Abstract
The ubiquitin interaction motif-containing protein RAP80 plays a key role in DNA damage response signaling. Using genomic and functional analysis, we established that the expression of the RAP80 gene is regulated in a DNA damage-responsive manner by the master regulator p53. This regulation occurs at the transcriptional level through a noncanonical p53 response element in the RAP80 promoter. Although it is inducible by p53, RAP80 is also able to regulate p53 through an association with both p53 and the E3 ubiquitin ligase HDM2, providing HDM2-dependent enhancement of p53 polyubiquitination. Depletion of RAP80 by small interfering RNA stabilizes p53, which, following DNA damage, results in an increased transactivation of several p53 target genes as well as greater apoptosis. Consistent with these observations, exogenous expression of RAP80 selectively inhibits p53-dependent transactivation of target genes in an mdm2-dependent manner in MEF cells. Thus, we identify a new DNA damage-associated role for RAP80. It can function in an autoregulatory loop consisting of RAP80, HDM2, and the p53 master regulatory network, implying an important role for this loop in genome stability and oncogenesis.
To assure genome integrity, all cellular organisms contain systems that can monitor and repair a variety of DNA lesions. The DNA damage response (DDR)4 in mammals is a highly dynamic and coordinated network that involves a plethora of proteins that sense damage and transduce signals to execute cellular responses, including cell cycle checkpoints, DNA repair mechanisms, cellular senescence, and apoptosis (1–4). Deregulation of components in these processes contributes to genomic instability, which can lead to tumorigenesis (5–7).
Recognition of DNA damage and propagation of the DDR signal involves the recruitment and assembly of many DDR mediators and effectors, including BRCA1, at sites flanking damage (2, 8). Recruitment occurs in a hierarchical manner and is dependent on a number of post-translational modifications including phosphorylation, ubiquitination, and acetylation (2, 9, 10). RAP80 (receptor-associated protein 80 or UIMC1) is associated with the BRCA1-BARD1-ccdc98(Abraxas) complex and plays a key role in the translocation of this complex to DNA damage sites (10–14). This translocation involves recognition of K63-linked polyubiquitin chains of histones H2A and H2AX by the ubiquitin interaction motifs (UIMs) within RAP80 (10, 15–18).
The tumor suppressor p53 plays a key part in DDR signaling. It functions as a master regulator that controls a broad transcriptional network activated in response to various types of cellular and environmental stress (19). Activation of p53, along with the subsequent induction of its target genes, plays a critical role in the regulation of cell cycle control and apoptosis to assure genome integrity (20). Disruption of p53 can compromise repair of DNA damage resulting in chromosome abnormalities, ultimately leading to oncogenesis. Mutations in the p53 gene have been associated with more than half of human cancers (21). Under normal physiological conditions, p53 levels are kept low because of its ubiquitination by the E3 ubiquitin ligase HDM2 (corresponding to mouse double-minute 2 protein mdm2), resulting in its rapid turnover by proteasomes. In response to DNA damage, p53 becomes stabilized through processes that include post-translational modification of p53. HDM2 is itself a p53 target gene that can become activated after stress and lead to p53 destabilization (22, 23). The resulting p53-HDM2 auto-regulatory loop is of vital importance in controlling the level of p53 and its activity.
In this study, we identify a new role for RAP80 as both a modulator of p53 activity and as a direct transcription target of p53 following DNA damage, mainly through a noncanonical response element (RE) sequence in its promoter. RAP80 is able to form a complex with p53 and increase HDM2-dependent polyubiquitination of p53. RAP80, therefore, expands the p53-HDM2 relationship to a DNA damage-responsive, autoregulatory RAP80-p53-HDM2 loop.
EXPERIMENTAL PROCEDURES
Plasmids
pLXIN and pEGFP were purchased from BD Biosciences. pCMV-HA-Ub, pCMV-Myc-p53, pCMV-Myc-HDM2, and pCMV-HDM2 were gifts from Dr. Yue Xiong (University of North Carolina at Chapel Hill). pGEX-p53 was kindly provided by Dr. Yang Shi (Harvard University). Plasmids pC53-SN3 coding for human p53 cDNA under the control of cytomegalovirus promoter and pCMV-Neo-Bam were provided by Dr. Bert Vogelstein (Johns Hopkins University). Luciferase reporter constructs containing the p53-REs were created in pGL4.26 (luc2/miniP/Hygro) reporter vector (Promega). pRL-SV40 is a reporter plasmid coding for Renilla reniformis luciferase (Promega). More detailed information of plasmids and constructs used in this study are described in the supplemental material.
Cell Cultures
Detailed information of the cell lines used is provided in the supplemental material. Where indicated, the cells were treated with doxorubicin (Sigma) (0.3 μg/ml), γ-irradiation (0.5 or 4 Gy), or UV radiation (10 or 15 J/m2) at ∼70% confluence; 24 h later cells were harvested for protein and RNA extraction. Evaluation of cellular death was assessed by annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit (BD Pharmigen) following the manufacturer's protocol. Protein analysis and Western blot protocols are described in the supplemental material.
siRNA Transfection
For RNA interference, U2OS cells were transfected with control or RAP80 siRNAs (Dharmacon and Invitrogen) following the manufacturer's suggestions.
Luciferase Reporter Assays
For transcriptional assays, the cells were transfected with the reporter gene in the absence or presence of expression vectors for the indicated proteins or empty expression vector (pCMV-Neo-Bam) as described previously (24). A more detail protocol is provided in the supplemental material.
GST Pulldown Assay
The methods for purifying GST or GST-p53 fusion proteins and their binding to [35S]methionine-labeled RAP80 were described previously (14).
Co-immunoprecipitation Assay
U2OS cells were transfected with wild type pLXIN-3×FLAG-RAP80 or their mutants and pcDNA3-Myc-p53 as indicated; 48 h later, the cells were collected and processed as described previously (14).
Real Time PCR
Evaluation of RAP80 and p53 target genes mRNA levels was determined using TaqMan probe-based chemistry (Applied Biosystems), and the relative quantitative values were calculated based on the 2-ΔΔCt method following the manufacturer's instructions.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were done as described previously (24) using the ChIP kit (Millipore) following the manufacturer's instructions. A more detailed protocol is provided in the supplemental material.
RESULTS
RAP80 Is a Novel Target of p53-dependent Transcriptional Regulation
Because the p53 protein plays an important role in DDR signaling by activating the transcription of many DDR effectors, we investigated the possibility that RAP80 might also be a target of transcriptional regulation by p53. A 4-kb region surrounding the transcription start site of the human RAP80 promoter was scanned for the presence of p53 response element sequences (p53-REs). Although the commonly accepted consensus p53-RE consists of (RRRCWWGYYY followed by a 0–13-nt spacer followed by RRRCWWGYYY) with up to approximately five mismatches, we recently established that a fully functional RE has a spacer of ≤3 nt and that a single decamer, a “noncanonical half-site” (23, 25, 26), could mediate transcriptional activation by p53. The CWWG core is important for p53 responsiveness, where C and G are essential and CATG is the strongest responder. Using these criteria, nine potential p53-REs were identified.
Based on sequence similarities between the nine potential p53-REs with the p53-RE consensus and on observations that more than 80% of functional p53-REs are located in the proximal promoter region of target genes (23), the p53-RE3, -4, and -5 located in the ∼1.5-kb promoter region just upstream of the transcription start site of the RAP80 gene appeared the best candidates. As shown in Fig. 1A, the potential p53-RE3 (−1266 to −1237) has a 9-nt spacer and contains three mismatches in the first decamer, one of which was located at a critical position (27–29) in the CWWG core. The p53-RE4 (−1211 to −1188) contains only two mismatches in the first decamer followed by a 3-nt spacer and a perfect second decamer with a moderately responsive CWWG core (CTAG) (29). The p53-RE5 (−717 to −693) had a single mismatch at a critical position in the first decamer, a 4-nt spacer, and a perfect second half-site with a strong core signature (CATG). The remaining potential p53-REs have either a long spacer, mismatches in the CWWG core or a predicted weak CWWG. We therefore focused our further studies on the region containing REs 3 to 5. The complete list of potential p53-REs found in the analyzed region is presented in supplemental Table S1. Interestingly, neither of the potential p53REs found in the region analyzed of the human RAP80 are conserved in rodents (supplemental information). Consistent with this observation, the expression of RAP80 was not induced in WT p53 MEFs treated with UV radiation, and the levels remained comparable with those in treated p53−/− MEFs (Fig. 1E).
FIGURE 1.
p53 regulates RAP80 expression. A, potential p53-RE sites in the RAP80 promoter (±4 kb from the transcription start site). The p53-RE sites indicated by black arrows contain a perfect decamer (i.e. no mismatches). Sequences for p53RE3, -4, and -5 are shown. Modification by site-directed mutagenesis of the original RE5 to a true half-site RE is also presented. Mismatches with respect to consensus sequence are shown as lowercase letters. B, RAP80 promoter ChIP analysis in HCT116 p53+/+ to evaluate the p53 occupancy in the p53 RE3–4 and p53 RE-5 regions after different genotoxic stresses. C, luciferase transactivation assay in SaOS2 to test the p53 responsiveness of the RAP80 p53 RE5 in comparison with other established p53 REs. D, assessment of RAP80 expression after DNA damage (UV radiation, 10 J/m2; DOXO, 0.3 μg/ml; IR, 4 Gy) and exogenous WTp53 expression in isogenic HCT116 cells (p53+/+ and p53−/−); p21 served as an internal p53 target control. E, analysis of RAP80 expression in WT and p53−/− MEFs treated with UV radiation (20 J/m2) for 24 h. F, RAP80 protein levels after DNA damage in U2OS cells.
The ability of these p53-REs to function as p53 targets sequences was first investigated using ChIP assays. Binding was assessed with primer pairs that amplify the regions encompassing p53-RE3/RE4 (nt −1308 to −1058) or p53-RE5 (nt −609 to −885). Colon carcinoma HCT116 p53+/+ cells were treated with doxorubicin (DOXO, 0.3 μg/ml) for 24 h or with UV radiation (10 J/m2). After cross-linking with formaldehyde, DNA-protein complexes were immunoprecipitated with anti-p53 antibody; mouse IgG provided a negative control. p53 activated by UV radiation was found to be associated with both regions, whereas p53 activated by DOXO treatment did not bind (Fig. 1B). p53 bound the region containing p53-RE5 with a nearly 2-fold greater efficiency than that of p53-RE3/4. As expected, activation of p53 by both DNA-damaging agents resulted in high p53 occupancy of the p21 promoter, a positive control. Similar results were found with cell lines U2OS and A549 (supplemental Fig. S1). Using SaOS2 cell lines containing either TET-inducible WT or G279E mutant p53 (24), only the expressed WT p53 was able to bind the putative sites in the 1.5-kb promoter region of RAP80 (supplemental Fig. S1).
To establish that the p53-RE5 sequence is also functional in p53-mediated transactivation, the 24-bp sequence (AAGCTgGCCTccttGAACATGTCT) was cloned upstream of a minimal promoter containing a TATA box promoter element upstream of the pGL4.26 luciferase reporter. In addition, to assess the potential function of the perfect p53 half-site, 4-nt changes were introduced into the first decamer. Luciferase reporter constructs containing the p53-RE of p21, PUMA, or AIP were used as positive controls. Each of these constructs were co-transfected into p53 null SaOS2 cells with either a pCSN3 control vector or a vector that expresses WT or the G279E mutant p53. As shown in Fig. 1C, the WT p53 greatly increased transcriptional activation and most of the activation was supported by the perfect half-decamer of p53-RE5 because mutations in the first decamer had little impact on the transactivation. The level of RE5-dependent p53 transactivation was comparable with that of the moderately active PUMA and AIP p53-REs.
These results led us to investigate whether DNA damage alters the expression of RAP80 in cells that express endogenous WT p53. Following UV radiation and IR treatment, RAP80 mRNA was up-regulated in p53+/+ HCT116 cells but not in the isogenic p53−/− counterpart (Fig. 1D) or in p53-deficient H1299 cells (supplemental Fig. S1). Consistent with p53 promoter occupancy results, DOXO did not induce RAP80 expression, whereas all treatments induced the expression of p21 in p53+/+ cells. UV radiation and IR, but not DOXO, also induced RAP80 expression in human lung carcinoma A549 and osteosarcoma U2OS cells (supplemental Fig. S1) containing WT p53. Finally, overexpression of p53 in the p53 null HCT116 (Fig. 1D) and H1299 cells (supplemental Fig. S1) resulted in increased RAP80 mRNA expression. In agreement with these observations, RAP80 protein levels were increased after UV radiation and IR treatment reaching a maximum between 12 and 18 h post-treatment (Fig. 1F). Taken together, these results strongly suggest that RAP80 expression is controlled by p53 in a DNA damage-dependent manner through a noncanonical p53 half-site in the RAP80 promoter.
RAP80 Associates with p53
We investigated whether the regulation of RAP80 by p53 might be linked directly to p53 function. This was stimulated by the observation that RAP80 interacts with estrogen receptor α (ERα) affecting its stability/activity (30) and that HDM2, which is regulated by p53, also determines p53 stability. We first examined whether RAP80 and p53 interact. HeLa cells were co-transfected with pCMV-Myc-p53 and pLXIN-3×FLAG-RAP80 expression plasmids. An antibody against FLAG-RAP80 was able to co-immunoprecipitate p53 (Fig. 2A), suggesting that RAP80 and p53 are associated with the same protein complex. In vitro pulldown analysis was performed to further confirm this interaction. Moreover, GST-p53 fusion protein effectively pulled down in vitro translated 35S-labeled RAP80, whereas GST alone did not bind RAP80. ERα, previously shown to bind p53 (30), was used as a positive control (Fig. 2B). The interaction of RAP80 and p53 was further confirmed with an MCF-7 cell line stably expressing FLAG-RAP80 (14). As shown in supplemental Fig. S2, antibodies against FLAG or RAP80 could co-immunoprecipitate endogenous p53. Finally, we demonstrated that endogenous RAP80 was able to pull down endogenous p53 in U2OS (Fig. 2C) and 293T cells (supplemental Fig. S2).
FIGURE 2.
RAP80 associates with p53. A, HeLa cells were co-transfected with pLXIN-3×FLAG-RAP80 and pCMV-Myc-p53 expression plasmids as indicated. Cell lysates were prepared 48 h later and incubated with anti-FLAG M2 resin to isolate FLAG-RAP80 protein complexes, and complexes were examined by Western blot with the antibodies against Myc and FLAG. B, GST and GST-p53 fusion proteins were bound to glutathione-Sepharose 4B beads and incubated with [35S]methionine-labeled RAP80 for 2 h. The beads were washed, and bound proteins were solubilized and analyzed by SDS-PAGE. The radiolabeled proteins were visualized by autoradiography. C, lysates prepared from U2OS cells were incubated with anti-RAP80 antibody or normal IgG (control). The immunoprecipitated protein complexes were examined by Western blot analysis with antibodies against p53. D, schematic view of the region of RAP80 that interacts with p53. ZF, putative zinc finger. E, schematic view of p53 protein and the interaction region with RAP80. TA, transactivation; DBD, DNA-binding domain; OD, oligomerization domain; RD, regulatory domain; IB, immunoblot.
To determine the region(s) important for RAP80 interaction with p53, RAP80 carboxyl-terminal deletion mutants were examined by co-immunoprecipitation. As shown in Fig. 2D and supplemental Fig. S3, the region between amino acids 122 and 204 that lacks the UIMs and two potential zinc fingers is essential for the interaction. Based on in vitro pulldown analysis with p53 deletion mutants (Fig. 2E), the DNA-binding domain (amino acids 100–200) of p53 is necessary and sufficient to bind RAP80. The p53 fragments containing the transactivation domain, oligomerization domain, and regulatory domain failed to interact with RAP80 (supplemental Fig. S4).
RAP80 Promotes HDM2-mediated Ubiquitination of p53
Given that RAP80 affects the stability of another protein, ERα (30) and that p53 ubiquitination and stability is controlled by HDM2, the major E3 ubiquitin ligase of p53, we examined whether RAP80 was associated with HDM2. Co-immunoprecipitation analysis using U2OS cells transfected with FLAG-RAP80 and Myc-HDM2 expression plasmids demonstrated that RAP80 was able to pull down HDM2 (Fig. 3A). Subsequently, we assessed the effect of RAP80 on HDM2-mediated p53 ubiquitination and proteasome-mediated degradation. U2OS cells were transfected with the plasmids indicated in Fig. 3B and treated with the proteasome inhibitor MG132 before collection. Ubiquitination was determined in immunoprecipitated p53. As shown in Fig. 3B, overexpression of RAP80 enhanced HDM2-mediated ubiquitination of p53. However, RAP80 had no effect on HDM2-p53 interaction based on reprobing the same filter with anti-HDM2 antibody. Because HDM2 self-ubiquitination was greatly enhanced (Fig. 3B), the RAP80 enhancement of p53 ubiquitination appears to result from increased HDM2 ligase activity. Additionally, several slower migrating, ubiquitinated forms of RAP80 were identified, indicating that HDM2 is also a potential E3 ligase of RAP80 (Fig. 3B).
FIGURE 3.
RAP80 promotes HDM2-mediated ubiquitination of p53. A, RAP80 associates with HDM2. U2OS cells were co-transfected with pLXIN-3×FLAG-RAP80 and pCMV-Myc-p53 expression plasmids as indicated. 48 h after transfection, the cells were collected and processed as described in the legend to Fig. 2A. B, effect of RAP80 on p53 ubiquitination. U2OS cells were transfected with plasmids as indicated for 48 h. The cells were treated with 25 μm MG132 for 4 h before collection. The cell lysates were prepared, and Myc-p53 proteins were immunoprecipitated with an anti-Myc antibody. Western blotting was performed with anti-Myc (first panel) or anti-HDM2 (second panel) antibody to detect p53 ubiquitination and HDM2 pulled down by p53. The levels of HDM2 and RAP80 expression were determined with anti-HDM2 (third panel) or anti-FLAG antibody (fourth panel). C, the UIMs are not required for RAP80 effect on p53 ubiquitination. U2OS cells were transfected with the plasmids as indicated and processed as described in the legend to Fig. 3B.
Because the UIMs of RAP80 are required for its effect on ERα ubiquitination (30), we determined whether they are required for the regulation of p53 ubiquitination by RAP80. As shown in Fig. 3C, the UIMs are not needed because WT and RAP80ΔUIM enhanced p53 ubiquitination to similar extents (lanes 4 and 5). This effect of RAP80 on p53 ubiquitination was dependent on HDM2 expression (lane 2 versus lane 4). Moreover, RAP80 was not ubiquitinated in the absence of expressed HDM2 (lane 2 versus lane 4). However, ubiquitination of RAP80ΔUIM was greatly diminished (lane 4 and lane 5) compared with that of RAP80, indicating that the UIMs are important for optimal ubiquitination of RAP80, consistent with our previous findings (30).
RAP80 Modulates p53-dependent Transactivation via HDM2
Because RAP80 can mediate HDM2 ubiquitination of p53, the effect of RAP80 on p53-mediated transcriptional activation of p53 target genes and the role of HDM2 was examined. p53-deficient SaOS2 cells were co-transfected with WT p53 along with a plasmid expressing either 3×FLAG-RAP80 or the deletion mutant 3×FLAG-RAP80(N1–122) and various p53-RE transcription reporter plasmids. Western blot analysis demonstrated that all proteins were expressed at similar levels (supplemental Fig. S5). Expression of RAP80 but not mutant RAP80(N1–122) interfered with p53-dependent transcriptional activation. As shown in Fig. 4A, RAP80 inhibited p53-dependent transactivation at p53-RE(PUMA) and p53-RE(AIP) by ∼95%, and P21-RE and the artificial pG13 reporters by ∼50%; RAP80(N1–122) had no effect. In the absence of p53, there was no effect of RAP80 or RAP80 (N1–122) on the residual transcription. As expected, co-transfection of HDM2 with p53 led to reduced transactivation at target REs. Inhibition was directly related to the amount of RAP80 plasmid added (supplemental Fig. S5). Similar results were obtained when p53+/+ U2OS cells were transfected with RAP80 expression plasmid and endogenous p53 was activated by DOXO (supplemental Fig. S5).
FIGURE 4.
RAP80 modulates p53 transactivation activity. A, effect of RAP80 on p53 dependent transactivation measured by luciferase reporter assay in SaOS2 cells using p53 REs from P21, PUMA, and AIP genes and one artificial p53-responsive construct (PG13). The constructs were co-transfected in the presence or absence of WT p53, WT or mutant RAP80-FLAG, and HDM2. RAP80 expression vectors were co-transfected in a 2:1 ratio relative to p53 vector transfected. p53 transcriptional activity using P21 and PUMA p53 REs was evaluated in mdm2−/− p53−/− (B) and mdm+/+ p53−/− MEFs co-transfected with WT p53 along with WT or mutant RAP80 constructs (C). Reporter assays were evaluated 48 h post-transfection. Presented are the averages and S.D. of three independent biological experiments.
RAP80 alone was unable to mediate p53 degradation, suggesting that RAP80 interacts with both p53 and HDM2 to enhance p53 ubiquitination by HDM2. To further assess the role of HDM2, we determined the effect of RAP80 on transactivation driven by exogenous p53 in mdm2−/− p53−/− and mdm2+/+ p53−/− MEF cells. As shown in Fig. 4 (B and C), p53 transactivation at the P21 and PUMA REs was not affected by RAP80 expression in the mdm2−/− cells, whereas RAP80 expression in the mdm2+/+ MEFs resulted in 40 and 70% reductions in P21-RE- and PUMA-RE-dependent transactivation, respectively. Similar to results obtained with SAOS2 cells, the inhibition of p53 transactivation activity by RAP80 in MEFs was directly dependent on the amount of RAP80 added (supplemental Fig. S5). As expected, ectopic expression of mouse MDM2 resulted in a large reduction in p53 transactivation (Fig. 4, B and C). Thus, the RAP80 reduction of p53-mediated transactivation is dependent on MDM2.
DNA Damage-dependent Stabilization of p53 by RAP80 Depletion
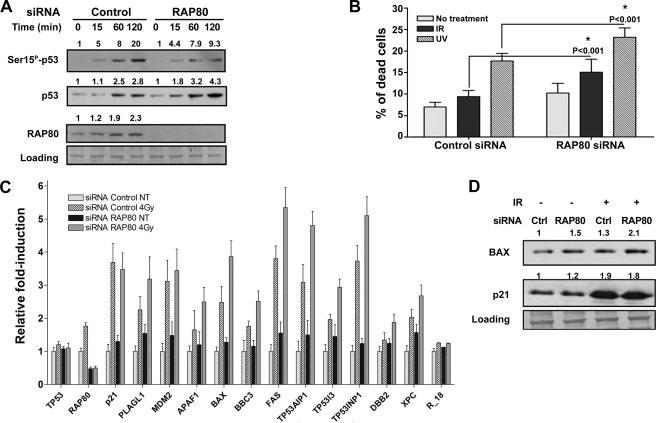
To examine the effect of RAP80 depletion on p53 protein levels, U2OS cells were transfected with RAP80 siRNA or scrambled siRNA (control) and 72 h later treated with 4 Gy (Fig. 5A) of γ-irradiation. The levels of p53 were determined during the subsequent 2 h. RAP80 siRNA greatly diminished the level of RAP80 protein without affecting p53 levels. However, after irradiation the p53 levels were considerably higher in RAP80-depleted cells as compared with post-irradiation controls, consistent with RAP80 enhancing HDM2-mediated ubiquitination and degradation of p53. Interestingly, knockdown of RAP80 inhibited the level of Ser(P)15-p53, which is commonly used as a marker for p53 activation after IR damage. As expected based on the results described above, there was a significant increase in RAP80 protein starting 1 h after treatment with IR for cells transfected with scrambled with siRNA. Qualitatively similar results were obtained at a much lower dose, 0.5 Gy, which is expected to result in only a small induction of p53 (supplemental Fig. S6).
FIGURE 5.
Reduction of RAP80 levels activates p53 and sensitizes cells to DNA-damaging agents. A, U2OS cells were treated with control or RAP80 siRNA. 72 h later the cells were irradiated (4 Gy) and collected at different times. Levels of p53, p53Ser15, and RAP80 were determined by Western blot analysis. Densitometric analysis for RAP80 and p53 protein levels, presented as relative-fold induction, are also shown above the lower Western blots. B, U2OS cells were transiently transfected with RAP80 siRNA, and 72 h later the cells were treated with IR (4 Gy) or UV radiation (15 J/m2) and apoptosis was evaluated 24 h later. *, statistical analysis with Tukey's multiple comparison test. The expression of p53 target genes by real time PCR in U2OS cells expressing RAP80 siRNA and treated with IR (C) is also presented. Shown are the averages and S.D. of three independent biological experiments. D, the protein levels of P21, BAX in U2OS cells transfected with RAP80-siRNA are shown together with densitometric analyses.
Because RAP80 overexpression inhibited p53-dependent transactivation through REs from the apoptosis-related genes PUMA and AIP to a greater extent than transactivation through the RE from the cell cycle arrest gene P21 (described above), depletion of RAP80 might particularly affect regulation of apoptosis-related genes. The effect of RAP80 depletion on p53-dependent apoptosis was therefore examined in U2OS cells treated with 4 Gy or 15 J/m2 UV radiation. As shown in Fig. 5B, apoptosis induced by IR was significantly enhanced in RAP80-depleted cells compared with cells transfected with scrambled siRNA. The difference in IR-induced (versus no IR) apoptotic cells was nearly twice that for RAP80-depleted (∼16%) as compared with scrambled siRNA cells (∼9%). Similarly, there was a significant increase in apoptosis following UV radiation for the RAP80-depleted cells.
The effect of RAP80 depletion on transactivation of p53 target genes after genotoxic stress was also examined in U2OS cells. Expression of the following well known p53 targets genes was evaluated by real time PCR: cell cycle arrest genes P21 and PLAGL1; apoptosis-related genes P53AIP1, APAF1, BAX, BBC3-PUMA, FAS, TP53I3-PIG3, and TP53INP1; DNA repair genes DDB2 and XPC; and the p53 gatekeeper gene HDM2. In general, the expression of most p53 target genes was elevated in RAP80-depleted cells treated with IR (Fig. 5C) or UV radiation (supplemental Fig. S6) relative to cells transfected with scrambled siRNA. Neither RAP80 knockdown or radiation affected p53 mRNA expression. At the doses used, IR had a larger effect than UV radiation. The effect was greatest for apoptosis genes, whereas the induction of P21 and HDM2 were not affected. The net impact of ionizing radiation on expression (exposure versus no exposure) was typically 1.5–3-fold greater in RAP80-depleted cells as compared with cells transfected with scrambled siRNA. At the protein level RAP80 siRNA led to no differences in p21, whereas there was a small increase in BAX in untreated and IR-treated U2OS cells (Fig. 5D).
DISCUSSION
In previous studies we and other groups demonstrated that RAP80 is part of a BRCA1-BARD1-ccdc98(Abraxas) complex and promotes the translocation of these complexes to DNA damage sites and as such is involved in DSB repair and cell cycle checkpoint control following IR exposure (10–14). Knockdown of RAP80 inhibits the translocation of the BRCA1 complex to sites of DNA damage, resulting in less efficient DNA repair through homologous recombination and compromise M phase checkpoint induced by IR. Because p53 interacts with several DDR proteins, including 53BP1 and BRCA1, and has a role in controlling cell cycle checkpoints, we were interested in examining possible links between RAP80 and p53. In this study, we identified a new RAP80-p53-HDM2 auto-regulatory loop that includes transcriptional regulation of RAP80 gene by p53 through a noncanonical p53 target sequence, interaction of RAP80 with p53 and HDM2, and negative modulation of p53 stability and activity by RAP80. A model summarizing our findings is presented in Fig. 6. It is well known that the p53-HDM2 auto-regulatory loop controls p53 level during DDR and is subject to fine-tuning through a number of proteins that include ARF (31), YY1 (32), gankyrin (33), nucleophosmin (34), WIP1 (35), and NUMB (36). These proteins regulate p53-HDM2 interaction and/or HDM2 localization, stability, and ligase activity, which in turn stimulate or prevent HDM2-mediated ubiquitination and p53 degradation. In our model, the p53-HDM2 auto-regulatory loop is augmented by another auto-regulatory loop in which RAP80 enhances p53 degradation by HDM2 and is itself under control of p53 (Fig. 6).
FIGURE 6.
Model for control of p53 stabilization by HDM2 and RAP80. In unstressed cells, p53 is kept inactive (green box) and at low levels (down arrow) mainly because of the action of HDM2 which promotes p53 ubiquitination through its ubiquitin-ligase activity and subsequent degradation by the proteasome; the HDM2 can also prevent p53 from recruiting transcriptional co-activators. UV radiation and IR exposure releases p53 from the HDM2 inhibitory interactions, resulting in activation and accumulation of p53. Activated p53 (indicated by a blue circle) induces the transcriptional activation of genes involved in cell cycle checkpoint, DNA repair, and apoptosis, as well HDM2. Induction of HDM2 by p53 results in a negative feedback loop, which switches off p53 activity, returning the system to baseline levels. In the present study, we demonstrate that RAP80 functions as a novel p53 target gene. RAP80 becomes part of an HDM2-p53 complex, enhances HDM2-dependent p53 ubiquitination and subsequently p53 degradation, resulting in RAP80 playing a central role in an additional negative feedback loop regulating p53 (red arrows). For simplicity, several other components involved in the regulation of p53 levels by HDM2 (see text for details) are not included.
Expression of RAP80 significantly increased p53 polyubiquitination (Fig. 3) in an HDM2-dependent manner that had a negative regulatory effect on p53 transactivation capabilities (Fig. 4). RAP80 itself does not have an intrinsic ubiquitin E3 ligase activity because it lacks a Ring or a HECT domain. Instead of strengthening the p53-HDM2 interaction, as described for YY1 and gankyrin (32, 33), RAP80 enhanced HDM2 ligase activity, as indicated by increased HDM2 ubiquitination, the mechanism of which remains to be determined. Unlike RAP80 regulation of ERα and BRCA1 signaling (11, 12, 16), the UIMs of RAP80 are not required for p53-regulation. Because the inhibition was totally abolished in mdm2 null MEFs (Fig. 4B), RAP80 functions as a co-factor of HDM2 to regulate p53 stability and activity. The relationship between RAP80 and various other modulators of the p53-HDM2 autoregulatory loop remains to be determined.
Although our experiments were designed to understand the biological implications of the RAP80-p53, our results indicate an interaction between RAP80 and HDM2 and suggest that HDM2 can also promote RAP80 ubiquitination. We are currently investigating what domains of both RAP80 and HDM2 are required for their interaction and whether the UIM domain of RAP80 is required for its ubiquitination process.
The RAP80-HDM2-p53 auto-regulatory loop could provide additional layers of target gene selectivity by affecting the amount of p53 available for transactivation. Reduced levels of RAP80 protein might be expected to have the greatest effect on weaker responding p53 targets. RAP80 depletion resulted in increased p53 stability following IR treatment and higher mRNA levels of p53 target genes, particularly pro-apoptotic genes (Fig. 5). Knockdown of RAP80 had little effect on the basal level of p53 protein, unlike YY1 and gankyrin, modifiers of the p53-HDM2 loop that inhibit basal p53 activity (32, 33). It is interesting that RAP80 knockdown resulted in reduced Ser(P)15-p53 phosphorylation after IR (Fig. 5 and supplemental Fig. S6). A previous study showed that BRCA1 is required for Ser(P)15-p53 phosphorylation by ATM or ATR after DNA damage (37). Because RAP80 is important for BRCA1 translocation and function after DNA damage, the impact of RAP80 depletion on Ser(P)15-p53 phosphorylation might relate to an effect of RAP80 depletion on BRCA1 translocation.
The effect of RAP80 knockdown on p53 protein level was similar to that found for reduction in Wip1 (wild type p53-induced phosphatase 1) (35). Wip1 interacts with and dephosphorylates HDM2 at serine 395, a site phosphorylated by ATM after IR treatment (38, 39). Dephosphorylation of HDM2 increases its stability and access to p53, increasing p53 degradation. Interestingly, both WIP1 and RAP80 are induced after IR treatment in a p53-dependent manner (39). The transcriptional activation of WIP1 and RAP80 by p53 after DNA damage and the negative effects of their encoded proteins on p53 stability and activity suggest that there are multiple mechanisms that control p53 via HDM2 following stress exposure. Possibly these autoregulatory loops are differentially responsive to various stress signals to assure that p53 levels return to normal levels after DNA damage repair. This is consistent with the reported differential activation of p53 targets following exposure to various DNA damage and stress inducers (23).
We found that p53 regulation of RAP80 expression was differentially dependent on induction of DNA damage (Fig. 1). Treatment with UV radiation and IR, but not DOXO, resulted in a p53-dependent induction of RAP80, suggesting that specific post-translational modifications on p53 might play a role in RAP80 regulation as reported for other p53 target genes (40, 41). Although several putative p53-REs were identified in the regulatory region of RAP80 surrounding the transcription start site, p53-RE3, -4, and -5 located in the ∼1.5-kb promoter region just upstream of the transcription start site appeared the best candidates. ChIP and promoter analysis suggested that transcriptional regulation of RAP80 by p53 was mainly driven by the p53-RE5 half-site in the upstream promoter region. This half-site (GAACATGTCT) has no mismatches relative to the established p53-RE consensus and contains a CATG core associated with higher p53-RE functionality (25, 27, 29, 42, 43). A second p53-RE decamer is located 4 nt away and contains one mismatch in the CWWG core, making it at most a very weak contributor to p53 transactivation. Most functional p53 target genes have 0–2-nt spacer (23, 27, 43–45), and we recently demonstrated that a spacer of ≥3 nt greatly impairs p53 transactivation (26). Thus, although RE-5 fits the generally acknowledged consensus, our results demonstrate that the half-decamer is likely responsible for most of the p53-mediated transactivation at p53RE-5. It is possible that other p53-REs, such as RE3/RE4 that weakly bind p53, might influence RAP80 transcription.
Interestingly, the regulatory region of human RAP80 is not conserved in rodents. We recently reported that most of the p53 target genes related to DNA repair/metabolism are not functionally conserved in the p53 transcriptional network in rodents (14). Similarly none of the nine putative REs in the human RAP80 region analyzed were conserved in rodents. Although alternative putative p53REs were found in the mouse and rat RAP80 proximal promoter region, none of these sites have the required characteristics expected for significant p53-mediated transactivation. In agreement with this, we also showed with MEF cells that RAP80 mRNA was not up-regulated by p53 in response to DNA-damaging agents. These findings are relevant to the use of animal models in studies of RAP80 and its relation with cancer and other diseases.
Also, additional factors could influence regulation. For example, we reported that a half-site p53-RE plus a nearby estrogen response element can mediate transactivation of the angiogenesis-related gene FLT1 (25). As part of a genome wide search for similar motifs,5 we found that the region around p53-RE5 as well as p53-RE3 and -4 is surrounded by several putative estrogen response elements. Interestingly, RAP80 has been reported to interact directly with ERα and to positively modulate ERα-mediated transactivation (30). Whether there is any functional link between the interactions of RAP80 with ERα and p53 requires further study.
Under normal conditions, p53 transactivates HDM2, which in turn targets p53 for degradation, thereby establishing a feedback control of p53 levels. DNA damage prevents HDM-2 from binding to p53, thus blocking degradation of p53. Here we show that RAP80 is an important negative regulator of the p53 tumor suppressor in response to genotoxic stress, modulating the p53-HDM2 autoregulatory loop. As a result RAP80 influences p53 stability and, therefore, its transactivation activities. This modulation could also be achieved through mechanisms not affecting p53 stability. We show that RAP80 can interact with the DNA-binding domain of p53, which also might compromise the ability of p53 to interact with target genes REs. The siRNA reduction of RAP80 protein levels sensitized cells to UV radiation and IR genotoxic stresses leading to an increase in the expression of pro-apoptotic p53 target genes. Along with this, there was a consistent modest increase in expression of the several pro-apoptotic genes tested (Fig. 5C), unlike for other nonapoptotic p53 targets such as p21 and HDM2. Possibly at low levels of DNA damage, the impact of reduced levels of p53 caused by the RAP80-HDM2-p53 interaction is greatest on weakly responding REs from p53 target genes such as those involving apoptosis. There might be less recruitment of p53-related transcriptional co-activators such as histone acetyltransferases, CREB-binding protein (CBP), p300, and P300/CBP-associated factor to the promoter-enhancer region of genes. At higher levels of DNA damage, p53 would be released from the HDM2 interaction because of site-specific post-translational modifications in both proteins so that the affinity of p53 for these co-activators, as well as for pro-apoptotic p53 co-factors such as ASPP1, ASPP2, JMY, and Tp53INP1, might enhance expression of weak p53 targets genes including those involved in apoptosis. Chromatin remodeling and the presence of such co-factors and its relation with RAP80 activities await further investigation. In addition to that, it is also possible that because RAP80 plays an important role in DNA repair by recruiting other proteins to the sites of DNA damage, such as the BRCA1-BARD1-ccdc98(Abraxas) complex, cells lacking RAP80 may experience increased levels of DNA damage because of reduced repair.
Mutations in a number of DDR proteins have been strongly linked to genome instability and cancer (1–4). The role of p53 in cancer is well established. Mutations in p53 are associated with ∼50% of human cancers, and ∼80% of human cancers have a defect in p53 signaling (46). Because RAP80 plays a key role in the DDR signaling, it might be implicated in cancer as well. Consistent with this concept are observations showing that RAP80-depleted cells exhibit impaired IR-induced CHK1 activation and, as a consequence, defective G2/M phase checkpoint control and reduction in the effectiveness to repair DNA damage (47). Moreover, recent studies by Shebzukhov et al. (47) reported that sera from 5–10% of patients with various types of cancer contained specific antibodies to RAP80/UIMC1 and proposed that RAP80 functions as a cancer-related antigen with limited tumor type specificity. Different studies analyzing RAP80 mutations in women with familial breast cancer, negative for BRCA1 and BRCA2 mutations, identified several novel haplotypes and rare missense mutations (48, 49). However, additional studies are needed to determine what role genetic changes in the RAP80 gene may have in influencing the susceptibility of humans to various cancers. Given the role that we have established for RAP80 regulation of p53, decreases in RAP80 might enhance protection provided by p53. This raises the possibility that blocking the p53-RAP80 interaction might provide a therapeutic strategy against human cancers with depressed p53 expression.
Supplementary Material
Acknowledgments
We thank Drs. Douglas Bell and Alex Merrick for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants Z01 ES101586 (to A. M. J.) and Z01 ES065079 (to M. A. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S6.
D. Menendez and M. A. Resnick, unpublished observations.
- DDR
- DNA damage response
- UIM
- ubiquitin interaction motif
- E3
- ubiquitin-protein isopeptide ligase
- siRNA
- small interfering RNA
- RE
- response element
- GST
- glutathione S-transferase
- ChIP
- chromatin immunoprecipitation
- nt
- nucleotide(s)
- WT
- wild type
- MEF
- mouse embryonic fibroblast
- DOXO
- doxorubicin
- ER
- estrogen receptor
- Gy
- gray
- IR
- ionizing radiation
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1.Bartek J., Lukas J. (2007) Curr. Opin. Cell Biol. 19, 238–245 [DOI] [PubMed] [Google Scholar]
- 2.Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 3.Kastan M. B., Bartek J. (2004) Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 4.Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 5.Hoeijmakers J. H. (2001) Nature 411, 366–374 [DOI] [PubMed] [Google Scholar]
- 6.McKinnon P. J., Caldecott K. W. (2007) Annu. Rev. Genomics Hum. Genet. 8, 37–55 [DOI] [PubMed] [Google Scholar]
- 7.Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 8.Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 9.Huen M. S., Chen J. (2008) Cell. Res. 18, 8–16 [DOI] [PubMed] [Google Scholar]
- 10.Yan J., Jetten A. M. (2008) Cancer Lett. 271, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H., Chen J., Yu X. (2007) Science 316, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 12.Sobhian B., Shao G., Lilli D. R., Culhane A. C., Moreau L. A., Xia B., Livingston D. M., Greenberg R. A. (2007) Science 316, 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., Matsuoka S., Ballif B. A., Zhang D., Smogorzewska A., Gygi S. P., Elledge S. J. (2007) Science 316, 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan J., Kim Y. S., Yang X. P., Li L. P., Liao G., Xia F., Jetten A. M. (2007) Cancer Res. 67, 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 19.Brooks C. L., Gu W. (2006) Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodier F., Campisi J., Bhaumik D. (2007) Nucleic Acids Res. 35, 7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 22.Wu X., Bayle J. H., Olson D., Levine A. J. (1993) Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 23.Riley T., Sontag E., Chen P., Levine A. (2008) Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 24.Menendez D., Inga A., Resnick M. A. (2006) Mol. Cell. Biol. 26, 2297–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menendez D., Inga A., Snipe J., Krysiak O., Schönfelder G., Resnick M. A. (2007) Mol. Cell. Biol. 27, 2590–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan J. J., Menendez D., Inga A., Nourredine M., Bell D., Ma R. (2008) PLoS Genet. 4, e1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inga A., Storici F., Darden T. A., Resnick M. A. (2002) Mol. Cell. Biol. 22, 8612–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menendez D., Inga A., Jordan J. J., Resnick M. A. (2007) Oncogene 26, 2191–2201 [DOI] [PubMed] [Google Scholar]
- 29.Jegga A. G., Inga A., Menendez D., Aronow B. J., Resnick M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J., Kim Y. S., Yang X. P., Albers M., Koegl M., Jetten A. M. (2007) Nucleic Acids Res. 35, 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Xiong Y., Yarbrough W. G. (1998) Cell 92, 725–734 [DOI] [PubMed] [Google Scholar]
- 32.Sui G., Affar el B., Shi Y., Brignone C., Wall N. R., Yin P., Donohoe M., Luke M. P., Calvo D., Grossman S. R., Shi Y. (2004) Cell 117, 859–872 [DOI] [PubMed] [Google Scholar]
- 33.Higashitsuji H., Higashitsuji H., Itoh K., Sakurai T., Nagao T., Sumitomo Y., Sumitomo H., Masuda T., Dawson S., Shimada Y., Mayer R. J., Fujita J. (2005) Cancer Cell 8, 75–87 [DOI] [PubMed] [Google Scholar]
- 34.Kurki S., Peltonen K., Latonen L., Kiviharju T. M., Ojala P. M., Meek D., Laiho M. (2004) Cancer Cell 5, 465–475 [DOI] [PubMed] [Google Scholar]
- 35.Lu X., Ma O., Nguyen T. A., Jones S. N., Oren M., Donehower L. A. (2007) Cancer Cell 12, 342–354 [DOI] [PubMed] [Google Scholar]
- 36.Colaluca I. N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., Di Fiore P. P. (2008) Nature 451, 76–80 [DOI] [PubMed] [Google Scholar]
- 37.Foray N., Marot D., Gabriel A., Randrianarison V., Carr A. M., Perricaudet M., Ashworth A., Jeggo P. (2003) EMBO J. 22, 2860–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maya R., Balass M., Kim S. T., Shkedy D., Leal J. F., Shifman O., Moas M., Buschmann T., Ronai Z., Shiloh Y., Kastan M. B., Katzir E., Oren M. (2001) Genes Dev. 15, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiscella M., Zhang H., Fan S., Sakaguchi K., Shen S., Mercer W. E., Vande Woude G. F., O'Connor P. M., Appella E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6048–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsson A., Manzl C., Strasser A., Villunger A. (2007) Cell Death Differ. 14, 1561–1575 [DOI] [PubMed] [Google Scholar]
- 41.Saito S., Yamaguchi H., Higashimoto Y., Chao C., Xu Y., Fornace A. J., Jr., Appella E., Anderson C. W. (2003) J. Biol. Chem. 278, 37536–37544 [DOI] [PubMed] [Google Scholar]
- 42.Veprintsev D. B., Fersht A. R. (2008) Nucleic Acids Res. 36, 1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., Ruan Y. (2006) Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 44.el-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B. (1992) Nat. Genet. 1, 45–49 [DOI] [PubMed] [Google Scholar]
- 45.Tomso D. J., Inga A., Menendez D., Pittman G. S., Campbell M. R., Storici F., Bell D. A., Resnick M. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6431–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S. V., Hainaut P., Olivier M. (2007) Hum. Mutat. 28, 622–629 [DOI] [PubMed] [Google Scholar]
- 47.Shebzukhov Y. V., Koroleva E. P., Khlgatian S. V., Belousov P. V., Sazykin A. Y., Kadachigova T. S., Pomerantseva E. A., Lagarkova M. A., Nedospasov S. A., Kuprash D. V. (2007) Cancer Lett. 255, 255–262 [DOI] [PubMed] [Google Scholar]
- 48.Akbari M. R., Ghadirian P., Robidoux A., Foumani M., Sun Y., Royer R., Zandvakili I., Lynch H., Narod S. A. (2009) Breast Cancer Res. Treat. 113, 377–381 [DOI] [PubMed] [Google Scholar]
- 49.Osorio A., Barroso A., García M. J., Martínez-Delgado B., Urioste M., Benítez J. (2009) Breast Cancer Res. Treat. 113, 371–376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.