HCs of IαI have been clearly demonstrated to be covalently coupled to HA (14, 18). However, TSG-6 has also been reported to form stable, possibly covalent, complexes with HA, either alone (19, 20) or when associated with HC (21).
The formation of covalent bonds between HCs and HA is mediated by TSG-6 (22–24) where its expression is often induced by inflammatory mediators such as tumor necrosis factor-α and interleukin-1 (25, 26). TSG-6 is also expressed in inflammatory-like processes, such as ovulation (21, 27, 28) and cervical ripening (29). TSG-6 interacts with both HA (17) and IαI (21, 24, 30–33), and is essential for covalently transferring HCs on to HA (22–24). The TSG-6-mediated formation of the HC·HA complex has been demonstrated to play a crucial role in female fertility in mice. The HC·HA complex is an integral part of an expanded extracellular “cumulus” matrix around the oocyte, which plays a critical role in successful ovulation and fertilization in vivo (22, 34). HC·HA complexes have also been found at sites of inflammation (35–38) where its pro- or anti-inflammatory role remain arguable (39, 40).
Immunostaining reveals abundant HA in the avascular stromal matrix of the AM (41, 42).5 In ophthalmology, cryopreserved AM has been widely used as a surgical graft for ocular surface reconstruction and exerts clinically observable actions to promote epithelial wound healing and to suppress inflammation, scarring, and angiogenesis (for reviews see Refs. 43–45). However, it is not clear whether HA in AM forms HC·HA complex, and if so whether such an HC·HA complex exerts any of the above therapeutic actions. To address these questions, we extracted AM with buffers of increasing salt concentration. Because HMW HA was found to form the HC·HA complex and was mainly extractable by isotonic solutions, we further purified it from the isotonic AM extract and reconstituted it in vitro from three defined components, i.e. HMW HA, serum IαI, and recombinant TSG-6. Our results showed that the HC·HA complex is an active component in AM responsible for the suppression of TGF-β1 promoter activity, linkable to the scarring process noted before by AM (46–48) and by the AM soluble extract (49), as well as for the promotion of macrophage death, linkable to the inflammatory process noted by AM (50) and the AM soluble extract (51).
EXPERIMENTAL PROCEDURES
Materials
The following reagents were from Sigma: anhydrous alcohol, cesium chloride, EDTA, guanidine hydrochloride, magnesium chloride, potassium acetate, sodium acetate, sodium chloride, sodium hydroxide, Tris base, Triton X-100, protease inhibitor mixture (including 4-(2-aminoethyl)benzenesulfonyl fluoride, aprotinin, bestatin hydrochloride, E-64, leupeptin, and pepstatin A), phenylmethanesulfonyl fluoride, Stains-all dye, λDNA-BstEII-digested restriction fragments, and a HMW HA purified from human umbilical cords (∼1000 kDa). Select-HA HiLadder (molecular mass of 1510, 1090, 966, 572, and 495 kDa) was purchased from AMS Biotechnology, UK. Hyaluronan-binding protein (HABP) and Streptomyces hyaluronidase (HAase) were from Seikagaku Biobusiness Corporation (Tokyo, Japan). A medical grade HMW HA, Healon® (∼4000 kDa) was purchased from Advanced Medical Optics (Santa Ana, CA). Agarose and phosphate-buffered saline (PBS) were from Invitrogen. Phosphatase inhibitors sodium fluoride and sodium vanadate, and dialysis tubes (Mr cutoff 3500) were from Fisher Scientific (Pittsburgh, PA). BCA Protein Assay Kit, Sulfo-NHS, and 1-ethyl-3-(3-dimethylaminopropyl)carbidodiimide were from Pierce. HA Quantitative Test Kit was from Corgenix (Westminster, CO). 4–15% gradient acrylamide ready gels and nitrocellulose membranes were from Bio-Rad. IαI were prepared in our laboratory from human plasma according to the published method (52). The antibody against human IαI and the secondary antibodies conjugated with the peroxidase were from DAKO (Carpinteria, CA). The recombinant human TSG-6 protein (TSG-6Q) was expressed in Drosophila S2 cells and purified to homogeneity as described previously (53) and an antibody against the C-terminal region of human TSG-6 (RAH-1) was as described in Ref. 29. Other antibodies to TSG-6 were from R&D Systems (mAb 2104) and Santa Cruz Biotechnology (sc-30140) (Santa Cruz, CA), respectively. Western LightingTM Chemiluminesence Reagent was from PerkinElmer Life Sciences. Luciferase assay reagents including the cell lysis buffer was from Promega (Madison, WI). Cell Proliferation Kit (MTT) was from Roche Applied Science. BioPulverizer was from Biospec Products, Inc. (Bartlesville, OK). The ultracentrifuge (LM8 model, SW41 rotor) was from Beckman Coulter, Inc. (Fullerton, CA).
Sequential AM Extraction by Buffers A, B, and C
Cryopreserved human AM, obtained from Bio-tissue, Inc. (Miami, FL), was sliced into small pieces, frozen in liquid nitrogen, and ground to fine powder by a BioPulverizer. The powder was mixed with Buffer A (100 mm Tris-HCl, pH 7.6, 150 mm NaCl, 4 mm EDTA, 1% (v/v) Triton X-100) at 1:1 (g/ml) at 4 °C for 1 h. The mixture was centrifuged at 48,000 × g for 30 min at 4 °C and the supernatant (Extract A) stored at −80 °C. The pellet was then washed three times with Buffer A before being extracted with Buffer B (100 mm Tris-HCl, pH 7.6, 1 m NaCl, 4 mm EDTA, 1% (v/v) Triton X-100) at 4 °C for 1 h. After the centrifugation, the supernatant (Extract B) was stored. The remaining pellet was washed with Buffer B before adding Buffer C (100 mm sodium acetate, pH 5.8, 4 m guanidine hydrochloride, 4 mm EDTA, 1% Triton X-100) at 4 °C for 24 h. Again after the centrifugation, the supernatant (Extract C) was stored. Buffers A–C were supplemented with the following protease and phosphatase inhibitors: protease inhibitor mixture (1:100 dilution according to manufacturer's suggestion), 0.5 mm phenylmethanesulfonyl fluoride, 50 mm sodium fluoride, and 0.2 mm sodium vanadate. Protein concentrations in AM extracts were determined by the BCA Protein Assay Kit, whereas HA concentrations by an enzyme-linked immunosorbent assay-based HA Quantitative Test Kit.
AM Extraction by PBS
The whole procedure for preparation of human AM extracts was carried out aseptically for subsequent cell culture-based experiments as recently reported (51). Most of preparation steps were the same as described above with the following modifications. The AM powder was mixed with the cold PBS buffer without protease and phosphatase inhibitors at 1:1 (g/ml). The mixture was centrifuged at 48,000 × g at 4 °C for 30 min. The supernatant was designated as Extract PBS.
Purification of Native HC·HA (nHC·HA) Complex
According to a reported method (36), Extract PBS (prepared in PBS) was dissolved in a CsCl, 4 m guanidine HCl mixture at the initial density of 1.35 g/ml, and centrifuged at 125,000 × g for 48 h at 15 °C. A total of 15 fractions (0.8 ml/fraction) were collected from the top to the bottom of each tube. Besides the density, the concentration of proteins and HA in each fraction were measured by the BCA Protein Assay and HA Quantitative Test Kit, respectively. Fractions 8–15, which contain HA but no detectable proteins, were pooled, adjusted with CsCl, 4 m guanidine HCl at the initial density of 1.40 g/ml, centrifuged, and fractionated in the same manner as described above. Fractions 3–15, which contained HA but no detectable proteins, were pooled and dialyzed against distilled water to remove CsCl and guanidine HCl. The dialysate was mixed with 3 volumes of 95% (v/v) ethanol containing 1.3% (w/v) potassium acetate at 0 °C for 1 h. After centrifugation at 15,000 × g, the pellet was washed with 70% (v/v) ethanol and centrifuged again. The pellet was briefly dried by air, stored at −80 °C, and designated as the nHC·HA complex.
Reconstitution of HC·HA (rcHC·HA) Complex in Vitro
HABP was covalently cross-linked to Covalink-NH 96-well plates using a reported method (54). In brief, Covalink-NH plates were sterilized in 70% alcohol for 2 h and dried before being added with 50 μl of 0.184 mg/ml Sulfo-NHS in distilled water containing 0.04 mg/ml HABP per 96-well plate. The cross-linking was performed by adding 1 μl of 0.123 mg/ml 1-ethyl-3-(3-dimethylaminopropyl)carbidodiimide in distilled water per well. The plate was incubated overnight at 4 °C or for 2 h at 25 °C before the coupling solution was removed, and washed 3 times with PBS containing 2 m NaCl and 50 mm MgSO4 followed by 3 washes with PBS. The HABP covalently cross-linked to the Covalink-NH 96-well plate was then incubated for 24 h at 25 °C with 50 μl of 1.5 to 200 μg/ml HMW HA (Healon) in PBS containing 5 mm MgCl2 with or without 40 μg/ml human IαI and/or 6 μg/ml recombinant human TSG-6Q (e.g. to form rcHC·HA complex (24)); the unbound component was removed by washing with PBS 4 times. Bound HA was released by incubation in 100 mm sodium acetate, pH 5.8, 4 m guanidine hydrochloride, 4 mm EDTA for 15 min, and quantitated using the HA Quantitative Test Kit after dialysis into distilled water. It was then subjected to HAase digestion or NaOH (see below) treatment before Western blotting.
HAase Digestion and NaOH or Heat Treatment
For biochemical characterization, AM extracts denoted as A, B, and C (with an equal volume) were dialyzed against the buffer D (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5 mm phenylmethanesulfonyl fluoride) at 4 °C for 6 h. After dialysis, the volume of each extract was measured and adjusted to the same volume with buffer D. Extracts A, B, C, PBS (e.g. 100 μl), and HMW HA (Sigma) were digested with 20 units/ml HAase in the reaction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% (v/v) Triton X-100, 0.1% (w/v) bovine serum albumin, with protease and phosphatase inhibitors) for 2 h at 37 °C. The enzymatic digestion was stopped by adding 2× Laemmli sample buffer (62.5 mm Tris-HCl, pH 6.8, 25% glycerol, 2% (w/v) SDS, 0.01% (w/v) bromphenol blue, 710 mm β-mercaptoethanol) to the reaction mixture (1:1). For HA size analysis, Extracts A, B, and C were treated with 0.2 n NaOH for 4 h at 25 °C to completely remove ester bonds between HA and proteins (36). For optimizing NaOH treatment, i.e. to determine the association between HA and proteins, a series of NaOH concentrations (0–0.2 n) were used to treat Extract A for 1 h at 25 °C (18). For the cell-based assay, HMW HA (Healon®) and Extract PBS were digested with 20 units/ml HAase in PBS, pH 7.5, for 1 h at 37 °C. For heat treatment, HMW HA (Healon) and Extract PBS were incubated into a water bath at 95 °C for 10 min followed by cooling on ice.
Determination of HA Sizes by Agarose Gel Electrophoresis
HA sizes in AM extracts were analyzed by an agarose gel electrophoresis according to a published method (55). Briefly, Extracts A, B, and C with or without treatment by HAase or NaOH were separated on 0.5% (w/v) agarose gels followed by staining with 0.005% (w/v) Stains-all dye in 50% (v/v) ethanol. The gels were stained overnight at 25 °C with light protection and HA visualized as bluish bands after de-staining in water and exposure to the room light for 6 h. The HA sizes were estimated by comparing to the Select-HA HiLadder and HMW HA (Healon).
Western Blot
Samples were electrophoresed on 4–15% (w/v) gradient acrylamide ready gels under denaturing and reducing conditions. Proteins were transferred to the nitrocellulose membrane. The membrane was then blocked with 5% (w/v) fat-free milk in TBST (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% (v/v) Tween 20) followed by sequential incubation with specific primary antibodies against IαI, TSG-6, and their respective secondary antibodies. Immunoreactive proteins were detected with Western LightingTM Chemiluminesence Reagent.
Construction of TGF-β1 Promoter/Luciferase Reporter Gene in Adenovirus
The plasmid containing the TGF-β1 promoter-luciferase reporter gene was kindly provided by Dr. Seong-Jin Kim (National Institutes of Health, Bethesda, MD) and the TGF-β1 promoter-luciferase reporter gene was inserted at KpnI and HindIII sites of pGL3-basic as previously reported (56). The adenovirus containing the TGF-β1 promoter-luciferase reporter gene was constructed by the DNA Core facility at the University of Michigan (Ann Arbor, MI), which also kindly provided adeno-CMV-β-galactosidase as previously described (48, 56).
TGF-β1 Promoter Assay with Luciferase Reporter Gene
Expression of the TGF-β1 gene was measured by a promoter assay in human corneal fibroblasts cultured to 80% confluence (∼1.0 × 106 cells/10-cm dish) in Dulbecco's modified Eagle's medium, 10% (v/v) fetal bovine serum as previously reported (48). In short, after cells were washed twice with the medium, adeno-TGF-β1 promoter-luciferase (multiplicity of infection = 37.5) and adeno-CMV-β-galactosidase (multiplicity of infection = 30) were added to cells and incubated at 37 °C for 4 h. Cells were then trypsinized for 5 min and collected by centrifuged at 1500 × g (∼600 × g) for 5 min. Cells were re-suspended in Dulbecco's modified Eagle's medium, 10% (v/v) fetal bovine serum and cell viability was measured by trypan blue stain. For experiments to determine the effect of Extract PBS, human corneal fibroblasts were seeded at 3 × 104/per well in a 24-well plastic plate before adding 1–125 μg/ml HMW HA (Healon), 0.04–125 μg of protein/ml Extract PBS or PBS (both HA and Extract PBS were dissolved in PBS). For experiments to determine the effect of nHC·HA and rcHC·HA, the same HA concentration of 25 μg/ml different components described under “Reconstitution of HC·HA (rc HC·HA) Complex in Vitro” or nHC·HA was added to the 96 wells with HABP cross-linked and incubated at 25 °C for 24 h. After the wash with PBS, human corneal fibroblasts were seeded at 3 × 103/per well in a 96-well plate, and incubated at 37 °C for 48 h. The medium was then removed and cells were rinsed twice with PBS. After 100 μl of lysis buffer (25 mm Tris phosphate, pH 7.8, 2 mm dithiothreitol, 2 mm 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% (v/v) glycerol, 1% (v/v) Triton X-100) was added, cells were scraped and transferred to a microcentrifuge tube placed on ice. Cell lysates were collected by vortexing for 10–15 s and centrifuged at 12,000 × g for 15 s at 25 °C. The supernatant was assayed for both luciferase and β-galactosidase activities. The relative luciferase activity was calculated by dividing the luciferase activity by the corresponding β-galactosidase activities.
Macrophage Viability Assay (MTT)
RAW264.7 cells were seeded at 1 × 104/per well in 96-well plates with HABP cross-linked and coated with various components (Extract PBS, HA, IαI, TSG-6, and nHC·HA) as described above. The cell viability was measured with the MTT as reported previously (51).
Statistical Analysis
Data are represented as the mean ± S.D. of 4 independent experiments with a sample size of 4 for each condition. Student's t test was performed to test statistical significance from controls. A p value equal to or less than 0.05 was considered statistically significant.
RESULTS
HMW HA in AM Is Readily Extractable by an Isotonic Buffer
Previous reports (41, 42) and immunostaining data5 showed that HA and IαI were present in AM stroma, respectively. To further determine the quantity and sizes of HA as well as its potential covalently coupled with HCs in AM, we extracted AM sequentially with Buffers A, B, and C, which consisted of increasing salt concentrations (0.15 m NaCl, 1.0 m NaCl, and 4 m guanidine HCl, respectively). As shown in Table 1, Buffer A extracted the highest percentage (48.8 ± 10.1%) of total proteins (combined protein amounts in Extracts A–C), followed by Buffer C (38.1 ± 8.8%) and Buffer B (13.2 ± 1.3%). Buffer A also extracted more HA (70.1 ± 6.0%) than either Buffer C (23.8 ± 3.9%) or Buffer B (6.2 ± 2.1%). As a result, the ratio of HA concentrations divided by protein concentrations (HA/protein) was much higher in Extract A (0.20 ± 0.02) as compared with Extract C (0.07 ± 0.01) or Extract B (0.06 ± 0.02). AM was also extractable with PBS, and yielded a similar ratio of HA/protein to that of Extract A although Extract PBS had overall lower concentrations of proteins and HA as compared with Extract A (Table 1). A higher ratio of HA/proteins indicated that HA present in AM could be preferentially extracted by an isotonic buffer such as Buffer A and PBS, and would be used to guide subsequent biochemical purification of HA-associated protein complex (see below).
TABLE 1.
Quantitation of proteins and HA in AM extracts
The whole AM from one human placenta was sequentially extracted with buffers A, B, and C, or separately extracted by PBS. Four independent experiments were performed (n = 4). The amount of proteins and HA was determined by BCA Protein Assay and an enzyme-linked immunosorbent assay-based HA Quantitative Test Kit, respectively, and reported as mean ± S.D. A higher ratio of HA/proteins was noted in Extracts A and PBS.
| AM extract | Protein |
HA |
HA/protein ratio | ||
|---|---|---|---|---|---|
| AM | Total | AM | Total | ||
| mg protein/g wet | % | mg HA/g wet | % | mg/mg | |
| A | 2.780 ± 1.661 | 48.8 ± 10.1 | 0.442 ± 0.139 | 70.1 ± 6.0 | 0.20 ± 0.02 |
| B | 0.707 ± 0.232 | 13.2 ± 1.3 | 0.033 ± 0.011 | 6.2 ± 2.1 | 0.06 ± 0.02 |
| C | 1.980 ± 0.387 | 38.1 ± 8.8 | 0.147 ± 0.012 | 23.8 ± 3.9 | 0.07 ± 0.01 |
| PBS | 1.493 ± 0.892 | 26.2 ± 5.4 | 0.236 ± 0.075 | 37.7 ± 3.2 | 0.22 ± 0.02 |
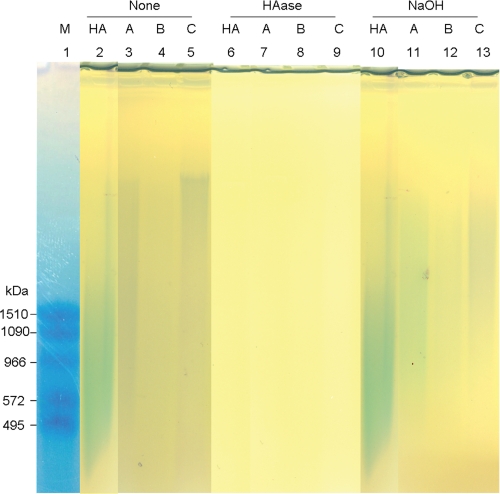
To determine the HA size in AM extracts, we used agarose gel electrophoresis as previously described (55). Notably, HA in all AM extracts appeared as a bluish smear and had a higher average molecular mass than that of the control HA (∼3000 kDa, Fig. 1, lanes 2–5). The bluish bands in Extracts A–C were shown to correspond to HA because they could be removed by digestion with a HAase from Streptomyces hyalurolyticus (Fig. 1, lanes 6–9). This HAase specifically digests HA, but not other glycosaminglycans such as chondroitin and chondroitin sulfate. Interestingly, 0.2 n NaOH treatment did not reduce the size of the control HA, but reduced those of HA in Extracts A–C (Fig. 1, lanes 10–13), suggesting that HA in these extracts might form aggregates with some proteins linked by NaOH-sensitive bonds, e.g. ester bonds. A similar HMW HA associated with proteins was also present in Extract PBS (see below, Fig. 5C).
FIGURE 1.
Presence of HMW HA in AM extracts. Extracts A, B, and C with or without HAase digestion (HAase) or NaOH treatment (NaOH) were resolved by 0.5% agarose gel electrophoresis and stained with Stains-all dye. The Mr of the Select-HA HiLadder (M, lane 1) is marked at the left of the gel. Five μg of HMW HA, with an average molecular mass of ∼1000 kDa (Sigma), was loaded as a comparison.
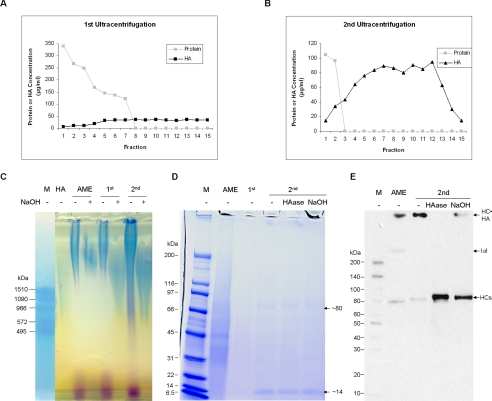
FIGURE 5.
Purification of HC·HA (nHC·HA) complex from Extract PBS by ultracentrifugation. Fractions 8–15 from the first CsCl, 4 m guanidine HCl ultracentrifugation (1st) started at the initial density of 1.35 g/ml (A) and fractions 3–15 from the second ultracentrifugation (2nd) started at the initial density of 1.40 g/ml (B) were pooled according to the presence of HA but the absence of proteins. The latter fraction was dialyzed in distill water to remove CsCl and guanidine. Extract PBS, 1st and 2nd pooled fractions were then treated with or without 0.05 n NaOH at 25 °C for 1 h (2nd pooled fractions was also digested with 20 units/ml HAase at 37 °C for 2 h) and analyzed on 0.5% agarose gel before being stained with Stains-all dye (C), stained with the Coomassie Blue dye (D), or on Western blot using an anti-IαI antibody (E). The results confirmed the nHC·HA complex was formed by HMW HA and HC of IαI via a NaOH-sensitive bond. Please note the pooled fractions from the second ultracentrifugation (labeled as 2nd) in D were concentrated ∼20-fold by lyophilization before loading to enhance the detection by the Coomassie Blue dye. AME, amniotic membrane extract.
HA in AM Extracts Is Covalently Linked with HCs of IαI
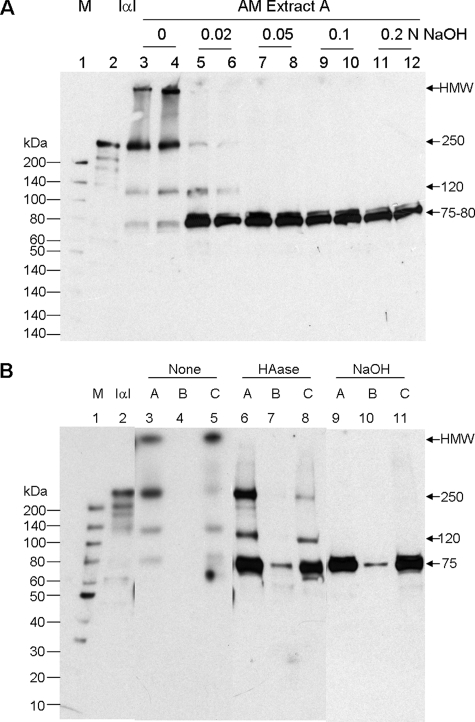
One prime protein candidate that can become covalently linked with HA is HCs of IαI (14, 15, 18). It has been shown that ester bonds are formed between two HCs and the chondroitin sulfate chain of bikunin in the intact IαI molecule (57), and also link HCs to HA (18). To investigate whether IαI is covalently linked with HMW HA in AM extracts, we used either HAase to digest HA into small fragments (that would run on SDS-PAGE) or weak NaOH to hydrolyze any ester bonds between HCs and HA (18). As shown in Fig. 1, HMW HA in these extracts was completely digested by 20 units/ml HAase at 37 °C for 2 h, but was not hydrolyzed by 0.2 n NaOH at 25 °C for 4 h. However, we found the amount of total proteins visualized by Coomassie Blue staining in Extracts A–C after 0.2 n NaOH treatment appeared to be less than those without such treatment (data not shown). To optimize NaOH treatment so as not to cause protein hydrolysis, we subjected Extract A to a range of NaOH concentrations at 25 °C for 1 h. Similar to what had been reported (18, 58), purified IαI (Fig. 2A, lane 2) yielded a major band at ∼250 kDa when analyzed by Western blot, representing the intact IαI. Other bands with smaller Mr were also seen, which are presumably degradation products of IαI. Untreated Extract A contained a band corresponding to IαI, but also had a HMW band just at the bottom of the loading well and two other major bands of 75 and 120 kDa (Fig. 2A, lanes 3 and 4). The HMW band is likely to be IαI components covalently linked with HMW HA (see below), where their size precludes them from entering the gel. The 75-kDa band is presumed to correspond to a free HC and the 120-kDa band is likely to be one HC covalently coupled to either the bikunin or TSG-6 (24, 33). Treatment with 0.02 n NaOH caused a large reduction of IαI-immunoreactive bands, with the exception of the 120-kDa species, and dramatically increased the intensity of the 75-kDa band and the emergence of an 80-kDa band, where the 75- and 80-kDa species are likely to correspond to HC1 and HC2, respectively. Treatment with 0.05–0.2 n NaOH led to complete removal of all bands except for the 75- and 80-kDa bands, where the highest concentrations of NaOH had a somewhat lower intensity of these bands, presumably due to protein hydrolysis. Therefore, in the subsequent experiments 0.05 n NaOH was chosen to treat AM extracts and the results were compared with those digested with HAase. Coomassie Blue staining showed that the sample loading of Extracts A–C was similar for non-treated (None), HAase-digested (HAase), and NaOH-treated (NaOH) samples (supplemental Fig. S1). Therefore, the same samples were then used for Western blot analysis with anti-IαI antibody. As shown in Fig. 2B, non-treated Extract C (lane 5) had similar band profiles to that of Extract A described above (Fig. 2, A, lane 3, and B, lane 3), whereas no IαI was detected in Extract B (lane 4). HAase digestion (lanes 6–8) completely removed the HMW band (retained in the well) in Extracts A and C, suggesting that this band is a HMW IαI·HA complex. For Extracts A and C, the 75-kDa band was increased after HAase digestion, where it also became visible in Extract B. These results clearly demonstrated that HCs and HA were linked and present in both isotonic (Extract A and PBS) and high-salt extractable Extracts B and C. Noticeably, the 250- and 120-kDa bands became much sharper and more intense following HAase digestion (Fig. 2B, lanes 6 and 8) indicating that some of these species may be released from HA. Because both 250- and 120-kDa bands were completely eliminated by 0.05 n NaOH (Fig. 2A), resulting in the most increase of the 75-kDa band (the 80-kDa band was difficult to see at most times) (Fig. 2B, lanes 9–11), this indicated that the 250- and 120-kDa bands are complexes of HCs and other components linked by ester bonds. These results are therefore consistent with the conclusion that the 250- and 120-kDa species correspond to intact IαI and a HC-containing complex (e.g. HC·bikunin or HC·TSG-6), respectively.
FIGURE 2.
Covalent linkage between HA and HCs of IαI in AM extracts. Extract A was treated (in duplicate) with a series of NaOH concentrations (0, 0.02, 0.05, 0.10, and 0.2 n) before Western blot with an anti-IαI antibody to determine the optimal NaOH concentration for cleaving the linkage between HA and HCs (A: M, protein ladder markers, and IαI, purified from the human plasma). Extracts A–C with or without HAase digestion or 0.05 n NaOH treatment were analyzed by Western blot with anti-IαI antibody (B).
TSG-6 Was Present in AM Extracts
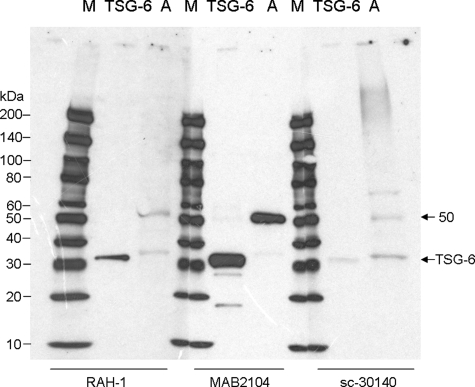
Because HMW HA was covalently linked with HCs of IαI in AM extracts and TSG-6 has been shown previously to be required for transferring HCs to HA by forming a unique covalent bond with IαI as an intermediate (22, 24, 59), we wanted to know whether TSG-6 was also present in AM extracts. To make sure the detection of TSG-6 in AM extracts was genuine, we obtained three anti-TSG-6 antibodies (RAH-1, mAb 2104, and sc-30140) from three different sources. RAH-1 (29) and sc-30140 are rabbit polyclonal antibodies raised against a C-terminal peptide and the full-length human TSG-6 pre-protein (amino acids 1–277), respectively, whereas mAb 2104 is a mouse monoclonal antibody raised against the mature human TSG-6 protein (amino acids 18–277). All three antibodies detected a single ∼32-kDa band in a preparation of recombinant human TSG-6Q protein (53) and a ∼35-kDa species in non-treated Extract A by Western blot (Fig. 3A). Such a difference in Mr between recombinant TSG-6Q and TSG-6 in Extract A might be due to the different sizes of N-linked glycans present in the Drosophila expressed material compared with the native protein.6 Noticeably, all three antibodies also detected a strong 50-kDa band in Extract A.
FIGURE 3.
TSG-6 was present in AM extracts. TSG-6 was found to be present in Extract A using three different antibodies that recognized the control TSG-6Q (25 ng) as a ∼32 kDa protein (A, bands of ∼35 and ∼50 kDa were seen in Extract A).
Suppression of TGF-β1 Promoter Activation by AM Extract PBS
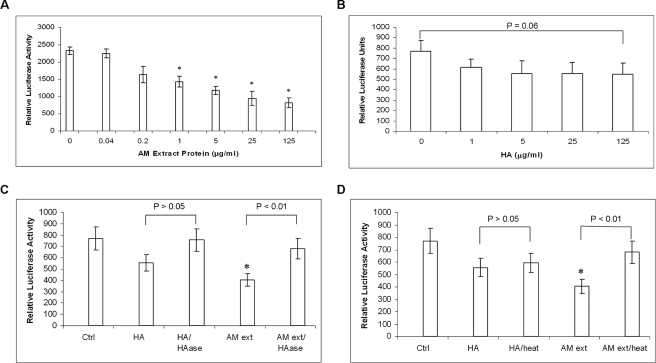
Down-regulation of TGF-βs signaling is a strategy for preventing scarring during wound healing. It had been shown that AM suppresses transcription of TGF-βs and their receptors in human corneal and limbal fibroblasts and hence downstream expression of α-smooth muscle actin, the EDA-spliced form of fibronectin, and integrin α5 (46). In mouse keratocytes, suppression of TGF-β by AM was essential for maintaining expression of keratocyte-specific keratocan and preventing differentiation into myofibroblasts (48). To test whether Extract PBS suppressed TGF-β transcription, we used a luciferase-based TGF-β1 promoter assay as previously reported (48). The suppression of TGF-β1 promoter activity in human corneal fibroblasts was dose-dependent over the range of protein concentrations from 0.2 to 125 μg/ml Extract PBS (Fig. 4A). As low as 1 μg/ml proteins significantly suppressed TGF-β1 promoter activation and there was a greater than 50% suppression when 125 μg/ml protein (containing ∼5 μg/ml HA) was added (p = 0.008). In contrast, 1 μg/ml HMW HA (Healon) did not significantly suppress TGF-β1 promoter activation when compared with the control (PBS added only) (p = 0.20, Fig. 4B). The suppression of the promoter activity by 5, 25, and 125 μg/ml HMW HA was further enhanced (p = 0.10, 0.09, and 0.06, respectively, Fig. 4B) but not as potent as Extract PBS. To further test whether this activity was related to HA alone or HA-protein complex, both HMW HA and Extract PBS were digested with HAase or heated at 95 °C for 10 min before testing. The results showed that both treatments abolished the significant suppressive effect of Extract PBS (p = 0.06 and 0.12 for HAase and heat treatment, respectively, Fig. 4, C and D). In contrast, the digestion of HMW HA by HAase did not cause a significant change in suppression of TGF-β1 promoter activity (p = 0.31) (Fig. 4C), and even less change when HMW HA was heat-treated (p = 0.70) (Fig. 4D). These data indicated both HA and proteins in Extract PBS were necessary for suppressing the TGF-β1 promoter activity.
FIGURE 4.
Suppression of TGF-β1 promoter activation by Extract PBS. A dose-dependent relationship was noted in the suppression of the TGF-β1 promoter activity by a series of concentrations of Extract PBS (A). In contrast, HMW HA (Healon), with an average molecular mass of HA ∼4000 kDa, showed some suppression of TGF-β1 promoter activity but not statistically significant (B). The suppressive effect of the TGF-β1 promoter activity was lost when HMW HA (125 μg/ml) or Extract PBS (125 μg/ml proteins) was digested with hyaluronidase (C). Heat treatment (95 °C for 10 min) also eliminated the suppressive activity by Extract PBS but this was not significant for HMW HA (D). Data are represented as the mean ± S.D. of four independent experiments (n = 4) with a sample size of 4 for each condition. In A–D, an asterisk indicated p < 0.05.
Purification and in Vitro Reconstitution of HC·HA Complex
Having known that suppression of TGF-β1 promoter activity by the Extract PBS required both HA and proteins and that HA in Extract PBS was covalently linked with HCs, we speculated that the suppressive activity was derived from the HC·HA complex. To test this hypothesis, we first purified this HC·HA complex away from free proteins by submitting Extract PBS to two successive rounds of CsCl ultracentrifugation in the presence of 4 m guanidine HCl followed by ethanol precipitation, according to a reported method (36). After the first ultracentrifugation step, the sample in each tube was subdivided into 15 fractions (from low to high density). The density and concentrations of proteins and HA in each fraction were measured. As the density increased there was a corresponding increase in HA and decrease in protein concentrations. From fraction 8 onwards, the protein concentration was below the detection limit (Fig. 5A). Therefore, fractions 8–15 (which contained the HC·HA complex, but little free protein) were pooled and subjected to a second ultracentrifugation. Samples were fractionated in the same manner, and fractions 3–15 in which the protein concentration was below the detection limit were pooled (Fig. 5B). After dialysis against water, this HC·HA containing material was precipitated by ethanol. Compared with the Extract PBS, which had 1370 μg/ml protein and 62 μg/ml HA, the purified HC·HA complex did not contain proteins detectable by the BCA Protein Assay (the minimum detection is ∼25 μg/ml) even when it was 10-fold concentrated. We thus estimated that there was at least 550-fold (1370 × 10/25) purification of the HA.
Agarose gel analysis demonstrated that HA in purified HC·HA complex was the HMW species, having an average molecular mass of greater than 3000 kDa, and was coupled with proteins via a NaOH-sensitive bond (Fig. 5C). Purified HC·HA was concentrated ∼20-fold prior to SDS-PAGE analysis. Coomassie Blue staining showed a prominent band likely corresponding to HC1 and/or HC2 (∼75–80 kDa) and a prominent 14-kDa band as well as few other minor protein bands in the range between 30- and 60-kDa (Fig. 5D). Western blot analysis using an anti-IαI antibody confirmed the presence of the HC·HA complex, which was detected as a high Mr band in the loading well and abolished or greatly reduced after HAase digestion or NaOH treatment, respectively (Fig. 5E). At the same time, the intensity of the HC band (∼75–80 kDa) was markedly enhanced due to its release from HMW HA by HAase or because the ester bond formed between HC and HA was hydrolyzed by NaOH. Western blot analysis using an anti-TSG-6 antibody did not detect any TSG-6 in the HC·HA complex purified from Extract PBS (data not shown).
Biological Function of HC·HA Complex
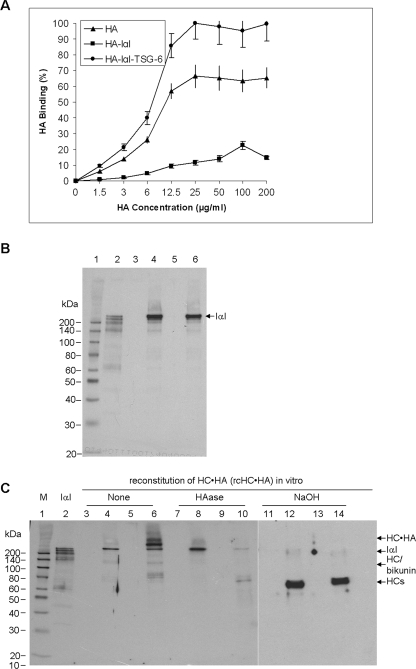
To further define the biological function of purified HC·HA complex, we followed the reported methods to reconstitute this complex in vitro using HMW HA (Healon), IαI purified from serum, and recombinant TSG-6 (24). In a 96-well Covalink-NH plastic plate, we first covalently cross-linked HABP to each well via amide bonds formed between the carboxylates of HABP and NH groups on the plate surface. We then added serial concentrations of 1.5 to 200 μg/ml HMW HA in the absence (Fig. 6A, ▴) or presence of either 40 μg/ml IαI alone (Fig. 6A, ■) or 40 μg/ml IαI and 6 μg/ml TSG-6 together (Fig. 6A, ●). After washing, the enzyme-linked immunosorbent assay-based HA Quantitative Test showed that the amount of bound HA was greatly decreased when added with IαI, but increased when added with both IαI and TSG-6. In the latter case the increase in HA binding is likely to result from its cross-linking by HC (36) giving rise to a greater avidity for the surface immobilized HABP, i.e. in a manner analogous to the increased adhesion of HA for HA receptors on leukocytes reported previously (40). However, the reason for the reduced binding of HA to HABP in the presence of IαI is likely due to a direct interaction between HABP and IαI (Fig. 6B), which competes for the interaction of HABP with HA. The above result indicated that there was maximal HA immobilization onto the plate surface when 25 μg/ml HMW HA was used, at least for HA alone and when in the presence of IαI and TSG-6.
FIGURE 6.
In vitro reconstitution of HC·HA (rcHC·HA) complex. The HA binding capacity (%) on HABP cross-linked wells was determined to be maximal at 25 μg/ml HMW HA (Healon) by addition of both human IαI and recombinant human TSG-6 (A, ●) when compared with HMW HA alone (A, ▴) or HMW HA with IαI (A, ■). Four independent experiments (n = 4) with a sample size of 4 for each condition were performed and data were represented as the mean ± S.D. IαI directly bound to HABP, which was covalently cross-linked to the 96-well surface, as shown by Western blot with anti-IαI antibody (B). Lane 1, protein marker; lane 2, purified IαI; lane 3, HABP only; lane 4, HABP + IαI; lane 5, HABP + HA; lane 6, HABP + IαI + HA. Western blot using an anti-IαI antibody (C) revealed that the bound HMW HA on HABP cross-linked wells formed the HC·HA complex when added with both IαI and TSG-6 (HA + IαI + TSG-6, lanes 6, 10, and 14) when compared with HMW HA alone (lanes 3, 7, and 11), with IαI alone (HA + IαI, lanes 4, 8, and 12) or TSG-6 alone (HA + TSG-6, lanes 5, 9, and 13) either without (lanes 3-6) or with HAase digestion (lanes 7–10) or NaOH treatment (lanes 11–14).
To verify the formation of rcHC·HA complex, we applied 25 μg/ml HMW HA to each well under the various conditions described above (Fig. 6A). After extensive washing to remove unbound components, each well containing the bound HA was subjected to HAase digestion or NaOH treatment before being solubilized in the Laemmli sample buffer for Western blotting with an anti-IαI antibody. As compared with IαI (Fig. 6C, lane 2) and HMW HA alone (lane 3), intact IαI, but not its degraded fragments, was present in the HABP/HA-coated wells and retained after extensive washing (lane 4). As expected, there was no IαI-immunoreactive band in lane 5 where HA was added with TSG-6 alone. Importantly, when HA, IαI, and TSG-6 were incubated together (lane 6), an additional HMW band (labeled as HC·HA) was seen at the bottom of the loading well, whereas the intensity of the IαI band was reduced, presumably because some IαI had been consumed in the transfer of HC to HMW HA by TSG-6. This HC·HA band and IαI were eliminated by HAase digestion (lane 10) or NaOH treatment (lane 14), resulting in the release (appearance) of a ∼75–80-kDa HC band. By a comparison, intact IαI (lane 4) was resistant to HAase digestion (lane 8), but cleavable by NaOH into at least two bands including higher 120- and ∼75–80-kDa bands (lane 12). These data verified that the HC·HA complex could be effectively reconstituted in vitro from HA and IαI in the presence of TSG-6 (24). Consistent with our earlier studies, once the HC·HA complex was formed, TSG-6 was released (24) and could be washed away (i.e. it does not associate with HC·HA and has low affinity for HA at physiological pH (60)), a finding that was supported by Western blot using an anti-TSG-6 antibody (data not shown).
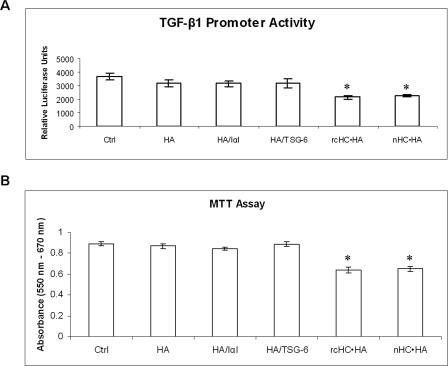
To determine whether suppression of the TGF-β1 promoter activity shown by Extract PBS was derived from the HC·HA complex, we seeded 3 × 104/ml of human corneal fibroblasts, which had been transfected with both adenoviruses containing TGF-β1 promoter-luciferase and the CMV-β-galactosidase gene for 48 h before the TGF-β1 promoter activity assay, onto HABP cross-linked plates coated with HA and HA/protein mixtures as described above. Compared with the control (Fig. 7A, Ctrl), the TGF-β1 promoter activity was not significantly suppressed by HA, HA/IαI, or HA/TSG-6 (p = 0.07, 0.06, and 0.10, respectively, Fig. 7A). In contrast, the rcHC·HA and nHC·HA complexes showed significant suppression (p = 0.004 and 0.005, respectively, Fig. 7A). The extent of suppression of the TGF-β1 promoter activity exerted by the nHC·HA complex was not significantly different from the rcHC·HA complex (p = 0.20, Fig. 7A). The same result was obtained by adding the above components as a solution in the well (without being bound to HABP-coated dish) (data not shown).
FIGURE 7.
Suppression of TGF-β1 promoter activity and promotion of macrophage death by rcHC·HA and nHC·HA complexes. As compared with the control (A, Ctrl, PBS only), rcHC·HA complex or nHC·HA significantly suppressed TGF-β1 promoter activity, i.e. as measured by the TGF-β1 promoter assay (A, p = 0.004 and 0.005, respectively), and promoted macrophage death as measured by the MTT assay (B, p = 0.0003 and 0.0007, respectively). In contrast, Healon HA alone (HA) or with additional IαI (HA + IαI) or TSG-6 (HA + TSG-6) did not show a statistically significant effect (all p > 0.05). Four independent experiments (n = 4) with a sample size of 4 for each condition were performed and data were represented as the mean ± S.D.
Recently, we demonstrated that Extract PBS suppressed cell viability in murine macrophages by inducing cell death via apoptosis (51), an effect that was also observed with AM (50). To further confirm that such an inhibitory activity on macrophage viability also resided in the HC·HA complex, we compared nHC·HA to rcHC·HA using the MTT assay. Compared with the control cultured on the HABP cross-linked wells (Fig. 7B, Ctrl), addition of HMW HA alone (HA) and addition of HMW HA with either 40 μg/ml IαI (HA/IαI) or 6 μg/ml TSG-6 (HA/TSG-6) did not cause a significant difference in macrophage viability (p = 0.30, 0.19, and 0.08, respectively, Fig. 7B). In contrast, the rcHC·HA complex as well as the nHC·HA significantly reduced macrophage viability (p = 0.0003 and 0.007, respectively, Fig. 7B). It should be noted that the rcHC·HA complex and nHC·HA exhibited a similar inhibitory activity (p = 0.64, Fig. 7B). The same result was obtained by adding the above components as a solution in the well (without being bound to HABP-coated dish) (data not shown). Collectively, these results indicated that nHC·HA as well as rcHC·HA exhibited similar suppression of TGF-β1 promoter activity and promotion of macrophage death, which have been correlated with anti-scarring and anti-inflammatory actions observed in the AM soluble extract, respectively (49, 51).
DISCUSSION
HA is a multifunctional non-sulfated glycosaminoglycan that is a known component of extracellular matrices. Although immunostaining has been used previously to show the presence of HA in the AM (41, 42),5 this study is the first providing the baseline biochemical characterization of HA in this unique AM with an avascular stromal matrix. Although HA could be extracted by buffers with different salt concentrations, the majority (∼70%) of HA in the AM was extractable by an isotonic Buffer A (0.15 m NaCl) (Table 1). Increasing NaCl concentration to 1 m (as in Buffer B) only slightly enhanced the efficiency of HA extraction. Because AM extracted by PBS yielded a similar HA/protein ratio (Table 1) and the extract is compatible with the functional assays because it does not contain Triton X-100, Extract PBS was used for further purification of HA complexes and cell-based assays. Importantly, HA in both isotonic and high salt-extractable extracts was of HMW with an average of ∼3000 kDa (Fig. 1).
Interestingly, HMW HA in the AM was associated with protein components, which were identified as HCs of IαI based on results by HAase digestion and immunoreactivity with anti-IαI antibodies (Fig. 2). Furthermore, we noted that the HC appeared to be covalently associated with HA, probably linked via ester bonds given that the HCs could be released by mild NaOH treatment (Fig. 2). This finding is consistent with previously published studies on HC·HA complexes, also referred to as SHAP-HA (18), and on this basis it seems likely that HMW complexes of HA in both isotonic and high salt extractable AM stromal matrices extracts are cross-linked via noncovalent HC-HC interactions as described for HC·HA from other tissues (36). Because the resultant free individual HA chain in AM extracts still had a molecular mass greater than ∼1500 kDa (supplemental Fig. S2) after NaOH treatment (Fig. 1), HA must be produced and maintained as HMW in the AM. Collectively, these results indicated that HA in the AM is another example utilizing ester bonds to link with HCs of IαI, a scenario resembling ovulated cumulus matrix (61), synovial fluids from individuals with inflammatory disease (61), and certain pericellular matrices (14, 62).
TSG-6 is a HA-binding protein (17) that has been associated with the formation of HC·HA complexes (21, 22, 24). Our data revealed that TSG-6 and TSG-6-related species (e.g. complexes with other components) were present in AM extracts. Besides the 35-kDa band, which closely matched the reported size of TSG-6 run on SDS-PAGE, a predominant 50-kDa was also detected in extracts (Fig. 3). Whether this 50-kDa band represents an isoform of TSG-6 uniquely present in the human AM or a complex formed between TSG-6 and one chain of IαI awaits further studies.
We and others have demonstrated that cryopreserved AM as a surgical graft exerted anti-inflammatory, anti-scarring, and anti-angiogenic actions when transplanted to diseased eye surfaces (for review see Refs. 43–45). Herein, we showed that Extract PBS suppressed TGF-β1 promoter activation, consistent with our recent report (49), which is similar to the effect of AM on human corneal (46, 48) and conjunctival (63) fibroblasts and murine corneal keratocytes (48). As shown by our biochemical analysis, Extract PBS contained HMW HA that is likely to be associated with proteins (Fig. 5C). HMW HA (∼2000 kDa) has been shown to down-regulate TGF-β1 signaling by binding to the CD44 receptor, thus affecting TGF-β1 receptor trafficking (64) and attenuating downstream SMAD activation (65). Based on the TGF-β1 promoter assay, our study showed that HMW HA suppressed TGF-β1 promoter activation but was not as potent as Extract PBS (Fig. 4B). In contrast, the significant suppressive activity of Extract PBS was dependent on both HMW HA and proteins (Fig. 4, C and D), where such an effect was also seen with nHC·HA and rcHC·HA complexes. Furthermore, we noted that the nHC·HA and rcHC·HA complexes also have a similar capacity to promote macrophage death (Fig. 7) as have recently been reported in Extract PBS (51). These results collectively indicated that besides a well established role in the fertility of female mice (22, 34), the HC·HA complex is likely to be one of the active components in AM responsible for the anti-inflammatory and anti-scarring actions that have been clinically observed in ocular surface reconstruction. Further studies into the mechanism of how the HC·HA complex exerts its actions at the cellular and molecular levels might lead to the development of therapeutics that have the ability to direct postnatal wound healing toward scarless tissue regeneration as seen in fetal wound healing (66, 67).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 EY06819 and EY15735 (to S. C. G. T.) from the NEI. This work was also supported by a research grant from TissueTech, Inc. and an unrestricted grant from Ocular Surface Research & Education Foundation, Miami, FL.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
W. Li, H. He, and S. C. G. Tseng, unpublished data.
M. S. Rugg, D. C. Briggs, and A. J. Day, unpublished data.
- HA
- hyaluronan
- AM
- amniotic membrane
- HMW
- high molecular weight
- IαI
- inter-α-inhibitor
- TSG-6
- tumor necrosis factor-αstimulated gene-6
- HC
- heavy chain
- TGF
- transforming growth factor
- HABP
- hyaluronan-binding protein
- PBS
- phosphate-buffered saline
- HAase
- Streptomyces hyaluronidase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- rcHC
- reconstitution of HC
- nHC
- native HC
- CMV
- cytomegalovirus.
REFERENCES
- 1.Tammi M. I., Day A. J., Turley E. A. (2002) J. Biol. Chem. 277, 4581–4584 [DOI] [PubMed] [Google Scholar]
- 2.Hascall V. C., Majors A. K., De La Motte C. A., Evanko S. P., Wang A., Drazba J. A., Strong S. A., Wight T. N. (2004) Biochim. Biophys. Acta 1673, 3–12 [DOI] [PubMed] [Google Scholar]
- 3.Holmes M. W., Bayliss M. T., Muir H. (1988) Biochem. J. 250, 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKee C. M., Penno M. B., Cowman M., Burdick M. D., Strieter R. M., Bao C., Noble P. W. (1996) J. Clin. Invest. 98, 2403–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooney P., Wang M., Kumar P., Kumar S. (1993) J. Cell Sci. 105, 213–218 [DOI] [PubMed] [Google Scholar]
- 6.Morrison H., Sherman L. S., Legg J., Banine F., Isacke C., Haipek C. A., Gutmann D. H., Ponta H., Herrlich P. (2001) Genes Dev. 15, 968–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slevin M., Krupinski J., Gaffney J., Matou S., West D., Delisser H., Savani R. C., Kumar S. (2007) Matrix Biol. 26, 58–68 [DOI] [PubMed] [Google Scholar]
- 8.Day A. J., Prestwich G. D. (2002) J. Biol. Chem. 277, 4585–4588 [DOI] [PubMed] [Google Scholar]
- 9.Hardingham T. E., Muir H. (1972) Biochem. J. 126, 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goetinck P. F., Stirpe N. S., Tsonis P. A., Carlone D. (1987) J. Cell Biol. 105, 2403–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeBaron R. G., Zimmermann D. R., Ruoslahti E. (1992) J. Biol. Chem. 267, 10003–10010 [PubMed] [Google Scholar]
- 12.Aruffo A., Stamenkovic I., Melnick M., Underhill C. B., Seed B. (1990) Cell 61, 1303–1313 [DOI] [PubMed] [Google Scholar]
- 13.Banerji S., Wright A. J., Noble M., Mahoney D. J., Campbell I. D., Day A. J., Jackson D. G. (2007) Nat. Struct. Mol. Biol. 14, 234–239 [DOI] [PubMed] [Google Scholar]
- 14.Yoneda M., Suzuki S., Kimata K. (1990) J. Biol. Chem. 265, 5247–5257 [PubMed] [Google Scholar]
- 15.Sandson J., Hamerman D., Schwick G. (1965) Trans. Assoc. Am. Physicians 78, 304–313 [PubMed] [Google Scholar]
- 16.Lee T. H., Wisniewski H. G., Vilcek J. (1992) J. Cell Biol. 116, 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohda D., Morton C. J., Parkar A. A., Hatanaka H., Inagaki F. M., Campbell I. D., Day A. J. (1996) Cell 86, 767–775 [DOI] [PubMed] [Google Scholar]
- 18.Zhao M., Yoneda M., Ohashi Y., Kurono S., Iwata H., Ohnuki Y., Kimata K. (1995) J. Biol. Chem. 270, 26657–26663 [DOI] [PubMed] [Google Scholar]
- 19.Lesley J., Gál I., Mahoney D. J., Cordell M. R., Rugg M. S., Hyman R., Day A. J., Mikecz K. (2004) J. Biol. Chem. 279, 25745–25754 [DOI] [PubMed] [Google Scholar]
- 20.Wisniewski H. G., Snitkin E. S., Mindrescu C., Sweet M. H., Vilcek J. (2005) J. Biol. Chem. 280, 14476–14484 [DOI] [PubMed] [Google Scholar]
- 21.Mukhopadhyay D., Hascall V. C., Day A. J., Salustri A., Fülöp C. (2001) Arch. Biochem. Biophys. 394, 173–181 [DOI] [PubMed] [Google Scholar]
- 22.Fülöp C., Szántó S., Mukhopadhyay D., Bárdos T., Kamath R. V., Rugg M. S., Day A. J., Salustri A., Hascall V. C., Glant T. T., Mikecz K. (2003) Development 130, 2253–2261 [DOI] [PubMed] [Google Scholar]
- 23.Jessen T. E., Ødum L. (2003) Reproduction 125, 27–31 [DOI] [PubMed] [Google Scholar]
- 24.Rugg M. S., Willis A. C., Mukhopadhyay D., Hascall V. C., Fries E., Fülöp C., Milner C. M., Day A. J. (2005) J. Biol. Chem. 280, 25674–25686 [DOI] [PubMed] [Google Scholar]
- 25.Wisniewski H. G., Vilcek J. (1997) Cytokine Growth Factor Rev. 8, 143–156 [DOI] [PubMed] [Google Scholar]
- 26.Milner C. M., Day A. J. (2003) J. Cell Sci. 116, 1863–1873 [DOI] [PubMed] [Google Scholar]
- 27.Fülöp C., Kamath R. V., Li Y., Otto J. M., Salustri A., Olsen B. R., Glant T. T., Hascall V. C. (1997) Gene 202, 95–102 [DOI] [PubMed] [Google Scholar]
- 28.Carrette O., Nemade R. V., Day A. J., Brickner A., Larsen W. J. (2001) Biol. Reprod. 65, 301–308 [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto T., Savani R. C., Watari M., Day A. J., Strauss J. F., 3rd (2002) Am. J. Pathol. 160, 1495–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisniewski H. G., Burgess W. H., Oppenheim J. D., Vilcek J. (1994) Biochemistry 33, 7423–7429 [DOI] [PubMed] [Google Scholar]
- 31.Wisniewski H. G., Hua J. C., Poppers D. M., Naime D., Vilcek J., Cronstein B. N. (1996) J. Immunol. 156, 1609–1615 [PubMed] [Google Scholar]
- 32.Getting S. J., Mahoney D. J., Cao T., Rugg M. S., Fries E., Milner C. M., Perretti M., Day A. J. (2002) J. Biol. Chem. 277, 51068–51076 [DOI] [PubMed] [Google Scholar]
- 33.Sanggaard K. W., Karring H., Valnickova Z., Thøgersen I. B., Enghild J. J. (2005) J. Biol. Chem. 280, 11936–11942 [DOI] [PubMed] [Google Scholar]
- 34.Ochsner S. A., Day A. J., Rugg M. S., Breyer R. M., Gomer R. H., Richards J. S. (2003) Endocrinology 144, 4376–4384 [DOI] [PubMed] [Google Scholar]
- 35.Wisniewski H. G., Maier R., Lotz M., Lee S., Klampfer L., Lee T. H., Vilcek J. (1993) J. Immunol. 151, 6593–6601 [PubMed] [Google Scholar]
- 36.Yingsung W., Zhuo L., Morgelin M., Yoneda M., Kida D., Watanabe H., Ishiguro N., Iwata H., Kimata K. (2003) J. Biol. Chem. 278, 32710–32718 [DOI] [PubMed] [Google Scholar]
- 37.Forteza R., Casalino-Matsuda S. M., Monzon M. E., Fries E., Rugg M. S., Milner C. M., Day A. J. (2007) Am. J. Respir. Cell Mol. Biol. 36, 20–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshihara Y., Plaas A., Osborn B., Margulis A., Nelson F., Stewart M., Rugg M. S., Milner C. M., Day A. J., Nemoto K., Sandy J. D. (2008) Osteoarthritis Cartilage 16, 1343–1355 [DOI] [PubMed] [Google Scholar]
- 39.Day A. J., de la Motte C. A. (2005) Trends Immunol. 26, 637–643 [DOI] [PubMed] [Google Scholar]
- 40.Zhuo L., Kanamori A., Kannagi R., Itano N., Wu J., Hamaguchi M., Ishiguro N., Kimata K. (2006) J. Biol. Chem. 281, 20303–20314 [DOI] [PubMed] [Google Scholar]
- 41.Meinert M., Eriksen G. V., Petersen A. C., Helmig R. B., Laurent C., Uldbjerg N., Malmström A. (2001) Am. J. Obstet. Gynecol. 184, 679–685 [DOI] [PubMed] [Google Scholar]
- 42.Higa K., Shimmura S., Shimazaki J., Tsubota K. (2005) Cornea 24, 206–212 [DOI] [PubMed] [Google Scholar]
- 43.Bouchard C. S., John T. (2004) The Ocular Surface 2, 201–211 [DOI] [PubMed] [Google Scholar]
- 44.Tseng S. C., Espana E. M., Kawakita T., Di Pascuale M. A., Li W., He H., Liu T. S., Cho T. H., Gao Y. Y., Yeh L. K., Liu C. Y. (2004) The Ocular Surface 2, 177–187 [DOI] [PubMed] [Google Scholar]
- 45.Dua H. S., Gomes J. A., King A. J., Maharajan V. S. (2004) Surv. Ophthalmol. 49, 51–77 [DOI] [PubMed] [Google Scholar]
- 46.Tseng S. C., Li D. Q., Ma X. (1999) J. Cell Physiol. 179, 325–335 [DOI] [PubMed] [Google Scholar]
- 47.Espana E. M., Kawakita T., Liu C. Y., Tseng S. C. G. (2004) Invest. Ophthalmol. Vis. Sci. 45, 2985–2991 [DOI] [PubMed] [Google Scholar]
- 48.Kawakita T., Espana E. M., He H., Hornia A., Yeh L. K., Ouyang J., Liu C. Y., Tseng S. C. (2005) J. Biol. Chem. 280, 27085–27092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W., He H., Chen Y. T., Hayashida Y., Tseng S. C. (2008) J. Cell Physiol. 215, 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W., He H., Kawakita T., Espana E. M., Tseng S. C. G. (2006) Exp. Eye Res. 82, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He H., Li W., Chen S. Y., Zhang S., Chen Y. T., Hayashida Y., Zhu Y. T., Tseng S. C. (2008) Invest. Ophthalmol. Vis. Sci. 49, 4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blom A. M., Mörgelin M., Oyen M., Jarvet J., Fries E. (1999) J. Biol. Chem. 274, 298–304 [DOI] [PubMed] [Google Scholar]
- 53.Nentwich H. A., Mustafa Z., Rugg M. S., Marsden B. D., Cordell M. R., Mahoney D. J., Jenkins S. C., Dowling B., Fries E., Milner C. M., Loughlin J., Day A. J. (2002) J. Biol. Chem. 277, 15354–15362 [DOI] [PubMed] [Google Scholar]
- 54.Frost G. I., Stern R. (1997) Anal. Biochem. 251, 263–269 [DOI] [PubMed] [Google Scholar]
- 55.Lee H. G., Cowman M. K. (1994) Anal. Biochem. 219, 278–287 [DOI] [PubMed] [Google Scholar]
- 56.Kawakita T., Espana E. M., He H., Smiddy R., Parel J. M., Liu C. Y., Tseng S. C. (2006) Invest. Ophthalmol. Vis. Sci. 47, 1918–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Enghild J. J., Salvesen G., Hefta S. A., Thøgersen I. B., Rutherfurd S., Pizzo S. V. (1991) J. Biol. Chem. 266, 747–751 [PubMed] [Google Scholar]
- 58.Huang L., Yoneda M., Kimata K. (1993) J. Biol. Chem. 268, 26725–26730 [PubMed] [Google Scholar]
- 59.Sanggaard K. W., Sonne-Schmidt C. S., Krogager T. P., Kristensen T., Wisniewski H. G., Thøgersen I. B., Enghild J. J. (2008) J. Biol. Chem. 283, 33919–33926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heng B. C., Gribbon P. M., Day A. J., Hardingham T. E. (2008) J. Biol. Chem. 283, 32294–32301 [DOI] [PubMed] [Google Scholar]
- 61.Jessen T. E., Odum L., Johnsen A. H. (1994) Biol. Chem. Hoppe-Seyler 375, 521–526 [DOI] [PubMed] [Google Scholar]
- 62.Selbi W., Day A. J., Rugg M. S., Fülöp C., de la Motte C. A., Bowen T., Hascall V. C., Phillips A. O. (2006) J. Am. Soc. Nephrol. 17, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 63.Lee S. B., Li D. Q., Tan D. T., Meller D. C., Tseng S. C. (2000) Curr. Eye Res. 20, 325–334 [PubMed] [Google Scholar]
- 64.Ito T., Williams J. D., Fraser D. J., Phillips A. O. (2004) J. Biol. Chem. 279, 25326–25332 [DOI] [PubMed] [Google Scholar]
- 65.Ito T., Williams J. D., Fraser D., Phillips A. O. (2004) Am. J. Pathol. 164, 1979–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adzick N. S., Longaker M. T. (1991) Clinical and Experimental Approaches to Dermal and Epidermal Repair: Normal and Chronic Wounds, Wiley-Liss, New York [Google Scholar]
- 67.Mast B. A., Diegelmann R. F., Krummel T. M., Cohen I. K. (1992) Surg. Gynecol. Obstet. 174, 441–451 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.